Abstract

Many physiological and pathophysiological processes, including Mycobacterium tuberculosis (Mtb) cell division, may involve fuzzy membrane association by proteins via intrinsically disordered regions. The fuzziness is extreme when the conformation and pose of the bound protein and the composition of the proximal lipids are all highly dynamic. Here, we tackled the challenge in characterizing the extreme fuzzy membrane association of the disordered, cytoplasmic N-terminal region (NT) of ChiZ, an Mtb divisome protein, by combining solution and solid-state NMR spectroscopy and molecular dynamics simulations. While membrane-associated NT does not gain any secondary structure, its interactions with lipids are not random, but formed largely by Arg residues predominantly in the second, conserved half of the NT sequence. As NT frolics on the membrane, lipids quickly redistribute, with acidic lipids, relative to zwitterionic lipids, preferentially taking up Arg-proximal positions. The asymmetric engagement of NT arises partly from competition between acidic lipids and acidic residues, all in the first half of NT, for Arg interactions. This asymmetry is accentuated by membrane insertion of the downstream transmembrane helix. This type of semispecific molecular recognition may be a general mechanism by which disordered proteins target membranes.
Keywords: intrinsically disordered proteins, NMR spectroscopy, fuzzy association, molecular dynamics simulations, acidic lipids, membrane association, semispecific molecular recognition
Introduction
Upon binding to their partners, intrinsically disordered proteins span a continuum in the extent of order, from fully folded to partially ordered to fully disordered. The complexes in which disordered proteins remain disordered are termed “fuzzy”. The fuzziness reaches an extreme when the partners are another disordered protein or nucleic acid and both subunits remain fully disordered.1−5 A third class of partners for disordered proteins comprise membranes.6−9 In a well-characterized case, membrane association of α-synuclein is accompanied by the formation of amphipathic α-helices.9 A large fraction of transmembrane and peripheral membrane proteins contain disordered regions,10 but there is little knowledge on any extreme fuzzy complexes with membranes. Here, we tackle the challenge of characterizing the extreme fuzzy membrane association of the disordered cytoplasmic N-terminal region of the transmembrane protein ChiZ, a member of the Mycobacterium tuberculosis (Mtb) divisome complex, by combining solution and solid-state NMR spectroscopy with molecular dynamics (MD) simulations.
Many disordered proteins are enriched in charged residues,11 and interactions between oppositely charged residues are crucial features of extreme fuzzy complexes between disordered proteins.1,2 Likewise, the interactions between basic residues of proteins and acidic phosphate groups of nucleic acids are crucial for their high-affinity, fuzzy association.3−5 The inner leaflet of the plasma membrane is highly acidic due to the asymmetric distribution of charged lipids, including phosphatidylserine, phosphatidylinositol, and the latter’s phosphorylated variants,12 and thus forms a target for polybasic proteins, including signaling molecules.7 The Mtb inner membrane contains an abundance of acidic lipids, with phosphatidylglycerol, cardiolipin, phosphatidylinositol, and phosphatidylinositol mannosides present at roughly a 7:3 ratio to the neutral phosphatidylethanolamine (based on the composition in Mycobacterium smegmatis, a nonpathogenic model13). This acidic surface provides ample opportunities for association by ChiZ and other Mtb divisome proteins with disordered cytoplasmic regions that are enriched in basic residues (Figure 1a and Figure S1).
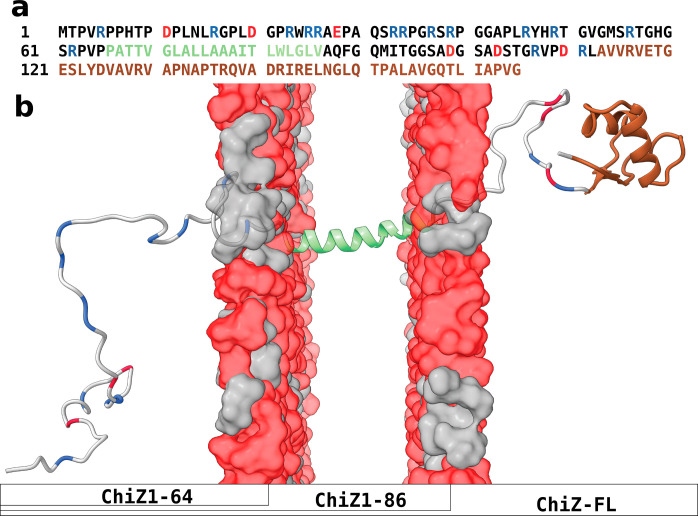
Figure 1.
Sequence and structure of ChiZ. (a) Amino-acid sequence. Disordered regions, transmembrane helix, and LysM domain are indicated by black, green, and brown letters, respectively; acidic and Arg residues in disordered regions are shown as red and blue letters, respectively. (b) Disposition of different regions or domains with respect to the membrane. Colors match those in panel (a). The compositions of the three ChiZ constructs are also indicated.
Very few fuzzy complexes between disordered proteins and membranes have been characterized at the residue level. The most intensely studied protein in this regard is α-synuclein, which forms amphipathic α-helices in the first 100 residues upon membrane association.9 α-Synuclein preferentially binds to vesicles containing acidic lipids,14 but membrane curvature also plays an important role. A disease-associated charge reversal, E46K, strengthened membrane binding but weakened selectivity for membrane curvature.15 Conversely, increasing negative charges in the C-terminal tail weakened membrane association but enhanced curvature selectivity.16 The entire 100 residues apparently do not bind to the same vesicle all the time; while the first 30 or so residues stably bind to a vesicle, the remaining segments can dissociate and even bind to a different vesicle, leading to vesicle clustering.17 Even when the 100 residues were membrane-bound, MD simulations showed significant conformational heterogeneity for α-synuclein, although the helices remained intact.18 By contrast, no information is available for how a basic region of the Wiscott–Aldritch Syndrome protein interacts with acidic lipids of the plasma membrane, even though the fuzzy interaction activates this protein for stimulating Arp2/3-mediated initiation of actin polymerization.6 Likewise, the disordered intracellular region of the prolactin receptor, known to interact with inner leaflet-specific lipids via conserved basic clusters and hydrophobic motifs,8 was modeled without considering membrane association due to lack of information.19
Here, we report residue-level characterization of the fuzzy association of the ChiZ 64-residue N-terminal region (NT) with acidic membranes. In full-length ChiZ (ChiZ-FL), NT is followed by a 21-residue transmembrane helix; on the periplasmic side, a C-terminal LysM domain (residues 113–165) is connected to the transmembrane helix by a 26-residue linker (Figure 1). In a previous study,20 we showed that, in solution, the NT-only construct ChiZ1-64 is fully disordered without detectable α-helix or β-sheet formation, but with polyproline II (PPII) formation and intramolecular interactions including salt bridges concentrated in the first half of the sequence. Here, we investigated NT-membrane association by solution NMR in the context of ChiZ1-64 and by solid-state NMR on both ChiZ1-64 and ChiZ-FL. In addition, extensive MD simulations of these two constructs and ChiZ1-86 associating with membranes (Figure 1b) were carried out, encompassing 16–20 trajectories and 20.6–38 μs of simulation time for each system. The conformation and pose of NT and the composition of the proximal lipids are all highly dynamic, making their association extremely fuzzy.
