Abstract
In Metazoa, four out of five complexes involved in oxidative phosphorylation (OXPHOS) are formed by subunits encoded by both the mitochondrial (mtDNA) and nuclear (nuDNA) genomes, leading to the expectation of mitonuclear coevolution. Previous studies have supported coadaptation of mitochondria-encoded (mtOXPHOS) and nuclear-encoded OXPHOS (nuOXPHOS) subunits, often specifically interpreted with regard to the “nuclear compensation hypothesis,” a specific form of mitonuclear coevolution where nuclear genes compensate for deleterious mitochondrial mutations due to less efficient mitochondrial selection. In this study, we analyzed patterns of sequence evolution of 79 OXPHOS subunits in 31 bivalve species, a taxon showing extraordinary mtDNA variability and including species with “doubly uniparental” mtDNA inheritance. Our data showed strong and clear signals of mitonuclear coevolution. NuOXPHOS subunits had concordant topologies with mtOXPHOS subunits, contrary to previous phylogenies based on nuclear genes lacking mt interactions. Evolutionary rates between mt and nuOXPHOS subunits were also highly correlated compared with non-OXPHO-interacting nuclear genes. Nuclear subunits of chimeric OXPHOS complexes (I, III, IV, and V) also had higher dN/dS ratios than Complex II, which is formed exclusively by nuDNA-encoded subunits. However, we did not find evidence of nuclear compensation: mitochondria-encoded subunits showed similar dN/dS ratios compared with nuclear-encoded subunits, contrary to most previously studied bilaterian animals. Moreover, no site-specific signals of compensatory positive selection were detected in nuOXPHOS genes. Our analyses extend the evidence for mitonuclear coevolution to a new taxonomic group, but we propose a reconsideration of the nuclear compensation hypothesis.
Keywords: mitonuclear coevolution, compensatory evolution, OXPHOS, bivalvia, evolutionary rates
Introduction
Mitochondria are the product of an ancient endosymbiotic event between an Archaea-like prokaryote and an alpha-proteobacterium (reviewed in Martin et al. 2015) that led to the evolution of eukaryotes and morphologically complex life as we know it today (Martin and Müller 1998; Martin and Koonin 2006; Lane and Martin 2010; Hill 2015; Zachar and Szathmáry 2017). The mitochondrial genome is a genetic relic of complex evolutionary processes that resulted in an extensive reduction of the alpha-proteobacterium genome, involving both gene loss and transfer to the nuclear genome (Gray et al. 1999; Timmis et al. 2004; Martin and Koonin 2006; Gray 2012).
At present, different eukaryotic lineages have variable mitochondrial genome sizes, organization, and gene content (Kolesnikov and Gerasimov 2012; Sloan et al. 2018). However, beside genes involved in translation, one consistent pattern is the maintenance of a limited set of protein-coding genes (PCGs) involved in the oxidative phosphorylation (OXPHOS) metabolic pathway, the main mechanism of ATP production in aerobic eukaryotes. OXPHOS is carried out by four protein complexes that produce a proton gradient across the internal mitochondrial membrane (Complexes I–IV or CI–IV), and an ATPase that exploits this gradient to produce ATP (Complex V or CV). In almost all bilaterian animals, 13 PCGs encoding components of CI and CIII–V are found in the mitochondrial genome. In Metazoa, the number of nuclear-encoded subunits is variable but ranges around 70, with CII being composed entirely of nuclear-encoded proteins.
One of the consequences of this binary genome delegation for such a critical metabolic process is that mitochondrial and nuclear genomes products must physically interact for proper OXPHOS functioning. However, these two genomes experience different evolutionary dynamics: for instance, mitochondria have a small effective population size, are uniparentally inherited, and often experience higher substitution rates (see Ballard and Whitlock 2004). This has led to a general prediction of mitonuclear coevolution: evolution in one genome should select for complementary changes in the other to ensure correct mitochondrial functions (Rand et al. 2004; Bar-Yaacov et al. 2012; Hill 2019, 2020). Probably, the most persuasive evidence of the tight coevolution of mitochondrial and nuclear OXPHOS genes comes from experiments with cytoplasmic hybrids. In these experiments, divergent mitochondrial genomes are expressed against foreign nuclear backgrounds via experimental crossing designs or nuclear transfer, often causing OXPHOS inefficiency and lowered fitness (McKenzie et al. 2003; Niehuis et al. 2008; Burton and Barreto 2012; Barreto and Burton 2013; Barreto et al. 2018; Healy and Burton 2020).
Signatures of mitonuclear coevolution are also present in the molecular evolution of OXPHOS genes. In insects, rates of evolution are strongly correlated in mitochondria-encoded and nuclear-encoded OXPHOS (mtOXPHOS and nuOXPHOS, respectively) genes, but not between mitochondrial genes and nuclear-encoded genes lacking mitochondrial interactions (Yan et al. 2019). Such evolutionary rate correlation (ERC) in mitochondrial genes and their nuclear-encoded counterparts generally extends across eukaryotes: Lineages with fast-evolving mitochondrial genes also have fast-evolving mitochondria-interacting nuclear genes (Havird and Sloan 2016).
However, why mitonuclear coevolution is common and whether it is adaptive are less thoroughly understood. Some have argued that increased dN/dS ratios (i.e., ratio between nonsynonymous substitutions per nonsynonymous site and synonymous substitutions per synonymous site, also known as ω) in nuOXPHOS genes of animals are due to relaxed functional constraints on peripheral nuOXPHOS subunits, not positive selection in response to mitochondrial changes (Nabholz et al. 2013; Popadin et al. 2013; Zhang and Broughton 2013). Closely related taxa in the angiosperm genus Silene with highly divergent mitochondrial mutation rates have proven valuable in addressing these hypotheses. In taxa with fast mitochondrial mutation rates, nuOXPHOS subunits show elevated dN/dS ratios as a result of positive selection, despite still acting as peripheral subunits (Sloan et al. 2014; Havird et al. 2015, 2017). Structural information has also been used to show that nuOXPHOS changes tend to occur in areas that interact with mitochondria-encoded residues (Osada and Akashi 2012; Havird et al. 2015). These results are consistent with the most popular hypothesis stemming from mitonuclear coevolution: nuclear compensation, which posits that inefficient selection in mitochondrial genomes causes mildly deleterious mutations to accumulate, which are offset by compensatory changes in interacting nuclear-encoded genes. However, direct evidence for nuclear compensation over other forms of mitonuclear coevolution remains scarce, especially in invertebrates.
Here, we examine mitonuclear coevolution in Bivalvia, a class of sedentary molluscs. These animals represent an interesting observational unit for such studies for several reasons. First, bivalve phylogenies inferred with mitochondrial DNA (mtDNA) show discordance with nuclear ones, mainly with regard to deep relationships between Pteriomorphia, Palaeoheterodonta, and Heterodonta (Doucet-Beaupré et al. 2010; Bieler et al. 2014; González et al. 2015; Plazzi et al. 2016). However, phylogenies based on nuOXPHOS subunits are lacking and phylogenetic concordance in these specific nuclear-encoded genes could be a consequence of mitonuclear coevolution. Moreover, bivalves include a unique and evolutionarily stable exception to the strictly maternal inheritance (SMI) of mitochondria in animals: more than 100 species (Gusman et al. 2016) show doubly uniparental inheritance (DUI), with a maternally transmitted mtDNA (F-type) and a paternally transmitted mtDNA (M-type) (see Zouros 2013 for a review). The amino acid p-distance between F- and M-type mtOXPHOS proteins can be >50% (Doucet-Beaupré et al. 2010; Zouros 2013) and both F- and M-type mtDNA and their products (RNAs and proteins) are present in females and males (i.e., heteroplasmy; see Ghiselli et al. 2019 for a thorough discussion). Such peculiar mitochondrial inheritance implies that the same nuclear background has to cofunction with two different mitochondrial genomes, adding another layer of complexity to mitonuclear coevolution. Bivalves also show variation in rates of mitochondrial evolution, but their sedentary nature suggests maintaining highly efficient OXPHOS may be under weaker selection compared to taxa with higher metabolic requests. Moreover, it appears that bivalve mitochondrial mutation rates are not dramatically higher than the nuclear ones (see, e.g., Allio et al. 2017), therefore potentially representing a different mitonuclear coevolutionary landscape respect to deeply investigated taxa like vertebrates (where mitochondrial mutation rates can be ∼30 times as high as the nuclear ones). Coherently, a recent study by Iannello et al. (2019) observed that mtOXPHOS and nuOXPHOS subunits did not show significantly different dN/dS ratios in two congeneric species of bivalves, one of which has DUI.
To explore mitonuclear coevolution in bivalves, we investigated phylogenetic signals of mtOXPHOS and nuOXPHOS proteins and dN/dS ratios in the OXPHOS complexes spanning the whole phylogenetic tree of Bivalvia, including both SMI and DUI species. We also examined ERCs between mtOXPHOS and nuOXPHOS subunits, as well as nuclear-encoded genes with no mitochondrial interactions as a negative control. Furthermore, we investigated signals of site-specific positive selection in the context of protein structures, mitonuclear interactions, and functional sites.
Results
Data Set and Annotation
Out of the 40 bivalve transcriptomes, we selected and assembled (based on proportional and wide phylogenetic sampling; supplementary table 1, Supplementary Material online), nine were excluded either because of low quality of the data (Nuculana pernula, Yoldia limatula, and Pteria colymbus), or because of massive contamination (Astarte sulcata—the only Archiheterodonta available—Anadara trapezia, Cerastoderma edule, Cyrenoida floridana, Hiatella arctica, and Nucula tumidula). Out of the seven DUI species included in the present study, we obtained M-type mtOXPHOS subunits for four of them, namely Cristaria plicata, Hyriopsis cumingii, Mytilus edulis, and Ruditapes philippinarum.
