Abstract
MiR-543 and Numb are associated with various malignancies, including prostate cancer (PCa). However, whether miR-543 regulates Numb in PCa development remains unclear. In this study, we identified Numb as a direct target of miR-543. The role of miR-543 was examined both in vitro and in vivo. The in vivo effects of miR-543 were investigated using tumor transplantation experiments and a lung metastasis model. The in vitro effects of miR-543 on proliferation, migration, invasion, and cancer stem-like cell (CSC)-associated properties were also examined. The binding sites of Numb were predicted using bioinformatics tools and confirmed by luciferase and rescue assays. QRT-PCR and western blot analyses were used to detect target expression levels. Expression levels of both miR-543 and Numb were manipulated in CD44+ and CD44-PCa cells followed by a series of functional assays. The results demonstrated that miR-543 promoted PCa growth and metastasis both in vivo and in vitro. Luciferase reporter assays, qRT-PCR, and western blot analyses revealed Numb as a direct target of miR-543. The function of miR-543 was abolished by Numb, as shown in rescue experiments. Moreover, miR-543 was verified to promote CSC properties, whereas Numb elicited the opposite effects. MiR-543 also influenced the expression of several stem-like factors, including Dll4, NF-κB, c-myc, and Oct-4, and the Numb/p53 signaling pathway. Taken together, these results demonstrate that miR-543 plays an oncogenic role by negatively controlling Numb, revealing the existence of an miR-543/Numb/p53 regulatory pathway in PCa tumorigenesis and development.
Keywords: miR-543, Numb, prostate cancer, CSC, proliferation
Introduction
Prostate cancer (PCa) is the most common malignancy worldwide and the second-leading cause of cancer-related deaths in males [1]. PCa first manifests as an androgen-dependent disease. Although patients with PCa initially respond to endocrine therapy, most will eventually develop hormone resistance, which leads to the progression of PCa relapse, metastasis, and even death. It has been reported that hormone therapy only maintains a median progression-free duration of two years [2]. Once a patient is considered androgen independent (AI), there are no effective treatments. Hence, there is extreme urgency to understand the molecular mechanisms of the AI phenotype of PCa (AI-PCa) and identify new targets for its treatment.
MicroRNAs (miRNAs) are a class of small (20-24 nucleotides in length), endogenous, noncoding RNAs expressed widely in eukaryotes. These miRNAs can regulate different biological processes by post-transcriptionally binding to the 3’-untranslated region (UTR) of a target gene to inhibit its translation. Various miRNAs have been shown to participate in cell proliferation, apoptosis, metabolism, differentiation, morphogenesis, stem cell maintenance, and other cellular activities. A previous report suggested that miRNAs may play a role in the diagnosis and prognosis of PCa, as well as in its potential therapeutic interventions [3]. For example, miR-1291 is reported to suppress cell proliferation and tumorigenesis via the downregulation of MED1 [4]; miR-34a attenuates paclitaxel resistance in PCa cells via the direct suppression of the JAG1/Notch1 axis [5]; and miR-30e regulates the adhesion, migration, invasion, and cell cycle progression of PCa cells via the downregulation of CHRM3 [6]. Recently, miR-543 was identified as an oncogenic miR that promoted migration, invasion, and the epithelial-mesenchymal transition (EMT) in esophageal cancer cells [7]. In addition, miR-543 was shown to enhance drug resistance by downregulating phosphatase and tensin homolog (PTEN) [8]. However, the mechanism by which miR-543 elicits its effects on AI-PCa and PCa stem cells (PCSCs) has not been elucidated.
Numb, an evolutionarily conserved developmental protein, plays a vital role in cell-fate determination and differentiation. Numb was originally identified as a mediator of tissue morphogenesis and patterning in Drosophila development [9]. Recent research has revealed an association between Numb and cancer development. The expression of Numb is frequently attenuated in solid tumors [10,11]. In PCa, Numb levels are deficient and correlate with aggressive disease and poor prognosis [10]. Numb was shown to inhibit the growth of prostate tumor xenografts and the development of castration-resistant PCa (CRPC) [10]. Numb-/low PCa cells are quiescent, preferentially express cancer stem-like cell (CSC)-associated genes, and are associated with resistance to androgen deprivation therapy [10,12]. Numb is involved in various functions connected with signaling (e.g., it regulates both Notch- and TP53-activated pathways) [13]. However, the upstream regulation of Numb is currently unclear. Peng et al. reported that Numb is targeted by miR-31-5p, which promotes the growth, migration, and invasion of colorectal cancer cells [14]. Colaluca et al. demonstrated that Numb is modulated by the KRT19/beta-catenin/RAC1 complex in breast cancer [15]. However, a causal relationship between miRNA and Numb has not been previously reported in PCa.
In the current study, we found that miR-543 serves as an oncogene in two classic AI-PCa cell lines: DU145 and PC3. Downregulation of miR-543 inhibited cell proliferation, clonogenic survival, self-renewal of PCSCs, metastasis, and tumorigenic ability both in vitro and in vivo. Bioinformatics analyses and luciferase reporter assays revealed that Numb is a direct target of miR-543. Restoring the expression of Numb partially reversed the effects of miR-543 on the clonality, invasiveness, and self-renewal of PCSCs. These results demonstrate a novel mechanism by which miR-543 regulates PCa progression by targeting Numb, which may represent a novel target for AI-PCa.
Materials and methods
Cells, animals, and reagents
PC3 cells were cultured in DMEM, and DU145 cells were cultured in 1640 medium. Both media contained 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. BALB/c mice were purchased from the Beijing Weitong Lihua Company. All animal experiments were approved by the local ethics committee. Antibodies were acquired against Numb (polyclonal, 1:1,000, Abcam), Notch1 (polyclonal, 1:1,000, Abcam), p53 (polyclonal, 1:1,000, Abclonal), E-cadherin (polyclonal, 1:1,000, Abclonal), vimentin (polyclonal, 1:1,000, Abclonal), Ki-67 (polyclonal, 1:1,000, Servicebio), caspase-3 (polyclonal, 1:1,000, Servicebio), and GAPDH (polyclonal, 1:5,000, Abclonal).