Results
ChiZ1-64 Associates with Acidic Membranes but Terminal Residues Remain Free
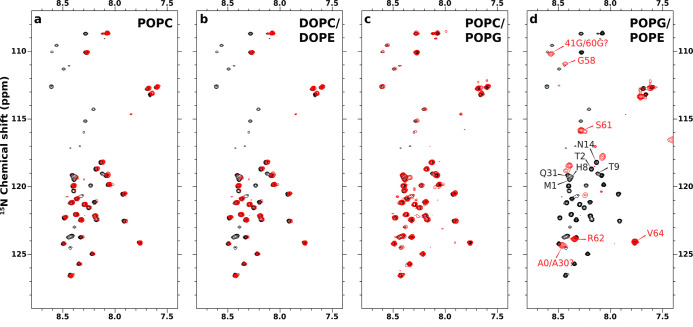
1H–15N HSQC spectra of ChiZ1-64 in the presence of liposomes with four lipid compositions were acquired to assess membrane association (Figure 2). Association is indicated by loss of crosspeaks due to line broadening from slow tumbling when a residue binds to a liposome. Relative to the 1H–15N HSQC spectrum of ChiZ1-64 in solution (hereafter “unbound” ChiZ1-64),20 no major loss of NMR signals was detected when the liposomes contained POPC only (Figure 2a), 4:1 DOPC:DOPE (Figure 2b), or 4:1 POPC:POPG (Figure 2c). These spectra show that ChiZ1-64 does not associate significantly with the neutral POPC and DOPC:DOPE membranes, or even with a membrane containing 20% acidic lipids. In contrast, the HSQC spectrum in the presence of 7:3 POPG:POPE liposomes, which mimic the charge composition of Mtb membranes,13 shows that the crosspeaks of most of the residues are broadened beyond detection (Figure 2d). Of the remaining crosspeaks, based on the overlap with the counterparts in unbound ChiZ1-64, assignments could be made for Gly58, Ser61, Arg62, and Val64, all located at the C-terminus. Due to slight shifts, the few other crosspeaks could not be unambiguously assigned, but appear to be N-terminal residues, including Met1, Thr2, His8, Thr9, and Asn14 as well as possibly Gln31. So, the HSQC spectra demonstrate that ChiZ1-64 associates with membranes containing 70% acidic lipids, but residues at the two termini remain free.
Figure 2.
Solution 15N–1H HSQC spectra of ChiZ1-64 in the presence and absence of liposomes. ChiZ1-64 spectra without and with liposomes are shown as black and red contours, respectively. ChiZ1-64 was mixed with (a) POPC, (b) DOPC:DOPE (4:1 molar ratio), (c) POPC:POPG (4:1 molar ratio), and (d) POPG:POPE (7:3 molar ratio) liposomes at a protein to lipid ratio of 1:100 in 20 mM phosphate buffer (pH 7.0) containing 25 mM NaCl. All spectra were collected with 100 μM protein at 25 °C.
Membrane Association Is Fuzzy But There Is Hint for a Subpopulation with a Stable Binding Motif
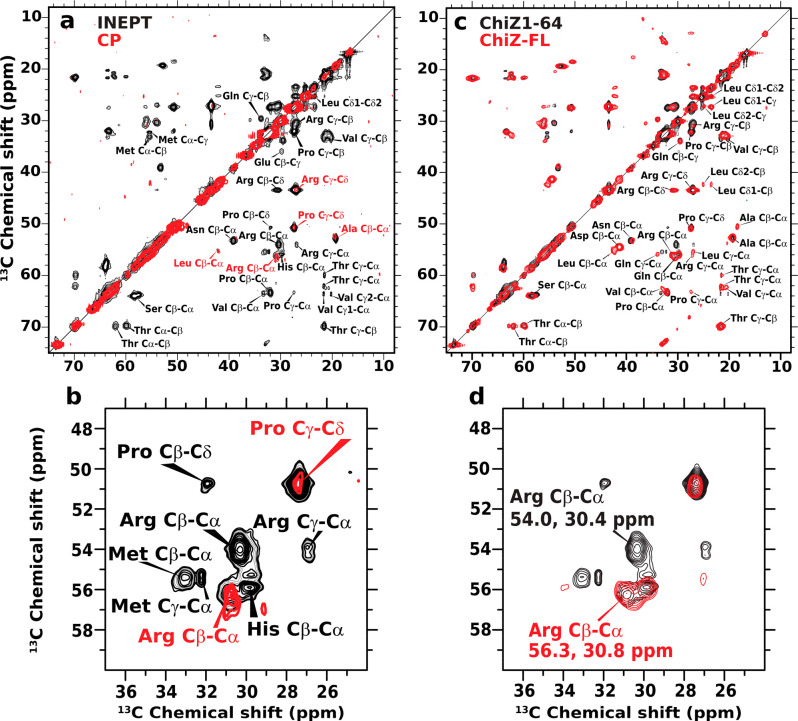
The solution NMR HSQC experiment is useful for indicating membrane association, but the loss of crosspeaks precludes further characterization of the association. We thus turned to magic-angle spinning (MAS) 13C solid-state NMR experiments: insensitive nuclei enhanced by polarization transfer (INEPT) and cross-polarization (CP). The former is sensitive to dynamic sites, whereas the latter is sensitive to static sites. The INEPT spectrum of ChiZ1-64 bound to POPG:POPE liposomes shows an abundance of crosspeaks (Figure 3a, black contours), indicating that most of the residues remain highly dynamic, and hence, the membrane association is extremely fuzzy. In fact, the dynamics apparently rival those in unbound ChiZ1-64 and result in the same, undispersed chemical shifts for a given pair of carbon–carbon sites (e.g., Arg Cβ-Cδ) at different positions along the amino-acid sequence. This spectral overlap is a strong indication that ChiZ1-64 does not fold upon membrane association and allowed the assignment of INEPT crosspeaks to types of carbon–carbon sites but not to specific residue positions.
Figure 3.
Solid-state NMR data of ChiZ1-64 bound to and ChiZ-FL reconstituted into POPG:POPE liposomes. The protein to lipid ratios were 1:50 and 1:80, respectively, for the two constructs. (a) 13C–13C correlation spectra of ChiZ1-64 using INEPT and CP magnetization transfer, shown in black and red, respectively. The INEPT spectrum was acquired with 512 transients and 128 scans per transient. For the CP spectrum, the PARIS pulse sequence was used with 100 ms mixing time and 400 transients with 268 scans per transient. (b) Zoom into a region centered around the Arg Cβ–Cα crosspeaks. (c) Comparison of ChiZ1-64 and ChiZ-FL INEPT spectra, shown in black and red, respectively. (d) Zoom into the region centered around the Arg Cβ–Cα crosspeaks. All experiments were carried out at 25 °C and at a 12.2 kHz spinning rate.
Both in unbound ChiZ1-6420 and in the INEPT spectrum of POPG:POPE-bound ChiZ1-64, the Arg Cα–Cβ pair has two distinct crosspeaks, one at (56.3, 30.8) (chemical shifts in ppm), and the other at (54.0, 30.4) (Figure 3b, black contours). With the help of MD simulations (Figure S2), we were able to recognize that, in unbound ChiZ1-64, these two crosspeaks were assigned to Arg residues with one distinction: whether the succeeding residue along the sequence is a Pro. We refer to these two groups of residues as RP Arg and non-RP Arg, respectively. The (56.3, 30.8) crosspeak belongs to nine non-RP Arg residues, whereas the (54.0, 30.4) crosspeak belongs to the four RP Arg residues: Arg5, Arg34, Arg39, and Arg62.
Corroborating the INEPT result that most residues in POPG:POPE-bound ChiZ1-64 are dynamic, the CP spectrum shows only a few crosspeaks (Figure 3a, red contours). They largely overlap with crosspeaks in the INEPT spectrum and accordingly can be assigned to Arg Cα–Cβ and Cγ–Cδ, Pro Cγ–Cδ, Ala Cα–Cβ, and Leu Cα–Cβ. Interestingly, Arg Cα–Cβ appears as a single crosspeak in the CP spectrum (Figure 3b, red contours); its overlap with the non-RP crosspeak in the INEPT spectrum suggests that this most prominent CP crosspeak comes from one or more non-RP Arg residues. Rather than appearing at isolated positions along the sequence, it is far more likely that the apparently static residues detected by CP form a contiguous stretch for overall stability. There is only a single such stretch, A43PLR46, and the Arg involved is indeed non-RP. The CP experiment thus hints at a stable motif, A43PLR46, that may form in a subpopulation of POPG:POPE-bound ChiZ1-64. Our MD simulations sampled a structure for this putative stable binding motif (Figure S3).
