SUMMARY
The objective of this study was to evaluate the association between aberrant right subclavian artery (ARSA) and chromosomal abnormalities. The study included 5211 women having attended our unit for fetal anatomic screening and fetal echocardiography from August 2016 until February 2019. After diagnosing ARSA, prenatal invasive testing was discussed with the patients. ARSA affected fetus was determined in 57 cases; of these, there were 38 cases of isolated ARSA and 19 cases of non-isolated ARSA but associated with soft markers and fetal anomalies. Nineteen patients underwent amniocentesis; Down syndrome was determined in two women, both of them from the non-isolated ARSA group, with fetal hydrops, atrioventricular septal defect and esophageal atresia. Fifteen of 38 patients who declined prenatal diagnostic testing, accepted karyotype analysis after delivery and none of these 15 cases had chromosomal abnormalities. Identification of ARSA should be followed by detailed ultrasound examination to ensure that there are no accompanying soft markers and/or structural defects. Isolated ARSA may not be an indication for karyotype analysis or 22q11.2 microdeletions. Non-ARSA implies a strong predictor of aneuploidy, and when additional findings are detected, invasive testing should be offered to the parents. The association between isolated ARSA and genetic disease should be evaluated in large powered prospective studies.
Key words: Aberrant right subclavian artery, 22q11.2 microdeletion, Aneuploidy, Trisomy 21, Prenatal diagnosis
Introduction
Aberrant right subclavian artery (ARSA) is identified by prenatal ultrasound scan in some fetuses. ARSA originates from the aortic arch distally instead of brachiocephalic artery as the fourth supra-aortic vessel and follows a retrotracheal course towards the right arm in the transverse 3-vessel tracheal view. The importance of this finding has been a source of debate in recent years, as ARSA is claimed to be a marker of some chromosomal abnormalities. ARSA is found in approximately 1%-2% of healthy people according to autopsy series (1). Its prevalence is estimated to range from 2.9% to 100% in cases of Down syndrome (2, 3). Besides, ARSA is also considered to be associated with 22q11.2 deletions (4). However, evidence is insufficient regarding the association between ARSA and these chromosomal conditions. Thus, counseling couples to perform further investigation with invasive prenatal diagnostic tests may be a difficult task in clinical practice.
A meta-analysis examining second trimester soft markers for Down syndrome reports on the positive and negative likelihood ratio (LR) of ARSA for trisomy 21 of 21.4 and 0.71, respectively, and the estimated LR for isolated ARSA of 3.94 (5). A study including patients with ARSA undergoing microarray analysis did not find significant differences between ARSA and Down syndrome or 22q11.2 deletions, irrespective of whether ARSA was an isolated finding or not (6). This descriptive study aimed to reflect a clinical standpoint for ARSA by assessing outcomes of prenatally diagnosed ARSA cases at a single institution.
Materials and Methods
This was a retrospective, descriptive cohort study of prenatally diagnosed ARSA cases, which was performed at the Department of Perinatology, Health Science University Van Training and Research Hospital, from August 2016 through February 2019. Our institution serves as a tertiary referral center in Van province, which is one of the low-income regions in Turkey. The study was approved by the institutional Ethics Committee (ethics approval number: 2019/06 21/03/2019).
Data on study subjects were obtained from electronic medical records in which all data are prospectively collected. All ultrasound exams were carried out using a Voluson E8 ultrasound machine (GE Healthcare, Milwaukee, USA) with a 4-8MHz transabdominal probe by a single operator (R. A.), who is a maternal-fetal specialist with extensive experience in performing fetal anatomy scan, as well as fetal echocardiography. The study included only ARSA cases diagnosed between 16 and 26 weeks of gestation. All cases underwent anatomy assessment and fetal echocardiography according to the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) guidelines (7, 8). ARSA cases with no if any soft markers or structural defects were assigned as isolated cases, whereas ARSA cases with at least one soft marker or structural defect were assigned as non-isolated cases.
Identification of ARSA
Aberrant right subclavian artery is one of the non-specific soft markers for aneuploidies and can be detected on the second-trimester ultrasound. Visualization of the right subclavian artery was adopted from the technique described by Chaoui et al. (9). The aortic arch was carefully evaluated to identify ARSA at the level of three vessel trachea view in the upper mediastinum where it courses between the trachea and the spine, and runs towards the right arm on color Doppler.
