Abstract
The aryl hydrocarbon receptor (AHR) is a transcription factor activated by exogenous halogenated polycyclic aromatic hydrocarbon compounds, including the environmental toxin TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin, and naturally occurring dietary and endogenous compounds. The activated AHR enhances transcription of specific genes including phase I and phase II metabolism enzymes and other targets genes such as the TCDD-inducible poly(ADP-ribose) polymerase (TiPARP). The regulation of AHR activation is a dynamic process: immediately after transcriptional activation of the AHR by TCDD, the AHR is exported from the nucleus to the cytoplasm where it is subjected to proteasomal degradation. However, the mechanisms regulating AHR degradation are not well understood. Here, we studied the role of two enzymes reported to enhance AHR breakdown: the cullin 4B (CUL4B)AHR complex, an E3 ubiquitin ligase that targets the AHR and other proteins for ubiquitination, and TiPARP, which targets proteins for ADP-ribosylation, a posttranslational modification that can increase susceptibility to degradation. Using a WT mouse embryonic fibroblast (MEF) cell line and an MEF cell line in which CUL4B has been deleted (MEFCul4b-null), we discovered that loss of CUL4B partially prevented AHR degradation after TCDD exposure, while knocking down TiPARP in MEFCul4b-null cells completely abolished AHR degradation upon TCDD treatment. Increased TCDD-activated AHR protein levels in MEFCul4b-null and MEFCul4b-null cells in which TiPARP was knocked down led to enhanced AHR transcriptional activity, indicating that CUL4B and TiPARP restrain AHR action. This study reveals a novel function of TiPARP in controlling TCDD-activated AHR nuclear export and subsequent proteasomal degradation.
Keywords: 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin); aryl hydrocarbon receptor (AHR); cullin 4B (CUL4B); cytochrome P450 (CYP450); E3 ubiquitin ligase; interferon beta 1 (Ifnb1); mouse embryonic fibroblasts (MEFs); nuclear export; protein proteasomal degradation; TCDD-inducible poly(ADP-ribose) polymerase (TiPARP, PARP7, ARTD14)
Abbreviations: +siTiPARP, silencing TiPARP by siRNA; 3pRNA, 5'-triphosphate RNA; AHR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon receptor nuclear translocator; CUL4B, cullin 4B; FBS, fetal bovine serum; Infb1, interferon beta 1; LMB, leptomycin B; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which the Cullin 4B gene had been knocked out; NES, nuclear export signal; RT-qPCR, real-time quantitative PCR; sgRNA, single-guide RNA; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TiPARP, TCDD-inducible poly(ADP-ribose) polymerase
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD, dioxin), a byproduct of incineration and other industrial processes, is a ubiquitous environmental contaminant whose toxic effects, widely observed in different species, include developmental defects, cancer, a wasting syndrome, hepatosteatosis, thymus involution, and dysregulation of immune responses (1, 2, 3, 4). TCDD is the best-known ligand of the aryl hydrocarbon receptor (AHR) (5), but other halogenated and polycyclic aromatic hydrocarbon compounds as well as naturally occurring dietary and endogenous compounds, such as tryptophan and indole metabolites, have also been identified as AHR ligands (6, 7).
AHR transcriptional activation is a dynamic process: an inactive AHR that is not bound to a ligand (either endogenous or exogenous) resides in the cytoplasm in a complex with several different proteins (i.e., heat shock protein 90, AHR-interacting protein, and AHR-activated 9 protein (8)). Upon ligand binding, the AHR moves into the nucleus where it dimerizes with the aryl hydrocarbon receptor nuclear translocator (ARNT) protein to produce an AHR/ARNT heterodimer that activates gene transcription by binding to dioxin-responsive elements, specific DNA sequences in the promoter regions of AHR-responsive genes, that is, phase I (oxidation) and phase II (conjugation) drug-metabolism enzymes (including cytochrome P450 enzymes in the CYP1A and CYP1B families (mammalian cyp1a1, cyp1a2, and cyp1b1), and many other targets including TCDD-inducible poly(ADP-ribose) polymerase (TiPARP) (also known as PARP7 and ARTD14)), and the aryl hydrocarbon receptor repressor (8, 9).
Simultaneously with activation of AHR-mediated gene transcription, the AHR begins to be degraded and AHR levels decline (10). Proteolytic degradation of transcription factors is a known mechanism for regulating signaling pathways (11), and although the phenomenon of AHR degradation has been known for many years, the process by which it occurs is not well understood. Through the late 1990s and early 2000s, a series of articles by Richard Pollenz et al. made inroads into the understanding of the mechanism, revealing that, after AHR ligand activation and translocation to the nucleus, a nuclear export signal (NES) on the N-terminal region of the AHR promotes its movement from the nucleus to the cytoplasm where the AHR is degraded via the 26S proteasome machinery (10, 12, 13). Inhibition of the ubiquitin–proteasome pathway was shown to increase the levels of the AHR–ARNT complex in the nucleus leading to ‘superinduction’ of TCDD-induced Cyp1a1 mRNA levels (14), thus indicating that AHR degradation is a mechanism for restraining the transcriptional activity of the AHR.
E3 ubiquitin ligase complexes catalyze ubiquitination of specific proteins, targeting them for proteasomal degradation and are involved in many cellular functions and biological processes (15). In 2007, Ohtake et al. (16) identified the ligand-activated AHR as an E3 ubiquitin ligase, part of an atypical cullin–RING ligase 4B complex (named CUL4BAHR), where CUL4B functions as a scaffold mediating interaction among the different components and AHR as the substrate-specific adaptor providing specificity for targeting proteins for ubiquitination (16, 17, 18). CUL4B interacts with the AHR via its N-terminal extension (16). Cullin 4A, which shares a mostly identical amino acid sequence with CUL4B but lacks the N-terminal extension (15), does not interact with the AHR (16). Other targets of the CUL4BAHR complex include the estrogen receptor (16), androgen receptor (AR) (16), β-catenin (17), peroxisome proliferator-activated receptor γ (18), and the AHR (16). This mechanism could contribute to regulate activated AHR levels. More recently, the AHR target gene TiPARP, a mono-ADP-ribosyltransferase, was shown to affect stability of the activated AHR, thus knocking out TiPARP increased AHR protein levels in both a mouse embryonic fibroblast (MEF) cell line and liver, an effect accompanied by enhanced AHR transcriptional activity by TCDD (19, 20) and supporting a role for TiPARP in regulating AHR levels and activity.
In the studies reported here, we sought to investigate the effects of loss of CUL4B and TiPARP on activated AHR degradation, cellular localization, and activity to better understand how these two factors regulate the AHR after its ligand activation. To this end, we used TCDD as a prototype ligand to activate the AHR, immortalized MEF cell line in which the Cullin 4B gene had been knocked out (MEFCul4b-null), and CRISPR/Cas9 technology to knock down TiPARP.
