Significance
The development of lymphocytes is critical for host immunity and relies on a series of developmental checkpoints regulated by key transcription factors such as Ikaros. We hypothesized that nonprotein-coding loci might represent an additional layer of control in lymphocyte development. We identified a noncoding region (Daedalus) whose absence leads to a profound loss of Ikaros protein and a severe reduction in early lymphocyte progenitors. In contrast to Ikaros deletion, removal of Daedalus also led to an increase in red-blood-cell colony formation, suggesting that Daedalus functions as a lineage-specific stabilizer of Ikaros activity, thus acting as a “gatekeeper” of a newly identified lymphoid-erythroid checkpoint. This finding presents a paradigm potentially applicable to the control of all developmental programs.
Keywords: lymphocytes, Ikaros, noncoding, hematopoiesis
Abstract
Haematopoiesis relies on tightly controlled gene expression patterns as development proceeds through a series of progenitors. While the regulation of hematopoietic development has been well studied, the role of noncoding elements in this critical process is a developing field. In particular, the discovery of new regulators of lymphopoiesis could have important implications for our understanding of the adaptive immune system and disease. Here we elucidate how a noncoding element is capable of regulating a broadly expressed transcription factor, Ikaros, in a lymphoid lineage-specific manner, such that it imbues Ikaros with the ability to specify the lymphoid lineage over alternate fates. Deletion of the Daedalus locus, which is proximal to Ikaros, led to a severe reduction in early lymphoid progenitors, exerting control over the earliest fate decisions during lymphoid lineage commitment. Daedalus locus deletion led to alterations in Ikaros isoform expression and a significant reduction in Ikaros protein. The Daedalus locus may function through direct DNA interaction as Hi-C analysis demonstrated an interaction between the two loci. Finally, we identify an Ikaros-regulated erythroid-lymphoid checkpoint that is governed by Daedalus in a lymphoid-lineage–specific manner. Daedalus appears to act as a gatekeeper of Ikaros’s broad lineage-specifying functions, selectively stabilizing Ikaros activity in the lymphoid lineage and permitting diversion to the erythroid fate in its absence. These findings represent a key illustration of how a transcription factor with broad lineage expression must work in concert with noncoding elements to orchestrate hematopoietic lineage commitment.
Haematopoiesis relies on tightly controlled gene expression patterns as development proceeds through a series of progenitors (1–8). These gene expression patterns are controlled by lineage-specifying transcription factors; however, these transcription factors are often found to have broad expression patterns and activities across lineages. Given their role in regulating cell-type–specific gene expression, noncoding loci are thought be important in the lineage-specific regulation of this complex process (6, 9–16).
Noncoding loci are a broad category of genetic elements that function independently of their protein-coding potential, although they are frequently marked by transcribed RNAs. The mechanisms through which noncoding loci exert their activity are diverse and can include direct DNA–DNA interactions (e.g., enhancer elements), recruitment of protein factors which modulate the surrounding epigenetic landscape, or the production of a functional RNA such as a microRNA or long noncoding RNA. Noncoding elements are now well appreciated as a primary mechanism by which gene expression across multiple, disparate cell types can be regulated in a lineage-specific manner.
To identify transcripts marking such loci, we performed RNA-sequencing (RNA-seq) in murine lymphocyte populations. We identified a variety of loci containing dynamically expressed transcripts and tested their role in hematopoiesis using an in vivo CRISPR knockout (KO) mouse screen.
Deletion of a locus proximal to Ikaros, here named Daedalus, led to a cell-autonomous reduction in lymphoid progenitor populations, exerting control over the earliest fate decisions during lymphoid lineage commitment. Congruent with this we observe a loss of Ikaros protein in Daedalus-deficient cells, although transcript levels appear consistent with wild type (WT). Using Hi-C, we found that the Daedalus locus interacts with that of Ikaros, suggesting that the Daedalus element may function through direct DNA interaction.
Finally, we demonstrate how loss of Daedalus leads to an increase in erythroid potential that is not seen in Ikaros-deficient progenitors. Using colony assays and single cell analyses we identify an Ikaros-regulated erythroid-lymphoid checkpoint that is governed by Daedalus in a lymphoid-lineage–specific manner. Daedalus appears to act as a gatekeeper of Ikaros’s broad lineage-specifying functions, selectively stabilizing Ikaros activity in the lymphoid lineage and permitting diversion to the erythroid fate in its absence.
Results
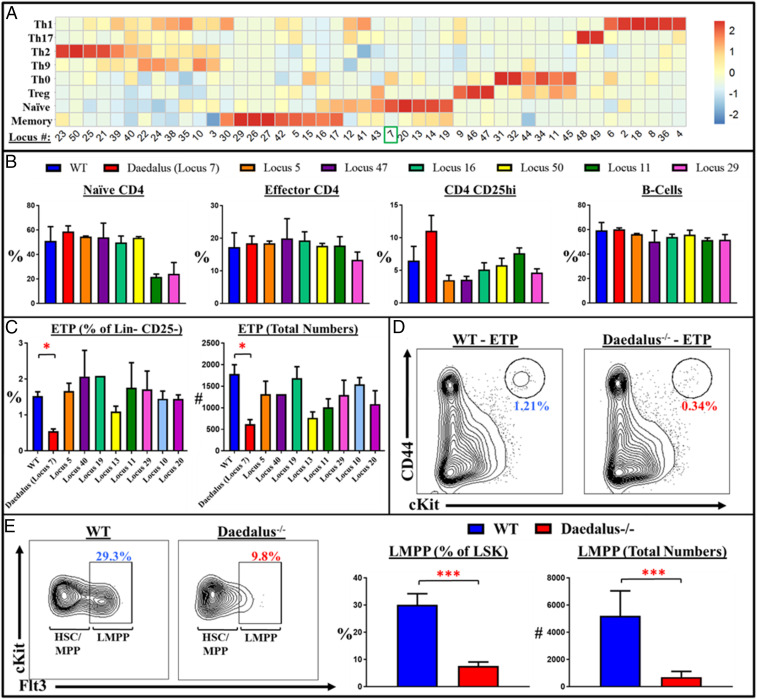
In order to uncover noncoding loci with the potential to regulate lymphoid cell biology, we performed an RNA-seq analysis on murine naive and effector CD4 T cells. We discovered 154 dynamically expressed unannotated RNAs and prioritized candidates that contained splice acceptor sites and had been established to lack coding potential in any frame, resulting in a list of 46 spliced, polyadenylated RNAs (Fig. 1A). In order to establish the function of these loci, we performed an in vivo knockout animal screen. We used CRISPR/Cas9 technology to generate 25 transgenic mouse lines lacking these loci. In order to investigate the impact of these loci on the lymphoid compartment, we evaluated steady-state hematopoietic development in the spleen (Fig. 1B and SI Appendix, Fig. S1), lymph nodes, and thymus (Fig. 1C and SI Appendix, Fig. S1) in these mice. While peripheral cell numbers appeared largely normal (Fig. 1B), we observed that loss of an ∼30-kb locus upstream of Ikaros (Chr11:11554730–11582873) (SI Appendix, Fig. S2), here named Daedalus, resulted in a severe reduction of early thymic progenitors (ETPs) (Fig. 1 C and D). Since these cells represent the earliest population of thymic-colonizing bone marrow progenitors (17, 18), we examined the bone marrow of these mice and identified a marked loss of Flt3+ LSK lymphoid progenitors (Fig. 1E), as well as canonical common lymphoid progenitor (CLP) and lymphoid-primed multipotent progenitor (LMPP) populations (SI Appendix, Fig. S3).