Genetic counseling and prenatal diagnostic testing
Following detailed genetic counseling, the parents of all ARSA cases were offered to undergo a prenatal diagnostic test to examine whether Down syndrome and/or 22q11.2 microdeletions accompanied current pregnancy. Amniocentesis procedure was performed in those who consented to do so. The choice of genetic test was conventional karyotyping in conjunction with fluorescence in situ hybridization (FISH). The option of postnatal genetic testing was offered to those who declined to have a prenatal diagnostic test. Blood samples of neonates were used for postnatal genetic assessment.
Descriptive statistics was used on data processing and data were expressed as number (percentage) or median (minimum-maximum).
Results
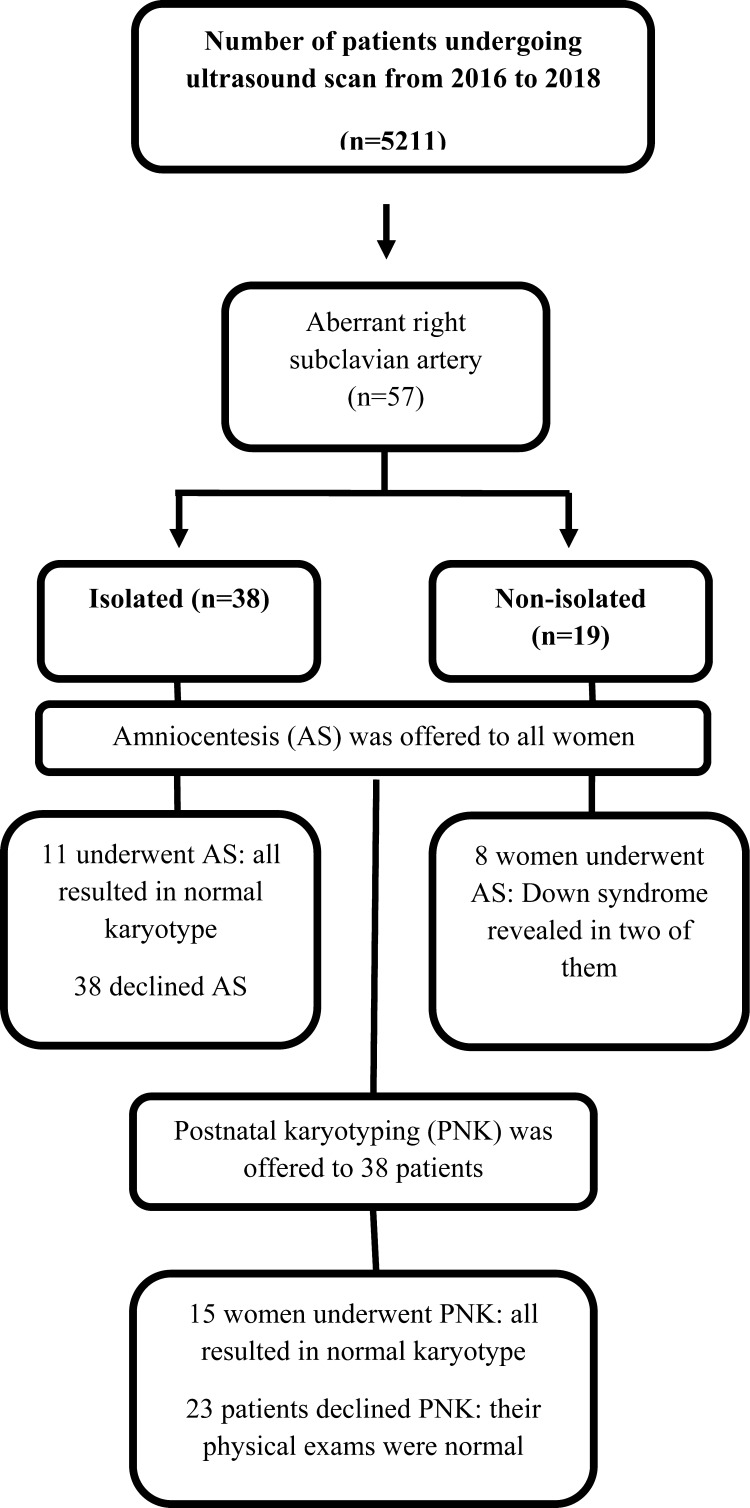
During the study period, 5211 women underwent ultrasound scan for fetal anatomy survey between 16 and 26 weeks of gestation. Figure 1 shows flowchart of the study. The median gestational age and median maternal age at diagnosis (min-max) was 21 weeks (range, 16-26) and 28 years (range, 17-44), respectively. Of these, 57 women had an ARSA affected fetus. Of these 57 cases, 38 had isolated ARSA and 19 had non-isolated ARSA. The rate of ARSA in our study population was 1.1% (57/5211).