Results
Loss of CUL4B increases AHR protein levels and TCDD-activated AHR transcriptional activity
To study the contribution of the CUL4BAHR E3 ligase complex to AHR degradation by TCDD, we used an MEFCul4b-null. Immortalized MEFCul4b-null cell line was derived from a CUL4B KO mouse described in ref (21). We treated WT and MEFCul4b-null cells with several doses of TCDD (0.3, 1, and 10 nM) or solvent (control) for 6 h. The mean decrease of AHR levels by TCDD at 1 nM (n = 5 independent experiments with 1 or 2 replicates per treatment group) was 84% in WT cells and 65% in MEFCul4b-null cells (Fig. 1A), indicating that loss of CUL4B diminished but did not prevent AHR degradation. Figure 1A also shows that CUL4B is detected and not affected by TCDD in the WT MEF cell line while CUL4B protein is absent in MEFCul4b-null cell line. To study the changes in AHR abundance after its activation by TCDD in correlation with AHR transcriptional activity, we conducted a time course experiment in WT MEF and MEFCul4b-null cells treated with TCDD for 2, 4, 6, and 15 h and we measured both AHR levels and Cyp1a1 mRNA levels, a hallmark of TCDD–AHR activation (22, 23). Left panels in Figure 1B show that after TCDD (T) treatment, levels of the AHR decreased with time in WT MEF cells by −43% (T, 2 h), −77% (T, 4 h), −89% (T, 6 h), and −91% (T, 15 h), consistent with AHR degradation occurring once the AHR is activated by TCDD. In MEFCul4b-null cells, AHR protein diminished less after TCDD treatment with time, −7%, (T 2 h), −64% (T 4 h), −68% (T 6 h), and −77% (T 15 h), suggesting that there is less AHR degradation with the loss of CUL4B (for each cell line, percentages were calculated comparing relative densitometry units of AHR levels at each time point with AHR levels before TCDD treatment). Figure 1B, right bar graph, shows that Cyp1a1 mRNA levels were increased by TCDD also in a time-dependent manner and were higher at each time point in MEFCul4b-null cells than WT MEF cells: MEFCul4b-null versus WT cells, +4.3-fold (T 2 h), +61% (T 4 h), +2.16-fold (T 6 h), +2.21-fold (T 15 h). These data show that diminished AHR degradation with loss of CUL4B leads to increased AHR transcriptional activity. Figure 1C shows that the AHR dimerization partner, ARNT, was not decreased by TCDD treatment in agreement with a previous report by Song and Pollenz (24), nor was ARNT affected by CUL4B KO.
Figure 1.
Loss of CUL4B increases AHR protein levels and AHR transcriptional activity in MEF cells.A, WT and MEFCul4b-null cells were treated with solvent dioxane or 0.3, 1, and 10 nM TCDD for 6 h. Left panels, cell homogenates were used for Western blotting (WB) analysis with antibodies against AHR, CUL4B, and β-actin; right bar graph shows densitometry analysis of AHR protein levels from n = 5 experiments using TCDD (1 nM) with 1 or 2 replicates per treatment group for each experiment (p ≤ 0.0001, T versus C for both WT MEF and MEFCul4b-null; p = 0.0004, T (WT MEF) versus T MEFCul4b-null). B, WT MEF and MEFCul4b-null cells treated with TCDD (0.3 nM) for 0, 2, 4, 6, and 15 h were used to prepare homogenates for WB analysis with antibodies indicated in the figure (left panels) or to extract RNA and analyze Cyp1a1 mRNA levels (an index of AHR transcriptional activity) by RT-qPCR (right bar graph); WT versus MEFCul4b-null: p = 0.0024 (T 2 h), p = 0.0005 (T 4 h), p < 0.0001 (T 6 h), p < 0.0001 (T 15 h). C, WB using homogenates of WT MEF and MEFCul4b-null cells (TCDD, 10 nM) with antibodies against ARNT and β-actin. Results are representative of n = 2 independent experiments. D, representative results for WB using homogenates of WT MEF and MEFCul4b-null cells treated with TCDD (1 nM, 6 h) ± MG132 (50 μM). For bar graphs in panels A and B, one-way ANOVA was used to calculate differences among the means and Tukey's honestly significant difference (HSD) test was used as a post hoc test. For this and other figures, bar graphs represent means ± SE with experimental replicates shown as black-filled circles; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. AHR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon receptor nuclear translocator; C, control (solvent); CUL4B, cullin 4B; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which CUL4B has been knocked out; n.s., not significant; RDU, relative densitometry units; RT-qPCR, real-time quantitative PCR; T, TCDD; T, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
We next asked whether the decrease in AHR protein levels observed in MEFCul4b-null cells by TCDD was attributable to proteasomal degradation. Figure 1D shows that cotreatment with TCDD and the proteasome inhibitor MG132 not only prevented the decrease of AHR levels by TCDD in WT cells but also prevented AHR degradation in MEFCul4b-null cells, indicating that the 26S proteasome is responsible for the decrease of AHR protein levels by TCDD also in the absence of the CUL4BAHR ubiquitin ligase.
TiPARP promotes AHR protein degradation in the absence of CUL4B. Loss of both TiPARP and CUL4B completely prevented TCDD-induced AHR degradation
The AHR target gene TiPARP has been reported to promote AHR degradation via its ADP-ribosylation activity (20), and PARP-mediated ADP-ribosylation has been shown to signal for ubiquitination and proteasomal degradation of proteins (25). Thus, we asked whether TiPARP can lead to AHR degradation after TCDD treatment in the absence of CUL4B or whether CUL4B is required for TiPARP action. Figure 2A shows that silencing TiPARP by siRNA (+siTiPARP) increased AHR protein levels in both TCDD treated WT MEF and MEFCul4b-null cells compared with cells treated with TCDD + scrambled siRNA, indicating that TiPARP can promote degradation of the AHR even in the absence of CUL4B. The combination of TiPARP knockdown and loss of CUL4B in MEFCul4b-null produced higher AHR protein levels than TiPARP knockdown or loss of CUL4B alone, indicating that TiPARP and CUL4B have combined effects in promoting AHR degradation. Higher AHR protein levels after silencing TiPARP in TCDD-treated WT MEF and MEFCul4b-null cells were accompanied by increased transcriptional activity of the AHR, as shown by increased mRNA levels of the AhR target gene Cyp1a1 (Fig. 2B, left bar graph).
Figure 2.
