Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) currently lacks sensitive approaches to detect cancer‐related traits in body fluid.
Methods
Methylation of tumor suppressor genes (TSGs) (PAX5, EDNRB, and DCC) were measured in the oral rinses from 50 HNSCC and 58 control subjects using droplet digital PCR (ddPCR). Diagnostic accuracies in detecting HNSCC and the detection rate of recurrence in the post‐treatment monitoring were analyzed.
Results
ddPCR TSG methylation detection in oral rinses for diagnosis of HNSCC had an AUC of 0.892 for PAX5, 0.753 for EDNRB, and 0.729 for DCC. Significant drop of TSG methylation was observed after completion of surgery (p < 0.01). 76.9% of the relapse cases had a pre‐emptive rebound of methylation above presurgery levels in at least one of the tested markers before confirmed recurrence.
Conclusions
Utilizing ddPCR for TSG methylation detection in oral rinses shows potential for detection and monitoring of HNSCC.
Keywords: droplet digital PCR, head and neck cancer, head and neck squamous cell carcinoma, liquid biopsy, methylation
1. INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer type worldwide with more than 700 000 new cases recorded annually. 1 , 2 Primary treatment of HNSCC has made advancements to reduce mortality and morbidity. 3 , 4 However, the 5‐year survival rate remains stagnant at approximately 40%–50%. Post‐treatment monitoring of patients with HNSCC is important for the early detection and treatment of the recurrent diseases. The current approach for post‐treatment monitoring utilizes physical examination and imaging like F‐18‐Fluorodeoxyglucose Positron Emission Tomography (F‐18‐FDG‐PET), for structural and functional analysis for recurrence. 5 , 6 , 7 The imaging approaches have been proposed and adopted in practice, but other approaches are required to address the possible concerns in both cost and radioactive exposures. 8 , 9 , 10 , 11
Molecular diagnostics for the detection of HNSCC offers the potential for early detection. Aberrant DNA methylation of CpG‐rich sequences within promoter regions of many tumor suppressor genes (TSGs) has been detected in tumor tissues, blood, and oral rinses from patients with HNSCC. 12 , 13 , 14 , 15 Previous attempts demonstrated successful detection of TSG methylation in the oral rinses and plasma of patients with HNSCC that were absent in normal patients but with relatively low sensitivity. 13 , 16 , 17 , 18 , 19 , 20 , 21 The development droplet digital PCR (ddPCR) technology, which facilitates partitioning of input DNA into 20 000 droplets, allowing PCR reactions in each individual droplet, and offering a distinct advantage in the ability to directly quantify reactions. 22 , 23 , 24 Hayashi et al. reported the change from qPCR to ddPCR for Paired Box 5 (PAX5) methylation detection increased the detection rate from 29% to 71% in surgical margin imprint specimens. 25 This demonstrated the potential of adapting ddPCR technology may enhance the detection of finite methylated signal found in patients with HNSCC. In this study, we developed ddPCR methylation detection optimized for fragmented DNA and sought to evaluate the feasibility of using ddPCR methylation analysis on a panel of TSG methylation markers in oral rinses for the noninvasive precise detection and monitoring of HNSCC.
2. MATERIALS AND METHODS
2.1. Patients recruitment
Patients who presented with a HNSCC to the Prince of Wales Hospital and United Christian Hospital in Hong Kong and agreed with written informed consent were included in the study. Subjects with the tumor type other than squamous cell carcinoma were excluded from the study. The study was approved by The Joint Chinese University of Hong Kong – New Territories East Cluster Clinical Research Ethics Committee (The Joint CUHK‐NTEC CREC) and Kowloon East Cluster Research Ethics Committee. Control subjects were recruited from Prince of Wales Hospital and Alice Ho Miu Ling Nethersole Hospital with no history of malignancies and above 18 years of age.
2.2. Sample collection
2.2.1. HNSCC cohort
Fifty patients with HNSCC were recruited with the collection of 30 mL oral rinse before surgery. Tumor and paired normal tissues (5 cm away from the tumor) were collected during surgery.
2.2.2. Control cohort
Fifty‐eight control cases were recruited in this study. Thirty‐two patients with head and neck cancer that required tonsillectomy for treatment of nonmalignant head and neck disease were recruited with the oral rinse collected before surgery and control tissue specimen collected during the surgery. Additional 26 healthy subjects were recruited with oral rinses alone. Two groups were combined for analysis in the oral rinse experiments.