Fig. 1.
An in vivo CRISPR screen identifies a noncoding locus regulating lymphoid development. (A) Heatmap of differentially expressed noncoding transcripts identified in RNA-seq of CD4 effector T lymphocytes. The Daedalus locus was originally termed as Locus #7 and is denoted here by the number 7 and highlighted with a green box. (B) Representative plots from splenic FACS data show peripheral lymphocyte populations largely unaffected in noncoding RNA locus-deficient mice. CD4 T cell populations gated off CD4+ TCRb+ splenocytes: Naive CD4: CD62L+CD44–; Effector CD4: CD44+CD62L–; CD25hi: CD4+ CD25+. B-cells: gated on TCRb- NK1.1– CD19+ splenocytes; n = 3, representative of more than three pooled experiments. (C) Representative plots from thymic screening data showing early thymic progenitor populations (ETP: lineage-negative [lineage mixture: B220, CD11b, CD11c, CD19, Gr1, NK1.1, TCRgd, Ter119], CD4–, CD8–, CD25–, CD44+, cKit+). (D) Representative FACS plot of wild-type and Daedalus−/− littermates showing relative ETP populations. (E) Representative FACS plots and bar graphs of WT and Daedalus−/− littermate mice showing the LMPP population (Lineage negative [lineage mixture: B220, CD4, CD8, CD11b, CD11c, CD19, Gr1, NK1.1, TCRgd, Ter119], cKit+, Sca1+ Flt3hi); n = 3, representative of 10+ experiments. In this figure, unless otherwise specified, all statistics result from comparisons to WT using Student’s t test. *P < 0.05, ***P < 0.001.
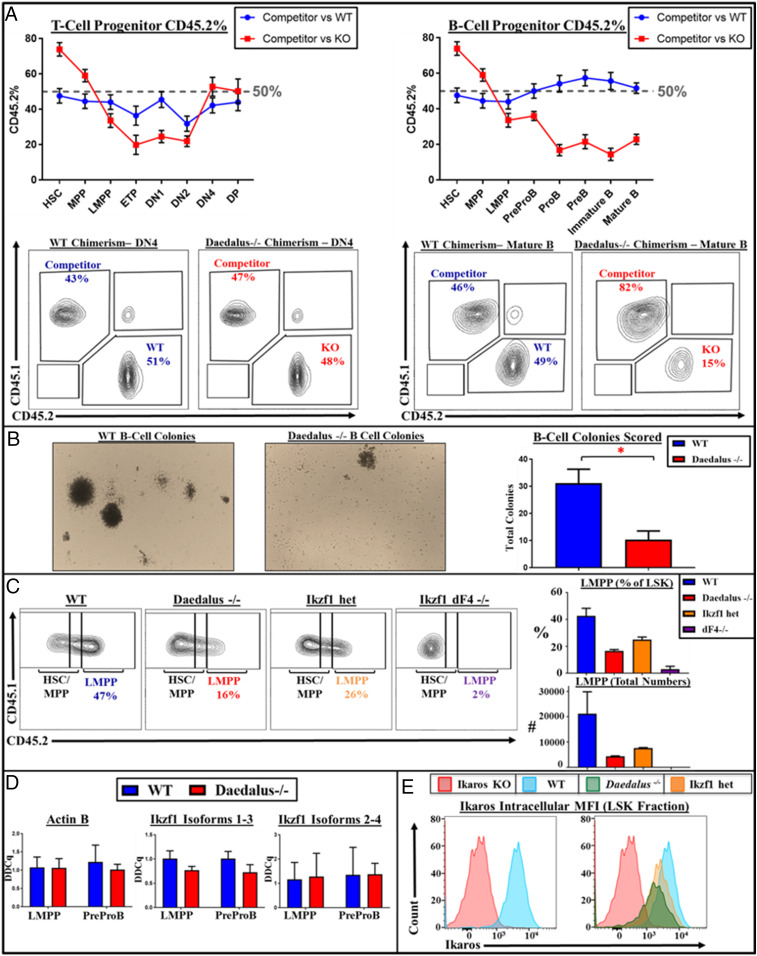
To distinguish between hematopoietic and nonhematopoietic contributions to the phenotype observed in Daedalus-deficient animals, we created bone marrow chimeras. We observed that the hematopoietic compartment is solely responsible for the phenotype of our Daedalus−/− mice. Irradiated donor mice injected with Daedalus−/− bone marrow exhibited a significant decrease in Flt3+ LSK lymphoid progenitors relative to wild type and a thymic phenotype consistent with that observed in Daedalus−/− mice (SI Appendix, Fig. S4). We next sought to determine whether these defects in lymphoid development were cell-autonomous by generating mixed-bone marrow chimeras. We found that, while the ratio of Daedalus−/− bone marrow to competitor is high in the HSC compartment, this significantly decreases in early lymphoid populations demonstrating that the defect originates at the LMPP stage (Fig. 2A). Furthermore, our mixed chimeras showed that, while this observed defect in early lymphoid progenitors recovers late in T-lymphocyte development, the ratio remains decreased in B-lineage commitment. This indicates that the lack of the Daedalus locus may impair B-lymphocyte development in our knockout mice. To confirm that lymphoid progenitors from Daedalus-deficient mice have diminished B-lineage potential, we performed B-cell colony-forming assays. Consistent with bone marrow chimera results, we found that Daedalus-deficient progenitors display impaired capacity to form B-cell colonies, compared to controls (Fig. 2B). Altogether, these data indicate that Daedalus is essential for normal lymphoid lineage development and likely acts around the LMPP/CLP stage.
