Abstract
The C1 (reductase) subunit of 4-hydroxy-phenylacetate (4-HPA) 3-hydroxylase (HPAH) from the soil-based bacterium Acinetobacterbaumannii catalyzes NADH oxidation by molecular oxygen, with hydrogen peroxide as a by-product. 4-HPA is a potent allosteric modulator of C1, but also a known urinary biomarker for intestinal bacterial imbalance and for some cancers and brain defects. We thus envisioned that C1 could be used to facilitate 4-HPA detection. The proposed test protocol is simple and in situ and involves addition of NADH to C1 in solution, with or without 4-HPA, and direct acquisition of the H2O2 current with an immersed Prussian Blue–coated screen-printed electrode (PB-SPE) assembly. We confirmed that cathodic H2O2 amperometry at PB-SPEs is a reliable electrochemical assay for intrinsic and allosterically modulated redox enzyme activity. We further validated this approach for quantitative NADH electroanalysis and used it to evaluate the activation of NADH oxidation of C1 by 4-HPA and four other phenols. Using 4-HPA, the most potent effector, allosteric activation of C1 was related to effector concentration by a simple saturation function. The use of C1 for cathodic biosensor analysis of 4-HPA is the basis of the development of a simple and affordable clinical routine for assaying 4-HPA in the urine of patients with a related disease risk. Extension of this principle to work with other allosteric redox enzymes and their effectors is feasible.
Keywords: allosteric regulation, biosensor, oxidase, reductase, redox signaling, reduced nicotinamide adenine dinucleotide (NADH), hydroxylase, disease biomarker, electroanalysis, amperometry
Abbreviations: 4-HPA, 4-hydroxy-phenylacetate; ITC, Isothermal titration calorimetry; NADH, nicotinamide adenine dinucleotide; PB-SPE, Prussian Blue–coated screen-printed electrode; RE, reference electrode; WE, working electrode
Allosteric up- or downregulation of enzyme activity is the result of protein evolution toward more efficient physiological function (1, 2, 3, 4). Binding of an activator or inhibitor to the enzyme and a subsequent conformational change affect substrate/protein interaction either through changes in binding affinity (“K-systems”) or in turnover rate (“V-systems”). Allosteric enzymes thereby synchronize complex cellular metabolic pathways through positive or negative feedback and mediate neuronal and endocrine chemical cell signaling (5, 6, 7, 8, 9). Recently, technological biocatalysis, using enzymes for green and sustainable chemical synthesis, has begun to explore allosteric features of native or genetically engineered protein catalysts in industrial process design, with enhanced yields, purity, and structural integrity of the product (10, 11, 12). Similarly, allosteric enzymes have been suggested as signal-generating elements of biosensors in the analysis of the products of enzymic processes, disease biomarkers and soil, water, air, and food pollutants (13).
Electroanalysis of reactants is the basis of the widely used clinical and home-care blood glucose meters that use strip-like sensor platforms with glucose oxidase on the working electrodes for amperometric sample testing (14). However, even 60 years after Clark and Lyon’s influential launch of amperometric glucose biosensing (15), the potential of this simple methodology has not yet exploited allosteric enzymes. The potential advantage of using allosteric enzymes as chemically tunable biological recognition elements on electrodes is that the activity of the immobilized signalling protein, and thus the sensor current, is altered by an activating or inhibiting modulator. The dependence of the allosteric effect on modulator concentration also permits assay of the allosteric modulator, widening the range of analytical applicability. Despite this useful practical feature, electrochemical biosensing with allosteric enzymes remains an underdeveloped practice, and the few published examples either work with rather complex designs of the sensor readout or require specially synthesized nanomaterials as components of the immobilization matrix in order to generate an adequate signal (16, 17, 18, 19, 20, 21). Thus, the utilization of allosteric enzymes on electrode surfaces and the design of simple and sustainable strategies for allosteric electrochemical assays of substrate or effector is a topical issue, with many methodological opportunities. Cheap, computerized devices for electroanalysis are now on the market and, in the case of compact cigarette-box-sized amplifiers, have the advantages of portability and a small bench footprint. Combining miniaturized apparatus with amperometric biosensing by natural or genetically modified allosteric enzymes may lead to novel analytical solutions for biomarker, drug, and environmental pollutant testing.
With the above considerations in mind, we studied the allosteric reductase subunit C1 of a hydroxylase from soil-based Acinetobacter baumannii (22, 23, 24, 25). This has an FMN prosthetic group and oxidizes NADH to NAD+; in the holo-enzyme it is reoxidized by the C2 subunit, which hydroxylates 4-HPA, but in the absence of C2, the reduced prosthetic group can be reoxidized by molecular oxygen, a reaction that is allosterically activated by 4-HPA. C1 was used as a model allosteric enzyme in the nonoptical assay of NADH oxidation by cathodic H2O2 amperometry, in a high-sensitivity, interference-free assay. This simple electroanalytic procedure could also be used to measure the enhancement of the rate of enzymic NADH oxidation by known allosteric activators of C1, to assess the relative strength of allosteric modulation by 4-HPA and structurally similar phenolic compounds, and provide a simple assay for the effector. 4-HPA is recognized as a urinary indicator for dysbiosis (26, 27, 28), a critical imbalance of the human microbiota, and for certain types of brain degeneration (29), diabetic nephropathy (30), and cancer (31, 32). The proven quality of the proposed amperometric C1 activity assay is thus an important advance toward simple and cheap 4-HPA biomarker analysis in urine samples, with a potential for monitoring clinical health and personal well-being. Apart from the possibility of an effective 4-HPA assay, the novel bioelectrochemical methodology may also serve as a complementary means of NADH detection, an optional screening tool for performance tests with genetically engineered allosteric C1 variants, and with suitable adaptations, a tactic for work with other allosteric redox enzymes.
Results
Adaptation of H2O2 amperometry for analytical readout of C1 activity
The substrate of C1 is the reduced form of nicotinamide adenine dinucleotide (NADH), which is oxidized to NAD+. As the reaction typically takes place in an aerobic environment, redox recycling of C1 occurs through electron transfer to dissolved oxygen, with production of hydrogen peroxide, which, due to its ambivalent redox character, can be quantified by anodic oxidation or cathodic reduction.
