Abstract
Objectives
This systematic review focuses on the use of the in vitro hollow fibre infection model (HFIM) for microbial culture. We summarize the direction of the field to date and propose best-practice principles for reporting of the applications.
Methods
Searches in six databases (MEDLINE®, EMBASE®, PubMed®, BIOSIS®, SCOPUS® and Cochrane®) up to January 2020 identified 129 studies meeting our inclusion criteria. Two reviewers independently assessed and extracted data from each publication. The quality of reporting of microbiological and technical parameters was analysed.
Results
Forty-seven out of 129 (36.4%) studies did not report the minimum pharmacokinetic parameters required in order to replicate the pharmacokinetic profile of HFIM experiments. Fifty-three out of 129 (41.1%) publications did not report the medium used in the HFIM. The overwhelming majority of publications did not perform any technical repeats [107/129 (82.9%)] or biological repeats [97/129 (75.2%)].
Conclusions
This review demonstrates that most publications provide insufficient data to allow for results to be evaluated, thus impairing the reproducibility of HFIM experiments. Therefore, there is a clear need for the development of laboratory standardization and improved reporting of HFIM experiments.
Introduction
The hollow fibre infection model (HFIM) is an in vitro system that offers a solution to culturing cells continuously at high density with flexibility and reproducibility.1 Applications range from the propagation of cell lines and the production of monoclonal antibodies and recombinant proteins to mimicking long-term physiologically relevant in vivo profiles.2–5 The HFIM is a preclinical closed system that allows the culturing of microbial cultures in an enclosed compartment. This compartment is usually a discrete cartridge that in turn is threaded with semi-permeable fibres.6,7 These fibres are attached to a circuit connecting to a central reservoir where the contents are rapidly circulated via a pump and nutrients and drugs equilibrate freely between the central reservoir circuit and the inoculum-containing compartment of the cartridge (Figure 1). Fresh medium is supplemented into the central compartment at a fixed rate, with central compartment contents removed via a pump at an identical rate. Through this, the drug in the central compartment is cleared at a rate determined by the supplementation rate. Adjustment of input/output rates allows simulation of clearance of the drug(s) added to the central reservoir, mimicking pharmacokinetic (PK) profiles seen in vivo. Samples can be taken from the enclosed compartment of the hollow fibre cartridge for quantification of the bacterial inoculum and determination of antimicrobial resistance. Samples can be taken from the central compartment to quantify the concentration of drug via bioanalysis to confirm recapitulation of the mimicked PK profile.
Figure 1.
Schematic of the HFIM. This schematic shows the hollow fibre compartment model. The hollow fibres in the cartridge are attached to a circuit connecting to a central reservoir (shown in blue). Test organisms are retained in the hollow fibre cartridge. The contents of the central reservoir can be topped up with fresh medium from the diluent compartment (shown on the left) and through the use of a waste removal tube the volume of the central reservoir is kept constant. Drug is administered directly into the central reservoir through the diluent tubing. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Over recent years, the HFIM has been increasingly used to characterize in vitro pharmacodynamics (PD) of antimicrobial agents and to determine PK-PD indices, such as AUC/MIC, T>MIC and Cmax/MIC.8,9 The model allows physiological PK drug profiles to be simulated with serial evaluation of the interplay between drug exposure and microbial response. In contrast, conventional in vitro infection models have generally relied on static time–kill experiments or dynamic one-compartment models with the microbial load in the central compartment.10,11 These latter models mimic drug concentrations by supplementation and removal of medium, such as in the HFIM, but cause continual dilution of the bacterial population. Furthermore, drug concentrations may be inaccurate due to difficulties of bioanalysis in infected medium and consumption of the drug by the microbial load.12,13 In contrast, in an HFIM, the microbial cell cultures remain contained in a separated compartment, whilst mimicking physiological PK profiles, allowing for accurate determination of drug concentration and no continual dilution of bacteria.14,15
The HFIM also has advantages over in vivo animal models. These in vivo models, particularly the thigh infection model, are well established and frequently used as preclinical models for anti-infective drug development.16–18 The advantages and disadvantages of both methods are well documented in publications; here we highlight the main advantages of the HFIM. Importantly, the life of an animal limits the duration of animal experiments and multiple animals need to be offered in each arm for each timepoint.19 Therefore, animal experiments are rarely long enough to quantify the emergence of resistance; however, the HFIM allows for higher sampling frequency over longer time periods, which enables the understanding of the PD of the emergence of resistance.20–23 In vivo animal models are also limited to bacterial loads that the animals can maintain over a period of time. In contrast the HFIM allows for high bacterial inocula to be maintained without issue. Furthermore, in vivo animal models rely on humanized doses, whilst needing to account for differences in elimination and distribution characteristics, such as metabolism and protein binding, whereas any PK parameter can be quite precisely mimicked in the HFIM.24 Animal models raise ethical concerns and require ethical approval of the experimental protocol, whereas culturing microbes in the HFIM does not. Ultimately, the HFIM provides more flexibility in experimental design and sampling.