TiPARP leads to TCDD-activated AHR degradation in the absence of CUL4B.A, WB using homogenates of WT MEF and MEFCul4b-null cells transfected with dsRNAs targeting mouse TiPARP (+siTiPARP) or nontargeted control dsRNAs (+scr) and treated with solvent or TCDD (10 nM) for 6 h; antibodies against AHR and β-actin were used. B, RT-qPCR for Cyp1a1 and Tiparp using RNA extracted from WT MEF and MEFCul4b-null cells treated as in panel A (n = 3 independent experiments). C, i, scheme showing the sequence in Tiparp exon 2 targeted by single-guide RNA (sgRNA) used for the CRISPR/Cas9-mediated deletion of Tiparp in MEFCul4b-null cells to abolish TiPARP. ii, panels, WB using homogenates of WT MEF, MEFCul4b-null, and MEFCul4b-null/TiparpK.D. cells treated with solvent or TCDD (1 nM, 6 h) and antibodies for AHR and β-actin. D, RT-qPCR for Cyp1a1 and Cyp1b1 using RNA extracted from WT MEF, MEFCul4b-null, and MEFCul4b-null with TiPARP knocked down cells treated with TCDD (1 nM, 6 h) (n = 3). For bar graphs in panels B and D, one-way ANOVA was used to calculate differences among the means and Tukey's honestly significant difference (HSD) test was used as a post hoc test. +scr, scrambled siRNA; +siTiPARP, silencing TiPARP by siRNA; AHR, aryl hydrocarbon receptor; C, control (solvent); CUL4B, cullin 4B; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which the Cullin 4B gene had been knocked out; n.s., not significant; PAM, protospacer adjacent motif; RDU, relative densitometry units; RT-qPCR, real-time quantitative PCR; T, TCDD, T, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TiPARP, TCDD-inducible poly(ADP-ribose) polymerase; WB, Western blotting.
+siTiPARP achieved only about a 40% decrease in Tiparp mRNA levels (Fig. 2B, right bar graph), thus the residual TiPARP could still account for the lower AHR protein levels in TCDD + siTiPARP–treated MEFCul4b-null cells than AHR protein levels in control (scrambled siRNA) MEFCul4b-null cells (Fig. 2A, upper right panel). To eliminate TiPARP more completely, we used CRISPR/Cas9 technology with a single-guide RNA (sgRNA) targeting exon 2 of the TiPARP gene (Fig. 2Ci) in MEFCul4b-null cells as described in Experimental procedures. To assess the loss of TiPARP, we performed Western blot analysis using several anti-TiPARP antibodies (including several commercially available and a custom-made antibody that successfully recognized chicken TiPARP (26)), but their lack of specificity did not allow the validation of the loss of TiPARP protein by this method. Therefore, we assessed the presence of mutations in the TiPARP gene sequence by ultra-deep sequencing of PCR amplicons of the TiPARP gene region containing exon 2. The sequence analysis revealed that we obtained a 100% homozygous mutant cell line that harbors 97% of frameshift mutations leading to a premature stop codon within the ORF of the TiPARP gene (Fig. S1). WT MEF, MEFCul4b-null cells, and MEFCul4b-null cells in which TiPARP was knocked down (MEFCul4b-null/Tiparp K.D.) were treated with TCDD (1 nM) or solvent for 2 or 6 h and analyzed by Western blotting to assess TCDD effects on AHR protein levels in these three cell lines. Figure 2Cii shows that after 2-h of treatment, TCDD decreased AHR protein levels by 70% in the WT MEF cells, while decreasing the AHR by 20% in the MEFCul4b-null cells; in MEFCul4b-null/Tiparp K.D. cells, TCDD did not decrease AHR protein levels. After 6 h of treatment, the AHR protein was barely detected in TCDD-treated WT MEF cells and was decreased by about 55% in TCDD-treated MEFCul4b-null cells. Strikingly, in MEFCul4b-null/Tiparp K.D. cells, AhR degradation by TCDD was completely prevented. Higher AHR protein levels correlated with higher AHR transcriptional activity as Cyp1a1 and Cyp1b1 mRNAs were higher in MEFCul4b-null/Tiparp K.D. cells than MEFCul4b-null cells after TCDD treatment (Fig. 2D). Together, these results indicate that AHR degradation by TCDD is completely abolished in the absence of both CUL4B and TiPARP and that TiPARP promotes AHR degradation in the absence of CUL4B.
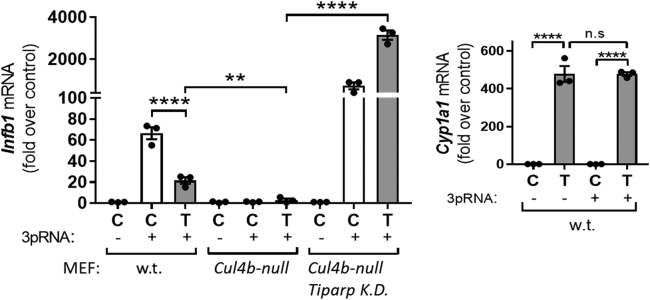
Loss of TiPARP and CUL4B affects the induction of Infb1 mRNA by a viral motif
We next asked whether the increased AHR protein levels with the loss of CUL4B and TiPARP could have biological consequences other than enhancing induction of AHR target genes. The induction of the cytokine interferon beta 1 (Infb1) is part of the first line of defense against viral infection in the host cell (27). It has been recently reported that AHR activation curtails virus-induced Infb1 mRNA, an effect mediated by TiPARP induction in the host cell (28). Thus, TiPARP was shown to ADP-ribosylate and diminish the activity of the TANK-binding kinase 1, which is a stimulator of Infb1 mRNA induction (28). Thus, we examined the effects of TCDD on the virus-induced Infb1 mRNA in WT MEF, MEFCul4b-null, and MEFCul4b-null/Tiparp K.D. cells. We transfected the three cell lines with 5'-triphosphate RNA (3pRNA, a viral motif that leads to the induction of Infb1 mRNA) followed by treatment of the cells with TCDD or solvent for 6 h. Figure 3 (left bar graph) shows that in WT cells, 3pRNA increased the induction of Infb1 mRNA levels (control + 3pRNA versus control - 3pRNA) and TCDD treatment decreased 3pRNA-induced Infb1 mRNA. Strikingly, 3pRNA did not induce Infb1 mRNA in both control and TCDD-treated MEFCul4b-null cells. Loss of both TiPARP and CUL4B (MEFCul4b-null/Tiparp K.D. cells) greatly increased the induction of Ifnb1 mRNA by 3pRNA in both control and TCDD-treated cells, consistent with a role for TiPARP in suppressing 3pRNA-induced Ifnb1 mRNA by TCDD-activated AHR. WT MEF cells transfected with 3pRNA had similar TCDD-induced Cyp1a1 mRNA compared with cells transfected with a control RNA motif (Fig. 3, right bar graph), indicating that 3pRNA did not alter TCDD-activated AHR transcriptional activity in these experimental conditions. The findings reveal that loss of CUL4B or TiPARP can have significant biological consequences as exemplified here by changes in virally induced Infb1 mRNA.
Figure 3.
Effects of loss of CUL4B and TiPARP on viral induced Infb1 mRNA.Left bar graph, RT-qPCR of Infb1 mRNA using RNA extracted from WT MEF, MEFCul4b-null, and MEFCul4b-null/TiparpK.D. transfected with 5'-triphosphate RNA (+3pRNA) or control RNA (−3pRNA) and treated with solvent or TCDD (1 nM, 24 h). Right bar graph, RT-qPCR analysis of Cyp1a1 mRNA in WT MEF cells transfected with ±3pRNA and treated with the solvent or TCDD (1 nM, 24 h). Bar graphs represent the means ± SEM (n = 3). One-way ANOVA was used to calculate differences among the means, and Tukey's honestly significant difference (HSD) test was used as a post hoc test. C, control (solvent); CUL4B, cullin 4B; Infb1, interferon beta 1; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which the Cullin 4B gene had been knocked out; n.s., not significant; RDU, relative densitometry units; RT-qPCR, real-time quantitative PCR; T, TCDD, T, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TiPARP, TCDD-inducible poly(ADP-ribose) polymerase.