Fig. 2.
The locus Daedalus controls lymphoid development through modulation of Ikaros protein levels. (A) WT and Daedalus−/− littermate mixed chimera studies, representative plots of chimerism in Mature B and DN4 cells, plots of percentage chimerism vs. CD45.1 competitor bone marrow in B and T lymphocyte developmental pathways; n = 3 representative of three experiments. (B) Representative pictures and bar graphs from B-cell methocult colony-forming assays; n = 3, representative of five or more experiments. (C) Representative FACS plots and bar graphs of LMPP populations from Ikaros heterozygous, Daedalus−/−, and Ikaros dF4−/− mice and WT littermates. LMPP: lineage-negative (lineage mixture: B220, CD4, CD8, CD11b, CD11c, CD19, Gr1, NK1.1, TCRgd, Ter119), cKit+, Sca1+ Flt3hi; n = 3, representative of three experiments. (D) qPCR experiment of Ikzf1 isoforms from mouse lineage-negative cKit+, Sca1+ hematopoietic progenitor cells; n = 1; data representative of multiple experiments with different sets of Ikzf1 primers (SI Appendix, Table S3 and Fig. S7). (E) Intracellular staining of Ikzf1 in Ikzf1 KO escape mutant (n = 1) and Daedalus−/− and WT littermate mice; Daedalus−/− data are representative of more than six experiments. In this figure, unless otherwise specified, all statistics result from comparisons to WT using Student’s t test. *P < 0.05.
Due to the proximity of Daedalus to the Ikzf1 locus and the role of Ikzf1 in lymphopoiesis, we hypothesized that Daedalus exerted its effects through Ikzf1 regulation. To test this, we compared hematopoiesis in Ikzf1 knockout heterozygotes, mice lacking zinc finger 4 (19) of the Ikaros protein (dF4−/−), and a homozygous Ikzf1 knockout escape mutant to Daedalus-deficient mice. Mice lacking Ikaros function (Ikzf1−/− and dF4−/−) exhibited an almost complete absence of Flt3+ LSK lymphoid progenitors and correspondingly large decreases in early B-lymphocyte progenitor numbers, consistent with prior studies (19, 20) (Fig. 2C and SI Appendix, Figs. S3 and S5). The phenotype of Daedalus−/− is most similar to that of Ikzf1+/− mice, with a significant reduction in lymphoid progenitor numbers, but not as few as observed for the Ikzf1−/− or dF4−/− mice (Fig. 2C). These analyses demonstrate that the Daedalus-locus–deficient mice exhibit an intermediate lymphoid phenotype quantitatively similar to Ikzf1 heterozygous mice.
Having observed a loss-of-function Ikzf1 phenotype in Daedalus-deficient animals, we next sought to evaluate how Daedalus impacts Ikaros expression. To test whether the Daedalus locus directly interacts with the Ikzf1 locus, we performed global run-on sequencing (GRO-seq) and Hi-C analysis in B-cell progenitors. We observed bidirectional transcription occurring at the Daedalus locus as well as interactions between Daedalus and the Ikzf1 locus and promoter regions (SI Appendix, Figs. S6 and S7), suggesting that Daedalus may function as an enhancer. To assay the impact Daedalus had on Ikzf1 transcript expression, we performed real-time qPCR on early hematopoietic progenitors from control and Daedalus knockout mice. We found that the levels of Ikaros transcript were largely comparable between control and Daedalus-deficient cells (Fig. 2D). In contrast, evaluation of Ikaros protein by intracellular flow cytometry analysis of these progenitors revealed that Daedalus knockout cells express reduced levels of Ikaros, comparable to what is observed in Ikzf1+/− cells (Fig. 2E). To confirm these findings, we performed intracellular flow cytometry for Ikaros protein using a separate antibody raised using a different Ikaros immunogen and again observed diminished Ikaros expression in Daedalus knockout cells (SI Appendix, Fig. S8). Altogether, these results indicate that Daedalus promotes lymphoid development by positively regulating Ikaros protein levels without marked changes in total Ikzf1 transcription.