Whilst there are established CLSI guidelines for static time–kill experiments, no comprehensive recommendations or standards (CLSI or otherwise) for performance of hollow fibre microbial experiments exist.25,26 Laboratory manuals are available, but consensus guidelines are currently lacking.27,28 Furthermore, the current literature lacks a systematic review detailing the experimental details of HFIM applications reported in original research papers. This, along with the high running costs and infrastructure requirements, forms a barrier to entry for groups new to the field, thus limiting the evaluation and reproducibility of published results. The aim of this work was therefore to undertake a systematic review of HFIM publications, describing the current state of the art with the objective of reviewing the reporting of data.
Methods
We included publications that reported use of the HFIM, describing the experimental aims as well as the methodology and outcomes. This review represents the direction of the field of HFIM research to date and focuses on in vitro infection models for microbial cell culture with the following objectives: (i) to describe the primary aims of the use of hollow fibre systems; (ii) to evaluate the experimental settings and parameter reporting; and (iii) to evaluate the microbiological outcome measure reporting.
Search strategy
PRISMA 2020 guidelines29 were followed and the following databases were searched for relevant records: MEDLINE®, EMBASE®, PubMed®, BIOSIS®, SCOPUS® and Cochrane®. The search strategy included three concepts: microbes, antimicrobials and the hollow fibre system. A detailed breakdown of the PICO framework alongside search concepts can be found in Table S1 (available as Supplementary data at JAC Online). All six databases were searched with our predefined search terms and our search strategy for each database is captured in Table S2. We included records published in English from January 1980 to January 2020.
Inclusion and exclusion criteria
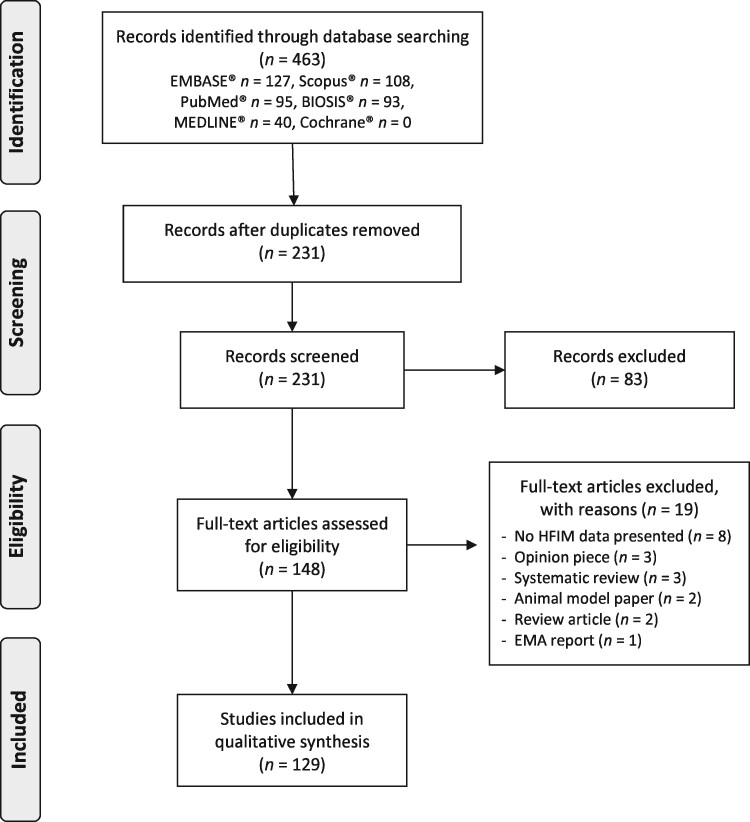
Search results were de-duplicated in the referencing software Mendeley© and all unique records were screened for the relevance of their title and abstract. We defined the inclusion criteria as records that: (i) presented primary hollow fibre data; (ii) studied microorganisms; and (iii) studied antimicrobials. Publications were excluded at the screening stage if their title and abstract did not meet the inclusion criteria. This included studies that did not use the HFIM, did not investigate microbial species and did not investigate antimicrobials. Screened abstracts were then assessed for their full-text eligibility by two independent reviewers in the data extraction phase. At full-text screening further papers were excluded if they didn’t fulfil the inclusion/exclusion criteria. All records excluded from the review at full-text screening are presented in the PRISMA flowchart diagram with details of their exclusion (Figure 2).