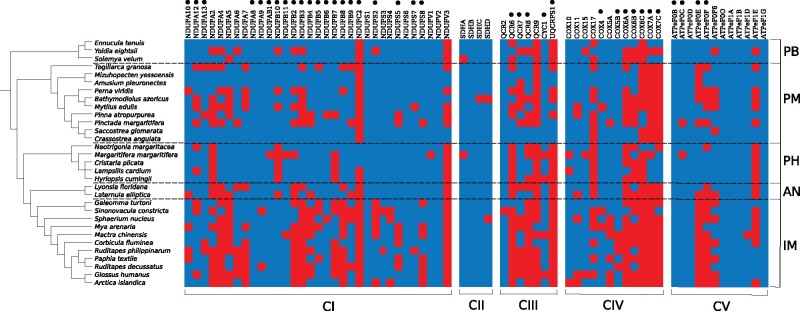
Out of 403 expected mtOXP HOS sequences (13 subunits for 31 species), 24 transcripts were completely missing from our data set, and 36 had only partial sequences (respectively, 4.73% and 7.10% of the total number of expected genes). However, 13 of the missing sequences were ATP8, which is known to be difficult to annotate because of a high rate of divergence (to the point of considering it missing in many bivalve lineages at first, see Dreyer and Steiner 2006; Breton et al. 2010). For annotation of nuOXPHOS subunits, seven subunits (namely QCR10, COX8, COX7B, NDUFA1, NDUFA3, NDUFB1, and NDUFC1) were not found in any of the species included in our data set; transcripts of COX6C and NDUFC2 were found in three species only, so they were excluded from the analyses; QCR1 was found in all the species but it is likely a misannotation so it was excluded as well; COX15 was excluded by BMGE (Criscuolo and Gribaldo 2010) during the alignment trimming phase. Out of the 66 remaining nuOXPHOS subunits, roughly 27.2% of the total expected sequences (66 subunits for 31 species = 2,046 sequences) were missing (fig. 1).
Fig. 1.
Annotation of nuDNA-encoded OXPHOS subunits. Presence and absence of each subunit in each species are depicted in blue and red, respectively. Left: species tree as built recovering data from literature (see Evolutionary Rate Correlations subsection of Materials and Methods for details). Top: protein nomenclature; black dots indicate subunits in contact with mitochondria-encoded proteins. Right: taxonomic clades (PB: Protobranchia; PM: Pteriomorphia; PH: Palaeoheterodonta; AN: Anomalodesmata; IM: Imparidentia). Bottom: respective OXPHOS complex.
Given the uneven, mtDNA-biased distribution of reference sequences for OXPHOS components, and the wide phylogenetic span that the species of the present study covered, we consider this data set to be adequate. The implementation of iterative intra-data set runs with the PSIBlast tool only moderately improved recovery of nuOXPHOS subunits. Crassostrea angulata (the most closely related species to C. gigas, the bivalve reference we used to annotate nuOXPHOS transcripts) was on average the most complete and Imparidentia (among the most diverged from C. gigas) were the most incomplete (fig. 1). However, the presence/absence patterns of nuOXPHOS subunits were not random in regard to the position of the nuclear-encoded subunits within the complexes. Subunits predicted to contact mtOXPHOS subunits tended to have lower annotation rates than “noncontact” subunits. To summarize, the protein sequence evolution analyses were conducted on 31 bivalve species for a total of 1,864 sequences (379 mitochondrial and 1,485 nuclear).
Concordance between mtOXPHOS and nuOXPHOS Phylogenetic Inferences
PartitionFinderProtein estimated LG + G as the best-fitting model for all partitions (LG+G + F for mitochondrial partitions). Maximum likelihood (ML) trees were inferred for both the mtOXPHOS and the nuOXPHOS concatenated data sets (see Materials and Methods for details). The two tree topologies mostly agreed between each other (fig. 2), with some minor exceptions. For instance, Yoldia eightsii clustered within Pteriomorphia in the mitochondrial tree (accordingly with Gusman et al. 2016), whereas in the nuclear tree, it represented a single taxon branch of a trichotomy with the branch of Autobranchia and a branch including the remaining Protobranchia samples. In any case, it never clustered with Opponobranchia (Solemya velum and Ennucula tenuis), as it usually does in nuclear-based phylogenies (González et al. 2015).
Fig. 2.
ML tree inference of mitochondrial and nuclear data sets. Trees were inferred with RAxML v8.2.11 on the two concatenated data sets. Only topologies are depicted in the figure. Bootstrap supports are depicted over each branch (supports lower than 70 were collapsed; 1,000 bootstrap replicates were performed). Left: mitochondrial topology (the star represents the omitted branching of unionids male mitochondria-encoded subunits. Other DUI species with both genomes available diverged terminally and the splits were collapsed in triangles, that is, Ruditapes philippinarum and Mytilus edulis). Right: nuclear topology. Clade acronyms as in fig. 1.
Paleoheterodonta clustered separately from all other Autobranchia in both data sets (bootstrap proportions: 97.7% and 70.9% for mitochondrial and nuclear data sets, respectively). This pattern is common for mitochondrial phylogenies of bivalves, which show topologies in which Euheterodonta (Imparidentia and Anomalodesmata) clusters together with Pteriomorphia (Doucet-Beaupré et al. 2010; Plazzi et al. 2016). However, such relationships have always been a matter of debate, since no phylogenetic analyses based on nuclear markers or genomewide data had obtained that topology so far, but rather displayed Euheterodonta clustering with Palaeoheterodonta (Kocot et al. 2011; Sharma et al. 2012; Stöger and Schrödl 2013; González et al. 2015).
Another difference between trees obtained with mtOXPHOS and nuOXPHOS data sets was the position of Anomalodesmata. In the nuclear data set, they figured as a well-supported sister group of Imparidentia (bootstrap: 99.1%), concordant with other nuclear data sets (González et al. 2015). Conversely, in the mitochondrial tree, they clustered separately from the clade Pteriomorphia + Imparidentia (although with only low bootstrap support: 70.6%). Inner relationships among Paleoheterodonta, Imparidentia, and Pteriomorphia are mainly concordant between the two trees and with the literature, with minor difficulties in solving an inner Pteriomorphia node for the mtOXPHOS tree and an inner Imparidentia node for the nuOXPHOS one.
Strong Correlation between Evolutionary Rates of mtOXPHOS and nuOXPHOS Proteins
In order to examine coevolutionary signals in mitochondrial and nuclear genes, we performed an ERC analysis. We obtained a “species tree” from the literature (based on genomic and transcriptomic data: see Materials and Method for details) and optimized branch lengths on that topology for the mtOXPHOS data set, the nuOXPHOS one, and a third data set of randomly chosen nuclear orthologs that share no roles in OXPHOS assembly or functioning (all values used for ERC analyses are in Supplementary Material). We then investigated the correlations between the branch lengths (root-to-tip, representing amino acid substitutions) of these three subsets of proteins.
There was a much stronger correlation between the branch lengths of mtOXPHOS and nuOXPHOS subunits compared with the other ERCs (fig. 3A). In particular, nuOXPHOS branch lengths had an almost perfect positive linear correlation of 0.967 with mtOXPHOS branch lengths (95% confidence interval: 0.931–0.984; p < 2.2e−16), while the random nuclear orthologues were only mildly correlated with the mtOXPHOS subunits (ρ = 0.437; 95% confidence interval: 0.098–0.686), although still with statistical significance (p = 1.39e−2, fig. 3B). Moreover, the correlation between nuOXPHOS and random nuclear orthologues was also statistically significant, but much lower than the correlation between nuOXPHOS and mtOXPHOS subunits (ρ = 0.548; 95% confidence interval: 0.240–0.756, p = 1.42e−3; supplementary fig. 1, Supplementary Material online). For the four DUI species with both sex-specific subsets of mtOXPHOS proteins available (out of seven DUI species overall), we used branch lengths of F-type subunits to perform ERCs. We reran the analyses by using the M-type branch lengths and the correlations with nuOXPHOS genes held strong (ρ = 0.943). The slightly lower ρ value was exclusively driven by the two Unionida M-type branch lengths that were significantly higher than F-type ones.
Fig. 3.
Evolutionary rate correlations analysis. (A, B) Correlation graphs between normalized branch lengths (per cumulative length of each tree, that is, cumulative sum of branch lengths for each tree = 1) of mtOXPHOS subunits versus nuOXPHOS subunits (ρ = 0.967; 95% confidence interval: 0.931–0.984; p = 2.2e−16), and mtOXPHOS subunits versus random orthologues (ρ = 0.437; 95% confidence interval: 0.098-0.686 p = 1.39e−2), respectively. (C) The black line represents the values of normalized mtOXPHOS branch lengths for each species in both graphs, the red line follows the values of normalized branch lengths on the same species for nuOXPHOS (average difference = 0.00219), and the blue line represents the branch lengths of random orthologues (average difference = 0.00819). This graph is useful to visualize the greater average difference in random orthologues’ branch lengths with respect to mtOXPHOS ones, compared with the differences between the latter and nuOXPHOS subunits. The lines that link the species are virtual and their purpose is simply to highlight the differences in the three relative trends of branch lengths.
The uniformity in the “nuclear signal” represented by the random orthologues was checked by dividing them in two random subsets (1,000 random divisions were performed) and assessing that there is a strong correlation between them (median ρ value for the 1,000 random subsets = 0.886, median p = 3.36e−11). Moreover, the strong correlation between mtOXPHOS and nuOXPHOS subunits held after normalizing the two distributions for each subset of random orthologues as an attempt to control for variation in overall rates of nuclear genome evolution among species (median ρ value for the 1,000 iterations = 0.925; median p = 9.99e−14; supplementary fig. 2, Supplementary Material online). The mitonuclear OXPHOS correlation was also robust after calculating it only using the terminal branches of each species (therefore avoiding any possible within-distribution bias): ρ = 0.937, p = 9.43e−15.
After normalizing the branch lengths of each tree for the total length of the trees themselves, it became clear that the curve trend of the nuOXPHOS proteins was more similar to that of the mtOXPHOS ones than to that of random orthologues (fig. 3C). For each species, the difference between the normalized branch lengths of mtOXPHOS proteins and nuOXPHOS proteins was on average 3.7 times lower than the difference between mtOXPHOS and random nuclear orthologues.
Another interesting coevolutionary signal resulted from correlation analyses performed for each component of each complex (i.e., when data sets of mtOXPHOS and nuOXPHOS subunits within each complex were correlated, fig. 4). All components, whether mitochondria- or nuclear-encoded, of all complexes were highly correlated with each other (ρ ranging from 0.819 to 0.950), with the exception of CII, which correlated to a very low extent with all other components (the highest ρ is 0.376). Moreover, all correlations with the branch lengths of random orthologues shared low values, and this was true also for CII subunits. With regard to CI, CIII, and CIV, the within-complex mitochondria- and nuclear-encoded components represented reciprocal best correlations. Nevertheless, these ρ values differed for at most 0.02.
Fig. 4.

Evolutionary rate correlations between each complex component. Graphical correlation matrix (Pearson’s r) between each component of each complex and the random orthologues. CII and the random orthologues data set shared lower correlation values with all other complex components, which were generally all consistently correlated with each other.