Oligonucleotides (oligos), plasmids, and transfection
The chemically synthesized miR-543 mimic was used to enhance the function of endogenous miRNAs. The miR-543 inhibitor is a chemically synthesized substance that was used to inhibit cellular miR-543 expression. Both the miR-543 mimic and inhibitor were purchased from Gemma. Lipofectamine RNAiMAX (Invitrogen) was utilized to transfect PCa cells with 30 nM miR-543 mimic and an inhibitor, as well as the corresponding negative control (NC), according to the manufacturer’s instructions. Small interfering RNA (siRNA), which silences complementary target RNA, was used to knockdown Numb expression. Numb siRNA and its NC were transfected into cells using Lipofectamine RNAiMAX. Lipofectamine 2000 (Invitrogen) was utilized to assist in the transfection of plasmids. The Numb-overexpressing plasmid (pBABE-Numb), which contains the full-length Numb cDNA, and its empty vector (pBABE) were prepared for functional experiments. Co-transfection of the miR-543 mimic and PmirGLO-Numb, lacking the Numb 3’-UTR, or its control PmirGLO (Con) was performed for the rescue experiments. The pGL3-control vector, pGL3-NUMB-WT, and pGL-NUMB-MUT plasmids were transfected with the miR-543 mimic or a NC for luciferase reporter assays.
Tumor transplantation experiments
Basic procedures pertaining to these experiments have been previously described [14]. For subcutaneous tumorigenesis experiments, PC3 and DU145 cells were digested, washed, and filtered 48 hours after transfection with the miR-543 mimic or an inhibitor. After counting, the cell concentration was adjusted to 5 × 106 cells/mL. One hundred microliters of cells (5 × 105 cells) were injected subcutaneously into each BALB/C nude mouse (6-8 weeks of age). The extent of tumorigenesis and weights of mice were measured every 3-4 days. When the tumor size approached 80-100 mm, the mice were sacrificed.
Lung metastasis model
To establish a lung metastasis model, the cell concentration was adjusted to 4 × 107 cells/mL. After fully resuspending the cells, 100 µL cells (4 × 106 cells) were injected into the tail veins of 5-week-old BALB/C nude mice. Changes in the mice were observed every 3-4 days. After 40 days, the mice were sacrificed, their lungs isolated, and the number of metastatic nodules was observed and counted.
Transwell migration and invasion
For the Transwell invasion assay, 70 µL of a mixture consisting of glue and serum (BD matrix gel:serum-free medium = 1:9) were added to each pore operated on ice on the previous day. The mixture was applied evenly to avoid bubble formation and placed into an incubator overnight. The migration assay did not require the aforementioned steps, but are consistent with the Transwell invasion assay. Forty-eight hours after transfection, the cells were counted. The cell concentration was adjusted to 1 × 106 cells/mL; 600 µL media containing 10% FBS were added to the lower chamber, and 200 µL of the serum-free cell suspension was added to the upper chamber. The Transwell plate was placed into an incubator for 36-48 hours, after which the cells were washed with PBS, fixed with polyformaldehyde for 30 min, and stained with crystal violet. The cells were then blotted carefully using a cotton swab. When the chambers had dried, photos were taken using an inverted microscope, and the cells counted using image J software.
Clonal, clonogenic, and sphere-formation assays
The procedures associated with these experiments were previously described [16]. For clonal experiments, PCa cells were inoculated into a 6-well plate at an appropriate density of 500 or 300 cells/well. When obvious clones were visible, cells were counted within 14 days. For clonogenic assays, the cell concentration was adjusted to 5 × 103 cells/mL. Sixty microliters of 4°C Matrigel were mixed with 60 µL of the cell suspension and added carefully around the edge of a 24-well dish. After solidification at 37°C for 20 min, 400 µL media containing serum were added, and cells were counted after 7-14 days in culture. For sphere-formation assays, 300-500 cells/well were inoculated into an ultralow attachment 6-well plate containing F12/DMEM, 1 × B27, fibroblast growth factor, and epidermal growth factor. Cells were counted and photographed within 14 days.
Immunohistochemical analysis
Immunohistochemical analysis was performed after cells were immobilized in polyformaldehyde. To determine the extent of subcutaneous tumorigenesis, the tumors were embedded, sectioned, and stained. Ki67 was located in the nucleus, and caspase-3 was located in the cytoplasm. The positive stains were brown and yellow. Sections of lung metastases were subjected to HE staining and photography.
Western blot
PIPA buffer (containing a protease inhibitor cocktail) was used to lyse the cells. Total cellular protein was extracted, and the protein concentration was measured. Polyacrylamide gel electrophoresis was carried out to separate cellular proteins based on their molecular weights, and proteins were then transferred to a polyvinylidene fluoride membrane. The membranes were blocked and incubated with primary antibodies at 4°C overnight. Membranes were washed three times and incubated with anti-rabbit HRP-linked secondary antibody (1:3,000, Servicebio) at room temperature for 2 hours. GAPDH was used as an internal control, and the relative quantification of proteins was carried out according to this protein.
Real-time PCR
Quantitative real-time RT-PCR (qRT-PCR) was performed according to a standard protocol. Total RNA was extracted from cells and tumor samples using the TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. Quantitative real-time PCR primers were synthesized by Genecreate (Wuhan, China). RNA was reverse-transcribed into cDNA, which was then amplified, according to the kit instructions. GAPDH and U6 were used as internal controls. Relative gene expression was calculated using the 2-ΔΔCT method. Each experiment was performed in triplicate. The primers used for the studies are listed in Table 1.
Table 1.