In an initial test, CI in solution was assayed using a platinum disk working electrode (WE) operated in amperometric anodic oxidation mode with a detection potential of +600 mV versus the reference electrode (RE) and challenged with successive additions of small aliquots of the C1 effector 4-HPA, the C1 substrate NADH, and the reaction product H2O2. Increased anodic currents were detected not only with addition of the diagnostic target H2O2 but also with 4-HPA and NADH (Fig. 1A). The initial test was then repeated with a Prussian Blue (PB)-modified screen-printed carbon disk electrode (PB-SPE) WE as the H2O2 sensor with a detection potential of –100 mV versus RE. In contrast to the signal pattern seen in the anodic test with a Pt-disk WE, a measurable current, now for cathodic reduction, was seen with the analyte H2O2, but not with the effector 4-HPA or the substrate NADH (Fig. 1B). With response times below 10 s, single H2O2 concentration increases as low as 2 μM led to reproducible and easily analyzed stepwise increases in cathodic sensor current (Fig. 1C). The response of cathodic PB-SPEs to H2O2 was reliably linear up to around 2000 μM, with a sensitivity of 0.01 μA μM-1, while at higher analyte levels, the curve approached a plateau, due to signal saturation (Fig. 1D).
Figure 1.
Anodic and cathodic electrochemical detection of H2O2with test of interference.A and B are the amperometric current responses of a Pt disk and a PB-SPE WE to serial additions of 50 μM of 4-HPA, NADH, and H2O2 at detection potentials of +600 and –100 mV versus RE, respectively. C, is a typical amperogram from a H2O2 calibration trial with a PB-SPE at –100 mV vs. RE. D, is the H2O2 calibration plot derived from the measurement shown in (C). The electrolyte in (A), (B), and (C) was 50 mM sodium phosphate buffer, pH 7. Data points and error bars represent the means and standard deviations of triplicate analyses.
The analytical performance of PB-SPE-based H2O2 amperometry was further tested with quantitative assays of the target in model samples. In triplicate repetitions of measurements in the standard addition mode, the concentration of 100 μM H2O2 in the model samples showed an acceptable recovery of (109.0 ± 2)% (Fig. 2).
Figure 2.
A, Cathodic H2O2amperometry in the standard addition mode for quantification of the analyte in a model sample of 100 μM concentration. The target, H2O2, was measured with a PB-SPE at a potential of –100 mV versus RE. The electrolyte was 50 mM sodium phosphate buffer, pH 7. B, Standard addition plot constructed with the data presented in (A).
C1 activity screening with cathodic H2O2 readout: allosteric activation monitoring
Cathodic H2O2 amperometry with PB-SPEs polarized at –100 mV versus RE was expected to reliably report production of H2O2 from O2, on oxidation of NADH to NAD+ by the allosterically-activated C1 subunit. A set of four amperometric PB-SPE recordings was carried out to confirm that the electrochemical method could also report the marked difference between the turnover rates of C1 with and without addition of the effector 4-HPA. In initial control runs with only enzyme or only NADH added to the electrolyte, no rise of cathodic current above residual level was detected (Fig. 3, solid and dotted black traces). On the other hand, a constantly increasing cathodic sensor current was observed when 50 μM NADH was added to buffer containing 1 μM C1 (Fig. 3, blue trace). The immediate signal increase on addition of the substrate visualized the onset and continuation of enzymic H2O2 production. The final steady current indicated complete consumption of NADH, with a stable H2O2 solution concentration corresponding to the added substrate concentration. Establishing a screen for allosteric enzyme activation was a further goal, and the last test was for a measurable increase in the rate of change of the the H2O2 current when the effector 4-HPA was added to the trial buffer before the substrate NADH (Fig. 3, red trace). The slopes of the traces indicate the rate of H2O2 production and thus correspond to the normal (Fig. 3, blue trace) and accelerated (Fig. 3, red trace) rates of NADH oxidation by C1. Under the conditions of the trial allosteric activation by 4-HPA produced an almost 12-fold increase in the turnover rate of C1.
Figure 3.
Measurement of NADH oxidation by C1, with and without allosteric activation, by cathodic H2O2amperometry. The blue trace is the amperometric current response of a PB-SPE at –100 mV versus RE in phosphate buffer, pH 7 containing 1 μM C1, with addition of 50 μM NADH about 360 s after the start of the trial. The red trace is the amperometric current response of a PB-SPE at –100 mV versus Ag/AgCl in sodium phosphate buffer containing 1 μM C1 and 100 μM 4-HPA with addition of 50 μM NADH about 360 s after the start of the trial. The steady signal increase after addition of NADH shows the development of the cathodic current for the reduction of enzymically produced H2O2; the curve slopes (ΔI/Δt) thus are indicators of normal (blue trace) and accelerated (red trace) NADH oxidation by C1. The plateau signifies full consumption of the substrate in each case.
Utilization of C1 reductase and cathodic H2O2 amperometry for quantitative NADH analysis
The observation of a linear correlation between NADH oxidation by C1 and the increase in H2O2 concentration in the reaction buffer solution allowed cathodic H2O2 amperometry at PB-SPEs to be used for the quantification of NADH in the media. In test trials, 1 μM C1 was dissolved in sodium phosphate buffer (50 mM, pH 7) and 100 μM 4-HPA was added to produce allosteric activation of the enzyme. Sequential additions of NADH to the test solution produced a series of stepped increases in the cathodic WE current (Fig. 4A). Plots of the background-corrected current step magnitudes against the actual NADH concentration were linear up to about 600 μM, while above this limit the current traces approached a maximum through C1 saturation with substrate (Fig. 4B). Using additions of 50 μM NADH, quantification was performed in the standard addition mode (Fig. 4, C and D). Triplicate repetition of the analysis of the 50 μM model sample revealed an acceptable recovery (109.2 ± 2.5%, n = 3) of the adjusted concentration. The practical limit of detection (LOD) of amperometric NADH biosensing with dissolved C1 was 2 μM.
Figure 4.