Last, we introduced positional information in the analysis by dividing the nuOXPHOS data set into subunits that share contact sites with mitochondria-encoded proteins, and subunits that are predicted to lack mitochondrial contact sites. The branch lengths of “contact” nuOXPHOS subunits showed a slightly higher correlation with mtOXPHOS branches compared with “noncontact” subunits (ρ = 0.969 vs. ρ = 0.951). On the other hand, the correlation with random orthologues was slightly higher for noncontact subunits (ρ = 0.572 vs. ρ = 0.533). These differences are very small, but a clearer signal is visible when normalizing branch lengths (fig. 5).
Fig. 5.
Evolutionary rate correlations with positional information of nuOXPHOS subunits. (A, B) Correlations between normalized branch lengths (per cumulative length of each tree) of mtOXPHOS subunits against contact nuOXPHOS subunits (A; ρ = 0.969; p < 2.2e−16) and noncontact ones (B; ρ = 0.951; p = 2.77e−16). (C, D) Correlations between normalized branch lengths (per cumulative length of each tree) of random orthologues against contact nuOXPHOS subunits (C; ρ = 0.533; p = 2.01e−3) and noncontact ones (D; ρ = 0.572; p = 7.83e−4).
Comparable dN/dS in mtOXPHOS and nuOXPHOS Subunits
For 48 out of the 79 OXPHOS subunits, the free-ratio model of Codeml (branch model 1, i.e., a different dN/dS value for each branch of the tree) fitted the data significantly better (likelihood ratio test [LRT]) than a single-ω model for all the branches of the species tree (i.e., branch model 0; supplementary table 2, Supplementary Material online). The results of the tests between the single-ω and the free-ratio models against the branch-specific model with tagged DUI branches (branch model 2, i.e., a different dN/dS value for each tagged branch) showed few sequences for which a DUI-specific calculation could be considered better-fitting (see Material and Methods for details on this branch-specific analysis, and supplementary fig. 3, Supplementary Material online, for a graphic summary). For six subunits (namely NADH3, NDUFA13, NDUFB11, NDUFB4, NDUFB8, and COX5B), the branch-specific model was better than the single-ω one, even when the free-ratio model was not significantly better than the latter. Moreover, for the other five subunits (namely COX7C, NADH4L, NDUFA4, NDUFA7, and NDUFS8), when both the branch-specific and the free-ratio models were better than the single-ω one, the free-ratio model was not significantly better than that with DUI-specific tags. Such results were, however, confined to few genes of the data set and did not allow us to consider the DUI phenomenon as a source of bias for the analysis. To confidently exclude this possibility, we removed those subunits from our data set and ran the same statistical tests we made for the whole data set. We did not observe any shift from any of the following results.
The distributions of dN/dS varied widely across OXPHOS gene products (supplementary fig. 4, Supplementary Material online). Interestingly, we observed significantly higher dN/dS values in contact nuOXPHOS subunits (median = 0.25) compared with noncontact ones (median = 0.1665; Wilcoxon–Mann–Whitney test: p < 2.2e−16; fig. 6A). Moreover, mtOXPHOS subunits had values of dN/dS almost one order of magnitude higher than what was previously observed in most metazoa (see, e.g., Nabholz et al. 2013; Havird and Sloan 2016), with a median value of 0.2241 (fig. 6A). The dN/dS values of mtOXPHOS subunits were significantly higher than those of noncontact nuOXPHOS subunits (Wilcoxon–Mann–Whitney test: p < 2.2e−16), but similar to those of contact nuOXPHOS ones (Wilcoxon–Mann–Whitney test: p = 0.183; fig. 6A).
Fig. 6.
dN/dS distributions of mtOXPHOS subunits, contact nuOXPHOS subunits, and noncontact nuOXPHOS. MtOXPHOS distributions are depicted in orange, nuOXPHOS ones in blue. Black lines within the boxes are the medians; the two hinges of the boxes approximate the first and the third quartile; whiskers extend to a roughly 95% confidence interval. Outliers are represented as black dots. Stars represent statistical significance of the relationship highlighted (single stars indicate statistically significant differences with all other distributions). Top: number of outliers not depicted in the figure. Bottom: Number of subunits included in the distributions. (A) Overall distributions. Noncontact nuOXPHOS subunits had a distribution statistically lower than both the other distributions (indicated by the star). MtOXPHOS and contact nuOXPHOS subunits shared statistically similar distributions. (B) Distributions of dN/dS for each complex and each compartment. Contact subunits are displayed as “cont” while noncontact subunits are displayed as “ncont.”
Variable evolutionary dynamics were observable when considering each OXPHOS complex separately (summary of the statistical relationships between all complexes distributions is in supplementary table 3, Supplementary Material online). Overall, CV displayed the fastest evolution among all complexes, both regarding its mitochondria-encoded components (0.2692 median dN/dS, significantly higher than all other mtOXPHOS subunits) and its nuclear-encoded ones (0.3836 median of contact subunits, significantly higher than all other nuOXPHOS subunits, except for nuCIV; fig. 6B). On the other hand, the slowest evolving complex was by far CII, which had lower dN/dS values compared with all other nuclear-encoded components (fig. 6B).
Within each complex, contact nuOXPHOS subunits showed dN/dS values on average higher than noncontact components (with an exception represented by the high dN/dS of noncontact nuCIII subunits; fig. 6B). However, rates of evolution of mtOXPHOS and contact nuOXPHOS components within a complex were never significantly different (reflecting the relationships observed for the overall data sets, see fig. 6A), in stark contrast to other animals, where dN/dS of nuOXPHOS subunits were higher. The only case where contact nuOXPHOS subunits had significantly higher values of dN/dS with respect to their mitochondria-encoded counterparts was in CIV.
By counting the normalized frequencies of branch-specific dN/dS values higher than 1, we were able to detect signatures of any historical or ongoing accelerated sequence evolution in the different complexes and in the different genomes. We observed that such signals were more frequent for mitochondria-encoded subunits (with mtCIII having the highest frequency, supplementary fig. 5A, Supplementary Material online). The only component that did not show any of such signatures was CII, which held the highest frequency of very low branch-specific dN/dS (considered as those values lower than 1/10 of the overall median; supplementary fig. 5B, Supplementary Material online).
No Clear Site-Specific Signature of Nuclear Compensation
For all subunits, the LRTs evaluated M3 as a better model compared with M0, meaning that a uniform rate of protein evolution across all sites would not represent the data set as well as variable rates. We then tested the likelihoods of models that implement distributions of sites under positive selection. M2a resulted to be better than M1a for 33 subunits, whereas M8 resulted better than M7 for 69 subunits (for model descriptions, see Materials and Methods; Yang 2007). We limited our data set to 33 subunits in which both M2a and M8 resulted as better models: 16 belonging to CI (two of which were mtOXPHOS: NADH1 and NADH5), 3 to CII, 2 to CIII, 5 to CIV (including all the three mitochondria-encoded subunits: COX1, COX2, and COX3), and 7 to CV. All these subunits were also analyzed for site-specific dN/dS under the “mechanistic-empirical combination” (MEC) model that includes empirical weights of the different amino acid substitutions. In all cases, no site under putative positive selection was detected. However, when comparing the corrected Akaike Information Criteria scores of MEC against M8, the latter still resulted in a better fit of the data for 9 of the 33 subunits. These nine subunits with site-specific signatures of positive selection included COX1 alongside nuclear-encoded subunits of CI (NDUFA2, NDUFB2, NDUFS2, and NDUFV1), CII (SDHA and SDHB), and CV (ATPeF1A and ATPeF1B) (summary of LRTs in supplementary table 2, Supplementary Material online; see Materials and Methods for details on this site-specific analysis and supplementary fig. 6, Supplementary Material online, for a graphic summary).
We analyzed these nine subunits with TreeSAAP (see Materials and Methods). We found that all the subunits that we analyzed except for NDUFA2 possessed regions that fell into the same category 8, which was “Equilibrium Constant (Ionization of COOH)”, meaning that such regions underwent amino acid substitutions that implied radical changes on the equilibrium constants of the constituting amino acids. However, when comparing the positively selected sites predicted by M8 with the regions under radical physicochemical divergence (i.e., that are the most likely candidate for positive selection) predicted by TreeSAAP, they did not always correspond. Moreover, when comparing these results with the annotated sites of catalysis, substrate binding, or subunits interface, there was no correlation. The only exceptions of annotated functional sites that were identified to be under positive selection were observed in COX1, and they represented interaction sites with other subunits: codons 357 (interaction with COX2) and 524 (interaction with COX5B) of the C. angulata sequence. Another interesting region consisted in positions 406–410 of NDUFS2, which were positively selected and close to the start of the C-t tail that makes contact with the mitochondria-encoded subunits of the complex. However, these did not represent actual contact sites. For both the NDUFS2 C-t tail, and the two positions of COX1, we manually checked amino acid composition in a phylogenetic frame, looking for concordant changes that could reflect compensatory-driven fixations in independent lineages, but we found no clear signal of putative convergent compensations. Unfortunately, the lack of high-quality 3D models and precise information on site-interactions in bivalves did not allow us to look for concordant changes in contact residues sited in other subunits.
All other sites showing putative positive selection were either physically close to a functional catalytic site (rather than subunit interfaces) or, much more often, located in distant regions of the protein. Positively selected positions were mostly associated to residues on the surface of complexes, and never buried ones (results of structural alignments with references in supplementary table 4, Supplementary Material online, predicted 3D structures with putative positively selected sites in the Supplementary Material). Amino acid frequencies of the sites under putative positive selection in the nine subunits were calculated and compared with the frequencies throughout all the alignments positions (see Supplementary Material). The amino acid frequencies of the positively selected sites were significantly different from the frequencies of the overall alignments. To assess whether such biased distributions were reflected in the enrichment of some specific quantitative property, we performed correlation tests between frequencies and 38 physicochemical amino acidic properties. Only four of them were significantly correlated (molecular weight, polarizability, refractive index, and average mass), but all four properties were also correlated among each other and were significantly correlated with the amino acid frequencies of the whole sequences as well, implying no specific physicochemical signal enriched for amino acids belonging to positively selected sites.
