Abstract
Key message
Reproductive stage salinity tolerance is most critical for rice as it determines the yield under stress. Few studies have been undertaken for this trait as phenotyping was cumbersome, but new methodology outlined in this review seeks to redress this deficiency. Sixty-three meta-QTLs, the most important genomic regions to target for enhancing salinity tolerance, are reported.
Abstract
Although rice has been categorized as a salt-sensitive crop, it is not equally affected throughout its growth, being most sensitive at the seedling and reproductive stages. However, a very poor correlation exists between sensitivity at these two stages, which suggests that the effects of salt are determined by different mechanisms and sets of genes (QTLs) in seedlings and during flowering. Although tolerance at the reproductive stage is arguably the more important, as it translates directly into grain yield, more than 90% of publications on the effects of salinity on rice are limited to the seedling stage. Only a few studies have been conducted on tolerance at the reproductive stage, as phenotyping is cumbersome. In this review, we list the varieties of rice released for salinity tolerance traits, those being commercially cultivated in salt-affected soils and summarize phenotyping methodologies. Since further increases in tolerance are needed to maintain future productivity, we highlight work on phenotyping for salinity tolerance at the reproductive stage. We have constructed an exhaustive list of the 935 reported QTLs for salinity tolerance in rice at the seedling and reproductive stages. We illustrate the chromosome locations of 63 meta-QTLs (with 95% confidence interval) that indicate the most important genomic regions for salt tolerance in rice. Further study of these QTLs should enhance our understanding of salt tolerance in rice and, if targeted, will have the highest probability of success for marker-assisted selections.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00122-021-03890-3.
Introduction
Biotic and abiotic stresses adversely affect crop growth and productivity. In crops, these abiotic stresses are generated by environmental factors such as drought, salinity and alkalinity, nutrient toxicity or deficiency, flooding and poor drainage, high or low soil pH, high and low temperatures and heavy metals; all are complex and often interacting phenomena and limit crop production worldwide (Shahbaz and Ashraf 2013; Almeida et al. 2016). Of these abiotic stresses, drought and salinity have a major impact on the productivity of a number of crops, including rice. For rice, salinity is next only to drought in limiting its productivity. Indeed, frequent occurrences of the combination of drought, due to declining water resources, and salinity, often due to poor irrigation management (Raes et al. 1995; Glick et al. 2007), have created a situation where rice ecosystems are now highly vulnerable to climate change. In addition, intrusion of sea water in coastal areas is converting arable lands to saline soils, while climatic conditions such as air humidity also affect the severity of salinity (Asch et al. 1995, 1997a, b). In this review, we summarize recent advances in understanding salinity tolerance in rice with particular emphasis on stage-specific tolerance. This review emphasises recent developments in available phenotyping methods for salt stress screening at different crop growth stages. Emphasis has been placed on a phenotyping protocol for reproductive stage salinity tolerance, as this has been most problematic for researchers. We have also reviewed QTL mapping studies and hotspots for effective introgressions of candidate genes, together with the application of marker-assisted selection (MAS) for developing commercial rice varieties suitable for salt-affected areas across the world. Potential candidate genes associated with salinity tolerance in the identified meta-QTL regions are also discussed.
Salinity, as far as soils are concerned, refers to the presence of soluble salts above an arbitrary limit, commonly defined by the electrical conductivity (EC) of a saturated soil paste. Agronomically, soil salinity is defined as the presence of sufficient soluble salts in the soil to reduce normal crop growth (Bockheim and Gennadiyev 2000), but this concentration varies from one crop to another and for different varieties within a species. The excess salts are commonly in the form of chlorides and sulfates of sodium and magnesium. Generally, problem soils due to salts are referred to as saline, sodic and saline-sodic based on their EC, exchangeable sodium percentage (ESP) and pH. Soils are termed ‘saline’ if the EC is more than 4 dS m−1 (see Ghassemi et al. 1995). If sodium (Na+) predominates with bicarbonate and carbonate anions, the soils are termed ‘sodic’ and are characterized by very poor soil structure that dramatically reduces water infiltration and drainage. Saline soils will have an EC > 4 dS m−1 and ESP < 15 with pH < 8.8; sodic soils have an EC < 4 dS m−1 and ESP > 15 percent with pH 8.5 to 10.7 while ‘saline-sodic’ soils will have characteristics of saline and sodic soils: EC > 4 dS m−1 and ESP > 15 percent with variable pH (USSL Staff 1954; Eynard et al. 2005).