Primer sequence
| Primer name | Sequence |
|---|---|
| Numb Forward primer | AACGCCAACTATCCCTAGG |
| Numb Reverse primer | ACTGGTTTGGTCATCGGAG |
| Dll4 Forward primer | CTTGCCATGACCTGGAGAA |
| Dll4 Reverse primer | ATGGATGTCCGCACCTCA |
| NF-κB Forward primer | GCACCCTGACCTTGCCTATT |
| NF-κB Reverse primer | AGCTGCTTGGCGGATTAG |
| c-Myc Forward primer | AGGCTCCTGGCAAAAGGT |
| c-Myc Reverse primer | CTGCGTAGTTGTGCTGATGTG |
| Oct-4 Forward primer | GCCAGAGGAAAGCACACT |
| Oct-4 Reverse primer | CAGATCAGCCACARCGC |
| Sox2 Forward primer | GAACCCCAAGATGCACAAC |
| Sox2 Reverse primer | GCTTAGCCTCGTCGATGAA |
| NANOG Forward primer | TCTTCTGCTGAGATGCCTCAC |
| NANOG Reverse primer | GGTTGTTTGCCTTTGGGAC |
| CD44 Forward primer | CCAAGTGGACTCAACGGAGA |
| CD44 Reverse primer | TGGTCTGGAGTTTCTGACGAC |
| GAPDH Forward primer | AGAAGGCTGGGGCTCATTTG |
| GAPDH Reverse primer | AGGGGCCATCCACAGTCTTC |
Statistical analysis
All experiments were repeated three times independently, and the weight of tumor xenografts were expressed as the mean ± S.D. Throughout the research, an unpaired, two-tailed Student’s t test was used to compare the differences between the experimental and control groups (the tumor weight, ki67+ and caspase3+ percentage in subcutaneous xenograft tumor model, lung metastasis nodules in tail vein injection assay, invasion and migration ability, clonal, clonogenic, and sphere-forming ability, the luciferase reporter assay and gene expression levels in RT-PCR). A P value less than 0.05 was considered statistically significant. Data were analyzed and drawings were made using Graphpad Prism software, version 7.0.
Results
MiR-543 promotes the growth of tumor xenografts in vivo
Previous studies [17] have reported that the expression of miR-543 in PCa is higher than that in adjacent normal prostate tissue; we found that miR-543 inhibited both proliferation and EMT in androgen receptor-expressing PCa cells. To further investigate the detailed function of miR-543 in AI-PCa cells, we transfected the synthetic miR-543 mimic, inhibitor, or their respective NC oligos (30 nM, 48 hours) into PC3 and Du145 cells. The transfected cells were subcutaneously implanted into BALB/C nude mice. Strikingly, exogenously introduced miR-543 significantly promoted tumor growth in both PC3 cells (P = 0.031) and DU145 cells (P = 0.0168) (Figure 1A), whereas the growth of tumor xenografts was significantly inhibited by the overexpression of anti-miR-543 using miR-543 inhibitors in both PC3 cells (P = 0.0176) and DU145 cells (P = 0.0263) (Figure 1A). Immunohistochemical staining was used to detect the expression of Ki67 and caspase-3 in the tumor xenografts. The results revealed increased numbers of Ki-67+ cells in two miR-543-mimic-transfected tumors and reduced numbers of caspase-3+ cells in miR-543-overexpressing DU145 and PC3 cell tumors (Figure 1B and 1C). These results suggest that the carcinogenic effects of miR-543 may be associated with the promotion of proliferation and the suppression of apoptosis. Taken together, these experiments provide in vivo evidence that miR-543 possesses PCa-promoting effects.
Figure 1.
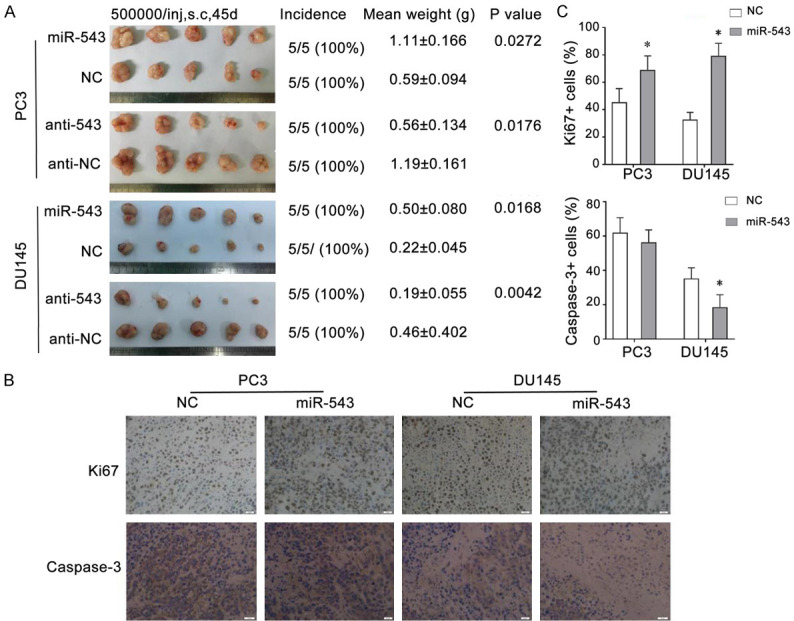
Overexpression of miR-543 promotes the growth of tumor xenografts in vivo. A. The images show tumors harvested from BALB/C nude mice that were subcutaneously injected with PC3 or DU145 cells transfected with the miR-543 mimic, mimic NC, miR-543 inhibitor (anti-543), or inhibitor NC (anti-NC). The tumor image, incidence (tumors/injections), final weights of the harvested tumors (mean ± S.D.), and their corresponding P values are shown on the left. B. Representative images of the immunohistochemical staining for Ki-67 and caspase-3 in harvested tumor tissues are shown on the bottom (magnification, × 200). C. Percentage analysis of cells staining positive for Ki67 and caspase-3 is shown on the right. Results are expressed as the means ± S.D. from three independent replicates, *P < 0.05.
MiR-543 increases pulmonary metastasis in a tail vein injection model
We asked whether the aberrant expression of miR-543 would also affect tumor metastatic behavior in vivo. To address this possibility, PC3 cells transfected with either the miR-543 mimic or NC for 48 hours were inoculated into BALB/C nude mice through the lateral tail vein. After six weeks, the number of metastatic lung nodules after lung isolation were analyzed (Figure 2A), and HE staining was performed (Figure 2C). As expected, the number of pulmonary metastatic nodules in the miR-543-overexpressing group was significantly higher than in the corresponding NC-transfected group (Figure 2B). These results demonstrate that miR-543 promotes lung tumor metastases of PCa in vivo.
