The natural history of SARS-CoV-2 infection is highly variable, ranging from asymptomatic infection on one hand to pneumonia, septic shock, multiple organ failure, and death on the other.1 The physiologic pathways that influence prognosis after infection are still incompletely understood; this represents a key barrier to optimal management.
The rs738409 G variant in PNPLA3 (patatin-like phospholipase domain containing 3) is a prominent genetic risk factor for steatosis, cirrhosis, and hepatocellular carcinoma.2 For 2 reasons, we hypothesized that rs738409 G may also affect COVID-19 severity. First, rs738409 G is associated with retinoid storage levels in liver mesenchymal cells,3 which may have a bearing on the ability to mount an effective immune response after viral infection.4 Second, previous data suggest that rs738409 G increases the cellular abundance of omega-3 polyunsaturated fatty acids, such as alpha linolenic acid,5 which may modulate inflammatory process levels during infections.
Our goal was to test a possible association of rs738409 G in PNPLA3 with outcomes of COVID-19 using data from the United Kingdom Biobank (UKB) study.
Methods
Our hypothesis was tested in the UKB, a population-based cohort of approximately 500,000 middle-aged individuals in the United Kingdom. Latterly, data on participants testing positive for SARS-CoV-2 in England after March 16, 2020, have been released.
All UKB unrelated participants with (1) a positive PCR test result for SARs-CoV-2 between March 16, 2020, and August 15, 2020, and (2) data available for the PNPLA3:rs738409 genotype were analyzed.
The primary endpoints in this study were (1) hospital admission for COVID-19 and (2) death from COVID-19.
Logistic regression was used to assess the association between the rs738409 G allele and each endpoint under additive and recessive genetic models. All models were adjusted for potential confounding factors.
We performed sensitivity analyses to check assumptions. First, we tested whether the association between rs738409 G and each outcome changed when individuals with liver disease were excluded. We also assessed if the association varied according to ethnicity, coronary artery disease, obesity, age, and liver disease. Third, we determined the rs738409 G association with 2 specific types of COVID-19 hospitalization, namely, (1) hospital admission for COVID-19 with pneumonia recorded and (2) hospital admission for COVID-19 entailing advanced respiratory support. Fourth, we calculated the association between rs738409 G and prevalent liver disease to ensure concordance with previous research.2
Further information, including detailed covariate and outcome definitions, is provided in the Supplementary Methods.
Validation Analysis
We meta-analyzed the association between rs738409 G and COVID-19 morbidity from 3 studies/sources comparing patients with COVID-19 morbidity to SARS-CoV-2–positive patients with no/minimal morbidity. These data sources were (1) the FinnGen biobank, (2) Geisinger Health Study, and (3) data on COVID-19 patients hospitalized in Palermo, Italy.6
Results
The primary analysis included 1585 UKB participants. The mean age was 67.9 years (standard deviation, 9.1), 52.7% were male, and three quarters were White British (78.1%). The rs738409 G allele frequency was 20.9% (Supplementary Table 1). The numbers of patients with a COVID-19 hospital admission, a COVID-19 admission with pneumonia, and a COVID-19 admission requiring advanced respiratory support were 759 (47.9%), 450 (28.4%), and 76(4.8%), respectively. Approximately one sixth of the sample died of COVID-19 (16.9%; n = 267).