Quantitative detection of NADH by C1 with cathodic H2O2amperometry.A, is the amperometric H2O2 current response of a PB-SPE at –100 mV versus RE in trial buffer with 1 μM C1 and 100 μM 4-HPA with sequential additions of small aliquots of NADH. B, is the NADH calibration plot derived from the measurement shown in (A). Data points and error bars represent the means and standard deviations of triplicate analyses. C, is an amperometric recording of NADH quantification in the standard addition mode for a sample with 50 μM NADH and three additions of 50 μM NADH. D, standard addition curve constructed with the data of the trace in C. Electrolyte for the measurements in (A) and (C) was 50 mM sodium phosphate buffer, pH 7.
Evaluation of allosteric activation of C1 by 4-HPA, using cathodic H2O2 amperometry
The previous section verified that the proposed electrochemical methodology was sufficiently fast and sensitive to monitor in situ generation of H2O2 by C1-catalyzed oxidation of NADH and to differentiate reaction rates with and without allosteric activation. Because the allosteric effect is produced by binding of the effector to a regulatory site on the enzyme, it was expected to show concentration dependence and saturation, and the rate of H2O2 generation, reflected in the slopes of the H2O2 current recordings, increased with increased concentration of 4-HPA in the measuring buffer (Fig. 5A). Increasing concentrations of the allosteric effector produced a linear increase in rate up to about 100 μM and then a gradual approach to a plateau as the effector-binding sites became saturated (Fig. 5B). The data were fitted with the Michaelis–Menten function, the maximum rate of H2O2 production being 37 nA s-1 and the apparent Kd (effector concentration for half-maximal activation) 103 μM. The lowest 4-HPA concentration to produce a measurable increase in the rate of NADH oxidation was 1 μM.
Figure 5.
Evaluation of concentration-dependent allosteric C1 activation by 4-HPA by cathodic H2O2amperometry.A, the amperometric H2O2 current responses of a PB-SPE at –100 mV versus RE in sodium phosphate buffer, pH 7 containing 1 μM C1 and 5–500 μM 4-HPA, with 50 μM NADH added about 360 s after the start of recording. B, changes of the slopes (ΔI/Δt) of the H2O2 current traces (ΔS = S4-HPA – Sno 4-HPA) in (A) plotted against the concentration of the allosteric effector, 4-HPA. Data points and error bars represent the means and standard deviations from triplicate analyses. The line through the data points is a fit based on the Michaelis–Menten function. The maximum rate change of allosterically enhanced enzymatic H2O2 production and the effector concentration for half-maximal activation (apparent dissociation constant, Kd) are 37 nA s-1 and 103 μM, respectively. The inset shows the first six points of the plot on an expanded scale.
Cathodic H2O2 amperometry as a tool for the assessment of C1 activators
The discovery of effective allosteric regulators of an enzyme such as the reductase studied here and a comparison of their activatory or inhibitory effects are important considerations when the protein is being exploited for biotechnological, biomedical, or analytical purposes. Cathodic H2O2 amperometry at PB-SPEs has therefore been used to assess the allosteric interaction with C1 of four phenolic compounds with structural similarity to the most potent known activator, 4-HPA (22, 23, 24, 25). This trial involved the 2- and 3-isomers of 4-HPA, 4-hydroxy-3-methoxy-phenylacetate, and 3(4-hydroxy-phenyl)-propionate.
As shown in the amperometric recordings in Figure 6, the compounds studied can be divided into three groups, with weak, moderate, or strong ability to activate NADH oxidation by C1, compared with the activity in the absence of any modulator. 4-HPA was identified as the most effective activator, followed by 3-HPA. 2-HPA, on the other hand, had almost no effect on the turnover rate, and 4-hydroxy-3-methoxy-phenylacetate and 3-(4-hydroxy-phenyl)-propionate were moderately effective.
Figure 6.
Allosteric activation of C1 by hydroxyphenyl analogues, measured by cathodic H2O2amperometry. The plots show amperometric H2O2 current responses of a PB-SPE at –100 mV versus RE in sodium phosphate buffer, pH 7 containing 1 μM C1 plus 100 μM allosteric effector, with 50 μM NADH added about 360 s after the start of recording. The chemical structures of the phenolic allosteric effectors are shown on the right.
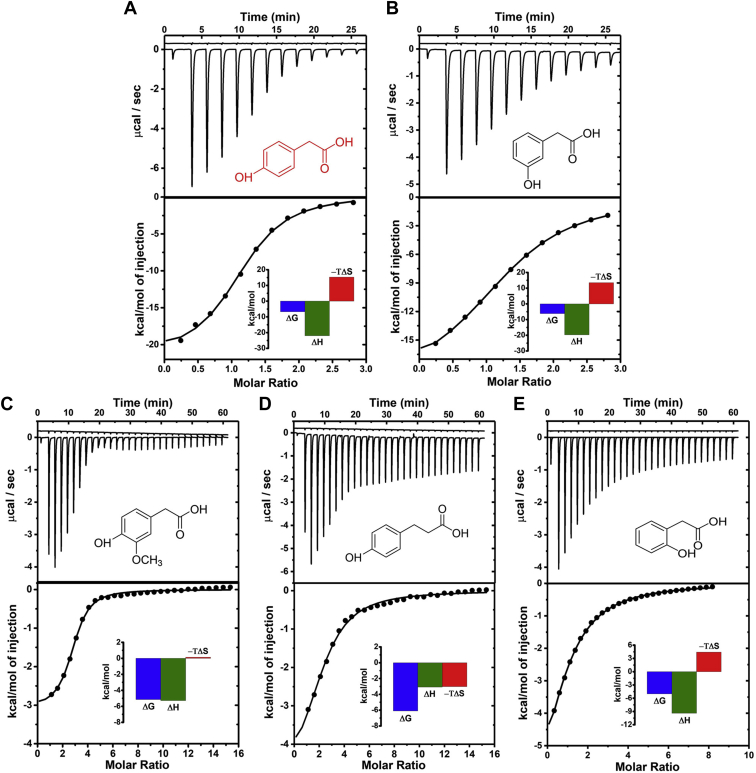
Isothermal titration calorimetry (ITC)-based determination of the affinities of the phenol derivatives for C1
The molecular affinities for C1 of the five phenolic compounds that had been electrochemically screened were also evaluated by ITC (33). As a routine, the phenol derivatives were used to fill the ITC syringe and to titrate C1 solution in the ITC sample cell. Figure 7 shows examples of thermograms obtained during the ITC trials, together with the corresponding plots of the integrated heat of the thermogram troughs for particular injections versus the molar ratio between the phenolic ligand and its binding partner C1.