We further performed INEPT on ChiZ-FL reconstituted into POPG:POPE liposomes to determine whether NT remained dynamic when the protein was tethered to the membrane via the transmembrane helix. The INEPT spectra of ChiZ1-64 (black contours) and ChiZ-FL (red contours) essentially overlap (Figure 3c), showing that, in the context of the full-length protein, NT also does not fold upon membrane association. However, one clear distinction emerges for the Arg Cα–Cβ crosspeaks (Figure 3d). Whereas ChiZ1-64 Arg Cα–Cβ has both an RP crosspeak at (54.0, 30.4) and a non-RP crosspeak at (56.3, 30.8) (red contours), only the latter crosspeak is observed in the ChiZ-FL INEPT spectrum. The disappearance of the RP crosspeak means that the corresponding Arg residues (more precisely, their Cα and Cβ atoms) become more static upon membrane insertion of the transmembrane helix. Of the four RP Arg residues, rigidification is expected for Arg62, which is right next to the transmembrane helix in ChiZ-FL. The most N-terminal Arg residue, Arg5, could become static because the N-terminal His-tag present in ChiZ-FL (but absent in ChiZ1-64) might attach to the membrane.21 That still leaves two RP residues, Arg34 and Arg39, in the midsection unaccounted for. As the data from the next experiment indicate, even in the ChiZ1-64 construct, these two residues, along with other midsection Arg residues, interact with lipids, and the resulting loss in dynamics potentially prevented their detection by INEPT, but the loss in dynamics was incomplete so Arg34 and Arg39 were not detectable by CP either. Upon membrane insertion of the transmembrane helix, Arg34 and Arg39 in ChiZ-FL may interact more strongly with lipids and further lose some dynamics (see below), thereby evading detection by INEPT.
Arg Residues Engage in Direct Interactions with Lipid Headgroups
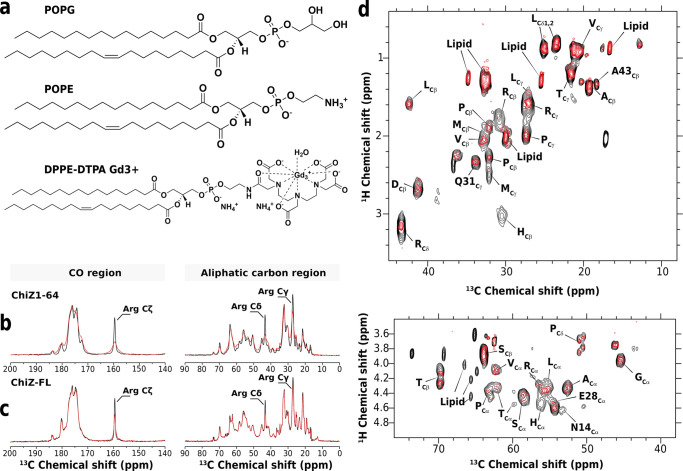
We used paramagnetic relaxation enhancement to identify NT residues that interact with membranes. By doping liposomes with lipids chelating the paramagnetic ion Gd3+ (Figure 4a), neighboring ChiZ nuclei would relax much faster due to increased dipolar interactions with the spin label, resulting in line broadening and loss of signal intensity. One-dimensional 13C direct-excitation spectra show that resonances of Arg side-chain carbons experience significant intensity loss in the presence of Gd3+-chelated lipids, while other resonances are largely unaffected (Figure 4b, c). This observation applies to both ChiZ1-64 bound to POPG:POPE liposomes and ChiZ-FL reconstituted into these liposomes and reveals that Arg residues are the major players in mediating NT association with membranes.
Figure 4.
Paramagnetic relaxation enhancement data of ChiZ1-64 bound to and ChiZ-FL reconstituted into POPG:POPE liposomes. The protein to lipid ratios were 1:50 and 1:80, and Gd3+-chelated lipids were at 2 and 1%, respectively, for the two constructs. (a) Molecular structure of POPG, POPG, and PE-DTPA (Gd). One-dimensional 13C direct-excitation spectra of (b) ChiZ1-64 and (c) ChiZ-FL in the absence (black) and presence (red) of Gd3+-chelated lipids. 128 scans were collected on each sample. (d) 1H–13C INEPT-based spectra of ChiZ-FL in the absence (black) and presence (red) of Gd3+-chelated lipids. The aliphatic and α-carbon regions are shown in two panels. All experiments were carried out at 25 °C and at a 12.2 kHz spinning speed.
To characterize NT-lipid interactions in more detail, we investigated paramagnetic relaxation enhancement in reconstituted ChiZ-FL by 1H–13C correlation experiments with INEPT magnetization transfer. We took advantage of the spectral overlap between the solid-state INEPT and solution HSQC spectra (Figure S4) and assigned the INEPT crosspeaks to types of carbon sites (e.g., Val Cγ; Figure 4d). In a few cases, assignment could be made to specific residues, either because there was only a single residue of a given type (Asn14, Glu28, or Gln31) in NT, or because it was the only NT residue of a given type (Ala43) that preceded a Pro. A comparison of the 1H–13C correlation spectra between ChiZ-FL samples without (black contours) and with (red contours) the Gd3+ spin label provides a global picture of the NT residues that are in contact with lipid headgroups. An immediate observation is that NT experiences a general loss in 1H–13C signals in the presence of Gd3+. As the relaxation enhancement effect of the spin label may reach protons as far as 20 to 25 Å away, we interpret the general loss in signal as an indication that the spin label senses the entire NT sequence. In other words, when ChiZ-FL is reconstituted into POPG:POPE liposomes, no portion of NT appears to dissociate constantly from membranes.
In the aliphatic region of the 1H–13C correlation spectra, upon adding the spin label, Arg Cδ sites experience the strongest loss of intensity. In addition, the His Cβ crosspeak disappears altogether. The considerable intensity loss for Arg side chains indicates direct interaction with lipids; the signal disappearance of His side chains likely can be attributed to membrane attachment of the N-terminal His-tag. Similar effects of the spin label on Arg and His residues are also observed in the Cα region of the spectra.
Together, the data from the different NMR experiments indicate that Arg residues away from the NT termini are the major mediators of the association with acidic membranes. The association is extremely fuzzy as NT remains highly dynamic and does not fold, apart from some hint for a subpopulation with A43PLR46 as a stable binding motif.
NT is Anchored to Membranes by Arg Residues in the Midsection
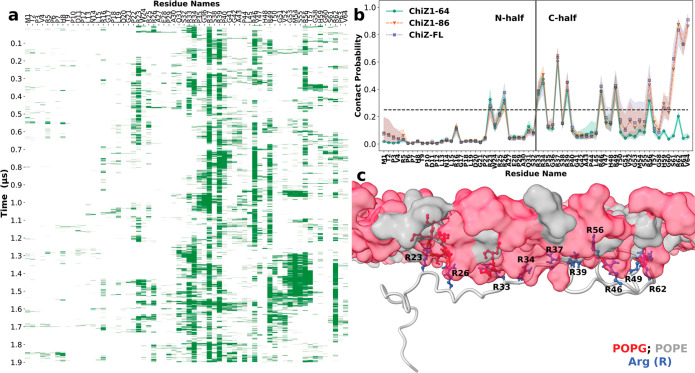
As is clear from the foregoing presentation, our MD simulations were crucial in the interpretation of the NMR data. More importantly, the simulations reveal atomistic details about the extreme fuzzy membrane association of NT, which we now describe. As a first step, we calculated the probabilities that individual NT residues in the three ChiZ constructs are in contact with POPG:POPE membranes (i.e., <3.5 Å between heavy atoms; Figure 5a, b). We denote the membrane-contact probability of residue i by Ci. In ChiZ1-64, the residues that contact membranes with relatively high probabilities (i.e., Ci > 0.25, indicated by a horizontal dashed line in Figure 5b) are all Arg residues, in accord with the paramagnetic relaxation enhancement data in Figure 4b. There are nine such Arg residues, including Arg23, Arg26, Arg33, Arg34, Arg37, Arg39, Arg46, Arg49, and Arg56. In complete agreement with the 1H–15N HSQC spectra of ChiZ1-64 reported in Figure 2d, the extreme N- and C-terminal residues do not frequently form contacts with POPG:POPE membranes. Indeed, except for Arg56, the frequent-contact Arg residues are limited to the midsection of NT with Arg37 having the highest contact probability at 60%. Furthermore, the distribution of the frequent-contact Arg residues along the sequence gives the first indication that the two halves of NT (denoted as N- and C-half) are not equal in membrane association, with C-half playing a more prominent role. We will further explore this asymmetry below. A representative snapshot illustrating the membrane anchoring of NT by midsection Arg residues is shown in Figure 5c.