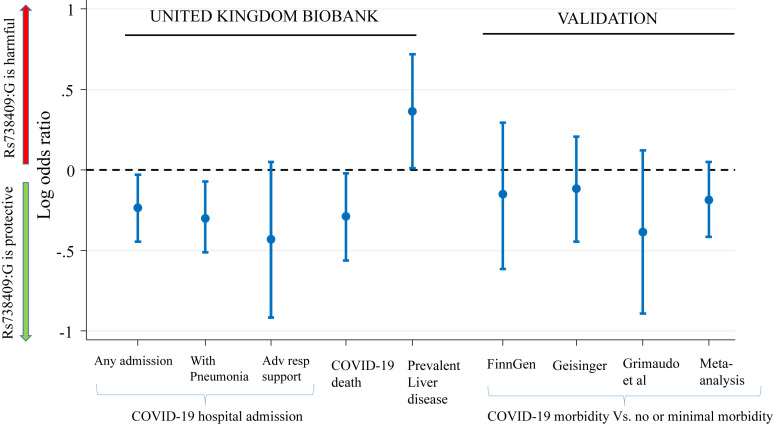
The rs738409 G allele was independently associated with a reduced risk of COVID-19 hospitalization and mortality (Figure 1 and Supplementary Table 2). On average, each additional G allele carried was associated with a 21% lower odds of COVID-19 hospitalization (adjusted odds ratio [aOR]: 0.79; 95% confidence interval [CI], 0.64–0.97; P = .027) and a 25% lower odds of COVID-19 death (aOR, 0.75; 95% CI, 0.57–0.98; P = .037). The aORs for the recessive model were 0.47 (95% CI, 0.25–0.88; P = .018) for COVID-19 hospitalization and 0.19 (95% CI, 0.06–0.64; P = .007) for COVID-19 mortality.
Figure 1.
Association of the rs738409 G allele with COVID-19 severity and prevalent liver disease. Adv resp, advanced respiratory.
In sensitivity analyses, the association between the rs738409 G allele and COVID-19 mortality/hospitalization did not attenuate after exclusion of individuals with liver disease (eg, adjusted additive OR for hospitalization, 0.78; 95% CI, 0.63–0.97; P = .027). There was a suggestion that the rs738409 G effect size was greater for younger individuals. For example, the additive OR for COVID-19 hospitalization was 0.59 (95% CI, 0.41–0.84) for those aged <65 years vs 0.92 (95% CI, 0.71–1.19) for individuals aged ≥65 years (P interaction = .040). Finally, as expected, the rs738409 G allele was associated with an increased risk of liver disease (aOR, 1.44; 95% CI, 1.01–2.05; P = .045) (see Figure 1).
Validation Analysis
In a meta-analysis of 3 independent data sources, rs738409 G showed a trend for association with a reduced risk of COVID-19 hospitalization/severe disease vs mild disease; but the result did not reach statistical significance (additive OR: 0.83; 95% CI, 0.66–1.05; P = .12; P heterogeneity = .68) (see Figure 1).
Discussion
These data show a lower risk of COVID-19 hospitalization and death in carriers of the rs738409 G allele in PNPLA3, which is athwart to its risk-increasing effect on liver disease (Figure 1). However, although this association was robust in the UKB study against a broad set of COVID-19–related endpoints and with adjustment for a comprehensive set of covariates, it was only moderately supported by independent validation data (ie, the effect was directionally concordant but not statistically significant in the validation data). Further data from larger well-defined cohorts, including data on mortality, are therefore required to verify the association of rs738409 G with COVID-19 sequelae. Functionally, this association could reflect the influence of lipid metabolism on the immune response to COVID-19; for example, retinoids are stored as retinyl esters in hepatic mesenchymal cells and also in adipose tissue where PNPLA3 is expressed. When required, retinoids are mobilized to extrahepatic tissues where they can stimulate interferon type 1 production as a potent cytokine response to viral infections.4 PNPLA3 has retinyl-palmitate lipase activity, which stimulates the release of retinol into the systemic circulation.7 Accordingly, individuals with the I148M loss-of-function variant (encoded by rs738409 G) exhibit lower circulating retinoid levels7 and increased retinoid storage levels in the liver.3 In contrast, some risk factors for severe COVID-19 (eg, obesity and liver disease8) are associated with diminished retinoid levels and/or impaired retinoid signaling, which may limit retinoid availability during infection. This finding may also reflect a lower ratio of omega-6 to omega-3 polyunsaturated fatty acids in individuals with the rs738409 G allele,5 which may temper inflammation and safeguard against cytokine storm syndrome.
In summary, we report a putative association between PNPLA3:rs738409 and COVID-19 severity. In doing so, we provide an example of how pleiotropic effects of certain genetic risk loci affect disease endpoints differently.