Figure 7.
ITC assessment of the interaction between reductase C1 and four allosteric activators: the top panels show the raw data for (A) 4-HPA, (B) 3-HPA, (C) 4-hydroxy-3-methoxy phenylacetate, (D) 3-(4-hydroxyphenyl) propionate and (E) 2-HPA and, as insets, the chemical structures of the five organic compounds. The bottom panels show the binding isotherms created by plotting the integrated heat peaks against the molar ratio between enzyme and activator. Data points were fitted for a one-site model with variable stoichiometry, N. The insets of the bottom panels of A–E are the thermodynamic profiles for binding of the various phenols to C1.
The values of the related stoichiometries (N), reaction enthalpy (ΔH°), the Gibb’s free energy (ΔG°), the entropy term (-TΔS°), and the dissociation constant (Kd) were extracted from the heat plots of phenol/C1 interactions, using the One Set of Sites Model. In the insets to Figure 7, A–E, ΔH°, ΔG°, –TΔS° are shown as thermodynamic profile plots, and the numerical values of Kd are provided in Table 1, which summarizes the outcome of triplicate ITC analyses of 4-HPA and 3-HPA and duplicate analyses of 2-HPA, 4-hydroxy-3-methoxy phenyl-acetate, and 3-(4-hydroxyphenyl) propionate. The conclusions were (1) 4-HPA had the lowest value of Kd, followed by 3-HPA and 4-hydroxy-3-methoxy phenylacetate, (approximately fourfold higher) and 2-HPA and 3-(4-hydroxyphenyl) propionate (20-fold higher) (2), the ΔG° values were negative for all five phenols and favorable to binding in all five cases (3), ΔH° was negative for all phenol/C1 interactions but heat release was greater for the 4- and 3-HPA isomers than for the other compounds, and (4) the –TΔS° term was markedly positive for the three HPA isomers but for 4-hydroxy-3-methoxy-phenylacetate was moderately negative and for 3-(4-hydroxyphenyl)-propionate only slightly positive.
Table 1.
Summary of the thermodynamic parameters Kd, ΔH°, -TΔS°, and ΔG° and the binding stoichiometry N for the interaction of hydroxyphenyl derivatives with the C1 subunit of 4-hydroxyphenylacetate (4-HPA) 3-hydroxylase from Acinetobacter baumannii
| Activator analogues | Kd (μM) | ΔH° (kcal/mol) | ΔG° (kcal/mol) | -TΔS° (kcal/mol) | N (site #) |
|---|---|---|---|---|---|
| 4-Hydroxyphenyl -acetate |
10.22 ± 0.27 | −20.27 ± 1.11 | −6.66 ± 0.20 | 13.47 ± 1.11 | 1.22 ± 0.12 |
| 3-Hydroxyphenyl -acetate |
36.43 ± 6.90 | −18.90 ± 2.21 | −6.07 ± 0.09 | 12.87 ± 2.10 | 1.20 ± 0.09 |
| 4-Hydroxy-3-methoxy –phenylacetate | 39.45 ± 5.75 | −3.03 ± 0.05 | −6.02 ± 0.12 | −2.99 ± 0.03 | 2.51 ± 0.34 |
| 3-(4-Hydroxyphenyl) -propionate | 173.50 ± 6.50 | −5.28 ± 0.01 | −5.13 ± 0.02 | 0.14 ± 0.04 | 2.41 ± 0.21 |
| 2-Hydroxyphenyl -acetate | 220.50 ± 13.44 | −8.78 ± 0.91 | −4.99 ± 0.04 | 3.79 ± 0.87 | 0.95 ± 0.04 |
Values are the means of triplicate (4-HPA, 3-HPA) and duplicate (4-hydroxy-3-methoxy phenylacetate, 3-(4-hydroxyphenyl) propionate and 2-HPA) determinations by isothermal titration calorimetry (ITC). Mean values and standard deviations from three experiments are listed.
Discussion
Redox enzymes fulfill essential physiological functions through their involvement in cellular metabolism, cell proliferation, growth, and apoptosis and also in cell-to-cell communication, while some isolated native or genetically engineered redox enzymes are used in the food, pharmaceutical, and biotechnology industries for chemical synthesis. Redox enzymes are also used as specific biosensors in clinical diagnostics and environmental and pharmaceutical analysis. For such applications, redox enzymes with allosteric binding sites that modulate their activity may offer strategies for optimization of process efficiency or biosensor performance. Because redox enzyme activity results in the appearance of electroactive chemical species, electrochemical analysis is a convenient and cost-effective alternative to spectroscopic biochemical assays for testing the potency of allosteric effectors of a biocatalyst and for assessing the concentration dependence of allosteric effects.
