Abstract
Fructus Rosae Roxburghii (FRR) has been considered as edible and medicinal fruit possessing antiatherosclerotic effect [1], [2], [3], [4], [5], but the mechanism is still unclear. Hyperlipidemia (HLP) is the material basis for atherosclerosis (AS) formation [6,7]. In this study, total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), lower high-density lipoprotein (HDL) and atherosclerotic index (ASI) in mice were analyzed under the action of FRR juice. Then differentially expressed proteins in liver were further analyzed by using TMT labeling and LC-MS/MS for better understanding the effect and molecular mechanism of FRR on diet-induced hyperlipidemic mice [8]. After the protein extraction and trypsin digestion, TMT labeling proteomic analysis were performed. The functions and KEGG signaling pathways of differentially expressed proteins were analyzed by bioinformatics methods. Hence, the potential antiatherosclerotic mechanism of FRR regulating blood lipids from protein level has great significance to explore new drug targets for AS.
Keywords: Fructus Rosae Roxburghii, Hyperlipidemia, Atherosclerosis, TMT proteomic analysis, KEGG signaling pathway, Antiatherosclerotic mechanism
Specifications Table
| Subject | Biotechnology |
| Specific subject area | Proteomic analysis has long been powerful and reliable technology for identification of proteins expression in tissues or cells [9,10]. Proteomics can provide understanding for cellular activities including the growth, development and metabolism in organisms, particularly in exploring potential mechanism of nutrients [11]. In this