Soil salinity is known to influence about 20% of the earth’s land and is relatively more widespread in arid and semi-arid climates compared to humid regions. The association with aridity leads to a link with irrigation: salinization affects about 50% of irrigated land worldwide, which includes about 30% of the rice areas (Wang et al. 2012). Globally one-fifth of the world’s arable land and one-third of irrigated agricultural area is salt-affected and has been estimated to be increasing at a very rapid pace (Machado and Serralheiro 2017; Collins 2014). About 30% of the world’s rice growing land is affected by soil salinity (Ahmad and Prasad, 2011; Wang et al. 2012; Hopmans et al. 2021).
Effects of salinity on various growth stages of rice
Following investigation of a few genotypes, rice was categorized as a ‘sensitive’ crop with a threshold salinity of 3 dS m−1 (Maas and Hoffman 1977) among the four categories of ‘tolerant,’ ‘moderately tolerant,’ ‘moderately sensitive’ and ‘sensitive’ to salinity. However, we now know that rice possesses a large variability for salt tolerance (Singh and Flowers 2010; Munns et al. 2006; Sabouri and Biabani 2009; Negrao et al. 2011; De Leon et al. 2015), variability that can be accessed in collections of rice germplasm that exist throughout world (e.g., the International Rice Research Institute (IRRI) alone has more than 129,000 accessions stored in its Germplasm Resource Centre). So we now classify rice as moderately tolerant to salinity since numerous rice genotypes fall into this category (see Table 1). Similarly, sodicity stress can be classified as low, moderate and high (Table 2) with rice again showing a considerable variation between cultivars. Under sodic conditions at pH 9.8, grain yield reduction of 25%, 37%, and 68% has been reported for tolerant, semi-tolerant and sensitive rice cultivars, respectively (Rao et al. 2008).
Table 1.
List of lines of rice tolerant to salinity that have been used in research studies and developmental breeding programs
| Salt-tolerant line | References |
|---|---|
| Pokkali, Nona Bokra, Bicol | Xie et al. (2000) |
| TCCP 266-2-49-B-B-3, IR51500-AC11-1, IR51500-AC17, IR51485-AC6534-4, IR72132-AC6-1, IR69997-AC1, IR69997-AC2, IR69997-AC3, R69997-AC4, | Senadhira et al. (2002) |
| Cuom | Hien et al. (2003) |
| IR 65195, PSBRC 50, Nona Bokra, At 401, BW 451, At354, Pokkali, Jhona 349, IR 4630, BW 351, IR 51500, Kombila, BW 302, Kharamana, IR 1721, Matarawee, Moddikarruppan, Pokkalian, Baticoloa, OB 678, SR 26B, Lankasamurdi, Uvarkarruppan | Safeena et al. (2003) |
| Cheriviruppu (IRGC 19928), Kalimekri 77-5 (IRTP 14213), TKM6 (IRTP 11703), Bhura Rata (IRGC 28590), Mushkan 41 (IRGC 6828), Kalarata 1-24 (IRGC 26913), Bhirpala (IRGC 37015), IR4630-22-2-5-1-3 (IRGC72958), Kajalsail, IR69502-6-SRN-3-UBN-1-B, IR65483-118-25-31-7-1-5, IR65483-141-2-4-4-2-5, IR77298-14-1-2, IR63262-AC201-1-7-2, IR73689-76-2 | Adorada et al. (2005) |
| Pokkali, Dang Dawk Kok, Luang Ta Moh, Supanburi 2 | Theerakulpisut et al. (2005) |
| SAL 187 (IR65209-3b-6-3-1), SAL 271 (IR65858-4B-11-1-2), SAL 345 (IR69588-4R-P-11-3), SAL 518 (IR72046-B-R-7-3-1-2), SAL 534 (IR71832-3R-2-2-1), SAL 543 (IR71899-2-1-1), SAL 546 (IR71991-3R-2-6-1), SAL 547( IR71995-3R-1-2-2), SAL 669 (IR74099-3R-3-3), SAL 699(IR74105-3R-2-1), SAL 729 (IR70023-4B-R-12-3-1), IRGC 19928(Chervirrupu), IRGC 26913(Kalarata 1-24), IRGC 108921(Pokkali), FL 478 (IR66946-3R-178-1-1), SAL 411 (IR72046-B-R-4-3-2-1-2B-1), ShahPasand (Iranian Variety) | Mohammadi-Nejad et al. (2008) |