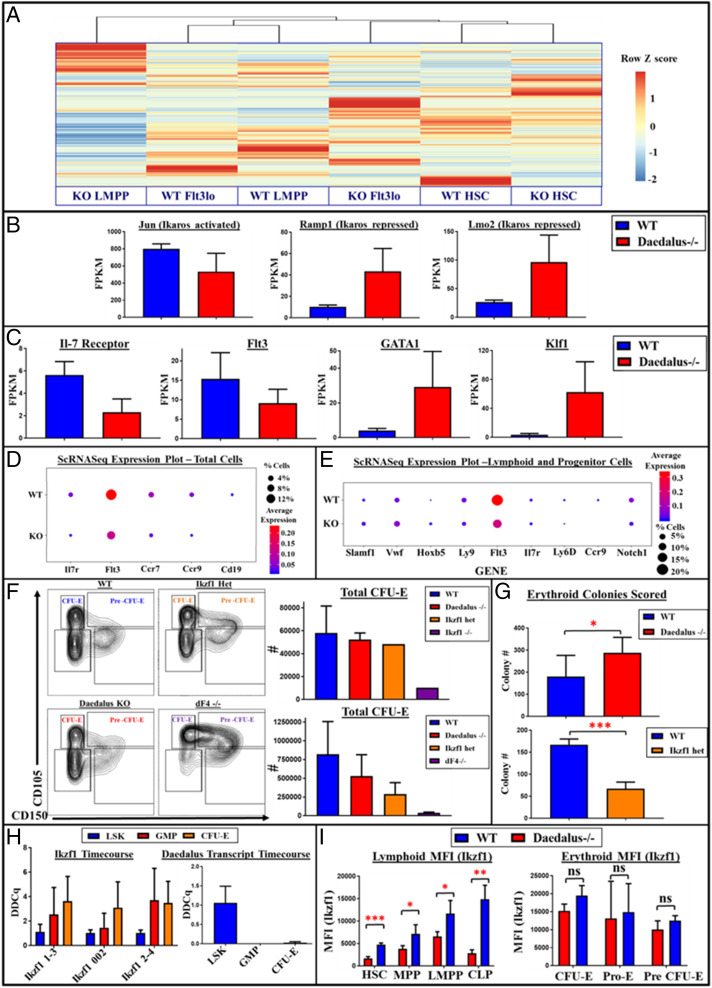
Ikaros is a transcription factor that has been implicated in the control of a wide array of developmental processes and transcriptional networks (19, 21–25). In order to better assess the changes resulting from ablation of Daedalus, we performed an RNA-seq analysis on progenitors from Daedalus KO and WT littermates (Fig. 3A). Consistent with a role for Daedalus in early lymphoid-lineage development, we found that expression patterns in early lymphoid progenitors (Flt3hi LSK) were most divergent from those of corresponding wild-type cells (Fig. 3A). As observed in qPCR experiments, total Ikaros transcript levels remained consistent between Daedalus-deficient and WT progenitors (SI Appendix, Fig. S9), although there were significant changes in the expression of some Ikaros isoforms (SI Appendix, Fig. S9), particularly in the LMPP compartment. In keeping with a role for Daedalus in regulating Ikaros function, our analysis revealed dysregulated expression of a number of important Ikaros target genes (26) (Fig. 3B). This included decreased expression of several genes previously reported as activated by Ikaros, including the transcription factor Jun, and increased expression of genes reported as repressed by Ikaros including Ramp1 and Lmo2 (26). In addition, consistent with our phenotypic finding of lymphoid defects, expression of lymphoid developmental markers such as Flt3 and the Il7 receptor decreased in Daedalus−/− progenitors (Fig. 3C). Strikingly, a number of important erythroid developmental regulators were also differentially expressed in these cells, including Klf1 (27, 28), Gata1 (29), and Gfi1b (30) (Fig. 3C and SI Appendix, Fig. S10). Genes associated with “erythrocyte differentiation” and “myeloid cell differentiation” were also overrepresented in a gene-ontology analysis of differentially expressed genes between these populations (SI Appendix, Fig. S10).
Fig. 3.
Daedalus controls Ikaros-dependent lymphoid erythromyeloid checkpoint. (A) Heatmap and dendrogram of total gene expression across hematopoietic progenitor subsets from whole-transcriptome analysis. (B) Fragments per kilobase of transcript per million mapped reads (Fpkm) expression levels of Ikaros target genes in WT and KO Flt3+ LSK progenitors. (C) Fpkm expression levels of key developmental regulators in WT and KO Flt3+ LSK progenitors. (D) Dot plots of marker genes across all cells for lymphoid-associated genes. The size of the dot illustrates the percentages of cells expressing a given gene, while the color indicates the level of expression. (E) Dot plots of marker genes across all cells in the clusters containing early progenitor and lymphoid progenitor markers for nonlymphoid and lymphoid genes. The size of the dot illustrates the percentages of cells expressing a given gene, while the color indicates the level of expression. (F) FACS plots of erythromyeloid progenitors from WT and Ikaros KO escape mutant littermates. (Top) Gated-off lineage-negative (lineage mixture: B220, CD4, CD8, CD11b, CD11c, CD19, Gr1, NK1.1, TCRgd), cKit+, Sca1–, and CD16/32– cells. (Bottom) Gated-off lineage-negative cKit+, Sca1–, CD16/32–, CD105+, and CD150 cells. (G) Bar graphs of total colonies from erythroid colony-forming assay results from equal numbers (2 × 106) of bone marrow from Daedalus–/– and WT littermate mice (Left); n = 3; data are representative of two experiments from Ikaros heterozygous mice and WT littermates (n = 2) (Right). (H) qRT-PCR experiment of Ikzf1 and Daedalus RNA expression in LSK progenitors, GMP progenitors (lineage-negative cKit+, Sca1–, CD16/32+), and erythroid progenitors (lineage-negative cKit+, Sca1–, CD16/32–, CD105+). (I) Intracellular Ikaros protein levels by median fluorescence intensity (MFI) in subsets of lymphoid, myeloid, and erythroid progenitors; n = 4, representative of two experiments. In this figure, unless otherwise specified, all statistics result from comparisons to WT using Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
In light of reports that Ikaros acts primarily as a part of the chromatin-modifying NuRD complex (31, 32), we next performed ATAC-sequencing (ATAC-seq) (33) to assess changes in chromatin accessibility to see if loss of Daedalus similarly disrupted histone remodeling at these known Ikaros-regulated loci. We determined that, in the absence of Daedalus, there is an increase in accessibility at loci of Ikaros target genes and erythroid-specific genes (SI Appendix, Fig. S11), consistent with the idea that these transcriptomic changes are due to an Ikaros insufficiency.
