Abstract
Objective
Feature engineering is a major bottleneck in phenotyping. Properly learned medical concept embeddings (MCEs) capture the semantics of medical concepts, thus are useful for retrieving relevant medical features in phenotyping tasks. We compared the effectiveness of MCEs learned from knowledge graphs and electronic healthcare records (EHR) data in retrieving relevant medical features for phenotyping tasks.
Materials and Methods
We implemented 5 embedding methods including node2vec, singular value decomposition (SVD), LINE, skip-gram, and GloVe with 2 data sources: (1) knowledge graphs obtained from the observational medical outcomes partnership (OMOP) common data model; and (2) patient-level data obtained from the OMOP compatible electronic health records (EHR) from Columbia University Irving Medical Center (CUIMC). We used phenotypes with their relevant concepts developed and validated by the electronic medical records and genomics (eMERGE) network to evaluate the performance of learned MCEs in retrieving phenotype-relevant concepts. Hits@k% in retrieving phenotype-relevant concepts based on a single and multiple seed concept(s) was used to evaluate MCEs.
Results
Among all MCEs, MCEs learned by using node2vec with knowledge graphs showed the best performance. Of MCEs based on knowledge graphs and EHR data, MCEs learned by using node2vec with knowledge graphs and MCEs learned by using GloVe with EHR data outperforms other MCEs, respectively.
Conclusion
MCE enables scalable feature engineering tasks, thereby facilitating phenotyping. Based on current phenotyping practices, MCEs learned by using knowledge graphs constructed by hierarchical relationships among medical concepts outperformed MCEs learned by using EHR data.
Keywords: embedding, representation learning, phenotyping, knowledge graph, electronic health records
LAY SUMMARY
Phenotyping is a task of identifying a patient cohort’s underlying specific clinical characteristics and has been considered as one of important research challenges. Among steps of phenotyping, identifying phenotype-relevant medical concepts, which is called feature engineering, is critical but labor-intensive step.
Neural embedding (ie, embedding), which transforms features into low-dimensional vector representations, has widely adopted to many tasks in various domains including medical research since efficiently trained embeddings can capture complex relationships of the features leading to improve the performance of downstream tasks.
In this study, we implemented several embedding methods to obtain embeddings of medical concepts and comparatively evaluated obtained medical concept embeddings on a task of identifying phenotype-relevant concepts.
INTRODUCTION
Phenotyping is a task of identifying a patient cohort’s underlying specific clinical characteristics. With the widespread adoption of electronic health records (EHR) data, phenotyping is one of the most fundamental research challenges encountered when using the EHR data for clinical research.1 As learned from the electronic medical records and genomics (eMERGE) network, the process of developing and validating a phenotype requires a large amount of manual effort and time, typically up to 6–10 months.2,3 A phenotype typically contains thousands to tens of thousands of relevant medical concepts. For example, type 2 diabetes mellitus (T2DM) phenotype developed by eMERGE network contains about 12 000 relevant medical concepts.4,5 Thus, identifying phenotype-relevant medical concepts (eg, diagnosis, laboratory test, medication, and procedure concepts), which is called feature engineering, is an essential but often labor-intensive step in developing a phenotype. Additionally, feature engineering for rule-based phenotyping heavily relies on domain experts, can be error-prone, and is not generalizable or portable. Recently, data-driven phenotyping methods have been proposed to readily extract relevant features from external knowledge sources.6–10 Existing methods, however, often required text mining techniques that are not easy to implement, thus making it difficult to implement in real-world phenotyping tasks.
Neural embedding, which was originally invented to learn low-dimensional vector representations of words,11,12 has recently been adopted to learn the representations of medical concepts since efficiently trained embeddings can capture complex relationships of input features. Since well-trained medical concept embeddings (MCEs) can capture underlying semantics of medical concepts, MCEs have been used to improve the performance of various downstream tasks,13,14 such as patient visit prediction,15 patient outcome and risk prediction,16 and phenotyping.17
Knowledge graphs are widely used resources to learn MCEs. A knowledge graph contains medical concept nodes connected via various relationships defined from domain knowledge. Commonly used knowledge graphs include Unified Medical Language System (UMLS), Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), International Classification of Disease (ICD), and Human Phenotype Ontology (HPO). Graph embedding (ie, network embedding) methods have been leveraged to capture the connectivity patterns and structure of a knowledge graph to learn the embedding of medical concept nodes.18 For example, Agarwal et al19 learned MCEs using SNOMED CT with several graph embedding methods and achieved impressive performance in various healthcare applications, including multi-label classification and link prediction tasks.. A knowledge graph can be enriched by introducing other kinds of relationships to increase connectivity of the nodes in a knowledge graph. Shen et al20 learned the embedding of HPO concepts by enriching an HPO knowledge graph using heterogeneous vocabulary resources.
EHR data are also commonly used resources to learn MCEs. Generally, EHR data are sliced into bags-of-medical-concepts by applying different sizes of context windows (eg, visit window, 1-year window) and then embedding methods that leverage co-occurrence information, such as GloVe21 and skip-gram,12 are applied to learn MCEs. Since complex relationships among co-occurring medical concepts are captured during training, well-trained MCEs improve the performance of various downstream tasks. For example, Med2vec used EHR data to learn non-negative MCEs and showed strong performance in predicting medical codes in future visits.15
In this study, we evaluated how MCEs learned by using various data sources and embedding methods can facilitate feature engineering for phenotyping. We trained MCEs using 5 different embedding methods with 2 different data sources—knowledge graphs and patient level data obtained from EHR. Thirty-three phenotypes developed and validated by the eMERGE network2 with their corresponding medical concept lists were used as benchmark data that reflect the current phenotyping practices to evaluate different MCEs. MCEs were evaluated on retrieving phenotype-relevant concepts given seed concept(s) selected from each phenotype. Additionally, we provided a concept recommender application operating on the learned MCEs to leverage the utility of MCEs and catalyze future studies.