| Mantaro rice, Guandong 51, Binre, Zhuziqing, Lansheng, IR 46, IR 4422-28-5, Pokkali, Kalarata 1-24 (IRGC 26913), Bhura Rata (IRGC 28590), BR 1, BR 203-26-2, Sail, Jingnuo 8, Linyi tangdao, Bairizao, Xiaojingdao, Cun-an lengshuibai, 80-85, Zhuxi 26, Sunuo 1, Zhengxian 139, Nanjing 570, Haoanxie, Zhuguang 23, Zhuguang 29, Taihuzao, Aijiaolaolaiqing, Jiucaiqing, Maxiangu, Maodao, Hongmangxiangjingnuo, Erzaobaigu, Hongkenuo, Meimanggui, Longjianghong, Dahonggu, Huangjingnuo, Dayanggu, Yingyang 1, Xigu, Wanmanzao, Shengshuilian, Xianzhan, Damangdao, Laohuangdao, Gaoliangdao, Liaoyan 2, Changbai 7 | Hu et al. (2012) |
| Ketumbar, Khao Seetha, Soc Nau, Damodar (CSR 1), Dasal (CSR 2), Getu (CSR 3), Pokkali, Vytilla 1, Vytilla 2, Vytilla 3, Vytilla 4, Vytilla 5 | Amaranatha et al. (2014) |
| SADRI (IRGC acc. 32329), FL478 (IR66946-3R-178-1-1), CSR28, IR4630-22-2-5-1-3, R70023-4B-R-12-3-1, SADRI (IRGC acc. 32329) | Mohammadi et al. (2014) |
| Cheriviruppu | Hossain et al. (2015) |
| Tarome-Molaei, Nona Bokra, Moroberekan | Khan et al. (2016) |
The results of these studies (i.e., developed varieties) are shown in Table 7
Table 2.
Relative salt tolerance of different growth stages of rice
| Kind of salt stress | Growth stage | Low | Moderate | High |
|---|---|---|---|---|
| Salinity—ECe (dS m−1) | Seedling | < 6 | 6–10 | > 10 |
| Reproductive | < 6 | 6–8 | > 8 | |
| Sodicity (pH1:2) | Seedling | < 9.2 | 9.2–9.8 | > 9.8 |
| Reproductive | < 9.2 | 9.2–9.6 | > 9.6 |
The figures in the table show the electrical conductivity of a saturated soil paste and the pH of a 1:2 soil water paste that define different levels of tolerance (low, moderate and high) to salinity and sodicity
Here, ECe is the electrical conductivity of a saturated soil paste; and pH1:2 is the pH of a stirred mixture of 1 part of soil and 2 parts of distilled water
Rice plants respond differently to salt stress at different growth stages (see Moradi and Ismail 2007; Singh and Flowers 2010). Over the entire growth period, rice is relatively tolerant at germination, but growth becomes very sensitive during the early seedling stage (1–3 weeks), and then more tolerant during active tillering. The most sensitive stage as far as overall grain yield is concerned is from panicle initiation to flowering and fertilization. The plants are relatively more tolerant at maturity (Khatun and Flowers 1995; Khan et al. 1997; Folkard and Wopereis 2001; Singh et al. 2004, 2008; Shereen et al. 2005; Agnihotri et al. 2006; Hakim et al. 2010; Ologundudu et al. 2014; Sajid et al. 2017). Hence, tolerance at the seedling and reproductive stages in rice are critical issues for breeding a salt-tolerant rice variety and for the management of rice productivity in the field (Zeng et al. 2001; Ahmadizadeh et al. 2016; Sajid et al. 2017).
Effects of salinity at germination, seedling and vegetative stages
Although rice is very sensitive at its seedling and reproductive stages, it is relatively tolerant at other growth stages including one of the shortest germination. In some cases, germination, which last for 2–3 d, is reportedly not significantly affected up to 16 dS m−1 (Khan et al.1997). Ologundudu et al. (2014) reported that salinity did not affect germination (80% germinating) up to 10 dS m−1 in rice genotypes tolerant at the seedling stage, while in sensitive genotypes germination was only reduced to 50 percent. At 5 dS m−1, both seedling-stage tolerant and sensitive genotypes recorded up to 90 percent germination (Ologundudu et al. 2014). However, irrespective of genotypes, salt stress reduced the rate of germination (Khan et al. 1997; Folkard and Wopereis 2001; Hakim et al. 2010; Ologundudu et al. 2014).
