Significance
Glioblastoma is a highly lethal form of brain cancer with no current treatment options that substantially improve patient outcomes. A key therapeutic challenge is the identification of methods that reduce tumor burden while leaving normal cells unaffected. We show that TERT-promoter mutations, common in glioblastoma, lead to TERT reactivation through increased binding of GABPB1L-isoform–containing transcription factor complexes. In turn, we find that cancer-cell–specific inhibition of TERT through GABPB1L reduction results in near-term anti-growth effects and an impaired DNA damage response that profoundly increase the sensitivity of glioblastoma tumors to frontline chemotherapy. Our results thus provide rationale for GABPB1L inhibition combined with temozolomide chemotherapy treatment as a promising therapeutic strategy for glioblastoma.
Keywords: TERT, cancer, CRISPR, glioblastoma, temozolomide
Abstract
Most glioblastomas (GBMs) achieve cellular immortality by acquiring a mutation in the telomerase reverse transcriptase (TERT) promoter. TERT promoter mutations create a binding site for a GA binding protein (GABP) transcription factor complex, whose assembly at the promoter is associated with TERT reactivation and telomere maintenance. Here, we demonstrate increased binding of a specific GABPB1L-isoform–containing complex to the mutant TERT promoter. Furthermore, we find that TERT promoter mutant GBM cells, unlike wild-type cells, exhibit a critical near-term dependence on GABPB1L for proliferation, notably also posttumor establishment in vivo. Up-regulation of the protein paralogue GABPB2, which is normally expressed at very low levels, can rescue this dependence. More importantly, when combined with frontline temozolomide (TMZ) chemotherapy, inducible GABPB1L knockdown and the associated TERT reduction led to an impaired DNA damage response that resulted in profoundly reduced growth of intracranial GBM tumors. Together, these findings provide insights into the mechanism of cancer-specific TERT regulation, uncover rapid effects of GABPB1L-mediated TERT suppression in GBM maintenance, and establish GABPB1L inhibition in combination with chemotherapy as a therapeutic strategy for TERT promoter mutant GBM.
Primary glioblastoma (GBM) is the most common and lethal form of malignant brain cancer in adults. Current treatment strategies are limited, with GBM progression leading to death within 2 y of diagnosis in 90% of cases (1–3). In GBM, as well as the vast majority of other cancers, transcriptional activation of the telomerase reverse transcriptase (TERT) gene, which is normally silenced in somatic cells, is a key step in tumorigenesis (4, 5). TERT encodes the catalytic subunit of telomerase, and its reactivation in cancer is thought to contribute to cell survival and immortalization (6–8). TERT ablation thus has the potential to directly affect both short- and long-term cell viability through telomere-length–dependent and independent pathways (9–15). Prior research has demonstrated that inhibition of TERT expression enhances sensitivity of cells to DNA damage by radiation and chemotherapy, suggestive of a possible combination therapy for cancer treatment (16, 17). However, telomerase inhibition is toxic to normal telomerase-dependent stem and germline cells, which has led to the failure of such approaches clinically (18–20). Therefore, understanding genetic contributors to aberrant TERT expression, as well as consequences of cancer-specific TERT ablation, are critical to develop novel therapeutic avenues for GBM.
A major mechanism by which TERT is reactivated in cancer involves the acquisition of somatic mutations in its promoter, which represent the most common noncoding mutations, and the third most common mutations overall, in cancer (21–26). In particular, ∼80% of primary GBMs contain one of two common single-nucleotide mutations that are associated with re-expression of TERT messenger RNA (mRNA), referred to as G228A and G250A (22, 24, 27). Both G-to-A transitions generate an identical 11-base-pair sequence (plus strand CCCGGAAGGGG) that creates a binding site for GA binding protein A (GABPA), an E26 transformation-specific (ETS)-family transcription factor (28, 29). Interestingly, these de novo ETS binding sites occur within three (G228A) or five (G250A) complete helical turns of two overlapping native ETS binding sites (ETS 195 and ETS 200) in the TERT promoter (TERTp), one or the other of which are also required for TERT reactivation (28). GABP transcription factors are obligate multimers that consist of the DNA-binding GABPA subunit (GeneID: 2551) and a transactivating GABPB subunit (30). Humans have two paralogues encoding different beta subunits, GABPB1 (GeneID: 2553) and GABPB2 (GeneID: 126626). Reduced function of the long protein isoform of GABPB1 (GABPB1L) via indel mutations has previously been linked to down-regulation of TERT mRNA and long-term telomere attrition (9). This could provide a cancer-specific way to target TERT, particularly given that GABPB1L is dispensable for normal murine development while GABPA and total GABPB1 are not (31, 32). However, the extended time period required to induce cell death via progressive telomere shortening limits the therapeutic potential of this approach for high-grade GBM (33). To further our understanding of the GABPB1L-TERT axis and its possible therapeutic benefit, here we examined the specificity and binding affinity of GABPB1L for the mutant TERT promoter, as well as functional effects of GABPB1L loss in a near-term, clinically relevant timeframe. Notably, we observed a dramatic synergistic effect between GABPB1L reduction and standard-of-care temozolomide chemotherapy, mediated through TERT down-regulation and a resulting attenuation of the DNA damage response (DDR) in TERTp mutant cells. These rapid effects of targeting the cancer-specific GABPB1L-TERT axis in combination with frontline chemotherapy in established intracranial GBM xenografts led to substantially increased survival, promising exciting avenues for TERTp-mutant GBM therapy.
Results
GABPB1L-Containing Complexes Bind and Regulate the Mutant TERT Promoter.
GABPB1 encodes two main transcript variants, a short isoform (GABPB1S) and a long isoform (GABPB1L). GABPB1S functions as a heterodimer with GABPA (GABPA1B1), while GABPB1L forms a heterotetramer (GABPA2B2) due to its unique terminal exon that contains a leucine zipper-like domain (30, 34) (SI Appendix, Fig. S1A). Previous work demonstrated that TERTp mutations create a binding site for the GABPA-B transcription factor complex and linked reactivation of TERT in this context to GABP complexes containing GABPB1L (9, 28). Indeed, the fact that two GABP binding sites distanced at complete helical turns from one another are required to enable TERT reactivation is suggestive of the recruitment of a GABPB1L-containing heterotetramer (9, 28, 35) (SI Appendix, Fig. S1A). However, GABPB1 isoform specificity in mutant TERT promoter regulation has not been examined. In order to test this, we made use of a doxycycline-inducible, microRNA-embedded short hairpin RNA (shRNA) system with a green fluorescent protein (GFP) marker reporting shRNA induction (36–38). We used a machine learning algorithm (38) to design shRNAs targeting either GABPB1L or GABPB1S specifically (SI Appendix, Fig. S1B). Doxycycline-mediated expression of GABPB1L or GABPB1S shRNAs led to reduction of the respective mRNA and protein levels in TERTp mutant (U-251) and TERTp wild-type (WT) (LN-18) GBM cells (Fig. 1A and SI Appendix, Fig. S1 C and D). Interestingly, GABPB1L—but not GABPB1S—knockdown led to reduced TERT mRNA and telomerase activity exclusively in TERTp mutant cells (Fig. 1 B and C and SI Appendix, Fig. S1E), suggesting that TERTp mutant regulation of TERT expression is dependent on GABPB1L-containing complexes specifically. Of note, residual TERT expression and telomerase activity are observed in TERTp mutant cells following GABPB1L knockdown when compared with a cancer cell line that is negative for TERT expression (U2OS) (Fig. 1 B and C and SI Appendix, Fig. S1E). This may be due to incomplete shRNA-mediated knockdown (Fig. 1A and SI Appendix, Fig. S1C) or to regulation of TERT by other factors in conjunction with, or in the absence of, GABPB1L (39–41).