Fig. 1.
Study flowchart.
The following markers or anomalies were identified in the non-isolated group: pyelectasis (n=4), hyperechogenic bowel (n=4), hyperechogenic intracardiac focus (n=4), single umbilical artery (n=1), nuchal fold thickness (n=1), short femur plus short humerus (n=1), hyperechogenic bowel plus persistent left superior vena cava (n=1), ventricular septal defect (VSD) plus single umbilical artery (n=1), atrioventricular septal defect (AVSD) plus esophageal atresia (n=1) and fetal hydrops (n=1).
Nineteen of 57 patients underwent amniocentesis after detailed genetic counseling, including 11 in the isolated group and eight in the non-isolated group. Karyotyping result was normal in the 11 isolated cases, while karyotyping resulted in trisomy 21 in two of eight non-isolated cases, one having AVSD plus esophageal atresia and another one having fetal hydrops.
Thirty-eight patients declined to have prenatal invasive test via amniocentesis. Of these, 15 patients accepted postnatal genetic testing, which showed 12 of them to have isolated ARSA and three of them to have non-isolated ARSA. None of these 15 cases had karyotype abnormalities based on conventional karyotyping in conjunction with FISH. On physical examination, the remaining 23 cases that did not undergo any genetic testing did not show any signs indicative of chromosomal abnormalities including Down syndrome and 22q11.2 microdeletions.
The overall rate of Down syndrome was 3.5% (2/57) in the presence of the finding of ARSA in our series. When only cases undergoing genetic testing (34 out of 57 cases) were included in the analysis, the rate of Down syndrome was 5.8% (2/34). Both cases with Down syndrome had major structural abnormalities. Maternal age in these two cases with trisomy 21 was 39 and 26 years. Their pregnancies were terminated. There was no case with Down syndrome in the isolated group. The cases tested did not have 22q11.2 microdeletions. Table 1 shows clinical and demographic characteristics of these cases.
Table 1. Ultrasonographic findings, measurement of nuchal translucency, results of screening tests and pre-postnatal genetic test in cases with aberrant right subclavian artery.
| Maternal age (yrs) | GW | Cardiac anomaly | Extracardiac anomaly | Soft markers | NT (mm) |
Screening test | Prenatal genetic test | Postnatal genetic test |
|---|---|---|---|---|---|---|---|---|
| 21 | 19 | - | - | - | 2.1 | Negative | N | - |
| 27 23 25 19 37 33 41 25 26 21 21 29 38 28 20 34 39 32 27 37 26 17 29 22 22 27 28 17 25 22 28 34 32 |
20+5 22 22 22 23 20 21 21 21 25 21 21 20 21 20 18 16 17 22 23 23 22 21 21 19 20 21 20 22 19 22 21 26 |
- - - - - - - VSD - - - - - - - PLSV AVSD - - - - - - - - - - - - - - - - |
- - - - - - - SUA - - - - - - - - - Fetal hydrops - - - - - - - - - - - - - - - |
Short FL Pyelectasis - Pyelectasis - - - Pyelectasis - - - - - - - - Hyperechogenic bowel Esophageal atresia - - - - Hyperechogenic bowel - - Pyelectasis - - - - - HIF - - |
1.7 2.1 1.5 1.9 1.3 - 1.1 1.6 - - 2.2 1.4 1.9 - 1.4 - 2.3 3.6 - - - 1.6 1.5 - 2.2 - 1.5 1.2 1.9 - 1.6 2.1 - |
Negative Negative Negative Negative Negative - Negative Negative Negative - Negative Negative Negative - Negative Trisomy 21:1/28 Trisomy 21:>1/50 Trisomy 21:>1/50 - Negative Negative Negative Negative Negative Negative - Negative Negative Negative - Negative Negative - |
N N N N N N N N N N N N N N N N Abnormal Abnormal - - - - - - - - - - - - - - - |
- - - - - - - - - - - - - - - - - - N N N N N N N N N N N N N N N |
GW = gestational week; NT = nuchal translucency; FL = femur length; VSD = ventricular septal defect; SUA = single umbilical artery; PLSV = persistent left superior vena cava; AVSD = atrioventricular septal defect; HIF = hyperechogenic intracardiac focus; N = Normal
Of 57 cases, 44 had the first or second trimester screening test and the remaining 13 cases did not undergo it. The risk of aneuploidy was high in eight of 44 cases; however, only three of them underwent invasive test and Down syndrome was found in two of these three cases.