The effects of salinity at the seedling and early vegetative stages are well documented (see Singh and Flowers 2010), with wide variability existing among germplasm lines (Negrao et al. 2013; Islam et al. 2012; Al-Amin et al. 2013; Babu et al. 2014). Most of the tolerant varieties were developed utilizing a limited number of resistant donors such as Nona Bokra or Pokkali or varieties derived from parents such as CSR 28 (Negrao et al. 2011): many of the identified sources of tolerance are landraces. Additionally, wild species can be explored for salinity tolerance mechanisms and new donors (Solis et al. 2020). Investigations of the variability among wild and cultivated rice species in response to salinity found that cultivars derived from crosses of O. glaberrima and O. sativa confer low Na+ and high K + concentrations in roots and shoots. Among the wild sources, O. rufipogon with its high compatibility with O. sativa is widely used for breeding salt-tolerant lines. Lines developed from O. rufipogon and O. sativa hybridization showed nine quantitative trait loci (QTL) and candidate genes (e.g., HKT1;5, HAK6) controlling salt tolerance at the seedling stage (Quan et al. 2018). Among other wild species, O. coarctata was found to be the most tolerant wild relative, followed by O. latifolia and O. alta (Prusty et al. 2018); all can be targeted in genetic improvement programs to develop salt-tolerant cultivars (Solis et al. 2020). New donors identified in various studies for seedling or egetative-stage tolerance listed in Table 1 will help to broaden the gene pool and hasten the pace of breeding of rice for salinity tolerance. This will also provide an opportunity to utilize the broad spectrum of available genetic resources in various region-specific breeding programs to develop tailor-made rice varieties designed for specific locations.
Effects of salinity at the reproductive stage: effects on yield and yield components
At the field level, the effects of salt stress during the reproductive stage are more important than at the vegetative stage (Rao et al. 2008) with the most deleterious effect on yield being stress during panicle initiation (PI) before booting. A significant reduction in tiller number per plant is observed if plants are exposed to salt stress before PI (Zeng et al. 2001); salinity at PI reduced yield by 50% (Zeng et al. 2002) to 80% (Asch and Wopereis 2001). Asch and Wopereis (2001) reported a yield loss for sensitive genotypes of 1 t ha−1 per unit EC (dS m−1) with water EC levels > 2 dS m−1, while yield loss for tolerant cultivars was less than 0.6 t ha−1 per unit increase in EC (genotypes used had yields of approximately 8 t ha−1 when irrigated with fresh water). In other reports, a 12% yield reduction per dS m−1 has been observed at salinities above a threshold level of 3 dS m−1 (Zeng et al. 2002).
Reductions in grain yield (Asch and Wopereis 2001; Abdullah et al. 2001; Zeng et al. 2002; Kiani et al. 2006; Motamed et al. 2008; Rao et al. 2008; Clermont-Dauphin et al. 2010; Singh et al. 2010; Mojakkir et al. 2015; Raghavendra et al. 2018) are particularly influenced by the number of panicles (Asch and Wopereis 2001) and panicle length (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015). Within-panicle characteristics affected by salinity include:
Spikelet number per panicle regardless of season and development stage (Asch and Wopereis 2001),
Number of primary (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015) and secondary branches per panicle (Mojakkir et al. 2015),
Number of grains per panicle (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015),
Number of filled grains per panicle (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015),
Grain weight per panicle (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015),
1000 grain weight (Asch and Wopereis 2001; Abdullah et al. 2001; Motamed et al. 2008; Clermont-Dauphin et al. 2010; Mojakkir et al. 2015),
Increased spikelet sterility (Asch and Wopereis 2001; Clermont-Dauphin et al. 2010) and
Increased unfilled spikelets per panicle (Abdullah et al. 2001; Motamed et al. 2008; Mojakkir et al. 2015).
There is a strong relationship between salt tolerance at the reproductive stage and grain yield as maintaining a high number of fertile florets contributes to high seed set and thus grain yields as seen in tolerant genotypes. Contrastingly, higher spikelet sterility leads to poor seed set and lower grain yields in sensitive genotypes, due to significantly higher uptake of Na+ by anthers of sensitive compared to tolerant genotypes. For example, in sensitive IR64, Na+ was 21 mmol/g dry weight (dwt) in the anthers, in comparison with the more tolerant Cheriviruppu where Na+ was just 0.35 mmol/g dwt (Sarhadi et al. 2012). The Na+/K+ ratio in the anthers of IR64 under salt stress was more than 1.7 times higher than in plants grown under normal conditions, but in the tolerant genotype, Cheriviruppu, no significant change was observed for the Na+/K+ ratio. Since there was no significant change in K+ concentration in the anthers of either IR64 or Cheriviruppu under stress, the increase in the Na+/K+ ratio could clearly be attributed to an increase in Na+ uptake in IR64 under stress (Sarhadi et al. 2012). The presence of Na+ reduces pollen fertility, an important parameter for salinity tolerance at the reproductive stage and a direct determinant of yield. Although pollen fertility has been commonly accepted as a reliable phenotyping method, it is quite cumbersome and time-consuming to determine (Sarhadi et al. 2012). In addition to pollen fertility, stigmatic receptivity, also cumbersome to assess, is related to percent seed set; this was reduced by 38%, 75% and 100% when female plants of IR36 were grown in 10, 25 and 50 mM Na+ concentrations, respectively (Khatun and Flowers 1995).