2.3. Sample preparation
2.3.1. Oral rinse
Pretreatment 30 mL of oral rinses were obtained with normal saline gargled twice under supervision, for 20 and 10 s, respectively, as previously described. 13 , 18 The samples were centrifuged at 1600g for 10 min at 4°C. Supernatants were discarded and the cellular pellet will be stored at −80°C until further use.
2.3.2. Tumor and paired normal tissue
At the time of surgery, approximately 5 mm3 tumor samples were excised from resected specimens within the tumor mass, without involving the margin. Paired normal tissue more than 5 cm away from the margin of the tumor was excised. Tissue specimens were stored at −80°C until further use.
2.3.3. DNA extraction and bisulfite modification
Approximately 2 mm3 of tissue samples were used for DNA extraction with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The DNA concentration and quality were assessed with spectrophotometry (Nanodrop; Thermo Fisher Scientific, Waltham, MA). One microgram of extracted DNA from each specimen was used for bisulfite conversion with EpiTect Plus DNA Bisulfite Kit (Qiagen), according to the manufacturer's protocol.
2.4. Methylation panel
TSG methylation markers panel were chosen with established markers that were shown differentially methylated in HNSCC and detected previously in tissue and showing higher detection rate. PAX5, Endothelin Receptor β (EDNRB), and Deleted in Colorectal Cancer (DCC) were selected as potential markers for the detection of HNSCC. 17 , 25 , 26 The primer and the probe sequences were listed in Table 1.
TABLE 1.
Oligonucleotide sequences used in the methylation‐specific droplet digital PCR (ddPCR) assays
| Target | Name of oligonucleotide | Sequence (5′‐3′) a |
|---|---|---|
| PAX5 promoter region (Chr 9: 37002661–37002595) | Forward primer | TAAGAGAGAY GAAGGTAAGAGAGG |
| Reverse M primer | AAACAAACCCCGTAAAACG | |
| Reverse UM primer | AAAAACAAACCCCATAAAACA | |
| M probe | (FAM)‐TCGCGTAGTTTCGTCGG‐(MGB) | |
| UM probe | (VIC)‐TTTGTGTAGTTTTGTTGGGGA‐(MGB) | |
| EDNRB promoter region (Chr 13: 77919140–77919056) | Forward prime | GGGAGTTGTAGTTTAGTTAGTTAGGGAG |
| Reverse M primer | TACCCCGCGATTAAACTCG | |
| Reverse UM primer | CCTCTATACCCCACAATTAAACTCA | |
| M probe | 6FAM‐TAGCGGTTTTTATTCGTCGG‐MGBNFQ | |
| UM probe | VIC‐TAGTGGTTTTTATTTGTTGGGA‐MGBNFQ | |
| DCC promoter region (Chr 18: 52340708–52340791) | Forward primer | TGTGTATGY GTGTGTGAGTGTATGTG |
| Reverse M primer | CCATATTTCAACCAACACCTTCG | |
| Reverse UM primer | CCATATTTCAACCAACACCTTCA | |
| M probe | 6FAM‐TCGTTGTTCGCGATTT‐MGBNFQ | |
| UM probe | VIC‐TTGTTGTTGTTTGTGATTT‐MGBNFQ |
Abbreviations: DCC, Deleted in Colorectal Cancer; EDNRB, Endothelin Receptor Beta; M, methylated specific; PAX5, Paired Box 5; UM, unmethylated specific.
The differentially methylated nucleotides are listed as underlined. The degenerate base in the designed sequence denoted as italic Y.
2.5. Methylation‐specific ddPCR
The ddPCR assays were designed with one hydrolysis probe labeled with FAM dye that targeted methylated sequences (from the tumor), with all the differentially methylated CpG sites remained unconverted a cytosine after bisulfite conversion. The other hydrolysis probe in the assays was labeled with VIC dye (from normal tissue), with differentially methylated CpG sites all converted uracil and amplified into thymidine. The assay also contained a nondifferentiating forward primer and a set of reverse primers with one targeting methylated sequences and the other targeted unmethylated sequences. Each reaction was performed in duplicate.