Figure 2.
PRISMA flowchart outlining the flow of information through the different phases of the systematic review, including the number of records identified, included and excluded.
Data extraction
Data extraction was performed on full-text articles in duplicate by two independent authors, Z.S. extracted data from all publications and the duplicate extractions were shared amongst all collaborating authors. A password-protected Rshiny form was designed using shiny version 1.4.0 to enable robust data extraction.30 The form was separated into three sections: (i) descriptive analysis of the records; (ii) engineering parameters; and (iii) the microbiological parameters. We defined the engineering parameters as settings or equipment that are not directly related to the microbiological measurements, but which impact the experimental dynamics. These included settings that determine the PK profile to be mimicked (e.g. pump settings and PK parameters), cartridge/fibre type and the tubing length and bore size. The microbiological section included parameters that influence the growth of the microorganisms or measures that arise from the sampling of the microorganism to determine the PD response to the antimicrobial. Technical repeats were defined as repeat testing of endpoints, for example bacterial quantification or MIC. Biological repeats were defined as repeat experiments of the organism investigated. Details of the parameters extracted alongside the definitions we used can be viewed in Table S3.
Results
The results are separated into four sections: (i) PRISMA flowchart; (ii) a descriptive analysis of records included; (iii) engineering parameters; and (iv) the microbiological parameters.
Section one: PRISMA flowchart
A PRISMA flowchart (Figure 2) outlines the number of publications at each stage of the systematic review.
Following deduplication, a total of 231 independent publications for title and abstract screening remained. Of these 83 were excluded and 148 were eligible for full-text screening. Upon full-text screening, 19 records were excluded and a total of 129 records were included in the data extraction and analysis.
Section two: Description of records included
The three most common aims of the HFIM experiments were investigating drug combinations [38.8% (50/129)], investigating antimicrobial resistance [26.4% (34/129)] and determining PK-PD indices [21.7% (28/129)] (Table 1). A few publications modelled intravascular time course [3.9% (5/129)] and the remaining 9.3% (12/129) ranged from investigating intracellular pathogens to investigating PK assay development. Further information and breakdown of the microbial species and antimicrobials investigated in the publications included is provided in Table S4.
Table 1.
HFIM publication aims frequency
| Primary publication aims | Number | Percentage |
|---|---|---|
| Drug combinations | 50 | 38.8 |
| Antimicrobial resistance | 34 | 26.4 |
| Determining PK-PD indices (drug development) | 28 | 21.7 |
| Modelling intravascular time course | 5 | 3.9 |
| Intracellular pathogens | 4 | 3.1 |
| Determine conditions for drug production | 2 | 1.6 |
| Dose finding | 2 | 1.6 |
| Other | 4 | 3.1 |
| Total | 127 | 100 |
Section three: Technical specifications
Simulating a PK profile is achieved by adjusting the HFIM settings, namely the pump settings. Technical settings play a vital role for interpreting the results and enabling reproducibility. Cmax, Tmax and t½ are required in order to replicate a PK profile of an extravascularly administered drug or intravenously administered drug by infusion (in which case Tmax is usually the infusion duration). For intravenous bolus administration only Cmax and t½ are required.
Twenty-seven out of 129 (20.9%) publications did not report any PK parameters. Cmax and t½ were the most reported PK parameters with 37.0% and 36.6% of total publications reporting them, respectively. Antimicrobial dose mimicked was reported by 45.7% of publications. Eighty-two out of 129 (63.6%) reported both Cmax and t½ - the minimum required to replicate the PK profile of intravenous bolus administration. Only one publication reported all of Cmax, Tmax and t½, the minimum required to replicate the PK profile of drugs administered extravascularly, or intravascularly via infusion. In summary, 47/129 (36.4%) publications did not meet the minimum criteria for reproducibility of PK profiles (Figure 3).
Figure 3.