experiment, Tandem Mass Tag (TMT) labeling proteomic analysis was used to quantify the dynamic changes of liver proteome of mice. To characterize the functional consequences of proteins in liver, GO functional enrichment and KEGG pathway analysis of differentially expressed proteins were performed. |
| Type of data | Figure |
| How data were acquired | Reverse-phase HPLC using Agilent 300Extend C18 column (5 µm particles, 4.6 mm ID, 250 mm length); reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific); Q Exactive™ Plus hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific). |
| Data format | Secondary Data |
| Parameters for data collection | The peptides were first separated with a gradient of 2% to 60% acetonitrile in 10 mM ammonium bicarbonate (pH 10) over 80 min into 80 fractions. Then, the peptides were combined into 18 fractions and dried by vacuum centrifugation. Peptides were dissolved in 0.1% FA, directly loaded onto a reversed-phase pre-column. Peptide separation was performed using a reversed-phase analytical column at constant flow rate on EASY-nLC 1000 UPLC system. The gradient was comprised of an increase from 9% to 25% solvent B (0.1% FA, 98% ACN, 1.9% H2O) over 40 min, 25% to 36% in 12 min, climbing to 80% in 4 min and then holding at 80% for last 4 min. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q Exactive™ Plus coupled to the UPLC online. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using NCE setting as 32. The ion fragments were detected at a resolution of 35,000. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 1E4 in MS survey scan with 30 s dynamic exclusion. The electrospray voltage was 2.0 kV. 5E4 ions were accumulated for generation of MS/MS spectra. The m/z scan range was 350 to 1800. Fixed first mass was set as 100 m/z. The resulting MS/MS data were processed using Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against Swissprot Mus musculus database. Trypsin/P was specified as cleavage enzyme allowing up to 2 missing cleavages. Mass error was set to 20 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys-were specified as fixed modification and oxidation on Met-was specified as variable modifications. For protein quantification, TMT 10plex was selected in Maxquant. FDR was adjusted to < 1% and peptide ion score was set ≥ 20. The distribution of mass error for identified peptides was mostly less than 0.02 Da. The length of most peptides distributed between 8 and 16 were in agreement with the property of tryptic peptides. |
| Description of data collection | Identified proteins were classified into three categories by Gene Ontology (GO) annotation: biological process, cellular component and molecular function. A two-tailed Fisher's exact test was employed to test the enrichment of differentially expressed protein against identified proteins. Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to annotate enriched protein pathways by a two-tailed Fisher's exact test. The KEGG pathway enrichment-based clustering analysis and enrichment pathway in the groups of Diet II vs I, III vs I and II vs III were performed. |
| Data source location | The proteomics data have been submitted to proteomexchange repository. Data are available via ProteomeXchange with identifier (PXD019962). |