Figure 2.
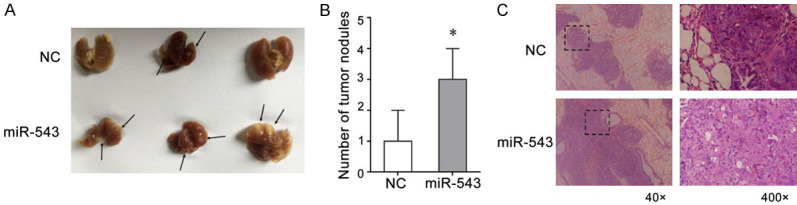
MiR-543 is significantly associated with pulmonary metastasis in a tail vein injection model. A. Images show lung tissues harvested 42 days after injecting PC3 cells transfected with the miR-543 mimic or mimic NC; arrows indicate visible metastatic lung nodules. B. The middle image shows the quantitative analysis of the number of metastatic lung nodules. C. HE staining of metastatic lung nodules from mice 42 days after injection with miR-543 mimic- and NC-transfected PC3 cells. Results are expressed as the means ± S.D. from three independent replicates, *P < 0.05.
MiR-543 facilitates PCa cell migration and invasion in vitro
To further study the mechanism of miR-543 function, we conducted a series of biological experiments in vitro. Exogenously introduced miR-543 significantly promoted the invasiveness of PC3 and DU145 cells, whereas cells transfected with the miR-543 inhibitor were unable to demonstrate invasiveness in vitro (Figure 3A and 3C). Consistent with this finding, the overexpression of miR-543 also promoted the migration of PCa cells, whereas the reduced expression of miR-543 had the opposite effects (Figure 3B and 3D). Considering that the EMT is closely related to the migration and invasion of tumor cells, E-cadherin and vimentin protein expression levels were measured by western blotting. Upregulated miR-543 expression levels enhanced vimentin protein expression but inhibited E-cadherin protein expression (Figure 3E). These results demonstrate that miR-543 may regulate tumor invasion and migration via the EMT in PCa cells. Taken together, our results suggest that miR-543 is a positive regulator of PCa metastasis.
Figure 3.
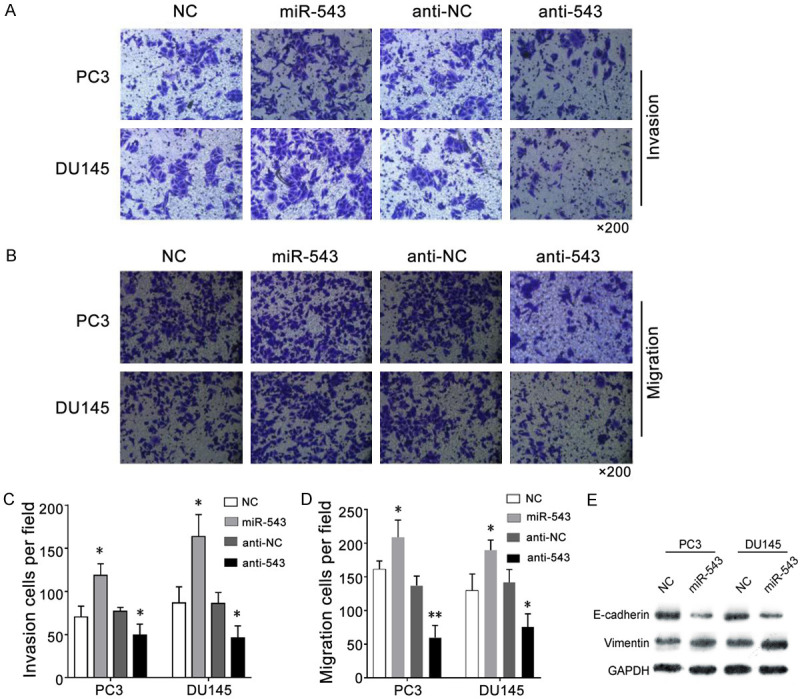
MiR-543 overexpression promotes PCa cell invasion and migration in vitro. A. A Transwell invasion assay was performed to compare the invasive abilities of different groups in PC3 and DU145 cells transfected with the miR-543 mimic, mimic NC, miR-543 inhibitor (anti-543), or inhibitor NC (anti-NC). B. A Transwell migration assay was performed to compare the migration abilities of the different groups. C. The invasion of cells from the different groups were analyzed. D. The migration of cells from the different groups were analyzed. E. Western blotting was used to detect differences in E-cadherin and vimentin protein levels in PCa cells transfected with the miR-543 mimic, miR-543 mimic, or their corresponding NC. Results are expressed as the means ± S.D., *P < 0.05.
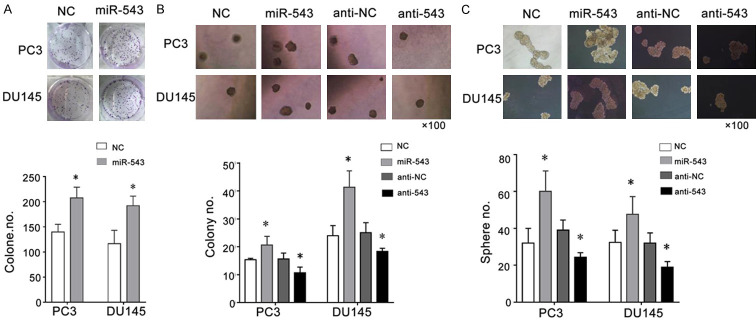
MiR-543 promotes the clonal, clonogenic, and sphere-forming activities of PCa cells in vitro
In addition to our study, a previous study showed that a fraction of PCa cells possess CSC properties [16], such as the ability to form spheres in a 3D matrix gel culture and floating spheres on ultralow adhesion culture plates. These CSC-related self-renewal properties can be regulated by certain miRs, such as miR-34a [18] and miR-128a [16]. We thus performed clonal, 3D Matrigel clonogenic, and sphere-formation assays to detect the self-renewal ability of CSCs in miR-543-overexpressing and miR-543-knockdown PCa cells. Compared with the NC group, PC3 and DU145 cells transfected with the miR-543 mimic showed stronger clonal, clonogenic, and sphere-forming activities in vitro (Figure 4A-C). When PC3 or DU145 cells were transfected with the miR-543 inhibitor, their clonogenic and sphere-forming activities were noticeably decreased (Figure 4B and 4C). These experiments confirm that miR-543 promotes the clonal and clonogenic abilities of PCa cells, as well as their CSC-related properties.
