Abstract
Machilus robusta W. W. Smith is an evergreen plant distributed in the Yangtze River Basin and the south regions of China. Here we analyzed the complete chloroplast (cp) genome sequence of M. robusta to determine its structure and evolutionary relationship to other Lauraceae. The cp genome is 152,737 bp in length and has an overall GC content of 39.2% The genome includes a large single-copy (LSC) region of 93,706 bp and a small single-copy (SSC) region of 18,885 bp, and these are separated by a pair of inverted repeats (IRs) of 20,073 bp. The cp genome contains 128 genes, including 83 protein-coding, 37 tRNAs, and 8 rRNAs. Phylogenetic analysis based on complete cp genome sequences fully resolved M. robusta in a clade with M. balansae. This work provides new molecular data for evolutionary studies of the Lauraceae.
Keywords: Complete chloroplast genome, Lauraceae, Machilus robusta W. W. Smith, phylogeny
Machilus robusta, an evergreen plant of the genus classified in the Lauraceae, is mainly distributed in the Yangtze River Basin and the south regions of China, with Guangdong, Guangxi, Guizhou, Hainan, Xizang, Yunnan as the main growing area. Its leaves are rich in bioactive metabolites with diverse structures, and are known to relaxmuscles, promoteblood circulation, reduceswelling, and relieve pain (Liu et al. 2007). The plant is also widely used as a folk herb for the treatment of gastric dullness, vomiting, and diarrhea in China (Li et al. 2011; Bu et al. 2013). To date, only genetic marker data are publicly available for M. robusta. In this study, the M. robusta cp genome was sequenced and assembled to document its plastid chromosomal content and structure, as well as confirm its relationship to other Lauraceae.
Fresh leaves of M. robusta were collected from Jiangxi Academy of Forestry, Nanchang, Jiangxi, China (28°44′41″N, 115°48′37″E). The voucher specimen was deposited in the laboratory of Camphor Engineering Technology Research Center for National Forestry and Grassland Administration (Yanfang Wu, yanfangwu2012@163.com) under accession number of WYF202001. Genomic DNA was extracted using the DNeasy plant mini kit (Qiagen). Paired-end reads were generated by using the Illumina NovaSeq system (Illumina, San Diego, CA). In total, ∼1.4 Gb of raw data (9,223,296 reads) were obtained. Quality control was performed to remove adapters and low-quality reads using fastp (Chen et al. 2018). The cp genome was de novo assembled by NOVOPlasty (Dierckxsens et al. 2017; Wang et al. 2018) and annotated by GeSeq (Tillich et al. 2017).
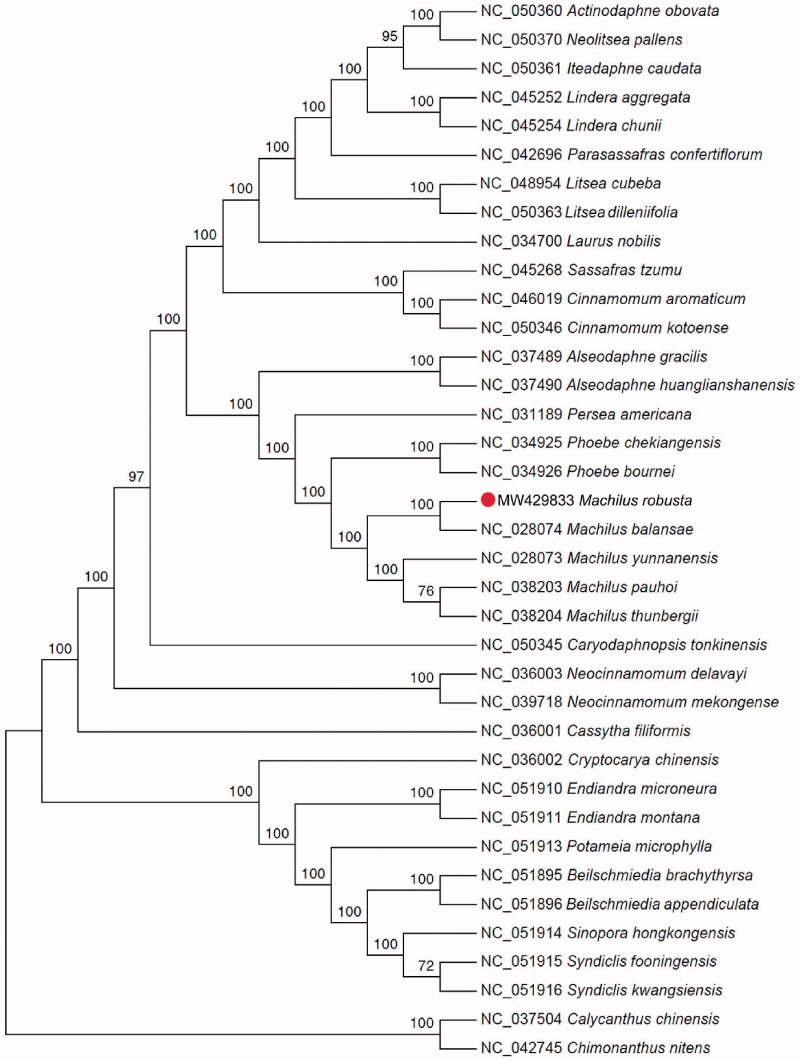