The ddPCR was performed using the QX200 Droplet Digital PCR system (Bio‐Rad, Hercules, CA). Bisulfite‐treated DNA (8 μL) were added to complete the reaction mix (20 μL). The forward primer was prepared at a final concentration of 900 nmol/L, with the set of reverse primers prepared at a final concentration each at 450 nmol/L. The probes were prepared at the concentration of 250 nmol/L. The thermal profile started with the initial activation step at 95°C for 10 min and then 45 cycles of the following two steps, 94°C for 15 s as the denaturing step and an anneal/extension step with temperature set differently for the markers, 52°C for 60 s (PAX5), 54°C for 60 s (EDNRB), 60°C for 60 s (DCC), followed by 98°C for 10 min of final incubation. Universal methylated human DNA and universally unmethylated human DNA (EpiTect Control DNA Set, Qiagen) was used in each run as the positive and negative control, respectively. QuantaSoft v1.7.4 (Bio‐Rad) was used in the system or machine control and subsequent analysis. Optimization of the assay is shown in Figure 1.
FIGURE 1.

Optimization of the droplet digital quantitative methylation specific PCR (ddqMSP) assays. (A) Testing of the ddqMSP assay of Deleted in Colorectal Cancer (DCC) using universally methylated DNA and universally unmethylated DNA, the assay showed specific identification of only methylated signal and unmethylated signal. (B) Quantification performances of the assays were evaluated under low methylation concentration testing, showing the assays could correctly identify the methylated sequence in low concentration conditions. Pearson correlation tests showed close correlation between the droplet digital PCR (ddPCR) results with the expected methylation density of the tested concentrations [Color figure can be viewed at wileyonlinelibrary.com]
In the digital PCR analysis, methylation density which refers to the fraction of molecules being methylated is calculated as
2.6. Statistical analysis
Statistical analyses were performed using MedCalc Statistical Software v18.5 and R software. 27 , 28 Differences of methylation markers between different groups are assessed with Student's t test, Mann–Whitney U test, and Wilcoxon signed‐rank test. The sensitivity and the specificity of the markers are determined by receiver‐operating characteristics (ROC) analysis and the area under the curve (AUC) of the ROC curves are compared.
3. RESULTS
3.1. Patient characteristics
Fifty patients with HNSCC were recruited in the study (Table 2). Their mean age was 66 years. Thirty‐seven (74%) were males and 13 (26%) were females. The group predominantly consisted of stage III/IV, 34 (68%). Fifty‐eight normal subjects were recruited, with a median age of 37.5 years, including 31 (53%) males and 27 (47%) females.
TABLE 2.
Demographics of recruited patients with head and neck squamous cell carcinoma (HNSCC) and control subjects
| Patient characteristics | Patients with HNSCC recruited, n = 50 (%) | Control subjects recruited, n = 58 (%) | p‐value | |
|---|---|---|---|---|
| Age (years) |
Median: 66 Range: 31–92 |
Median: 37.5 Range: 19–77 |
p < 0.001 | |
| Gender | M | 37 (74%) | 31 (53%) | p < 0.03 |
| F | 13 (26%) | 27 (26%) | ||
| Smoking status | Non‐smoker | 19 (38%) | 41 (71%) | p < 0.001 |
| Smoker | 31 (62%) | 13 (22%) | ||
| Unknown | 0 (0%) | 4 (7%) | ||
| Alcohol consumption | Non‐drinker | 32 (64%) | 45 (78%) | p < 0.01 |
| Drinker | 18 (36%) | 7 (12%) | ||
| Unknown | 0 (0%) | 6 (10%) | ||
| Primary site | Oral cavity | 37 (74%) | ||
| Oropharynx | 3 (6%) | |||
| Hypopharynx | 5 (10%) | |||
| Larynx | 4 (8%) | |||
| Paranasal sinus | 1 (2%) | |||
| Pathological T classification | T1/T2 | 24 (48%) | ||
| T3/T4 | 26 (52%) | |||
| Pathological N classification | N0 | 29 (58%) | ||
| N+ | 21 (42%) | |||
| Overall stage | I/II | 16 (32%) | ||
| III/IV | 34 (68%) | |||
Note: Comparison of the demographic feature between patients with HNSCC and control subjects were completed with Student's t test.
Abbreviations: F, female; HNSCC, head and neck squamous carcinoma; M, male.
3.2. TSG methylation analysis in tissue specimens
We first analyzed the methylation status of the tumor in the patients with HNSCC to assess the degree of the methylation of the selected TSG methylation markers, PAX5, EDNRB, and DCC. Selected markers showed significantly higher methylation density in the tested 50 tumor specimens, compared with paired normal tissues from patients (p < 0.001) and 32 control tissues from control subjects (p < 0.001). The comparison of the methylation density between tumor and control tissues of the individual markers is shown in Figure 2.