Discussion
The incidence of ARSA was 1.1% in our patient population, which resides in a low-income region. This rate is similar to the reported incidence rate of ARSA in the general population (10). There were two Down syndrome cases among 57 ARSA cases, translating into a rate of 3.5% for Down syndrome. As described above, both Down syndrome cases had structural defects, with a high risk first trimester combined screening (a risk of 1/50). None of the isolated cases had Down syndrome. We did not diagnose any cases with 22q11.2 microdeletions in our series. It seems that further genetic investigation does not contribute to clarifying the isolated ARSA cases. Thus, ARSA may be considered as a variant of normal if found in isolation.
The finding of isolated ARSA implies that there are no other identifiable soft markers and structural defects during anatomy survey. It has also been proposed that the background risk of aneuploidy in a patient carrying an ARSA-affected fetus should be evaluated before assigning ARSA as an isolated finding. It remains elusive whether the finding of isolated ARSA is an indication for performing prenatal invasive test. Although earlier studies found an association between ARSA and chromosomal abnormalities, they did not differentiate isolated ARSA cases from non-isolated ones (11). Esmer et al. report on 10 Down syndrome cases with ARSA, which was in isolation in six of these 10 cases (12). However, of these six cases, five had advanced maternal age (≥35) and four had a high-risk screening test. It is known that the risk of having an aneuploid baby was significantly higher in older women compared with younger women, with a 4-fold increase in those aged 35-40 years compared with those aged 30-35 years (13). In their study conducted in a high-risk population, Borenstein et al. estimated the identification of ARSA in the second trimester to imply a 20-fold increase in the risk of Down syndrome and 13-fold increase in the risk of trisomy 18 (14). However, their study did not provide data whether any of other ultrasound-based soft markers accompanied ARSA. Paladini et al. suggest that detection of ARSA is the third most important marker for offering prenatal invasive test after nasal bone hypoplasia/absence and increased nuchal fold. Furthermore, they report ARSA to be the only finding in eight of 27 (29.6%) fetuses with Down syndrome; however, maternal age and the results of first trimester screening test were not provided in their study (15).
The finding of isolated ARSA has been reported to be a poor maker of Down syndrome in several studies (6, 15-18). Zalel et al. identified Down syndrome in three of 16 fetuses with ARSA; all three cases had at least one additional soft marker or structural defect (16). Sagi-Dain et al. identified only one Down syndrome among 246 isolated ARSA cases according to microarray analyses, and the first trimester screening test result was 1/65 in this case, with maternal age of 39 years (6). De León-Luis et al. found 60 cases with ARSA in unselected 8781 pregnant women, with ARSA being in isolation in 39 cases and non-isolated in 21 cases. They did not detect any Down syndrome cases in the isolated group, whereas there were seven Down syndrome cases in the non-isolated group (19). Similar to our findings, some recent studies also report no aneuploidies and pathogenic copy number variants in cases of isolated ARSA based on conventional karyotyping and microarray analysis (16, 18). In support of the existing literature, both Down syndrome cases had at least one additional marker and a high-risk screening result in our study. It seems reasonable to define ARSA as an isolated finding if there are no other ultrasound-based soft markers and structural defects in conjunction with a low-background risk. Relevant literature suggests that ARSA in isolation as an ultrasound marker may not be a finding requiring further work-up. Thus, the decision-making process to proceed with prenatal invasive testing should not only hinge on whether the finding of ARSA is present or not.