Discussion
Strong Signals of Coevolution between mtOXPHOS and nuOXPHOS Genes by Phylogenetic Inference and ERCs
One of the most contradictory findings in opposition to mitonuclear coevolution is the prevalence of mitonuclear phylogenetic discordance in animals—mitochondrial genes yield one topology, whereas nuclear genes produce another (Sharma et al. 2012; Toews and Brelsford 2012). Bivalves are no exception, and the major difference between the previous phylogenies inferred with the mitochondrial versus nuclear markers lies in the deep relationships. This may be due to incomplete lineage sorting, mitochondrial introgression, or errors in reconstructing phylogenies. One resolution to this contradiction may be that nuclear phylogenies are often based on anonymous loci (e.g., single nucleotide polymorphisms obtained by RAD-Seq) or genes that lack mitochondrial interactions. Accordingly, it has been suggested that nuOXPHOS genes should show more similar phylogenetic signals to mtOXPHOS genes under mitonuclear coevolution compared with noninteracting nuclear genes (Sloan et al. 2017). The results of our phylogenetic analysis (fig. 2) were consistent with such predictions, since our nuOXPHOS phylogeny was more similar to the mtOXPHOS phylogeny than previous topologies based on either a handful of nuclear markers or transcriptome-wide analyses (Bieler et al. 2014; González et al. 2015).
Another strong signal of mitonuclear coevolution was the almost perfect positive linear correlation between branch lengths calculated on the same species tree for mtOXPHOS and nuOXPHOS subunits (ρ = 0.967; fig. 3A). Random nuclear orthologues lacking mitochondrial interactions were only mildly correlated to mtOXPHOS subunits (ρ = 0.437; fig. 3B), suggesting that genomewide changes in evolutionary rates only partially explain the strong ERC between mt and nuOXPHOS genes. Similar results were previously found for insects (e.g., Yan et al. 2019) and between plastid-encoded and plastid-interacting genes in angiosperms (Williams et al. 2019). Such strong ERCs between mitochondrial and mitochondria-interacting nuclear genes represent some of the strongest evidence of shared evolutionary dynamics between the mitochondrial and the nuclear genomes. This approach has also been used to find novel nuclear-encoded genes that likely play an important role in mitochondrial dynamics, as such genes can show similar ERCs as nuOXPHOS genes (Williams et al. 2019; Yan et al. 2019). The lack of high-quality genomic data in many invertebrates is currently a hindrance to such studies but will likely not be so for long.
Because CII, the only OXPHOS complex exclusively formed by nuclear-encoded subunits, did not show a strong ERC with either mtOXPHOS or nuOXPHOS subunits of chimeric complexes (fig. 4), it is most likely that mitonuclear coevolution, not relaxation of constraints for OXPHOS function in general, is driving the strong ERC between mtOXPHOS and nuOXPHOS genes. Supporting this, ERCs were generally stronger within a complex compared with across complexes (fig. 4) and the strongest ERCs were found when using contact nuOXPHOS that directly interact with mitochondria-encoded residues (fig. 5). Although categorizing nuclear-encoded proteins into contact versus noncontact is likely an oversimplification that ignores allosteric effects, all of these observations are consistent with mitonuclear coevolution in bivalves.
Our finding that mtOXPHOS rates are correlated with nuOXPHOS rates, but not those of other nuclear genes, has interesting ramifications on bivalve phylogenetic inference. For example, the ERC may be driving the pattern of mitonuclear concordance described above. If mt and nuOXPHOS genes show similar rates of protein evolution compared with other nuclear genes, then long-branch attraction issues may affect nuclear phylogenies based on different genes differently. Such a scenario is consistent with the disagreement at deep nodes between mitochondrial and previous nuclear phylogenies. More focused analyses involving other nuclear markers and finer phylogenetic methods might be worthwhile.
Limited Signals of Nuclear Compensation in Bivalves
Many studies of bilaterian animals show that dN/dS ratios are extremely low in mitochondrial genes, despite low effective population size and higher mutation rate, suggesting strong selective constraints acting on mtOXPHOS subunits (distribution of values from 1 to 3 quantile < 0.05; see, e.g., Nabholz et al. 2013; Popadin et al. 2013; Havird and Sloan 2016). In the present study, dN/dS for mtOXPHOS subunits was an order of magnitude higher than those previously reported for most Metazoa, with a median value of 0.2241. This value is consistent with recent work that compared the congeneric bivalve species R. philippinarum and R. decussatus (Iannello et al. 2019) and with values calculated among mitochondrial genomes across Bivalvia (Plazzi et al. 2016). A possible explanation could be that lower metabolic needs of bivalves (due to a sedentary lifestyle) result in relaxed selection on mtOXPHOS proteins (as observed for loss of flight: Mitterboeck and Adamowicz 2013; and swimming performances: Strohm et al. 2015). Another, mutually nonexclusive, hypothesis could be that adaptations to stress tolerance (Sokolova 2018; Sokolova et al. 2019) increased the robustness of the OXPHOS system to nonsynonymous substitutions without relevant consequences in terms of fitness.
Although the biological reasons for high dN/dS in bivalve mtOXPHOS proteins are unclear, they may provide insights into mitonuclear coevolutionary dynamics. According to the “nuclear compensation hypothesis,” nuOXPHOS subunits are the prime sites for compensatory changes that maintain proper functioning of OXPHOS complexes in the face of deleterious mitochondrial mutations (Dowling et al. 2008; Gershoni et al. 2010; Osada and Akashi 2012; Havird and Sloan 2016). Some support for this hypothesis was provided in our study by the entirely nuclear-encoded CII, which had significantly lower dN/dS compared with all other, mt-interacting nuclear components (fig. 6B, supplementary fig. 5, Supplementary Material online). Similarly, the dN/dS of noncontact nuOXPHOS subunits was significantly lower than those of contact subunits (fig. 6A), which are the most obvious sites for potential compensatory changes (although this may be an oversimplification).
However, in bivalves, overall dN/dS of contact nuOXPHOS subunits was not elevated compared with mtOXPHOS subunits (fig. 6A), unlike in most animals (Nabholz et al. 2013; Havird and Sloan 2016). Under nuclear compensation, it is generally assumed that dN/dS should be elevated in nuOXPHOS subunits, reflecting positive selection for compensatory changes. When considering each complex separately, this signal is not uniform (fig. 6B). CIV does show the expected trend under nuclear compensation of nuclear contact proteins that appear to evolve significantly faster than mitochondrial ones. However, CIV is constituted by some of the slowest-evolving mtOXPHOS subunits. Therefore, nuclear compensation might be expected to show the weakest signal in CIV. One possibility is that each complex and each set of subunits are undergoing different evolutionary dynamics that are driven by specific selective pressures, rather than all complexes being primarily shaped by coevolutionary constraints (see, e.g., Zhang and Broughton 2013; Iannello et al. 2019). Future studies might benefit by examining each complex and each subunit separately to reveal different selective pressures associated with different functional constraints. Another possibility is that the elevated dN/dS ratios in mitochondria- and nuclear-encoded contact subunits could be due to different reasons. Relaxed selection on mitochondrial genes coupled with positive selection on nuclear-encoded contact subunits could result in similarly high dN/dS ratios and would be consistent with nuclear compensation. Phylogenetic and population genetic tools to explicitly test for positive selection may be useful in exploring this possibility (Wertheim et al. 2015; Havird et al. 2017).
We also examined signatures of nuclear compensation in site-specific signals of positive selection, predicting that contact nuOXPHOS subunits should be enhanced for positive selection. However, out of eight nuOXPHOS subunits in which positively selected sites were inferred, only NDUFB2 and NDUFS2 were predicted to physically contact mitochondria-encoded subunits. All other proteins represent key subunits involved in catalysis and are located in regions of the complexes that are distant to mitochondria-encoded proteins. We acknowledge that in order to be tied by coevolutionary constraints, residues do not necessarily need to be in physical contact, since perturbations in the tertiary structure due to an amino acid mutation can compromise stability also in distant residues. However, two subunits of CII were among the putative positively selected sequences and comparable numbers of positively selected sites were found in mtOXPHOS subunits, further reducing the possibility that these results were a reflection of compensatory nuclear evolution. It could be possible that these signals of positive selection were the result of false positives due to the higher rates of sequence conservation of these proteins (see supplementary fig. 4, Supplementary Material online; Anisimova et al. 2002). These sites may simply represent residues under loose purifying selection due to their exposition in the mitochondrial matrix (therefore not involved in catalysis nor structural conformation). Regardless, our site-specific analyses do not support nuclear compensation as in the dN/dS analyses.
Heterogeneity of Mitonuclear Evolutionary Dynamics across Metazoa
The extent of nuclear compensatory evolution may vary among taxa. For example, in corals, dipterans, and some fungi mitochondrial and nuclear dS values are fairly similar (Havird and Sloan 2016), whereas Vertebrata show the highest values of mutation rate ratios between mitochondrial and nuclear genomes calculated so far (up to an average ratio per gene of 32.5 in primates: Allio et al. 2017). In our samples, mitochondrial dS values are not high on average (median is ∼0.3; supplementary fig. 7, Supplementary Material online), however, these values are still noticeably higher than the nuclear ones, and the same is observed for nonsynonymous substitution rates (dN; supplementary fig. 7, Supplementary Material online). Precisely, the ratio of dS between mtOXPHOS and nuOXPHOS genes in our samples was ∼2.5 (ratio between the two medians), similar to the ratio recently calculated in Bivalvia based on comparisons between mutation rates of mitochondrial genes and 398 nuclear nonmitochondria-interacting genes (median = 1.8; Allio et al. 2017). Under these conditions, we should nevertheless expect relatively higher nuclear dN/dS for our data set under nuclear compensation (like observed in fast-mutating mtDNA taxa; Havird and Sloan 2016), but that is not the case. In other words, the high mitochondrial dN/dS observed in bivalves is not likely due entirely to a low mitochondrial mutation rate but also due to increased rates of nonsynonymous fixations.
In our opinion, there is an important caveat to comparing dN/dS values in different genomes that may have widely differing mutation rates. Correlation between the ratio of mitonuclear dN/dS and mitonuclear dS observed in Havird and Sloan (2016) may have been misleading, since one of the variables is nested within the other and would automatically be expected to result in a negative correlation. Havird and Sloan attempted to control for this by examining genes without mitochondrial interactions as a control, which showed different patterns than nuOXPHOS genes. If nuclear compensation is predominantly responsible for the types of correlations observed in Havird and Sloan (2016), then amino acid substitution rate (dN) in the nuclear genes should be driving the trend. However, by reanalyzing the Havird and Sloan (2016) data set (one of the few works with a wide phylogenetic sampling across eukaryotes), we found that the mitonuclear dN/dS ratio is only mildly correlated with the mitonuclear dN ratio, and the correlation is driven mainly by the plant/animal dichotomy (supplementary fig. 8, Supplementary Material online). Moreover, when considering also Bivalvia values as calculated in the present study, the correlation is even weaker (supplementary fig. 8, Supplementary Material online). Other meaningful correlations, like mitochondrial dN/dS against nuclear dN/dS, or mitochondrial dN against nuclear dN, are not significant (neither excluding nor including bivalves; supplementary fig. 9, Supplementary Material online), even though they may represent more direct predictions of nuclear compensation (all correlation tests were performed with R on the data set of Havird and Sloan 2016; data in Supplementary Material). Therefore, although mitonuclear coevolution may drive some of the observed differences between dN/dS in mtOXPHOS and nuOXPHOS genes in many metazoans, the large difference in underlying mutation rates between the two genomes certainly also contributes. In a case where mutation rate is high (e.g., high mitochondrial dS) but purifying selection is very strong, the need for compensation may not be high, since few protein residues actually change (e.g., low mitochondrial dN). Disentangling such nuances of dN/dS analyses should be a goal of future work.