While tolerance at the vegetative stage increases biomass for later stages, there is a poor association between seedling and reproductive stage salinity tolerance and it has been reported that there are different QTLs/genes independently controlling the tolerance at the two different stages (Moradi and Ismail 2007; Singh and Flowers 2010; Mohammadi et al. 2014).
Phenotyping for salt stress at different stages
Accuracy in phenotyping is very important, and at the reproductive stage, a precise treatment at a specific growth stage is the key for phenotypic repeatability. The results of screening depend on the ambient conditions, particularly temperature and relative humidity, which play a vital role under salinity. Under controlled conditions (29 °C / 21 °C D/N at 70% RH), 50–120 mM NaCl is adequate to discriminate tolerant and sensitive genotypes of rice at the seedling stage, and 30–100 mM for the reproductive stage. However, under high temperature and low RH (34 °C / 25 °C D/N at 50% RH), the rate of transpiration increases, thus carrying more salt into the plant tissues and ultimately leading to severe injury or death (Singh et al. 2005). Hence, knowledge of ambient conditions, an optimum level of stress to use and the correct stage of crop growth along with standard tolerant and sensitive checks are vital in phenotyping for salinity tolerance. Most of the required conditions can be achieved in a controlled environment, unlike field-based screening techniques. However, comparing the results from the controlled conditions with those from field conditions is important in the final selection of desirable plants with a high level of tolerance (Kranto et al. 2016).
Seedling and early vegetative stage
Phenotyping protocols for screening at the seedling and early vegetative stage are very well standardized and repeatable. Screening is mostly based on morphological parameters and relatively easy to achieve. Hydroponics is the best culture method available and ensures a uniform stress with ample nutrients, so that genotypic differences can be attributed to inherent differences of tolerance. The Yoshida culture-based method proposed by Gregorio et al. (1997) has been extensively used as a rapid method for screening large number of genotypes/populations. To counter the adverse effects of Na+ on other nutrients in Yoshida culture solution (Yoshida et al. 1976), a modified Yoshida solution was devised (modified by making the minor nutrients in neutral rather than acid solution, thus avoiding high concentrations of Na+, K+ or NH4+ required to adjust the pH; see Flowers and Yeo 1981) and is considered as the most appropriate for rice growth (Singh et al. 2010). While there are many variants in the way to screen in hydroponics, at IRRI, the use of perforated Styrofoam sealed with net, worked well as the Styrofoam platforms float on culture solution. Four-day-old, germinated seeds are grown on floats for three more days on nutrient solution under stress (usually NaCl of 100–120 mM, i.e., 10–12 dS m−1) before scoring. To validate the screening, every tray must include a tolerant genotype (like IR63307-3R-178-1-1 also known as FL 478) and a sensitive check (e.g., IRRI 154 or IR29). The scoring for seedling injury (Standard Evaluation System or SES score; Table 3) is recorded after 2 weeks based on the damage to the entry (see IRRI 2013; Singh et al. 2010).
Table 3.
Average number of filled grains/plant and 100-grain weight (g) of rice varieties IR64 and IR4630-22-2-5-1-3 grown with different pruning regimes under non-saline conditions, from Calapit-Palao et al. (2013)
| Trait | Variety | Control | Regime 1 | Regime 2 | Regime 3 | LSD (5%) |
|---|---|---|---|---|---|---|
| Filled grains | IR64 | 451a | 208b | 428a | 388a | 62.8 |
| IR4630-22-2-5-1-3 | 349a | 247b | 325a | 346a | 60.7 | |
| 100-grain weight(g) | IR64 | 2.23a | 2.08a | 2.15a | 2.18a | 0.16 |
| IR4630-22-2-5-1-3 | 2.19a | 1.91b | 2.24a | 2.35a | 0.27 |
Lowercase letters indicate grouping (a,b) based on Duncan Multiple Range Test (DMRT) for significant difference from control at P = 0.05
Control—untrimmed plant/no leaf cut
Regime 1—only the flag leaf was left in the plant
Regime 2—two leaves left (penultimate and flag leaves)
Regime 3—top three leaves left
Reproductive stage
There is a good correlation between reproductive stage salinity tolerance and grain yield, but not always with seedling stage tolerance (Singh et al. 2004; Moradi and Ismail 2007; Singh and Flowers 2010). The best example is genotype FL478 which is used as a highly tolerant check for the seedling stage salinity screening. In studies at IRRI, FL478 shows a very high degree of sterility under salinity stress during its reproductive stage (Ahmadizadeh et al. 2016). Contrastingly, Sadri, an Iranian genotype, is very sensitive to salinity at the seedling stage but moderately tolerant at the reproductive stage (Mohammadi et al. 2014).