Inhibition of AHR nuclear export leads to nuclear accumulation of the AHR in TCDD-treated WT cells
Davarinos and Pollenz (12) showed, using HepG2 and Hepa-1 cell lines, that the TCDD-activated AHR requires export from the nucleus to the cytoplasm to be degraded by the 26S proteasome. We assessed the effect of inhibiting protein export from the nucleus on AHR stability using WT MEF cells. We treated the cells with solvent or TCDD and different concentrations of leptomycin B (LMB), an inhibitor of chromosome maintenance region 1 (also known as exportin 1) (29), which inhibits the nuclear export of proteins including the AHR (12). After 6-h treatment, LMB led to accumulation of the AHR in the nucleus in TCDD-treated WT MEF cells in a dose-dependent manner (Fig. 4A, upper panels). LMB decreased AHR protein levels in the cytoplasm, consistent with the inhibition of AHR nuclear export (Fig. 4A, lower panels). To validate these findings, we studied the subcellular localization of the AHR by immunofluorescence in WT MEF cells treated with the solvent or TCDD (1 nM, 6 h) with or without LMB (50 nM, a dose that did not alter cell morphology). Figure 4B shows that the AHR (green signal) resides largely in the cytoplasm of control cells and was decreased by TCDD by about 80% (bar graph). The remaining AHR protein localized in the nucleus, as shown by the overlapping of the AHR signal and the nuclear stain 4′,6-diamidino-2-phenylindole (blue). Cotreatment with TCDD + LMB increased the AHR signal in the nucleus 2.4-fold, consistent with the results shown in Figure 4A and by Davarinos and Pollenz (12) who reported that inhibiting nuclear export leads to accumulation of the ligand-activated AHR in the nucleus.
Figure 4.
Inhibition of the nuclear export of the TCDD-activated AHR by LMB leads to inhibition of AHR degradation and AHR nuclear accumulation; loss of CUL4B and TiPARP leads to TCDD-activated AHR accumulation in the nucleus.A, WB using nuclear- and cytoplasmic-enriched subcellular fractions of WT MEF cells treated with the solvent or TCDD (1 nM) with or without LMB (25, 50, and 100 nM) for 6 h. Antibodies against AHR, histone 3 (H3, nuclear marker), and α/β tubulin (cytoplasmic marker) were used. B, left panels, representative images of immunofluorescence analysis in WT MEF cells treated with the solvent or TCDD (1 nM, 6 h) with or without LMB (50 nM, overnight) using an AHR antibody (FITC, green). DAPI, nuclear marker (blue). Merge, visualization of both AHR and nuclear signals. Right bar graph, means ± SEM (n = 15 cells per treatment group) of AHR fluorescence signal (green). The white scale bar represents 20 μm. C, WB using nuclear- and cytoplasmic-enriched subcellular fractions of WT MEF, MEFCul4b-null, and MEFCul4b-null/TiparpK.D. cells treated with the solvent or TCDD (1 nM, 6 h). Antibodies against AHR, histone 3 (H3, nuclear marker), and α/β tubulin (cytoplasmic marker) were used. D, left panels, representative images of immunofluorescence analysis in MEFCul4b-null/TiparpK.D. treated with the solvent or TCDD (1 nM, 6 h) with or without LMB (50 nM, overnight) using an AHR antibody (FITC, green). DAPI, nuclear marker (blue). Merge, visualization of both AHR and nuclear signals. Right bar graph, means ± SEM (n = 15 cells per treatment group) of AHR fluorescence signal (green). One-way ANOVA was used to calculate differences among the means, and Tukey's honestly significant difference (HSD) test was used as a post hoc test. The white scale bar represents 20 μm. AHR, aryl hydrocarbon receptor; C, control (solvent); CUL4B, cullin 4B; DAPI, 4′,6-diamidino-2-phenylindole; LMB, leptomycin B; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which the Cullin 4B gene had been knocked out; n.s., not significant; RDU, relative densitometry units; T, TCDD, T, 2,3,7,8-tetrachlorodibenzo-p-dioxin; WB, Western blotting.
Loss of CUL4B and TiPARP increases nuclear AHR levels
To study the roles of CUL4B and TiPARP in AHR nuclear localization, we treated MEFCul4b-null and MEFCul4b-null/Tiparp K.D. cells along with WT MEF cells with solvent (control) or TCDD (1 nM) for 6 h and prepared nuclear and cytoplasmic fractions. Figure 4C shows that AHR protein levels were increased by about 2.7-fold in the nuclear fraction of TCDD-treated MEFCul4b-null cells compared with the control MEFCul4b-null cells, and the loss of TiPARP in addition to the loss of CUL4B in MEFCul4b-null/Tiparp K.D. cells produced a greater accumulation of the AHR in the nucleus (TCDD versus C, +7.8-fold). Figure 4C shows that in the cytoplasmic fraction of TCDD-treated WT cells, the AHR was hardly detectable, whereas there was a faint band in the cytoplasmic fractions of TCDD-treated MEFCul4b-null and MEFCul4b-null/Tiparp K.D. cells (TCDD versus C, 80% decrease within each cell line). We then validated the findings obtained by Western blotting by immunofluorescence studies. Figure 4D (panels and bar graph) shows that TCDD treatment did not lead to AHR degradation in MEFCul4b-null/Tiparp K.D and that the AHR signal was detected in the nucleus (while it was detected mainly in the cytoplasm in solvent-treated cells), consistent with the Western blot results shown in Figures 2C and 4C, which show that the AHR is not degraded and accumulates in the nucleus after TCDD treatment in MEFCul4b-null/Tiparp K.D. cells. Inhibition of nuclear export by LMB treatment had no further effect on AHR levels after TCDD treatment in MEFCul4b-null/Tiparp K.D. cells, indicating that the loss of CUL4B and TiPARP is sufficient to inhibit the nuclear export of the AHR. In summary, these results show that in the absence of CUL4B and TiPARP, the AHR is not exported from the nucleus to the cytoplasm and accumulates in the nucleus.