Is DUI Compatible with Nuclear Compensation?
Mitonuclear coevolution is particularly interesting in bivalves because of the frequent occurrence of DUI. In the present work, given the low representativeness of DUI species in online database, and given the additional difficulties in extracting both F- and M-type mtDNAs within a species, we could not include more DUI-specific analyses. The only signals we could get from the present sampling were represented by a handful of genes for which a specific tagging of DUI branches resulted in a better fitting Codeml model, and a slight lowering in the mtOXPHOS–nuOXPHOS branch length correlation when considering M-type mitochondria-encoded subunits for the four available species (almost exclusively driven by the two Unionid species).
However, the DUI system presents some interesting considerations for mitonuclear coevolution and nuclear compensation. In DUI species, two highly divergent mitochondrial genomes have to cofunction with the same nuclear background. This introduces a potential challenge for the nuclear compensation theory because nuclear changes must offset changes happening in the two lineages of mitochondrial genes. For instance, if a mutation arises in an F-type subunit, the nuclear compensatory mutation might disrupt the coassembly with the corresponding M-type subunit, lowering the efficiency of M-type mitochondria. However, M-type genomes, despite being usually rare in somatic tissues (but with exceptions, see Ghiselli et al. 2011), are still functionally important, since the whole male germline relies exclusively on them (Ghiselli et al. 2013; Milani and Ghiselli 2015).
One explanation for maintenance of DUI along with nuclear compensation could be the presence of two separate sets of nuOXPHOS genes that underwent duplication and evolved sex-specific expression. Such male-biased nuOXPHOS orthologues are common in mammals and Drosophila (Gallach et al. 2010; Eslamieh et al. 2017; Havird and McConie 2019). While this explanation cannot be completely excluded and future studies should examine it more thoroughly, no clues of duplicated sets of nuOXPHOS genes have been found so far in DUI species (Maeda G, Iannello M, McConie HJ, Ghiselli F, Havird JC, unpublished), and we found only a single transcript per gene in all DUI species in the present study, with the exception of M. edulis COX4. Another possibility is sex-specific splice variants or sex-specific nuclear-encoded OXPHOS expression, which has been found in humans (Barshad et al. 2018).
A second explanation for the stable presence of DUI could lie in mitochondrial compensatory evolution, an underexplored version of mitonuclear coevolution. In such scenario, an amino acid change in a nuclear gene could be independently compensated in both M- and F-type mitochondrial genomes. The fact that these two highly divergent lineages have been kept evolutionarily stable for millions of years without disrupting respiratory capacity may be explained by considering the “mitochondrial compensation hypothesis” as the primary coevolutionary force. The production of more DUI-specific data in the future will allow us to properly address such questions.
Considerations on the Directions of Compensatory Mitonuclear Coevolution
Others have highlighted that mitonuclear coevolution could take many forms and deleterious compensatory changes are only one class (Sloan et al. 2017). The nuclear compensation hypothesis has been favored because classic evolutionary theory suggests nonrecombining genomes such as mitochondrial genomes are likely to suffer from mutational meltdown (Lynch 1996; Lynch and Blanchard 1998; Neiman and Taylor 2009). Both empirical and modeling work has challenged this assumption (Cooper et al. 2015; Christie and Beekman 2017) and the assumption that mitochondrial genomes never recombine is also being undermined (Havird et al. 2019).
Mitochondrial genomes usually mutate faster and many variants of mtDNA are constitutively present in a heteroplasmic state (Burr et al. 2018). In the heteroplasmic pool, there might be some mtDNA copies that present a compensatory mutation for a novel amino acid change that occurred in a nuclear subunit. In this case, mitochondria that contain higher amounts of this “compensatory” mtDNA would present better functioning OXPHOS complexes with respect to the wild-type ones. Such mitochondria would have higher fitness than the others and might eventually be fixed (Milani and Ghiselli 2015; Burr et al. 2018; Zhang et al. 2018). The mechanisms that allow this selection are yet to be clarified, however, the fact that better-performing mtDNA variants are favorably transmitted (Wilding et al. 2001; Zhou et al. 2010; Ghiselli et al. 2013; Hill et al. 2014; Milani 2015; Milani and Ghiselli 2015; Tworzydlo et al. 2016; Bilinski et al. 2017; Marlow 2017) could represent a coherent mechanism for mitochondrial compensation of nuOXPHOS mutations in very short evolutionary times.
Referring to this interpretation, it should be noted that almost all observations previously associated and explained in terms of nuclear compensation could be equally explained as mitochondrial compensations. For example, the fact that nuOXPHOS genes have higher dN/dS than nuclear non-OXPHOS genes, and nuOXPHOS genes without mitochondrial counterparts (Havird and Sloan 2016; Havird et al. 2017; Li et al. 2017; Yan et al. 2019) could be due to mitochondrial compensation. NuOXPHOS genes can indeed be more variable because they can be efficiently compensated by a fast-mutating mitochondrial genome. When no compensation is possible, a structural deleterious mutation should simply be selected against. The same holds true for nuclear-encoded ribosomal proteins that form mitochondrial ribosomes (Barreto and Burton 2013; Sloan et al. 2014; Weng et al. 2016) and for aminoacyl-tRNA-synthetases that act on mt-tRNAs (Adrion et al. 2016). Also, many site-specific coevolutionary signals do not specifically favor nuclear compensation because they lack temporal data that could discern the order of appearance of the mutations (inter alia Gershoni et al. 2010, 2014; Levin and Mishmar 2017).
Little direct evidence supports nuclear compensation in contrast to other forms of mitonuclear coevolution, with one notable exception being the observation that nuclear changes tended to occur later in time than mitochondrial ones at contact residues in primates (Osada and Akashi 2012). However, a recent study by Wernick et al. (2019), showed in vivo evidence of direct mitochondrial compensation in Caenorhabditis elegans. In gas-1 mutated lines, they directly observed functional recovery of OXPHOS efficiency through 60 generations in populations under food competition driven by novel mutations in nadh1 and nadh6 genes, which are mitochondria-encoded subunits in contact with the nuclear-encoded gas-1. It is therefore possible that in some cases, the mitochondrial genome is responsible for compensatory mutations. Future studies focusing on specific residues and the temporal order of changes are needed.
Conclusions
Overall, a clear signal of mitonuclear coevolution in bivalves emerges from our data.
Both the phylogenetic analysis and the ERC analyses showed strong evidence of shared evolutionary trajectories for mtOXPHOS and nuOXPHOS subunits in contrast to nuclear genes that do not interact with mitochondria. However, mitochondrial dN/dS in our samples were almost an order of magnitude higher than previously recorded bilaterian data and similar to nuclear dN/dS ratios, calling into question the idea of nuclear compensation as the driving force of mitonuclear coevolution in bivalves. Similar results were obtained in previous analyses of bivalves (Iannello et al. 2019). However, contact nuOXPHOS subunits displayed higher rates of evolution than noncontact and nonchimeric nuclear proteins, again supporting a general observation of mitonuclear coevolution. No site-specific signal of accelerated compensatory evolution was found in any of the nuclear OXPHOS subunits. Overall, support for nuclear compensation as the specific form of mitonuclear coevolution was scarce. This pattern is in contrast to other metazoans, possibly due to different reasons, including relaxed selection on OXPHOS proteins in sedentary living bivalves, increased selection on stress-tolerance pathways, or a combination of these factors. Examining a diverse sample of bivalve taxa, we extend the evidence for mitonuclear coevolution to a novel taxonomic group, but question the ubiquity of the nuclear compensation hypothesis.
Materials and Methods
Data Set
We downloaded the RNA-Seq raw reads for a total of 40 bivalve species from the short read archive (SRA) of NCBI (www.ncbi.nlm.nih.gov/sra; supplementary table 1, Supplementary Material online). Species were selected to evenly represent the biodiversity of the class: 14 Imparidentia, 2 Anomalodesmata, the only Archiheterodonta available, 5 Palaeoheterodonta, 12 Pteriomorphia, and the 6 available species of Protobranchia. We included Haliotis tubercolata (Gasteropoda), Octopus bimaculoides (Cephalopoda), Graptacme eborea (Scaphopoda), and Acanthochitona crinita (Polyplacophora) as outgroups for the inference of phylogenetic trees.
When multiple RNA-Seq data sets were available for the same species, we downloaded reads from different tissues to limit the effect of potential tissue-specific transcription of OXPHOS genes. Even though we did not consider expression data in the present work, tissue-specific and/or sample-specific transcription patterns could affect the de novo assembly of less abundant transcripts, preventing the detection of the target sequences or compromising their quality. When DUI species were considered, we downloaded reads from both sexes in order to retrieve both mitochondrial genomes. When sex was not specified, we pooled multiple SRA experiments, increasing the chances of obtaining the M-type genome as it is abundant in male gonads but sometimes rare/absent in the somatic tissues.
We removed sequencing primers and filtered out low-quality and unpaired reads using Trimmomatic-0.36 (Bolger et al. 2014) with the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:25:33 MINLEN:75. Transcriptomes were then assembled de novo using Trinity-v2.4.0 (Haas et al. 2013) with default parameters. To assess quality and completeness of the transcriptomes, we used BUSCO v2 (Simão et al. 2015), as implemented in the gVolante website (https://gvolante.riken.jp/analysis.html; Nishimura et al. 2017). Gene content (complete and fragmented) was assessed comparing the transcripts with the “Metazoa” core ortholog database.