MATERIALS AND METHODS
Data description and processing
Medical concepts used in this study are defined by the Observational Health Data Science and Informatics (OHDSI) Observational Medical Outcomes Partnership common data model (OMOP CDM). OHDSI is a multi-stakeholder, interdisciplinary collaborative that aims to bring out the value of health data through large-scale analytics.22 The OMOP CDM harmonizes several different medical coding systems, including but not limited to ICD-9-CM, ICD-10-CM, SNOMED CT, and LOINC, to achieve standardized vocabularies while minimizing information loss, thus provides a unifying data format for various analysis pipelines.23 The standard vocabularies were built by the OMOP CDM and defined the meaning of a clinical entity uniquely across all databases and independent from the coding system. Non-standard concepts that have the equivalent meaning to the standard concept were then mapped to the standard concept. Tables and identifiers (ID) from OMOP were styled in italics (eg, concept_relationship table, visit_occurrence_id) throughout this article. In this study, we focused on condition (ie, diagnosis) concepts, which play a critical role in phenotyping.
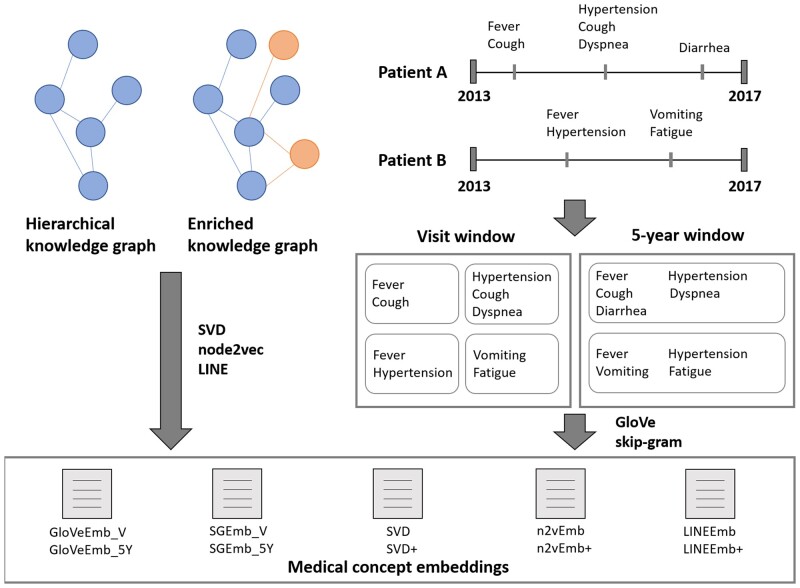
We used 2 kinds of knowledge graphs in this study, hierarchical and enriched knowledge graphs (Figure 1). The hierarchical knowledge graph was constructed by using “is-a” and “subsume” relationships between standard condition concepts obtained from the concept_relationship table in the OMOP CDM. The enriched knowledge graph expanded upon the hierarchical knowledge graph by adding non-standard condition concepts to the hierarchical knowledge graph. Those non-standard concepts were connected to the existing nodes of the hierarchical knowledge graph with additional hierarchical relationships obtained from the OMOP concept_ancestor table.23 As a result, the enriched knowledge graph has more nodes and increased connectivity in comparison with the hierarchical knowledge graph (Table 1). A subgraph of the hierarchical knowledge graph and the enriched knowledge graph are depicted in Supplementary Figure S1 as examples.
Figure 1.
The entire process to obtain medical concept embeddings (MCEs) from knowledge graphs and electronic health record (EHR) data. EHR data were sliced into the collection of bags-of-medical-concepts by applying visit windows and 5-year windows. Sliced bags-of-medical-concepts containing only a single concept were excluded. Blue nodes and edges of the knowledge graphs represent standard concepts and “is-a” or “subsume” relationships, respectively. Orange nodes and edges of the knowledge graphs represent non-standard concepts and additional hierarchical relationships that are used to enrich hierarchical knowledge graph, respectively. Processed data were used as input to obtain MCEs. SVD, node2vec, and LINE were employed to generate MCEs from the knowledge graphs. GloVe and skip-gram were used to generate MCEs from the EHR data.
Table 1.
Summary statistics of the knowledge graphs
| Hierarchical knowledge graph | Enriched knowledge graph | |
|---|---|---|
| # of unique condition concepts (medical concept nodes) | 306 266 | 312 089 |
| # of edges (relationships) | 588 298 | 671 591 |
| Avg. degree of nodes | 1.93 | 2.15 |
EHR data were obtained from the Columbia University Irving Medical Center (CUIMC) EHR database containing inpatient and outpatient data from more than 5 million patients. To ensure consistent data quality, we used the recent 5-year EHR data from January 1, 2013 to December 31, 2017.24,25 We sliced the EHR data into bags-of-medical-concepts by applying 2 different context windows: visit-window; and 5-year window (Figure 1). The visit-window was defined for each patient by distinct and unique visits and identified by visit_occurrence_id, an OMOP identifier assigned to each patient’s inpatient and outpatient visit. It is worth noting that visit window corresponding to each visit has different length since visits vary in length, generally less than a few hours. Five-year window aggregated all visits of each patient. We excluded the bags-of-medical-concepts containing only a single concept since they do not provide any meaningful co-occurrence information. Summary statistics of the sliced EHR data are provided in Table 2.