FIGURE 2.

Methylation density of tumor suppressor gene (TSG) markers in head and neck squamous cell carcinoma (HNSCC) tissue specimens and tissues from recruited control subjects. Paired Box 5 (PAX5) (A), Endothelin Receptor Beta (EDNRB) (B), and Deleted in Colorectal Cancer (DCC) (C). All three markers showed aberrant methylation in tumor, compared with the paired normal tissues (p < 0.001, Mann–Whitney U test) and with the control tissues (p < 0.001, Mann–Whitney U test). Each dot in the three groups represents individual patients. Median methylation level of each group was indicated [Color figure can be viewed at wileyonlinelibrary.com]
3.3. TSG methylation analysis in the oral rinse for HNSCC diagnosis
After evaluation in the tumor tissue methylation status, the markers were tested in oral rinses for their diagnostic performances. The methylation status of the three markers was examined in both the preoperative oral rinses of the 50 patients with HNSCC and 58 controls. All three markers including PAX5 (p < 0.001), EDNRB (p < 0.001), DCC (p < 0.02) showed a significantly higher methylation density in patients with HNSCC. Compared with the tumor methylation results, 38 (88%) cases showed PAX5 methylation in the tumor also recorded aberrant methylation in their oral rinse specimens, whereas for EDNRB and DCC, the number of cases that detected methylation in both tumor and oral rinse was 22 (82%) and 28 (78%), respectively. The diagnostic performances of the three TSG markers in differentiating patients with HNSCC from control subjects were compared using ROC analysis (Figure 3(A)). The sensitivity of PAX5 methylation analysis in oral rinse to detect HNSCC was 84.0% (95%CI: 70.9–92.8) with a specificity of 87.9% (95%CI: 76.7–95.0). The cut‐off for the PAX5 methylation density (PAX5 M%) in oral rinse was set at M% > 0.5%. The cut‐off for EDNRB hypermethylation in oral rinse was set at EDNRB M% > 0.1%. The sensitivity and specificity for EDNRB methylation analysis were 78.0% (95%CI: 64.0–88.5) and 72.4% (95%CI: 59.1–83.3), respectively. The cut‐off for DCC hypermethylation in oral rinse was set at DCC M% > 0.08%. While the sensitivity and specificity of DCC methylation analysis were 76.0% (95%CI: 61.8–86.9) and 72.4% (95%CI: 59.1–83.3). The AUC of the ROC analysis of PAX5 for HNSCC diagnosis was 0.892. While the AUC for the ROC of EDNRB and DCC were 0.753 and 0.729, respectively (Figure 3(A)).
FIGURE 3.

Comparison of the HNSCC diagnostic accuracy of TSG methylation markers. (A) Diagnostic accuracy of the individual TSG methylation markers. The cut‐off selected in the model was marked by a dot in each ROC curve. Diagnostic accuracy of Paired Box 5 (PAX5) methylation in oral rinse is significantly better compared with Endothelin Receptor Beta (EDNRB) and Deleted in Colorectal Cancer (DCC) (p < 0.004, log‐rank test). The cut‐off for PAX5 methylation was set at methylation density (M%) >0.5%. The cut‐off for EDNRB and DCC methylation was set at EDNRB M% >0.1% and DCC M% >0.08%, respectively. (B) Comparison between the marker combinations and the best‐performed TSG marker (PAX5). In the trial of the combination of the TSG methylation markers, the best performed three marker combination did not show significant improvement compared with PAX5 (p = 0.306, log‐rank test) [Color figure can be viewed at wileyonlinelibrary.com]
The markers were also evaluated of possible improvement in diagnostic accuracy for HNSCC through the marker combinations. A combination of all three markers had an AUC of 0.897, which showed no significant improvement when compared to using only PAX5 for the HNSCC diagnosis in oral rinse (p = 0.31). The ROC comparison is illustrated in Figure 3(B). There is no significant difference in the methylation level of the markers in the oral rinse of patients with HNSCC and the control cases, regardless of their smoking and alcohol consumption status (Tables S1–S10 and Figures S1 and S2, Supporting Information). In addition, there was no significant effect on the methylation level of the oral rinse with regard to the tumor load based on the T classifications and the site of the tumors (Tables S5–S10).