Acknowledgments
This research has been conducted using the UKB resource (application number: 8764). The authors also acknowledge the participants and investigators of the FinnGen study.
CRediT Authorship Contributions
Hamish Innes, BSc, MSc, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Methodology: Equal; Resources: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Stephan Buch, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal); Eleanor Barnes, PhD (Conceptualization: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Writing – review & editing: Equal) Jochen Hampe, M.D; PhD (Conceptualization: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Writing – review & editing: Equal); Thomas Majot, PhD (Conceptualization: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Writing – review & editing: Equal); Felix Stickel, MD, PhD (Conceptualization: Equal; Data curation: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by the Swiss National Science Foundation grant (no. 310030_169196 to Felix Stickel). Hamish Innes is a Medical Research Foundation Viral Hepatitis Fellow (grant no: C0825). Eleanor Barnes is an NIHR Senior Investigator. Thomas Marjot has received a registry grant from the European Association for Study of the Liver (2020RG03) and is supported by the Wellcome Trust as a Clinical Research Fellow (MSDTC_868356).
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2021.02.059.
Supplementary Methods
Inclusion Criteria
The present analysis included all UKB participants with a positive test result for SARs-CoV-2 in the 3-month period between March 16, 2020, and 15 August 15, 2020. National Health System (NHS) England Hospital Episode Statistics (HES) and mortality data were complete until September 30, 2020, at the time of analysis. Thus, the August 15 cutoff date ensured there was a minimum of 6 weeks of follow-up data between the first positive test for SARs-CoV-2 and any subsequent COVID-19 hospitalization or death.
Participants who were first- or second-degree relatives with another participant in the sample were excluded. We identified first- or second-degree relations via a kinship coefficient of ≥0.10, as recommended by the UKB. We excluded participants with unreliable genetic data, using the UKB field identifier (ID) 22010.
Definition of Outcome Events
A COVID-19 death was defined through the presence of 1 or more of the following 3 International Classification of Diseases–10th Revision (ICD-10) codes in any cause of death position: U071 (COVID-19 virus identified) and/or U072 (COVID-19 virus not identified) and/or B342 (coronavirus infection, unspecified site). Of note, the U072 ICD-10 code is used to denote clinical or epidemiologic cases of COVID-19 where laboratory data are inconclusive or not available.
COVID-19 hospital admissions were identified using NHS England HES data. The same 3 ICD-10 codes (U071 and/or U072 and/or B342) in any diagnostic position were used to identify a COVID-19–related admission. Instances of COVID-19–related pneumonia were identified using the J12.8 ICD-10 code (other viral pneumonia, not classified elsewhere) in combination with U071 and/or U072 and/or B342.
Information on advanced respiratory support is collected on the HES critical care data set. It has been defined as: “i) Invasive medical ventilatory support applied via a trans-laryngeal tracheal tube or applied via a tracheostomy; ii) Bi-level positive airway pressure applied via a trans-laryngeal tracheal tube or applied via a tracheostomy; iii) Continuous positive airway pressure via a trans-laryngeal tracheal tube; iv) Extracorporeal respiratory support.”1 We used the NHS England HES critical care data set available for UKB participants to identify individuals receiving this intervention.
Prevalent liver disease was defined through 1 or more liver-related hospital admissions before the first SARs-CoV-2–positive test. Liver-related hospital admissions were identified using the ICD-10 K70–K77 “diseases of the liver” chapter or the equivalent ICD–Ninth Revision (ICD-9) codes (571–573).
Adjustment for Confounding Factors
All regression models were adjusted for a broad range of potential confounding factors. These were age, month of positive SARS-CoV-2 test result, body mass index, sex, coronary artery disease, chronic liver disease/liver cirrhosis, current smoker, type 2 diabetes, rs657152, rs11385942, ethnic group, and the top 5 principal components of genetic ancestry. Individuals with missing data for 1 or more of these variables were excluded from the final sample.