Figure 5.
Membrane-contact probabilities of NT residues. (a) Contact status of individual residues in snapshots along a 1.9-μs molecular dynamics trajectory of ChiZ1-64. Green bars and blanks indicate that a residue either is or is not in contact with the membrane. (b) Membrane-contact probabilities of NT residues in the three constructs. The shaded bands represent standard deviations among the snapshots analyzed. The extreme N-terminal residues that show high membrane-contact probabilities in ChiZ1-86 and ChiZ-FL are from two MD trajectories where Met1 was started as nearly embedded in the headgroup region, mimicking in a small way potential membrane attachment of the N-terminal His-tag; Met1 eventually dissociated from the membrane. For these two constructs, residues 49–56 penetrated into the membrane in two trajectories. These events led to relatively large standard deviations in membrane-contact probability. (c) A snapshot of ChiZ1-64 at 1.56 μs from the same trajectory as in (a), illustrating the membrane anchoring of NT by Arg residues in the midsection.
Comparing the membrane-contact probabilities of ChiZ1-64 with those of the longer constructs (Figure 5b), the most obvious effect of membrane tethering of the NT C-terminus is the near 100% contact probabilities of the three most C-terminal residues, R62PV64. The effect of the membrane tethering is apparent up to residue Thr50, and small increases in membrane-contact probabilities are seen all the way to the start of C-half. These changes accentuate the asymmetry between the two halves of NT in membrane association. Additional evidence below will show that the effect of the membrane tethering even propagates into N-half. The resulting further loss in dynamics for Arg34 and Arg39 in ChiZ-FL explains why they, along with Arg5 and Arg62, are not detectable by INEPT (Figure 3b).
Lastly, we note that while the membrane-contact probabilities of NT residues are very similar between ChiZ1-86 and ChiZ-FL, there are subtle differences. A majority (11 out of 16) of the frequent-contact residues have slightly higher contact probabilities in ChiZ-FL than in ChiZ1-86 (Figure S5a). This difference will also be further addressed below.
Competition between Acidic Residues and POPG Contributes to Asymmetry between the Two Halves of NT in Membrane Association
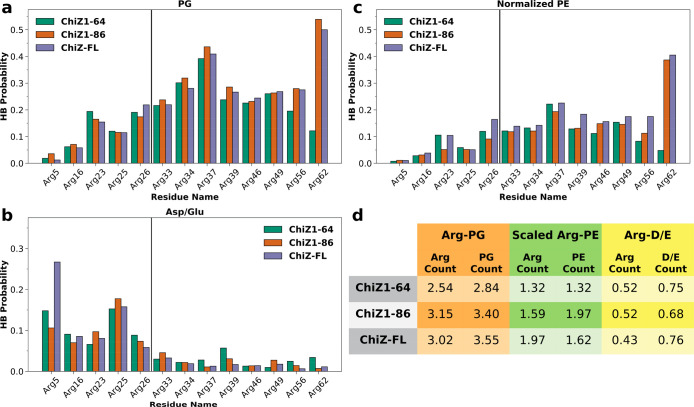
The scant involvement in membrane association by Arg residues in ChiZ1-64 N-half stands in contrast to their deep involvement in intramolecular salt bridges when ChiZ1-64 is unbound20 (Figure S6). The latter result has been explained by the fact that the salt-bridge partners, i.e., acidic residues (Asp11, Asp20, and Glu28), are all in N-half. Apparently, acidic residues and acidic lipids compete for interactions with Arg residues; when Arg residues (in particular, in N-half) engage in intramolecular interactions with acidic residues, they lose the ability to engage in intermolecular interactions with POPG lipids. Indeed, with the partners being either POPG lipids or acidic residues, the profiles of hydrogen bonding probabilities of Arg residues are mirror images of each other, with POPG lipids favored by C-half residues whereas acidic residues favored by N-half residues (Figure 6a, b).
Figure 6.
Hydrogen bonding probabilities of NT Arg residues. (a) Hydrogen bonding probabilities of Arg residues with POPG lipids. (b) Hydrogen bonding probabilities of Arg residues with Asp and Glu residues. (c) Hydrogen bonding probabilities of Arg residues with POPE lipids, scaled up by a factor of 7/3. (d) Average number of Arg residues that hydrogen bond with a particular type of partner at a given moment, and the counterpart for the partner hydrogen bonding with Arg residues. The partners are either POPG or POPE lipids or Asp and Glu residues.
Expectedly, the probabilities that Arg residues hydrogen bond with POPG lipids (Figure 6a) track closely the corresponding membrane-contact probabilities (Figure 5b). Indeed, these two sets of data are highly correlated, with a slope of approximately 0.62 (Figure S7). In other words, each time an Arg residue comes into contact with membranes, there is a 2/3 chance that it forms hydrogen bonds with POPG lipids, therefore indicating that Arg-POPG hydrogen bonds are the main driving force for membrane association. Non-Arg residues in ChiZ1-64 have minimal probability for hydrogen bonding with POPG (Figure S8a). Seven of the nine Arg residues that most frequently hydrogen bond with POPG lipids are in C-half.
In contrast, Arg residues that frequently hydrogen bond with acidic residues are all in N-half (Figure 6b). The most prevalent of these Arg residues are Arg5 and Arg25. The prevalence of Arg5 can be attributed to its proximity to Asp11 along the sequence, while that of Arg25 to its proximity to both Asp20 and Glu28. The frequent hydrogen bonding with Asp20 and Glu28 explains why Arg25 has lower probabilities than both of its neighbors, Arg23 and Arg26, for hydrogen bonding with POPG lipids and for membrane contact. Compared to unbound ChiZ1-64 (Figure S6), Arg5 and Arg16 near the N-terminus have increased probabilities of hydrogen bonding with acidic residues upon membrane association, but Arg23, Arg26, and Arg33 have reduced probabilities of hydrogen bonding with acidic residues, showing that, for these latter Arg residues, acidic residues lose their competition against POPG lipids.
Besides the acidic POPG, Arg residues can also hydrogen bond with the zwitterionic POPE, though at much lower probabilities (Figure 6c). Even after compensating for the fact that POPE is at a lower mole fraction in the membranes, Arg residues are still 1.5 to 2.0 times less likely to hydrogen bond with POPE than with POPG (Figure 6d). On average, 2.5 NT Arg residues in ChiZ1-64 hydrogen bond with POPG lipids at each moment. This number increases to 3.2 in ChiZ1-86 and 3.0 in ChiZ-FL, mostly from C-half Arg residues starting at position 37 (Figure 6a). In comparison, the average numbers of NT Arg residues that hydrogen bond with POPE lipids at each moment, after scaling up by a factor of 7/3, are only 1.3, 1.6, and 2.0, respectively, in ChiZ1-64, ChiZ1-86, and ChiZ-FL. Therefore, POPG lipids preferentially distribute around the membrane-associated NT (see Figure 5c). Such preferential distribution of acidic lipids around basic groups of membrane-associated proteins have been observed in previous MD simulation studies.22,23 On average, each Arg residue engages with 1.1 to 1.2 POPG lipids in their hydrogen bonding. The average numbers of NT Arg residues that hydrogen bond with acidic residues range from 0.52 to 0.43 in the three ChiZ constructs, slightly less than the counterpart, 0.62, in unbound ChiZ1-64.
The two halves of unbound ChiZ1-64 are asymmetric not only in salt-bridge formation but also in PPII propensity (there are very low propensities for helices and β-strands; Figure S9).20 Three PPII stretches form with high probabilities (>50%), all in N-half: V4RP6, P10DP12, and A27EP29. In C-half, residues that sample the PPII region with the highest probabilities are P44L45 at 35% and S38R39 at 32%. In agreement with the NMR data, ChiZ1-64 does not gain any secondary structure upon membrane association (Figure S9). In fact, while N-half largely preserves its PPII probabilities upon membrane association, P44L45 in C-half suffers a modest reduction in its PPII probability, down to 31%. In ChiZ1-86 and ChiZ-FL, this probability further deteriorates to 26 and 25%, respectively. Similar losses in PPII probability are also seen for S38R39. So, NT sacrifices PPII formation in C-half to gain stability in membrane association.