Before gene annotation, we filtered the transcriptomes with a sequence-similarity Blast-based method in order to avoid the presence of contaminant reads (this filter consists in the first step of an RNA-Seq annotation pipeline developed for nonmodel organisms, complete protocol available at: https://osf.io/cdkb9/?view_only=f0b2cde926db43719f3d705012c4eeaa). Whole transcriptomes were aligned with DIAMOND v0.9.19.120 (Buchfink et al. 2015) against NCBI nonredundant protein database (nr), retrieving taxon ID annotation for each hit. Through E-fetch (NCBI E-utilities package) we extracted the taxonomic lineage for each hit, and we retained only those transcripts for which the best hit was against a lophotrochozoan. We did not choose a stricter filtering method (e.g., Mollusca or Bivalvia) because the underrepresentation of bivalve and mollusc nuclear sequences in the online databases could have led to false negatives.
OXPHOS Subunits Annotation
MtOXPHOS transcripts were identified with a BlastX (Blast v2.6.0+; Camacho et al. 2009) search of each transcriptome against a custom database containing all molluscan mtOXPHOS PCGs (downloaded from NCBI). Due to the higher representation of mollusc mitochondrial genes, a stricter filter could be applied for mitochondrial genes: We kept only those transcripts whose best hit against nr was a mollusc sequence. We then manually extracted open reading frames (ORFs) using the NCBI ORFfinder online tool (www.ncbi.nlm.nih.gov/orffinder), validating the results with a BlastP against nr. When both F- and M-type mitochondrial gene products were annotated in DUI species, we considered them as separate operational taxonomic units throughout the whole analysis. To solve the problem of a high occurrence of partial subunits, we implemented annotation with MitoRNA (available at: https://github.com/mozoo/mitoRNA; Forni et al. 2019), a software used to assemble mitochondrial genes from raw reads through iterations of Bowtie2 mapping (Langmead and Salzberg 2012) and Trinity assembling, based on a reference genome. Such a step was possible only for those species for which the mtDNA—or at least a congeneric one—was present in the databases.
ORFs of nuOXPHOS subunits were retrieved using the Findorf tool (Krasileva et al. 2013), that uses a BlastX search against a user-defined database, and an HMMER (Mistry et al. 2013) search against the Pfam database 30.0 (Finn et al. 2016). To build the user-defined Blast database, we downloaded nuOXPHOS protein sequences of seven reference species from the KEGG database (https://www.genome.jp/kegg/; Kanehisa and Goto 2000): Crassostrea gigas, Octopus bimaculoides, Lottia gigantea, Helobdella robusta, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens. The complete set consisted of 78 subunits (38 of CI, 4 of CII, 9 of CIII, 14 of CIV, and 13 of CV; see fig. 1). We implemented the annotation with the PSI-Blast tool that consists of a series of consecutive BlastP iterations (from which a position-specific score matrix is built) in which each newly detected sequence is included in the database for the subsequent cycles. We used the protein sequences positively annotated with the Findorf tool as databases to complete the annotation in the species with missing genes. ORFs annotated this way were verified with a BlastP against nr, checking that the first hit corresponded to the presumptive gene.
We performed an additional check for the possible presence of nuOXPHOS paralogs in the data set by back-Blasting the annotated subunits against each species transcriptome. In almost all cases, only a single transcript for each gene was present. In some cases, multiple transcripts were present but they simply represented fragments with identical sequence composition (de novo assembly constructs or length isoforms). However, for a few genes in a few species (therefore not bivalve-wide duplications), we found two plausible transcripts, and reciprocal Blasts with other species were performed to ensure that we did not include the duplicated genes in the analyses. For three sequences (out of 1,485), our annotation pipeline extracted an ORF for which the correct orthology could not be defined, assessing the high predictive power of the pipeline but confirming the need for paralogy check.
A further additional step was performed for nuOXPHOS subunits. The first N-terminal amino acids of these proteins constitute the mitochondrial targeting signal (MTS), necessary for recognition by the mitochondrial import machinery. However, the MTS sequence is cleaved once the protein enters the organelle, and therefore, it is not included in the final functional product. Such sequences were excluded from our study, since they do not participate in the coevolutionary dynamics of the mitonuclear OXPHOS complexes and are subjected to rather different evolutionary forces (e.g., ligand–receptor interactions). To exclude MTSs from the retrieved ORFs, we used MitoFates (Fukasawa et al. 2015), a software that predicts both the presence of an MTS and the heterodimer mitochondrial processing peptidase (MPP) cleavage sites. We consider in the subsequent analyses only the sequence from the MPP cleavage site to the stop codon for those proteins, where the presence of MTS was assessed with default MitoFates cutoffs.
Phylogenetic Inference
Two ML phylogenies were inferred for the concatenated sets of mtOXPHOS and nuOXPHOS subunits. We aligned the amino acid sequences with PSICOFFEE (Floden et al. 2016) and trimmed the alignments with BMGE v1.12 (BLOSUM30 -h 0.75 -b 3; Criscuolo and Gribaldo 2010). We concatenated the sequences with PhyUtility (Smith and Dunn 2008), and we inferred partitions and best-fitting models with PartitionFinderProtein (Lanfear et al. 2017). ML trees were built with RAxML v8.2.11 (Stamatakis 2014) with 1,000 bootstrap replicates, forcing bivalve monophyly, and setting Acanthochitona crinita as outgroup. Nodes with a bootstrap support value lower than 0.7 were collapsed.
Evolutionary Rate Correlations
We also examined ERCs (a useful test to investigate protein coevolutionary dynamics, see de Juan et al. 2013; Williams et al. 2019; Yan et al. 2019) between the two mt and nuOXPHOS proteins. We built a species tree and optimized the branch lengths of the concatenated alignments with RAxML v8.2.11 (Stamatakis 2014). The species tree of our sample species was built manually using data from the literature (see the tree in fig. 1). We mostly referred to the phylotranscriptomic analysis of González et al. (2015): relationships between and within the orders (with the exception of Pteriomorphia) were obtained from the ML tree of all genes found in 37.5% or more of the species included in their work. Some of our species were included in that phylogeny: for those that were not, we considered the genus or the family. The inner relationships among the three Unionidae species considered here (Cristaria plicata, Lampsilis cardium, and Hyriopsis cumingii) could not be solved, because the only work with solved relationships that included all three families was based only on COX1 and 28S (Lopes-Lima et al. 2017). For this reason, we chose to keep them as a polytomy. The relationships within Pteriomorphia were based on the phylogenomic work of Lemer et al. (2016).
A set of random nuclear proteins was used as control for the ERC; for this purpose, we used Proteinortho v6.0.7 (Lechner et al. 2011) to obtain ortholog transcripts from the 31 bivalve transcriptomes of our study. We selected 24 orthologue clusters from the output (maximizing the species representation) for a total of 605 transcripts (139 missing sequences). We extracted ORFs with TransDecoder (https://github.com/TransDecoder/TransDecoder), and through a BlastP search against the nr database we ensured that no mitochondria-interacting proteins were included in these clusters. Alignments, trimming, partitioning, branch length optimization on the species tree, and distance to the root calculations were performed as for the OXPHOS proteins. We then performed correlation tests (cor.test function in R) on the distances to the root (patristic method of distRoot function in R, adephylo package) of every species in the three different sets of proteins as a proxy for coevolutionary dynamics. Correlating distributions of distances to the root introduces nonindependence among within-distribution values because of shared branches, and this could bias the calculations. For this reason, we also tested for correlation between the lengths of the terminal branches (i.e., species-specific). Since the correlation analyses shared the same trend, we kept root-to-tip distances (that represent a more precise evolutionary history of each species) for the subsequent analyses.
Another possible bias that may affect ERC is the potential nonrandom representativeness of the 24 nuclear control orthologue clusters. To test this, we randomly divided the cluster in two subsets of 12 proteins and tested for ERC between the two. This was performed 1,000 times to obtain a median correlation coefficient and its confidence intervals for each ERC. Moreover, we divided both the mtOXPHOS and the nuOXPHOS for the branch lengths of both orthologues subsets to check if the correlation held after a normalization to control for variation among species in overall rates of nuclear evolution (normalization performed for all 1,000 iterations). To check for more specific coevolutionary signals, we calculated branch lengths for separate data sets of both mitochondria- and nuclear-encoded subunits of each complex and tested for correlations between the different components. Last, we divided the nuOXPHOS subunits in two clusters: those predicted to be in physical contact to mitochondria-encoded subunits and those without any supposed direct interaction with mtOXPHOS proteins (CIV: Richter and Ludwig 2003; CV: Jonckheere et al. 2012; CI: Zhu et al. 2016; CII: Amporndanai et al. 2018). We then performed correlation tests for these two subsets against branch lengths of mtOXPHOS subunits and random orthologues.
Rates of Protein Evolution
For the analyses on the rates of protein evolution, we followed the same alignment procedure for the phylogenetic analyses, except that we excluded the four outgroups from the data set. We optimized branch lengths of the species tree (see above) for each alignment of our analyses with RAxML v8.2.11 (Stamatakis 2014). Best-fitting models for RAxML were inferred with ProtTest v3.4 (Darriba et al. 2011). We calculated for each backtranslated alignment dN/dS using Codeml (PAML v4.9 package; Yang 2007; supplementary fig. 3, Supplementary Material online).
Each alignment was tested for a free-ratio model of dN/dS calculation over the tree (each branch associated to a different value, i.e., branch model 1; model = 1) against a uniform rate model (a single value averaged for all branches, i.e., branch model 0; model = 0). The best-fitting model was estimated through LRTs, comparing log-likelihood values for each model (maximized over six replicates). In the cases where branch model 1 was the best-fitting model, we pooled together dN/dS values for all branches of each subunit tree, therefore associating a distribution to each gene product, rather than a single value. We considered this as the best way to represent the evolutionary dynamics of the subunits, since a single value averaged for all the sites of the sequences and for all the branches of the tree would have been an extreme approximation, also taking into account that some species in our data set have been separated for hundreds of millions of years and have underwent extensive diversification. In order to be able to compare the single-ω subunits with the others, we replicated the single dN/dS value for all the branches of their trees, therefore equally weighing the two sets of subunits in the overall distribution.
Statistical group analyses were conducted with Wilcoxon–Mann—Whitney and Dunn tests (with Bonferroni correction) as implemented in R v3.4.4. Zero values of dN or dS, which resulted in calculations of dN/dS of either 0 or 999 in Codeml, were excluded. We plotted distributions of dN/dS values higher than 1 and lower than 1/10 of the overall median to detect signals of fast-evolving and slow-evolving sequences, respectively. We performed chi-squared tests to test whether these distributions differed significantly from the expected ones.