Approximately 10%-15% of all pregnant women experience a variety of emotional changes that increase the risk of anxiety and depression, which can adversely affect both pregnant women and developing fetuses, and detection of soft markers during the second trimester fetal anatomic screening examination causes anxiety in pregnant women (20, 21). During prenatal genetic counseling of a couple having a fetus with ARSA, the pre-existing risk factors including maternal age, result of first trimester combined screening test, and other second trimester soft markers should be taken into consideration. Isolated soft marker findings except for thickened nuchal fold, nasal bone agenesis/hypoplasia, ARSA and ventriculomegaly, which have a high likelihood ratio, are rarely associated with chromosomal or other abnormalities (5). Otherwise, the risk of aneuploidy is higher whenever more than one marker is found. In 2013, a meta-analysis calculated the finding of isolated ARSA to increase the risk of Down syndrome about 3- to 4-fold, which was based on only two studies. In addition, the majority of studies included in this meta-analysis examined high-risk populations (5). In 2014, another meta-analysis concluded that the likelihood ratio of isolated and non-isolated ARSA was 0 and 35.3 for Down syndrome, respectively. It seems that the conclusion of the latter meta-analysis more accurately reflects actual situation regarding the association between ARSA and Down syndrome, as it included a higher number of studies compared with the former one (19). In 2015, Scala et al. suggested in their meta-analysis that the background risk and other ultrasound-based markers should be assessed before making decision to perform invasive testing for chromosomal abnormalities (10).
As regards other chromosomal aberrations, 22q11.2 microdeletions are considered to be associated with ARSA. In particular, concern has been raised regarding a link between conotruncal anomalies secondary to 22q11.2 microdeletions and ARSA. However, it remains speculative whether there is a need for further work-up to investigate 22q11.2 microdeletions (4, 18, 22). McElhinney et al. report that chromosome 22q11.2 deletions were found in 2/7 (29%) patients with isolated ARSA and noassociated intracardiac defects (4). Pico et al. and Rembouskos et al. report on ARSA to accompany two cases with 22q11.2 deletions; however, both had additional cardiac and extra-cardiac findings (23, 24). Sagi-Dain et al. analyzed 246 isolated ARSA cases for chromosome 22q11.2 deletions by microarray analysis and there was no case with 22q11 deletions among them (6). Maya et al. demonstrated that ARSA was strongly associated with 22q11.2 deletion syndrome if accompanying enlarged nuchal translucency (NT; >4 mm), VSD, right aortic arch, clubfoot, echogenic intracardiac focus, and high-risk screening for trisomy 21 (17). Our cohort did not have any cases with 22q11.2 microdeletions and none of the patients with ARSA had conotruncal anomalies. The fact that the incidence of DiGeorge syndrome is approximately 3 per 10,000 plays a limiting factor in revealing its association with ARSA. Thus, more work is needed to explore this association.
Cell-free DNA (cfDNA) screening is the most accurate form of screening testing for trisomy 21 (25). However, it is not recommended for microdeletion syndromes (26). As reducing the risk of miscarriage depends on invasive tests, cfDNA in maternal blood might be offered for common aneuploidies but not for 22q11.2 microdeletion.
Our study had several limitations. One of the limitations was that it was retrospectively conducted. The second limitation was the inability to perform microarray analysis for further genetic investigation because the Ministry of Health in Turkey only covered conventional karyotyping and FISH as diagnostic genetic tests, and the study included a low-income patient population. Third, the follow up was based on physical examination. Fourth, our sample size did not allow us to make definitive recommendation for or against performing an invasive test in case of isolated ARSA.In addition, genetic information could not be obtained for 23 fetuses with ARSA as their parents denied genetic testing. Although physical examinations of all neonates that did not undergo genetic testing were within the normal range, 22q11.2 microdeletions could not be ruled out with certainty as the typical features of 22q11.2 microdeletions, such as facial dysmorphism and neurodevelopmental disorder, may not be seen in the neonatal period.