Table 2.
Summary statistics of the sliced EHR data based on visit-window and 5-year window
| EHR data sliced with visit-window | EHR data sliced with 5-year window | |
|---|---|---|
| # of patients | 1 292 369 | 1 370 787 |
| # of bags-of-medical-concepts | 8 865 691 | 1 370 787 |
| # of unique concepts | 17 175 | 17 288 |
| Avg. (SD) # of concepts per bag-of-medical-concepts | 4.3 (3.3) | 12.4 (17.2) |
Learning medical concept embeddings from EHR data
GloVe
GloVe was originally developed to learn word representations by using global co-occurrence statistics of the words in an input corpus.21 Although GloVe was designed to learn word embeddings, we can apply GloVe to learn MCEs by treating medical concepts as words and sliced EHR data as the collection of bags-of-medical-concepts to calculate co-occurrence statistics. We obtained 2 sets of MCEs, GloVeEmb_V and GloVeEmb_5Y, by implementing GloVe on the global co-occurrence statistics of the EHR data sliced by visit window and 5-year window, respectively.
Skip-gram
Similar to GloVe, skip-gram was developed to learn word representations. Skip-gram learns word representations by maximizing occurrence probabilities of the context words given a target word. Context words are the words around the target word based on the pre-defined context window size, therefore skip-gram tries to make the distance between the words that appear together in the same context window closer in an embedding space.12 We can apply skip-gram to learn MCEs by slicing the EHR data by visit window and 5-year window, creating 2 sets of MCEs, SGEmb_V and SGEmb_5Y, respectively.
Learning medical concept embeddings from knowledge graphs
Singular value decomposition
Singular value decomposition (SVD) is one of the commonly used traditional matrix factorization methods, which factorizes a data matrix into a lower dimensional matrix. Two sets of MCEs were obtained by implementing SVD on the adjacency matrix of the hierarchical and enriched knowledge graphs, named SVD and SVD+, respectively.
node2vec
node2vec26 learns embeddings of the nodes in a graph using random walk. Since node2vec has 2 hyperparameters that govern breadth-first and depth-first search, the resulting node embeddings have information regarding homophily and structural equivalence of the nodes. Two sets of MCEs, n2vEmb and n2vEmb+, were obtained by implementing node2vec on the hierarchical and enriched knowledge graphs, respectively.
Large-scale information network embedding
Large-scale information network embedding (LINE) learns embedding of the nodes in a graph by approximating first-order proximity and second-order proximity of the nodes.27 The first-order proximity is the local pairwise proximity between the nodes in the graph and the second-order proximity is the context proximity among the nodes in the graph. We obtained 2 sets of MCEs, LINEEmb and LINEEmb+, by implementing LINE on the hierarchical and enriched knowledge graphs, respectively.
Implementation details
All MCEs were trained on a machine equipped with 2 × Intel Xeon Silver 4110 CPUs with 192GB RAM and using 1 Nvidia GeForce RTX 2080 TI GPU. SVD was implemented using Numpy 1.18.5. GloVe and skip-gram were implemented using Tensorflow 2.2.0.28 LINE and node2vec were implemented using OpenNE,29 a python package for graph embedding. Python 3.7.1 was used for implementation. Hyperparameter settings are provided in Supplementary Table S1. Source codes are publicly available at https://github.com/WengLab-InformaticsResearch/mcephe.
Evaluation strategy
To evaluate the performance of learned MCEs on retrieving relevant medical concepts for phenotypes, we first obtained 33 available phenotyping algorithms generated and validated by the eMERGE Network and built evaluation concept set by using those phenotyping algorithms. We obtained all available phenotyping algorithms from the eMERGE Network’s Phenotype Knowledgebase30 (PheKB) as of September 2018. PheKB was created by the eMERGE Network to facilitate phenotyping and sharing of phenotyping knowledge. Phenotypes are shared in PheKB as descriptive texts, workflow charts, and code books of medical concepts. The Columbia eMERGE team converted code books of the 33 phenotyping algorithms into OMOP standard concepts, on which the evaluation concept set for the 33 phenotypes are based. In total, the evaluation concept set for the 33 phenotypes contained 20 640 unique condition concepts. A list of all 33 phenotypes and the number of concepts in the evaluation concept set for each phenotype are provided in Supplementary Table S2. We excluded the concepts related to exclusion criteria of each phenotype.
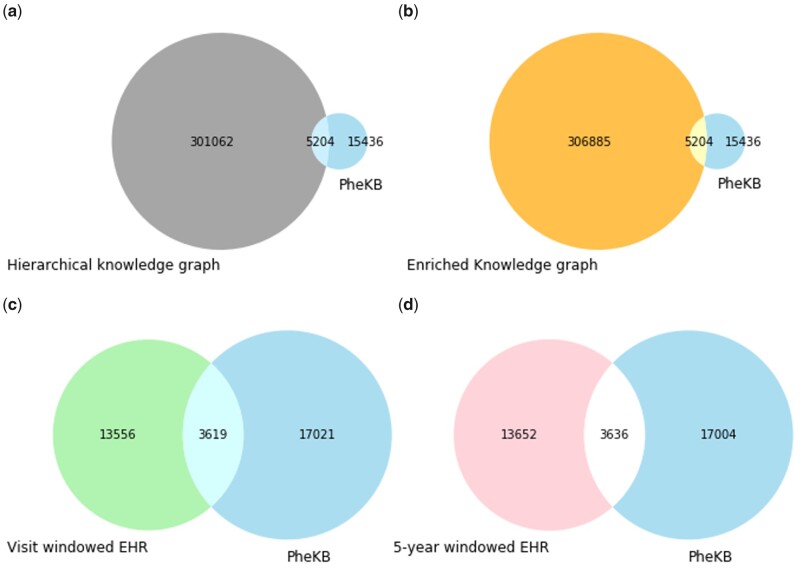
A concept must be included in both the input data (ie, knowledge graphs or EHR data) and the evaluation concept set to be trained and evaluated. Since the unique concepts included in each kind of input data are different across all 4 data sources (hierarchical knowledge graph, enriched knowledge graph, EHR data sliced by visit window, and 5-year window), we built an evaluation set for each data source for comparable evaluation. The evaluation set for each data source only contains the concepts that lies in the intersection of the evaluation concept set and the input data (Figure 2).
