Abstract
The p75 neurotrophin receptor (p75NTR) is a critical mediator of neuronal death and tissue remodeling and has been implicated in various neurodegenerative diseases and cancers. The death domain (DD) of p75NTR is an intracellular signaling hub and has been shown to interact with diverse adaptor proteins. In breast cancer cells, binding of the adaptor protein TRADD to p75NTR depends on nerve growth factor and promotes cell survival. However, the structural mechanism and functional significance of TRADD recruitment in neuronal p75NTR signaling remain poorly understood. Here we report an NMR structure of the p75NTR-DD and TRADD-DD complex and reveal the mechanism of specific recognition of the TRADD-DD by the p75NTR-DD mainly through electrostatic interactions. Furthermore, we identified spatiotemporal overlap of p75NTR and TRADD expression in developing cerebellar granule neurons (CGNs) at early postnatal stages and discover the physiological relevance of the interaction between TRADD and p75NTR in the regulation of canonical NF-κB signaling and cell survival in CGNs. Our results provide a new structural framework for understanding how the recruitment of TRADD to p75NTR through DD interactions creates a membrane-proximal platform, which can be efficiently regulated by various neurotrophic factors through extracellular domains of p75NTR, to propagate downstream signaling in developing neurons.
Keywords: NMR, protein structure, cell signaling, TRADD, p75 neurotrophin receptor, death domain
Abbreviations: CGN, cerebellar granule neuron; DD, death domain; ECD, extracellular cysteine-rich domain; ES, embryonic stem; ICD, intracellular domain; NGFR, nerve growth factor receptor; NMR, nuclear magnetic resonance; NT, neurotrophin; PEI, polyethylenimine; p75NTR, p75 neurotrophin receptor; RMSD, root mean square deviation; TMD, transmembrane domain; TNFRSF, tumor necrosis factor receptor superfamily; TRADD, TNF receptor associated death domain
p75 neurotrophin receptor (p75NTR), also known as tumor necrosis factor receptor superfamily (TNFRSF) 16 and nerve growth factor receptor (NGFR), is the first receptor identified for a family of neurotrophic growth factors known as the neurotrophins (NTs) (1, 2, 3, 4). p75NTR is a critical mediator of neuronal death and tissue remodeling and has been implicated in a variety of neurodegenerative diseases (5). In the early development of mammals, p75NTR is highly expressed in the nervous system and plays an essential role in promoting nerve growth. Its expression is significantly downregulated in the adult and only observed in a small number of neuronal subpopulations; however, lesions to the nervous system strongly stimulate p75NTR re-expression (5, 6, 7, 8). In many cancers, such as melanoma and breast cancer, p75NTR is also expressed as a tumor promoter or suppressor depending on cancer types (9). Similar to other TNFRSF members, p75NTR does not have intrinsic catalytic activity, and its signaling depends on the recruitment of intracellular interactors (10).
p75NTR consists of an extracellular cysteine-rich domain (ECD), a single-pass transmembrane domain (TMD) involved in the formation of an intermolecular disulfide bridge, and an intracellular domain (ICD), containing an unstructured juxtamembrane domain (JMD) and a helical death domain (DD) (10, 11). Upon binding extracellular signals, such as the neurotrophin nerve growth factor (NGF), the p75NTR-ECD undergoes significant conformational changes from an open to a closed state (12). This movement further propagates to the p75NTR-DD through the disulfide-bonded p75NTR-TMD, leading to separation of the p75NTR-DD homodimer and exposure of active sites on the p75NTR-DD surface for recruitment of DD interactors (13). As a signaling hub, the p75NTR-DD has been shown to interact with a variety of intracellular molecules to regulate different signaling cascades, including NF-κB, RhoA, and JNK/caspase pathways in diverse cellular contexts (10, 14, 15). Among the many proteins that interact with the p75NTR-ICD, TRADD (TNF receptor associated death domain) is rather unique due to its ability to create a platform on the membrane for recruitment of additional proteins for downstream signaling (16). TRADD is a 34 kDa protein with two functional domains (N- and C-terminal domains) connected by an unstructured peptide of ∼38 amino acids (17, 18). The C-terminal domain of TRADD contains a novel DD. TRADD was initially identified as an adaptor protein for tumor necrosis factor receptor 1 (TNFR1) signaling since its DD can interact with the TNFR1-DD in response to TNF activation (16, 19). It has also been reported that TRADD interacts with p75NTR and promotes NGF-dependent signaling that opposes cell death in breast cancer cells (20). Aside from that first report, there are no other studies on the role and physiological relevance of TRADD in p75NTR signaling. In particular, it remains unknown whether and, if so, how TRADD participates in p75NTR signaling in neurons. Here, we report the high-resolution NMR complex structure of p75NTR-DD:TRADD-DD and reveal the specific mechanism of TRADD-DD recognition by the p75NTR-DD. We also identify spatiotemporal overlap of p75NTR and TRADD expression in developing cerebellar granule neurons (CGNs), a physiologically relevant neuronal subpopulation, the development of which is strongly regulated by p75NTR signaling (15, 21, 22, 23).