The complete cp genome sequence of M. robusta is 152,737 bp in length, and contains a large single-copy region (LSC) of 93,706 bp, a small single-copy region (SSC) of 18,885 bp, and a pair of inverted repeats (IR) regions of 20,073 bp. A total of 128 genes were annotated, including 83 protein-coding genes, 37 tRNAs, and 8 rRNAs. The GC content of the cp genome is 39.2%. To reveal the phylogenetic position of M. robusta with other members in Lauraceae, a phylogenetic analysis was performed based on 37 complete cp genomes, and two taxa from the Calycanthaceae, Calycanthus chinensis and Chimonanthus nitens to serve as outgroups. The sequences were aligned by MAFFT v7.271 (Katoh and Standley 2013). The maximum likelihood (ML) bootstrap analysis with 1000 replicates was performed using IQ-TREE v1.6.12 (Minh et al. 2020). TVM + F+R3 was selected as the best-fit model according to the built-in ModelFinder. The phylogenetic tree showed that M. robusta is closely related to M. balansae (Figure 1). Machilus was fully resolved in a sister position to the genus Phoebe in Lauraceae. These results were similar to those presented by Kong et al. (2014) based on matK sequence analysis and Chen et al. (2017) based on cp gene analysis. The cp genome sequence of M. robusta provides new molecular data for evolutionary studies of Lauraceae.
Figure 1.
Maximum likelihood phylogenetic tree based on the complete chloroplast genome sequences of 35 plant species from Lauraceae and two outgroup plant species from the Calycanthaceae.
Funding Statement
This work was supported by the Science and Technology Planning Project of Jiangxi Province [20171BBF60014 and 20203BBF62W010], the Forestry Science and Technology Innovation Special Project of Jiangxi Forestry Department [JXTG(2021)16]. There was no additional external funding received for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW429833. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA714487, SRR13962180, and SAMN18310375 respectively.
References
- Bu PB, Li YR, Jiang M, Wang XL, Wang F, Lin S, Zhu CG, Shi JG.. 2013. Lignans from Machilus robusta. Zhongguo Zhong Yao Za Zhi. 38(11):1740–1746. [PubMed] [Google Scholar]
- Chen C, Zheng Y, Liu S, Zhong Y, Wu Y, Li J, Xu LA, Xu M.. 2017. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ. 5:e3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Ma C, Zhang Q, Li L, Chen X, Zeng H, Guo D.. 2014. Leading dimensions in absorptive root trait variation across 96 subtropical forest species. New Phytol. 203(3):863–872. [DOI] [PubMed] [Google Scholar]
- Li Y, Cheng W, Zhu C, Yao C, Xiong L, Tian Y, Wang S, Lin S, Hu J, Yang Y, et al. 2011. Bioactive neolignans and lignans from the bark of Machilus robusta. J Nat Prod. 74(6):1444–1452. [DOI] [PubMed] [Google Scholar]
- Liu MT, Lin S, Wang YH, He WY, Li S, Wang SJ, Yang YC, Shi JG.. 2007. Two novel glycosidic triterpene alkaloids from the stem barks of Machilus yaoshansis. Org Lett. 9(1):129–132. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N.. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW429833. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA714487, SRR13962180, and SAMN18310375 respectively.