It is now known that sorted progenitors represent heterogenous populations that may contain many different short-lived cell types (34). Since our phenotype showed a highly significant reduction specifically in lymphoid progenitors sorted by fluorescence-activated cell sorting (FACS), we sought to confirm that Daedalus deficiency exerts a unique effect upon these populations using an unbiased approach. We performed single-cell transcriptomic analysis on total bone marrow progenitors from Daedalus locus-deficient mice and WT littermates in order to determine the changes in progenitor populations that occur in these mice (SI Appendix, Fig. S12). Unbiased clustering analyses revealed separation of total bone marrow progenitor populations into discrete groups that could be identified through the expression of known marker genes (SI Appendix, Fig. S13). Analysis of marker gene expression across all cells showed that, while myeloid and erythroid genes appear unaffected, there are significant differences in lymphoid gene expression between WT and Daedalus−/− cells both in terms of the percentages of cells expressing these genes and the level at which they are expressed (Fig. 3D and SI Appendix, Fig. S14). Differences in early progenitor markers as well as more pronounced differences in multiple lymphoid markers can be observed when taking into account only those clusters containing these markers (Fig. 3E). These results conclusively demonstrate that the effects of Daedalus locus deletion are highly lineage specific, affecting only early lymphoid progenitor and HSC populations without affecting other lineages.
It has been previously reported that defects in Ikaros lead to a reduction in erythroid cell potential (35, 36) and that Ikaros null mice on the B6 background suffer from embryonic lethality due to absence of erythroid potential (19). In our hands, B6 mice with an Ikaros null mutation exhibit embryonic lethality with incomplete penetrance, with fewer than 10% of Ikaros null homozygotes surviving postnatally (SI Appendix, Fig. S15). The phenotyping of one such escape mutant demonstrated that this mouse did indeed exhibit severe defects in erythroid development and terminal differentiation at several stages that were not shared by Daedalus-deficient mice (SI Appendix, Fig. S16). The Ikaros KO escape mutant exhibited severe anemia and an almost complete absence of early erythroid progenitors (SI Appendix, Figs. S15 and S16), congruent with previously published results suggesting defects in these areas (35, 36). While the dF4−/− bone marrow did not exhibit anemia as severe as that observed for the Ikaros null mutant, there were significant defects in early erythroid development in these mice. (Fig. 3F). Ikaros heterozygote bone marrow showed an intermediate but significant phenotype in the erythroid compartment, whereas Daedalus−/− bone marrow did not show a significant difference from wild type in terms of the numbers or percentages of these populations (Fig. 3F).
During hematopoietic development, opposing lineage programs often act to repress one another in order to firmly commit a cell to a single terminal differentiation pathway, including in initial specification of myeloerythroid and myelolymphoid lineages (37–39). Disruption of the lineage master regulators can lead to aberrant differentiation or to an increase in proliferation in upstream stem-cell populations (40, 41). Consistent with this observation, and with the increased expression of erythroid markers observed in Daedalus-deficient bone marrow, these cells displayed a higher potential to form erythroid colonies compared to cells from WT littermates (Fig. 3G). While the phenotype of the Daedalus−/− mice closely mirrored that of the Ikaros heterozygous animals in the lymphoid compartment, erythroid colony-forming potential significantly decreases in Ikzf1+/− bone marrow, contrasting with the increase in potential observed for Daedalus deletion.
To test the per-cell erythroid potential of precursors lacking the Daedalus locus, we sorted pure populations of hematopoietic progenitors and performed erythroid colony-forming assays (SI Appendix, Fig. S17). Consistent with our data from whole bone marrow, Daedalus-deficient progenitor cells displayed a slight increase in colony-forming potential relative to bone marrow from wild-type littermates, demonstrating that Daedalus not only impacts the frequency of lymphoid progenitors but also the capacity of individual hematopoietic progenitors to engage the lymphoid versus erythroid programs.
Our observation that Daedalus deletion phenocopies Ikzf1 deficiency in the lymphoid lineage, but leads to an increase in erythroid- and myeloid-associated genes, led us to hypothesize that Daedalus might regulate Ikaros biology in a lineage-specific manner. To test this, we performed qPCR to look at expression patterns of Daedalus and Ikzf1 in these populations and determined that, while Ikzf1 messenger RNA (mRNA) is expressed, unlike in lymphoid progenitors, Daedalus transcript does not appear to be present in early erythroid and myeloid progenitors (Fig. 3H). These data suggest that Daedalus plays a critical role in enforcing lymphoid-specific Ikaros expression and that, in the absence of Daedalus, erythroid-specific developmental programs become de-repressed. Deletion of a 3.5-kb region comprising a single predicted exon of a predicted Daedalus transcript was sufficient to reproduce the Daedalus null phenotype (SI Appendix, Fig. S18), suggesting that the defects observed are likely not due to compound effects of multiple regulatory elements in the larger region.
In order to test the effect of Daedalus locus deletion on Ikaros in nonlymphoid development, we performed intracellular Ikaros flow cytometry analysis in their respective early erythroid and myeloid progenitor populations. This analysis demonstrated that per-cell Ikaros protein expression was unaffected in Daedalus-deficient mice in erythroid (Fig. 3I) and myeloid progenitor populations unlike the Ikaros deficiency in the lymphoid lineage in these animals.
Discussion
In this study we identify a noncoding locus expressed during hematopoietic development whose absence leads to a significant reduction in lymphoid gene expression and lymphoid progenitor numbers via an Ikaros-dependent mechanism. This decrease in lymphoid lineage factors leads to a functional decrease in lymphoid lineage potential and a concomitant increase in development into erythroid lineages. Daedalus-deficient mice largely phenocopy Ikzf1 heterozygous null mice in the lymphoid compartment, but additionally display increased erythroid colony-forming potential (Fig. 3G). As evidence that these phenotypes require Daedalus to act as a modifier of Ikaros, epistasis studies using Ikzf1+/−Daedalus+/− mice found that compound heterozygote cells displayed decreased erythroid colony-forming potential compared to controls (SI Appendix, Fig. S14). Collectively, our findings demonstrate the following: 1) Ikaros is required for both normal erythroid and lymphoid development; 2) Daedalus expression and activity are restricted to the lymphoid lineage; 3) the absence of Daedalus yields dysregulated Ikaros expression that is lineage restricted to lymphoid progenitors, revealing an Ikaros-dependent erythromyeloid-lymphoid checkpoint.