Results and discussion
Solution structure of the complex between the p75NTR-DD and the TRADD-DD
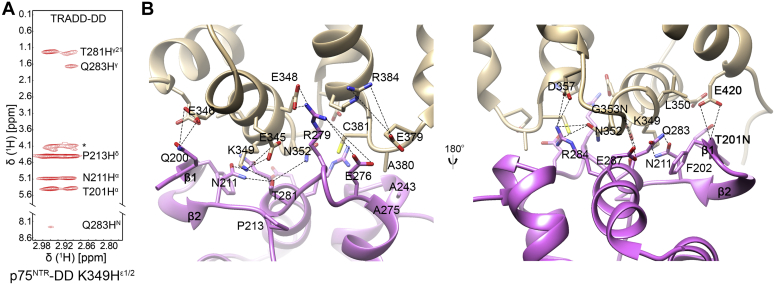
Our recent NMR structural studies have identified a direct binding of the TRADD-DD to the p75NTR-DD in vitro and characterized a novel structural fold of the TRADD-DD in the DD superfamily (18). The TRADD-DD contains a unique β-hairpin motif, which is not found in any other known DDs. However, it was unclear if this unusual structural element has a specific role in DD-DD interactions. To better understand the structural mechanisms of DD recognition in the p75NTR/TRADD interaction, we first determined the solution structure of the complex between the p75NTR-DD and the TRADD-DD by multidimensional nuclear magnetic resonance (NMR) spectroscopy (Table S1). Due to the fully and partially exposed hydrophobic residues on the surface, the TRADD-DD exhibits aggregation-prone property in solution. Thus, the protein complex of the p75NTR-DD and TRADD-DD for NMR structural studies was prepared in salt-free water, where TRADD-DD aggregation can be minimized and the protein still retains its function for binding to the p75NTR-DD (18, 24, 25) (Fig. S1). The ensemble of ten lowest-energy structures of the p75NTR-DD:TRADD-DD complex and a representative cartoon structure are depicted in Figure 1, A and B, respectively. Multiple refinements converged to a mean backbone root mean square deviation (RMSD) of 0.32 ± 0.09 Å in the structural region of the complex (G334-E420 of the p75NTR-DD and T201-L302 of the TRADD-DD). The representative slice of the detected intermolecular nuclear Overhauser effects (NOEs) shows that the residue K349 from the p75NTR-DD is critical for binding the TRADD-DD (Fig. 2A). The structure of the p75NTR-DD in the complex was close to that in the p75NTR-DD homodimer with a pairwise RMSD of ∼1.1 Å. A small observable movement of helix orientation occurs in helixes ⍺1 and ⍺4 (Fig. S2). In comparison, the TRADD-DD in the complex is nearly the same as TRADD-DD monomer with a pairwise RMSD of ∼0.5 Å. Therefore, the recruitment of TRADD to p75NTR does not impose noticeable conformational changes on either of these death domains.
Fig. 1.
Solution structure of p75NTR-DD:TRADD-DD complex.A, superposition of backbone heavy atoms of the ten lowest-energy complex structures of the p75NTR-DD:TRADD-DD. N- and C-termini of DDs are indicated. B, Ribbon drawing of p75NTR-DD:TRADD-DD.
Fig. 2.
Binding interface of p75NTR-DD:TRADD-DD complex.A, representative slice from the 13C,15N-filtered 3D NOESY spectrum. ∗, ambiguous NOE peaks. The p75NTR-DD was labeled with 13C and 15N, and the TRADD-DD was unlabeled. B, detail of binding interface in the p75NTR-DD:TRADD-DD complex. Key residues at the binding interface are labeled and depicted as stick models. Close distances between nitrogen and oxygen atoms (~5 Å or less) are showed in dash lines.
DD interaction interface
High resolution of the NMR structure allows us to closely inspect the binding interface in the complex. Figure 2B shows that the DD interaction interface is mainly formed by helices α1, α6, and α3-α4 loop of the p75NTR-DD and β-hairpin (β1 and β2), helices α5, α2-α3, and α4-α5 loops of the TRADD-DD. Hydrophobic interactions between residues L350 and A380 of the p75NTR-DD and F202, A243, and A275 of the TRADD-DD also contribute to the binding of two DDs. We have previously shown that the unique β-hairpin motif is indispensable for the TRADD-DD to fold correctly in water solution while its structural role in DDs assembly was not determined (18). The interactions observed in the p75NTR-DD:TRADD-DD complex highlight the structural importance of this β-hairpin motif in specific DD recognition. Electrostatic interactions were found to play a crucial role in the binding interface, and an extensive set of charged and polar residues, including Arg, Lys, Glu, Asp, Gln, Asn, were identified (Fig. 2B). Hydrogen bonding between E348 in the p75NTR-DD and R279 in the TRADD-DD as well as between N352 in the p75NTR-DD and R284 in the TRADD-DD was detected. Since electrostatic interactions significantly contribute to the binding interface, we selected fully surface-exposed charge residues for mutagenesis studies. Importantly, isothermal titration calorimetry (ITC) assays as well as coimmunoprecipitation experiments verified the functional relevance of six key interface residues, namely E345, K349, and E379 in the p75NTR-DD and E276, R279, and R284 in the TRADD-DD. Single-point mutation of these charged residues to Ala significantly diminished TRADD interaction with p75NTR in ITC assays using purified proteins and in transfected HEK cells (Fig. 3, Figs. S3 and S4). One of the most critical residues in the binding interface is K349 from the p75NTR-DD, which shows a number of interactions with residues N211, T281, Q283, and P213 from the TRADD-DD (Fig. 2, A and B). A single K349A mutation without disturbing its global helical folding can nearly completely disrupt the interaction between the TRADD-DD and the p75NTR-DD, leading to undetectable binding affinity measured by ITC assay (Figs. 3B and S5A). We further confirmed this data in transfected HEK cells where the interaction between WT TRADD-DD and p75NTR-DD in the K349A mutation was undetectable (Fig. 3C). Similar results were obtained using E276A mutation in the TRADD-DD (Figs. 3D and S5B). Taken together, these mutagenesis studies highlight the critical contribution of the charged surface to DD interaction between p75NTR and TRADD.