Figure 4.
MiR-543 overexpression promotes clonal, clonogenic, and sphere-forming activities of PCa cells in vitro. A. PC3 and DU145 cells transfected with the miR-543 mimic or NC were plated onto a six-well plate for 48 hours, and clonal experiments were performed. B. The two cell lines were transfected with the miR-543 mimic, mimic NC, inhibitor (anti-543), or inhibitor NC (anti-NC) for 48 hours, and the 3D Matrigel clonogenic assay was performed. C. The sphere-formation assay was performed in transfected PC3 and DU145 cells to determine the self-renewal ability of the CSCs. All results are expressed as the means ± S.D. from three independent replicates. *P < 0.05.
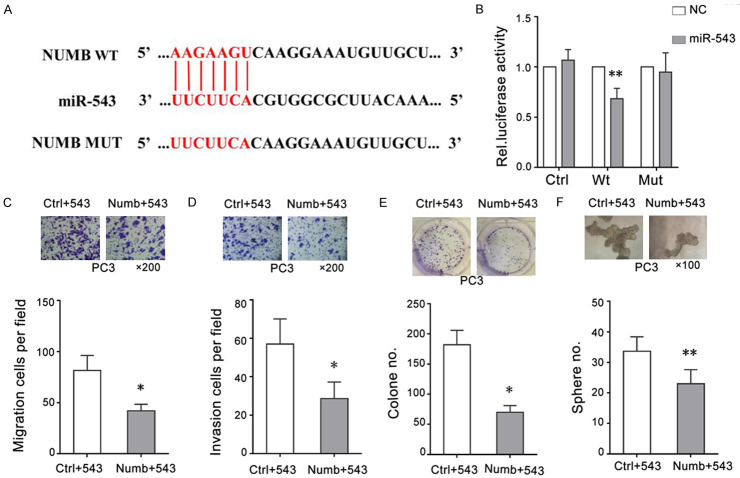
Numb is a direct and functional target of miR-543 in PCa cells
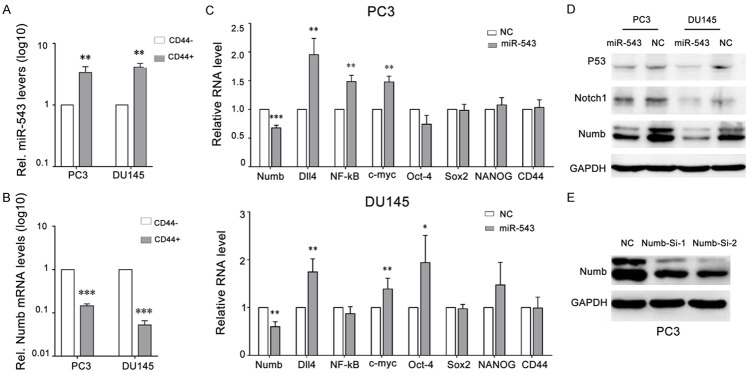
To identify the target of miR-543, we performed a prediction analysis using PIPA/miRanda/TarBase software (v.8) and found that Numb was a potential target of miR-543 (Figure 5A). Western blot and qRT-PCR analyses indicated that Numb protein and mRNA were negatively regulated by miR-543 in PCa cells (Figure 6C and 6D). In CD44+ PCSCs (Figure 6B), Numb expression levels were reduced compared to levels in CD44- non-PCSC cells, in contrast to miR-543 expression levels (Figure 6A). Further, miR-543 overexpression reduced the mRNA levels of Dll4, NF-κB, c-myc, and Oct-4 (Figure 6C), all of which are known oncogenes and stem cell regulators implicated in PCa initiation and progression. We also found that miR-543 overexpression reduced p53 expression but did not affect Notch1 expression (Figure 6D). When we used two siRNAs (Numb-si-1 and Numb-si-2) to knockdown Numb protein levels (Figure 6E), there were no effects on Notch1 expression in DU145 and PC3 cells (data not shown).
Figure 5.
Numb is a direct and functional target of miR-543 in PCa cells. (A) The schematic diagram shows the region where miR-543 is believed to bind the Numb 3’-UTR and the corresponding mutation of Numb in the luciferase reporter assay. (B) Dual luciferase assays showed that luciferase activity was inhibited by miR-543. Cells were co-transfected with miR-128 or NC, and reporter vectors containing empty (Ctrl), wild-type (Wt), or mutant (Mut) Numb for 48 hours. (C-F) PC3 cells were co-transfected with the miR-543 mimic and Numb cDNA lacking the 3’-UTR to restore the expression of Numb for 48 hours, and then the migration (C), invasion (D), and clonal (E) and sphere-formation abilities (F) of PC3 cells were measured. The results are represented as the means ± SEM of three independent assays, *P < 0.05, **P < 0.01.
Figure 6.
MiR-543 regulates the expression of Numb and several stem-like regulators. A. A qPCR assay was performed to assess miR-543 levels in CD44+ DU145 and CD44+ PC3 cells compared to CD44- PCa cells. B. A qPCR assay was performed to assess Numb levels in CD44+ DU145 and CD44+ PC3 cells compared to CD44- PCa cells. C. The effects of miR-543 overexpression are shown on Numb expression and the expression of several stem cell regulators in DU145 and PC3 cells. D. Western blotting was performed to assess the effects of miR-543 overexpression on Numb, p53, and Notch1 levels. E. Numb protein was knocked down using Numb siRNA-1 and Numb siRNA-2. The results are represented as the means ± SEM of three independent assays, *P < 0.05, **P < 0.01, ***P < 0.001.