Fig. 1.
GABPB1L-containing complexes bind and regulate the mutant TERT promoter. (A) Representative immunoblots of GABPB1 in U-251 and LN-18 cells expressing doxycycline-induced shRNAs targeting GABPB1S (shGB1S.82, shGB1S.77, shGB1S.32, and shGB1S.110) and GABPB1L (shGB1L.377, shGB1L.699, shGB1L.968, and shGB1L.1202) compared with negative control (olfactory receptor OR2B6, shOR2B6.83) and nontargeting (renilla luciferase, shRen.713) shRNAs. Cells were incubated with doxycycline for 6 d prior to harvest. The lower bands represent GABPB1S, and upper bands represent GABPB1L. (B) TERT mRNA expression measured via qRT-PCR in cell lines from A compared with a control cell line (U2OS) lacking TERT expression. (C) Representative gels of telomerase activity measured via TRAP assay in cell lines from A compared with a control cell line (U2OS) lacking TERT expression. The control condition reflects no lysate. (D and E) Representative gels (D) and quantification (E) of electrophoretic mobility shift assays (EMSAs) comparing binding affinity of GABPA-B1L heterodimers to the mutant (G228A) TERT promoter (additional ETS binding site), WT TERT promoter (native ETS binding sites), and control WT TERT promoter sequences lacking native ETS binding sites (G201T: native ETS single mutant; G201T and A197T: native ETS double mutant).
Though binding of GABP to the TERT promoter has previously been demonstrated (28), possible GABPA2B1L2 heterotetramer formation at the mutant promoter has not directly been examined. To determine the nature of the GABPA-B1L interaction with the TERT promoter, we titrated purified heterodimer (SI Appendix, Fig. S1 F and G) against a fixed concentration of radiolabeled wild-type TERTp DNA, mutant TERTp DNA (G228A), and control probes with single (G201T) or double (A197T and G201T) mutations in the native ETS sites contained within the WT promoter. We observed that GABPB1L-containing complexes bind with ∼twofold increased affinity to the mutant TERTp relative to wild type. However, calculating Kd values that accurately reflect GABP binding was challenging due to the observation of multiple intermediate states, which may be reflective of heterodimer binding, as has been previously suggested (42, 43) (Fig. 1D and SI Appendix, Fig. S1H). Thus, we segregated observed states into unbound, intermediate bound, and fully bound states. The comparison of signal ratios in each state for each target DNA tested across multiple GABP concentrations showed that the TERTp mutation (G228A) leads to increased formation of the fully bound state, which we attribute to a likely tetrameric complex (42, 43) (Fig. 1 D and E). We saw that GABP has the ability to bind all TERT promoter sequences tested in this study. However, binding to WT and native ETS site mutants (G201T or A197T and G201T) showed an increased number of intermediate states when compared with the G228A-bearing promoters and a decrease in the fully bound state (Fig. 1 D and E and SI Appendix, Fig. S1H). Together, these results demonstrate that TERTp mutations result in increased binding affinity for probable GABPB1L-containing heterotetramer complexes, which help drive TERT reactivation.
GABPB1L Knockout Impairs Near-Term Growth of TERT Promoter Mutant GBM Cells.
For GBM therapy, short-term effects of TERT reduction, rather than long-term consequences of canonical telomere attrition, would increase the clinical relevance of targeting the cancer-specific GABP-TERT axis (33). Given that GABPB1L regulates TERT in the context of TERTp mutations, our data raise the question of possible early functional consequences of GABPB1L inhibition on TERTp-mutant cell growth and viability. CRISPR-Cas methods enable the precise dissection of genetic dependencies through the generation and analysis of specific heterozygous and homozygous genetic deletions in isogenic cell lines (44). Therefore, to assess near-term consequences of complete GABPB1L deletion, we used CRISPR-Cas9 and single-guide RNAs (sgRNAs) targeting the intron and 3′ untranslated region (UTR) flanking the unique exon 9 of GABPB1L (Fig. 2A and SI Appendix, Fig. S2 A and B and Table S1). We compared a set of three TERTp mutant GBM cell lines (U-251, LN-229, and T98G) and four TERTp WT cell lines (HEK293T, HAP1, NHA-S2, and LN-18) by generating monoclonal isogenic derivatives. The TERTp WT lines include an immortalized human astrocyte line (NHA-S2) as well as a GBM line (LN-18). Sequencing of the TERT promoter confirmed TERTp mutation status (SI Appendix, Fig. S2 C and D).
Fig. 2.
Inhibition of GABPB1L rapidly prevents growth of TERT promoter mutant cells. (A) GABPB1L exon 9 excision strategy. A diagram of the major human GABPB1 transcript variants, encoding either a short (GABPB1S) or long (GABPB1L) isoform. Red rectangles represent the coding sequence, and green rectangles represent the 3′ UTR. For clarity, the first exon containing the 5′ UTR is not shown. GABPB1L sgRNA targeting sites are highlighted in purple. (B) The editing strategy for scarless knockout of the long isoform of GABPB1 (GABPB1L). In each case, a pair of sgRNAs were used targeting GABPB1L intron number 8 and the 3′ UTR of GABPB1L, respectively. (C and D) Genotyping analysis of GABPB1L edited monoclonal TERT promoter mutant U-251, LN-229, and T98G GBM cell lines (C) and TERT promoter WT HEK293T, HAP1, NHA-S2, and LN-18 cell lines (D). The superscript characters indicate the sgRNA pair used for editing. WT: parental WT cell line. C, PCR no template control. Unedited, full-length genotyping band; Δ2-12, band after excision with sgGB2 and sgGB12; Δ3-14, band after excision with sgGB3 and sgGB14. (E) A comparison of GABPB1L editing outcomes between TERTp WT (blue) and TERTp mutant (yellow) lines. The bars represent the percentage of each editing outcome averaged over three TERTp mutant and four TERTp WT lines. Retained refers to clones where the GABPB1L exon 9 was overall retained, even though they may contain indels at either or both of the sgRNA target sites flanking exon 9. p, P value (unpaired, two-tailed Student’s t test). n.s., not significant (alpha level = 0.05). (F and G) Representative bioluminescence images (F) and Kaplan–Meier survival curve (G) of mice injected with U-251 cells expressing a control shRNA (shRen.713) or GABPB1L-targeting shRNAs (shGB1L.968, shGB1L.1202). Mice were fed doxycycline chow to induce shRNA expression starting at day 7 postinjection. n = 10 to 12 mice per condition. **P < 0.01 compared with shRen.713 (Kaplan–Meyer log-rank test). M, median survival.