Summary of PK parameter reporting in HFIM studies, presented in table format (a) and as a Euler proportional diagram (b).
Seventy-two out of 129 (55.8%) publications reported the manufacturer of the cartridge used in the hollow fibre experiments and 37/129 (28.7%) publications reported the type of fibre used by either reporting the cartridge catalogue number or explicitly mentioning the fibre (e.g. cellulosic, polysulfone or polypropylene). Only 6/129 (4.7%) publications reported monitoring of pH in the system. Bore size of the tubing used was reported by 3/129 (2.3%) publications. No publications reported the length of the tubing used in any part of the HFIM system (Table 2).
Table 2.
Percentage and raw number of publications reporting hollow fibre settings
| Hollow fibre setting | % Reported (n) | % Not reported (n) | % Not applicable (n) |
|---|---|---|---|
| Cartridge source | 55.8 (72) | 44.2 (57) | 0.0 (0) |
| Fibre type | 28.7 (37) | 71.3 (92) | 0.0 (0) |
| Pump dynamics | 6.2 (8) | 93.0 (120) | 0.8 (1) |
| pH | 4.7 (6) | 94.6 (122) | 0.8 (1) |
| Tubing bore size | 2.3 (3) | 96.9 (125) | 0.8 (1) |
| Tubing length | 0 (0) | 99.2 (128) | 0.8 (1) |
| Dose administration | 74.4 (96) | 24.8 (32) | 0.8 (1) |
| Dose mimicked | 45.7 (59) | 54.3 (70) | 0 (0) |
Section four: Microbiology
One hundred and eighteen out of 129 (91.5%) publications reported the results for a control experiment—often an antimicrobial-free growth control. Seventy-five out of 129 (58.2%) publications reported the medium used in the HFIM. None of the 129 publications reported sampling and monitoring for contamination. One hundred and eight out of 129 publications (83.7%) quantified the bacterial population by cfu to determine the antimicrobial killing effect. Only 6/129 (4.7%) measured markers of cell viability beyond growth (e.g. using flow cytometry). Antimicrobial resistance was measured and reported by 95/129 (73.6%) publications; however, only 27/129 (20.9%) publications reported genotypic analysis of samples taken from the hollow fibre cartridge (Table 3).
Table 3.
Percentage and raw number reporting of microbiological outcome measures
| Outcome measure | % Reported (n) | % Not reported (n) | % Not applicable (n) |
|---|---|---|---|
| Medium | 58.14 (75) | 41.09 (53) | 0.78 (1) |
| Contamination | 0.0 (0) | 99.22 (128) | 0.78 (1) |
| Control | 91.47 (118) | 5.43 (7) | 3.10 (4) |
| Inoculum | 76.74 (99) | 20.16 (26) | 3.10 (4) |
| cfu | 83.72 (108) | 13.95 (18) | 2.33 (3) |
| Viability | 4.65 (6) | 91.47 (118) | 3.88 (5) |
| Resistance | 73.64 (95) | 23.26 (30) | 3.10 (4) |
| Genotyping | 20.93 (27) | 75.97 (98) | 3.10 (4) |
The overwhelming majority of publications did not perform any technical repeats [107/129 (82.9%)] or biological repeats [97/129 (75.2%)] (Table 4). Duplicate testing (i.e. one original test and one repeat test) were performed by 18/129 (14.0%) publications for technical repeats and by 23/129 (17.8%) publications for biological repeats. Four publications (3.1%) reported technical repeats in triplicate and nine publications (7.0%) reported biological repeats in triplicate.
Table 4.