3.4. Methylation analysis in the oral rinse for patient monitoring
Forty‐eight recruited patients with HNSCC had oral rinse specimens collected 4 weeks after completion of surgical treatment. The methylation density of PAX5 (p < 0.001), EDNRB (p < 0.001), and DCC (p < 0.01) in oral rinse showed a significant drop after the surgical excision of the tumor (Figure 4). These 48 patients were serially monitored with a mean follow‐up period of 323 days (range: 75–447 days). For the cases that reached and remained in remission, the methylation densities of the three TSG markers remained at a low level after completion of the tumor resection. Seventy‐five percent of nonrecurring cases recorded PAX5 methylation density remained below presurgery level. While for EDNRB and DCC, 66% and 75% of those cases remained below presurgery methylation density, respectively (Figure 5(A)). Thirteen patients with disease recurrence were able to complete longitudinal study after treatment completion. Four (80%) patients with local recurrence have shown a significant rebound in at least one of the markers. Six (86%) patients with regional spread showed a significant rebound in at least one of the markers. Interestingly, two (100%) with distant metastasis showed a rebound in all three TSG markers. Both cases were diagnosed with lung metastasis. The variation of the TSG methylation in oral rinse has shown a correlation with the clinical events as illustrated with the cases below (Figure 5(B)).
FIGURE 4.
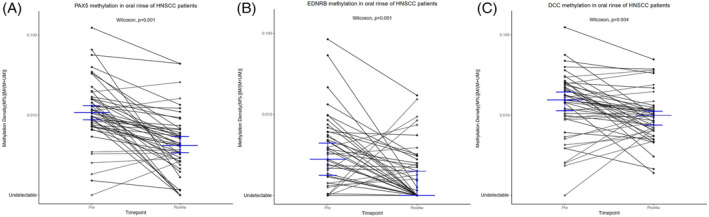
Change in methylation density of tumor suppressor gene (TSG) markers in oral rinse specimens before and after surgical treatment. The figure showed the paired comparison of the methylation densities of Paired Box 5 (PAX5) (A), Endothelin Receptor Beta (EDNRB) (B), and Deleted in Colorectal Cancer (DCC) (C) in oral rinse collected on presurgery and post 4‐week (first postsurgery timepoint) timepoint. Median level in each group was indicated by the blue mark. Three markers all shown significant drop in the methylation densities after completion of the surgery [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 5.

Longitudinal variation of the oral rinse tumor suppressor gene (TSG) methylation in head and neck squamous cell carcinoma (HNSCC) patients. (A) The methylation densities of TSG markers in serial collected oral rinse samples from non‐relapse patients showed significant drop after completion of the treatment and remained at low levels till final followup time point. (B) Some relapse cases showed close correlation between the variation of TSG methylation in oral rinse and the disease progression and treatment [Color figure can be viewed at wileyonlinelibrary.com]
Case HN2 was diagnosed with stage II buccal SCC. The methylation density of PAX5 in oral rinse dropped from 1.4% before surgery to 0% on Day 32 after tumor resection. PAX5 methylation remained at 0.3% on Day 111. The level rose to 0.8% on Day 181 and to 1.6% on Day 384. A parallel increase was also observed in EDNRB M% and DCC M% at 1.6% and 2.5%, respectively, on Day 384. The rise in the methylation level of the three tumor markers was 152 days ahead of the confirmed presence of lung metastasis.
Case HN6 was diagnosed with stage I buccal SCC. PAX5 M% dropped from 0.9% before surgery to 0.5%, 25 days after tumor resection. EDNRB M% also observed a drop from 0.2% before surgery to 0% at the same timepoint. PAX5 and EDNRB M% showed rebound reaching 1.1% and 0.5%, respectively, on 223 days after surgery. The patient was confirmed to have a regional recurrence on Day 265. The patient received second surgery on Day 336 and the methylation density of PAX5, EDNRB, and DCC all rose further to 2.7%, 1.4%, and 2.8% on Day 363 after the first surgery, 27 days after the second surgery. The patient eventually showed distant metastasis on Day 514 and died on Day 550.