To identify ideal guide RNAs, all possible combinations of three intronic and three 3′ UTR targeting guides were compared in HEK293T and U-251 cells to assess editing efficiency (Fig. 2B and SI Appendix, Fig. S2E). Next, the two most efficient pairs of sgRNAs were used to edit all seven cell lines, and 169 monoclonal cell line derivatives were established, genotyped, and subjected to further analysis (Fig. 2 C and D and SI Appendix, Figs. S3 A–H and S4 A–C). Strikingly, we observed that while full homozygous knockout of GABPB1L occurred at an appreciable rate across all TERTp WT cell lines (13 to 36%), we were able to generate only one full GABPB1L knockout line, LN229-C19, among the 76 TERTp mutant clones that were screened (Fig. 2E and SI Appendix, Fig. S3 A–H). Importantly, the discrepancy between TERTp WT and mutant cell lines did not appear to result from differences in overall editing efficacy between cell lines, as equal numbers of heterozygous knockouts were observed (Fig. 2E and SI Appendix, Fig. S3H). Thus, loss of GABPB1L may have near-term effects on growth and survival of TERTp mutant cells specifically that prevent the generation of full knockout clones. Prior studies from our laboratory demonstrated that impaired GABPB1L protein function through indel-based editing led to long-term telomere attrition over a more than 90 d timeframe (9). However, the present data suggest that full knockout of GABPB1L causes TERTp mutant cells to become either slow growing or nonviable over the ∼30 d editing and selection process, a timeframe that is not consistent with gradual telomere shortening (9, 15, 45).
GABPB1L Knockdown Slows Growth of Established TERTp Mutant Tumors.
To more directly assess the therapeutic potential of GABPB1L reduction, we next examined its effect on growth of established GBM tumors in vivo, using the above-described doxycycline-inducible shRNA system (Fig. 1A and SI Appendix, Fig. S1B). We first tested the system for in vivo efficacy using a bona fide essential gene, Replication Protein A1 (RPA1; GeneID: 6117), along with controls. Addition of doxycycline to U-251 cells stably transduced with an inducible vector encoding RPA1-targeting shRNAs resulted in reduced RPA1 mRNA in vitro (SI Appendix, Fig. S5A) and prolonged animal survival in vivo (SI Appendix, Fig. S5 B and C). Because the doxycycline-inducible vectors encode an shRNA within the 3′ UTR of a GFP construct (Fig. 1A), the percentage of GFP-positive cells was used as a proxy readout for shRNA expression. Postmortem tumor analysis showed that in vivo expression of the RPA1-targeting shRNA construct was reduced compared with controls (SI Appendix, Fig. S5D), consistent with a survival advantage for loss of RPA1 knockdown (37).
We then made use of our GABPB1L-targeting shRNAs to determine the effect of GABPB1L knockdown on growth of existing GBM tumors. Two separate GABPB1L-targeting shRNAs tested in orthotopic intracranial xenografts of U-251 cells slowed tumor growth within 12 d of shRNA induction when induced post tumor implantation (Fig. 2F and SI Appendix, Fig. S5E) and increased median survival by 70 to 90% (Fig. 2G). GABPB1L shRNA-mediated target knockdown and linked GFP expression were maintained in vivo via analysis of ex vivo dissociated tumor cells postmortem (SI Appendix, Fig. S5 F and G). A third tested GABPB1L-targeting shRNA (shGB1L.699) did not impact survival (SI Appendix, Fig. S5H). Correspondingly, postmortem analysis demonstrated minimal GABPB1L protein reduction in tumors with this shRNA and showed that only ∼30% of tumor cells retained GFP expression (SI Appendix, Fig. S5 F and G), indicating that sustained GABPB1L knockdown is necessary to slow tumor growth. A short-term cell-culture–based competitive proliferation assay with the same shRNAs did not show any toxicity (SI Appendix, Fig. S5I), pointing toward a specific in vivo effect of sustained GABPB1L knockdown. Together, these data demonstrate that shRNA-mediated GABPB1L suppression in established tumors significantly prolongs animal survival, highlighting its therapeutic value.
GABPB2 Promotes Resistance to GABPB1L Loss in TERTp Mutant GBM Cells.
Given the potential therapeutic benefit of GABPB1L inhibition for TERTp mutant GBM, we chose to analyze heterozygous knockout clones generated from TERTp mutant lines compared with full and heterozygous knockouts from TERTp WT lines in order to better understand the molecular impact of GABPB1L loss (Fig. 2 C–E). Full knockout of GABPB1L led to a complete loss of GABPB1L mRNA and protein in most cell lines, whereas heterozygous knockout led to various degrees of GABPB1L reduction, consistent with the expected effects of total versus partial removal of GABPB1L loci (SI Appendix, Fig. S6 A and B). We also observed unchanged or up-regulated total GABPB1 mRNA (all transcript variants) and substantially up-regulated GABPB1S mRNA and protein across all cell lines (SI Appendix, Fig. S6 A–F), supporting the possibility that all GABPB1 transcripts from a GABPB1L knockout locus are now GABPB1S. Notably, GABPB1L reduction was associated with a decrease in TERT mRNA expression and reduced telomerase activity in TERTp mutant cells only (SI Appendix, Fig. S6 G–J), confirming regulation of TERT by GABPB1L in a TERTp mutant-specific manner.
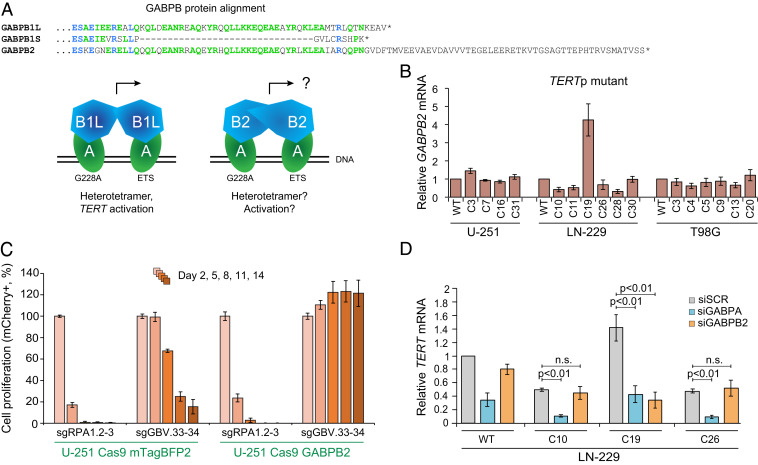
The single TERTp mutant GABPB1L full knockout cell line (LN229-C19) that we obtained did not exhibit reduction of either TERT mRNA or telomerase activity (SI Appendix, Fig. S5 G, I, and J), suggesting a compensatory mechanism to regulate TERT independent of GABPB1L. GABPB1 and GABPB2 are distinct genes with high sequence conservation that perform similar but separate functions, though GABPB2 is thought to function exclusively as a heterotetramer with GABPA (30, 34), mediated by the presence of a leucine zipper-like domain (Fig. 3A and SI Appendix, Fig. S7A). Since GABPB2 is expressed at lower levels than GABPB1 in GBM based on data from the TCGA Research Network (https://www.cancer.gov/tcga) (SI Appendix, Fig. S7B), we probed for possible GABPB2 up-regulation in our GABPB1L knockout clones. While deletion of GABPB1L did not affect GABPB2 mRNA in any other line, the LN229-C19 clone showed a 4.5-fold elevation in GABPB2 transcript levels (Fig. 3B and SI Appendix, Fig. S7C). In support of GABPB2 up-regulation as a potential compensatory mechanism, stable GABPB2 overexpression in U-251 cells rescued cell dropout following CRISPR editing of the GABPB1 locus (Fig. 3C). In addition, small interfering RNA–mediated inhibition of GABPB2 showed that TERT mRNA levels are responsive to GABPB2 knockdown in LN229-C19 but not in WT LN-229 cells or heterozygous knockout clones (Fig. 3D). These data suggest that GABPB2 up-regulation can compensate functionally for GABPB1L deletion in TERTp mutant cells and indicate that TERTp mutant GBM can tolerate total loss of GABPB1L in the context of alternative mechanisms rescuing TERT expression. This finding reveals a possible resistance mechanism for GABPB1L-targeting therapies.