To verify whether Numb is a functional target of miR-543, we cloned a fragment sequence containing the miR-543-binding site (wild-type Numb) and also mutated the miR-543-binding site (mutant-type Numb) to construct luciferase reporter gene vectors (Figure 5A). Results of the luciferase reporter assay showed that luciferase activity was significantly decreased when cells were co-transfected with the wild-type Numb vector and the miR-543 mimic, whereas co-transfection with the mutant-type Numb vector and the miR-543 mimic did not significantly alter the luciferase activity (Figure 5B). These data illustrate that a mutation in the region where miR-543 binds to the Numb 3’-UTR blocks the function of miR-543 and highlights that Numb is the target of miR-543. We then constructed a Numb-overexpressing plasmid that contains the entire Numb cDNA without the 3’-UTR (Numb-ORF), which does not bind miR-543. A rescue assay demonstrated that cells co-transfected with the Numb-ORF plasmid and the miR-543 mimic were noninvasive and could not form tumor spheres compared to cells co-transfected with empty vector and the miR-543 mimic (Figure 5C-F). These results suggest that Numb is a functional target gene of miR-543, as miR-543 was overexpressed in CD44+ PCa cells. Taken together, these data support that Numb is a direct and functional target that mediates the proliferative, metastatic, and tumorigenic effects of miR-543 in PCa cells.
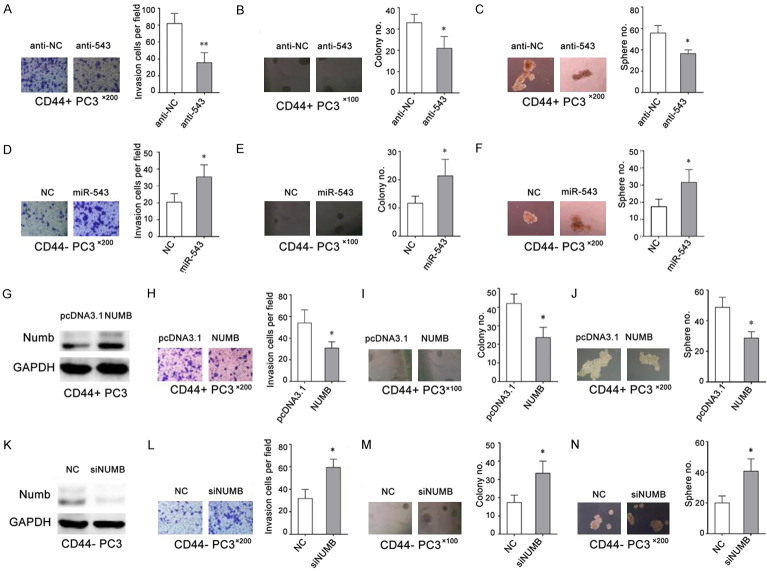
MiR-543 and Numb exert opposite effects in CD44+ PCSCs
As miR-543 and its target Numb have inversely proportional expression levels in CD44+ PCSCs, we speculated that miR-543 and Numb may play different roles in PCSCs. To verify whether miR-543 positively regulates PCSCs, we manipulated miR-543 expression in CD44+ PCSCs and CD44- non-PCSCs followed by cell clonal, clonogenic, and sphere-forming assays. As shown in Figure 7A-C, the downregulation of miR-543 in purified CD44+ PC3 cells significantly inhibited tumor cell clonal, clonogenic, and sphere-forming activities in that fewer clones were regenerated and the developed spheres were much smaller. In contrast, the overexpression of miR-543 in purified CD44- PC3 cells promoted tumor cell clonal, clonogenic, and sphere-forming activities (Figure 7D-F).
Figure 7.
MiR-543 and Numb exert opposite effects in CD44+ PCSCs. A-C. Transwell invasion assays, 3D Matrigel clonogenic assays, and sphere-formation assays were performed in CD44+ PC3 cells transfected with the miR-543 inhibitor (anti-543) or inhibitor NC (anti-NC). D-F. Transwell invasion assays, 3D Matrigel clonogenic assays, and sphere-formation assays were respectively performed in CD44- PC3 cells transfected with the miR-543 mimic or mimic NC. G. Numb protein levels in CD44+ PC3 cells transfected with Numb vector containing full-length Numb. H-J. Transwell invasion assays, 3D Matrigel clonogenic assays, and sphere-formation assays were performed in pcDNA3.1-Numb- and pcDNA3.1-Ctrl-transfected CD44+ PC3 cells. K. Numb protein levels in CD44- PC3 cells transfected with Numb siRNA. L-N. Transwell invasion assays, 3D Matrigel clonogenic assays, and sphere-formation assays were performed in CD44- PC3 cells transfected with Numb siRNA or NC siRNA. The results are represented as the means ± SEM of three independent assays, *P < 0.05.
Because Numb was expressed at lower levels in CD44+ PCa stem cell populations, we further verified whether Numb negatively regulate PCSCs. To address this question, we manipulated Numb expression in CD44+ and CD44- PC3 cells followed by tumor invasion, 3-D clone formation, and sphere-formation assays. As expected, the overexpression of PCDNA3.1-Numb upregulated Numb protein levels in CD44+ Du145 cells (Figure 7G) and reduced invasion capacity (Figure 7H), 3-D clone establishment (Figure 7I), and sphere-formation (Figure 7J). In contrast, Numb-1 downregulation by siNumb (Figure 7K) promoted invasion capacity (Figure 7L), 3-D clone establishment (Figure 7M), and sphere-formation (Figure 7N). These results suggest that miR-543 and Numb exert opposite functions in CD44+ PCSCs and that the PCSC-promoting effects of miR-543 are mediated, at least in part, through Numb.