TiPARP is a major factor promoting AHR nuclear export and degradation after TCDD treatment
To deepen the understanding of the role of TiPARP in regulating AHR nuclear export and degradation, we used WT MEF cells and CRISPR/Cas9 technology as described for Figure 2C to abolish TiPARP. Ultra-deep sequencing analysis showed that we obtained a 100% homozygous mutant cell line that harbors 99.8% of frameshift mutations leading to premature stop codons within the ORF of the TiPARP gene (Fig. S1). We treated MEFTiparp K.D. and WT MEF cells with solvent (control) or TCDD (1 nM, 6 h). Figure 5Ai shows that TCDD-treated MEFTiparp K.D. cells had higher AHR levels (about 2-fold) than TCDD-treated WT cells. Furthermore, immunofluorescence studies (Fig. 4Aii) showed that AHR protein accumulated in the nucleus of TCDD-treated MEFTiparp K.D. cells. Similar AHR levels in TCDD-treated MEFTiparp K.D. cells were obtained by Western blotting and immunofluorescence (TCDD versus C, −30%; bar graphs, Fig. 5Ai and ii). We also assessed AHR levels and localization in the absence of TiPARP by reintroducing CUL4B in MEFCul4b-null/Tiparp K.D. cells. MEFCul4b-null/Tiparp K.D. cells were transfected with a CUL4B construct (0.5 or 5 μg) and treated with the solvent or TCDD (1 nM, 6 h). Figure 5B shows that overexpressing CUL4B produced a minor decrease of AHR levels in both homogenates (left panels) and nuclear fractions (right panels) of MEFCul4b-null/Tiparp K.D. cells after TCDD treatment, even when overexpressed CUL4B (+5 μg CUL4B-HA) was about 80-fold higher than the levels of endogenous CUL4B in WT cells (Fig. 5Bi lower panels, comparing bands in lanes 2 and 4). In summary, these results show that TiPARP has a greater effect than CUL4B in promoting TCDD-activated AHR nuclear export and proteasomal degradation.
Figure 5.
TiPARP is a major factor promoting AHR nuclear export and degradation after TCDD treatment.A, i, WB using homogenates of WT MEF and MEFTiparpK.D. treated with the solvent (C) or TCDD (1 nM, 6 h). Antibodies against AHR and β-actin were used. The bar graph represents densitometry analysis of WB bands (means ± SEM). ii, representative images of immunofluorescence analysis in MEFTiparpK.D. treated with the solvent or TCDD (1 nM, 6 h) using an AHR antibody (FITC, green). DAPI, nuclear marker (blue). Merge, visualization of both AHR and nuclear signals. The bar graph represents the means ± SEM (n = 15 cells per treatment group) of AHR fluorescence signal (green). The white scale bar represents 20 μm. B, i, representative WB using homogenates of MEFCul4b-null/TiparpK.D. transfected with CUL4B-HA construct (0.5 or 5 μg) and treated with the solvent or TCDD (1 nM, 6 h); antibodies against AHR and β-actin were used. Lower panels, WB using homogenates of WT MEF, MEFCul4b-null, and MEFCul4b-null/TiparpK.D. transfected with the CUL4B-HA construct (0.5 or 5 μg) and treated as above; antibodies against CUL4B and β-actin were used. ii, WB using nuclear- and cytoplasmic-enriched fractions of MEFCul4b-null/TiparpK.D. transfected and treated as described in panel Bi. Antibodies against AHR, CUL4B, and H3 were used. AHR, aryl hydrocarbon receptor; C, control (solvent); CUL4B, cullin 4B; DAPI, 4′,6-diamidino-2-phenylindole; MEF, mouse embryonic fibroblast; MEFCul4b-null, MEF cell line in which the Cullin 4B gene had been knocked out; n.s., not significant; RDU, relative densitometry units; T, TCDD, T, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TiPARP, TCDD-inducible poly(ADP-ribose) polymerase; WB, Western blotting.
Discussion
AHR degradation, which occurs after TCDD activation, is an important event in regulating AHR signaling (10, 12, 13). Using genetically engineered MEF cell lines, we studied the roles of two factors that affect AHR protein stability: the E3 ubiquitin ligase CUL4BAHR complex and the AHR target gene, TiPARP. We report here the discoveries that are as follows: (1) CUL4B and TiPARP collaborate to promote AHR protein degradation; (2) TiPARP has a greater effect than CUL4B in promoting AHR proteasomal degradation; and (3) loss of both CUL4B and TiPARP completely prevent TCDD-induced AHR degradation. We further showed that loss of CUL4B and TiPARP can have major effects on ligand-activated AHR transcriptional activity and action.
Export of the AHR from the nucleus to the cytoplasm is required for ligand-activated AHR proteasomal degradation (10). We showed here that loss of TiPARP caused accumulation of the TCDD-activated AHR in the nucleus, indicating that TiPARP participates in the regulation of ligand-activated AHR nuclear export. The AHR contains a nuclear localization signal in the N-terminal region and NESs both in the N-terminal region and in other domains (30, 31, 32, 33). Conformational changes of the activated AHR could reveal the NES leading to AHR nuclear export (34), and posttranslational modifications of the AHR could play a role in this process. TiPARP is a member of the PARP family, enzymes which target proteins for ADP-ribosylation, leading to different biological effects (25, 35), including inhibition or promotion of protein nuclear export (36, 37). Interestingly, Gomez et al. (38) recently reported, using in vitro studies, that TiPARP can mono-ADP-ribosylate the AHR at multiple sites (aa 430–848), including sites within the AHR Q-rich domain, which has been reported to contain a motif regulating the N-terminal NES and the AHR nuclear export to the cytoplasm (39, 40). It will be of interest to validate the role of the specific TiPARP-mediated ADP-ribosylation sites in promoting AHR nuclear export.
ADP-ribosylation by PARP enzymes has been reported to signal proteins for ubiquitination and proteasomal degradation (41, 42, 43) by the recruitment of E3 ubiquitin ligases to the ADP-ribosylated site of the proteins (25). Interestingly and relevant to our findings, a recent report (44) showed that TiPARP ADP-ribosylates and targets for proteasomal degradation HIF1A and other nuclear transcriptional factors, that is, c-MYC and the estrogen receptor, by forming nuclear complexes with the E3 ubiquitin ligase HUWE1 and several other E3 ubiquitin ligases. We showed here that AHR degradation still occurs via the 26S proteasome in the absence of the E3 ubiquitin ligase CUL4B as MG132 treatment abolished AHR degradation in TCDD-treated MEFCul4b-null cells (Fig. 1D), supporting the role for another E3 ubiquitin ligase besides CUL4BAHR in AHR degradation. Further studies are needed to understand whether HUWE1 or another reported TiPARP-binding E3 ubiquitin ligase is involved in activated AHR proteasomal degradation.
We showed also that TiPARP and CUL4B have additive effects in curtailing AHR transcriptional activity as increased AHR protein levels caused by the loss of CUL4B or both CUL4B and TiPARP lead to enhanced TCDD induction of Cyp1a1 and 1b1 mRNAs (Figs. 1B and 2, B and D). These findings are consistent with previous reports that pharmacological inhibition of AHR proteasomal degradation (10) and silencing TiPARP (20, 45) led to increased ligand-activated AHR levels and AHR transcriptional activity. It will be worthwhile in the future to study whether SNPs that affect the function of TiPARP (and CUL4B) can exacerbate or protect against the effects of TCDD and other environmental toxins that activate the AHR. In vitro studies using human TiPARP constructs harboring identified human SNPs showed that TiPARP SNPs can affect AHR activation (Ahmed 2015). No known human CUL4B SNPs have been identified so far.