While Ikaros protein levels correlate with active transcription at the Daedalus locus and deletion of Daedalus results in a significant reduction in Ikaros protein, transcription at Daedalus may simply mark regulatory element activity rather than the presence of a functional RNA. Although the transcript detected appears to be polyadenylated and spliced, GRO-seq in B-cell progenitors found bidirectional transcription occurring at the Daedalus locus, and a Hi-C analysis demonstrated interactions between this locus and that of Ikaros (SI Appendix, Figs. S6 and S7), consistent with a role for Daedalus as an enhancer, marked by a transcribed enhancer-associated RNA. It is therefore likely that this effect is mediated through direct DNA–DNA interactions and that the RNA produced is a by-product of active transcriptional machinery recruited to this region of euchromatin. Daedalus may act as a lineage-specific enhancer that modulates the isoform expression of Ikaros in the lymphoid lineage, and, in the absence of which, differential Ikaros isoform expression occurs. It is possible that these observed changes in isoform balance lead to destabilization of the Ikaros transcript, which may explain the severe reduction in Ikaros protein that we observe. A less parsimonious hypothesis could ascribe an incremental function to the transcripts as well as the enhancer or, as has been previously reported, a requirement for transcription or splicing of the transcript for cis-regulatory functions of the DNA element (42, 43).
These findings illustrate how noncoding loci can act as lineage-specific gatekeepers of transcription factors with broad lineage-specifying activities. While Ikaros is broadly expressed and required in both lymphoid and erythroid progenitors, the Daedalus locus is active only in early and lymphoid progenitors, where it stabilizes Ikaros activity and, in turn, lymphopoiesis. While further study is required to fully elucidate the molecular mechanism in the absence of Daedalus, Ikaros activity is destabilized in the lymphoid lineage, but preserved in erythroid progenitors, resulting in developmental diversion. The regulation of Ikaros by Daedalus represents a paradigm, demonstrating how noncoding loci can imbue transcription factors that exert a function in multiple lineages with lineage-specific activity that likely exists not just in hematopoiesis but more broadly throughout all developmental programs.
Materials and Methods
Mice.
Mice deficient in specific loci marked by noncoding RNAs were generated by injection of Cas9 RNA and chimeric guideRNAs flanking the locus (SI Appendix, Table S1) into C57/B6N embryos as previously reported (44). All mice were housed and maintained under pathogen-free conditions at Yale University animal facilities. Female and male littermates from heterozygous crossings between 4 and 12 wk old were used for all experiments unless otherwise indicated. All animal experimentation was performed in strict compliance with Yale Institutional Animal Care and Use Committee protocols.
We thank the Hilde Schjerven laboratory at the University of California, San Francisco (UCSF), for its generosity in providing us with samples from Ikaros DF4−/− mice (19) to use in our experiments.
Flow Cytometry Staining, Analysis, and Cell Sorting.
Cells were isolated from the tissues indicated from euthanized mice. For splenic and bone marrow analysis, red blood cells were first lysed using ACK lysis buffer. DAPI and eBioscience Fixable Viability Dye eFluor 780 were used for flow cytometric assessment of live/dead cells. Cells were stained in antibody mixtures containing 5% normal rat serum (NRS) at 4 °C for 30 to 60 min in phosphate-buffered saline (PBS) + 2% fetal calf serum (FCS). For intracellular staining, the eBioscience Foxp3/transcription factor staining kit (catalog #00–5523-00) was used according to the manufacturer’s specifications. Intracellular staining was performed with antibodies in 1x permeabilization wash buffer (eBioscience) with 2% NRS and antibodies for 60 min at room temperature or at 4 °C overnight. Antibodies used for Ikaros intracellular staining were monoclonal antibodies raised against different Ikaros epitopes: 2A9-mIkaros (eBioscience, catalog #14–5780-82) and 4E9 (ThermoFisher, catalog # MA5-28613). The epitope recognized by the 2A9 antibody or the immunogen used to produce it was not disclosed by the company. The immunogen for the 4A9 antibody was the recombinant C-terminal part of human Ikaros (encoded by exon 7), and the epitope was not determined (this antibody is cross-reactive with mouse and human Ikaros protein). All flow cytometric analyses and cell-sorting procedures were performed at the Yale Flow Cytometry core using BD LSRII cell analyzers and BD FACSAria cell sorters running FACSDiva software (BD Biosciences). Data analysis, table generation, and graphic rendering of flow cytometry plots was performed using FlowJo (version 10). SI Appendix, Table S3, lists all antibodies used for flow cytometry. Unless otherwise specified, lineage mixtures consist of antibodies against B220, CD4, CD8, CD11b, CD11c, CD19, Gr1, NK1.1, TCRgd, and Ter119 (for B-cell progenitor populations, CD19 and B220 were omitted; for specific erythroid progenitor populations, Ter119 was not included; other more tailored lineage mixtures are listed in the figure legends of respective figures).
ATAC-seq Preparation, Sequencing, and Analysis.
Purified populations of hematopoietic progenitors were sorted from WT and KO littermate mice. The ATAC-seq preparation was performed as outlined previously (33). Briefly, cells were washed once and resuspended in PBS, centrifuged again, and resuspended in 50 μL of lysis buffer at 4 °C. The cells were centrifuged at 500 x g for 10 min at 4 °C, the supernatant was removed, and the sample was resuspended in 5 μL of Nextera transposition reaction mix and incubated at 37 °C for 30 min. Library amplification was performed using Nextera PCR primers and NEBNext High-Fidelity 2x PCR Master Mix, and thermocycling was performed at 72 °C for 5 min for initial primer extension, followed by 98 °C for 30 s and then five cycles of 10 s at 98 °C, 30 s at 63 °C, and 1 min at 72 °C. The appropriate number of additional cycles was determined via qPCR using an aliquot of the PCR mix and iTaq Universal SYBR Green Supermix (BioRad). Additional amplification via PCR was performed for a number of cycles calculated from the qPCR amplification plot as previously described (33). Library concentration and quality was assessed using a Bioanalyzer, and sequencing was performed on an Illumina HiSEq. 2500. All sequencing was performed at the Yale Center for Genomic Analysis.