Figure 2.
Set diagrams between the unique concepts in the evaluation concept set of 33 phenotypes and in each data source: (A) hierarchical knowledge graph; (B) enriched knowledge graph; (C) EHR data sliced by visit window; and (D) EHR data sliced by 5-year window. The intersection of each set diagram forms the evaluation set for the MCE learned by using the corresponding data source. Since we excluded bags-of-medical-concepts that had less than 2 concepts, there are slight differences in the total number of unique concepts between the EHR data sliced by visit window and EHR data sliced by 5-year window.
In practice, feature engineering is often started by generating seed concepts or features. Inspired by this seed generation step, we quantitatively evaluated the performance of MCEs on retrieving phenotype-relevant concepts given varying numbers of seed concepts, from a single seed concept to multiple seed concepts. In addition to quantitative evaluation, we also visualized MCEs in two-dimensional space.
Evaluation based on a single seed concept
Hits@k is a commonly used metric in information retrieval to assess how well the retrieved results satisfy a user’s query intent. Given a single seed concept selected from a phenotype, each MCE retrieved the topmost candidate concepts based on the cosine similarity to the seed concept from the evaluation set. Since the number of relevant concepts in each of 33 phenotype varies, we used a modified version of Hits@k—Hits@k% as our evaluation metric to provide a more consistent comparison across different phenotypes. Specifically, Hits@k% for phenotype p based on MCE is defined as Eq (1):
| (1) |
where t is the number of unique concepts in phenotype p, is the number of relevant concepts retrieved for the embedding of the seed concept (ie, true positives), and k% is the percentage that determines the number of candidate concepts retrieved for the seed concept. The seed concept was selected by iterating over all concepts in phenotype p. The number of retrieved candidate concepts is k% of t, which is proportional to the number of unique concepts in phenotype p. Therefore, Hits@k% provides a consistent comparison across different phenotypes regardless of the number of unique concepts in the phenotypes. We reported the average Hits@k% for each MCE by averaging individual Hits@k% of all phenotypes.
Evaluation based on multiple seed concepts
We again calculated Hits@k% using multiple seed concepts selected from each of the 33 phenotypes. Given n randomly selected seed concepts from a phenotype, we generated a “conceptual” single seed embedding by summing the embeddings of n seed concepts. Each MCE then retrieved the topmost candidate concepts based on the cosine similarity to the conceptual seed embedding. Retrieval for the evaluation was repeated t times for a given phenotype, where t is the number of unique concepts in the given phenotype, yielding the same number of retrievals as in the single seed concept case. We conducted the evaluation with n = 5. Similar to the single concept seed case, Hits@k% for phenotype p based on MCE are defined as Eq (2):
| (2) |
where is the conceptual single seed embedding generated by summing the embeddings of n randomly selected concepts from phenotype p and is the number of relevant concepts retrieved for the . Average Hits@k% was reported for each MCE.
Visualization of MCEs
We visualized the embeddings of the 1221 concepts which lie in the intersection between all 4 standard evaluation sets in two-dimensional space using t-SNE.31 For clear visualization, we excluded the concepts that were included in multiple phenotypes, resulting in 26 phenotypes.
EXPERIMENT RESULTS
Overall performance
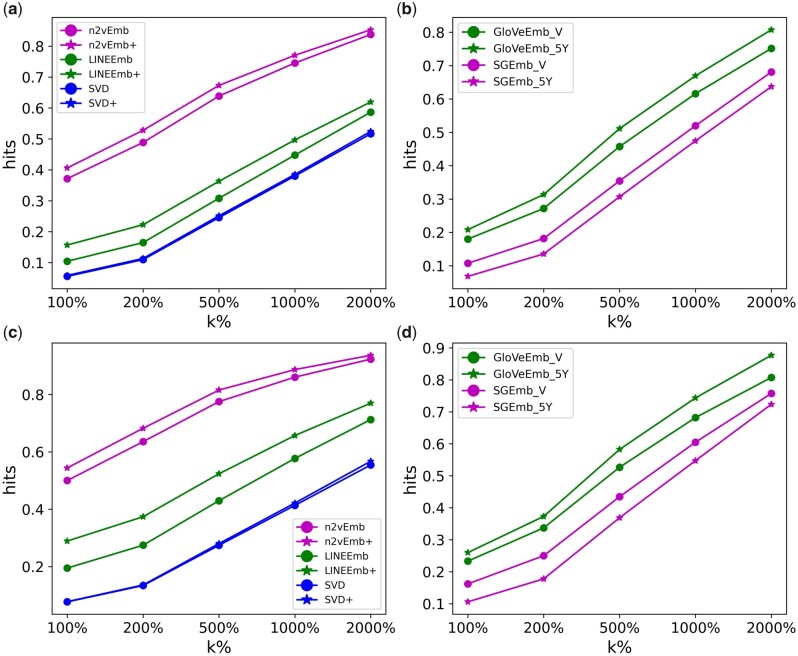
Figure 3 shows the average Hits@k% of all MCEs based on a single seed concept and 5 seed concepts. Of all MCEs based on both single seed concept and multiple seed concepts scenarios, n2vEmb+ outperformed all other MCEs. n2vEmb and n2vEmb+ outperformed others among MCEs learned by using knowledge graphs. GloVeEmb_V and GloVeEmb_5Y outperformed others among MCEs learned by using EHR data.