Percentage and raw number of publications reporting repeat testing in HFIM
| Number of repeats | % Technical repeats (n) | % Biological repeats (n) |
|---|---|---|
| Single | 82.9 (107) | 75.2 (97) |
| Duplicate | 14.0 (18) | 17.8 (23) |
| ≥Triplicate | 3.1 (4) | 7.0 (9) |
Discussion
To the best of our knowledge, this is the first systematic review focusing on the reproducibility of the microbial applications of the HFIM. In this systematic review of in vitro hollow fibre PK-PD studies of antimicrobials, we found wide variability in reporting. Most studies did not provide enough information for their results to allow comparison, reproduction or modification of HFIM studies. This review demonstrates that the reproducibility of published HFIM work remains impaired by the practice of inadequate reporting of the hollow fibre settings. We found many publications outsourced their methodology to previously published papers.31–34 This practice leaves too much interpretation and creates a further barrier for the sourcing of important information, as the specifics are often under several layers of prior publications with the referenced publications often in a different context. We would recommend that study design and methodology is restated in each individual paper to provide clarity. A systematic review of clinical PK-PD methodologies reported similar findings in lack of conduct and reporting, further highlighting the need for the field to standardize reporting.35
Future HFIM studies should be designed and reported carefully so that confidence can be given to the relationships (or lack of) demonstrated by these studies. This would improve clarity in the field and eliminate ambiguity in interpretation of results. This systematic review further highlights the need for laboratory methodology standards to be developed for the use of the HFIM and has identified the key areas of inconsistency and we believe there is a need for a consensus standard checklist for minimum reporting. We have identified criteria we consider necessary for other researchers to be able to evaluate and reproduce an experiment, which we offer as a recommendation for reporting HFIM experiments (Table 5). We hope these recommendations will spark interest in setting a common standard to all HFIM studies investigating antimicrobial activity, similar to antimicrobial susceptibility guidelines (e.g. CLSI and EUCAST). These standards of reporting will also help reduce the barrier to entry for new laboratories setting up HFIM experiments. These recommendations can also pave the way for further conversations that the field could delve into. For instance, exploring thresholds for varying degrees of observed antimicrobial activity.
Table 5.
Recommendations for suggested reporting of specifications in the HFIM experiments
| Section | HFIM feature | Further explanation |
|---|---|---|
| Descriptive specifications | primary study aim | main research question of the HFIM study |
| microbial species | microbial species inoculated into the hollow fibre cartridge | |
| antimicrobial(s) | antimicrobial(s) administered to the HFIM system | |
| duration | duration of the HFIM experiment in days | |
| Technical specifications | mimicked dosea | dose being mimicked in the HFIM system in mg/L |
| C max | peak concentration of antimicrobial(s) in mg/L | |
| t ½ | elimination half-life | |
| T max a | time taken to reach Cmax in h | |
| C min a | lowest concentration of the drug in a dosing interval | |
| Τ | time between antimicrobial dose administrations | |
| cartridge source | manufacturer of the cartridge used and catalogue number | |
| fibre type | cartridge fibre type (e.g. cellulosic, polysulfone or polypropylene) | |
| flow rate | rate between the central compartment and the hollow fibre cartridge and rate from the diluent compartment | |
| pump model | pump models used | |
| tubinga | bore size and length of tubing used in the central compartment | |
| administration | route and details of administration, e.g. bolus or infusion, with details of rate and volume | |
| Microbiological specifications | medium | name and manufacturer of medium used in the HIFM |
| control | details of control experiment, e.g. drug-free arm | |
| contamination | measurement of sterility in the cartridge and the central compartment | |
| inoculum | method used to determine inoculum quantification stated | |
| cfu | cfu of the microbe quantified from the cartridge sampling | |
| viabilitya | cell viability markers beyond cfu (e.g. flow cytometry) | |
| resistancea | resistance of microbial sample phenotypically quantified | |
| genotypinga | molecular testing of the microbial sample | |
| biological repeat | number of repeat testing of single microbial species | |
| technical repeat | number of repeat testing of endpoint measures, e.g. cfu |
Additional parameters not required for minimal reporting.
We appreciate reporting of some parameters may be challenging and therefore propose some solutions.
For example, suitability of cartridge types (cellulosic, polysulfone etc.) for specific antimicrobials may not be available. For these situations, we suggest it is more important to check equilibration and binding for the chosen cartridge fibre type in these novel studies. We appreciate, with a wide variety of pump brands, that reporting the specific pump model and settings could get complex. We propose a standardized method of reporting flow rates (e.g. mL/min) that were used in the experiment that can be replicated by others. Factors that affect the time taken for equilibrium to be reached between the central reservoir and hollow fibre compartment should be reported, as often there can be a significant delay between input of the antimicrobial to the central reservoir and distribution to the cartridge. For example, the volume of the dose administered relative to the volume of the central compartment, or the flow rate of the dose infused relative to the volume of the central compartment, is also a critical parameter that affects the attainment of Cmax at Tmax. Although tubing length from the drug medium and waste compartment is not important, the length and bore size of tubing between the central compartment and the cartridge has an impact on the distribution and equilibration rate of the drug.