Fig. 3.
Mutagenesis studies.A, ITC binding curves of human TRADD to human p75NTR. Dissociation constants (Kd) are the mean ± SEM from three independent experiments. B, binding affinities expressed as Kd of WT and point mutants of p75NTR and TRADD derived from ITC data. Dissociation constants are the mean ± SEM from three independent experiments. C, coimmunoprecipitation of wild-type (WT) and point mutants of HA-tagged human p75NTR with Myc-tagged human TRADD in transfected HEK 293T cells. The immunoblots shown are representative of three independent experiments. D, coimmunoprecipitation of WT and point mutants of Myc-tagged human TRADD with HA-tagged human p75NTR in transfected HEK 293T cells. The immunoblots shown are representative of three independent experiments. IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysate.
p75NTR-DD interaction specificity
In p75NTR-mediated signaling pathways, the p75NTR-DD functions as a central hub for recruitment of various intracellular interactors or domains, including RhoGDI (Rho guanine nucleotide dissociation inhibitor), RIP2 (Receptor-interacting protein 2) kinase, TRADD, p45-DD, etc., in different cellular contexts. An important question is what could confer the interaction specificity of the p75NTR-DD. Although the p75NTR-DD exhibits a similar structural fold or 3D topology to other members in the DD superfamily, it has distinctive lengths and orientations of six ⍺- and one 310 helices, leading to unique surface features, such as protrusion or concave surfaces with different charge distribution, which are crucial for determining binding specificity of the p75NTR-DD (Fig. 4) (18). Structural investigation of the complexes between the p75NTR-DD and its interactors reveals that the p75NTR-DD takes advantage of specific surfaces for recruiting different interactors. Mutagenesis studies on the charged or hydrophobic residues involved in the binding interfaces further provide functional validation of the structural insights obtained on the specific recognition of different interactors by the p75NTR-DD (Fig. 3) (26, 27).
Fig. 4.
p75NTR-DD with specific binding sites for intracellular interactors. Charged surface of the p75NTR-DD and the binding sites for intracellular interactors. Unstructured regions are not shown. Color code is blue for positive charges, red for negative charges, and white for neutral surface. The patches on the surface of the p75NTR-DD responsible for binding TRADD, RhoGDI, RIP2, and the p75NTR-DD itself are circled in pink, cyan, green, and brown, respectively.
The interaction specificity of the p75NTR-DD is not compromised by overlapping binding sites. Partially overlapping binding sites on the p75NTR-DD can lead to competitive binding of two or more proteins to the p75 NTR-DD if they are coexpressed in a cell context (26). For example, the TRADD-DD-binding site on the p75NTR-DD partially overlaps with p75NTR-DD homodimerization and RhoGDI-binding sites (Figs. S6 and 4). Therefore, recruitment of TRADD to p75NTR requires separation of p75NTR-DD homodimer, which is consistent with previous observation showing NGF-dependent binding of TRADD to p75NTR (20). Although coexpression of RhoGDI and TRADD in the same neuron cell is unknown, it is conceivable that signaling pathways regulated by RhoGDI and TRADD could not be activated at the same time.
Expression of TRADD in developing cerebellum
For TRADD to have a physiological role in p75NTR signaling, it needs to be present in the same cells that express the receptor. We and others have previously reported on the expression and function of p75NTR in developing CGNs (15, 21, 22, 23), the most abundant neuron subtype in the mammalian brain. In contrast, the specific expression of TRADD in distinct neuronal subpopulations, including CGNs, is not well understood. We therefore assessed whether TRADD is expressed in developing CGNs at early postnatal stages, coincident with the height of p75NTR expression. Immunohistochemical analysis revealed that TRADD and p75NTR were highly coexpressed in proliferating CGN precursors in the external granule layer (EGL) at postnatal day 2 (P2) (Fig. 5A). Their expression levels were significantly lower at P5 (Fig. 5B). By P7, the EGL is reduced, as CGNs mature and migrate to the internal granule layer (IGL), and both proteins were expressed at very low levels (Fig. 5C). Thus, the spatiotemporal overlap of p75NTR and TRADD expression is compatible with TRADD being involved in p75NTR signaling during early stages of CGN development. Moreover, their predominant coexpression in early, proliferating CGN precursors suggests roles in cell-cycle progression and/or withdrawal.
Fig. 5.
Expression of TRADD in developing cerebellum. Micrographs of a representative mid-sagittal section through the developing cerebellum of P2 (A/B), P5 (C/D), and P7 (E/F) wild-type mouse stained with anti-TRADD (red) and anti-p75NTR (green) antibodies, and counterstained with DAPI (blue). White outlines in C and E indicate no tissue in these areas. Higher magnification panels of P2, P5, and P7 are shown in B, D, and F, respectively. Scale bars: A, 300 μm; B, 50 μm (upper) and 100 μm (lower); C, 300 μm; D, 100 μm; E, 300 μm; F, 100 μm.