Figure 3.
Average Hits@k% of medical concept embedding learned from (A and C) knowledge graphs and (B and D) EHR data based on a single seed concept and 5 seed concepts, respectively.
Performance on individual phenotypes
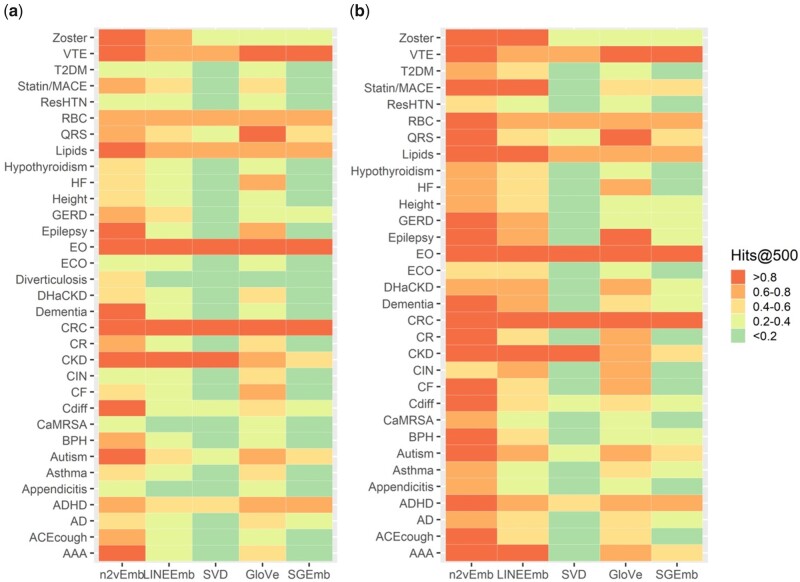
Figure 4 shows average Hits@500% of all individual phenotypes for each MCE based on a single seed concept (4A) and 5 seed concepts (4B). Hits@k% when k = 100, 200, 500, 1000, 2000 based on a single and 5 seed concept(s) are provided in Supplementary Tables S3–S22. We excluded 1 phenotype (Diverticulosis), which contains less than 10 concepts, from the evaluation based on 5 seed concepts. In both evaluations based on a single concept seed and 5 seed concepts, n2vEmb+ showed the best performance among the MCEs in more than half of the phenotypes.
Figure 4.
Average Hits@500% of all individual phenotypes based on (A) a single seed concept and (B) 5 seed concepts for MCEs. Full names for abbreviated phenotypes are provided in Supplementary Table S2.
Visualization of learned MCEs
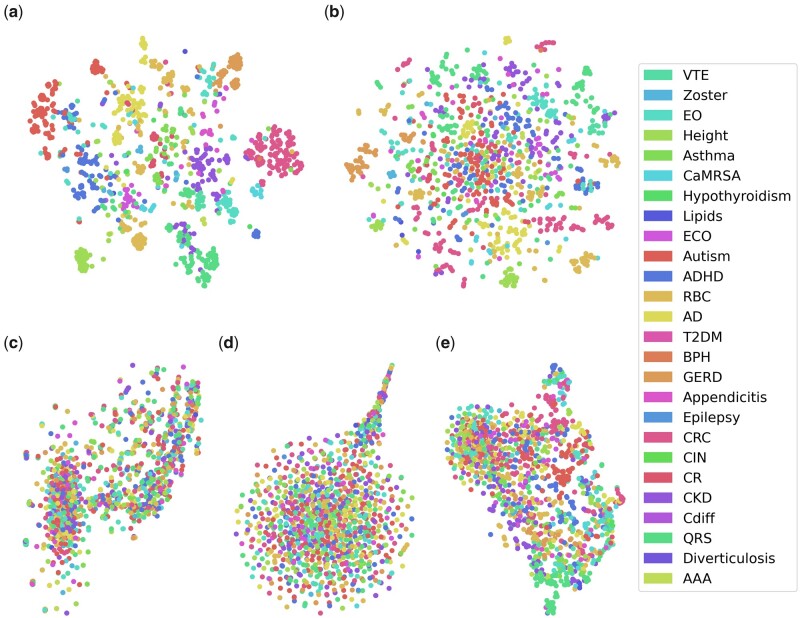
t-SNE scatterplots of the embeddings of the 1221 concepts for all MCEs are shown in Figure 5. The color of each marker represents the phenotype that the concept belongs to.
Figure 5.
t-SNE scatterplots of the 1221 concepts which lie in the intersection between the evaluation set of all MCEs, for (A) n2vEmb+, (B) LINEmb+, (C) SVD+, (D) SGEmb_5Y, and (E) GloVeEmb_5Y. Since we excluded concepts that were included in multiple phenotypes, there were only 26 phenotypes included in the scatterplots. Full names for abbreviated phenotypes are provided in Supplementary Table S2.