A novel phenotyping methodology for reproductive stage salinity
The very poor correlation between tolerance at the seedling and reproductive stages in some genotypes suggests that tolerance at these two stages is controlled by a different set of genes (Moradi et al. 2003; Moradi and Ismail 2007; Singh et al. 2008; Singh and Flowers 2010; Mohammadi et al. 2014). Of late, the importance of addressing the reproductive stage tolerance has been realized as it ultimately determines grain yield (Hossain et al. 2015). However, progress in phenotyping has been slow due to time-consuming and laborious protocols for the reproductive stage screening as compared with the relatively easy phenotyping protocols for the seedling stage (Jena and Mackill 2008; Calapit-Palao 2010). Screening for reproductive-stage tolerance in micro-plots filled with soil irrigated with saline water or soil preparations in pots or in natural field conditions have been proposed (Mishra 1996; Singh and Mishra 2004; Singh et al. 2008). However, under field conditions, controlling spatial variability in the soil and the imposition of uniform stress to a population consisting of genotypes with different phenology has proved difficult (Hossain 2014; Ahmadizadeh et al. 2016). Since the development of a precise and accurate phenotyping approach for the reproductive stage is both critical and very challenging, a technique has been devised at IRRI to salinize an individual genotype at the appearance of the flag leaf, which is about 1 week before the most sensitive gametophytic stage and pollen formation. In this way, each genotype can grow normally without any stress until the start of the most sensitive reproductive stage, irrespective of their growth duration.
The method developed at IRRI is based on the fact that in rice plants older leaves act as sinks where Na+ is accumulated so there is a cascade of loading from lower to upper leaves (Yeo and Flowers 1982) and ultimately the flag leaf, whose contribution to grain development is greater than other leaves (see Box 1). The method developed at IRRI (see Box 1 and Box 2) addresses the two major challenges for reproductive stage screening: (1) imposition of salinity stress exactly at the reproductive stage without stressing the plants at the seedling or late vegetative stages, and (2) imposition of stress on different genotypes or mapping populations at the same stage of development—the appearance of the flag leaf (Calapit-Palao et al. 2013; Ahmadizadeh et al. 2016). In this method, salt translocation at the reproductive stage is accelerated by the pruning of old leaves so salt moves faster to the developing panicle than in control plants without pruning (Box 1; Table 3). The protocol, which is described in Box 2 and involves screening at 10–12 dS m−1, has enabled genotypes displaying clear differences in tolerance at the reproductive stage to be identified (Table 4) and the independence of salinity tolerance at the seedling and reproductive stages to be established. For example, FL478, which is used as tolerant check (score 1–3) for seedling-stage-salinity screening, is sensitive (score 7) at the reproductive stage. Contrastingly, Sadri, an Iranian rice variety, is highly sensitive at the seedling stage (7–9) but moderately tolerant (5) at the reproductive stage (Mohammadi et al. 2014; Singh et al. 2021).
| Box 1: Phenotyping protocol developed for screening for reproductive stage salinity tolerance |
| The phenotyping technique was developed based on two small sub-experiments |
| Experiment 1 |
|
In the first experiment, the yield data for control (no leaf blade pruning) were compared with (1) rice plants where only the flag leaf remained, (2) plants where the flag leaf and penultimate leaf were left and (3) plants where the flag leaf and two preceding leaves remained. Salt translocation at the reproductive stage is accelerated by the pruning of old leaves immediately after the first appearance of the flag leaf so that the sink for toxic ion compartmentation in treatment 2 is limited to two leaves (flag leaf and penultimate leaf) and leaf sheaths. Consequently, salt moves faster to the developing panicle than in control plants without pruning. There is a question of yield reduction due to leaf clipping, but there are many studies that indicate that the top three leaves are those that make the major contribution to grain yield, with the flag leaf alone contributing 45–60% to grain yield (Enyi 1962; Yoshida 1981; Abou-khalifa et al. 2008) (Fig. 1). The result of a study from Calapit-Palao et al. (2013) indicated that there was only any significant effect of leaf pruning on yield when the only remaining leaf was the flag leaf (Table 3). Experiment 2 The second experiment was to compare the time taken by Na+ to reach the flag leaf when all leaves were intact with those where leaves had been pruned (only two leaves left). It was observed that Na+ reached the flag leaf/reproductive organs in about 9–10 days after treatment, if all the leaves were intact. However, if only the two top leaves were left and the rest clipped, Na+ took only 3–4 days to reach the flag leaf/reproductive organs (Fig. 1a and b). These two experiments fine-tuned the phenotyping protocol for salinity tolerance at the reproductive stage and ensured the stress was applied to all genotypes at a precise growth stage (Ahmadizadeh et al. 2016). |
Table 4.