Here, the allosterically activated reductase C1 subunit of 4-hydroxy-phenylacetate 3-hydroxylase from A. baumannii served as a model for which an amperometric readout of activity could be established. A hurdle for realization of the methodology was the shared tendency to oxidation of the C1 substrate NADH, its phenolic allosteric activators and H2O2, the product of C1 reoxidation by O2 during conversion of NADH to NAD+. Accordingly, useful H2O2 current signals following enzymic NADH oxidation could not be acquired using anodic detection (Fig. 1A). However, interference was avoided with cathodic determination of H2O2 at the surface of screen-printed carbon electrodes that carried a PB coating as a catalyst for H2O2 reduction. The complete absence of substrate and effector reduction (Fig. 1B) and a reducing current response proportional to H2O2 concentration (Figs. 1C and 2) allowed cathodic PB-SPEs to be used in electrochemical assays of C1 activity (Fig. 3). Their satisfactory performance and potential for sensitive C1 activity screening were verified with 1 μM C1, activated with 100 μM 4-HPA, in which the response of PB-SPEs to increasing concentrations of NADH was calibrated by NADH quantification in the standard addition mode (Fig. 4). The signal proportionality (up to 600 μM NADH) and recovery of 50 μM NADH in prepared model samples (better than 110%) replicated the analytical figures of merit of recently introduced amperometric biosensors with fully 4-HPA-activated C1 immobilized in a thin-film electrode redox polymer coating (20). The practical limit of NADH detection (LOD) of amperometric NADH biosensing with dissolved (here) and immobilized C1 (20) was 2 and 15 μM, respectively. The lower LOD for the electrochemical bioassay with dissolved C1 was likely to be because NADH had freer access to C1 in solution than with the reductase immobilized within the redox polymer. It is worth mentioning that use of C1 as an immobilized element of redox polymer-based biosensors, as proposed in earlier published work (20, 21), could form the basis of a personal urinary test device with screen-printed electrode strips modified with the enzyme-polymer layer. For urinary 4-HPA analysis in clinical laboratories and as an analytical tool for inspection of libraries of effector candidates and/or genetically engineered redox enzyme variants, work with C1 in solution is more practical because of its simplicity and speed and because it avoids the extra steps needed to prepare biosensors modified with a redox polymer.
Urine samples from healthy volunteers have 4-HPA contents of 55 to 110 μM (28, 32, 34) while those from subjects with dysbiosis (28) or breast cancer (32), for instance, have concentrations many times higher. As 4-HPA levels as low as 1 μM are detectable and the linear range of the signal response extends up 100 μM, the proposed methodology of 4-HPA biosensing is feasible as an alternative assay for urinary dysbiosis and cancer analysis. Urinary H2O2 concentrations up to about 10 μM have been reported (35). Though our PB-SPEs detect low concentrations of H2O2, its presence in urine should not affect urinary 4-HPA measurements as it would just cause a small offset in the cathodic signal recordings, with the enzymatically generated H2O2 signal appearing as an analyzable trace, enabling HPA quantification.
Cathodic H2O2 amperometry was used to evaluate activity changes of soluble C1 produced by several potential allosteric effectors, added to the reaction buffer before NADH. An alternative physicochemical technique for characterizing interactions between large and small molecules in solution is isothermal titration calorimetry (ITC) (33). In the present work, ITC identified favorable enthalpies and unfavorable entropies defining the binding of 4-, 3-, and 2-HPA to C1 (Fig. 7, A, B and E); N-values for these interactions were close to 1.0, suggesting a 1:1 stoichiometry of binding by HPAs. The observed dominance of the enthalpic over entropy contributions implied that the energetics of hydrogen bond formation and van der Waals cooperativity favored HPA/C1 interaction and outweighed the disadvantageous entropy loss in the process. Finally, 4-, 3-, and 2-HPAs bound to C1 with ΔG° values of –6.8, –6.0, and –5.0 kcal/mol (Table 1). This Gibb’s free energy ranking is consistent with the relative C1 activity data from the electrochemical work in this study and with spectroscopic analysis (22), both of which rank 4-HPA, 3-HPA, and 2-HPA as the strongest, median, and weakest allosteric activators of C1. Analysis of the ITC thermograms of the other two compounds tested, 4-hydroxy-3-methoxy phenylacetate (Fig. 7C) and 3-(4-hydroxyphenyl) propionate (Fig. 7D), showed C1 interaction with N-values greater than 2 (Table 1), suggesting multiple and independent binding sites with similar affinities. Data analysis using the “Two Sets of Sites” model produced unsatisfactory fits with higher Chi-squared values. Thus, we believe the data obtained with the “One Set of Sites” model are reliable. Possibly addition of a methoxy group to the aromatic phenylacetate structure and the lengthened aliphatic chain of the propionate species produced nonspecific binding to C1, as well as specific binding to the allosteric site. However, stimulation of C1 activity may be restricted to one binding site. A marked difference between the behavior of the three HPA isomers, the methoxy variant, and the propionate species is the entropy contribution to their interaction with C1. For the methoxy-modified 4-HPA, the –TΔS° term is negative and thus favorable to binding, while for the propionate –TΔS° is virtually zero (Table 1). Multiple binding of these two compounds may result in substantial release of bound water molecules and the entropy gain from desolvation would then be a stabilizing factor for effective complex formation with C1. Finally, the Gibb’s free energy changes are –6.1 and –5.2 kcal/mol, respectively (Table 1). The ITC results suggest they are less potent C1 activators than 4-HPA but stronger effectors than 2-HPA, consistent with electrochemical and spectroscopic testing.
We found good correlation between the electrochemical assessment of relative allosteric effectiveness and the binding strength determination by ITC. However, the concentration of 4-HPA producing half-maximal activation of C1 (103 μM) was about tenfold higher than the Kd measured by ITC (10.2 μM). This may reflect multiple steps in the allosteric activation and does not affect the usefulness of electrochemistry as a technique for assaying 4-HPA in biological samples. More generally, the potential of H2O2 amperometry as an analytical tool for allosteric redox enzyme assays has been demonstrated, as long as oxygen is the electron acceptor in recycling the enzyme. Amperometric identification and assessment of allosteric modulation by an effector candidate are easily perfomed, using a simple experimental arrangement without the involvement of extra signaling enzymes and/or chromophore supplementation. The proposed concept of allosteric amperometric NADH biosensing with dissolved C1 is new and, as its H2O2-based substrate signaling is amplified by 4-HPA, most importantly a realistic basis for the detection of the effector as medical biomarker in urine. An approved clinical trial on urine samples from patients with dysbiosis, brain disfunction, or cancer will be the next step.