In our data set, we included 7 DUI species that are known to possess two different mitochondrial genomes that are maintained separately by sex-specific segregation (see Introduction). Since we pooled all dN/dS values of the tree together, we tested whether the DUI species biased the overall signal, especially for the subunits where the free-ratio model was better than the single-ω model. In order to do so, we performed LRTs between single-ω model and the branch-specific model that allows to tag different branches or clades for which a specific dN/dS is calculated (in this case, we tagged the private branch of each DUI species—or the whole clade in the case of Unionida). Such a model was also tested against the free-ratio model. The two paired tests allowed us to evaluate whether the dN/dS calculation with the single-ω or the free-ratio models were biased by the presence of DUI species, and if it would have been better to consider such taxa separately (for a graphic summary of this branch-specific analysis, see supplementary fig. 3, Supplementary Material online).
Signatures of Positive Selection
We also used Codeml to investigate the site-specific evolutionary rate of gene products (graphic summary: supplementary fig. 6, Supplementary Material online). To test whether a model considering different dN/dS for different sites fit the data better than one implementing a uniform rate, we tested log-likelihood values (maximum values over six calculation replicates) of M0 (single dN/dS; NSsites = 0) and M3 (n categories of dN/dS: five in our case; NSsites = 3) with LRTs. When M3 was the best model, we tested for the presence of positive selection comparing two pairs of models. Each pair consisted in a model that included parameters admitting positively selected sites, and another that did not (the null model; Yang 2007): they were M1a (variable selective pressure but no positive selection; NSsites = 1) versus M2a (M1a plus positive selection; NSsites = 2) and M7 (beta-distributed variable selective pressure; NSsites = 7) versus M8 (M7 plus positive selection; NSsites = 8). When both the models that included sites with dN/dS > 1 were the best, we performed additional tests to evaluate whether we could actually consider positive selection as a possibility. We tested M8 against the MEC model, which takes into account the weight of each amino acid replacement (Doron-Faigenboim and Pupko 2007) in terms of radical and conservative modifications based on empirical replacement probability matrices (calculation performed on the Selection server; Stern et al. 2007). Those models are not nested within each other, therefore an LRT was not possible. Hence, we compared Akaike Information Criteria scores in order to evaluate the best model. When M8 was the best, we considered the sites under positive selection as predicted by the Bayes empirical Bayes method as implemented in Codeml.
We further analyzed subunits where sites under putative positive selection were found with TreeSAAP (Woolley et al. 2003), a software that estimates the selection dynamics of 31 physicochemical amino acid properties, predicting whether each property is subject to radical or conservative shifts, and in which regions. The parameters were set as follows: all properties, eight categories, sliding window length = 15. Codeml and TreeSAAP results were compared with the aid of IMPACT_S (Maldonado et al. 2014), a software that allows the comparison and combination of results of protein sequence evolution analyses obtained with different calculation methods. The results of the different analyses were then compared with annotated ligand and catalytic sites from the literature. In detail, we compared the protein sequences of C. angulata (as annotated in our data set) with the functional sites as predicted in C. gigas (NCBI protein database), or with annotated sites in Homo sapiens when such information was not available for any Crassostrea species. The comparison was implemented by plotting the sites under putative positive selection on the tertiary structures of the C. angulata proteins. Structural conformations were predicted for C. angulata subunits on the I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/; Zhang 2008) and we kept only those structures that had a C-score higher than 0 (C-score is typically in the range [−5,2], with higher values representing more confident model predictions). Structural alignment against known structures of the OXPHOS complexes (downloaded from the Protein Data Bank archives, https://www.rcsb.org) were performed in order to visualize the sites of interest in the context of the quaternary structure (CI: Homo sapiens, 10.2210/pdb5XTD/pdb; CII: Escherichia coli, 10.2210/pdb1NEK/pdb; CIV and CV: Bos taurus, 10.2210/pdb5XDX/pdb and 10.2210/pdb5ARA/pdb, respectively). Last, we tested whether the sites under putative positive selection were enriched for specific amino acids or for specific physicochemical properties. In order to do so, we calculated the frequencies of each of the 20 amino acids in all the positively selected sites of the final positively selected candidate subunits and compared it with the frequencies as calculated from the whole alignments (chi-square tests). Moreover, we tested for correlation among the amino acid frequencies and 38 physicochemical properties (31 of which were those included in TreeSAAP), exploring the possibility that those putatively positively selected sites included some overrepresentation of quantitative indexes (Spearman’s correlation as implemented in R).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We gratefully thank Daniel Sloan for his useful comments on the manuscript, and Liliana Milani for discussions and help with computational resources. We thank the anonymous Editor and two Reviewers for their useful comments. This work was supported by the Italian Ministry of Education, University and Research (MIUR) FIR2013 Program (grant no. RBFR13T97A to F.G.), “Ricerca Fondamentale Orientata” (RFO) funding from the University of Bologna (grant to F.G.), and the Canziani bequest (grant to F.G.).
Data Availability
All raw reads accession numbers are available in supplementary material, along with branch length calculations and codeml models tests. Sequence alignments and other results will be shared on reasonable request to the corresponding author.
References
- Adrion JR, White PS, Montooth KL.. 2016. The roles of compensatory evolution and constraint in aminoacyl tRNA synthetase evolution. Mol Biol Evol. 33(1):152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Donega S, Galtier N, Nabholz B.. 2017. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol Biol Evol. 34(11):2762–2772. [DOI] [PubMed] [Google Scholar]
- Amporndanai K, Johnson RM, O’Neill PM, Fishwick CWG, Jamson AH, Rawson S, Muench SP, Hasnain SS, Antonyuk SV.. 2018. X-ray and cryo-EM structures of inhibitor-bound cytochrome bc1 complexes for structure-based drug discovery. IUCrJ. 5(2):200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z.. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19(6):950–958. [DOI] [PubMed] [Google Scholar]
- Ballard JW, Whitlock MC.. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13(4):729–744. [DOI] [PubMed] [Google Scholar]
- Barreto FS, Burton RS.. 2013. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 30(2):310–314. [DOI] [PubMed] [Google Scholar]
- Barreto FS, Watson ET, Lima TG, Willett CS, Edmands S, Li W, Burton RS.. 2018. Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat Ecol Evol. 2(8):1250–1257. [DOI] [PubMed] [Google Scholar]
- Barshad G, Blumberg A, Cohen T, Mishmar D.. 2018. Human primitive brain displays negative mitochondrial-nuclear expression correlation of respiratory genes. Genome Res. 28(7):952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Blumberg A, Mishmar D.. 2012. Mitochondrial-nuclear co-evolution and its effects on OXPHOS activity and regulation. Biochim Biophys Acta. 1819(9–10):1107–1111. [DOI] [PubMed] [Google Scholar]
- Bieler R, Mikkelsen PM, Collins TM, Glover EA, González VL, Graf DL, Harper EM, Healy J, Kawauchi GY, Sharma PP, et al. 2014. Investigating the Bivalve Tree of Life: an exemplar-based approach combining molecular and novel morphological characters. Invert Syst. 28(1):32. [Google Scholar]
- Bilinski SM, Kloc M, Tworzydlo W.. 2017. Selection of mitochondria in female germline cells: is Balbiani body implicated in this process? J Assist Reprod Genet. 34(11):1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Hoeh WR.. 2010. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: mytilidae): identification of the “missing” ATPase 8 gene. Mar Genomics. 3(1):11–18. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Burr SP, Pezet M, Chinnery PF.. 2018. Mitochondrial DNA heteroplasmy and purifying selection in the mammalian female germ line. Dev Growth Differ. 60(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Barreto FS.. 2012. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol Ecol. 21(20):4942–4957. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JR, Beekman M.. 2017. Uniparental inheritance promotes adaptive evolution in cytoplasmic genomes. Mol Biol Evol. 34(3):677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper BS, Burrus CR, Ji C, Hahn MW, Montooth KL.. 2015. Similar efficacies of selection shape mitochondrial and nuclear genes in both Drosophila melanogaster and Homo sapiens. G3. 5:2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D, Pazos F, Valencia A.. 2013. Emerging methods in protein co-evolution. Nat Rev Genet. 14(4):249–261. [DOI] [PubMed] [Google Scholar]
- Doron-Faigenboim A, Pupko T.. 2007. A combined empirical and mechanistic codon model. Mol Biol Evol. 24(2):388–397. [DOI] [PubMed] [Google Scholar]
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR.. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 10(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J.. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol. 23(10):546–554. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G.. 2006. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica—and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslamieh M, Williford A, Betrán E.. 2017. Few nuclear-encoded mitochondrial gene duplicates contribute to male germline-specific functions in humans. Genome Biol. Evol. 9(10):2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floden EW, Tommaso PD, Chatzou M, Magis C, Notredame C, Chang J-M.. 2016. PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 44(W1):W339–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni G, Puccio G, Bourguignon T, Evans T, Mantovani B, Rota-Stabelli O, Luchetti A.. 2019. Complete mitochondrial genomes from transcriptomes: assessing pros and cons of data mining for assembling new mitogenomes. Sci Rep. 9(1):14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, Imai K.. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 14(4):1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betrán E.. 2010. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol. Evol. 2:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni M, Fuchs A, Shani N, Fridman Y, Corral-Debrinski M, Aharoni A, Frishman D, Mishmar D.. 2010. Coevolution predicts direct interactions between mtDNA-encoded and nDNA-encoded subunits of oxidative phosphorylation complex I. J Mol Biol. 404(1):158–171. [DOI] [PubMed] [Google Scholar]
- Gershoni M, Levin L, Ovadia O, Toiw Y, Shani N, Dadon S, Barzilai N, Bergman A, Atzmon G, Wainstein J, et al. 2014. Disrupting mitochondrial-nuclear coevolution affects OXPHOS complex I integrity and impacts human health. Genome Biol Evol. 6(10):2665–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Maurizii MG, Reunov A, Ariño-Bassols H, Cifaldi C, Pecci A, Alexandrova Y, Bettini S, Passamonti M, Franceschini V, et al. 2019. Natural heteroplasmy and mitochondrial inheritance in bivalve Molluscs. Integr. Comp Biol. 59(4):1016–1032. [DOI] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, Passamonti M.. 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol Evol. 5(8):1535–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M.. 2011. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: veneridae). Mol Biol Evol. 28(2):949–961. [DOI] [PubMed] [Google Scholar]