Physiological relevance of TRADD/p75NTR interaction in NF-κB signaling
It has been reported that interaction of TRADD with TNFR1 leads to activation of the NF-κB signaling pathway (28). In order to elucidate the functional relevance of the interaction between p75NTR and TRADD, we investigated the activation state of this pathway in cultured CGNs derived from p75NTR null mice after reconstitution with wild-type p75NTR, the p75NTR mutant K349A that is unable to bind TRADD or the p75NTR mutant R384A that has a decreased interaction with TRADD. Our previous studies indicated that p75NTR mediates tonic activation of NF-κB in cultured CGN neurons through endogenous production of NGF and autocrine stimulation of p75NTR in these cells (15, 21). We assessed NF-κB activation by quantification of the levels of the p65 NF-κB subunit in the nuclei of cultured CGNs, a key step in canonical NF-kB signaling. In agreement with our previous studies, we observed reduced levels of p65 NF-κB in the nuclei of CGNs derived from p75NTR knockout mice, compared with wild-type controls (Fig. 6, A and B). Importantly, transfection of knock-out CGNs with wild-type p75NTR, but not K349A mutant, restored the levels of nuclear p65 NF-κB (Fig. 6B). R384A mutant only partially restored NF-kB activation due to its weaker binding affinity to TRADD. This assessment indicates that interaction of TRADD with p75NTR is required for normal activation of the NF-κB pathway in CGNs.
Fig. 6.
Functional role of TRADD/p75NTRinteraction in NF-κB signaling.A, representative micrographs of p75NTR knockout (KO) CGNs transfected with expression plasmids containing HA-tagged p75WT (wild-type) and HA-tagged p75K349A mutant. After 2 days in vitro, cultures were immunostained with anti-P65 NF-κB (green) and anti-HA (magenta) antibodies, and counterstained with DAPI (blue). Transfected cells expressing HA constructs are indicated with white arrows. Scale bar, 20 μm. B, quantification of nuclear p65 NF-κB (expressed as nuclear p65 fluorescence intensity/cytoplasmic p65 fluorescence intensity) in p75NTR wild-type (WT) and knockout (KO) CGNs transfected with HA-p75WTand HA-p75K349A plasmids as indicated. Results are expressed as mean ± SEM from three independent cultures (∗p < 0.05 and ∗∗p < 0.01, one-way ANOVA followed by Turkey's multiple comparison test).
As NF-κB activity has been shown to be essential for survival of CGNs (15, 21), we further assessed apoptotic cell death in cultured CGNs from p75NTR knockout mice after reconstitution with wild-type, K349A, or R384A mutant by immunostaining for cleaved caspase-3. We found twice as many apoptotic cells in cultures reconstituted with the mutant p75NTR K349A construct, compared with wild-type (Fig. S7), in agreement with the inability of the former to efficiently activate NF-κB. Together, our expression and functional studies indicate that TRADD binding to p75NTR is essential for the ability of the receptor to regulate NF-κB signaling and the balance between cell survival and cell death in CGNs and perhaps other neuronal subpopulations and cell types outside the nervous system.
Although regulation of the NF-κB pathway by p75NTR is often studied in the context of cell survival, there is also strong evidence for the importance of NF-κB signaling in cell cycle regulation, primarily through its effect on the expression of several key components of the cell cycle machinery, including cyclin D1 (29). Interestingly, recent work has indicated a role for p75NTR signaling in the regulation of cell cycle duration and withdrawal in CGN precursors (23, 30). Although those studies highlighted the role of the RhoA pathway in the effects of p75NTR on the cell cycle, the NF-κB and RhoA signaling pathways are known to intersect on p75NTR, through competition between RhoGDI and RIP2 effectors for binding to the receptor DD (26). Thus, it is possible that the interaction of TRADD with the p75NTR-DD described here plays a dual role in the early development of CGNs, both through direct regulation of the NF-κB pathway, as well as indirectly modulating RhoA signaling through competition for access to binding determinants in the DD.
Model for the initial stage of NF-κB signaling engaged by p75NTR and TRADD
Based on the present studies, we propose a structural model for the initial stage of p75NTR engagement with the TRADD/NF-κB pathway in developing cerebellar neurons (Fig. 7). p75NTR can form a disulfide-bound dimer at the plasma membrane, and homodimerization of two p75NTR-DDs closes their potential binding sites for downstream effectors. Since the N-terminal domain of TRADD (TRADD-NTD) can interact with the TRADD-DD (31), TRADD could also exist in a closed state before recruitment to the receptor, although the exact binding interface between the TRADD-NTD and the TRADD-DD is unclear. p75NTR signaling through TRADD depends on NGF, which is expressed by developing cerebellar neurons. In the unliganded state, the self-association of the p75NTR-DDs could be very strong since the movement of p75NTR-DDs is restricted by the transmembrane domain. NGF binding to the p75NTR-ECD leads to separation of intracellular p75NTR-DDs through disulfide-bonded p75NTR-TMD(26). Since p75NTR-DD homodimerization site partially overlaps with TRADD-DD-binding site on the p75NTR-DD (Fig. S6), dissociation of p75NTR-DD homodimer is required for the recruitment of the TRADD-DD to p75NTR, which could lead to the opening of two domains of TRADD. The free TRADD-NTD is envisioned to further recruit as yet unknown downstream molecules to activate the NF-κB pathway and promote neuron cell survival. In TNFR1-and DR3-mediated signaling pathways, the three molecules of TRADD-NTD were shown to associate with trimeric TRAF domains of TNF receptor-associated factor 2 (TRAF2) to initiate NF-κB signaling (17, 32, 33). Since the unstructured linker between the TRADD-NTD and the TRADD-DD is long (∼37 amino acid residues), it is conceivable that three p75NTR dimers are clustered together by two TRAF2 trimers on the membrane through TRADD and form a larger complex if TRAF2 is coexpressed with TRADD and p75NTR in developing CGNs. In this scenario, opening of two-domain TRADD is also necessary since key residues of the TRADD-NTD for binding the TRADD-DD are involved in the interaction with TRAF2 (Fig. S8) (31, 32).