The asymmetry in NT’s membrane association is dramatically illustrated by one of the ChiZ1-64 simulation runs (Movie S1). In this run, ChiZ1-64 initially binds to one leaflet via N-half. After only 20 ns, it dissociates but then quickly reassociates at 120 ns with another leaflet, this time via C-half. The association is stable for the rest of the 1.9-μs simulation. Apart from this brief episode in ChiZ1-64, NT in each of the three constructs is associated with membranes essentially all the time. When membrane contact is broken into N- and C-halves, we further find that C-half is membrane-bound constantly, whereas N-half is membrane-bound approximately 71% of the time in each of the three constructs. That both halves of NT spend at least 70% of the time on POPG:POPE membranes explains why the Gd3+ spin label senses the entire NT sequence (Figure 4d).
Both Transmembrane Helix and LysM Domain Contribute, Directly or Allosterically, to NT-Membrane Association
Several characteristics of NT-membrane association have emerged from the foregoing analyses of MD simulations. The association is largely maintained by Arg-POPG hydrogen bonding. For ChiZ1-64, these Arg residues are mostly located in the midsection of the sequence, but there is also an asymmetry that favors C-half. This intrinsic asymmetry is partly due to competition between acidic residues, all in N-half, and POPG lipids for interactions with Arg residues, and partly due to high PPII propensities in N-half. This asymmetry is accentuated by the membrane tethering of the NT C-terminus via the transmembrane helix. As illustrated by Movie S2 for ChiZ1-64 and Movie S3 for ChiZ-FL, NT-membrane association is highly dynamic. At each given moment, several Arg residues hydrogen bond with the membranes, but the identities of the Arg residues rapidly change (Figure 5a). As NT changes its conformation and hydrogen bond donors, the lipid acceptors, primarily POPG, also adapt to surround the Arg donors.
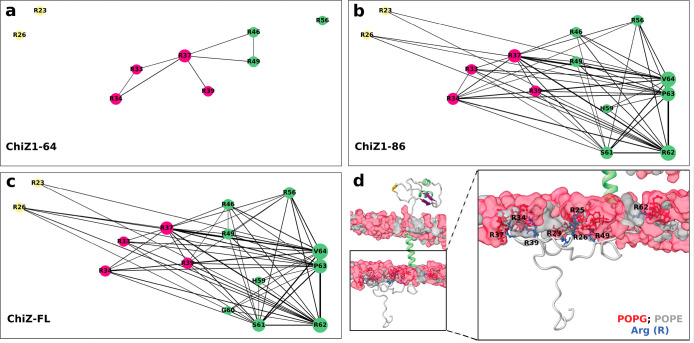
At a given moment, the numbers of NT residues in contact with membranes are 7.0 ± 1.4, 10.9 ± 1.9, and 11.1 ± 1.4, respectively, in ChiZ1-64, ChiZ1-86, and ChiZ-FL; of these, 74, 82, and 83% are in C-half. To gain a deeper sense of which residues contact membranes at the same time, we calculated the probability, Cij, that two residues, i and j, contact membranes simultaneously. Figure 7a–c displays the Cij networks of the three ChiZ constructs as graphs, where circular nodes (with radii proportional to Ci) represent residues with Ci > 0.25, and edge widths represent Cij (with Cij threshold at 0.20). It is clear that, relative the contact network of ChiZ1-64, the counterparts of ChiZ1-86 and ChiZ-FL are much more connected, with strong connections extending into N-half. The strengthened network connectivity of the longer constructs arises largely from the higher membrane-contact probabilities of the C-half residues (Figure 5b), which in turn can be attributed to the membrane insertion of the transmembrane helix. This is the basis of the assertion made above that the effect of membrane tethering propagates all the way into N-half. The direct effect of the membrane-contact probabilities can be removed by normalizing the co-occurrence probability: Ĉij ≡ Cij/CiCj, where CiCj is the expected probability that residues i and j would contact membranes at the same time by chance. A Ĉij that is greater than 1 indicates correlation between the two residues, and hence, we refer the Ĉij – 1 network as the contact correlation network. The contact correlation networks no longer show a clear-cut difference in connectivity among nine common residues for ChiZ1-86 and ChiZ-FL and for ChiZ1-64 (Figure S10a–c).
Figure 7.
Networks of membrane-contacting residues. (a–c) Contact networks of the three ChiZ constructs. Node radii are proportional to contact probabilities Ci; only nodes with Ci > 0.25 are shown. Edge widths are proportional to co-occurrence probabilities Cij; only edges with Cij > 0.20 are shown. (d) A snapshot of ChiZ-FL, illustrating residues that contact the membrane at the same time..
On the other hand, closer inspection reveals that the network connectivity of ChiZ-FL is stronger than that of ChiZ1-86, in line with the slightly but consistently higher membrane-contact probabilities of ChiZ-FL shown in Figure S5a. This difference is made clearer by comparing the degree, di, defined as the sum of Cij over all the partner (i.e., j) residues, of each node in ChiZ-FL and ChiZ1-86 (Figure S5b). Of the 16 frequent-contact residues, 13 have higher di in ChiZ-FL than in ChiZ1-86. As shown by the contact correlation networks (Figure S10b,c), membrane-contact residues in ChiZ-FL also have a higher level of correlation than in ChiZ1-86.
The stronger network connectivity of ChiZ-FL reveals that the periplasmic linker and LysM domain also contribute to the stability of NT-membrane association. Periplasmic residues only occasionally contact membranes (Figure S8b) and thus do not influence NT’s membrane association through their own membrane association on the opposite leaflet. Instead, we found that the positioning and tilting of the transmembrane helix are affected by the presence of the periplasmic linker and LysM domain (Figure S11a,b). In ChiZ-FL, the helix shifts toward the periplasmic side by approximately 1 Å, and the helix tilt samples a narrow range of angles. These make the transmembrane helix more deeply (from NT’s perspective) and more stably inserted in the membrane. By these changes in the transmembrane helix, the periplasmic linker and LysM domain allosterically strengthen NT-membrane association. Lastly, we display a snapshot from the MD simulations of ChiZ-FL in Figure 7d to illustrate the extreme fuzzy membrane association of NT in the full-length protein.
Discussion
By combining solution and solid-state NMR spectroscopy with molecular dynamics simulations, we have characterized the extreme fuzzy membrane association of the disordered N-terminal region of ChiZ. The association is largely driven by hydrogen bonding between Arg residues and acidic POPG lipids. Not only the conformation of NT but also the residues that contact the membrane at a given moment are highly dynamic. As NT frolics on the membrane, lipids quickly redistribute, with the acidic POPG lipids preferentially taking up Arg-proximal positions. We refer to membrane association represented by the disordered NT as “semispecific”, to be contrasted with specific binding between a protein and a macromolecular partner, with a defined interface, and nonspecific binding of proteins at high concentrations, where there is no clear demarcation between a bound state and an unbound state. Membrane association of the disordered NT is also distinct from that of folded domains such as C2 domains in synaptotagmin-1, which have one or more defined membrane-binding sites.23 For these reasons, ChiZ NT-membrane association represents a new paradigm of biomolecular binding. Other disordered proteins that engage in semispecific membrane association include α-synuclein9 and the Wiscott–Aldritch Syndrome protein.6
The term “semispecific” is also fitting in the sense that NT-membrane association has mixed random and nonrandom characteristics, similar to fuzzy association between two disordered proteins.1,2 While the random aspect is obvious from the highly dynamic nature of bound NT (see, e.g., Movies S2 and S3), the nonrandom aspect is also worth emphasizing. First, as already noted, it is largely Arg residues that drive the association. Second, for ChiZ1-64, the association-driving Arg residues are located in the midsection of the sequence. Third, the NT sequence codes for asymmetry between the two halves in membrane association. N-half contains all the acidic residues (which compete with POPG lipids for Arg interactions), and has high PPII propensities. N-half is therefore more recalcitrant while C-half is more adaptive to membrane association. Fourth, the intrinsic asymmetry between the two halves of NT is accentuated when its C-terminus is tethered to membranes via the subsequent transmembrane helix. Interestingly, NTs of ChiZ homologues in Mycobacterium species have a very conserved C-half, with 6–8 Arg residues (plus a rare Lys residue) and no acidic residues (other than a rare Asp), and a very variable N-half containing all the acidic residues (Figure S12). The characteristics of NT-membrane association determined here for Mtb ChiZ thus largely apply to other Mycobacterium species, and the conservation of the features important for membrane association argues for a functional role of membrane association.