Ethnic group was defined through a combination of genetic and self-reported information (field IDs 22006 and 21000). Type 2 diabetes was defined using field ID 2443: “Has a doctor ever told you that you have diabetes?” From here, we excluded participants with a history of gestational diabetes only (field ID 4041) and individuals with evidence of type 1 diabetes from hospital admission records (ICD-10 E10) or from detailed information collected on diagnosed medical conditions at UKB enrollment during the nurse interview (field ID 20002). The presence of coronary artery disease was inferred through hospital admissions occurring before the first positive test result for SARs-CoV-2 using ICD-10 120–125, and/or ICD-9 4120–4129, 4130–4139, 4140, 4141, or 4149. As a covariate, liver disease was divided into 2 categories: those with a hospital admission for cirrhosis and those with a noncirrhosis liver-related admission. As before, a liver-related admission was defined using the ICD-10 K70–K77 liver disease chapter (or equivalent ICD-9 codes). Cirrhosis was defined using ICD codes outlined previously by Ratib et al.2 Age was based on age at the time of the first positive test result for SARs-CoV-2. Current smoking was inferred only through information provided at UKB enrollment (field ID 20116). Body mass index was determined from each participant’s height and weight at the time of the initial UKB assessment visit. Standing height was measured via the Seca 202 height measure, and body weight was measured with the Tanita BC-418 MA body composition analyzer.
Assessing Variability in rs738409 Effect Size
We assessed if the rs738409 effect size varied according to 3 covariates: ethnic group, coronary artery disease, obesity, age <65 years, and history of prior liver disease. This was done by adding interactions terms into the model (ie, between rs738409 and the covariate of interest) and performing a likelihood ratio test to gauge whether the interaction model significantly improved model fit.
Validation Analysis
We retrieved the association between rs738409 G and COVID-19 morbidity (vs SAS-CoV-2 infection but no/minimal morbidity) from 3 independent studies. These were
-
•
the FinnGen population–based biobank study, comprising 83 patients with a COVID-19 hospital admission vs 274 SARS-CoV-2–positive patients without a hospital admission (Supplementary Table 2);
-
•
individuals in the Geisinger Health Study data set of European descent, comprising 165 individuals COVID-19 hospitalization vs 689 SARS-CoV-2–positive patients without a hospital admission (Supplementary Table 2);
-
•
data from a study by Grimaudo et al,3 comprising 52 patients admitted to the hospital for COVID-19 who either died or required intensive care therapy vs 314 patients admitted to the hospital for COVID-19 but without any complications.
The rs738409:G association from each study was then aggregated through fixed-effects meta-analysis, weighted according to the study sample size.
Supplementary Material
References
- 1.Huang C., et al. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trepo E., et al. J Hepatol. 2016;65:399–412. doi: 10.1016/j.jhep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Kovarova M., et al. J Clin Endocrinol Metab. 2015;100:E1568–1574. doi: 10.1210/jc.2015-2978. [DOI] [PubMed] [Google Scholar]
- 4.Trottier C., et al. Antiviral Res. 2008;80:45–53. doi: 10.1016/j.antiviral.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Peter A., et al. Diabetologia. 2014;57:2103–2107. doi: 10.1007/s00125-014-3310-0. [DOI] [PubMed] [Google Scholar]
- 6.Grimaudo S et al. 2020. https://www.researchsquare.com/article/rs-40510/v1
- 7.Mondul A., et al. J Nutr. 2015;145:1687–1691. doi: 10.3945/jn.115.210633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trasino S.E., et al. Sci Rep. 2015;5:15893. doi: 10.1038/srep15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.NHS Digital. HES Data Dictionary: Adult Critical Care. Page 7.
- 2.Ratib S., et al. Am J Gastroenterol. 2015;110:1149–1158. doi: 10.1038/ajg.2015.191. [DOI] [PubMed] [Google Scholar]
- 3.Grimaudo et al. 2020. Available at: https://www.researchsquare.com/article/rs-40510/v1. Accessed April 22, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.