We also present the unexpected finding that loss of CUL4B abrogated the induction of Infb1 mRNA both in control and TCDD-treated cells (Fig. 3), suggesting that CUL4B might affect Infb1 mRNA induction by 3pRNA independently of AHR activation. These interesting findings will require further studies.
Questions regarding AHR ubiquitination, nuclear export, and consequent degradation remain. For example, Is TiPARP-mediated ADP-ribosylation of the AHR required to signal for ubiquitination as has been reported for other proteins that undergo ubiquitination after being ADP-ribosylated by PARP enzymes? (25, 44). Where does AHR ubiquitination occur? The evidence that CUL4B has been reported to be predominantly localized in the nucleus (15, 21) and that Ohtake et al. (16) identified and characterized the CUL4BAHR protein complex from nuclear extracts support the hypothesis that the AHR is ubiquitinated before being exported from the nucleus to the cytoplasm.
Furthermore, although there is some evidence that the AHR can be degraded in the nucleus in the absence of a ligand (24), other reports from Pollenz (12, 13) and our findings (Fig. 4A) showing that the nuclear export inhibitor LMB prevents TCDD-activated AHR degradation support that the AHR degradation occurs by the 26S proteasome in the cytoplasm after TCDD-activated AHR nuclear export.
In summary, we provided evidence that the CUL4BAHR complex and TiPARP have important roles in AHR protein degradation and action, showing that (1) CUL4BAHR and TiPARP have collaborative effects on AHR protein levels, (2) CUL4BAHR has a lesser effect than TiPARP on AHR degradation, and (3) TiPARP is a major factor needed for TCDD-activated AHR translocation from the nucleus to the cytoplasm and consequent proteasomal degradation.
Experimental procedures
Cells, reagents, and constructs
MEFCul4b-null cell line was originally derived from the CUL4B KO mouse described in ref (21) in the laboratory of P.Z. The WT MEF cell line was also derived from the control mouse used in those studies (21). Cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (ATCC) and 1% Penicillin-Streptomycin solution (Invitrogen). Cells were cultured at least for 24 h before the addition of treatment compounds directly in the medium (without medium change) to avoid possible activation of the AHR in control samples (46). Other reagents and their sources were as follows: TCDD (MRIGlobal Chemical Carcinogen Repository); MG132, 10 mM ready-made solution in DMSO from Sigma; LMB, 25 μg/ml solution in ethanol from Abcam; mouse HA-CUL4B vector was provided by P. Z.
RNA extraction and real-time quantitative PCR
MEF cells were seeded at 0.3 × 106 cells/well in 6-well plates (Corning Inc) containing DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin solution. The next day, cells were treated for 6 h with TCDD or dioxane at the concentration indicated in each figure. RNA STAT-60 (Tel-Test “B”) was used for total RNA extraction, following the manufacturer’s directions. For preparation of cDNA, 0.8 to 1 μg of total RNA was mixed with 4 μl of qScript cDNA SuperMix (Quanta Biosciences) and nuclease-free water to 20-μl total reaction volume. The mixture was incubated sequentially at 25 °C for 5 min, at 42 °C for 40 min, and 85 °C for 5 min. The cDNA obtained was diluted in water 1:5. Real-time quantitative PCR (RT-qPCR) was carried out in a 20-μl reaction mixture containing 2 μl of cDNA, 10 μl PerfeCTa SYBR Green FastMix (Quanta Biosciences), 1 μl forward and reverse primers (10 μM), and the remaining volume of nuclease-free water. The primers used for qPCR amplification and their corresponding annealing temperatures are shown in the Table S1.
siRNA-mediated gene silencing
WT MEF or MEFCul4b-null cells were seeded at cell densities of 0.5 × 106 cells/well in 6-well plates. Cells were transfected the following day with 1.25-μg siGENOME Mouse TiPARP siRNA (M-060174-01-0005, Dharmacon) or 1.25-μg siGENOME Non-Targeting siRNA Pool #2 (D-001206-14-05, Dharmacon) in 7.5-μl Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. Twenty-four hours after transfection, cells were treated with 10 nM TCDD or the solvent dioxane (control). Six hours after treatment, cells were scraped in 1-ml RNA STAT-60 for RNA extraction and RT-qPCR analysis or in 300-μl 2× sample buffer for Western blotting analysis.
Preparation of cell lysates for SDS-PAGE/Western blotting
MEF cells were scraped in 300-μl 2× sample buffer (125 mM Tris HCl, 4% SDS, 16% glycerol, 10% β-mercaptoethanol, 0.002% bromophenol blue) and boiled for 5 min. Total protein concentrations were measured by Bio-Rad assay following the manufacturer’s instructions. For Western blotting, equal amounts of protein from each sample, up to 30 μg per lane, were separated on precast Tris-Glycine gels (Invitrogen) and transferred to nitrocellulose membranes. Primary antibodies and their dilutions were as follows: anti-AHR (BML-SA210; Enzo Life Sciences), 1:1000; anti-ARNT (sc-17811, Santa Cruz Biotechnology), 1:200; anti-CUL4B (12916-1-AP; ProteinTech), 1:1000; anti-β actin (A5441, Sigma) 1:50,000; anti-histone-3, H3 (H0164, Sigma), 1:30,000, and anti-α/β tubulin (2168, Cell signaling), 1:1000. Secondary antibodies were peroxidase-conjugated goat anti-rabbit (A6154, Sigma) or mouse IgG kappa binding protein (sc-516102S, Santa Cruz Biotechnology). Protein bands were detected with ECL Western Blotting Detection reagents (GE Healthcare). Band intensities were measured by densitometry and were normalized to β-actin, α/β tubulin, or histone 3 (H3) levels using GeneTools analysis software (Syngene).
CRISPR/Cas9–mediated suppression of TiPARP in WT MEF and MEFCul4b-null cell lines
WT MEF or MEFCul4b-null cells were resuspended in Mouse ES Cell Nucleofector Solution (Lonza Inc) at 1 × 106 cells/0.1 ml. Each 0.1-ml cell suspension was mixed with ribonucleoprotein particles consisting of 1.2 μmol of synthetic sgRNA + 150 pmol of Cas9 2NLS protein (Synthego) and 2 μg GFP plasmid (Lonza Walkersville, Inc) to control transfection efficiency. For TiPARP gene KO, the following sgRNA sequence recognizing the nucleotides from position 973 to 989 in the exon 2 of the mouse TiPARP gene was used: CTGCAAGGCACTTGCATTTA (CRISPRevolution sgRNA EZ Kit, modified; Synthego). Transfections were performed using program A023 with Amaxa Nucleofector device following the manufacturer's instructions. Cells were plated, and after 48 h, they were collected and re-seeded with serial dilutions in 96-well culture plates to achieve a cell population with enriched mutations. Cell populations were then analyzed for TiPARP mutations by AMPLICON-EZ next-generation sequencing at GENEWIZ Global Headquarters using the following primers: Forward, 5’-GCTTCCCTTGAGCTTGTGTT-3’; Reverse, 5’- TGGAAACACTCTGCCACTTCT-3’.