Discussion
PCa is one of the leading causes of morbidity and mortality among adult males worldwide, especially in western countries [19]. It is a biologically heterogeneous and multifactorial disease. Localized PCa can be successfully treated with radical prostatectomy or radiation therapy. However, about 40% of patients will eventually progress to an advanced, recurrent, or metastatic PCa stage [20]. Though androgen deprivation therapy can induce positive responses among patients, most eventually develop CRPC. Despite such efforts, CRCP is not an effective therapy. There is, therefore, an urgent need to identify new therapeutic targets for PCa. Although recent studies have provided additional insights into both the oncogenic and tumor suppressive functions of miRNAs in the development of PCa [21], the exact molecular mechanisms underlying the miRNA/target interaction in PCa are unclear. The in vitro and in vivo analyses included in the present study demonstrate that the tumor oncogene miR-543 promotes tumor growth and metastasis, including stem cell-like traits, by directly targeting Numb. Inhibiting the expression of miR-543 or overexpressing Numb can elicit antitumor effects in PCa.
MiR-543 is a member of the miRNA cluster located in the imprinted DLK1-DIO3 region on human chromosome 14 [22]. Some studies have demonstrated an association between miR-543 and several human cancers, but contrasting reports exist. Zhao et al. found that miR-543 facilitated the migration, invasion, and EMT of esophageal cancer cells by targeting phospholipase A2 group IVA members [7]. Yu et al. showed that miR-543 acted as an oncogene by targeting PAQR3 in hepatocellular carcinoma (HCC), promoting tumor proliferation and invasion [23]. In contrast, Xu et al. reported that miR-543 expression was downregulated in glioma and functioned as a tumor suppressor to induce apoptosis and inhibit growth, the cell cycle, and tumor invasion [24]. A similar study found that miR-543 inhibited colorectal cancer metastasis by restraining the expression of KRAS, MTA1, and HMGA2 [25]. These conflicting reports suggest that miR-543 may exert heterogeneous functions in different tumors or cell types at different developmental stages and/or under normal vs. pathological conditions. Our present research confirms that miR-543 is negatively correlated with the expression of Numb in AI-PCa cell lines (DU145 and PC3) and functions as an oncogene. At the cellular and whole-animal levels, miR-543 positively regulates PCa cells and PCSCs. Its overexpression also promotes the growth of tumor xenografts and pulmonary metastases in vivo, and increases clonal, clonogenic, and sphere-forming activities of PCa cells and the EMT in vitro, whereas its downregulation inhibits the malignant behavior of PCa cells both in vivo and in vitro. The results presented here fully indicate that in AI-PCa, miR-543 plays an oncogenic role.
Numb was first described as a regulator of cell-fate in neurons [26] and was consequently reported to be responsible for the anti-oncogenic properties in certain cancers [13]. Numb proteins are involved in various functions, such as the control of asymmetric cell division [27], cell migration [28], cell adhesion [29], and endocytosis [30]. The tumor suppressive functions of Numb have been observed in multiple cancers. As highlighted in a previous study [12] of a panel of PCa tissues with different Gleason scores, Numb protein expression decreased with increasing Gleason score, indicating a positive correlation with a poor prognosis. Numb exerts its biological function principally by negatively regulating a number of signaling pathways (i.e., Notch signaling [10] and p53 signaling [31]). As for its upstream regulatory mechanisms, Numb appears to be regulated by miR-31-5a [14] in colorectal cancer and by miR-31/96/182 in head and cancer [32]. However, which miRs can functionally regulate Numb in PCa remains unclear. In this study, we found that miR-543 can directly target Numb mRNA in PCa cells and reduce Numb protein expression. Numb represents a bona fide as well as a functional target of miR-543, as the expression of a Numb cDNA lacking the miR-543-binding site at the 3’-UTR partly reduced the migration, invasion, and clonal and sphere-forming activities of miR-543-overexpressing PCa cells. Further, Numb inhibited the self-renewal of PSCSs, in contrast to miR-543. These observations suggest that Numb is an important downstream target of miR-543 for exerting miR-543’s oncogenic functions.
We also examined the impact of miR-543 on potential downstream effectors of Numb (Notch1 and p53) in PCa. The results showed that p53 protein levels were significantly reduced, whereas miR-543 was overexpressed and Numb was downregulated compared with control cells; however, Notch1 protein levels were not notably changed. Although Notch1 is widely considered to be a downstream regulator of Numb, this presumption has not been fully proven in PCa, and the involved regulatory mechanism remains unclear. Recent studies revealed the dual tumor suppressing and promoting function of Notch1 in human PCa [33]. Carvalho et al. revealed that the expression of Notch1 can confer a survival advantage in PCa but can also antagonize the survival of both benign and malignant PCa cells [34]. In our present research, we found that Notch1 was not a downstream target of Numb in DU145 or PC3 cells, as when we used Numb siRNA to inhibit Numb expression, Notch1 was unaffected. Future studies are thus need to clarify the role of Notch signaling in different PCa cell lines with different levels of androgen receptor expression.
Conclusion
Our study defines Numb as a novel target of the oncogene miR-543 in PCa cells. Inhibition of miR-543 or overexpression of Numb decreases PCa cell proliferation, the clonogenic and sphere-forming activities of CSCs, the EMT, and metastasis, and induces the regression of PCa tumors. These results suggest that miR-543 may act as an oncogene and that the miR-543/Numb/p53 pathway may be a potential target for the diagnosis and treatment of PCa.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81602255). National Natural Science Foundation of China, Grant/Award Number: 81602255.
Disclosure of conflict of interest
None.