Based on the foregoing information on ChiZ NTs, we may speculate that 6–8 Arg residues, minimally interrupted by acidic residues and distributed in a sequence of 30 or so amino acids, may be required for stable fuzzy association with highly acidic membranes. Of course, not all acidic lipids are alike. Although we used POPG as a representative of acidic lipids, the actual composition of the M. smegmatis inner membrane is approximately 35% cardiolipin, 35% phosphatidylinositol, and 30% phosphatidylethanolamine.13 Our preliminary results from MD simulations of ChiZ1-64 binding to a membrane with this composition closely track those reported for POPG:POPE membranes (Figure S13). However, lipids with higher negative charges, in particular phosphatidylinositol 4,5-bisphosphate, may have increased propensities for interacting with polybasic proteins, and hence, the number of Arg residues required for extreme fuzzy association might be reduced. Additional disordered membrane proteins need to be studied before we can establish the sequence requirements.
For specific binding between two structured domains, the dogma is that sequence codes for structure, which in turn codes for specificity, but for fuzzy binding of intrinsically disordered regions including semispecific membrane association of ChiZ NT and others, how sequence codes for binding specificity is still an open question. Contrary to α-synuclein and other disordered proteins that associate with membranes through amphipathic helices, ChiZ NT does not gain any secondary structure upon membrane association (apart from some hint for an A43PLR46 binding motif in a subpopulation). In the former cases, a mechanism to code for binding specificity is through amino-acid patterning that favors amphipathic-helix formation, i.e., by positive design, as exemplified by the KTKEGV motifs in α-synuclein.9 Illustrated by the exclusion of acidic residues in C-half, the specificity of ChiZ NT-membrane association appears to be achieved partly by negative design. As found in our previous study,20 the NT sequence codes for correlated segments, mostly in N-half, that are stabilized by salt bridges, cation−π interactions, and high PPII propensities. Just as we speculated previously, these correlated segments lead to the recalcitrance of N-half toward membrane association. Conversely, lack of strongly correlated segments in C-half allows it to be more adaptive to membrane association.
Due to reduced dimensionality, membrane association increases the chances that proteins interact with each other. A main function of ChiZ is to halt cell division via overexpression under DNA damage conditions.24 Overexpression may present ChiZ at a level where NTs of different copies come into contact at the membrane. The work presented here characterizing the conformations and dynamics of membrane-bound NT in a single copy of ChiZ lays a solid foundation for understanding interactions between multiple NTs as well as interactions of ChiZ NT and membrane-bound disordered regions of partner proteins, including FtsI and FtsQ25 (Figure S1).
Materials and Methods
Protein Expression and Purification
Expression and purification of ChiZ1-64 was performed as previously described.2013C–15N labeled ChiZ-FL containing a noncleavable N-terminal 6× His-tag was expressed in Escherichia coli BL21 Codon Plus RP competent cells. Cells were grown at 37 °C in LB media until OD at 600 nm reached 0.7. Cells were pelleted and transferred to M9 media containing 1 g of 15N-ammonium chloride and 2 g of 13C uniformly label glucose (Cambridge Isotope Laboratories). After transfer, cells were incubated at 37 °C for 30 min before adding IPTG to a final concentration of 0.4 mM to induce protein expression for 5 h. Cells were then pelleted and resuspended in a lysis buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl) for cell lysis using a French press. n-Dodecylphosphocholine (DPC; Anatrace) was added to the lysate to a final concentration of 2% (wt/vol) and then incubated overnight at 4 °C with agitation. Cell lysate was centrifuged at 250 000g for 30 min. Protein purification was performed using Ni-NTA resin (Qiagen) equilibrated with the lysis buffer containing 20 mM imidazole. The column was washed using the lysis buffer containing 0.5% (wt/vol) DPC and 60 mM imidazole. Protein was eluted with the same buffer but containing 400 mM imidazole.
Mixing of ChiZ1-64 with Liposomes
ChiZ1-64 was mixed with liposomes containing: (i) pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); (ii) 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) at 4:1 molar ratio; (iii) POPC and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) at 4:1 ratio; or (iv) POPG and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) at 7:3 ratio (all lipids from Avanti Polar Lipids). The protein–liposome mixtures, at a protein to lipid molar ratio of 1:100, were loaded into an NMR tube for collecting 1H–15N HSQC spectra.
For solid-state NMR experiments, ChiZ1-64 was mixed with POPG:POPE (7:3) liposomes at a 1:50 protein to lipid ratio. The mixture was pelleted down by centrifugation at 15,000g for 15 min. The pellet was then loaded into a 3.2 mm MAS rotor. Liposomes in samples for paramagnetic relaxation enhancement experiments also contained 2% 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid gadolinium salt (PE-DTPA-GD; Avanti Polar Lipids) as a spin label.
Reconstitution of ChiZ-FL into Liposomes
ChiZ-FL samples in MAS solid-state NMR experiments were reconstituted into POPG:POPE (7:3) liposomes at a protein to lipid molar ratio of 1:80. Methyl-β-cyclodextrin (MβCD; Sigma-Aldrich) was used to remove the DPC detergent from the protein–detergent–lipid mixture. Specifically, POPG and POPE lipids in chloroform were mixed, and the solvent was removed using nitrogen stream and extensive vacuum. Lipid films were resuspended in 20 mM Tris-HCl (pH 8.0) and sonicated. DPC was added until the solution became clear. Then, ChiZ-FL was added, and the mixture was incubated for 1 h at room temperature. To remove DPC, a solution of MβCD in 20 mM Tris-HCl (pH 8.0) was added to the protein–detergent–lipid mixture at a DPC to MβCD molar ratio of 1:1.5. Proteoliposomes were collected by centrifugation at 250 000g for 3 h at 8 °C. The pellet was resuspended in 20 mM Tris-HCl (pH 8.0), and an MβCD solution containing 10% of the previous level was added to remove residual detergent. Proteoliposomes were finally collected by centrifugation at 100 000 rpm in a TLA-100 rotor at 8 °C for 16 h and washed with 20 mM Tris-HCl (pH 8.0) at least twice. ChiZ-FL proteoliposomes were packed into a 3.2 mm MAS rotor for solid-state NMR experiments. Samples for paramagnetic relaxation enhancement experiments were doped with 1% PE-DTPA-GD.
NMR Spectroscopy
Solution NMR experiments of ChiZ1-64 mixed with liposomes were performed in 20 mM sodium phosphate (pH 7.0) containing 25 mM NaCl, 50 μM sodium trimethylsilylpropanesulfonate (DSS; NMR standard) and 10% D2O. 1H–15N and 1H–13C heteronuclear single quantum coherence (HSQC) spectra were collected at 25 °C on an 800 MHz NMR spectrometer equipped with a cryoprobe. Chemical shift assignments of ChiZ1-64 have been reported previously (BMRB accession # 50115).20 MAS solid-state NMR experiments of reconstituted ChiZ-FL and liposome-bound ChiZ1-64 were performed at 25 °C on a 600 MHz NMR spectrometer equipped with a Low-E MAS probe with a spinning rate of 12.2 kHz. Glycine carbonyl carbon with a chemical shift frequency of 178.4 ppm was used as 13C chemical shift reference. One-dimensional 13C direct-excitation spectra were collected using a 13C 90° pulse of 62.5 kHz and proton decoupling at 75 kHz using the SPINAL64 decoupling sequence. 13C–13C (and 1H–13C) correlation spectra using cross-polarization (CP) and INEPT-based pulse sequences were collected using the same proton and carbon frequencies as for one-dimensional experiments. For CP-based experiments, the PARIS pulse sequence was used.26
Molecular Dynamics Simulations
Three ChiZ constructs (Figure 1) were modeled and simulated: (i) ChiZ1-64 bound to a 7:3 POPG:POPE bilayer; (ii) ChiZ1-86 with the 22-residue transmembrane helix inserted in a 7:3 POPG:POPE bilayer and NT bound to the inner leaflet; and (iii) ChiZ-FL, which extended the ChiZ1-86 system by the periplasmic linker and LysM domain. The simulations of the three systems consisted of 20, 20, and 16 replicate trajectories, respectively; the production lengths of these trajectories were 1.9, 1.8, and 1.29 μs, respectively. The production simulations were preceded by preparatory simulations. The force field combination was AMBER14SB27 for proteins, TIP4P-D28 for solvent (water plus ions), and Lipid1729 for membranes.