Hyperparameter sensitivity analysis
Besides using the hyperparameter settings suggested by the original publications of the methods to learn MCEs, we also empirically evaluated the sensitivities of the methods to hyperparameter choices. Table 3 lists important hyperparameters for each method and Figure 6 shows the average Hits@500% based on a single seed concept for each MCE in terms of different hyperparameter choices. We did not experiment with the different sizes of context window of skip-gram since the results were already shown in Figure 3.
Table 3.
Important hyperparameters for the embedding methods used in this study
| Method | Hyperparameters |
|---|---|
| GloVe |
|
| skip-gram |
|
| Singular value decomposition |
|
| node2vec |
|
| Large-scale information network embedding (LINE) |
|
Figure 6.
Average Hits@500% based on a single seed concept for MCEs trained with different hyperparameter choices. (A) Average Hits@500% based on a single seed concept with different embedding dimension for GloVeEmb_V, SGEmb_V, SVD, n2vEmb, and LINEEmb. (B) Hits@500% with different (maximum number of co-occurrences) of GloVeEmb_V. (C and D) Hits@500% with different p and q of n2vEmb, respectively. GloVeEmb_V and SGEmb_V were trained for 10 epochs.
DISCUSSION
In this study, we evaluated MCEs learned by using 5 methods with 2 different data sources on the task of retrieving phenotype-relevant medical concepts. MCEs learned by using node2vec with knowledge graphs achieved the best performance. In practice, feature engineering can be facilitated by retrieving relevant medical concepts based on the given query concepts along with well-trained MCEs. We provided a medical concept recommender application (https://github.com/WengLab-InformaticsResearch/concept-recommender) that can be used to find relevant medical concepts based on the given query concept(s) for future studies and a practical use of trained MCEs used in this study.
Among 3 different graph-based embedding methods, node2vec achieved the best performance. SVD showed the worst performance, which indicates feature engineering tasks often involve complex relationships among nodes rather than first-order proximity that can be captured by simple matrix factorization. While LINE and node2vec both consider the first- and second-order proximity to learn the embeddings of the nodes, node2vec is able to reuse the samples via a random walk strategy. Our results suggest that node2vec learns better MCEs for feature engineering tasks, indicating random walk is an important strategy to learn more efficiently from large knowledge graphs with low average degree of nodes. We admit that there are many other popular graph embedding methods in addition to the methods used in this study. Graph Convolutional Networks32 (GCN) and Graph Autoencoder33 (GAE) are graph embedding methods leveraging the power of neural networks. GCN and GAE, however, cannot be implemented on a large graph with currently available source codes. DeepWalk34 and VERSE35 are also widely used graph embedding methods. Since node2vec can be considered as a generalized version of DeepWalk and VERSE shares some similarities with node2vec and LINE depending on the choice of similarity function for learning, we did not include those 2 methods in this study.
The difference of the performance between MCEs learned from hierarchical knowledge graph and enriched knowledge graph showed that enriching a knowledge graph by introducing additional relationships connected to the existing nodes is beneficial for efficient learning of MCEs. This finding aligns with the result from Shen et al,20 where the authors obtained efficient embeddings of the concepts in HPO using an enriched knowledge graph. It is not always true, however, that enriching a knowledge graph will lead to more efficient learning of MCEs. For example, introducing a singular node that is connected to less than 1 existing node cannot improve learning since the singular node does not increase connectivity of the knowledge graph, which is crucial for learning efficient MCEs from a knowledge graph. This hinders MCEs from leveraging a knowledge graph that is built upon concepts from multiple domains but which lacks sufficient inter-domain relationships. The currently available knowledge graphs from OMOP CDM have this limitation. In contrast to the 2 148 636 and 23 435 796 relationships between condition–condition and drug–drug concept pairs, respectively, there are only 22 334 relationships between condition–drug pairs in the concept_relationship table.
Of EHR-based embedding methods, GloVe achieved the best performance. The better performance of GloVe can be explained by its ability to learn by using global co-occurrence statistics instead of using local context windows in skip-gram. Besides the methods used in this study, there are other powerful embedding methods that can be applied to learn MCEs from EHR data, such as ELMo36 and BERT,37 2 widely used state-of-the-art models to learn pre-trained word representations. Future efforts, however, will be needed to tailor those methods to learn MCEs.
Between GloVeEmb_5Y and GloVeEmb_V, GloVeEmb_5Y outperformed GloVeEmb_V. This is perhaps because phenotype-relevant condition concepts often appear across multiple visits with irregular time intervals between visits. For example, concepts related to heart failure, including initial presenting signs, symptoms, and complications, can appear in multiple visits with the progression of heart failure within a long time period. The 5-year window, which aggregates multiple visits within a 5-year time range, can capture the long-term relationships between concepts better than the visit window. The 5-year window also builds a less sparse co-occurrence matrix of the concepts than visit window, resulting in learning more efficient MCEs. Among SGEmb_5Y and SGEmb_V, however, SGEmb_V outperformed SGEmb_5Y. The disparity of the result in using GloVe and skip-gram is because skip-gram simply tries to minimize the distance between co-occurring concepts while GloVe utilizes global co-occurrence statistics to cancel out the noise from non-discriminative concepts.
The context window size to slice EHR data can be adjusted with consideration of data quality and downstream tasks. We decided to use visit and 5-year window to ensure data quality of our EHR data obtained from CUIMC.24 It is worth noting that a larger window size might introduce noise into the co-occurrence statistics for some acute diseases where intra-visit information between concepts is more important than inter-visit information. Therefore, if one aims to learn MCEs using EHR data for feature engineering of a specific phenotype, characteristics of the phenotype must be considered while selecting a context window size. For example, visit window can be used to learn MCEs for acute diseases such as clostridioides difficile and a larger context window (eg, lifetime window or 5-year window) can be used to learn MCEs for the diseases where symptoms appear in a long time period, such as heart failure and chronic kidney disease.