Fig. 7.
Structure model of p75NTRNF-κB signaling via the recruitment of TRADD. p75NTR model was built based on available structures of individual domains and domain complexes. The domain orientation and interface between TRADD-NTD and TRADD-DD are not defined in this cartoon.
Previous studies have demonstrated that the caspase recruitment domain (CARD domain) of RIP2 also interacts with the p75NTR -DD to activate NF-κB pathway in developing cerebellum as well as in Schwann cells (14, 21, 34). Interestingly, structural analysis demonstrates that the RIP2-CARD and the TRADD-DD do not share overlapping binding sites on the p75NTR -DD (Fig. 4C). This finding was further supported by coimmunoprecipitation experiments performed in HEK 293T cells, which shows that mutations of key residues at the binding interface of p75NTR-DD:TRADD-DD complex do not reduce the capacity of p75NTR to bind RIP2 (Fig. S9). However, it is still unknown if RIP2 is coexpressed with TRADD in developing CGNs. RIP2 was also shown to diminish the interaction between TRAF6 and p75NTR through a steric hindrance effect on the receptor itself. Nevertheless, whether TRADD binding to the death domain of p75NTR interferes with TRAF6 binding remains to be investigated. Given that neither TRADD binds to the same residues of p75NTR as TRAF6 does and the MW of RIP2 (≈60 kDa) is nearly twice as big as TRADD (≈34 kDa), TRADD could be too small to interfere with TRAF6 binding in the same way RIP2 is capable of.
In summary, here we report the structural mechanism underlying recruitment of TRADD to p75NTR through DD interactions and reveal that interaction specificity of the p75NTR-DD, an intracellular signaling hub, relies on its distinct surface patches. We also identify the physiological relevance of TRADD/p75NTR interaction in NF-κB signaling in developing CGNs. Our results provide a structural framework for understanding the mechanisms by which this adaptor protein creates a large complex on p75NTR to propagate downstream signaling.
Materials and methods
Protein expression and purification
The cDNAs of human p75NTR DD (330–427) and TRADD-DD (199–312) were amplified from total human embryonic stem (ES) cell cDNA and subcloned into pET32-derived expression vectors between BamH I and Xho I restriction sites. Unlabeled proteins were expressed in E.coli strain SoluBL21 (DE3) in M9 medium. All protein samples were purified by Ni-NTA affinity chromatography, ionic exchange (MonoQ or MonoS), and/or gel filtration (Superdex 75). Isotopic labeling of 15N and/or 13C was carried out by expressing the proteins in M9 minimal medium containing 15N-NH4Cl and/or 13C-labeled glucose as the sole source of nitrogen and carbon. For protein complexes, two double-labeled samples were prepared in pure water with 10 mM D10-DTT: (1) 0.8 mM 13C, 15N-labeled p75NTR-DD mixed with 1.0 mM unlabeled TRADD-DD; (2) 0.8 mM 13C, 15N-labeled TRADD-DD mixed with 1.0 mM unlabeled p75NTR-DD.
NMR spectroscopy experiments and structure determination
All NMR experiments were carried out on a Bruker 800 MHz NMR spectrometer (AVANCE) with a cryogenic probe at 301K. All the NMR spectra were processed with NMRPipe (35) and analyzed with NMRDraw and NMRView supported by an NOE assignment plugin (36). Sequence-specific assignments of backbone and side chains were obtained by using previously described methods (37, 38). The chemical shift values of backbone Cα and Cβ were analyzed by TALOS+ to predict backbone dihedral angles (39). Intramolecular NOEs were assigned from 4D time-shared 13C, 15N-edited NOESY spectra (40). Intermolecular NOEs were obtained from 13C, 15N-filtered 3D experiments NOESY spectra of two double-labeled samples as mentioned above. Ambiguous NOEs were obtained with iterated structure calculations by CYANA (41). Based on NOE peak volume, intramolecular NOE values were binned into short (1.8–2.8 Å), medium (1.8–3.4 Å), and long (1.8–5.5 Å) distances. All the intermolecular NOEs were binned into long (1.8–6.0 Å) distances. Final structure calculation was started from 100 conformers. Ten conformers with the lowest target function values were selected for energy minimization in AMBER force field (42). PROCHECK-NMR (43) was used to assess the quality of the structures. All the structural figures including charged surfaces were made using UCSF Chimera (44).
ITC binding assay
All protein samples used in the ITC binding assay were prepared in deionized water with 1.4 mM β-mercaptoethanol. ITC binding assay was performed via an ITC200 (GE Healthcare) equipment at 25 °C. To determine the binding affinity, 0.4 mM WT TRADD-DD was titrated into 0.04 mM WT p75NTR-DD. Mutagenesis studies were performed in a similar way. The thermograms were integrated by the Origin software and fitted to a single-site binding model. The standard deviation of each Kd value was calculated from three independent ITC titration experiments.
Animals
Mice were housed in a 12-h light/dark cycle and fed a standard chow diet. The transgenic mouse lines used were p75NTR knockout (KO) (45). Transgenic mice were maintained in a c57bl6J background. Mice of both sexes were used for the experiments. All animal experiments were conducted in accordance with the National University of Singapore Institutional Animal Care. The lab certificate/approval number is OSHM/PI/14/SOM-250.