Case HN15 was a patient with stage IVa buccal SCC. The methylation densities of the three TSG markers showed a significant drop 34 days after tumor resection. PAX5 methylation density dropped from 0.9% to 0.4%, EDNRB dropped from 0.3% to 0%, and DCC dropped from 1.4% to 0.1%. The patient was given subsequent concurrent chemoradiotherapy. After completion of the regiment, PAX5 and EDNRB showed an increase of methylation density to 0.5%, while DCC rose to 1%. The methylation density continued to rise till the last timepoint on Day 391, with PAX5 methylation density rose from 0.9% before surgery to 2.0% on Day 391 after tumor resection. While EDNRB methylation density rose from 0.3% pretreatment to 1.1% on Day 391, DCC methylation density rose from 1.4% before surgery to 1.5% on Day 391. The patient has subsequently diagnosed with Lung, adrenal and skull base metastasis on Day 658. The marked rebound of the three markers was recorded 267 days before confirmed relapse.
Case HN36 was a patient with stage II tongue SCC. PAX5 M% in oral rinse dropped from 0.8% to 0.3% on Day 27 after tumor resection. EDNRB methylation density dropped from 0.5% to 0.2% and DCC methylation density dropped from 1.4% to 0.7% at the same timepoint. The patient was found to have recurrence on Day 230 and salvage surgery was performed on Day 262. PAX5 methylation density rose from 0.6% on Day 188 to 1.7% at the time of salvage surgery. EDNRB and DCC M% also showed a parallel increase in the same period, EDNRB M% rose from 0.03% to 0.6%. DCC M% rose from 0.3% to 1.0%. PAX5, EDNRB, and DCC M% showed drop on Day 280, 18 days after salvage surgery. PAX5 M% dropped from 1.7% to 0.3%, EDNRB M% dropped from 0.6% to 0.3%, and DCC M% dropped from 1.0% to 0.7%. The patient was given concurrent chemoradiotherapy following the second surgery. The oral rinse TSG M% remained at a low level on Day 447, with PAX5 M% measured at 0.1%, EDNRB M% measured at 0.03%, and DCC M% measured at 0.5%. Follow‐up for the patient ended on Day 489 with the patient in remission.
4. DISCUSSION
This study illustrated the possible application of ddPCR methylation detection in oral rinses for the detection and monitoring of HNSCC. The evaluation of the markers in the study cohort confirmed that PAX5, EDNRB, and DCC were frequently methylated among patients with HNSCC. Such an observation was replicated in the pretreatment oral rinse specimens, showing aberrant methylation compared with the normal oral rinse specimens. This observation demonstrates the potential of ddPCR of methylation markers in the noninvasive detection of HNSCC. Compared to previous studies which utilized quantitative real‐time PCR approach, ddPCR‐based methylation detection achieved higher sensitivity, with EDNRB increased from 38% to 78% and DCC raised from 27% to 72%, respectively. AUC in the ROC analysis also increased from 0.58 to 0.75 for EDNRB and from 0.57 to 0.73 for DCC. 29
Previous studies demonstrated ddPCR provides enhanced resolution for the detection of finite targets in the pool of nontarget molecules. 22 , 30 , 31 This approach also allows for the absolute quantification of the target molecule without the need for an external reference standard. In this study, the ddPCR method allowed quantification and detection of methylated target that is common in the salivary rinse, with the dilution of unmethylated normal DNA to as low as 0.01%. This low detection limit allows the differentiation of background low methylation signals from the high tumor derived methylation signal and an improved sensitivity compared to previous studies.
The decrease in methylation density of TSG markers after treatment completion indicated the potential utility of the methylation analysis in oral rinse as a mean for disease monitoring. Utilizing oral rinse for analysis provided a noninvasive way to detect cancer‐specific aberrations. Wang et al. have previously shown that oral rinses have better detection of cancer‐specific aberrations and HPV infection in patients with oral and oropharyngeal cancer corroborating the use of oral rinses in the detection of HNSCC in these particular sites. 32 In our cohort of nonrelapse cases, the methylation density of the markers remained at consistently low levels throughout the study. In comparison, some of the relapse cases showed a substantial increase in TSG methylation density in oral rinses, which predates the confirmed diagnosis of disease recurrence. Such different patterns between relapse and nonrelapse patients suggest the possible use of the methylation analysis of serially collected oral rinse as a personalized tool for disease monitoring of patients with HNSCC. The methylation markers may provide molecular detection toward the subclinical residual disease before they manifest (Figure 5(B)).