From Figure 3, we can confirm that the performance improved with multiple seed concepts. This is natural since more seed concepts from a specific phenotype can provide more information to find the concepts relevant to that phenotype. In practice, however, as a trade-off for the improved performance, selection of seed concepts will require more efforts from domain experts.
We can see from Figure 5 that n2vEmb+ and LINEEmb+ showed better visualized embeddings that align with phenotypes than other MCEs. This result suggests that co-occurrence information from EHR data may not be sufficient for learning interpretable MCEs that are consistent with phenotypes. It is interesting to see that other existing studies also found that simple co-occurrence information cannot learn interpretable embeddings that align with medical knowledge, although they did not use phenotyping knowledge to assess interpretability.38,39 Nevertheless, co-occurrence information from EHR data can reflect daily clinical operations, providing complementary information to ontological knowledge for phenotype development.
From Figure 6, we can see the performance of MCEs for the feature engineering task is sensitive to hyperparameter choices. All MCEs showed saturated performance between 128 and 256 embedding dimensionality. For MCEs learned by using GloVe, low resulted in better performance. For MCEs learned by using node2vec, better performance was achieved with larger p and smaller q, perhaps because the knowledge graphs used in this study have low average degree of nodes.
Several limitations of the study warrant mention. First, since benchmark data for evaluating MCEs were obtained from the phenotypes based on rule-based algorithms, our findings may not be generalized well to the phenotypes based on machine learning or data-driven approaches. Second, considering that concepts from domains other than the condition domain (eg, drug and procedure) are often involved in phenotype development, future efforts will be required to expand the study using concepts from multiple domains. Finally, we admit that besides the embedding methods investigated in this study, there are other more powerful embedding methods. Although this study focused on evaluation of MCEs learned by using knowledge graphs and EHR data, our evaluation framework can be applied to a wide range of MCEs learned by using diverse data sources. Therefore, future works will include more extensive evaluation of MCEs learned by using more diverse data sources and embedding methods.
Analysis of false positive concepts
The above evaluation assessed the accuracy of recommended concepts from the perspective of using the recommended concepts in conventional rule-based phenotyping, which create inclusion and exclusion criteria based on medical concepts that are directly related to the phenotype. Data-driven and machine learning-based phenotyping approaches might leverage other concepts that are relevant but not necessarily directly related to the phenotype from the perspective of rule-based phenotyping approach. Concepts retrieved by using MCEs that were considered false positives relative to the PheKB evaluation sets could still be useful for feature engineering in data-driven phenotyping. We thus qualitatively investigated the false positive concepts in the retrieved results (ie, retrieved as candidate concepts but not included in the evaluation concept set of a phenotype) based on n2vEmb+ and GloVeEmb_5Y for a selected phenotype—the type 2 diabetes mellitus (T2DM). T2DM was selected because it is one of the phenotypes that have been validated across multiple clinical sites in the eMERGE Network.40 We first manually selected 5 seed concepts to create the conceptual seed embedding and obtained the top 50 retrieved concepts given the seed. The false positive concepts were then scored based on the relevance to the T2DM phenotype: 1 was assigned for concepts that were considered as strongly relevant to the T2DM and can directly be included in the phenotype; 0.5 was assigned for concepts that were considered as relevant to the T2DM and can be used a covariate concept or proxy concept for developing the phenotype; and 0 was assigned for concepts that were considered as irrelevant to the T2DM. The selection of the seed concepts and scoring of the false positive concepts were conducted with help of a clinically experienced researcher. Table 4 shows the average relevant score of the false positive concepts for T2DM based on n2vEmb+ and GloVeEmb_5Y. Among 27 and 40 false positive concepts for n2vEmb+ and GloVeEmb_5Y, respectively, 67% and 50% of them were confirmed to be strongly relevant or relevant to the T2DM phenotype. This result suggests that although there were some phenotypes that showed low Hits@k%, it does not necessarily mean the MCE is not useful for feature engineering tasks in developing phenotypes.
Table 4.
Average relevance score of the false positive concepts for type 2 diabetes mellitus based on n2vEmb+ and GloVeEmb_5Y
| n2vEmb+ | GloVeEmb_5Y | |
|---|---|---|
| # of false positive concepts in the 50 retrieved concepts | 27 | 40 |
| # of false positive concepts that can be used directly or used as covariate concepts | 18 | 20 |
| Average relevance score of the false positive concepts | 0.473 | 0.275 |
CONCLUSIONS
We assessed the potential of several different MCEs for feature engineering based on current phenotyping practices. MCEs learned by using knowledge graphs outperformed MCEs learned by using EHR data in a task of retrieving phenotype-relevant concepts. We also found that enriching a knowledge graph by adding relationships to increase connectivity of the knowledge graph improves the performance of MCEs in retrieving phenotype-relevant concepts. Future works include more extensive evaluations of MCEs using concepts from multiple domains and additional embedding methods.
FUNDING
This work was supported by National Library of Medicine grants R01LM009886-11 and 1R01LM012895-03, National Human Genome Research Institute grant 2U01-HG008680-05, and National Center for Advancing Translational Science grant 1OT2TR003434-01.
AUTHOR CONTRIBUTIONS
JL and CL implemented the methods and conducted all the experiments. JHK and AB contributed to conducting experiments and evaluation of the results. NS, CP, KN, and PR contributed to generating dataset and implementing OMOP CDM for PheKB phenotypes. CT and CW co-supervised the research and edited the manuscript. All authors were involved in developing the ideas and drafting the paper.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGEMENTS
This study received institutional review board approval (AAAD1873) with a waiver for informed consent. We would like to thank eMERGE phenotyping workgroup who inspired this study.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this study cannot be shared publicly due to the patient privacy.