In conclusion, a detailed ultrasound survey including fetal echocardiography is the crucial part of assessment in cases of ARSA. The finding of non-isolated ARSA seems to be a risk-modifying factor for Down syndrome. Thus, complete prenatal genetic work-up should be offered to the expectant parents in the presence of accompanying soft marker and/or structural defect, advanced maternal age, or a high-risk screening result. However, the decision to proceed with invasive diagnostic testing for aneuploidy is less certain in low-risk pregnancies if ARSA is found in isolation. Cell-free DNA test may be an alternative to invasive test in these cases. Further studies are still needed to fill the knowledge gap regarding the association between isolated ARSA and aneuploidy.
References
- 1.Evans PR. Cardiac anomalies in Mongolism. Br Heart J. 1950. July;12(3):258–62. 10.1136/hrt.12.3.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss L. Congenital cardiac anomalies with mongolism. Trans Am Coll Cardiol. 1953;3:214–21. [PubMed] [Google Scholar]
- 3.Zapata H, Edwards JE, Titus JL. Aberrant right subclavian artery with left aortic arch: associated cardiac anomalies. Pediatr Cardiol. 1993;14:159–61. 10.1007/BF00795645 [DOI] [PubMed] [Google Scholar]
- 4.McElhinney DB, Clark BJ, 3rd, Weinberg PM, et al. Association of chromosome 22q11 deletion with isolated anomalies of aortic arch laterality and branching. J Am Coll Cardiol. 2001;37:2114–9. 10.1016/S0735-1097(01)01286-4 [DOI] [PubMed] [Google Scholar]
- 5.Agathokleous M, Chaveeva P, Poon LC, Kosinski P, Nicolaides KH. Meta-analysis of second-trimester markers for trisomy 21. Ultrasound Obstet Gynecol. 2013;41:247–61. 10.1002/uog.12364 [DOI] [PubMed] [Google Scholar]
- 6.Sagi-Dain L, Singer A, Josefsberg S, Peleg A, Lev D, Samra NN, et al. Microarray analysis has no additional value in fetal aberrant right subclavian artery: description of 268 pregnancies and systematic literature review. Ultrasound Obstet Gynecol. 2019. June;53(6):810–5. 10.1002/uog.20208 [DOI] [PubMed] [Google Scholar]
- 7.Alfirevic Z, Berghella V, Bilardo C, Hernandez-Andrade E, Johnsen SL, Kalache K, et al. ISUOG Clinical Standards Committee . Practice guidelines for performance of the routine mid-trimester fetal ultrasound scan. Ultrasound Obstet Gynecol. 2011. January;37(1):116–26. 10.1002/uog.8831 [DOI] [PubMed] [Google Scholar]
- 8.Carvalho JS, Allan LD, Chaoui R, Copel JA, DeVore GR, Hecher K, et al. ISUOG Practice Guidelines (updated): Sonographic screening examination of the fetal heart. International Society of Ultrasound in Obstetrics and Gynecology. Ultrasound Obstet Gynecol. 2013. March;41(3):348–59. 10.1002/uog.12403 [DOI] [PubMed] [Google Scholar]
- 9.Chaoui R, Heling KS, Sarioglu N, Schwabe M, Dankof A, Bollmann R. Aberrant right subclavian artery as a new cardiac sign in second- and third-trimester fetuses with Down syndrome. Am J Obstet Gynecol. 2005. January;192(1):257–63. 10.1016/j.ajog.2004.06.080 [DOI] [PubMed] [Google Scholar]
- 10.Scala C, Leone Roberti Maggiore U, Candiani M, et al. Aberrant right subclavian artery in fetuses with Down syndrome: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015;46(3):266–76. 10.1002/uog.14774 [DOI] [PubMed] [Google Scholar]
- 11.Borenstein M, Cavoretto P, Allan L, Huggon I, Nicolaides KH. Aberrant right subclavian artery at 11 + 0 to 13 + 6 weeks of gestation in chromosomally normal and abnormal fetuses. Ultrasound Obstet Gynecol. 2008. January;31(1):20–4. 10.1002/uog.5226 [DOI] [PubMed] [Google Scholar]