Fig. 3.
GABPB2 up-regulation can compensate for loss of GABPB1L. (A) Similarity between GABP proteins. Partial protein alignment (MUSCLE) of GABPB1L, GABPB1S, and GABPB2. GABPB1L is the long isoform and GABPB1S the short isoform of the GABPB1 gene. GABPB2 is a distinct gene. Amino acids matching across all three proteins are highlighted in blue, and those matching across two proteins are highlighted in green. GABPA (alpha) subunits of the GABP complex bind to both the native ETS site (ETS) and the mutation-derived ETS sites (G228A or G250A) at the TERT promoter locus. The GABPB1 short (B1S) or long (B1L) isoform subunits bind to the alpha subunits to form either heterodimers (GABPA1B1S1) or heterotetramers (GABPBA2B1L2). Heterotetramer formation is presumably mediated through the leucine zipper-like domain of GABPB1L, which is also present in GABPB2. (B) GABPB2 mRNA expression measured via qRT-PCR in GABPB1L knockout clones, plotted relative to control (WT) cells, in TERTp mutant cell lines. The data represent mean ± SEM. (C) Competitive proliferation assay in U-251 cells using pairs of sgRNAs targeting a positive control locus (sgRPA1.2 and 3) and total GABPB1 (targeting exon 3, sgGBV.33 and 34) in the presence of a lentiviral vector expressing either GABPB2 or mTagBFP2 (control). The data represent mean ± SD of triplicates. (D) TERT mRNA expression measured via qRT-PCR following siRNA-mediated knockdown of GABPA or GABPB2 in LN-229 WT, full knockout, or heterozygous knockout cells. p, P value (unpaired, two-tailed Student’s t test). n.s., not significant (alpha level = 0.05). The data represent mean ± SEM.
Increased Response of GABPB1L-Reduced GBM Tumors to Chemotherapy.
Currently, most patients diagnosed with primary GBM are treated with a regimen that includes temozolomide (TMZ) chemotherapy, a DNA-alkylating chemotherapeutic agent that is most effective at eliminating cells that lack O-6-methylguanine-DNA methyltransferase (MGMT) through generation of single- and double-strand DNA breaks (46, 47). Prior studies have indicated that reduction of TERT in TERT-expressing cells may sensitize them to DNA damage from ionizing radiation and chemotherapy (16, 17). Reduction of TERT mRNA by RNA interference has been shown to prevent the repair of DNA breaks in vitro by inhibiting central components of the DDR, such as histone H2AX phosphorylation (yH2AX), through an incompletely understood mechanism (16). Loss of DNA damage signaling increases DNA damage-related senescence or cell death, possibly due to a reduction in normal cell cycle arrest (16, 48, 49). Therefore, we wondered whether a GABPB1L-mediated down-regulation of TERT mRNA in TERTp mutant GBM might promote TMZ response, enabling a possible path to GBM-cell–specific chemotherapy sensitization (Fig. 4A).
Fig. 4.
Reduction of GABPB1L potentiates anti-tumor response of TERTp mutant GBM to chemotherapy. (A) A schematic for possible sensitization of TERT promoter mutant GBM cells to TMZ through GABPB1L inhibition. Inhibiting GABPB1L leads to reduced TERT expression, resulting in a blunted DDR and lack of normal cell cycle arrest, ultimately reducing cancer cell viability following DNA damage. SSB, single-strand break; DSB, double-strand break; DDR, DNA-damage response. (B) Representative immunoblots of yH2AX in U-251 cells expressing doxycycline-induced shRNAs targeting TERT (shTERT.3795 and shTERT.3592) or GABPB1L (shGB1L.968 and shGB1L.1202) compared with a nontargeting (renilla luciferase, shRen.713) shRNA. (C) Representative immunoblots (Bottom) and quantification (Top, from triplicate blots) of yH2AX in U-251 cells engineered to stably express either BFP (control) or TERT (rescue) in the presence of shRNAs targeting TERT (shTERT.3795 and shTERT.3592) or GABPB1L (shGB1L.968 and shGB1L.1202) compared with a nontargeting shRNA (renilla luciferase, shRen.713). (B and C) Cells were incubated with doxycycline for 6 d prior to harvest and treated with specified dose(s) of TMZ 20 h prior to harvest. (D) Representative images (Left) and G2/G1 ratio quantification (Right) from flow cytometry–based cell cycle analysis of U-251 WT and GABPB1L heterozygous knockout cells treated with a dose titration of TMZ 72 h prior to harvest. p, P value (unpaired two-tailed Student’s t test). (E) A schematic of in vivo TMZ treatment. Mice were xenografted with U-251 cells expressing doxycycline-inducible shRNAs and placed on doxycycline chow 7 d postorthotopic xenograft. Mice were then treated with TMZ or a vehicle control by oral gavage starting 12 d postxenograft for 5 d. IHC, immunohistochemistry. (F) Bioluminescence imaging of mice injected with U-251 cells engineered to express shRNAs targeting TERT (shTERT.3592), GABPB1L (shGB1L.968), or a nontargeting control shRNA (shRen.713). Mice were placed on doxycline chow and per os (p.o.) dosed with either TMZ or a vehicle control as described in E. (G) Representative images of immunofluorescence staining in tumors from mice described in E, cohort 1. Mice were euthanized 30 d postorthotopic xenograft for analysis. Doxycycline-induced tumor cells are GFP positive. n = 5 images per mouse, three mice per condition. (Scale bar, 50 μm.) (H) Kaplan–Meier survival curves for mice described in E, cohort 2. For statistics, survival of mice with no tumor burden was set at the experimental endpoint (130 d). n = 5 to 7 mice per condition. **P < 0.01, relative to all vehicle conditions (Kaplan–Meier log-rank test). M, median survival. (I) A comparison of the relative fold increase in median survival based on overall survival in H. The dashed line represents a simple addition of effects of TMZ plus GABPB1L or TERT knockdown. The survival of mice xenografted with WT cells and treated with vehicle control was used for normalization.