Plasmids
Full-length HA-tagged p75NTR and full-length Myc-tag TRADD were expressed from a pcDNA3 vector backbone (Invitrogen). The HA-p75E345A, HA-p75K349A, HA-p75E379A, HA-p75R384A, Myc-TRADDE276A, Myc-TRADDR279A, Myc-TRADDR284A, and Myc-TRADDE287A mutant constructs were selected based on p75NTR-DD:TRADD-DD complex structure.
Neuronal cultures
Wild-type and p75NTR KO CGNs were trypsinized and plated at a density of 40,000 (for cell death assay) or 200,000 (for protein collection) cells per coverslip coated with poly-L- lysine (Sigma, Cat: P7280) in a 24-well (Starlab) in neuro basal medium supplemented with B27 (Gibco, Cat: 17504001), 25 mM KCl (Sigma, Cat: P9541), 1 mM glutamine (Gibco, Cat: 25030149), and 2 mg/ml gentamicin (Invitrogen, Cat: 15750060).
Transfection
Neurons were transfected with either HA-p75WT, HA-p75K349A, or HA-p75R384A plasmids using Neon transfection system (Thermo Scientific, Cat: MPK10025) prior to plating. In total, 250 ng plasmid per well in the 24-well plate was used.
Cell death
For assessing apoptosis, p75NTR KO neurons transfected with the different plasmids (see above) were cultured for 2 days, fixed with solution containing 4% paraformaldehyde and 4% sucrose. The fixed cells were labeled with cleaved caspase 3 (Cell signal, 9761, 1:400), anti HA-tag (Invitrogen, 71–5500, 1:250), and DAPI. For each experiment, neurons were culture in duplicates and at least 15 images were taken per coverslip.
Immunohistochemistry
P2, P5, and P7 wild-type animals were perfused first with PBS, followed by 4% paraformaldehyde. Harvested cerebellar were post fixed in 4% paraformaldehyde for 16 h and cryoprotected in 30% sucrose before freezing. OCT-embedded cerebellar were frozen at –80 °C overnight and serially sectioned at 30 μm in the sagittal plane using cryostat. Midline sections were mounted onto electrostatic charged slides (Leica Microsystems), blocked with 5% donkey serum (Fisher scientific) containing 0.3% Triton X-100 (Sigma) in PBS for 1 h at room temperature, and then incubated for 16 h at 4 °C with primary antibodies. The sections were washed in PBS before incubated with the appropriate secondary antibodies.
Immunocytochemistry
The cultures were fixed in 4% paraformaldehyde and 4% sucrose for 15 min and washed with PBS before blocking nonspecific binding and permeabilizing with blocking solution (5% donkey serum and 0.3% Triton X-100) in PBS for 1 h at room temperature. Neurons were incubated overnight with the primary antibodies in 1% blocking solution at 4 °C. After washing with PBS, the cultures were incubated with the appropriate secondary antibodies. The primary antibodies used in this study were: polyclonal anti-p75NTR (Neuromics, GT15057, 1:250), monoclonal anti-TRADD (Santa Cruz, sc-46653, 1:500), polyclonal anti-HA (Sigma, H3667, 1:250), polyclonal anti-P65 NF-κB (Santa cruz, sc-372, 1:500), and polyclonal anti-cleaved caspase 3 (Cell signal, 9761, 1:400). Secondary antibodies were Alexa Fluor–conjugated anti-immunoglobulin from Life Technologies, Invitrogen, used at 1:1000 (donkey anti-rabbit IgG Alexa Fluor 555, A31572, donkey anti-goat IgG Alexa Fluor 488, A11055, donkey anti-mouse IgG Alexa Fluor 555, A31570, and donkey anti-mouse IgG Alexa Fluor 647, A31571). Images were obtained using a Zeiss Axioplan confocal microscope.
Immunoprecipitation
HEK 293T cells were transfected with the polyethylenimine (PEI) method. Forty-eight hours post transfection, cells were lysed in lysis buffer (50 mM Tris/HCl pH 7.5, 1 mM EDTA, 270 mM Sucrose, 1% (v/v) Triton X-100, 0.1% (v/v) 2-mercaptoethanol, and 60 mM n-Octyl-β-D-Glucopyranoside) containing protease inhibitor (Roche). Total protein was collected and incubated with anti-p75NTR (Alomone, ANT-007, 1 μg) overnight at 4 °C and then incubated with Sepharose protein-G beads (GE Healthcare). Samples were then prepared for immunoblotting as described below.
Immunoblotting
Immunoblotting protein samples were prepared for SDS-PAGE in SDS sample buffer (Life Technologies) and boiled at 95 °C for 10 min before electrophoresis on 12% gels. Proteins were transferred to PVDF membranes (Amersham). Membranes were blocked with 5% nonfat milk and incubated with primary antibodies. The following primary antibodies were used at the indicated dilutions: mouse anti-Myc (Roche, 1:3000), goat anti-p75NTR (Neuromics, GT15057, 1:3000), and anti-GAPDH (Sigma, G9545, 1:15,000). Immunoreactivity was visualized using appropriate HRP-conjugated secondary antibodies. Immunoblots were developed using the ECL Advance Western blotting detection kit (Life Technologies) and exposed to Kodak X-Omat AR films.
Statistical analysis
Data are expressed as mean and standard errors. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those generally used in the field. Following normality test and homogeneity variance (Kolmogorov–Smirnov test or Brown–Forsythe test), group comparison was made using one-way ANOVA followed by Turkey's post-hoc test. Differences were considered significant for p < 0.05. The experiments were not randomized. Data from all experiments are included; none were excluded.
Data availability
The atomic coordinates of p75NTR-DD:TRADD-DD have been deposited at the Protein Data Bank with accession number 7CSQ.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this manuscript.