| Data accessibility | Repository name:proteomexchange Data identification number:PXD019962 Direct URL to data: Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD019962 FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2021/01/PXD019962 |
| Related research article | P.P. Song, X.C. Shen, Proteomic Analysis of Liver in Diet-induced Hyperlipidemic Mice under Fructus Rosa roxburghii Action, J. Proteomics. |
Value of the Data
-
•
From obtained data, much more valuble information could be deeply digged out. GO functional enrichment and KEGG pathway analysis showed that FRR juice could ameliorate the blood lipids and maintain metabolic homeostasis by regulating the expression of differentially co-expressed proteins in HLP mice. Various kinds of bioactive substances in FRR might regulate the lipid metabolism by synergistic effect. This is of great significance to provide theoretical basis for application of FRR and explore new drug targets for AS.
-
•
Some researchers in the field of atherosclerotic cardiovascular disease (CVD) could benefit from these data. A comparison of experimental data may facilitate the discovery of common factors that play the major roles in the development of a disease. It may narrow down the potential targets for future research. These potential drug targets could be further explored and verified according to the upregulation or downregulation of differentially expressed proteins. The people with HLP or early-stage AS could also try to supplement the intake of FRR juice, which would be beneficial for the prevention of CVD.
-
•
Based on the results, narrowing down interested proteins and performing functional study were recommended. These differentially expressed proteins with significant expression, specific functions, specific pathways and antibodies preparation and validation could be used for target selection. Once appropriate proteins targets are selected, functional study will be performed. Differentially expressed proteins between the control and experimental samples could be validated by using Western Blot. The results from bioinformatic analysis could be validated by the immnuoprecipiation (IP), for example, whether the target proteins are interacted with other signaling molecules or protein complexes. Overexpression or knockdown/knockout version of target proteins could be introduced to in vivo or in vitro system, and then the phenotype and functional changes could be examined.
1. Data Description
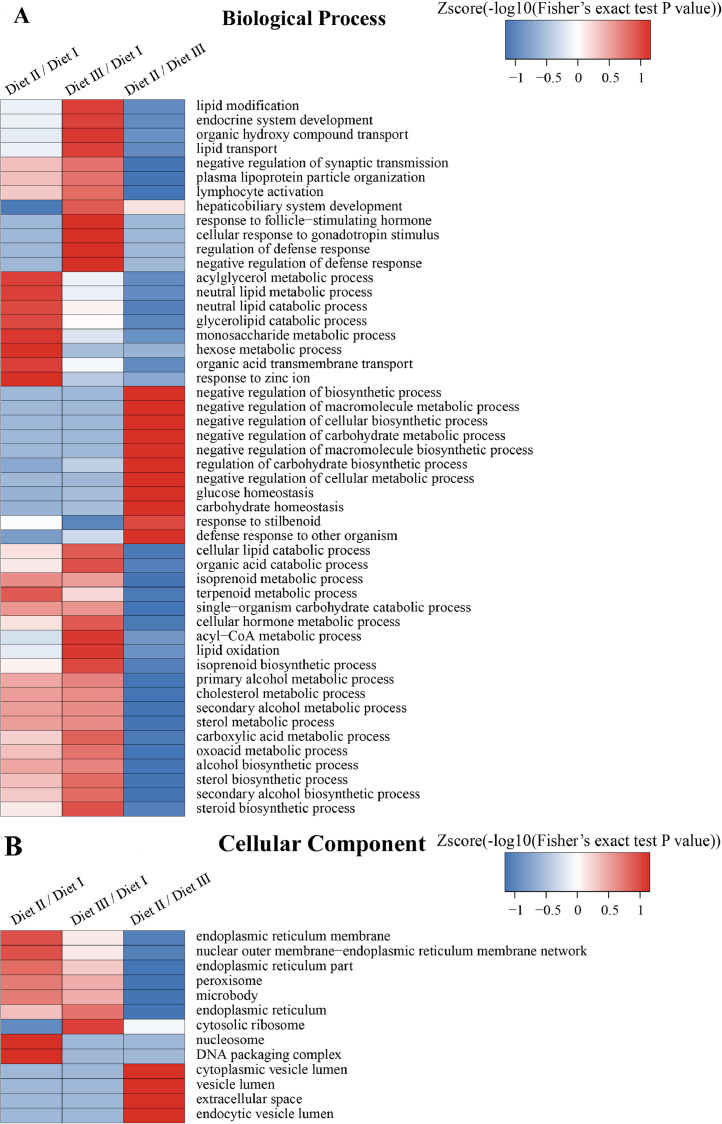
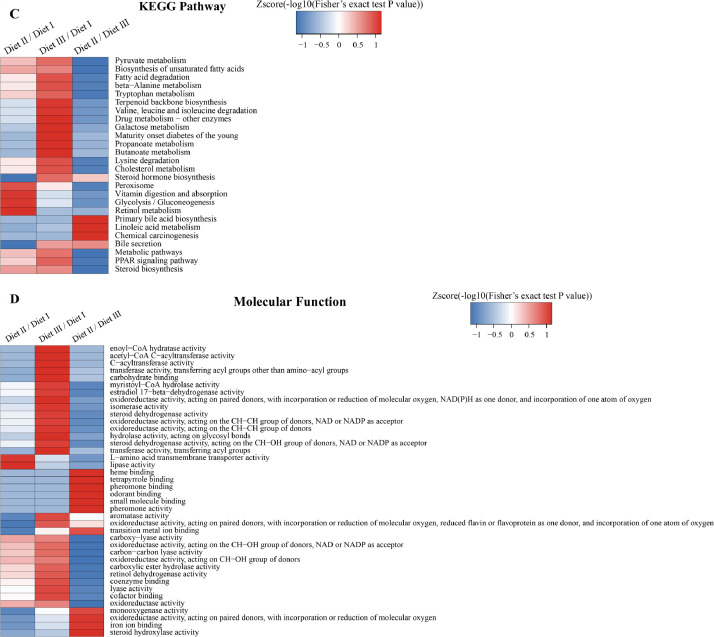
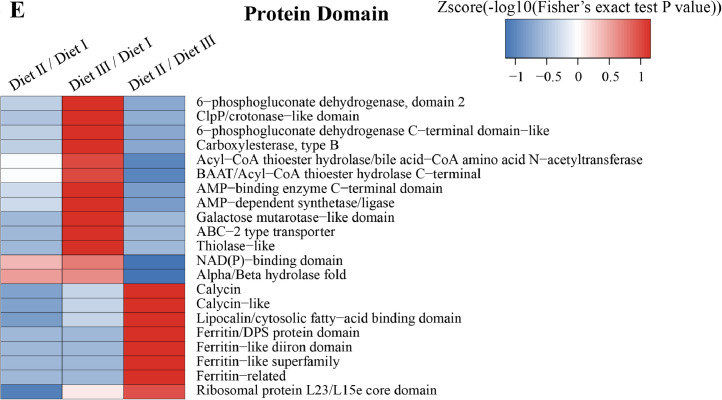
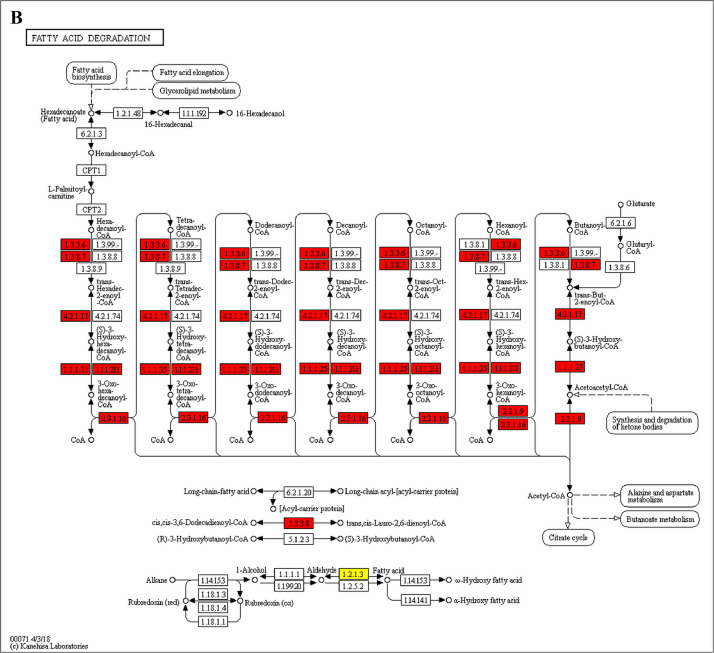
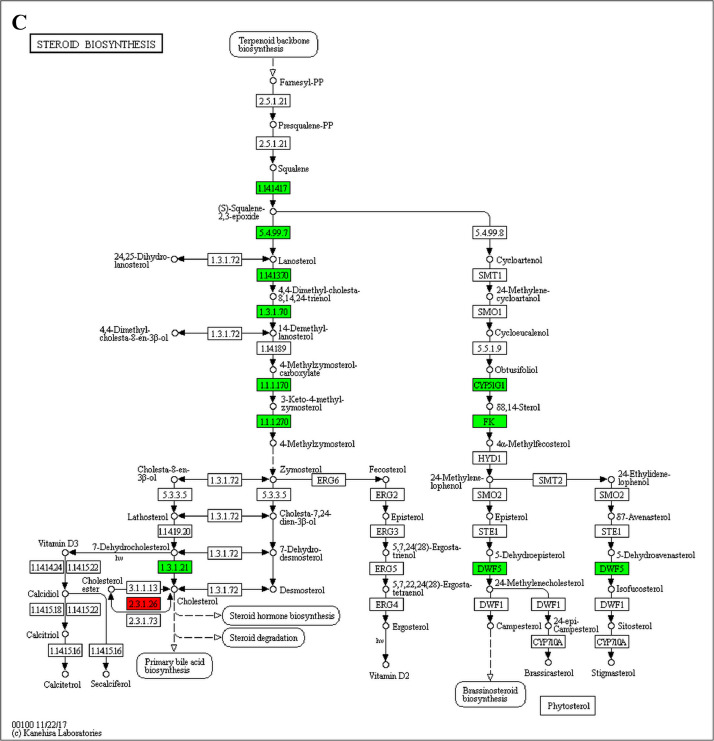
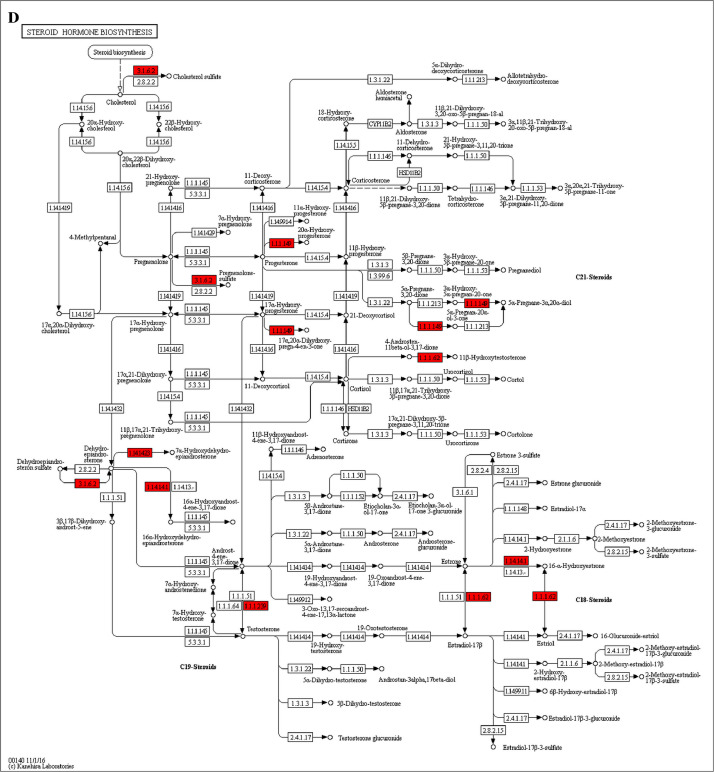
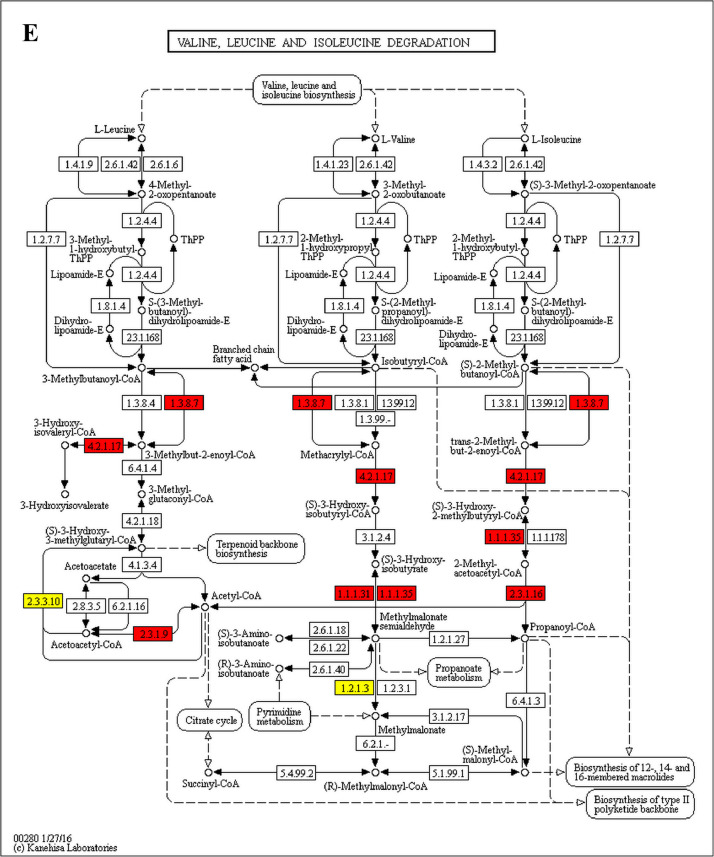
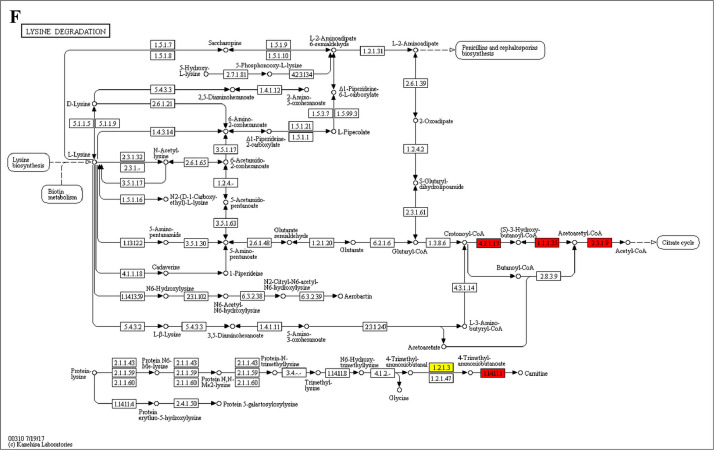
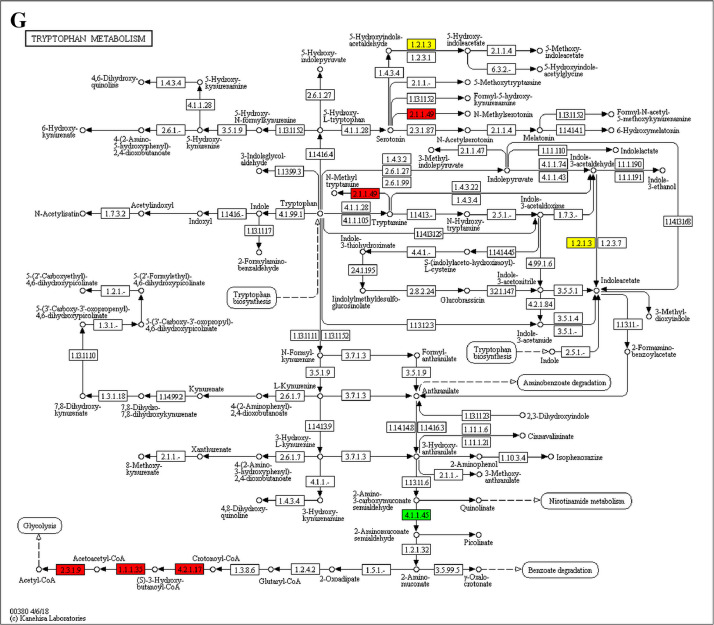
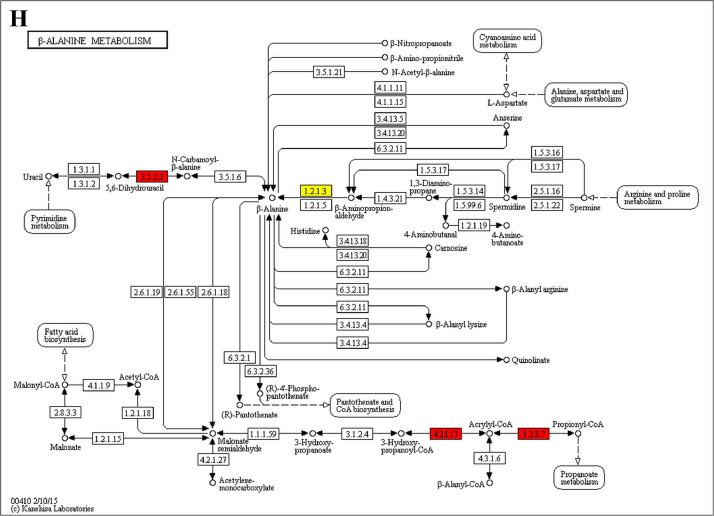
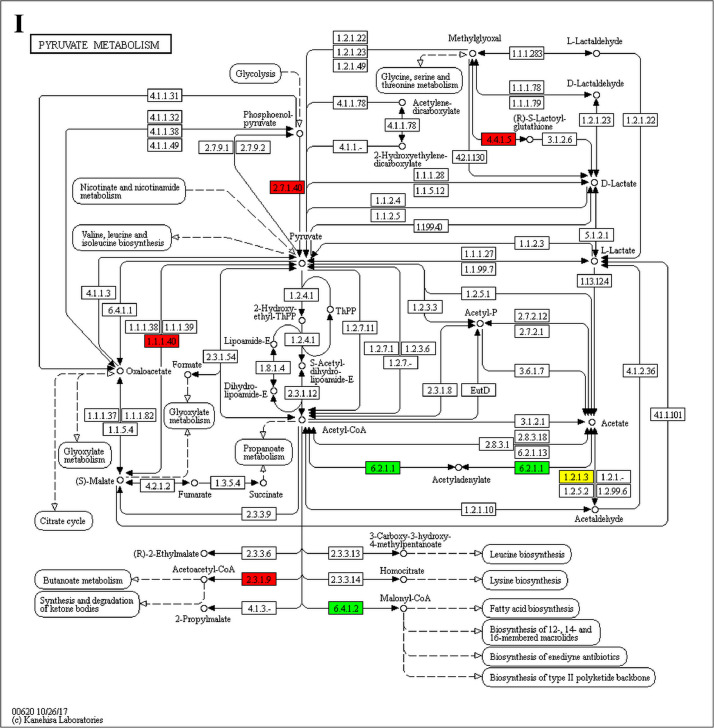
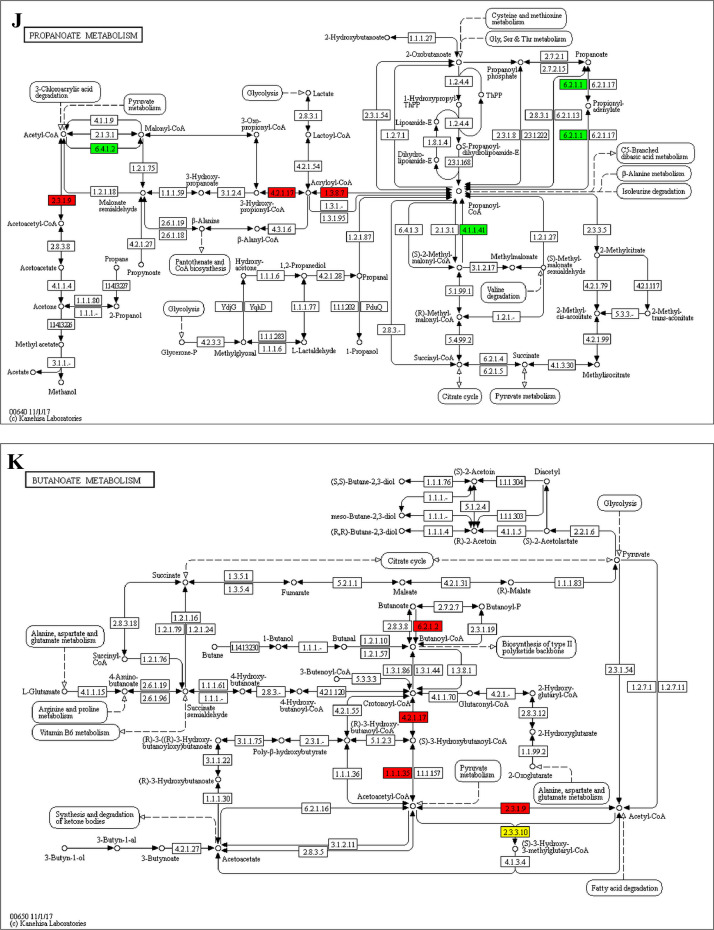
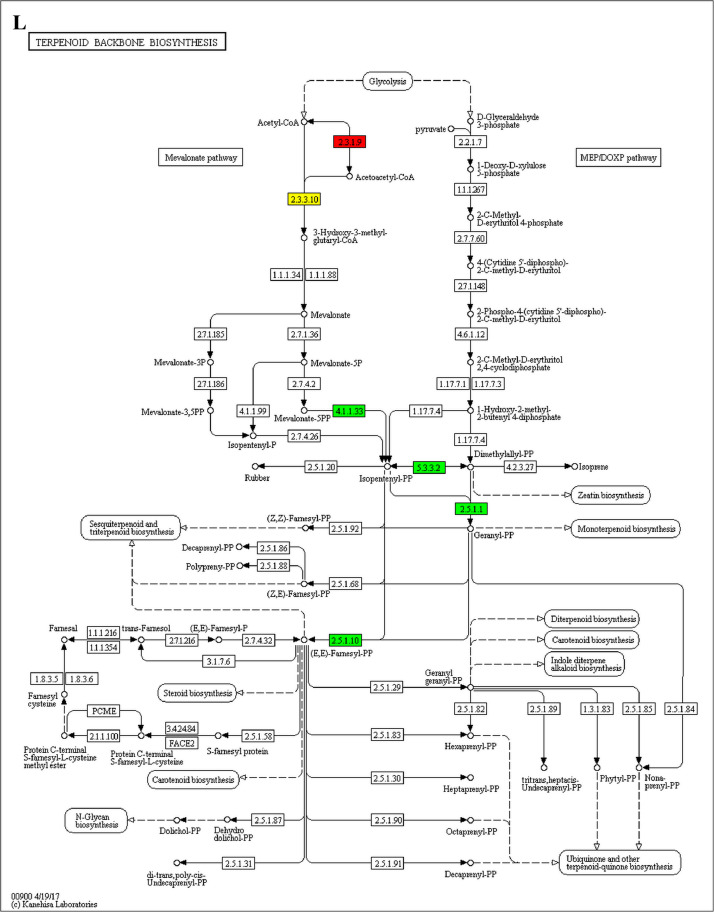
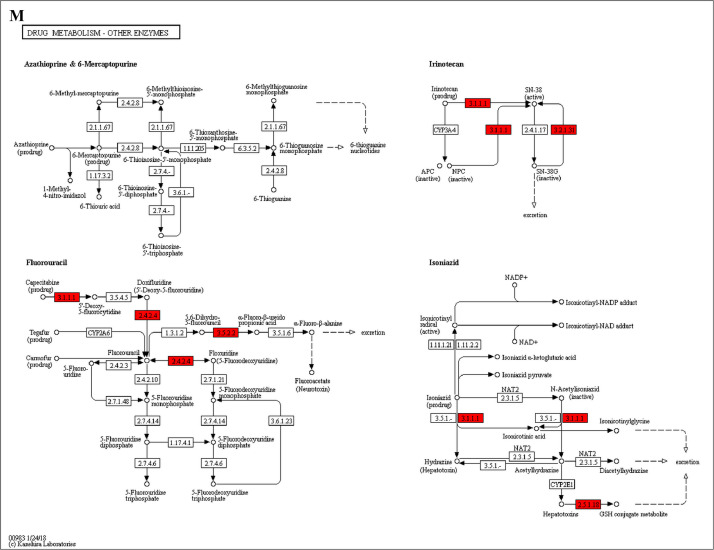
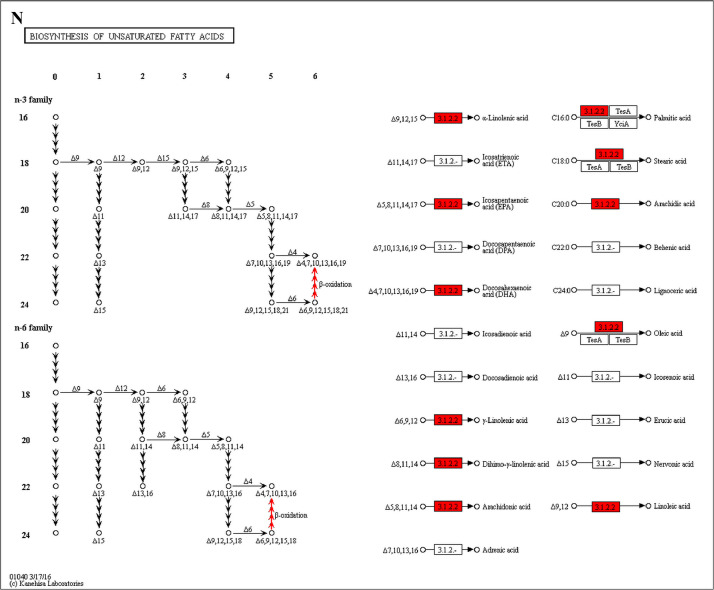
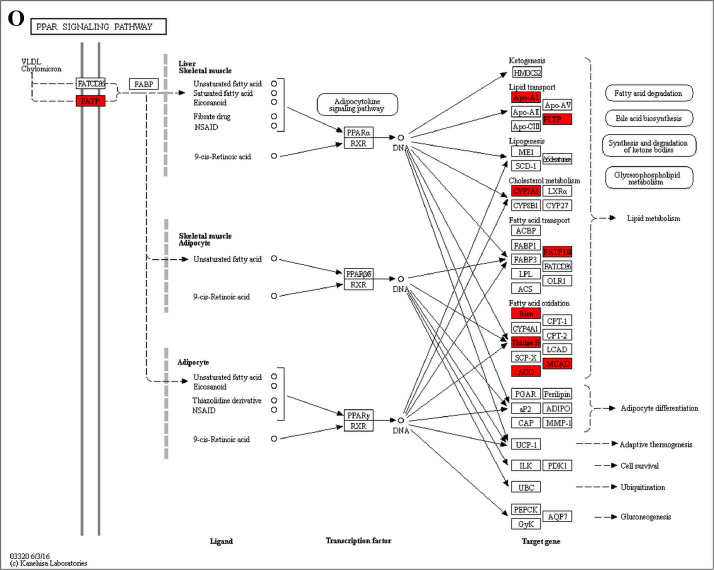
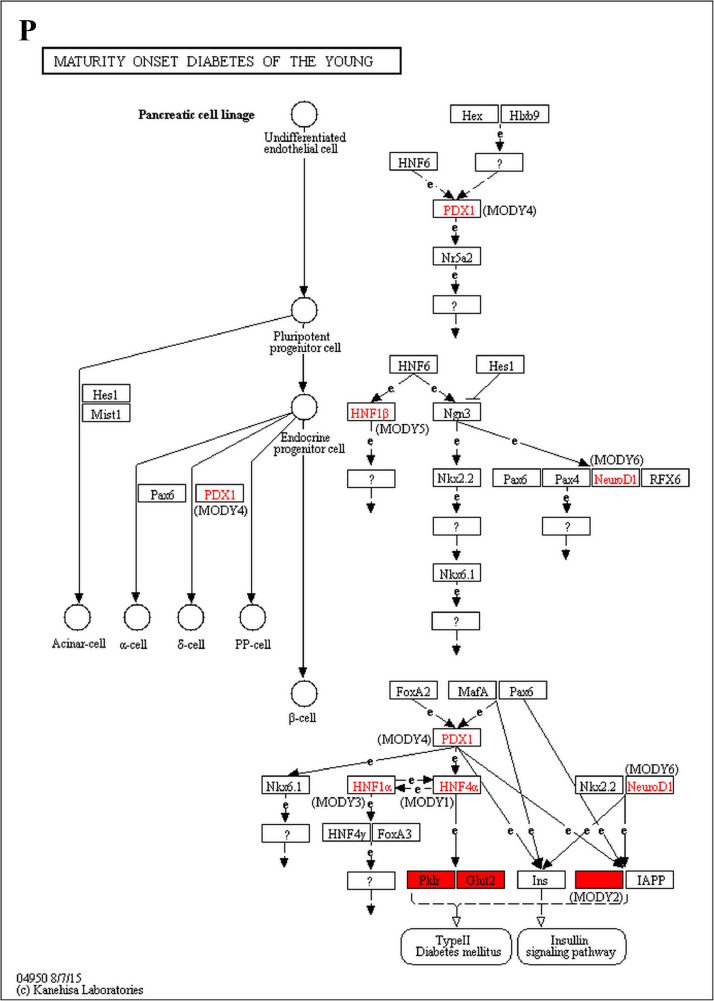