Conclusion
Cathodic H2O2 amperometry at screen-printed electrodes was demonstrated to be a successful alternative assay technique for redox enzymes that reduce dissolved oxygen during redox state recycling. Using the allosteric reductase C1 subunit from A. baumannii as model, the electrochemical bioassay allowed real-time monitoring of NADH conversion rates. Measurements in the absence and presence of allosteric C1 effectors offered an easy and quick method of assessing the potency of potential molecular stimulants. The relative strengths of five phenols as activators of C1-catalyzed NADH corresponded with those determined using optical assays, with 4-HPA the most potent. The proposed methodology is thus applicable to screening allosteric redox enzymes for practical applications, especially when combined with robotic electroanalysis (36, 37, 38, 39, 40, 41, 42) or electrochemical minicells for fluid testing (43, 44, 45). The results of this study may encourage consideration of voltammetric or amperometric detection as an economical and convenient alternative to spectroscopic methods, for initial screening of library members from engineered enzyme synthesis though not necessarily for detailed kinetic or mechanistic studies. Adaptation to enzymes other than the reductase C1, e.g., for pyranose and glucose oxidase genetic engineering studies, is feasible. Our future aims are further simplification of the electrochemical redox enzyme assay, its integration into a microplate-based electrochemical format for automated sample handling and minicells for sustainable allosteric biosensing, use for inspections of libraries of C1 mutants, and most importantly, exploration of the feasibility of its application to 4-HPA biomarker analysis in urine samples from dysbiosis, brain degeneration, and cancer patients, in an approved clinical study.
Experimental procedures
Reagents, materials, and solutions
The 4-hydroxyphenylacetate reductase subunit (C1-HPAH) from A. baumannii was isolated and purified as described in previously published reports [22–25]. All chemicals for solution preparation and flavin mononucleotide (FMN) were obtained as analytical grade Sigma Aldrich products through local distributor S.M. Chemical Supplies Co Ltd (Bangkok, Thailand). Reduced β-Nicotinamide adenine dinucleotide disodium salt hydrate (β-NADH), 2-hydroxyphenyl-acetate, 3-(4-hydroxyphenyl) propionate, 4-hydroxy-3-methoxy-phenyl-acetate, 3-hydroxy-phenylacetate, and 4-hydroxy-phenylacetate were from Acros Organics (Geel, Belgium). Commercial superstable PB-SPEs were products of RUSENS Ltd (Moscow, Russia).
Sodium phosphate buffer solution (50 mM, pH 7) was prepared from mixtures of NaH2PO4 and Na2HPO4 in ultrapure deionized water. In total, 10 mM stock solutions of phenolic compounds were prepared in 50 mM sodium phosphate buffer (pH 7), and 10 mM stock solution of NADH was prepared in 10 mM tris-buffer, pH 10.
All H2O2, NADH, and 4-HPA determinations were performed in triplicate on standard solutions in sodium phosphate buffer of pH 7, and results are reported as means ± standard deviations.
Instrumentation
Electroanalysis
Amperometry was carried out with a 910 PSTAT mini potentiostat from Metrohm Instruments (Herisau, Switzerland). Testing used two different three-electrode arrangements: (1) a common 3-mm-diameter Pt disk WE from Gamry Instruments (Warminster, PA, USA) with a coiled Pt wire as counter-electrode (CE) and an Ag/Ag wire RE. (2) The three integrated electrodes of the RUSENS PB- SPE platform. WE of the assembly was the centered PB electrocatalyst-coated carbon disk, CE a carbon semicircle surrounding the WE, and RE a small disk of Ag/AgCl next to the WE/CE assembly.
The electrolyte/buffer for all electrochemical C1 studies, 50 mM sodium phosphate buffer, pH 7, was held in a 10 ml glass beaker on a magnetic stir plate and all data were acquired at room temperature (25 °C). Amperometric data analysis was handled with Origin and 50-point FFT-based smoothing was applied prior to graph plotting to reduce the effect of electrical noise.
Isothermal calorimetry
The instrument for the ITC measurements of binding of potential phenolic effectors to C1 was a MicroCal PEAQ-ITC device from Malvern Panalytical (Malvern, UK). The binding reaction took place in a reaction cell that was initially filled with 280 μl of 100 μM C1 in 50 mM sodium phosphate buffer pH 7, containing 1 mM FMN, (for 4-HPA and 3-HPA), and with 280 μl of 200 μM C1 in the same buffer (for the other activator analogues). The binding reaction was initiated by injection of the phenolic compounds into the stirred C1-containing buffer in the reaction cell (3.2 μl for each of 13 injections of a 1.5 mM solution of 4-HPA or 3-HPA and 1.32 μl for each of 30 injections of a 10 mM solution of the other activators). Multiple injections were applied to allow saturation of available allosteric sites to take place and the peak sizes in the ITC heat plots became constant, as dilution, rather than molecular interaction, became the source of heat changes. The MicroCal PEAG-ITC software integrated the individual peaks of the original heat plots at the end of a trial and created graphs of the integrated peak area versus the molecular ratio of molecular binding partners (Wiseman plots). From isotherm fitting, the instrument software then calculated the dissociation constant Kd, binding enthalpy ΔH°, entropy change, ΔS°, and finally the Gibb’s free energy change, ΔG° and the stoichiometry N, for the C1:phenol complex. The means and standard deviations of three- (4-HPA, 3-HPA) and twofold (2-HPA, 4-Hydroxy-3-methoxy phenylacetate and 3-(4-Hydroxy-phenyl) propionate) repetitions of the determinations of the best-fit parameter values for ΔH°, ΔS°, ΔG°, and N are listed in the Table 1.
Data availability
All data are contained within this article.
Conflict of interest
The authors declare no conflicts of interest in regard to this article.
Acknowledgments
All authors thank their institutes and their sponsors from the industry and business section for the kind general study funding. In addition, ST appreciates the financial support from VISTEC through a Postdoctoral Research Fellow grant and from SUT and The Thailand Research Fund under the Royal Thai government (TRF) through a Royal Golden Jubilee (RGJ) PhD scholarship. WS, PC, and AS acknowledge the Thailand Science Research and Innovation (TSRI) public funding agency for complementary funding through a grant within the organization’s Global Partnership Program. And last but not least, all authors thank the critical manuscript reading and language improvements by Dr David Apps, Edinburgh Medical School, Edinburgh, Scotland.