Acknowledgments
We thank Dr Jing-song Fan for NMR data collection, and Miss Ket Yin Goh for technical assistance.
Author contributions
Z. Lin. conceptualization; N. Z. data curation; C. F. I. and Z. Lin. funding acquisition; N. Z., L. K., A. R., Z. Li., E. W. S., X. T., and W. Y. investigation; Z. Lin. project administration; C. F. I. and Z. Lin. supervision; L. K., C. F. I., and Z. Lin. writing-original draft; A. R. validation.
Funding and addditional information
This research was supported by funds from National Natural Science Foundation of China (Grant No. 21974093) to Z. L.; as well as Singapore National Medical Research Council (NMRC/CBRG/0107/2016), Singapore Ministry of Education (MOE2018-T2-1-129), and Swedish Research Council (VR-2016-01538) to C. F. I.
Edited by Wolfgang Peti
Supporting information
References
- 1.Chao M.V., Bothwell M.A., Ross A.H., Koprowski H., Lanahan A.A., Buck C.R., Sehgal A. Gene transfer and molecular cloning of the human NGF receptor. Science. 1986;232:518–521. doi: 10.1126/science.3008331. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D., Lanahan A., Buck C.R., Sehgal A., Morgan C., Mercer E., Bothwell M., Chao M. Expression and structure of the human NGF receptor. Cell. 1986;47:545–554. doi: 10.1016/0092-8674(86)90619-7. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Tebar A., Dechant G., Gotz R., Barde Y.A. Binding of neurotrophin-3 to its neuronal receptors and interactions with nerve growth factor and brain-derived neurotrophic factor. EMBO J. 1992;11:917–922. doi: 10.1002/j.1460-2075.1992.tb05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radeke M.J., Misko T.P., Hsu C., Herzenberg L.A., Shooter E.M. Gene transfer and molecular cloning of the rat nerve growth factor receptor. Nature. 1987;325:593–597. doi: 10.1038/325593a0. [DOI] [PubMed] [Google Scholar]
- 5.Ibanez C.F., Simi A. p75 neurotrophin receptor signaling in nervous system injury and degeneration: paradox and opportunity. Trends Neurosci. 2012;35:431–440. doi: 10.1016/j.tins.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Ernfors P., Henschen A., Olson L., Persson H. Expression of nerve growth factor receptor mRNA is developmentally regulated and increased after axotomy in rat spinal cord motoneurons. Neuron. 1989;2:1605–1613. doi: 10.1016/0896-6273(89)90049-4. [DOI] [PubMed] [Google Scholar]
- 7.Mobley W.C., Woo J.E., Edwards R.H., Riopelle R.J., Longo F.M., Weskamp G., Otten U., Valletta J.S., Johnston M.V. Developmental regulation of nerve growth factor and its receptor in the rat caudate-putamen. Neuron. 1989;3:655–664. doi: 10.1016/0896-6273(89)90276-6. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt S., Shooter E.M., Davies A.M. Expression of the NGF receptor gene in sensory neurons and their cutaneous targets prior to and during innervation. Neuron. 1990;4:421–427. doi: 10.1016/0896-6273(90)90054-j. [DOI] [PubMed] [Google Scholar]
- 9.Demir I.E., Tieftrunk E., Schorn S., Friess H., Ceyhan G.O. Nerve growth factor & TrkA as novel therapeutic targets in cancer. Biochim. Biophys. Acta. 2016;1866:37–50. doi: 10.1016/j.bbcan.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Yuan W., Ibanez C.F., Lin Z. Death domain of p75 neurotrophin receptor: A structural perspective on an intracellular signalling hub. Biol. Rev. 2019;94:1282–1293. doi: 10.1111/brv.12502. [DOI] [PubMed] [Google Scholar]
- 11.Liepinsh E., Ilag L.L., Otting G., Ibanez C.F. NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J. 1997;16:4999–5005. doi: 10.1093/emboj/16.16.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Y., Cao P., Yu H.J., Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454:789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 13.Vilar M., Charalampopoulos I., Kenchappa R.S., Simi A., Karaca E., Reversi A., Choi S., Bothwell M., Mingarro I., Friedman W.J., Schiavo G., Bastiaens P.I., Verveer P.J., Carter B.D., Ibanez C.F. Activation of the p75 neurotrophin receptor through conformational rearrangement of disulphide-linked receptor dimers. Neuron. 2009;62:72–83. doi: 10.1016/j.neuron.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charalampopoulos I., Vicario A., Pediaditakis I., Gravanis A., Simi A., Ibanez C.F. Genetic dissection of neurotrophin signaling through the p75 neurotrophin receptor. Cell Rep. 2012;2:1563–1570. doi: 10.1016/j.celrep.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Vicario A., Kisiswa L., Tann J.Y., Kelly C.E., Ibanez C.F. Neuron-type-specific signaling by the p75NTR death receptor is regulated by differential proteolytic cleavage. J. Cell Sci. 2015;128:1507–1517. doi: 10.1242/jcs.161745. [DOI] [PubMed] [Google Scholar]
- 16.Natoli G., Austenaa L.M. A birthday gift for TRADD. Nat. Immunol. 2008;9:1015–1016. doi: 10.1038/ni0908-1015. [DOI] [PubMed] [Google Scholar]
- 17.Tsao D.H., McDonagh T., Telliez J.B., Hsu S., Malakian K., Xu G.Y., Lin L.L. Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway. Mol. Cell. 2000;5:1051–1057. doi: 10.1016/s1097-2765(00)80270-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N., Yuan W., Fan J.S., Lin Z. Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily. Sci. Rep. 2017;7:7073. doi: 10.1038/s41598-017-07348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu H., Xiong J., Goeddel D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 20.El Yazidi-Belkoura I., Adriaenssens E., Dolle L., Descamps S., Hondermarck H. Tumor necrosis factor receptor-associated death domain protein is involved in the neurotrophin receptor-mediated antiapoptotic activity of nerve growth factor in breast cancer cells. J. Biol. Chem. 2003;278:16952–16956. doi: 10.1074/jbc.M300631200. [DOI] [PubMed] [Google Scholar]