As the HFIM can be used to understand the dynamic microbial response to antimicrobials, wherever possible we should take the opportunity to undertake further analyses of the microbial population sampled, for example genetic sequencing, transcriptomics or flow cytometry. We understand the cost of performing biological replicates for HFIM experiments is high, with several cartridges required per experiment. In instances where repurposed cartridges (e.g. dialysis fibres) are used instead of proprietary cartridges on cost grounds we strongly advise reporting this. In addition, cartridge pore size should also be reported; this can be captured by manufacturer and catalogue number. Further to this, we appreciate that for slow-growing organisms, such as mycobacteria, performing biological repeats under time constraints may be challenging. In these instances, we suggest performing static time–kill experiments in triplicate, to build a model hypothesis that can be simulated in the HFIM.
Conclusions
This systematic review found wide variability in reporting, with most HFIM studies not providing sufficient information for their results to be evaluated. This creates difficulty in data comparison and reproducibility of studies. We believe there is scope for developing standards of reporting widely accepted as the recommendation for future HFIM studies.
Funding
This systematic review was performed as part of Z.S.'s PhD studentship that was partially funded by an educational grant from Shionogi B.V. and by the University College London Institute for Global Health (IGH) and Centre for Clinical Microbiology (CCM). F.K. was supported by a United Kingdom Medical Research Council (MRC) Fellowship (Grant Number P014534) and a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 220587/Z/20/Z).
Transparency declarations
C.L. is an employee of Shionogi Europe and D.M. is an employee of QIAGEN and holds QIAGEN shares. All other authors: none to declare.
Supplementary data
Tables S1 to S4 are available as Supplementary data at JAC Online.
Supplementary Material
References
- 1.Cadwell JJS, Whitford WG, Three‐dimensional cell‐based assays in hollow fibre bioreactors. In: Technology Platforms for 3D Cell Culture. 2017; 327–50.
- 2.Handa-Corrigan A, Nikolay S, Fletcher D et al. Monoclonal antibody production in hollow-fiber bioreactors: process control and validation strategies for manufacturing industry. Enzyme Microb Technol 1995; 17: 225–30. [Google Scholar]
- 3.Gorter A, van de Griend RJ, van Eendenburg JDH et al. Production of bi-specific monoclonal antibodies in a hollow-fibre bioreactor. J Immunol Methods 1993; 161: 145–50. [DOI] [PubMed] [Google Scholar]
- 4.Chu L, Robinson DK. Industrial choices for protein production by large-scale cell culture. Curr Opin Biotechnol 2001; 12: 180–7. [DOI] [PubMed] [Google Scholar]
- 5.Ryll T, Lucki-Lange M, Jäger V et al. Production of recombinant human interleukin-2 with BHK cells in a hollow fibre and a stirred tank reactor with protein-free medium. J Biotechnol 1990; 14: 377–92. [DOI] [PubMed] [Google Scholar]
- 6.Griffith DC, Sabet M, Tarazi Z et al. Pharmacokinetics/pharmacodynamics of vaborbactam, a novel β-lactamase inhibitor, in combination with meropenem. Antimicrob Agents Chemother 2018; 63: e01659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose W, In vitro pharmacodynamic models to evaluate anti-infective pharmacodynamics. In: Rotschafer JC, Andes DR, Rodvold K, eds. Antibiotic Pharmacodynamics 2016; 29–57. [Google Scholar]
- 8.Singh R, Almutairi M, Alm RA et al. Ceftaroline efficacy against high-MIC clinical Staphylococcus aureus isolates in an in vitro hollow-fibre infection model. J Antimicrob Chemother 2017; 72: 2796–803. [DOI] [PubMed] [Google Scholar]
- 9.VanScoy BD, Trang M, McCauley J et al. Pharmacokinetics-pharmacodynamics of a novel β-lactamase inhibitor, CB-618, in combination with meropenem in an in vitro infection model. Antimicrob Agents Chemother 2016; 60: 3891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasso S, Meinardi G, De Carneri I et al. New in vitro model to study the effect of antibiotic concentration and rate of elimination on antibacterial activity. Antimicrob Agents Chemother 1978; 13: 570–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haag R, Lexa P, Werkhauser I. Artifacts in dilution pharmacokinetic models caused by adherent bacteria. Antimicrob Agents Chemother 1986; 29: 765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaumann R, Goldstein EJC, Forberg J et al. Activity of moxifloxacin against Bacteroides fragilis and Escherichia coli in an in vitro pharmacokinetic/pharmacodynamic model employing pure and mixed cultures. J Med Microbiol 2005; 54: 749–53. [DOI] [PubMed] [Google Scholar]