U-251 cells expressing doxycycline-inducible shRNAs targeting either TERT or GABPB1L were treated with a dose titration of TMZ, and the DDR was assessed through analysis of yH2AX levels. Compared with control, shRNA-mediated TERT knockdown significantly reduced the usual increase in yH2AX 20 h after treatment with TMZ (Fig. 4B and SI Appendix, Fig. S8 A and B). Strikingly, GABPB1L knockdown led to an equally blunted yH2AX increase following TMZ treatment (Fig. 4B and SI Appendix, Fig. S8B). Similarly, TMZ-mediated yH2AX iduction was reduced in both U-251 and LN-229 clones with heterozygous GABPB1L knockout, compared with WT cells (SI Appendix, Fig. S8 C and D). However, the full GABPB1L knockout clone LN229-C19 with GABPB2 up-regulation and maintained TERT expression exhibited a TMZ-induced yH2AX increase that mirrored WT, consistent with the impairment of the DDR being mediated by TERT down-regulation (SI Appendix, Fig. S8D).
To directly assess whether GABPB1L knockdown leads to a reduced yH2AX increase through reduced TERT, we generated U-251 cells that stably express either blue fluorescent protein (BFP; control) or TERT (rescue) constructs (SI Appendix, Fig. S8E). While the BFP construct showed no effects, overexpression of TERT completely rescued the loss of yH2AX induction following shRNA-mediated knockdown of both TERT and GABPB1L (Fig. 4C). In the control shRNA condition (shRen.713), TERT overexpression significantly increased yH2AX levels following TMZ treatment when compared with WT TERT levels, providing further evidence for regulation of the DDR by TERT (Fig. 4C and SI Appendix, Fig. S8E).
Ablated DDR signaling can lead to a blunting of the G2 cell cycle arrest that is commonly observed following treatment with DNA damaging agents such as TMZ, causing an increased loss of viability as cells continue to divide (48, 50, 51). In concordance, here we also observe reduced G2 arrest in both U-251 and LN-229 GABPB1L heterozygous knockout clones treated with TMZ, as compared with both WT cells and the LN-229 full GABPB1L knockout clone C19 (Fig. 4D and SI Appendix, Fig. S8 F and G). Lack of MGMT expression was confirmed in all cell lines and corresponding clones (SI Appendix, Fig. S8H).
Both partial and full loss of H2AX, or inhibition of its phosphorylation, have been shown to significantly impact cell survival following exposure to DNA damage (52–54). Similarly, inhibition of G2 arrest following TMZ administration is associated with decreased cell viability and growth, resulting in TMZ sensitization (50). Therefore, we hypothesized that TERTp mutant GBM cells with reduced GABPB1L would be more sensitive to TMZ chemotherapy. We first assessed the therapeutic implications of this finding in vivo through administration of a TMZ regimen following orthotopic intracranial xenograft of either U-251 or LN-229 WT and GABPB1L knockout cells. Initial analysis—in the absence of TMZ therapy—of GABPB1L heterozygous knockout cell tumors in vivo demonstrated that GABPB1L reduction slowed growth of both U-251 and LN-229 tumors and increased animal survival (SI Appendix, Fig. S8 I–L), while the full knockout LN229-C19 clone with maintained TERT expression formed tumors at a similar rate to WT cells. Strikingly, in vivo administration of chemotherapy in combination with GABPB1L heterozygous status radically ablated tumor growth of U-251 clones (SI Appendix, Fig. S9 A–D). A similar potentiation of treatment efficacy was also observed, though to a lesser extent, in GABPB1L-reduced LN-229 tumors, again with the expected exception of the full knockout LN229-C19 clone that maintained WT TERT mRNA levels (SI Appendix, Fig. S9 E–H).
Lastly, in order to assess the therapeutic potential of this combination therapy in a more clinically relevant manner, we performed orthotopic xenografts of U-251 cells engineered to express doxycycline-inducible shRNAs targeting either TERT (shTERT.3952) or GABPB1L (shGB1L.968) compared with a control (shRen.713). Following tumor engraftement, mice were placed on doxycycline and treated with a regimen of either TMZ or vehicle, then either assessed 30 d postxenograft for immunohistochemistry (Cohort 1) or followed to endpoint for survival analysis (Cohort 2) (Fig. 4E). We verified that doxycycline and TMZ do not interfere with each other to affect either tumor growth or shRNA induction in vivo (SI Appendix, Fig. S9 I and J). Interestingly, knockdown of TERT and GABPB1L both slowed growth of tumors and prolonged survival to a similar extent, which was accompanied by a significant reduction in the percentage of Ki-67 positive tumor cells in these conditions (Fig. 4 F–H and SI Appendix, Fig. S9 K–M). More importantly, both TERT and GABPB1L reduction in established intracranial xenografts resulted in a similar potentiation of TMZ efficacy, such that tumor growth was either dramatically slowed or eliminated (Fig. 4H and SI Appendix, Fig. S9 K and L). The median animal survival was increased beyond a simple addition of effects (Fig. 4I), showing the signature of a synergistic combination therapy. Ki-67 status was not affected by TMZ treatment, as assessed 14 d following the TMZ regimen (Fig. 4G and SI Appendix, Fig. S9M). Together, the dramatic anti-tumor effects of TMZ combined with GABPB1L reduction and the associated cancer-specific down-regulation of TERT mRNA are of significant clinical interest, as they provide a path to increased TMZ response for TERTp mutant GBMs. Indeed, our data suggest that MGMT-silenced TERTp mutant GBM could be effectively targeted through a combination of GABPB1L reduction and TMZ treatment.
Discussion
A major challenge in GBM therapy is finding efficient methods to reduce growth of established tumors while leaving normal cells unaffected. Here, our data directly demonstrate increased binding of GABPB1L-isoform–containing complexes to the mutant compared with WT TERT promoter. Additionally, we show that GABPB1L knockdown in vivo, and full knockout in vitro, result in near-term anti-growth effects on TERTp mutant cells and that up-regulation of GABPB2 can compensate for the loss of GABPB1L in both contexts. Mechanistically, the early appearance of these anti-growth effects hints toward noncanonical functions of TERT rather than telomere attrition or immediate vulnerability of cells with short telomeres, though this warrants further investigation (9–11, 15, 45). Additionally, these effects may be the direct result of TERT down-regulation following GABPB1L reduction, a combined effect between reduced TERT and other factors associated with GABPB1L loss, or involve other differences in gene expression specific to TERTp mutant cells (55). Regardless, our results provide evidence that even after tumor initiation, GABPB1L knockdown rapidly impairs growth of GBM tumors in vivo, which is of great therapeutic interest.
More strikingly, we also find that GABPB1L reduction combined with TMZ treatment dramatically potentiates anti-tumor effects in vivo through cancer-specific loss of TERT, emphasizing the clinical potential of this combination therapy. Specific targeting of TERT through the long isoform of GABPB1 (GABPB1L) is particularly promising given that both GABPA and total GABPB1 (the short and long isoform together) are required for normal murine development, while the GABPB1L isoform is dispensable (9, 31, 32). Previous studies have observed that TERT reduction sensitizes cells to chemotherapy and radiation by inhibiting the normal DDR, measured in part by a loss of yH2AX increase (16). Here, our data show that reducing TERT through knockdown of GABPB1L prevents both the induction of yH2AX and the G2 cell cycle arrest normally seen following exposure of TERTp mutant cells to standard-of-care TMZ chemotherapy. The mechanism of action of TMZ is largely through mismatch repair pathway–associated single-strand breaks but also results in DNA double-strand breaks, and yH2AX signaling may occur in response to single-strand breaks as well as canonically to double-strand breaks (46, 47, 56, 57). Loss of yH2AX signaling leads to reduced cell viability following DNA damage induced by radiation and chemotherapy (48, 49, 54). Our findings extend this by demonstrating that TERTp mutant GBM tumors with reduced TERT mRNA, either through knockdown of GABPB1L or by targeting TERT directly, show a dramatically increased response to TMZ in vivo.