- 21.Kisiswa L., Fernandez-Suarez D., Sergaki M.C., Ibanez C.F. RIP2 gates TRAF6 interaction with death receptor p75(NTR) to regulate cerebellar granule neuron survival. Cell Rep. 2018;24:1013–1024. doi: 10.1016/j.celrep.2018.06.098. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Suarez D., Krapacher F.A., Andersson A., Ibanez C.F., Kisiswa L. MAG induces apoptosis in cerebellar granule neurons through p75(NTR) demarcating granule layer/white matter boundary. Cell Death Dis. 2019;10:732. doi: 10.1038/s41419-019-1970-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanin J.P., Verpeut J.L., Li Y., Shiflett M.W., Wang S.S., Santhakumar V., Friedman W.J. The p75NTR influences cerebellar circuit development and adult behavior via regulation of cell cycle duration of granule cell progenitors. J. Neurosci. 2019;39:9119–9129. doi: 10.1523/JNEUROSCI.0990-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M., Liu J., Ran X., Fang M., Shi J., Qin H., Goh J.M., Song J. Resurrecting abandoned proteins with pure water: CD and NMR studies of protein fragments solubilized in salt-free water. Biophys. J. 2006;91:4201–4209. doi: 10.1529/biophysj.106.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N., Yuan W., Fan J.S., Lin Z. (1)H, (15)N and (13)C chemical shift assignments of the C-terminal domain of TRADD. Biomol. NMR Assign. 2017;11:281–284. doi: 10.1007/s12104-017-9763-6. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z., Tann J.Y., Goh E.T., Kelly C., Lim K.B., Gao J.F., Ibanez C.F. Structural basis of death domain signaling in the p75 neurotrophin receptor. Elife. 2015;4 doi: 10.7554/eLife.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilar M., Sung T.C., Chen Z., Garcia-Carpio I., Fernandez E.M., Xu J., Riek R., Lee K.F. Heterodimerization of p45-p75 modulates p75 signaling: Structural basis and mechanism of action. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pobezinskaya Y.L., Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11:871–876. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledoux A.C., Perkins N.D. NF-kappaB and the cell cycle. Biochem. Soc. Trans. 2014;42:76–81. doi: 10.1042/BST20130156. [DOI] [PubMed] [Google Scholar]
- 30.Zanin J.P., Abercrombie E., Friedman W.J. Proneurotrophin-3 promotes cell cycle withdrawal of developing cerebellar granule cell progenitors via the p75 neurotrophin receptor. Elife. 2016;5 doi: 10.7554/eLife.16654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D., Zhao H., Jin M., Zhu H., Shan B., Geng J., Dziedzic S.A., Amin P., Mifflin L., Naito M.G., Najafov A., Xing J., Yan L., Liu J., Qin Y. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature. 2020;587:133–138. doi: 10.1038/s41586-020-2757-z. [DOI] [PubMed] [Google Scholar]
- 32.Park Y.C., Ye H., Hsia C., Segal D., Rich R.L., Liou H.C., Myszka D.G., Wu H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell. 2000;101:777–787. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- 33.Bittner S., Ehrenschwender M. Multifaceted death receptor 3 signaling-promoting survival and triggering death. FEBS Lett. 2017;591:2543–2555. doi: 10.1002/1873-3468.12747. [DOI] [PubMed] [Google Scholar]
- 34.Khursigara G., Bertin J., Yano H., Moffett H., DiStefano P.S., Chao M.V. A prosurvival function for the p75 receptor death domain mediated via the caspase recruitment domain receptor-interacting protein 2. J. Neurosci. 2001;21:5854–5863. doi: 10.1523/JNEUROSCI.21-16-05854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J.Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 36.Johnson B.A., Blevins R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 37.Lin Z., Xu Y., Yang S., Yang D. Sequence-specific assignment of aromatic resonances of uniformly 13C,15N-labeled proteins by using 13C- and 15N-edited NOESY spectra. Angew. Chem. Int. Ed. 2006;45:1960–1963. doi: 10.1002/anie.200503558. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y., Lin Z., Ho C., Yang D. A general strategy for the assignment of aliphatic side-chain resonances of uniformly 13C,15N-labeled large proteins. J. Am. Chem. Soc. 2005;127:11920–11921. doi: 10.1021/ja053539b. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y., Long D., Yang D. Rapid data collection for protein structure determination by NMR spectroscopy. J. Am. Chem. Soc. 2007;129:7722–7723. doi: 10.1021/ja071442e. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 42.Case D.A., Cheatham T.E., 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Lee K.F., Davies A.M., Jaenisch R. p75-deficient embryonic dorsal root sensory and neonatal sympathetic neurons display a decreased sensitivity to NGF. Development. 1994;120:1027–1033. doi: 10.1242/dev.120.4.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of p75NTR-DD:TRADD-DD have been deposited at the Protein Data Bank with accession number 7CSQ.