The membrane-bound ChiZ1-64 simulations were prepared starting from nine ChiZ1-64 models selected from the simulations of the unbound system.20 A membrane plus solvent system (220 lipids per leaflet with POPG and POPE at 7:3 ratio) was built using the CHARMM-GUI server.30 The output was converted to AMBER-formatted coordinate and topology files using the charmmlipid2amber.py script and tleap in AmberTools17.31 Upon aligning N-half of ChiZ1-64 to the inner leaflet of the bilayer, ChiZ1-64 was inserted into the system using PARMED with clashing solvent removed. Using tleap, neutralizing ions plus 25 mM NaCl were added, and the combined system was built into AMBER topology. The final system size was 122 × 122 × 140 Å with 261 493 atoms.
Preparatory simulations starting from the nine ChiZ1-64 models were run in NAMD 2.1232 with AMBER topology. Energy minimization (10 000 cycles of conjugate gradient) was followed by the six-step CHARMM-GUI equilibration protocol30 with gradually decreasing restraints on the protein and lipids. Bond lengths involving hydrogens were constrained by the SHAKE algorithm.33 The time step was 1 fs in the first four of the six-step protocol but 2 fs in the last two. The durations of the six steps were 25, 25, 25, 200, 200, 2000 ps. van der Waals interactions were force-switched starting at 10 Å and cut off at 12 Å. The same cutoff was used for calculating short-range electrostatic interactions; long-range electrostatic interactions were treated by the particle mesh Ewald method.34 The first three steps were under constant temperature (300 K) and volume, whereas the last three were under constant temperature and pressure (1.0 atm). Temperature was regulated by the Langevin thermostat with a friction coefficient of 1.0 ps–1; pressure was regulated by the Langevin piston35 with an oscillation period of 50.0 fs and decay of 25.0 fs. Here, and below, whenever pressure was regulated, semi-isotropic scaling in the x–y plane was applied to maintain the constant ratio of the two dimensions, with no added surface tension. Following the six-step equilibration, the nine simulations continued under constant temperature and pressure for 40 ns. A total of 20 snapshots, i.e., the nine at the start and the nine at the end of 40 ns simulations, plus two in between, were restarted to run AMBER production simulations for 1.9 μs on GPUs (see below for further details).
ChiZ1-86 models were built using MODELER36 with residues 65–86 modeled as a helix. Ten models were selected for insertion into a POPG:POPE bilayer (at 7:3 ratio with a total of 220 lipids per leaflet) using CHARMM-GUI, with 25 mM NaCl and neutralizing ions added. The final system size was 135 × 135 × 251 Å with 446 510 atoms. Preparatory simulations of the 10 ChiZ1-86 models were the same as for ChiZ1-64 with the following exceptions. (i) Pressure was regulated by the Monte Carlo barostat; (ii) the durations of the last three steps of the equilibration were 100 ps each; (iii) the subsequent NAMD run was replaced by an AMBER GPU simulation of 1 ns. The 10 final snapshots were each restarted with two random seeds to run AMBER production simulations for 1.8 μs on GPUs.
ChiZ-FL models were built using MODELER by combining eight homology models of the LysM domain (residues 113–165) from SWISS-MODEL37 with eight of the ChiZ1-86 starting models. The rest of the ChiZ-FL preparations was the same as for ChiZ1-86. The system contained 300 lipids per leaflet (with POPG and POPE at 7:3 ratio) with a size of 147 × 149 × 235 Å and 522 825 atoms. Each of the eight final snapshots in the preparatory simulations was restarted with two random seeds to run AMBER production simulations for 1.29 μs on GPUs.
Production simulations were on GPUs using pmemd.cuda(38) in AMBER18. Temperature was held at 300 K using the Langevin thermostat with a friction coefficient at 1.0 ps–1. Pressure was held at 1.0 atm using the Berendsen barostat.39 For van der Waals interactions, the force-switch distance was 9 Å and cutoff was 11 Å. The latter was also used for dividing direct calculation of electrostatic interactions from a particle mesh Ewald treatment. Bond lengths involving hydrogens were constrained by the SHAKE algorithm. The time step was 2 fs. Snapshots were saved every 10 ps in the ChiZ1-64 simulations and every 20 ps in the ChiZ1-86 and ChiZ-FL simulations. The first 2000 saved snapshots for each system were discarded.
MD Trajectory Analyses
Heavy atom contacts, hydrogen bonds, distances along the z axis, and secondary structures were calculated with cpptraj.40 Further analyses and plotting were performed using in-house python scripts. Two heavy atoms between a protein and lipids were in contact if they were within 3.5 Å. Hydrogen bonds were defined as formed when the donor–acceptor distance was less than 3.5 Å and the donor–hydrogen–acceptor angle was greater than 135°.
The membrane-contact probability Ci and the probability Cij that two residues contact membranes at the same time were calculated after pooling all the saved snapshots of each system. From Ci, Cij, and Cij/CiCj – 1, the python module networkx was used to build the membrane contact networks and the contact correlation networks. The SHIFTX241 software was used to calculate the chemical shifts of all atoms on snapshots taken at 200 ps intervals. The seaborn plotting module in python3 was implemented to create the violin plots. Images of structures were rendered using ChimeraX,42 and movies were composed using Blender.43
Acknowledgments
A.H. would like to thank Dr. Sean Smrt and Dr. Rongfu Zhang for discussions on NMR. This work was supported by National Institutes of Health Grants R35 GM118091 and R01 AI119178. The NMR experiments were performed at the National High Magnetic Field Laboratory, funded by the National Science Foundation Division of Materials Research (DMR-1644779) and the State of Florida.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.0c00039.