Targeted cancer therapies have the potential to be highly impactful, but cancer cells commonly develop resistance to anti-tumor drugs (58). Uncovering mechanisms of resistance often allows development of second-generation therapies that stunt tumor relapse. Our work points to sporadic GABPB2 up-regulation as a possible resistance mechanism to loss of GABPB1L, providing preliminary evidence that GABPB2, which is typically expressed only at very low levels in GBM, could be targeted in conjunction with GABPB1L for increased treatment robustness. Multiple other transcription factors have also been linked to TERT regulation, both in normal and oncogenic contexts (59–62). Such factors could play an additional role in basal expression of TERT from the mutant promoter or compensate for loss of GABPB1L and merit further study. While clinical methods to inhibit GABPB1L do not currently exist, and transcription factors such as the GABP complex are generally considered difficult to target, viable pathways might include approaches such as anti-sense oligo therapy, which has been successful at targeting transcription factors through regulation of splicing (63), or development of small molecules that prevent GABPA2B2 heterotetramer formation. The possible clinical benefit of these approaches warrants further study. Together, our data suggest that GABPB1L inhibition combined with TMZ treatment can provide a tumor-specific path to improve disease outcomes for patients with TERTp mutant GBMs and possibly the many other cancers harboring TERTp mutations.
Materials and Methods
See SI Appendix, Methods for additional experimental details. See SI Appendix, Table S1 for sequence information for sgRNAs, shRNAs, oligonucleotides, and vectors.
Protein Purification.
GABPB1L and GABPA subunits were cloned into the pET-Duet-1 vector with Gibson assembly. The GABPB1L subunit was inserted after the first ribosome binding site and a plasmid-encoded HisTag. The HisTag and GABPB1L were separated by insertion of a tobacco etch virus (TEV) cleavage sequence. GABPA was inserted after the second ribosome binding site without any affinity purification tags.
Mammalian Cell Culture.
HEK293T human kidney cells (293FT; Thermo Fisher Scientific, #R70007; RRID: CVCL_6911), NHA-PC5 normal human astrocyte cells (a kind gift from Russell Pieper, University of California, San Francisco [UCSF]), and derivatives thereof were grown in Dulbecco’s Modified Eagle Medium (DMEM; Corning Cellgro, #10-013-CV) supplemented with 10% fetal bovine serum (FBS; Seradigm #1500-500), and 100 Units/mL penicillin and 100 µg/mL streptomycin (100-Pen-Strep; Gibco, #15140-122). U-251 human GBM cells (Sigma-Aldrich, #09063001; RRID: CVCL_0021), LN-229 human GBM cells (American Type Culture Collection [ATCC], #CRL-2611; RRID: CVCL_0393), T98G human GBM cells (ATCC, #CRL-1690; RRID: CVCL_0556), LN-18 human GBM cells (ATCC, #CRL-2610; RRID: CVCL_0392), and derivatives thereof were cultured in DMEM/Nutrient Mixture F-12 (DMEM/F-12; Gibco, #11320-033 or Corning Cellgro, #10-090-CV) supplemented with 10% FBS and 100-Pen-Strep. HAP1 cells (a kind gift from Jan Carette, Stanford University) and derivatives thereof were grown in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, #12440-053 or HyClone, #SH30228.01) supplemented with 10% FBS and 100-Pen-Strep. HAP1 cells had been derived from the near-haploid chronic myeloid leukemia cell line KBM7 (64).
Establishment of a Puromycin-Sensitive Normal Human Astrocyte Cell Line.
The normal human astrocyte cell line NHA-PC5 was previously described (45) and is puromycin resistant. For our experiments, we established a puromycin-sensitive monoclonal derivative, termed NHA-S2, through CRISPR editing of the previously inserted puromycin-resistance cassette.
Plasmid and Lentiviral Vectors.
To generate monoclonal knockout cell lines, sgRNAs and Cas9 were expressed from either the pCF120 or pCF123 vectors. In brief, the plasmid vector pCF120, expressing a U6-promoter–driven sgRNA and an EF1-alpha short (EFS)-promoter–driven Cas9-P2A-Hygro cassette was derived from pX459 (65) by replacing the chicken beta-actin promoter with an EFS promoter from pCF204 (66), swapping the T2A-Puro cassette with a P2A-Hygro cassette and exchanging the guide RNA scaffold for a more efficient variant (67). The plasmid vector pCF123, expressing a U6-driven sgRNA and an EFS-driven Cas9-T2A-Puro cassette, was generated based on pX459 by replacing the chicken beta-actin promoter with an EFS promoter and exchanging the guide RNA scaffold for a more efficient variant (67).
For CRISPR-Cas9–mediated competitive proliferation assays, sgRNAs were expressed from the previously established pCF221 lentiviral vector and Cas9 from the pCF226 lentiviral vector (66). For low-level stable overexpression of GABPB2 (NCBI gene ID: 126626) and mTagBFP2 (negative control), we first modified the pCF525-mTagBFP2 lentiviral vector (68), encoding an EF1a-Hygro-P2A-mTagBFP2 cassette, by replacing the strong EF1-alpha promoter with a weak EFS) promoter. This new vector, EFS-Hygro-P2A-mTagBFP2, was named pCF554. Next, we cloned the coding sequence of homo sapiens GA binding protein transcription factor subunit beta 2 (GABPB2), transcript variant 1, and mRNA (NCBI reference sequence: NM_144618.2) into pCF554 by replacing mTagBFP2, yielding lentiviral vector pCF553.
For doxycycline-inducible, microRNA-embedded miR-E shRNA expression, both in cell culture and in vivo, we used the lentiviral vector LT3GEPIR (36).
Lentiviral Transduction.
Lentiviral particles were produced in HEK293T cells using polyethylenimine (Polysciences #23966)-based transfection of plasmids, as previously described (66).
Design of sgRNAs for CRISPR-Cas9 Genome Editing.
Standard sgRNA sequences were either designed manually or using GuideScan (69). To generate specific genomic excisions rather than indels, pairs of sgRNAs were designed and used in tandem. This strategy was applied to remove sequences specific to the long isoform of GABPB1L while not affecting the short isoform.
CRISPR-Cas9 Competitive Proliferation Assay.
CRISPR-Cas9 competitive proliferation assays were used to assess whether Cas9-mediated editing of genes of interest leads to a change in proliferation speed compared with unedited cells. First, cell lines (U-251) were stably transduced with a lentiviral vector expressing Cas9 (pCF226) and selected on puromycin (1.0 to 2.0 µg/mL) as previously described (66). Subsequently, Cas9-expressing cell lines were further stably transduced with pairs of lentiviral vectors (pCF221) expressing various mCherry-tagged sgRNAs.
Design of shRNAs for Reversible Target Inhibition.