- González VL, Andrade SCS, Bieler R, Collins TM, Dunn CW, Mikkelsen PM, Taylor JD, Giribet G.. 2015. A phylogenetic backbone for Bivalvia: an RNA-seq approach. Proc R Soc B. 282(1801):20142332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 4(9):a011403–a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF.. 1999. Mitochondrial evolution. Science 283(5407):1476–1481. [DOI] [PubMed] [Google Scholar]
- Gusman A, Lecomte S, Stewart DT, Passamonti M, Breton S.. 2016. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. Peer J. 4:e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, , Whitehill NS, , Snow CD, , Sloan DB. 2015. Conservative and compensatory evolution in oxidative phosphorylation complexes of angiosperms with highly divergent rates of mitochondrial genome evolution. Evolution 69(12):3069–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Forsythe ES, Williams AM, Werren JH, Dowling DK, Sloan DB.. 2019. Selfish mitonuclear conflict. Curr Biol. 29(11):R496–R511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, McConie HJ.. 2019. Sexually antagonistic mitonuclear coevolution in duplicate oxidative phosphorylation genes. Integr Comp Biol. 59(4):864–874. [DOI] [PubMed] [Google Scholar]
- Havird JC, Sloan DB.. 2016. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial versus nuclear genomes. Mol Biol Evol. 33(12):3042–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Trapp P, Miller C, Bazos I, Sloan DB.. 2017. Causes and consequences of rapidly evolving mtDNA in a plant lineage. Genome Biol Evol. 9(2):323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy TM, Burton RS.. 2020. Strong selective effects of mitochondrial DNA on the nuclear genome. Proc Natl Acad Sci U S A. 117(12):6616–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. 2015. Mitonuclear ecology. Mol Biol Evol. 32(8):1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. 2019. Mitonuclear ecology. New York: Oxford University Press. [Google Scholar]
- Hill GE. 2020. Mitonuclear compensatory coevolution. Trends Genet. 36(6):403–414. [DOI] [PubMed] [Google Scholar]
- Hill GE, Havird JC, Sloan DB, Burton RS, Greening C, Dowling DK.. 2019. Assessing the fitness consequences of mitonuclear interactions in natural populations. Biol Rev. 94(3):1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H.. 2014. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 46(4):389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannello M, Puccio G, Piccinini G, Passamonti M, Ghiselli F.. 2019. The dynamics of mito-nuclear coevolution: a perspective from bivalve species with two different mechanisms of mitochondrial inheritance. J Zool Syst Evol Res. 57(3):534–547. [Google Scholar]
- Jonckheere AI, Smeitink JAM, Rodenburg RJT.. 2012. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 35(2):211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, Cannon JT, Todt C, Citarella MT, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, et al. Phylogenomics reveals deep molluscan relationships. Nature 477(7365):452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov AA, Gerasimov ES.. 2012. Diversity of mitochondrial genome organization. Biochemistry (Moscow) 77(13):1424–1435. [DOI] [PubMed] [Google Scholar]
- *Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, Tabbita F, Soria M, Wang S, Akhunov E, Uauy C, et al. 2013. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 14(6):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W.. 2010. The energetics of genome complexity. Nature 467(7318):929–934. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ.. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemer S, González VL, Bieler R, Giribet G.. 2016. Cementing mussels to oysters in the pteriomorphian tree: a phylogenomic approach. Proc R Soc B. 283(1833):20160857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin L, Mishmar D.. 2017. The genomic landscape of evolutionary convergence in mammals, birds and reptiles. Nat Ecol Evol. 1:0041. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang R, Liu S, Donath A, Peters RS, Ware J, Misof B, Niehuis O, Pfrender ME, Zhou X.. 2017. The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evol Biol. 17(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Lima M, Froufe E, Do VT, Ghamizi M, Mock KE, Kebapçı Ü, Klishko O, Kovitvadhi S, Kovitvadhi U, Paulo OS, et al. 2017. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes. Mol Phylogenet Evol. 106:174–191. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol Biol Evol. 13(1):209–220. [DOI] [PubMed] [Google Scholar]
- Lynch M, Blanchard JL.. 1998. Deleterious mutation accumulation in organelle genomes. Genetica 102/103:29–39. [PubMed] [Google Scholar]
- Maldonado E, Sunagar K, Almeida D, Vasconcelos V, Antunes A.. 2014. IMPACT_S: integrated multiprogram platform to analyze and combine tests of selection. PLoS One 9(10):e96243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow FL. 2017. Mitochondrial matters: mitochondrial bottlenecks, self-assembling structures, and entrapment in the female germline. Stem Cell Res. 21:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF, Garg S, Zimorski V.. 2015. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc B. 370(1678):20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV.. 2006. Introns and the origin of nucleus–cytosol compartmentalization. Nature 440(7080):41–45. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M.. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392(6671):37–41. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Chiotis M, Pinkert CA, Trounce IA.. 2003. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol. Biol. Evol. 20(7):1117–1124. [DOI] [PubMed] [Google Scholar]
- Milani L. 2015. Mitochondrial membrane potential: a trait involved in organelle inheritance? Biol Lett. 11(10):20150732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F.. 2015. Mitochondrial activity in gametes and transmission of viable mtDNA. Biol Direct . 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M.. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41(12):e121–e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterboeck TF, Adamowicz SJ.. 2013. Flight loss linked to faster molecular evolution in insects. Proc R Soc B. 280(1767):20131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Ellegren H, Wolf JBW.. 2013. High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol. Biol. Evol. 30(2):272–284. [DOI] [PubMed] [Google Scholar]
- Neiman M, Taylor DR.. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc R Soc B. 276(1660):1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Gadau J.. 2008. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 178(1):413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Hara Y, Kuraku S.. 2017. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics. 33(22):3635–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N, Akashi H.. 2012. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol. Biol. Evol. 29(1):337–346. [DOI] [PubMed] [Google Scholar]
- Plazzi F, Puccio G, Passamonti M.. 2016. Comparative Large-Scale Mitogenomics Evidences Clade-Specific Evolutionary Trends in Mitochondrial DNAs of Bivalvia. Genome Biol Evol. 8(8):2544–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin KY, Nikolaev SI, Junier T, Baranova M, Antonarakis SE.. 2013. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol. Biol. Evol. 30(2):347–355. [DOI] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19(12):645–653. [DOI] [PubMed] [Google Scholar]
- Richter O-MH, Ludwig B.. 2003. Reviews of Physiology, Biochemistry and Pharmacology. Cytochrome c oxidase — structure, function, and physiology of a redox-driven molecular machine. In: Berlin, Heidelberg: Springer Berlin Heidelberg. p. 47–74. [DOI] [PubMed] [Google Scholar]
- Sharma PP, González VL, Kawauchi GY, Andrade SCS, Guzmán A, Collins TM, Glover EA, Harper EM, Healy JM, Mikkelsen PM, et al. 2012. Phylogenetic analysis of four nuclear protein-encoding genes largely corroborates the traditional classification of Bivalvia (Mollusca). Mol. Phylogenet. Evol. 65(1):64–74. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Havird JC, Sharbrough J.. 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 26(8):2212–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Triant DA, Wu M, Taylor DR.. 2014. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol. Biol. Evol. 31(3):673–682. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Warren JM, Williams AM, Wu Z, Abdel-Ghany SE, Chicco AJ, Havird JC.. 2018. Cytonuclear integration and co-evolution. Nat Rev Genet. 19(10):635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Dunn CW.. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716. [DOI] [PubMed] [Google Scholar]
- Sokolova I. 2018. Mitochondrial Adaptations to Variable Environments and Their Role in Animals’ Stress Tolerance. Integr. Comp. Biol. 58(3):519–531. [DOI] [PubMed] [Google Scholar]
- Sokolova IM, Sokolov EP, Haider F.. 2019. Mitochondrial Mechanisms Underlying Tolerance to Fluctuating Oxygen Conditions: lessons from Hypoxia-Tolerant Organisms. Integr. Comp. Biol. 59(4):938–952. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T.. 2007. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35(Web Server):W506–W511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger I, Schrödl M.. 2013. Mitogenomics does not resolve deep molluscan relationships (yet?). Mol. Phylogenet. Evol. 69(2):376–392. [DOI] [PubMed] [Google Scholar]
- Strohm JHT, Gwiazdowski RA, Hanner R.. 2015. Fast fish face fewer mitochondrial mutations: patterns of dN/dS across fish mitogenomes. Gene. 572(1):27–34. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W.. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5(2):123–135. [DOI] [PubMed] [Google Scholar]
- Toews DPL, Brelsford A.. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 21(16):3907–3930. [DOI] [PubMed] [Google Scholar]
- Tworzydlo W, Kisiel E, Jankowska W, Witwicka A, Bilinski SM.. 2016. Exclusion of dysfunctional mitochondria from Balbiani body during early oogenesis of Thermobia. Cell Tissue Res. 366(1):191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Jansen RK.. 2016. Plastid-nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol Evol. 8(6):1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernick RI, Christy SF, Howe DK, Sullins JA, Ramirez JF, Sare M, Penley MJ, Morran LT, Denver DR, Estes S.. 2019. Sex and mitonuclear adaptation in experimental Caenorhabditis elegans populations. Genetics 211(3):1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrel B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding M, Carotenuto R, Infante V, Dale B, Marino M, Di Matteo L, Campanella C.. 2001. Confocal microscopy analysis of the activity of mitochondria contained within the “mitochondrial cloud” during oogenesis in Xenopus laevis. Zygote 9(4):347–352. [DOI] [PubMed] [Google Scholar]
- Williams AM, Friso G, van Wijk KJ, Sloan DB.. 2019. Extreme variation in rates of evolution in the plastid Clp protease complex. Plant J. 98(2):243–259. [DOI] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA.. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19(5):671–672. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yan Z, Ye G, Werren JH.. 2019. Evolutionary rate correlation between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects. Mol Biol Evol. 36(5):1022–1036. [DOI] [PubMed] [Google Scholar]
- Zachar I, Szathmáry E.. 2017. Breath-giving cooperation: critical review of origin of mitochondria hypothesis. Biol Direct. 12:19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Broughton RE.. 2013. Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol. 5(10):1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Burr SP, Chinnery PF.. 2018. The mitochondrial DNA genetic bottleneck: inheritance and beyond. Essays Biochem. 62(3):225–234. [DOI] [PubMed] [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou RR, Wang B, Wang J, Schatten H, Zhang YZ.. 2010. Is the mitochondrial cloud the selection machinery for preferentially transmitting wild-type mtDNA between generations? Rewinding Müller’s ratchet efficiently. Curr Genet. 56(2):101–107. [DOI] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, Hirst J.. 2016. Structure of mammalian respiratory complex I. Nature 536(7616):354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 40(1):1–31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw reads accession numbers are available in supplementary material, along with branch length calculations and codeml models tests. Sequence alignments and other results will be shared on reasonable request to the corresponding author.