References
- 1.Klemann N, Roder MA, Helgstrand JT, Brasso K, Toft BG, Vainer B, Iversen P. Risk of prostate cancer diagnosis and mortality in men with a benign initial transrectal ultrasound-guided biopsy set: a population-based study. Lancet Oncol. 2017;18:221–229. doi: 10.1016/S1470-2045(17)30025-6. [DOI] [PubMed] [Google Scholar]
- 2.Hellerstedt BA, Pienta KJ. The current state of hormonal therapy for prostate cancer. CA Cancer J Clin. 2002;52:154–179. doi: 10.3322/canjclin.52.3.154. [DOI] [PubMed] [Google Scholar]
- 3.Wen X, Deng FM, Wang J. MicroRNAs as predictive biomarkers and therapeutic targets in prostate cancer. Am J Clin Exp Urol. 2014;2:219–230. [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Q, Zhao A, Ren L, Chen J, Liao K, Wang Z, Zhang W. MicroRNA-1291 mediates cell proliferation and tumorigenesis by downregulating MED1 in prostate cancer. Oncol Lett. 2019;17:3253–3260. doi: 10.3892/ol.2019.9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Luo X, Wu Y, Xia D, Chen W, Fang Z, Deng J, Hao Y, Yang X, Zhang T, Zhou L, Wu Y, Wang Q, Xu J, Hu X, Li L. MicroRNA-34a attenuates paclitaxel resistance in prostate cancer cells via direct suppression of JAG1/notch1 axis. Cell Physiol Biochem. 2018;50:261–276. doi: 10.1159/000494004. [DOI] [PubMed] [Google Scholar]
- 6.Zheng XM, Zhang P, Liu MH, Chen P, Zhang WB. MicroRNA-30e inhibits adhesion, migration, invasion and cell cycle progression of prostate cancer cells via inhibition of the activation of the MAPK signaling pathway by downregulating CHRM3. Int J Oncol. 2019;54:443–454. doi: 10.3892/ijo.2018.4647. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhao H, Diao C, Wang X, Xie Y, Liu Y, Gao X, Han J, Li S. MiR-543 promotes migration, invasion and epithelial-mesenchymal transition of esophageal cancer cells by targeting phospholipase A2 group IVA. Cell Physiol Biochem. 2018;48:1595–1604. doi: 10.1159/000492281. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Zhou J, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39:BSR20190249. doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat Neurosci. 2002;5:1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Zhang K, Cheng C, Ji Z, Wang X, Wang M, Chu M, Tang DG, Zhu HH, Gao WQ. Numb(-/low) enriches a castration-resistant prostate cancer cell subpopulation associated with enhanced notch and hedgehog signaling. Clin Cancer Res. 2017;23:6744–6756. doi: 10.1158/1078-0432.CCR-17-0913. [DOI] [PubMed] [Google Scholar]
- 11.Zeng YL, Shao XM, Li HS. Numb expression in colon cancer and its significance. Sichuan Da Xue Xue Bao Yi Xue Ban. 2012;43:6–8. 14. [PubMed] [Google Scholar]
- 12.Sun J, Wang K, Teng J, Yu Y, Hua R, Zhou H, Zhong D, Fan Y. Numb had anti-tumor effects in prostatic cancer. Biomed Pharmacother. 2017;92:108–115. doi: 10.1016/j.biopha.2017.04.134. [DOI] [PubMed] [Google Scholar]
- 13.Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Peng H, Wang L, Su Q, Yi K, Du J, Wang Z. MiR-31-5p promotes the cell growth, migration and invasion of colorectal cancer cells by targeting NUMB. Biomed Pharmacother. 2019;109:208–216. doi: 10.1016/j.biopha.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 15.Saha SK, Choi HY, Kim BW, Dayem AA, Yang GM, Kim KS, Yin YF, Cho SG. KRT19 directly interacts with beta-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2017;36:332–349. doi: 10.1038/onc.2016.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV, Tang DG. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74:4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y, Liu XH, Zhu HC, Wang L, Ning JZ, Xiao CC. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roehrborn CG, Black LK. The economic burden of prostate cancer. BJU Int. 2011;108:806–813. doi: 10.1111/j.1464-410X.2011.10365.x. [DOI] [PubMed] [Google Scholar]
- 20.Beltran H, Beer TM, Carducci MA, de Bono J, Gleave M, Hussain M, Kelly WK, Saad F, Sternberg C, Tagawa ST, Tannock IF. New therapies for castration-resistant prostate cancer: efficacy and safety. Eur Urol. 2011;60:279–290. doi: 10.1016/j.eururo.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 21.Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21:126–144. doi: 10.1007/s12094-018-1910-8. [DOI] [PubMed] [Google Scholar]
- 22.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang J, Guo M, Liu N, Zhu L. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res. 2014;4:897–906. [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Yu J, Wang Z, Zhu Q, Wang W, Lan Q. miR-543 functions as a tumor suppressor in glioma in vitro and in vivo. Oncol Rep. 2017;38:725–734. doi: 10.3892/or.2017.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma H, Yu D, Maitikabili A, Xiao H, Zhang C, Liu F, Luo Q, Ouyang G. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7:21825–21839. doi: 10.18632/oncotarget.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chia W, Cai Y, Morin X, Tio M, Udolph G, Yu F, Yang X. The cell cycle machinery and asymmetric cell division of neural progenitors in the Drosophila embryonic central nervous system. Novartis Found Symp. 2001;237:139–151. doi: 10.1002/0470846666.ch11. discussion 151-163. [DOI] [PubMed] [Google Scholar]
- 27.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- 30.Dho SE, Trejo J, Siderovski DP, McGlade CJ. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: structural determinants of numb association with the cortical membrane. Mol Biol Cell. 2006;17:4142–4155. doi: 10.1091/mbc.E06-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 32.Chou CH, Tu HF, Kao SY, Chiang CF, Liu CJ, Chang KW, Lin SC. Targeting of miR-31/96/182 to the Numb gene during head and neck oncogenesis. Head Neck. 2018;40:808–817. doi: 10.1002/hed.25063. [DOI] [PubMed] [Google Scholar]
- 33.Lefort K, Ostano P, Mello-Grand M, Calpini V, Scatolini M, Farsetti A, Dotto GP, Chiorino G. Dual tumor suppressing and promoting function of Notch1 signaling in human prostate cancer. Oncotarget. 2016;7:48011–48026. doi: 10.18632/oncotarget.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvalho FL, Simons BW, Eberhart CG, Berman DM. Notch signaling in prostate cancer: a moving target. Prostate. 2014;74:933–945. doi: 10.1002/pros.22811. [DOI] [PMC free article] [PubMed] [Google Scholar]