MicroRNA-embedded miR-E shRNA sequences were predicted using SplashRNA (38) and cloned into LT3GEPIR (36), an all-in-one doxycycline-inducible miR-E shRNA expression vector. The shRNA numbers refer to the position of the shRNA, specifically the 3′ nucleotide of the guide strand, on the target transcript (38, 70). All shRNA sequences are shown (SI Appendix, Table S1) and were cloned as previously described (70).
Generation of Isogenic GABPB1L Knockout Cell Lines.
To generate isogenic monoclonal knockout cell lines of the long isoform of GABPB1 (NCBI gene ID: 2553), pairs of sgRNAs flanking the long isoform–specific exon 9 of GABPB1 were used.
GABPB1L locus editing was assessed by genotyping with the primers oCF862_GAL-fw1 (AAAAGTACAGGTGCCCAGTTTG) and oCF863_GAL-rev2 (GCCTAACCAACAACGATCAC), yielding an 808-base-pair band in WT cells. Where mentioned, Sanger sequencing of the GABPB1L locus and Tracking of Indels by Decomposition analysis (71) were also carried out using the primers oCF862_GAL-fw1 and oCF863_GAL-rev2.
Telomerase Repeated Amplification Protocol.
Cy5-based telomerase repeated amplification protocol (TRAP) assays were performed as previously described (72). Briefly, 150,000 cells per condition were collected via trypsinization and resuspended in Nonidet P-40 lysis buffer (10 mM Tris HCl pH 8.0, 1 mM MgCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Nonidet P-40, 0.25 mM sodium deoxycholate, 10% glycerol, 150 mM NaCl, 5 mM 2-mercaptoethanol, and 0.1 mM AEBSF) at a concentration of 2,500 cells/ul to maintain telomerase activity. A total of 1 μL of lysate (or 1 μL of Nonidet P-40 lysis buffer for the negative control) was added to a PCR master mix containing a Cy5-labeled telomeric substrate primer, an internal standard control, and an extension primer mix at the described ratios (72).
Orthotopic Xenografts and Bioluminescence Imaging.
Animal procedures conformed to care guidelines approved by the UCSF Institutional Animal Care and Use Committee (protocol numbers: AN179550, AN17599, and AN179565). U-251 and LN-229 cell lines were stably transduced with Firefly Luciferase Lentifect Purified Lentiviral Particles (Genecopoiea, #LPP-FLUC-Lv105) at a multiplicity of infection (MOI) of 3. All cell lines were verified to be stably expressing luciferase at similar levels in vitro 48 h prior to xenograft. Either 4 × 105 U-251 cells or 1 × 105 LN-229 cells were injected in a 4 µl total volume into the right frontal cortex (mediolateral [ML]: 1.5 mm, anteroposterior [AP]: 1 mm, and dorsoventral [DV]: -3.5 mm, relative to bregma) of 6 to 7 wk old female athymic nu/nu mice (inducible shRNA TMZ combination experiment: The Jackson Laboratory, RRID: IMSR_JAX:002019; all other experiments: Envigo, Hsd:Athymic Nude-Foxn1nu, #069).
Imaging and Analysis.
For immunofluorescence-based analysis of GFP, Ki-67, and glial fibrillary acidic protein (GFAP) expression in inducible shRNA-expressing U-251 tumors treated with vehicle or TMZ, animals were euthanized 30 d postxenograft to compare across conditions at identical timepoints. Animals were perfused and brains fixed in 4% paraformaldehyde (PFA) for 24 h. Brains were then washed in phosphate-buffered saline (PBS) and soaked in 30% sucrose prior to freezing in optimal cutting temperature (OCT) compound blocks for cryosectioning. Slides were washed in PBS to remove OCT, blocked in 5% bovine serum albumin + 0.3% Triton-X in PBS for 1 h, then incubated with primary antibodies toward Ki-67 (Thermo Fisher, #PA5-19462) or GFAP (Abcam, #ab4674-50) overnight, followed by secondary antibodies for 1 h (Alexa Fluor 647, Abcam #ab150171 or Alexa Fluro 555, and Thermo Fisher #A27039). Slides were incubated with Hoechst stain for 10 min prior to mounting. GFP immunofluorescence was detectable without the need for antibody staining. Imaging was performed using a Keyence digital microscope by a researcher blinded to experimental condition. Then, 2× representative images were taken of the entire tumor field for each of three mice per condition, and 40× images near the tumor edges were obtained for analysis of Ki-67 expression, since Ki-67 expression is highest at the tumor edge and is used as a pathology-based marker for tumor grade. Ki-67 image quantification was carried out using similar methods to previously published work (73–75).
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism 9 software. Two-tailed, unpaired Student’s t tests were used with alpha = 0.01 or alpha = 0.05, as specified. Statistical significance between Kaplan Meier survival curves was performed using a Kaplan Meier log-rank test with alpha = 0.01 or alpha = 0.05, as specified. The Wilcoxon signed-rank test was used for comparison of GABPB1 and GABPB2 expression levels from TCGA RNA sequencing datasets. Statistical analysis for qPCR data were performed among specific samples, rather than relative to WT control, due to normalization on WT control using the deltaCT method. Quantified data represent mean ± SD or mean ± SEM, as referenced in the figure legends. Values of “n” are listed in relevant methods and/or figure legends.
Supplementary Material
Acknowledgments
We thank Russell Pieper (UCSF) for generously sharing the immortalized astrocyte line, NHA-PC5, and for consultation on experiments involving temozolomide. We thank Mitchel Berger (UCSF) for insightful discussions and generous ongoing support. We thank Abdullah Syed (Gladstone Institutes) for assistance with image analysis. We thank the UCSF Preclinical Therapeutics Core and the UCSF Preclinical Animal Core for their assistance with animal experiments and maintenance of imaging equipment. We thank Mary West (University of California, Berkeley) and the University of California, Berkeley CIRM/QB3 Shared Stem Cell Facility/High-Throughput Screening Facility for support. We thank the Gladstone Institutes Histology and Microscopy core for assistance with cryosectioning. This work was supported by NIH Grant NCI F32CA228365 (A.M.A.), NIH K99/R00 Pathway to Independence Awards NIGMS K99GM118909 and R00GM118909 (C.F.), National Cancer Institute (NCI) fellowship F99 CA222987 (A.M.), NCI fellowship F31 CA243187 (A.M.M.), a generous gift from Mitchel Berger and the Fishgold Hurwitt Brain Tumor Research Fund (C.F. and J.A.D.), NIH Grant NCI P50CA097257 (J.F.C. and J.A.D.), funding from the GBM Precision Medicine Project (J.F.C. and J.A.D.), a generous gift from the Dabbiere family (J.F.C.), and a generous gift from the Hana Jabsheh Research Initiative (J.F.C.). J.A.D. is an investigator of the HHMI. We also thank all J.A.D. and J.F.C. laboratory members for insightful discussions.