REFERENCES
- 1. Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH.. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shang N, Liu C, Rasmussen LV, et al. Making work visible for electronic phenotype implementation: lessons learned from the eMERGE network. J Biomed Inform 2019; 99: 103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei W-Q, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc 2012; 19 (2): 219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012; 19 (2): 212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 22 (5): 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCoy TH Jr, Yu S, Hart KL, et al. High throughput phenotyping for dimensional psychopathology in electronic health records. Biol Psychiatry 2018; 83 (12): 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gronsbell J, Minnier J, Yu S, Liao K, Cai T.. Automated feature selection of predictors in electronic medical records data. Biometrics 2019; 75 (1): 268–77. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26 (11): 1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bengio Y, Ducharme R, Vincent P, Janvin C.. A neural probabilistic language model. J Mach Learn Res 2003; 3: 1137–55. [Google Scholar]
- 12. Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: advances in neural information processing systems; arXiv preprint arXiv:1310.4546; 2013.
- 13. Xiao C, Choi E, Sun J.. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc 2018; 25 (10): 1419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weng W-H, Szolovits P. Representation learning for electronic health records. arXiv preprint, arXiv:1909.09248; 2019.
- 15. Choi E, Bahadori MT, Searles E, et al. Multi-layer representation learning for medical concepts. In: proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining; San Francisco, CA; Aug 13–17, 2016.
- 16. Xiao C, Ma T, Dieng AB, Blei DM, Wang F.. Readmission prediction via deep contextual embedding of clinical concepts. PloS One 2018; 13 (4): e0195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miotto R, Li L, Kidd BA, Dudley JT.. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6 (1): 26094–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yue X, Wang Z, Huang J, et al. Graph embedding on biomedical networks: methods, applications and evaluations. Bioinformatics 2020; 36 (4): 1241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agarwal K, Eftimov T, Addanki R, Choudhury S, Tamang S, Rallo R. Snomed2Vec: random walk and poincaré embeddings of a clinical knowledge base for healthcare analytics. arXiv preprint arXiv:1907.08650; 2019.
- 20. Shen F, Peng S, Fan Y, et al. HPO2Vec+: leveraging heterogeneous knowledge resources to enrich node embeddings for the human phenotype ontology. J Biomed Inform 2019; 96: 103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pennington J, Socher R, Manning CD. Glove: global vectors for word representation. In: proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP); Doha, Qatar; Oct 25–29, 2014.
- 22. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 23. The Observational Health Data Sciences and Informatics (OHDSI). The Book of OHDSI; 2019.
- 24. Ta CN, Dumontier M, Hripcsak G, Tatonetti NP, Weng C.. Columbia open health data, clinical concept prevalence and co-occurrence from electronic health records. Sci Data 2018; 5: 180273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ta CN, Weng C.. Detecting systemic data quality issues in electronic health records. Stud Health Technol Inform 2019; 264: 383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grover A, Leskovec J. node2vec: scalable feature learning for networks. In: proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining; San Francisco, CA; Aug 13–17, 2016. [DOI] [PMC free article] [PubMed]
- 27. Tang J, Qu M, Wang M, Zhang M, Yan J, Mei Q. LINE: large-scale information network embedding. In: Proceedings of the 24th international conference on world wide web; Florence, Italy; May 20–22, 2015.
- 28. Abadi M, Barham P, Chen J, et al. Tensorflow: a system for large-scale machine learning. In: 12th {USENIX} symposium on operating systems design and implementation ({OSDI} 16); Savannah, GA; Nov 2–4, 2016.
- 29. OpenNE: an open source toolkit for network embedding. https://github.com/thunlp/OpenNEAccessed February, 2021.
- 30. The Phenotype Knowledgebase website. https://www.phekb.org/Accessed February, 2021.
- 31. Maaten L, Hinton G.. Visualizing data using t-SNE. J Mach Learn Res 2008; 9: 2579–605. [Google Scholar]
- 32. Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907; 2016.
- 33. Kipf TN, Welling M. Variational graph auto-encoders. arXiv preprint arXiv:1611.07308; 2016.
- 34. Perozzi B, Al-Rfou R, Skiena S. Deepwalk: online learning of social representations. In: proceedings of the 20th ACM SIGKDD international conference on knowledge discovery and data mining; New York, NY; Aug 24–27, 2014.
- 35. Tsitsulin A, Mottin D, Karras P, Müller E. Verse: versatile graph embeddings from similarity measures. In: proceedings of the 2018 world wide web conference; Lyon, France; Apr 23–27, 2018.
- 36. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. arXiv preprint arXiv:1802.05365; 2018.
- 37. Devlin J, Chang M-W, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805; 2018.
- 38. Choi E, Bahadori MT, Song L, Stewart WF, Sun J. GRAM: graph-based attention model for healthcare representation learning. In: proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining; Halifax, Canada; Aug 13–17, 2017. [DOI] [PMC free article] [PubMed]
- 39. Song L, Cheong CW, Yin K, Cheung WK, Fung BC, Poon J. Medical concept embedding with multiple ontological representations. In: IJCAI; 2019.
- 40. Hripcsak G, Shang N, Peissig PL, et al. Facilitating phenotype transfer using a common data model. J Biomed Inform 2019; 96: 103253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this study cannot be shared publicly due to the patient privacy.