Cell treatment and transfection with viral motifs for analysis of Infb1 gene expression
WT MEF, MEFCul4b-null, and MEFCul4b-null/Tiparp K.D. cells were seeded at cell densities of 0.3 × 106 cells/well in 6-well plates. The following day, cells were treated with 1 nM TCDD or dioxane. After 1 h, cells were transfected with 1 μg/ml of triphosphate dsRNA (3pRNA) or its negative control (3pRNAc) (InvivoGen) in 7.5-μl Lipofectamine 3000, following the manufacturer’s instructions. Twenty-four hours after transfection with 3pRNA, cells were scraped in 1-ml RNA STAT-60 for RNA extraction and RT-qPCR analysis to assess Infb1 mRNA expression using the primers shown in the Table S1.
Subcellular nuclear and cytoplasmic fraction preparation
Cytoplasmic- and nuclear-enriched fractions from MEF cells were obtained by using the NE-PER Nuclear and Cytoplasmic Extraction kit (78833; Invitrogen) following the manufacturer’s instructions with the following modifications: CER II buffer was added at a ratio of 0.68 μl per 10 μl of packed cell pellet, and vortexing was avoided at all times. After addition of buffer CER II, lysed cells were centrifuged for 10 min (1000g). The supernatant (cytoplasmic fraction) was snap-frozen, while the pellet (nuclear fraction) was washed once with 1× PBS before being snap-frozen. The subcellular fractions were used for Western blotting analysis following the same procedures indicated above. To assess the enrichment of the nuclei and cytoplasmic fractions, primary antibodies anti-histone-3 H3, nuclear marker, and anti-α/β tubulin, cytoplasmic marker, were used.
Immunofluorescence analysis
To visualize AHR protein in MEF cells by immunofluorescence, cells were seeded in 6-well plates on poly-D-lysine–coated glass coverslips at a cell density of 0.12 × 106 cells/well in DMEM supplemented with 10% dialyzed FBS (Invitrogen) and 1% Penicillin-Streptomycin solution (Invitrogen). The next day, cells were pretreated with LMB (50 nM, 16 h) or solvent ethanol, and followed by TCDD treatment (10 nM, 6 h) or solvent dioxane (control). After treatments, cells were fixed in a 1:1 mixture of cold MetOH:acetone at −20 °C for 10 min. Cells were then air-dried and blocked in 10% normal goat serum (50197Z, Invitrogen) + 1% BSA in 1× PBS for 1 h at room temperature (RT). Cells were then incubated with an AHR primary antibody (BML-SA210, Enzo Life Sciences; 1:100) overnight at 4 °C. For detection, cells were rinsed with 1× PBS two times and then incubated with a secondary antibody goat anti-rabbit conjugated with Alexa Fluor 488 green fluorescent dye (A-11008, Invitrogen; 1:500) for 1 h at RT. Glass coverslips were then rinsed two times with 1× PBS and mounted on microscope slides using an anti-fade solution (S36967, Invitrogen) and examined using a fluorescence microscope (Nikon Eclipse TE2000-E). 4′,6-Diamidino-2-phenylindole solution (62248, Invitrogen) was used as the nuclear counterstain. Fluorescence intensity measurements of AHR green signal were obtained using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD, http://imagej.nih.gov/ij/, 1997–2016).
Statistics
Statistical significance of the differences between group means was evaluated by one-way ANOVA using Tukey's honestly significant difference test as a post hoc test; p values ≤0.05 were considered statistically significant.
Data availability
All data are contained in the article and in the Supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Rana Mekawy (Weill Cornell Medicine, Department of Pharmacology) for technical assistance.
Author contributions
M. P. R., S. D.-M., P. Z., and A. B. R. conceptualization; M. P. R., S. D.-M., C. Y., and A. B. R. investigation; M. P. R., S. D.-M., and A. B. R. methodology; M. P. R., S. D.-M., and A. B. R. writing–original draft; M. P. R., S. D.-M., P. Z., and A. B. R. writing–review and editing; S. D.-M., C. Y., P. Z., and A. B. R. data curation; S. D.-M. formal analysis; S. D.-M. and A. B. R. supervision; A. B. R. resources; A. B. R. funding acquisition; A. B. R. project administration.
Funding and additional information
This work was supported by the National Institute of Environmental Health Sciences, National Institutes of Health, Grant # R01ES027488 to A. B. R. and S. D.-M.; National Cancer Institute, National Institutes of Health, Grants # R01CA213992 and R01CA221152 to P. Z. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by George DeMartino
Footnotes
In memory of Dr Arleen B. Rifkind who dedicated more than 40 years of her life to advance the understanding of the effects of the ligand-activated aryl hydrocarbon receptor.
Present address for Mercedes Paz Rijo: Institut Pasteur de Montevideo, Montevideo, Uruguay.
Supporting information
References
- 1.Poland A., Knutson J.C. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 2.Diani-Moore S., Shoots J., Singh R., Zuk J.B., Rifkind A.B. NAD(+) loss, a new player in AhR biology: Prevention of thymus atrophy and hepatosteatosis by NAD(+) repletion. Sci. Rep. 2017;7:2268. doi: 10.1038/s41598-017-02332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock K.W., Kohle C. Ah receptor: Dioxin-mediated toxic responses as hints to deregulated physiologic functions. Biochem. Pharmacol. 2006;72:393–404. doi: 10.1016/j.bcp.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Stockinger B., Hirota K., Duarte J., Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Semin. Immunol. 2011;23:99–105. doi: 10.1016/j.smim.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Whitlock J.P., Jr. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 6.Denison M.S., Soshilov A.A., He G., DeGroot D.E., Zhao B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez-Vazquez C., Quintana F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48:19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beischlag T.V., Luis Morales J., Hollingshead B.D., Perdew G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Q., Baldwin K.T., Renzelli A.J., McDaniel A., Dong L. TCDD-inducible poly(ADP-ribose) polymerase: A novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001;289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- 10.Pollenz R.S. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol. Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- 11.Pahl H.L., Baeuerle P.A. Control of gene expression by proteolysis. Curr. Opin. Cell Biol. 1996;8:340–347. doi: 10.1016/s0955-0674(96)80007-x. [DOI] [PubMed] [Google Scholar]
- 12.Davarinos N.A., Pollenz R.S. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J. Biol. Chem. 1999;274:28708–28715. doi: 10.1074/jbc.274.40.28708. [DOI] [PubMed] [Google Scholar]
- 13.Pollenz R.S., Barbour E.R. Analysis of the complex relationship between nuclear export and aryl hydrocarbon receptor-mediated gene regulation. Mol. Cell. Biol. 2000;20:6095–6104. doi: 10.1128/mcb.20.16.6095-6104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Q., Baldwin K.T. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced degradation of aryl hydrocarbon receptor (AhR) by the ubiquitin-proteasome pathway. Role of the transcription activaton and DNA binding of AhR. J. Biol. Chem. 2000;275:8432–8438. doi: 10.1074/jbc.275.12.8432. [DOI] [PubMed] [Google Scholar]