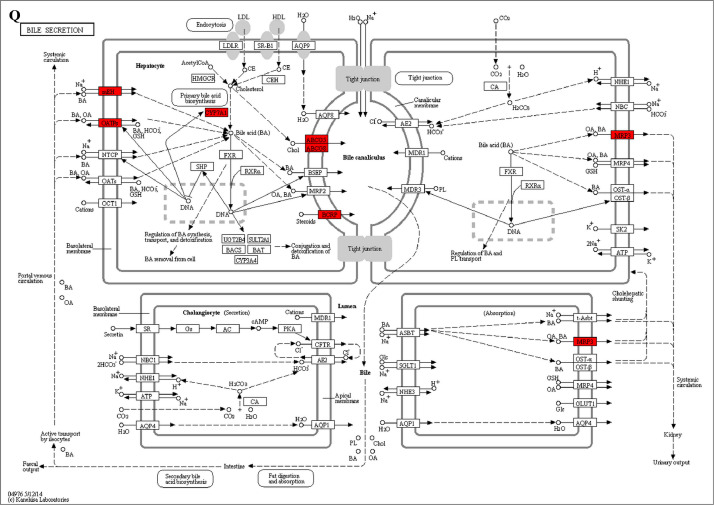
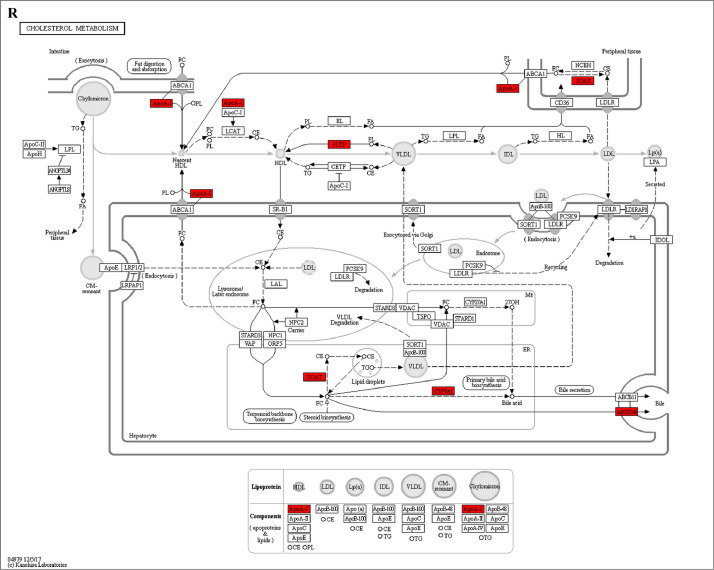
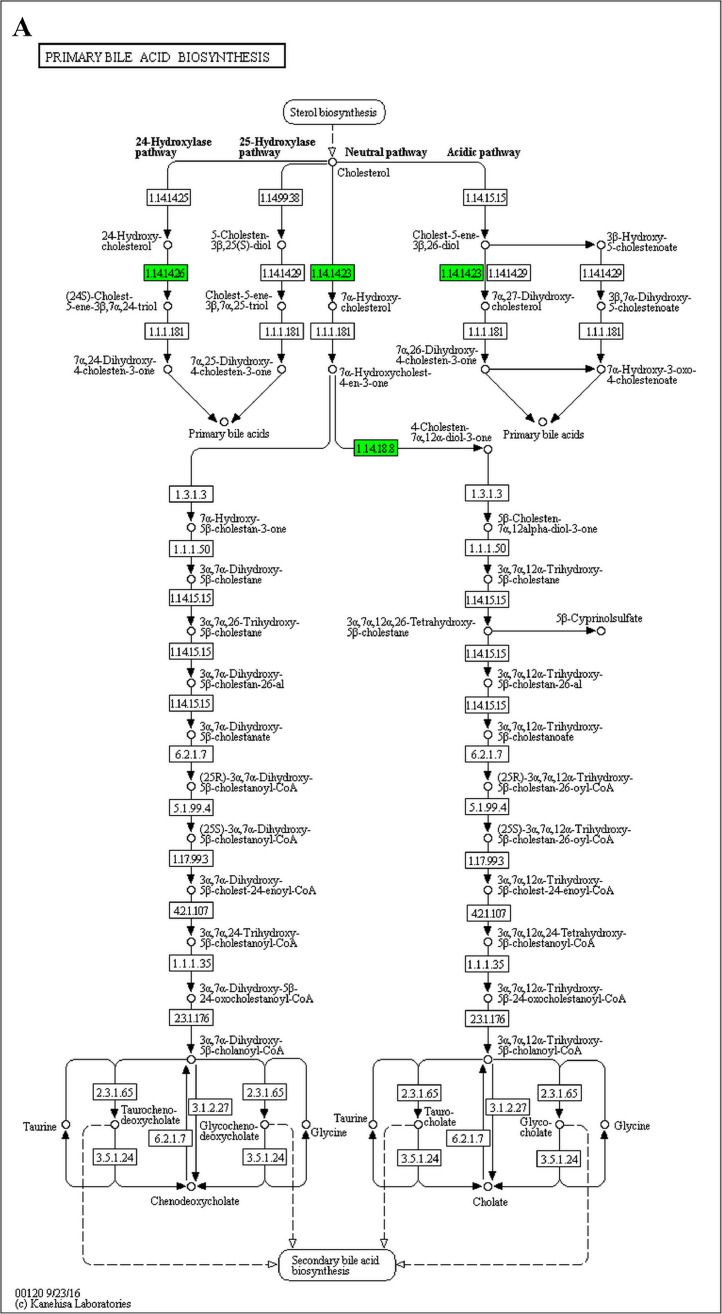
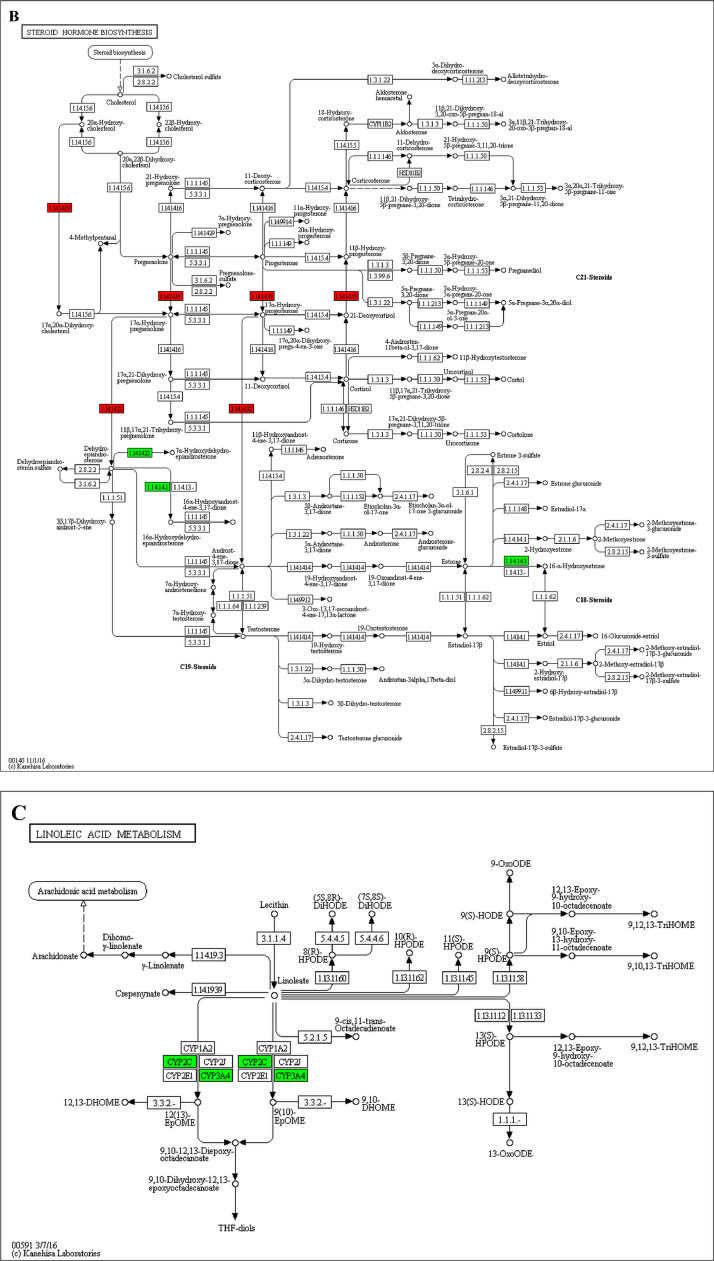
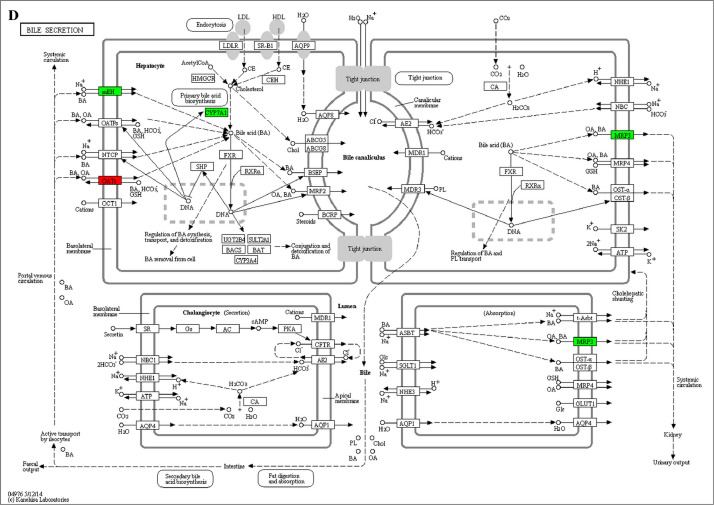
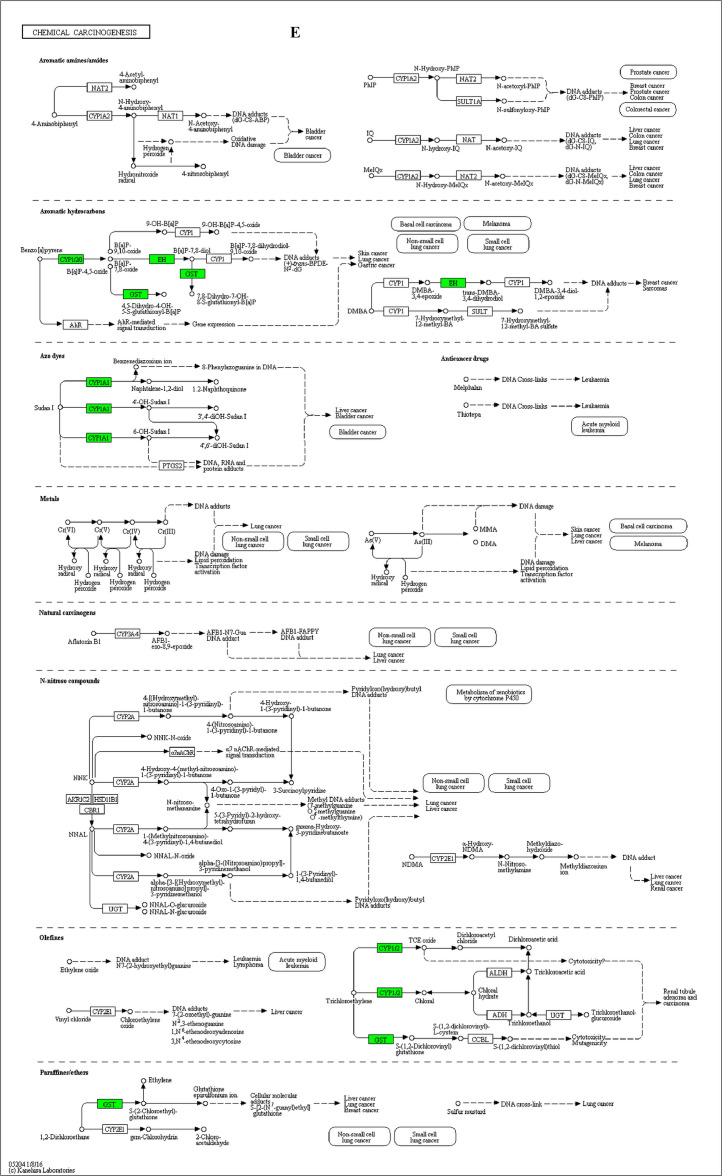
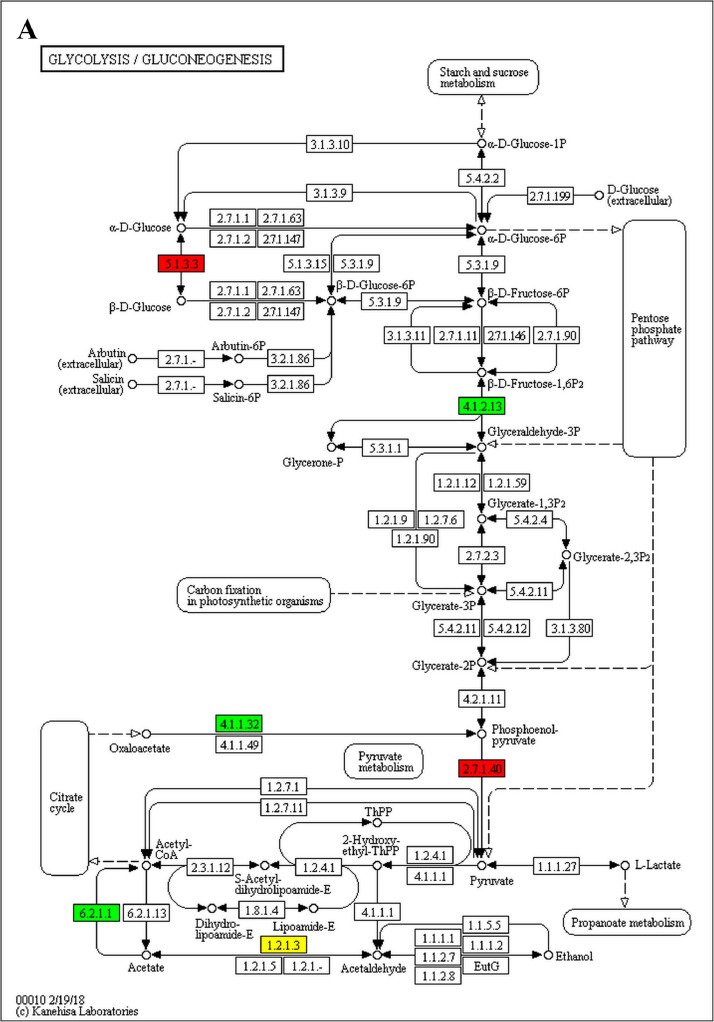
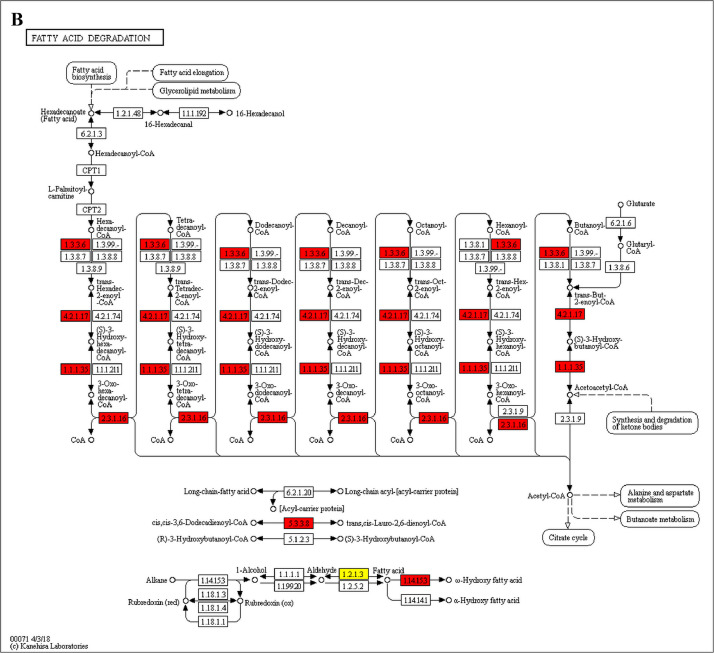
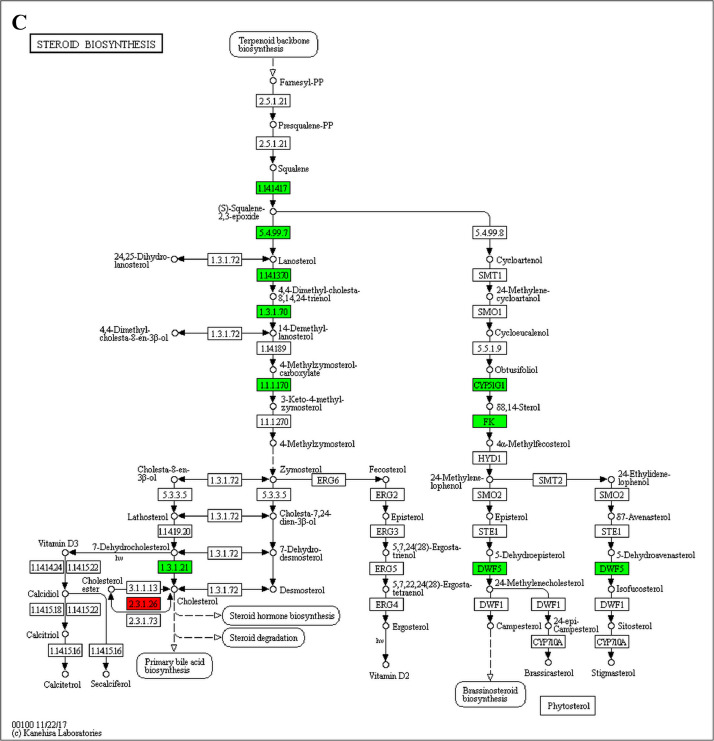
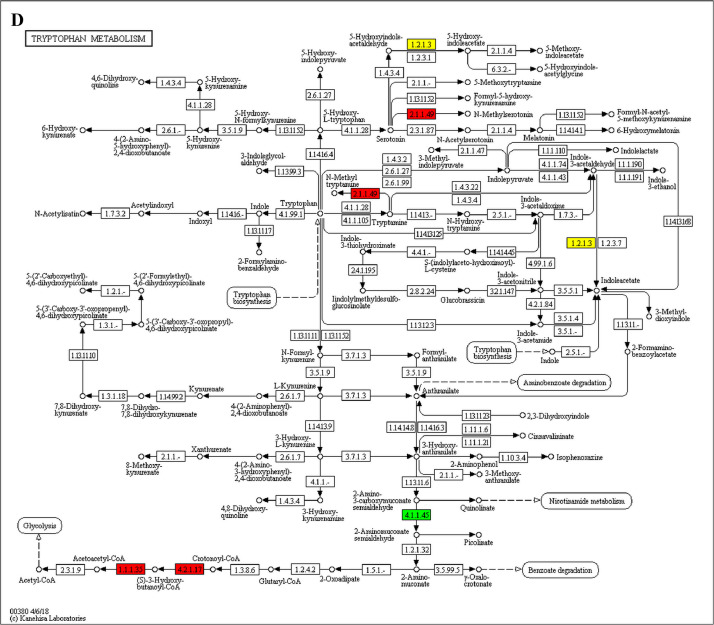
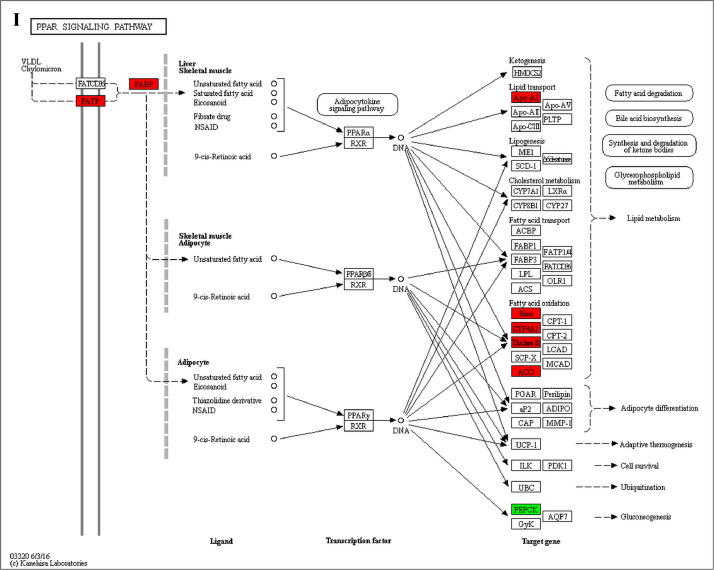
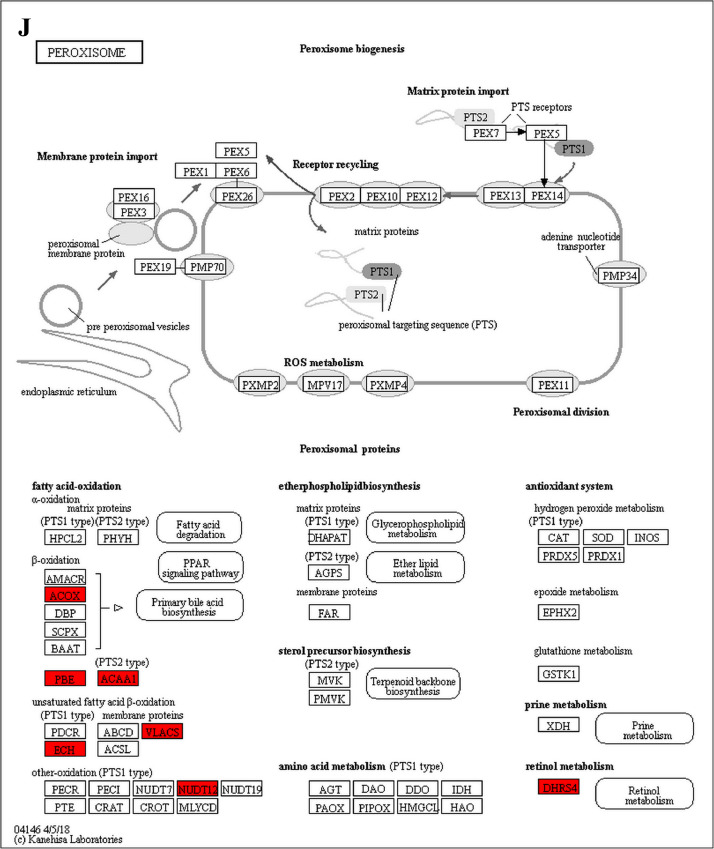
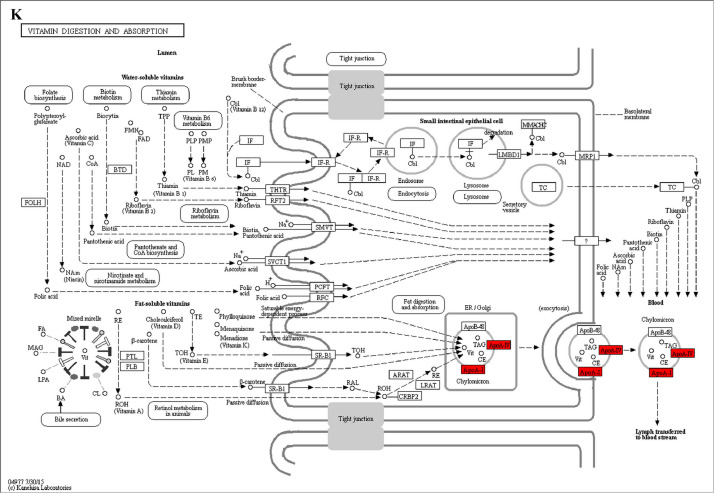
The GO enrichment and KEGG pathway analysis were used to analyze the differentially expressed proteins, which showed that many biological processes were significantly altered between the proteomics of three diet groups. The KEGG pathway enrichment-based clustering analysis were also performed in the groups of Diet II vs I, III vs I and II vs III (Fig. 1). Comparing Diet III and I groups, eighteen KEGG terms were significantly enriched (p<0.05, Fig. 2). The five KEGG terms between the groups of Diet II and III were significantly enriched (p<0.05, Fig. 3). The eleven KEGG terms between the groups of Diet II and I were significantly enriched (p<0.05, Fig. 4). The original data from KEGG pathway analysis have been uploaded in PRIDE (W8026TQ).
Fig. 1.
The KEGG pathway enrichment-based clustering analysis in the groups of Diet II vs I, III vs I and II vs III. A: Biological Process; B: Cellular Component; C: KEGG Pathway; D: Molecular Function; E: Protein Domain. The categories were at least enriched in one of protein groups with P<0.05. Filtered P matrix was transformed by the x=−log10 (P).