Standard Evaluation System (SES) scores for phenotyping for salinity tolerance at the reproductive Stage/maturity
| Score | Category | Symptoms | Examples (genotypes) |
|---|---|---|---|
| 1 | Highly Tolerant | Normal growth, spikelet sterility at ≤ 5% | Cheriviruppu, CSR28, Hasawi, Pokkali (IRGC Acc. No. 28689) |
| 3 | Tolerant | Growth slightly stunted, spikelet sterility at > 5 to 20% | IR4630-22-2-5-1-3, BRRI dhan 47 |
| 5 | Moderately Tolerant | Growth moderately stunted, ¼ of all leaves brown, panicles partially exerted, spikelet sterility at 21 to 40% | Sadri, CSR43 |
| 7 | Sensitive | Growth severely stunted with about ½ of all leaves become brown, panicles poorly exerted, high sterility at 41% to 70% | FL478, IR 64 |
| 9 | Highly Sensitive | Growth severely stunted with almost all the leaves becoming brown and affected, panicles not exerted, delayed heading or papery florets/chaffy panicle with very high sterility at > 70% | IRRI 154, IR29, Swarna |
Fig. 1.
Mechanism of salt movement in rice and comparison of time taken by Na+ to reach the flag leaf after salinization with (a) no leaf pruning and (b) when only top two leaves are left. Red arrows indicate the movement of Na+ from the root zone towards the flag leaf
| Box 2: Screening rice for salt tolerance at the reproductive stage |
| Protocol |
| The method involves sowing pre-germinated seeds in perforated plastic pots filled with fertilized soil (50 N, 25P and 25 K mg kg−1 soil), which are kept in concrete tanks filled with water. Two plants per plot are allowed to grow initially, thinned later to one plant per pot. A water level of 3 cm below the soil surface of the perforated pots is maintained in the tanks. All plants are grown under control conditions (using harvested rain water; EC < 0.2 dS/m) until the flag leaf appears when salt stress is applied—at the same growth stage for all genotypes. At the first appearance of the flag-leaf, individual pots are transferred to saline conditions with EC 10 dS m−1 (ca.100 mM NaCl) and are maintained under these saline conditions for 15–20 days, depending upon the ambient conditions (temperature and humidity as well as the degree of tolerance of the donor). Plants grown under similar conditions without salinization serve as controls. Leaf pruning, done at the first appearance of the flag leaf, is used to accelerate salt accumulation in the flag leaf by clipping old leaf blades. Consequently, only the flag leaf and penultimate leaf are available for salt accumulation and translocation to the reproductive organs. This accelerates, after 2 or 3 days, the effects of stress treatment and its effect on yield components as compared to that of control plants where all leaves are left untrimmed. Subsequently, all plants are transferred back to non-saline conditions. Yield and yield components including plant height, tiller number, panicle number, panicle length and panicle characteristics are estimated on a single plant basis. Samples for Na+ and K+ analysis can be taken from the flag leaf (small sample from base, middle and top of the leaf) as an indication of the degree of damage within the plant |
| The genotypes evaluated at the reproductive stage following the protocol can be scored based on the SES scoring system using a 1–9 scale where 1–3 are considered tolerant and 7–9 as sensitive—mainly based on flowering behavior and spikelet sterility (Table 4). This technique has greatly increased the efficiency of screening for the reproductive stage salinity tolerance and could be used as the basis of reproductive stage-specific screening for salinity tolerance in rice |
QTL mapping for seedling stage tolerance
Research on mapping quantitative trait loci, QTL, for salt tolerance in rice has advanced significantly in the last two decades. Several molecular markers in the form of isozymes and DNA markers (such as RFLP, RAPD, SSR, AFLP, VNTRs, CAPS, RAD-Seq) were designed and employed in QTL mapping studies. These methods have been used for the improvement of salt tolerance, utilizing wild rice genetic resources (Quan et al. 2018). Several studies have been undertaken to identify QTLs that quantify indices for plant survival and development under normal vis-a-vis stress conditions (Table 5).