Thirteen supporting figures, Figures S1–S13 (PDF)
Author Contributions
□ A.H. and C.A.E. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Wu S.; Wang D.; Liu J.; Feng Y.; Weng J.; Li Y.; Gao X.; Liu J.; Wang W. The Dynamic multisite interactions between two intrinsically disordered proteins. Angew. Chem., Int. Ed. 2017, 56 (26), 7515–7519. 10.1002/anie.201701883. [DOI] [PubMed] [Google Scholar]
- Borgia A.; Borgia M. B.; Bugge K.; Kissling V. M.; Heidarsson P. O.; Fernandes C. B.; Sottini A.; Soranno A.; Buholzer K. J.; Nettels D.; Kragelund B. B.; Best R. B.; Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555 (7694), 61–66. 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. L.; Watson M.; Wilkins O. G.; Cato L.; Travers A.; Thomas J. O.; Stott K. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (47), 11964–11969. 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira J. L.; Palomino-Schätzlein M.; Ricci C.; Ortore M. G.; Rizzuti B.; Iovanna J. L. Dynamics of the intrinsically disordered protein NUPR1 in isolation and in its fuzzy complexes with DNA and prothymosin α. Biochim. Biophys. Acta, Proteins Proteomics 2019, 1867 (11), 140252. 10.1016/j.bbapap.2019.07.005. [DOI] [PubMed] [Google Scholar]
- Meyer N. H.; Dellago H.; Tam-Amersdorfer C.; Merle D. A.; Parlato R.; Gesslbauer B.; Almer J.; Gschwandtner M.; Leon A.; Franzmann T. M.; Grillari J.; Kungl A. J.; Zangger K.; Falsone S. F. Structural fuzziness of the RNA-organizing protein SERF determines a toxic gain-of-interaction. J. Mol. Biol. 2020, 432 (4), 930–951. 10.1016/j.jmb.2019.11.014. [DOI] [PubMed] [Google Scholar]
- Higgs H. N.; Pollard T. D. Activation by Cdc42 and PIP2 of Wiskott-Aldrich Syndrome protein (WASP) stimulates actin nucleation by Arp2/3 complex. J. Cell Biol. 2000, 150 (6), 1311–1320. 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T.; Terebiznik M.; Yu L.; Silvius J.; Abidi W. M.; Philips M.; Levine T.; Kapus A.; Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science 2006, 313 (5785), 347–51. 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Haxholm G. W.; Nikolajsen L. F.; Olsen J. G.; Fredsted J.; Larsen F. H.; Goffin V.; Pedersen S. F.; Brooks A. J.; Waters M. J.; Kragelund B. B. Intrinsically disordered cytoplasmic domains of two cytokine receptors mediate conserved interactions with membranes. Biochem. J. 2015, 468 (3), 495–506. 10.1042/BJ20141243. [DOI] [PubMed] [Google Scholar]
- Das T.; Eliezer D. Membrane interactions of intrinsically disordered proteins: The example of α-synuclein. Biochim. Biophys. Acta, Proteins Proteomics 2019, 1867 (10), 879–889. 10.1016/j.bbapap.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B.; Li L.; Meroueh S. O.; Uversky V. N.; Dunker A. K. Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol. BioSyst. 2009, 5 (12), 1688–1702. 10.1039/b905913j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N.; Gillespie J. R.; Fink A. L. Why are “natively unfolded” proteins unstructured under physiologic conditions?. Proteins: Struct., Funct., Genet. 2000, 41 (3), 415–427. . [DOI] [PubMed] [Google Scholar]
- Yang Y.; Lee M.; Fairn G. D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293 (17), 6230–6240. 10.1074/jbc.R117.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.; Crick D. C.; Brennan P. J. Phosphatidylinositol Is an essential phospholipid of Mycobacteria. J. Biol. Chem. 2000, 275 (39), 30092–30099. 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- Rhoades E.; Ramlall T. F.; Webb W. W.; Eliezer D. Quantification of α-synuclein binding to lipid vesicles using fluorescence correlation spectroscopy. Biophys. J. 2006, 90 (12), 4692–4700. 10.1529/biophysj.105.079251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovere M.; Powers A. E.; Jiang H.; Pitino J. C.; Fonseca-Ornelas L.; Patel D. S.; Achille A.; Langen R.; Varkey J.; Bartels T. E46K-like α-synuclein mutants increase lipid interactions and disrupt membrane selectivity. J. Biol. Chem. 2019, 294 (25), 9799–9812. 10.1074/jbc.RA118.006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S. S. Y.; Taneva S. G.; Lee J. M. C.; Cornell R. B. The curvature sensitivity of a membrane-binding amphipathic helix can be modulated by the charge on a flanking region. Biochemistry 2014, 53 (3), 450–461. 10.1021/bi401457r. [DOI] [PubMed] [Google Scholar]
- Fusco G.; Pape T.; Stephens A. D.; Mahou P.; Costa A. R.; Kaminski C. F.; Kaminski Schierle G. S.; Vendruscolo M.; Veglia G.; Dobson C. M.; De Simone A. Structural basis of synaptic vesicle assembly promoted by α-synuclein. Nat. Commun. 2016, 7 (1), 12563. 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas J. V.; Tajkhorshid E. Conformational heterogeneity of α-synuclein in membrane. Biochim. Biophys. Acta, Biomembr. 2014, 1838 (12), 3107–17. 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge K.; Papaleo E.; Haxholm G. W.; Hopper J. T. S.; Robinson C. V.; Olsen J. G.; Lindorff-Larsen K.; Kragelund B. B. A combined computational and structural model of the full-length human prolactin receptor. Nat. Commun. 2016, 7 (1), 11578. 10.1038/ncomms11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks A.; Escobar C. A.; Cross T. A.; Zhou H.-X. Sequence-dependent correlated segments in the intrinsically disordered region of ChiZ.. Biomolecules 2020, 10 (6), 946. 10.3390/biom10060946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailescu M.; Sorci M.; Seckute J.; Silin V. I.; Hammer J.; Perrin B. S. Jr.; Hernandez J. I.; Smajic N.; Shrestha A.; Bogardus K. A.; Greenwood A. I.; Fu R.; Blazyk J.; Pastor R. W.; Nicholson L. K.; Belfort G.; Cotten M. L. Structure and function in antimicrobial piscidins: Histidine position, directionality of membrane insertion, and pH-dependent permeabilization. J. Am. Chem. Soc. 2019, 141 (25), 9837–9853. 10.1021/jacs.9b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb C. N.; Sansom M. S. Defining the membrane-associated state of the PTEN tumor suppressor protein. Biophys. J. 2013, 104 (3), 613–21. 10.1016/j.bpj.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R.; Zhou H. X. Membrane association and functional mechanism of synaptotagmin-1 in triggering vesicle fusion. Biophys. J. 2020, 119, 1255–1265. 10.1016/j.bpj.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A.; Lofton H.; Maloney E.; Moore J.; Fol M.; Madiraju M. V. V. S.; Rajagopalan M. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 2006, 62 (1), 132–147. 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- Vadrevu I. S.; Lofton H.; Sarva K.; Blasczyk E.; Plocinska R.; Chinnaswamy J.; Madiraju M.; Rajagopalan M. ChiZ levels modulate cell division process in mycobacteria. Tuberculosis 2011, 91, S128–S135. 10.1016/j.tube.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarth M.; Demco D. E.; Bodenhausen G.; Tekely P. Improved magnetization transfer in solid-state NMR with fast magic angle spinning. Chem. Phys. Lett. 2009, 469, 342–348. 10.1016/j.cplett.2008.12.084. [DOI] [Google Scholar]
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11 (8), 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana S.; Donchev A. G.; Robustelli P.; Shaw D. E. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 2015, 119 (16), 5113–5123. 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- Gould I. R.; Skjevik A. A.; Dickson C. J.; Madej B. D.; Walker R. C., Lipid17: A comprehensive AMBER force field for the simulation of zwitterionic and anionic lipids. In prep. 2019. [Google Scholar]
- Wu E. L.; Cheng X.; Jo S.; Rui H.; Song K. C.; Dávila-Contreras E. M.; Qi Y.; Lee J.; Monje-Galvan V.; Venable R. M.; Klauda J. B.; Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35 (27), 1997–2004. 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A.; Ben-Shalom I. Y.; Brozell S. R.; Cerutti D. S.; Cheatham I. T.E.; Cruzeiro V. W. D.; Darden T. A.; Duke R. E.; Ghoreishi D.; Gilson M. K.; Gohlke H.; Goetz A. W.; Greene D.; Harris R.; Homeyer N.; Izadi S.; Kovalenko A.; Kurtzman T.; Lee T. S.; Le Grand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Mermelstein D. J.; Merz K. M.; Miao Y.; Monard G.; Nguyen C.; Nguyen H.; Omelyan I.; Onufriev A.; Pan F.; Qi R.; Roe D. R.; Roitberg A. E.; Sagui C.; Schott-Vergudo S.; Shen J.; Simmerling C. L.; Smith J.; Salomon-Ferrer R.; Swails J.; Walker R. C.; Wang J.; Wei H.; Wolf R. M.; Wu X.; Xiao L.; York D. M.; Kollman P. A.. AMBER 2018; University of California: San Francisco, 2018.
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26 (16), 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23 (3), 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103 (19), 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Feller S. E.; Zhang Y. H.; Pastor R. W.; Brooks B. R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103 (11), 4613–4621. 10.1063/1.470648. [DOI] [Google Scholar]
- Webb B.; Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc Bioinformatics 2016, 54 (1), 5.6.1–5.6.37. 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.; Bertoni M.; Bienert S.; Studer G.; Tauriello G.; Gumienny R.; Heer F. T.; de Beer T. A. P.; Rempfer C.; Bordoli L.; Lepore R.; Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46 (W1), W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Ferrer R.; Götz A. W.; Poole D.; Le Grand S.; Walker R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013, 9 (9), 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81 (8), 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Roe D. R.; Cheatham T. E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9 (7), 3084–3095. 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Han B.; Liu Y.; Ginzinger S. W.; Wishart D. S. SHIFTX2: Significantly improved protein chemical shift prediction. J. Biomol. NMR 2011, 50 (1), 43. 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T. D.; Huang C. C.; Meng E. C.; Pettersen E. F.; Couch G. S.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27 (1), 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender Online Community. Blender - a 3D modelling and rendering package; Blender Foundation: Stichting Blender Foundation: Amsterdam, 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.