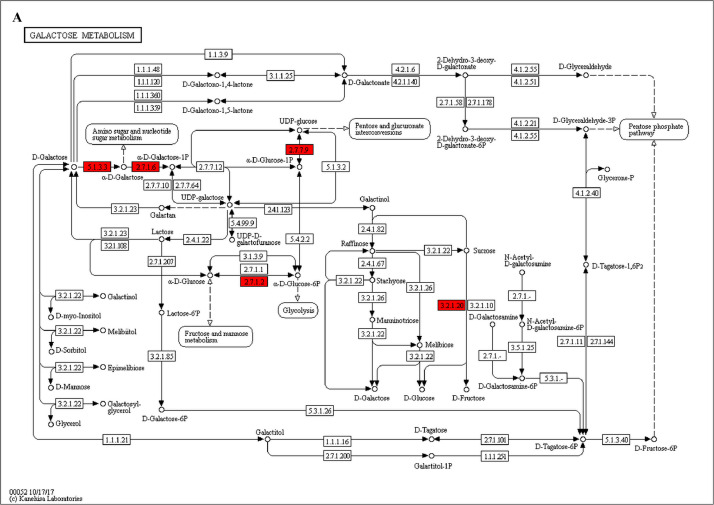
Fig. 2.
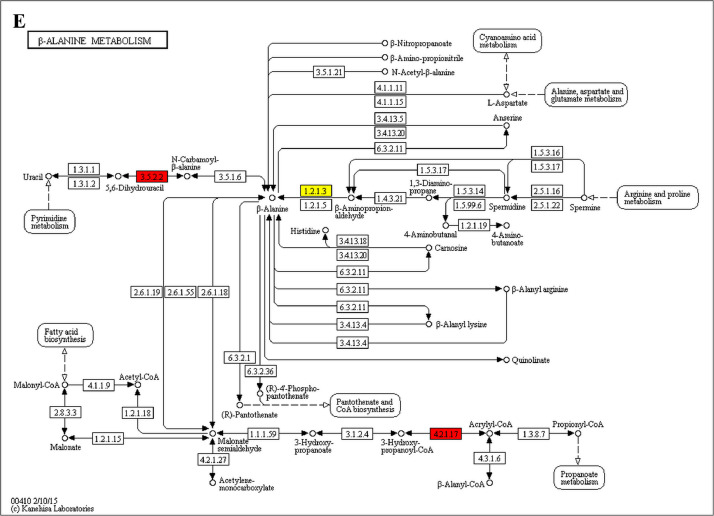
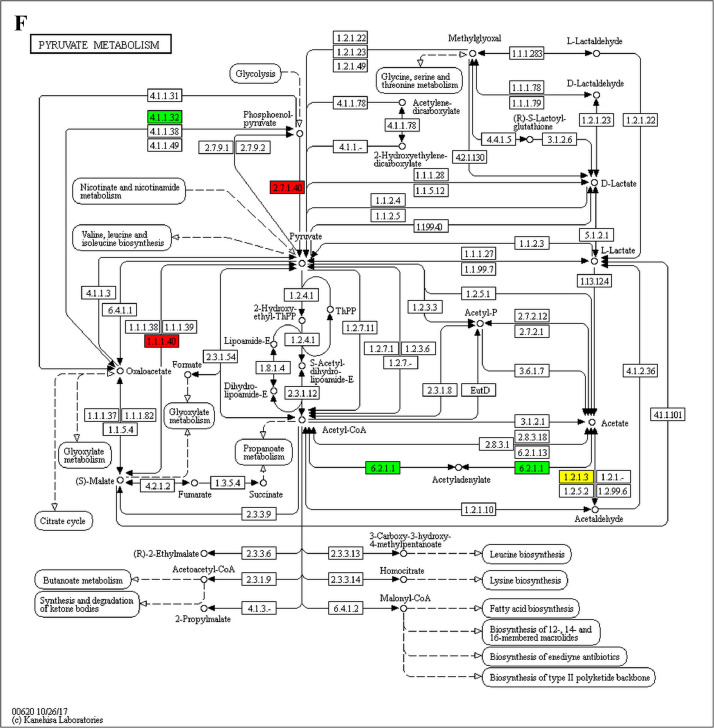
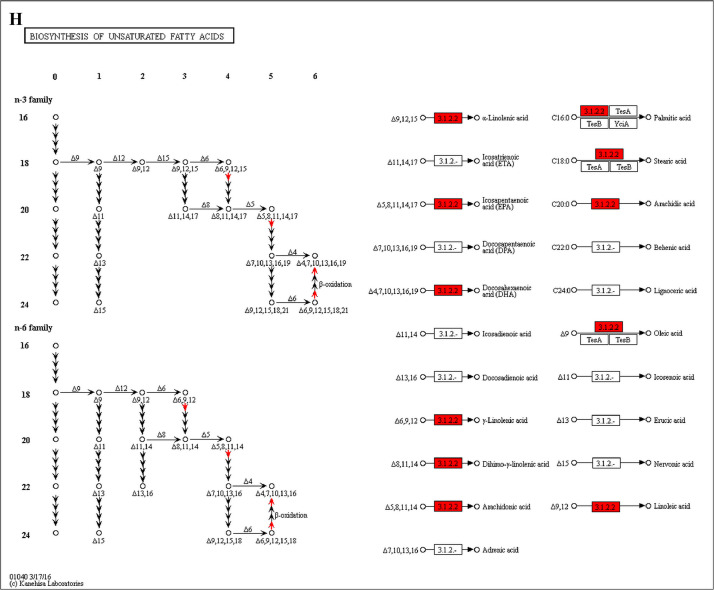
The analysis for enrichment pathway between Diet III vs I groups. A: Galactose metabolism; B: Fatty acid degradation; C: Steroid biosynthesis; D: Steroid hormone biosynthesis; E: Valine, leucine and isoleucine degradation; F: Lysine degradation; G: Tryptophan metabolism; H: β-alanine metabolism; I: Pyruvate metabolism; J: Propanoate metabolism; K: Butanoate metabolism; L: Terpenoid backbone biosynthesis; M: Drug metabolism-other enzymes; N: Biosynthesis of unsaturated fatty acids; O: PPAR signaling pathway; P: Maturity onset diabetes of the young; Q: Bile secretion; R: Cholesterol metabolism. The proteins in green were down-regulated, proteins in red were up-regulated and in yellow some proteins were up-regulated and some were down-regulated.
Fig. 3.
The analysis for enrichment pathway between Diet II vs III groups. A: Primary bile acid biosynthesis; B: Steroid hormone biosynthesis; C: Linoleic acid metabolism; D: Bile secretion; E: Chemical carcinogenesis. The proteins in green were down-regulated, proteins in red were up-regulated and in yellow some proteins were up-regulated and some were down-regulated.
Fig. 4.
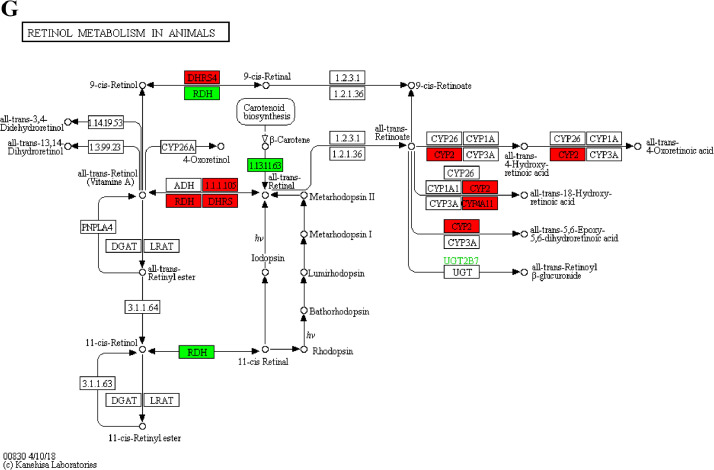
The analysis for enrichment pathway between Diet II vs I groups. A: Glycolysis/Gluconeogenesis; B: Fatty acid degradation; C: Steroid biosynthesis; D: Tryptophan metabolism; E: β-alanine metabolism; F: Pyruvate metabolism; G: Retinol metabolism in animals; H: Biosynthesis of unsaturated fatty acids; I: PPAR signaling pathway; J: Peroxisome; K: Vitamin digestion and absorption. The proteins in green were down-regulated, proteins in red were up-regulated and in yellow some proteins were up-regulated and some were down-regulated.
2. Experimental Design, Materials and Methods
2.1. The preparation of animal model and protein sample
2.1.1. Materials
Fresh FRR juice from Guinong No. 5 was prepared by automatic squeezer and preserved in a fridge (4 °C). The male Yunnan KM mice were purchased from Animal Center of Guizhou Medical University (No. SCXK Jing 2012–0001). All studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Guizhou Medical University. The mouse feeds were purchased from Xiaoshu Biotechnology Limited Company (Beijing, China). The components of normal diet were 45% corn, 20% bean, 20% wheat flour, 10% pear skin, 1% fish meal, 0.2% yeast, 1% salt, 0.58% vegetable oil, 2% bone meal, 0.2% methionine, 0.02% multivitamin. The high-fat diet were composed of 78.7% normal diet, 1% cholesterol, 0.3% cholic acid, 10% pork lard, 10% yolk
powder. All experimental chemicals were obtained from Kaixin Biotechnology Limited Company (Guizhou, China).
Thirty healthy male mice with 6 weeks were divided into three groups by average weight, and given a 12 h light/dark cycle and free access to purified water. The control mice were fed normal diet (Diet I), the model group was fed high-fat diet (Diet II), and the treatment group (Diet III) was fed 50% FRR juice based on high-fat diet daily. FRR juice was given to mice by gavage (0.25 ml/10 g). On day 30, the blood was collected by retro-orbital bleeding, finally these mice were euthanized by cervical dislocation. After mice anatomization, hepatic tissue was washed by 0.9% physiological saline, and preserved in liquid nitrogen for following proteomic analysis (Table 1).
Table 1.