Author contributions
S. T. conducted all the experiments, generated and analyzed the data, wrote the first article draft, and prepared all the figures. J. S. prepared the C1 reductase enzyme and guided its enzyme assay utilization. W. S. guided the ITC trials and performed with S. T. and T. F. the ITC data analysis and interpretation. A. S. designed the electrochemical dissolved C1-based 4-HPA biosensing methodology. P. C. and A. S. conceptualized the proposed enzyme assay. A. S. managed the project completion. All the authors discussed the results and contributed to draft revision and submitted the article for finalization. All the authors approved the final version of the study report.
Funding and additional information
This work was primarily supported by a faculty startup grant from the Vidyasirimedhi Institute of Science and Technology, Thailand (to A. S.), with additional funding support obtained from the Thailand Science Research and Innovation public funding agency, Thailand through a grant within the organization’s Global Partnership Program (to A. S., W. S., and P. C.). S. T. was supported by a Vidyasirimedhi Institute of Science and Technology, Thailand, Postdoctoral Researcher Fellowship and received support from Suranaree University of Technology, Thailand, and The Thailand Research Fund, Thailand, under the Royal Thai government (TRF) through a Royal Golden Jubilee (RGJ) PhD scholarship.
Edited by Ruma Banerjee
References
- 1.Fenton A.W. Allostery: An illustrated definition for the ‘second secret of life’. Trends Biochem. Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodey N.M., Benkovic S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 3.Motlagh H.N., Wrabl J.O., Li J., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campitelli P., Modi T., Kumar S., Ozkan S.B. The Role of conformational Dynamics and allostery in modulating protein evolution. Annu. Rev. Biophys. 2020;49:267–288. doi: 10.1146/annurev-biophys-052118-115517. [DOI] [PubMed] [Google Scholar]
- 5.Changeux J.-P., Edelstein S.J. Allosteric mechanism of signal Transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 6.Ma B., Nussinov R. Amplification of signaling via cellular allosteric relay and protein disorder. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6887–6888. doi: 10.1073/pnas.0903024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nussinov R., Tsai C.-J., Liu J. Principles of allosteric interactions in cell signaling. J. Am. Che. Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arey B.J. Allosteric modulators of glycoprotein hormone receptors: Discovery and therapeutic potential. Endocrinology. 2008;34:1–10. doi: 10.1007/s12020-008-9098-2. [DOI] [PubMed] [Google Scholar]
- 9.Khoury E., Clèment S., Laporte S.A. Allosteric and biased G protein-coupled receptor signaling regulation: Potentials for new therapeutics. Front. Endocrinol. 2014;5:1–8. doi: 10.3389/fendo.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fastrez J. Engineering allosteric regulation into biological catalysts. ChemBioChem. 2009;10:2824–2835. doi: 10.1002/cbic.200900590. [DOI] [PubMed] [Google Scholar]
- 11.Rocheville M., Garland S.L. An industrial perspective on positive allosteric modulation as a means to discover safe and selective drugs. Drug Discov. Today Technol. 2010;7:e87–e94. doi: 10.1016/j.ddtec.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis L.M. Drug hunters explore allostery’s advantages. Chem. Eng. News. 2019;97:39–42. [Google Scholar]
- 13.Villaverde A. Allosteric enzymes as biosensors for molecular diagnosis. FEBS Lett. 2003;554:169–172. doi: 10.1016/s0014-5793(03)01160-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang J. Electrochemical glucose biosensors. Chem. Rev. 2008;108:814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- 15.Clark L.C., Lyons C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962;102:29–45. doi: 10.1111/j.1749-6632.1962.tb13623.x. [DOI] [PubMed] [Google Scholar]
- 16.Wollenberger U., Scheller F.W. Enzyme activation for activator and enzyme activity measurements. Biosens. Bioelectron. 1993;8:291–297. doi: 10.1016/0956-5663(93)85009-d. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M., Zhang S., Chen Z., Zhao C., Wang L., Liu S. Allosteric kissing complex-based electrochemical biosensor for sensitive regenerative and versatile detection of proteins. Biosens. Bioelectron. 2018;105:42–48. doi: 10.1016/j.bios.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Bollela P., Edwardraja S., Guo Z., Alexandrov K. Control of allosteric protein electrochemical switches with biomolecular and electronic signals. J. Phys. Chem. Lett. 2020;11:5549–5554. doi: 10.1021/acs.jpclett.0c01223. [DOI] [PubMed] [Google Scholar]
- 19.Bollela P., Edwardraja S., Guo Z., Alexandrov K. Control of allosteric electrochemical protein switch using magnetic signals. Chem. Commun. 2020;56:9206–9209. doi: 10.1039/d0cc04284f. [DOI] [PubMed] [Google Scholar]
- 20.Teanphonkrang S., Janke S., Chaiyen P., Sucharitakul J., Suginta W., Khunkaewla P., Schuhmann W., Ruff A., Schulte A. Tuned amperometric detection of reduced β-nicotinamide adenine dinucleotide by allosteric modulation of the reductase component of the p-hydroxyphenylacetate hydroxylase immobilized within a redox polymer. Anal. Chem. 2018;90:5703–5711. doi: 10.1021/acs.analchem.7b05467. [DOI] [PubMed] [Google Scholar]
- 21.Teanphonkrang S., Ernst A., Janke S., Chaiyen P., Sucharitakul J., Suginta W., Khunkaewla P., Schuhmann W., Schulte A., Ruff A. Amperometric detection of the urinary disease biomarker p-HPA by allosteric modulation of a redox polymer-embedded bacterial reductase. ACS Sens. 2019;4:1270–1278. doi: 10.1021/acssensors.9b00144. [DOI] [PubMed] [Google Scholar]
- 22.Chaiyen P., Suadee C., Wilairat P. A novel two-protein component flavoprotein hydroxylase. Eur. J. Biochem. 2001;268:5550–5561. doi: 10.1046/j.1432-1033.2001.02490.x. [DOI] [PubMed] [Google Scholar]