Footnotes
Competing interest statement: The Regents of the University of California have patents issued and pending for CRISPR technologies on which J.A.D. is an inventor. C.F. is a co-founder of Mirimus, Inc. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi, Mammoth Biosciences, and Inari. J.A.D. is a Director at Johnson & Johnson and has sponsored research projects by Pfizer, Roche Biopharma, and Biogen. J.F.C. is a co-founder of Telo Therapeutics, Inc. and has ownership interests. The other authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008772118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Reardon D. A., Rich J. N., Friedman H. S., Bigner D. D., Recent advances in the treatment of malignant astrocytoma. J. Clin. Oncol. 24, 1253–1265 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Stupp R.et al.; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Louis D. N., et al., The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kim N. W., et al., Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Yuan X., Larsson C., Xu D., Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 38, 6172–6183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Tergaonkar V., Noncanonical functions of telomerase: Implications in telomerase-targeted cancer therapies. Cancer Res. 74, 1639–1644 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Leão R., et al., Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 25, 22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancini A., et al., Disruption of the β1L isoform of GABP reverses glioblastoma replicative immortality in a TERT promoter mutation-dependent manner. Cancer Cell 34, 513–528.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celeghin A., et al., Short-term inhibition of TERT induces telomere length-independent cell cycle arrest and apoptotic response in EBV-immortalized and transformed B cells. Cell Death Dis. 7, e2562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin E., et al., Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart S. A., et al., Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 99, 12606–12611 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y., Li H., Deb S., Liu J. P., TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 21, 3130–3138 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Anderson R., et al., Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. 38, e100492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Crothers J., Haqq C. M., Blackburn E. H., Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference-mediated depletion of telomerase RNA. J. Biol. Chem. 280, 23709–23717 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Masutomi K., et al., The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. U.S.A. 102, 8222–8227 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura M., et al., Efficient inhibition of human telomerase reverse transcriptase expression by RNA interference sensitizes cancer cells to ionizing radiation and chemotherapy. Hum. Gene Ther. 16, 859–868 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Tefferi A., et al., A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N. Engl. J. Med. 373, 908–919 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Shay J. W., Wright W. E., Telomerase therapeutics for cancer: Challenges and new directions. Nat. Rev. Drug Discov. 5, 577–584 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Jäger K., Walter M., Therapeutic targeting of telomerase. Genes (Basel) 7, E39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang F. W., et al., Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Killela P. J., et al., TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U.S.A. 110, 6021–6026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn S., et al., TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Zehir A., et al., Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arita H., et al., Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 126, 267–276 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Arita H., et al., TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol. 126, 939–941 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Chiba K., et al., Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 4, e07918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell R. J., et al., Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern J. L., Theodorescu D., Vogelstein B., Papadopoulos N., Cech T. R., Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 29, 2219–2224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosmarin A. G., Resendes K. K., Yang Z., McMillan J. N., Fleming S. L., GA-binding protein transcription factor: A review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol. Dis. 32, 143–154 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Xue H. H., et al., GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat. Immunol. 5, 1036–1044 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Xue H. H., et al., Targeting the GA binding protein beta1L isoform does not perturb lymphocyte development and function. Mol. Cell. Biol. 28, 4300–4309 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guterres A. N., Villanueva J., Targeting telomerase for cancer therapy. Oncogene 39, 5811–5824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Brousse F. C., Birkenmeier E. H., King D. S., Rowe L. B., McKnight S. L., Molecular and genetic characterization of GABP beta. Genes Dev. 8, 1853–1865 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Chiba K., et al., Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 357, 1416–1420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fellmann C., et al., An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Zuber J., et al., Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelossof R., et al., Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 35, 350–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X., et al., Role of Ets/Id proteins for telomerase regulation in human cancer cells. Exp. Mol. Pathol. 75, 238–247 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Xu D., Dwyer J., Li H., Duan W., Liu J. P., Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J. Biol. Chem. 283, 23567–23580 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., et al., Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 17, 1327–1338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinenov Y., Henzl M., Martin M. E., The α and β subunits of the GA-binding protein form a stable heterodimer in solution. J. Biol. Chem. 275, 7749–7756 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Carter R. S., Avadhani N. G., Cooperative binding of GA-binding protein transcription factors to duplicated transcription initiation region repeats of the cytochrome c oxidase subunit IV gene. J. Biol. Chem. 269, 4381–4387 (1994). [PubMed] [Google Scholar]
- 44.Fellmann C., Gowen B. G., Lin P. C., Doudna J. A., Corn J. E., Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 16, 89–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohba S., et al., Mutant IDH1 expression drives TERT promoter reactivation as part of the cellular transformation process. Cancer Res. 76, 6680–6689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegi M. E., et al., MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Ochs K., Kaina B., Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 60, 5815–5824 (2000). [PubMed] [Google Scholar]
- 48.Fragkos M., Jurvansuu J., Beard P., H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol. Cell. Biol. 29, 2828–2840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meador J. A., et al., Histone H2AX is a critical factor for cellular protection against DNA alkylating agents. Oncogene 27, 5662–5671 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Hirose Y., Berger M. S., Pieper R. O., Abrogation of the Chk1-mediated G(2) checkpoint pathway potentiates temozolomide-induced toxicity in a p53-independent manner in human glioblastoma cells. Cancer Res. 61, 5843–5849 (2001). [PubMed] [Google Scholar]
- 51.Hirose Y., Berger M. S., Pieper R. O., p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 61, 1957–1963 (2001). [PubMed] [Google Scholar]
- 52.Celeste A., et al., H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celeste A., et al., Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taneja N., et al., Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J. Biol. Chem. 279, 2273–2280 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Stern J. L., et al., Mesenchymal and MAPK expression signatures associate with telomerase promoter mutations in multiple cancers. Mol. Cancer Res. 18, 1050–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinner A., Wu W., Staudt C., Iliakis G., Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward I. M., Chen J., Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Sabnis A. J., Bivona T. G., Principles of resistance to targeted cancer therapy: Lessons from basic and translational cancer biology. Trends Mol. Med. 25, 185–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takakura M., Kyo S., Inoue M., Wright W. E., Shay J. W., Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol. Cell. Biol. 25, 8037–8043 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmeyer K., et al., Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336, 1549–1554 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Wu K. J., et al., Direct activation of TERT transcription by c-MYC. Nat. Genet. 21, 220–224 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Song Y. S., et al., Interaction of BRAF-induced ETS factors with mutant TERT promoter in papillary thyroid cancer. Endocr. Relat. Cancer 26, 629–641 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Aslesh T., Maruyama R., Yokota T., Systemic and ICV injections of antisense oligos into SMA mice and evaluation. Methods Mol. Biol. 1828, 455–465 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Carette J. E., et al., Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ran F. A., et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oakes B. L., et al., CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell 176, 254–267.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen B., et al., Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watters K. E., Fellmann C., Bai H. B., Ren S. M., Doudna J. A., Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 362, 236–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perez A. R., et al., GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35, 347–349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fellmann C., et al., Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell 41, 733–746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mender I., Shay J. W., Telomerase repeated amplification protocol (TRAP). Bio Protoc. 5, e1657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai Q., et al., Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors. ACS Nano 12, 8423–8435 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Kingston B. R., Syed A. M., Ngai J., Sindhwani S., Chan W. C. W., Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc. Natl. Acad. Sci. U.S.A. 116, 14937–14946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Syed A. M., et al., Liposome imaging in optically cleared tissues. Nano Lett. 20, 1362–1369 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.