The basic reagents for proteomic analysis.
| Name | Company |
|---|---|
| TMT Kit | Thermo |
| Sequencing Grade Modified Trypsin | Promega |
| EtOH (ethyl alcohol) | Fisher Chemical |
| ACN (acetonitrile) | Fisher Chemical |
| TFA (trifluoroacetic acid) | Sigma-Aldrich |
| FA (formic acid) | Fluka |
| IAA (iodoacetamide) | Sigma |
| DTT (dithiothreitol) | Sigma |
| 2-D Quant kit | GE Healthcare |
2.1.2. Protein extraction and trypsin digestion
Four volumes of lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) were added to protein powder, then sonicated three times on ice by using ultrasonic processor. After the centrifugation (12,000 g, 4 °C, 10 min), the supernatant was collected and protein concentration was determined by BCA kit. The protein solution was reduced with 10 mM DTT for 1 h at 37 °C and alkylated with 20 mM IAA for 45 min at room temperature in the darkness. Then protein sample was diluted to urea concentration less than 2 M by adding 100 mM TEAB. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion. ~100 µg protein for each sample was digested with trypsin for following experiments.
2.1.3. TMT labeling and HPLC fractionation
The peptide was desalted by Strata X C18 SPE column (Phenomenex) and vacuum-dried. Peptide was reconstituted in 0.5 M TEAB and processed for 10plex TMT kit. One unit of TMT reagent required to label 100 µg of protein were thawed and reconstituted in 24 µl ACN. Then peptide mixtures were incubated for 2 h at room temperature and pooled, desalted and vacuum-dried. The sample was fractionated into fractions by the reverse-phase HPLC using Agilent 300Extend C18 column (5 µm particles, 4.6 mm ID, 250 mm length). Peptides were separated with a gradient of 2% to 60% acetonitrile in 10 mM ammonium bicarbonate (pH 10) over 80 min into 80 fractions, then combined into 18 fractions and vacuum-dried.
2.2. Quantitative proteomic analysis by LC-MS/MS [12], [13], [14], [15]
Peptides were dissolved in 0.1% FA, directly loaded onto a reversed-phase pre-column (Acclaim PepMap 100, Thermo Scientific). Peptide separation was performed using a reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific). The gradient was comprised of solvent B (0.1% FA in 98% ACN) over 40 min at an increase from 9% to 25%, 25% to 36% in 12 min and climbing to 80% in 4 min, then holding at 80% for 4 min, all are at a constant flow rate by EASY-nLC 1000 UPLC system. The resulting peptides were analyzed by Q Exactive™ Plus hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific).
The peptides were subjected to NSI source, followed by the MS/MS in Q Exactive™ Plus (Thermo) coupled to the UPLC online. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using NCE setting as 32, ion fragments were detected in the Orbitrap at a resolution of 35,000. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans was applied for top 20 precursor ions above a threshold ion count of 1E4 in MS scan with 30 s dynamic exclusion. The applied electrospray voltage was 2.0 kV. Automatic gain control (AGC) was used to prevent the overfilling of ion trap, 5E4 ions were accumulated for generation of MS/MS spectra. For MS scans, m/z scan range was 350 to 1800. Fixed first mass was set as 100 m/z.
The resulting MS/MS data were processed using Maxquant search engine (v.1.5.2.8). MS/MS were searched against Swissprot M. musculus database. Trypsin/P was specified as cleavage enzyme allowing up to 2 missing cleavages. Mass error was set to 20 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys-was specified as fixed modification and oxidation on Met-was specified as variable modifications. For protein quantification, TMT 10plex was selected in Maxquant. FDR was adjusted to < 1% and peptide ion score was set ≥ 20. The distribution of mass error for identified peptides is near zero and most of them are less than 0.02 Da. The length of most peptides is distributed between 8 and 16, which agree with the property of tryptic peptides.
2.3. Bioinformatics methods
Gene Ontology (GO) annotation was derived from UniProt-GOA database (http://www.ebi.ac.uk/GOA/). Converting identified protein ID to UniProt ID and mapping to GO IDs by protein ID are necessary. If identified proteins were not annotated by UniProt-GOA database, InterProScan software would be used to annotate their GO function according to protein sequence alignment. Then proteins were classified into three categories by GO annotation: biological process, cellular component and molecular function.
Kyoto Encyclopedia of Genes and Genomes (KEGG) can connect known information with molecular interaction networks, such as pathways and complexes. KEGG pathways are mainly including the metabolism, genetic information processing, environmental information processing, cellular processes, rat diseases and drug development. KEGG database was used to annotate the protein pathway. KEGG online service tools KAAS is used to annotated protein's KEGG database description. Then the annotation for KEGG pathway database is mapped using KEGG mapper.
For the proteins of each category, KEGG database was used to identify enriched pathways by a two-tailed Fisher's exact test to test the enrichment of differentially expressed protein against identified proteins. Correction for multiple hypothesis testing was carried out using standard FDR. The pathway with a corrected P<0.05 was considered to be significant. These pathways were classified into hierarchical categories according to KEGG website.
Functional enrichment-based clustering for different protein groups was used to explore potential relationships between different protein groups from special protein functions (GO, Domain, Pathway). The protein groups after functional enrichment analysis were collated along with P-value, then filtered for those categories which were at least enriched in one of protein groups with P<0.05. Filtered P value matrix was transformed by the x=−log10 (P). These x values were z-transformed for each functional category, z scores were clustered by one-way hierarchical clustering. Cluster membership was visualized by a heat map using the “heatmap.2” function from “gplots” R-package.
Ethics Statement
All animal experiments comply with the ARRIVE guidelines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This work was supported by Scientific and Technological Fund in Guizhou Province of China (No. 2017–2843, 19NSP064). The author declared that no competing financial interests or personal relationships influenced the work reported in this article.
References
- 1.Lu X., Bao S. Effect of polysaccharides from Fructus Rosae Roxburghii on stress tolerance and immune function. J. GZ Univ. Tradit. Chin. Med. 2002;19:141–142. [Google Scholar]
- 2.Dai T.T., Yang X.S. Advances in chemical constituents and pharmacological activities of Rosa roxburghii. J. GY. Coll. TCM. 2015;37:93–97. doi: 10.19540/j.cnki.cjcmm.20191108.201. [DOI] [Google Scholar]
- 3.Zhang C., Liu X., Qiang H. Inhibitory effects of Rosa roxburghii tratt juice on in vitro oxidative modification of low density lipoprotein and on the macrophage growth and cellular cholesteryl ester accumulation induced by oxidized low density lipoprotein. Clin. Chim. Acta. 2001;313:37–43. doi: 10.1016/S0009-8981(01)00647-7. [DOI] [PubMed] [Google Scholar]
- 4.Xu J., Vidyarthi S.K., Bai W. Nutritional constituents, health benefits and processing of Rosa Roxburghii: a review. J. Func. Foods. 2019;60 doi: 10.1016/J.JFF.2019.103456. [DOI] [Google Scholar]
- 5.Burke D.S., Smidt C.R., Vuong L.T. Momordica cochinchinensis, Rosa roxburghii, wolfberry, and sea buckthorn-highly nutritional fruits supported by tradition and science. Curr. Top. Nutraceu. R. 2005;3:59. [Google Scholar]
- 6.Juźwiak S., Wójcicki J., Mokrzycki K., Marchlewicz M., Białecka M., Wenda-Różewicka L., Droździk M. Effect of quercetin on experimental hyperlipidemia and atherosclerosis in rabbits. Pharmacol. Rep. 2005;57:604–609. [PubMed] [Google Scholar]
- 7.Liu M.H., Zhang Q., Zhang Y.H. Chemical analysis of dietary constituents in Rosa roxburghii and Rosa sterilis fruits. Molecules. 2016;21:1204. doi: 10.3390/molecules21091204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.P.P. Song, X.C. Shen, Proteomic Analysis of Liver in Diet-induced Hyperlipidemic Mice under Fructus Rosa roxburghii Action, J. Proteomics. 2021, In Press. [DOI] [PubMed]
- 9.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic serum biomarkers for neuromuscular diseases. Expert Rev Proteomics. 2018;15:277–291. doi: 10.1080/14789450.2018.1429923. [DOI] [PubMed] [Google Scholar]
- 10.Aslam B., Basit M., Nisar M.A., Khurshid M., Rasool M.H. Proteomics: technologies and Their Applications. J Chromatogr Sci. 2017;5:182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- 11.Palma J.M., Corpas F.J., del Río L.A. Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J. Proteomics. 2011;74:1230–1243. doi: 10.1016/j.jprot.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Du C., Liang J.R., Chen D.D., Xu B., Zhuo W.H., Gao Y.H., Chen C.P., Bowler C., Zhang W. iTRAQ-based proteomic analysis of the metabolism mechanism associated with silicon response in the marine diatom Thalassiosira pseudonana. J. Proteome. Res. 2014;13:720–734. doi: 10.1021/pr400803w. [DOI] [PubMed] [Google Scholar]
- 13.Guerreiro A.C., Benevento M., Lehmann R., van Breukelen B., Post H., Giansanti P., Maarten Altelaar A.F., Axmann I.M., Heck A.J. Daily rhythms in the cyanobacterium synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol. Cell. Proteomics. 2014;13:2042–2055. doi: 10.1074/mcp.M113.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshishian H., Burgess M.W., Gillette M.A., Mertins P., Clauser K.R., Mani D.R., Kuhn E.W., Farrell L.A., Gerszten R.E., Carr S.A. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol.Cell. Proteomics. 2015;14:2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateos J., Landeira-Abia A., Fafian-Labora J.A., Fernandez-Pernas P., Lesende-Rodriguez I., Fernandez-Puente P., Fernandez-Moreno M., Delmiro A., Martin M.A., Blanco F.J., Arufe M.C. iTRAQ-based analysis of progerin expression reveals mitochondrial dysfunction, reactive oxygen species accumulation and altered proteostasis. Stem. Cell. Res. Ther. 2015;6:119. doi: 10.1186/s13287-015-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]