This main limitation of the study is that the sample cohort mainly comprised of patients with oral cancer. Other studies had shown a similar pattern with the oral cancer cases comprised of more than 60% of the recruited cohort. 32 , 33 This may have led to the skewed representation of the status of the markers more in line with mostly patients with oral cancer, with the study among the other subsites of the patients with head and neck cancer yet to be confirmed. In the combined TSG marker panel, the PAX5 methylation contributed to a majority of the prediction power in the ROC curve analyses. For the HNSCC cases, especially human papilloma virus (HPV) negative cases, TP53 mutations has been shown to be highly prevalent. 33 PAX5 methylation has been indicated to be prevalent among the cases that carry the TP53 mutations in another report from Gurrero‐Preston et al. 34 A close correlation of PAX5 methylation with TP53 mutations was shown to be prevalent among a large number of patients with HNSCC. For EDNRB and DCC methylation, markers were previously shown to be methylated among 33%–67% of patients with HNSCC. 29 , 35 , 36 In this study, the detection rate of oral rinse EDNRB methylation and DCC methylation was 78% and 76%, respectively, demonstrating that our method has a higher sensitivity toward detecting the finite methylation signal found in the oral rinse of the patients with HNSCC for the two markers. PAX5 demonstrated superior performances as a biomarker for the HNSCC in comparison with EDNRB and DCC. But with the relatively small cohort, the diagnostic accuracies and performances for the markers may require further validation.
5. CONCLUSION
This study highlights the diagnostic potential of ddPCR‐based detection of TSG methylation in the oral rinse for the HNSCC. PAX5 methylation analysis in the oral rinse could be a highly sensitive and specific marker for the diagnosis and disease monitoring after treatment completion for the detection of the residual disease and traces for relapse but requires a larger cohort study.
AUTHOR CONTRIBUTIONS
Study design: Sherwood Y. H. Fung, Jason Y. K. Chan, and K. C. Allen Chan. Materials support: Sherwood Y. H. Fung, Jason Y. K. Chan, Cherrie W. K. Ng, Eddy W. Y. Wong, Ryan Cho, and Zenon W. C. Yeung. Conduct of experiment: Sherwood Y. H. Fung and K. C. Allen Chan. Data interpretation and analysis: Sherwood Y. H. Fung, K. C. Allen Chan, Jacky W. K. Lam, and Jason Y. K. Chan. Manuscript writing: all authors. Approved of the final manuscript: all authors.
Supporting information
Appendix S1: Supporting information
ACKNOWLEDGMENTS
We thank the anonymous participants providing samples for this study. The work was supported by the Dr. Stanley Ho Medical Foundation (Jason Y. K. Chan), the Research Grants Council of the Hong Kong SAR government under the Theme‐based research scheme (T12‐401/16‐W) (K. C. Allen Chan) and General Research Fund (CUHK 14109716 and 14108818) (Jason Y. K. Chan). The funders had no role in study design, data collection, analysis, interpretation, or writing of the report. K. C. Allen Chan is a consultant of and holds equity in Grail. K. C. Allen Chan holds equity of and is a director of Take2. K. C. Allen Chan and Jacky W. K. Lam have filed patents/patent applications on molecular diagnostics and receive royalties from Grail and Take2. Jacky W. K. Lam holds equity in Grail. Jason Y. K. Chan is a consultant for Intuitive Surgical Inc.
Fung SYH, Chan KCA, Wong EWY, et al. Droplet digital PCR of tumor suppressor gene methylation in serial oral rinses of patients with head and neck squamous cell carcinoma. Head & Neck. 2021;43:1812–1822. 10.1002/hed.26647
Section Editor: Patrick Ha
Funding information Dr Stanley Ho Medical Foundation; Research Grants Council, University Grants Committee, Grant/Award Numbers: General Research Fund (CUHK 14108818), General Research Fund (CUHK 14109716), Theme‐based Research Scheme (T12‐401/16‐W)
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Economopoulou P, Psyrri A. Epidemiology, risk factors and pathogenesis of squamous cell tumours. In: Licitra L, Karamouzis MV, Ghielmini M, eds. Essentials for Clinicians: Head & Neck Cancers. Lugano, Switzerland: European Society for Medical Oncology; 2017:1‐2. [Google Scholar]
- 3. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386‐396. [DOI] [PubMed] [Google Scholar]
- 4. Chan JYK, Tsang RK, Holsinger FC, et al. Prospective clinical trial to evaluate safety and feasibility of using a single port flexible robotic system for transoral head and neck surgery. Oral Oncol. 2019;94:101‐105. [DOI] [PubMed] [Google Scholar]
- 5. Kawecki A, Krajewski R. Follow‐up in patients treated for head and neck cancer. Memory. 2014;7:87‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saba NF. Posttreatment surveillance of squamous cell carcinoma of the head and neck. In: Posner M, Brockstein B, Brizel D, Fried M, Shah S, eds. UpToDate. Waltham, MA: UpToDate; 2019. [Google Scholar]