Reads were mapped using Bowtie2 (45) after trimming and quality checking was made using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were processed using SamTools (46), BEDTools (47), and Picard JAVA toolkit (broadinstitute.github.io/picard/); peaks were called using MACS2 (48) and visualized using IGV Browser (49).
Bone Marrow Chimeras.
Bone marrow was extracted from age matched Daedalus−/− mice and WT littermates, subjected to red cell lysis using ACK lysis buffer (ThermoFisher catalog #A1049201) and resuspended in PBS (Gibco). For noncompetitive chimeras, a total of 1 to 2 million bone marrow cells were injected into age-matched B6.SJL-Ptprca Pepcb/Boy (BoyJ) mice obtained from Jackson Laboratories that had been irradiated with a dose of 1,000 cGy 3 to 5 h previously using an X-ray irradiator. For mixed chimeras, bone marrow was harvested from BoyJ mice obtained from Jackson Laboratories, subjected to red-blood-cell lysis using ACK buffer, and resuspended in PBS. Bone marrow from BoyJ competitor and WT or KO experimental mice was mixed at a 1:1 ratio and 2 million total bone marrow cells were injected into B6.BoyJ F1 mice deriving from a cross between C57/B6N and BoyJ mice, both derived from Jackson Laboratories, that had been irradiated with a dose of 1,000 cGy 3 to 5 h previously using an X-ray irradiator. Injected mice were cohoused and treated with a course of antibiotic food (sulfamethoxazole and trimethoprim) in order to limit potential pathogen infection. Bone marrow was harvested from these mice after 16 wk and used for flow cytometric analysis.
RNA Sequencing.
Purified populations of hematopoietic progenitors were sorted from WT and KO littermate mice as described above. Purified RNA was obtained using the Qiagen RNeasy Micro Kit or Zymo-research Direct-zol RNA Microprep Kit. Libraries for RNA sequencing were generated using Illumina TruSeq mRNA-Seq Sample Prep Kits. Library concentration and quality were assessed using a Bioanalyzer, and sequencing was performed on an Illumina HiSEq. 2500. All sequencing was performed at the Yale Center for Genomic Analysis.
Analysis was performed using the Tuxedo suite: splice junction mapping was performed using TopHat, and transcript quantification was performed using Cufflinks (50). Downstream analysis was performed in R using CummeRbund analysis software (http://compbio.mit.edu/cummeRbund/). Comparison pseudoalignment analysis was also performed using Kallisto and Sleuth (51).
RNA Extraction, Complementary DNA Synthesis, and Quantitative RT-PCR.
Purified populations of hematopoietic progenitors were sorted from WT and KO littermate mice as described above. Purified RNA was obtained using Qiagen RNeasy Micro Kit or TRIzol (ThermoFisher catalog #15596026) purification per manufacturer’s specifications. Complementary DNA synthesis was performed using Maxima RT minus reverse transcriptase according to the manufacturer’s specifications. Quantitative real-time PCR was performed using iTaq Universal SYBR Green Supermix (BioRad) and CFX96 Touch Deep Well Real-Time PCR Detection Systems. Primers used in this paper are outlined in SI Appendix, Table S2.
Hi-C Library Preparation and Analysis of Hi-C Data.
Hi-C libraries of pro-B cells were prepared, and Hi-C data were analyzed as described previously (52). In short, bone marrow pro-B cells were cultured for 5 d, and Hi-C libraries were prepared from 2 × 107 cells as described in detail (53). Sequencing was performed using the Illumina NextSeq system with a read length of 75 nucleotides in the paired-end mode, according to the manufacturer’s guidelines. Reads were truncated, aligned, and filtered using the HiCUP pipeline version 0.5.10 with the scorediff parameter set to “10.” The data of two Hi-C experiments were merged to produce contact matrix files with the Juicer tools 1.8.9. The HiCCUPS algorithm from the Juicer tools was used to call intrachromosomal loops. The contact matrix was calculated and visualized with Juicebox.
GRO-seq Analysis.
GRO-seq libraries were prepared, and GRO-seq data were analyzed as described previously (54). In short, bone marrow pro-B cells of Rag2−/− mice were cultured for 5 d, and nuclei from 10 × 107 cells were prepared and subjected to nuclear run-on. The reaction was stopped, and RNA was isolated and fragmented, followed by isolation and sequencing of the nascent transcripts.
B-Cell Precursor and Erythroid Colony Assays.
Total bone marrow was extracted from euthanized mice, red blood cells were lysed using ACK lysis buffer, and cell numbers were normalized. Colony assays were performed using Stem Cell MethoCult Medium according to the manufacturer’s specifications. Cells were resuspended in IMDM + 2% FCS and mixed at a ratio of 1:10 with Stem Cell MethoCult Medium for erythroid (including EPO–M3434, M3436) and B-cell progenitor (M3630) colony formation. Cells in colony medium were mixed thoroughly and then plated on 35-mm plates, incubated under controlled conditions at 37 °C for 7 to 12 d, and analyzed using a microscope as described in the manufacturer’s recommendations. For sorted cell colony assays, purified populations of hematopoietic progenitors were sorted from WT and KO littermate mice, resuspended at equal volumes in IMDM + 2% FCS, and mixed with Stem Cell MethoCult Medium; plating procedure was done according to the manufacturer’s recommendations as described above.
Single-Cell RNA Sequencing Analysis.
Purified populations of hematopoietic progenitors (lineage negative, cKit positive cells) were sorted from WT and KO littermate mice. Cells were sorted into PBS + 0.01% bovine serum albumin (ThermoFisher UltraPure BSA Cat#:AM2616), washed twice, and counted using a hemocytometer, and counts were checked using a Countess II Automated Cell Counter (ThermoFisher). Single-cell libraries were generated using a 10x Genomics Chromium Controller instrument and associated kits. Sequencing was performed on an Illumina HiSEq. 4000.
Demultiplexing, genome alignment, and gene counting were performed using 10x Genomics Cell Ranger software. Downstream analysis was performed in R using the Seurat R toolkit for single-cell genomics (55). In some cases, the ALRA algorithm was used to improve resolution of distinct cell populations; ALRA is an algorithm for imputation of technical zeroes based on low-rank approximation that emphasizes preserving biological zeroes from nonexpressed genes (56).