- 13.Rustige C, Wiedemann B. Antibacterial activity of lomefloxacin in a pharmacokinetic in vitro model. Antimicrob Agents Chemother 1990; 34: 1107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knazek RA, Gullino PM, Kohler PO et al. Cell culture on artificial capillaries: an approach to tissue growth in vitro. Science 1972; 178: 65–7. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd JR, Hirst TR, Bunch AW. Hollow-fibre bioreactors compared to batch and chemostat culture for the production of a recombinant toxoid by a marine Vibrio. Appl Microbiol Biotechnol 1997; 48: 155–61. [DOI] [PubMed] [Google Scholar]
- 16.Dandekar PK, Tessier PR, Williams P et al. Pharmacodynamic profile of daptomycin against Enterococcus species and methicillin-resistant Staphylococcus aureus in a murine thigh infection model. J Antimicrob Chemother 2003; 52: 405–11. [DOI] [PubMed] [Google Scholar]
- 17.Zuluaga AF, Agudelo M, Cardeño JJ et al. Determination of therapeutic equivalence of generic products of gentamicin in the neutropenic mouse thigh infection model. PLoS One 2010; 5: e10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertlein T, Sturm V, Kircher S et al. Visualization of abscess formation in a murine thigh infection model of Staphylococcus aureus by 19F-magnetic resonance imaging (MRI). PLoS One 2011; 6: e18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velkov T, Bergen PJ, Lora-Tamayo J et al. PK/PD models in antibacterial development. Curr Opin Microbiol 2013; 16: 573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos-Martin V, Johnson A, McEntee L et al. Pharmacodynamics of teicoplanin against MRSA. J Antimicrob Chemother 2017; 72: 3382–9. [DOI] [PubMed] [Google Scholar]
- 21.Ramos-Martin V, Johnson A, Livermore J et al. Pharmacodynamics of vancomycin for CoNS infection: experimental basis for optimal use of vancomycin in neonates. J Antimicrob Chemother 2016; 71: 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ly NS, Bulitta JB, Rao GG et al. Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J Antimicrob Chemother 2015; 70: 1434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Wang L, Zhang XJ et al. Evaluation of meropenem regimens suppressing emergence of resistance in Acinetobacter baumannii with human simulated exposure in an in vitro intravenous-infusion hollow-fiber infection model. Antimicrob Agents Chemother 2014; 58: 6773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulitta JB, Hope WW, Eakin AE et al. Generating robust and informative nonclinical in vitro and in vivo bacterial infection model efficacy data to support translation to humans. Antimicrob Agents Chemother 2019; 63: e02397-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CLSI. Methods for Determining Bactericidal Activity of Antimicrobial Agents: M26. 1999.
- 26.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100. 2020.
- 27.Hollow Fiber System of Tuberculosis (HFS-TB): A Laboratory Manual to Guide System Engineering, Study Design and Execution. 2018; 1–30.
- 28.EMA. Qualification Opinion: In-Vitro Hollow Fiber System Model of Tuberculosis (HSF-TB). 2015; 1–9.
- 29.Page MJ, McKenzie JE, Bossuyt PM et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021; 372: n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang W, Cheng J, Allaire JJ et al. shiny: Web Application Framework for R. Version 1.4.0. 2019.
- 31.Hirsch EB, Ledesma KR, Chang KT et al. In vitro activity of MK-7655, a novel β-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 2012; 56: 3753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuji BT, Brown T, Parasrampuria R et al. Front-loaded linezolid regimens result in increased killing and suppression of the accessory gene regulator system of Staphylococcus aureus. Antimicrob Agents Chemother 2012; 56: 3712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao M, Bulman ZP, Lenhard JR et al. Pharmacodynamics of colistin and fosfomycin: a ‘treasure trove’ combination combats KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 2017; 72: 1985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith NM, Bulman ZP, Sieron AO et al. Pharmacodynamics of dose-escalated ‘front-loading’ polymyxin B regimens against polymyxin-resistant mcr-1-harbouring Escherichia coli. J Antimicrob Chemother 2017; 72: 2297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAleenan A, Ambrose PG, Bhavnani SM et al. Methodological features of clinical pharmacokinetic-pharmacodynamic studies of antibacterials and antifungals: a systematic review. J Antimicrob Chemother 2020; 75: 1374–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.