- 23.Thotsaporn K., Sucharitakul J., Wongratana J., Suadee C., Chaiyen P. Cloning and expression of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii: Evidence of the divergence of enzymes in the class of two-protein component aromatic hydroxylases. Biochim. Biophys. Acta. 2004;1680:60–66. doi: 10.1016/j.bbaexp.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Sucharitakul J., Chaiyen P., Entsch B., Ballou D.P. The reductase of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii requires p-hydroxyphenylacetate for effective catalysis. Biochemistry. 2005;44:10434–10442. doi: 10.1021/bi050615e. [DOI] [PubMed] [Google Scholar]
- 25.Sucharitakul J., Phongsak T., Entsch B., Svasti J., Chaiyen P., Ballou D.P. Kinetics of a two-component p-hydroxyphenylacetate hydroxylase explain how reduced flavin is transferred from the reductase to the oxygenase. Biochem. 2007;46:8611–8623. doi: 10.1021/bi7006614. [DOI] [PubMed] [Google Scholar]
- 26.Sarosiek I., Schicho R., Blandon P., Bashashati M. Urinary metabolites as noninvasive biomarkers of gastrointestinal diseases: A clinical review. World J. Gastrointest. Oncol. 2016;8:459–465. doi: 10.4251/wjgo.v8.i5.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord R.S., Bralley J.A. Clinical applications of urinary organic acids. Part 2. Dysbiosis markers. Altern. Med. Rev. 2008;13:292–306. [PubMed] [Google Scholar]
- 28.Chalmers R., Valman H., Liberman M. Measurement of 4-hydroxyphenylacetic aciduria as a screening test for small-bowel disease. Clin. Chem. 1979;25:1791–1794. [PubMed] [Google Scholar]
- 29.An M., Gao Y. Urinary biomarkers of brain diseases. GPB. 2015;13:345–354. doi: 10.1016/j.gpb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stec D.F., Wang S., Stothers C., Avance J., Denson D., Harris R., Voziyan P. Alterations of urinary metabolite profile in model diabetic nephropathy. Biochem. Biophys. Res. Commun. 2015;456:610–614. doi: 10.1016/j.bbrc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung J., Jung Y., Bang E.J., Cho S.-i., Jang Y.-J., Kwak J.-M., Park S., Hwang G.-S. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann. Surg. Oncol. 2014;21:736–742. doi: 10.1245/s10434-014-3886-0. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y., Liu F., Wan Y. Simultaneous determination of 4-hydroxyphenyl lactic acid, 4-hydroxyphenyl acetic acid, and 3, 4-hydroxyphenyl propionic acid in human urine by ultra-high performance liquid chromatography with fluorescence detection. J. Sep. Sci. 2017;40:2117–2122. doi: 10.1002/jssc.201700014. [DOI] [PubMed] [Google Scholar]
- 33.Ladbury J.E. Application of isothermal titration calorimetry in the biological Sciences: Things are heating up! BioTechniques. 2004;37:885–887. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- 34.Zamora-Ros R., Achaintre D., Rothwell J.A., Rinaldi S., Assi N., Ferrari P., Leitzmann M., Boutron-Ruault M.-C., Fagherazzi G., Auffret A., Kuehn T., Katzke V., Boeing H., Trichopoulou A., Naska A. Urinary excretion of 34 dietary polyphenols and their associations with lifestyle factors in the EPIC cohort study. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep26905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen J.W.M., Benzie I.F.F. Hydrogen peroxide in urine as a potential biomarker of whole body oxidative stress. Free Radic. Res. 2003;37:1209–1213. doi: 10.1080/10715760310001616032. [DOI] [PubMed] [Google Scholar]
- 36.Intarakamhang S., Leson C., Schuhmann W., Schulte A. A novel automated electrochemical ascorbic acid assay in the 24-well microtiter plate format. Anal. Chim. Acta. 2011;687:1–6. doi: 10.1016/j.aca.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Intarakamhang S., Schulte A. Automated electrochemical free radical scavenger screening in dietary samples. Anal. Chem. 2012;84:6767–6774. doi: 10.1021/ac301292c. [DOI] [PubMed] [Google Scholar]
- 38.Intarakamhang S., Schuhmann W., Schulte A. Robotic heavy metal anodic stripping voltammetry: Ease and efficacy for trace lead and cadmium electroanalysis. J. Solid State Electrochem. 2013;17:1535–1542. [Google Scholar]
- 39.Theanponkrang S., Suginta W., Weingart H., Winterhalter M., Schulte A. Robotic voltammetry with carbon nanotube-based sensors: A superb blend for convenient high-quality antimicrobial trace analysis. Int. J. Nanomedicine. 2015;10:859–868. doi: 10.2147/IJN.S75237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teanphonkrang S., Schulte A. Automated quantitative enzyme biosensing in 24-well microplates. Anal. Chem. 2017;89:5261–5269. doi: 10.1021/acs.analchem.6b04694. [DOI] [PubMed] [Google Scholar]
- 41.Jaikaew W., Ruff A., Khunkaewla P., Erichsen T., Schuhmann W., Schulte A. Robotic microplate voltammetry for real-time hydrogel drug release testing. Anal. Chim. Acta. 2018;1041:33–39. doi: 10.1016/j.aca.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 42.Ruff A., Jaikaew W., Khunkaewla P., Schuhmann W., Schulte A. Drug Release from polymer thin films and gel pellets: Insights from programmed microplate electroanalysis. ChemPlusChem. 2020;85:627–633. doi: 10.1002/cplu.202000129. [DOI] [PubMed] [Google Scholar]
- 43.Sripirom J., Sim W.C., Khunkaewla P., Suginta W., Schulte A. Simple and economical analytical voltammetry in 15 μl volumes: Paracetamol voltammetry in blood serum as a working example. Anal. Chem. 2018;90:10105–10110. doi: 10.1021/acs.analchem.8b01135. [DOI] [PubMed] [Google Scholar]
- 44.Sim W.C., Kutrakul N., Khunkaewla P., Schulte A. Three-electrode 30-60 μl mini-cell for ecologically conscious analytical voltammetry with common macro- and microelectrodes. ACS Sustainable Chem. Eng. 2020;8:5082–5090. [Google Scholar]
- 45.Teanphonkrang S., Sripirom J., Schulte A. A Closed 50-u L Three-Electrode Minicell for green voltammetry without O2 interference. IEEE Sens. J. 2020;20:14050–14057. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within this article.