Supplementary Material
Acknowledgments
This work was supported by NIH National Institute of Allergy and Infectious Diseases (NIAID) Grant 1R21AI110776-01 (to A.W.) and the Howard Hughes Medical Institute (HHMI) (C.C.D.H. and R.A.F.). C.C.D.H. was supported by a Gruber Science Fellowship from the Yale Graduate School of Arts and Sciences and by an HHMI International Student Fellowship. W.B. was supported by a CRI Irvington Postdoctoral Fellowship and a K22 (NIAID 5K22AI141758). We thank Hilde Schjerven and her laboratory at UCSF for providing us with samples from Ikaros DF4−/− mice to use in our experiments.
Footnotes
Competing interest statement: R.A.F. is a consultant for GSK and Zai Lab Ltd.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918062118/-/DCSupplemental.
Data Availability.
All sequencing data for this study was uploaded to the Gene Expression Omnibus (GEO) repository under accession superseries GSE153879 unless otherwise specified. Hi-C sequencing data for this study is available at the GEO repository under the accession nos. GSM4350200 and GSM4350198 (in GSE140975) and was published in Hill et al. (52).
References
- 1.Boehm T., Design principles of adaptive immune systems. Nat. Rev. Immunol. 11, 307–317 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Greenberg P. D., Riddell S. R., Deficient cellular immunity: Finding and fixing the defects. Science 285, 546–551 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Buckley R. H., Primary cellular immunodeficiencies. J. Allergy Clin. Immunol. 109, 747–757 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Buckley R. H., Primary immunodeficiency diseases due to defects in lymphocytes. N. Engl. J. Med. 343, 1313–1324 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Cobaleda C., Schebesta A., Delogu A., Busslinger M., Pax5: The guardian of B cell identity and function. Nat. Immunol. 8, 463–470 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Ramírez J., Lukin K., Hagman J., From hematopoietic progenitors to B cells: Mechanisms of lineage restriction and commitment. Curr. Opin. Immunol. 22, 177–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye M., Graf T., Early decisions in lymphoid development. Curr. Opin. Immunol. 19, 123–128 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Bryder D., Sigvardsson M., Shaping up a lineage: Lessons from B lymphopoesis. Curr. Opin. Immunol. 22, 148–153 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Deniz E., Erman B., Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genomics 17, 135–143 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ransohoff J. D., Wei Y., Khavari P. A., The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez-Dominguez J. R., Lodish H. F., Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 130, 1965–1975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paralkar V. R., Weiss M. J., Long noncoding RNAs in biology and hematopoiesis. Blood 121, 4842–4846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satpathy A. T., Chang H. Y., Long noncoding RNA in hematopoiesis and immunity. Immunity 42, 792–804 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Rothenberg E. V., Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32, 283–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M., Rinn J. L., Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K. C., Chang H. Y., Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love P. E., Bhandoola A., Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 11, 469–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz B. A., et al. , Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 178, 2008–2017 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Schjerven H., et al. , Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 14, 1073–1083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J. H., et al. , Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Georgopoulos K., et al. , The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Marke R., van Leeuwen F. N., Scheijen B., The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica 103, 565–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng S. Y., Yoshida T., Georgopoulos K., Ikaros and chromatin regulation in early hematopoiesis. Curr. Opin. Immunol. 19, 116–122 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Tinsley K. W., et al. , Ikaros is required to survive positive selection and to maintain clonal diversity during T-cell development in the thymus. Blood 122, 2358–2368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsiö J. M., Tarchini B., Cayouette M., Livesey F. J., Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 110, E716–E725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwickert T. A., et al. , Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15, 283–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siatecka M., Bieker J. J., The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 118, 2044–2054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tumburu L., Thein S. L., Genetic control of erythropoiesis. Curr. Opin. Hematol. 24, 173–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M., et al. , GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells 18, 921–933 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Saleque S., Cameron S., Orkin S. H., The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16, 301–306 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida T., Georgopoulos K., Ikaros fingers on lymphocyte differentiation. Int. J. Hematol. 100, 220–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J., et al. , Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345–355 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu V. W., Scadden D. T., Heterogeneity of the bone marrow niche. Curr. Opin. Hematol. 23, 331–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichogiannopoulou A., Trevisan M., Neben S., Friedrich C., Georgopoulos K., Defects in hemopoietic stem cell activity in Ikaros mutant mice. J. Exp. Med. 190, 1201–1214 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papathanasiou P., et al. , Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 19, 131–144 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Arinobu Y., et al. , Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 1, 416–427 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Nerlov C., Querfurth E., Kulessa H., Graf T., GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95, 2543–2551 (2000). [PubMed] [Google Scholar]
- 39.Rekhtman N., Radparvar F., Evans T., Skoultchi A. I., Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes Dev. 13, 1398–1411 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Galen P., et al. , Reduced lymphoid lineage priming promotes human hematopoietic stem cell expansion. Cell Stem Cell 14, 94–106 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T., Ng S. Y., Georgopoulos K., Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr. Opin. Immunol. 22, 154–160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engreitz J. M., et al. , Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tippens N. D., et al. , Transcription imparts architecture, function and logic to enhancer units. Nat. Genet. 52, 1067–1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., et al. , Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Hill L., et al. , Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584, 142–147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bönelt P., et al. , Precocious expression of Blimp1 in B cells causes autoimmune disease with increased self-reactive plasma cells. EMBO J. 38, e100010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linderman G. C., Zhao J., Kluger Y., Zero-preserving imputation of scRNA-seq data using low-rank approximation. bioRxiv [Preprint] (2020). 10.1101/397588 (Accessed 4 January 2021). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data for this study was uploaded to the Gene Expression Omnibus (GEO) repository under accession superseries GSE153879 unless otherwise specified. Hi-C sequencing data for this study is available at the GEO repository under the accession nos. GSM4350200 and GSM4350198 (in GSE140975) and was published in Hill et al. (52).