- 7. Ng SP, Pollard C, Berends J, et al. Usefulness of surveillance imaging in patients with head and neck cancer who are treated with definitive radiotherapy. Cancer. 2019;125:1823‐1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner DJ, Hall EJ. Computed tomography—an increasing source of radiation exposure. N Engl J Med. 2007;357:2277‐2284. [DOI] [PubMed] [Google Scholar]
- 9. Smith‐Bindman R, Lipson J, Marcus R, et al. Radiation dose associated with common computed tomography examinations and the associated lifetime attributable risk of cancer. Arch Intern Med. 2009;169:2078‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Annunziata S, Caldarella C, Treglia G. Cost‐effectiveness of Fluorine‐18‐Fluorodeoxyglucose positron emission tomography in tumours other than lung cancer: a systematic review. World J Radiol. 2014;6:48‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yankevich U, Hughes MA, Rath TJ, et al. PET/CT for head and neck squamous cell carcinoma: Should we routinely include the head and abdomen? Am J Roentgenol. 2017;208:844‐848. [DOI] [PubMed] [Google Scholar]
- 12. Sanchez‐Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892‐895. [PubMed] [Google Scholar]
- 13. Rosas SL, Koch W, da Costa Carvalho MG, et al. Promoter hypermethylation patterns of p16, O6‐methylguanine‐DNA‐methyltransferase, and death‐associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939‐942. [PubMed] [Google Scholar]
- 14. Reed AL, Califano J, Cairns P, et al. High frequency of p16 (CDKN2/MTS‐1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630‐3633. [PubMed] [Google Scholar]
- 15. Dayyani F, Etzel CJ, Liu M, Ho CH, Lippman SM, Tsao AS. Meta‐analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC). Head Neck Oncol. 2010;2:15‐3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carvalho AL, Henrique R, Jeronimo C, et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res. 2011;17:4782‐4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carvalho AL, Chuang A, Jiang WW, et al. Deleted in colorectal cancer is a putative conditional tumor‐suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401‐9407. [DOI] [PubMed] [Google Scholar]
- 18. Carvalho AL, Jeronimo C, Kim MM, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97‐107. [DOI] [PubMed] [Google Scholar]
- 19. Viet CT, Jordan RC, Schmidt BL. DNA promoter hypermethylation in saliva for the early diagnosis of oral cancer. J Calif Dent Assoc. 2007;35:844‐849. [PubMed] [Google Scholar]
- 20. Ovchinnikov DA, Cooper MA, Pandit P, et al. Tumor‐suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Transl Oncol. 2012;5:321‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schrock A, Leisse A, de Vos L, et al. Free‐circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin Chem. 2017;63:1288‐1296. [DOI] [PubMed] [Google Scholar]
- 22. Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real‐time PCR. Nat Methods. 2013;10:1003‐1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol. 2015;26:1994‐1999. [DOI] [PubMed] [Google Scholar]
- 24. Gai W, Ji L, Lam WKJ, et al. Liver‐ and colon‐specific DNA methylation markers in plasma for investigation of colorectal cancers with or without liver metastases. Clin Chem. 2018;64:1239‐1249. [DOI] [PubMed] [Google Scholar]
- 25. Hayashi M, Guerrero‐Preston R, Sidransky D, Koch WM. Paired box 5 methylation detection by droplet digital PCR for ultra‐sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer Prev Res (Phila). 2015;8:1017‐1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pattani KM, Zhang Z, Demokan S, et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev Res (Phila). 2010;3:1093‐1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MedCalc Software bvba . MedCalc Statistical Software version 18.5. 2018.
- 28. R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 29. Schussel J, Zhou XC, Zhang Z, et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin Cancer Res. 2013;19:3268‐3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hindson BJ, Ness KD, Masquelier DA, et al. High‐throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604‐8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003‐1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guerrero‐Preston R, Michailidi C, Marchionni L, et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031‐1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun W, Zaboli D, Liu Y, et al. Comparison of promoter hypermethylation pattern in salivary rinses collected with and without an exfoliating brush from patients with HNSCC. PLoS One. 2012;7:e33642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Demokan S, Chang X, Chuang A, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127:2351‐2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