- 15.Hannah J., Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene. 2015;573:33–45. doi: 10.1016/j.gene.2015.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtake F., Baba A., Takada I., Okada M., Iwasaki K., Miki H., Takahashi S., Kouzmenko A., Nohara K., Chiba T., Fujii-Kuriyama Y., Kato S. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 17.Kawajiri K., Kobayashi Y., Ohtake F., Ikuta T., Matsushima Y., Mimura J., Pettersson S., Pollenz R.S., Sakaki T., Hirokawa T., Akiyama T., Kurosumi M., Poellinger L., Kato S., Fujii-Kuriyama Y. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou H., Duan Y., Zhang X., Yu Q., Di Q., Song Y., Li P., Gong Y. Aryl hydrocarbon receptor (AhR) regulates adipocyte differentiation by assembling CRL4B ubiquitin ligase to target PPARgamma for proteasomal degradation. J. Biol. Chem. 2019;294:18504–18515. doi: 10.1074/jbc.RA119.009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., Cho T., MacPherson L., Sugamori K.S., Yang Y., Grant D.M., Cummins C.L., Matthews J. Loss of the mono-ADP-ribosyltransferase, Tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J. Biol. Chem. 2015;290:16824–16840. doi: 10.1074/jbc.M115.660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacPherson L., Tamblyn L., Rajendra S., Bralha F., McPherson J.P., Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41:1604–1621. doi: 10.1093/nar/gks1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L., Yin Y., Li Y., Prevedel L., Lacy E.H., Ma L., Zhou P. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 2012;22:1258–1269. doi: 10.1038/cr.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mimura J., Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 23.Nebert D.W., Dalton T.P., Okey A.B., Gonzalez F.J. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 24.Song Z., Pollenz R.S. Ligand-dependent and independent modulation of aryl hydrocarbon receptor localization, degradation, and gene regulation. Mol. Pharmacol. 2002;62:806–816. doi: 10.1124/mol.62.4.806. [DOI] [PubMed] [Google Scholar]
- 25.Gupte R., Liu Z., Kraus W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diani-Moore S., Zhang S., Ram P., Rifkind A.B. Aryl hydrocarbon receptor activation by dioxin targets phosphoenolpyruvate carboxykinase (PEPCK) for ADP-ribosylation via 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TiPARP) J. Biol. Chem. 2013;288:21514–21525. doi: 10.1074/jbc.M113.458067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen G.C. Viruses and interferons. Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 28.Yamada T., Horimoto H., Kameyama T., Hayakawa S., Yamato H., Dazai M., Takada A., Kida H., Bott D., Zhou A.C., Hutin D., Watts T.H., Asaka M., Matthews J., Takaoka A. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 2016;17:687–694. doi: 10.1038/ni.3422. [DOI] [PubMed] [Google Scholar]
- 29.Kudo N., Wolff B., Sekimoto T., Schreiner E.P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 30.Petrulis J.R., Kusnadi A., Ramadoss P., Hollingshead B., Perdew G.H. The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J. Biol. Chem. 2003;278:2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- 31.Ikuta T., Eguchi H., Tachibana T., Yoneda Y., Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J. Biol. Chem. 1998;273:2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- 32.Berg P., Pongratz I. Differential usage of nuclear export sequences regulates intracellular localization of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 2001;276:43231–43238. doi: 10.1074/jbc.M105261200. [DOI] [PubMed] [Google Scholar]
- 33.Greb-Markiewicz B., Kolonko M. Subcellular localization signals of bHLH-PAS proteins: Their significance, current state of knowledge and future perspectives. Int. J. Mol. Sci. 2019;20:4746. doi: 10.3390/ijms20194746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seok S.H., Lee W., Jiang L., Molugu K., Zheng A., Li Y., Park S., Bradfield C.A., Xing Y. Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. U. S. A. 2017;114:5431–5436. doi: 10.1073/pnas.1617035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus W.L. PARPs and ADP-ribosylation: 60 years on. Genes Dev. 2020;34:251–253. doi: 10.1101/gad.336420.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abd Elmageed Z.Y., Naura A.S., Errami Y., Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): New roles in intracellular transport. Cell. Signal. 2012;24:1–8. doi: 10.1016/j.cellsig.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Grimaldi G., Corda D. ADP-ribosylation and intracellular traffic: An emerging role for PARP enzymes. Biochem. Soc. Trans. 2019;47:357–370. doi: 10.1042/BST20180416. [DOI] [PubMed] [Google Scholar]
- 38.Gomez A., Bindesboll C., Satheesh S.V., Grimaldi G., Hutin D., MacPherson L., Ahmed S., Tamblyn L., Cho T., Nebb H.I., Moen A., Anonsen J.H., Grant D.M., Matthews J. Characterization of TCDD-inducible poly-ADP-ribose polymerase (TIPARP/ARTD14) catalytic activity. Biochem. J. 2018;475:3827–3846. doi: 10.1042/BCJ20180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tkachenko A., Henkler F., Brinkmann J., Sowada J., Genkinger D., Kern C., Tralau T., Luch A. The Q-rich/PST domain of the AHR regulates both ligand-induced nuclear transport and nucleocytoplasmic shuttling. Sci. Rep. 2016;6:32009. doi: 10.1038/srep32009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollenz R.S., Wilson S.E., Dougherty E.J. Role of endogenous XAP2 protein on the localization and nucleocytoplasmic shuttling of the endogenous mouse Ahb-1 receptor in the presence and absence of ligand. Mol. Pharmacol. 2006;70:1369–1379. doi: 10.1124/mol.106.027672. [DOI] [PubMed] [Google Scholar]
- 41.Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G., Dawson V.L., Dawson T.M. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu K., Wu W., Li Y., Lin L., Chen D., Yan H., Xiao X., Chen H., Chen Z., Zhang Y., Xu S., Guo Y., Koeffler H.P., Song E., Yin D. Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep. 2019;20 doi: 10.15252/embr.201846166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C.J., Mickanin C., Myer V. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Cao J., Dong L., Lin H. TiPARP forms nuclear condensates to degrade HIF-1alpha and suppress tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 2020;117:13447–13456. doi: 10.1073/pnas.1921815117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacPherson L., Ahmed S., Tamblyn L., Krutmann J., Forster I., Weighardt H., Matthews J. Aryl hydrocarbon receptor repressor and TiPARP (ARTD14) use similar, but also distinct mechanisms to repress aryl hydrocarbon receptor signaling. Int. J. Mol. Sci. 2014;15:7939–7957. doi: 10.3390/ijms15057939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diani-Moore S., Labitzke E., Brown R., Garvin A., Wong L., Rifkind A.B. Sunlight generates multiple tryptophan photoproducts eliciting high efficacy CYP1A induction in chick hepatocytes and in vivo. Toxicol. Sci. 2006;90:96–110. doi: 10.1093/toxsci/kfj065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the article and in the Supporting information.