- 12.Esmer AC, Gul A, Nehir A, Yuksel A, Dural O, Kalelioglu I, et al. Detection rate of trisomy 21 in fetuses with isolated and non-isolated aberrant right subclavian artery. Fetal Diagn Ther. 2013;34(3):140–5. 10.1159/000354650 [DOI] [PubMed] [Google Scholar]
- 13.Hook EB. Rates of chromosome abnormalities at different maternal ages. Obstet Gynecol. 1981. September;58(3):282–5. [PubMed] [Google Scholar]
- 14.Borenstein M, Minekawa R, Zidere V, Nicolaides KH, Allan LD. Aberrant right subclavian artery at 16 to 23 + 6 weeks of gestation: a marker for chromosomal abnormality. Ultrasound Obstet Gynecol. 2010. November;36(5):548–52. 10.1002/uog.7683 [DOI] [PubMed] [Google Scholar]
- 15.Paladini D, Sglavo G, Pastore G, et al. Aberrant right subclavian artery (ARSA). Incidence and correlation with other markers of Down syndrome in 2ndtrimester fetuses. Ultrasound Obstet Gynecol. 2012;39(2):191–5. 10.1002/uog.10053 [DOI] [PubMed] [Google Scholar]
- 16.Zalel Y, Achiron R, Yagel S, Kivilevitch Z. Fetal aberrant right subclavian artery in normal and Down syndrome fetuses. Ultrasound Obstet Gynecol. 2008;31:25–9. 10.1002/uog.5230 [DOI] [PubMed] [Google Scholar]
- 17.Maya I, Kahana S, Yeshaya J, Tenne T, Yacobson S, Agmon-Fishman I, et al. Chromosomal microarray analysis in fetuses with aberrant right subclavian artery. Ultrasound Obstet Gynecol. 2017. March;49(3):337–41. 10.1002/uog.15935 [DOI] [PubMed] [Google Scholar]
- 18.Svirsky R, Reches A, Brabbing-Goldstein D, Bar-Shira A, Yaron Y. Association of aberrant right subclavian artery with abnormal karyotype and microarray results. Prenat Diagn. 2017. August;37(8):808–11. 10.1002/pd.5092 [DOI] [PubMed] [Google Scholar]
- 19.De León-Luis J, Gámez F, Bravo C, Tenías JM, Arias Á, Pérez R, et al. Second-trimester fetal aberrant right subclavian artery: original study, systematic review and meta-analysis of performance in detection of Down syndrome. Ultrasound Obstet Gynecol. 2014. August;44(2):147–53. 10.1002/uog.13336 [DOI] [PubMed] [Google Scholar]
- 20.Watson MS, Hall S, Langford K, Marteau TM. Psychological impact of the detection of soft markers on routine ultrasound scanning: a pilot study investigating the modifying role of information. Prenat Diagn. 2002. July;22(7):569–75. 10.1002/pd.373 [DOI] [PubMed] [Google Scholar]
- 21.Nakić Radoš S, Tadinac M, Herman R. Anxiety during pregnancy and postpartum: course, predictors and comorbidity with postpartum depression. Acta Clin Croat. 2018. March;57(1):39–51. 10.20471/acc.2018.57.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpe P, Tuo G, De Robertis V, et al. Fetal interrupted aortic arch: 2D-4D echocardiography, associations and outcome. Ultrasound Obstet Gynecol. 2010;35:302–9. 10.1002/uog.7530 [DOI] [PubMed] [Google Scholar]
- 23.Pico H, Mancini J, Lafouge A, Bault J-P, Gorincour G, Quarello E. Prenatal associated features in fetuses diagnosed with an aberrant right subclavian artery. Fetal Diagn Ther. 2016;40(3):187–94. 10.1159/000443524 [DOI] [PubMed] [Google Scholar]
- 24.Rembouskos G, Passamonti U, De Robertis V, Tempesta A, Campobasso G, Volpe G, et al. Aberrant right subclavian artery (ARSA) in unselected population at first and second trimester ultrasonography. Prenat Diagn. 2012;32(10):968–75. 10.1002/pd.3942 [DOI] [PubMed] [Google Scholar]
- 25.Gil MM, Akolekar R, Quezada MS, Bregant B, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for aneuploidies: meta‐analysis. Fetal Diagn Ther. 2014;35:156–73. 10.1159/000358326 [DOI] [PubMed] [Google Scholar]
- 26.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016. October;18(10):1056–65. 10.1038/gim.2016.97 [DOI] [PubMed] [Google Scholar]