Table 5.
Reported QTL studies in rice for salinity tolerance (details in Online Resource 1)
| Trait governing QTL | QTL study/ reference |
|---|---|
| Na+ accumulation in roots & shoots; K+ absorption, Na+ absorption, Na+/K+ ratio | Claes et al. (1990), Gregorio (1997), Lang et al. (2001b), Koyama et al. (2001), Bonilla et al. (2002), Niones (2004), Lin et al. (2004), Ren et al. (2005), Yao et al. (2005), Sabouri and Sabouri (2008), Zang et al. (2008), Thomson et al. (2010), Ahmadi and Fotokian (2011), Javed et al. (2011), Zheng et al. (2015), Qiu et al. (2015), Gimhani et al. (2016), De Leon et al. (2016), Dhanaya ka et al. (2018), Puram et al. (2017), Puram et al (2018) |
| Seedling survival, root dry weight, seedling dry matter, shoot dry weight, fresh weight shoot, fresh weight root, total biomass, seedling root length, shoot length | Gregorio (1997), Lang et al. (2001b), Koyama et al. (2001), Takehisa et al. (2004), Lin et al. (2004), Yao et al. (2005), Sabouri and Sabouri (2008), Zang et al. (2008), Sabouri et al. (2009), Thomson et al. (2010), Javed et al. (2011), Wang et al. (2012), Ghomi et al. (2013), Qiu et al. (2015), Gimhani et al. (2016), De Leon et al. (2016), Wang et al. (2017), Rahman et al. (2017), Bizimina et al. (2017), Puram et al. (2017), Dhanayaka et al. (2018), |
| Seedling stage salt injury, SES score, SSI for spikelet fertility at high salt concentration | Yao et al. (2005), Lee et al. (2006), Ammar et al. (2007), Zang et al. (2008), Sabouri et al. (2009), Thomson et al. (2010), Alam et al. (2011), Islam et al. (2011), Pandit et al. (2010), Tian et al. (2011), Javed et al. (2011), Ghomi et al. (2013), Zheng et al. (2015), De Leon et al. (2016), Wang et al.(2017), Rahman et al. (2017), Bizimana et al. (2017), Dahanayaka et al. (2017) |
| Leaf bronzing | Takehisa et al. (2006) |
| Plant stand, chlorophyll content, green leaf area | Sabouri and Sabouri (2008), Thomson et al. (2010), Ghomi et al. (2013), De Leon et al. (2016), Puram et al.(2017), Puram et al.(2018) |
| Relative germination energy, relative germination range, relative seedling height, relative root length, relative root number, relative vigor index, alkali damage rate in germination period, alkali damage rate at early seedling stage | Cheng et al. (2008), Tian et al. (2011), Puram et al. (2018) |
| Reduction of dry weight, reduction of fresh weight, reduction of leaf area, reduction of seedling height | Kim et al. (2009) |
| Na+ in leaves at reproductive stage, Cl− in leaf at reproductive stage, Na+/K+ ratio in leaf at reproductive stage, K+ in leaves at reproductive stage | Ammar et al. (2009), Pandit et al. (2010) |
| K+ concentration in leaves at vegetative stage, Na+ concentration in straw at high salt stress, Na+/K+ ratio in straw at high-salinity stress, Cl− ion concentration in leaves at vegetative stage, Na+ concentration in stem at vegetative stage | Ammar et al. (2009), Pandit et al. (2010), Fayed and Farid (2017) |
| Plant height, tiller number, panicle length, number of fertile spikelets, grain yield per plant, spikelet fertility, number of sterile spikelets, days to flowering, number of panicles, straw dry weight, number of fertile spikelets, total spikelets number, 1000-grain weight | Mohammadi et al. (2013), Zang et al. (2008), Gregorio (1997), Takehisa et al. (2004), Sabouri and Sabouri (2008), Thomson et al. (2010), Calapit-Palao et al. (2015), Hossain et al. (2015) |
| Germination rate, imbibition radicle length, coleoptile fresh weight, coleoptile dry weight, plumule fresh weight, radicle dry weight, germination percentage, radicle fresh weight, plumule dry weight, plumule length | Mardani et al. (2014), Wang et al. (2011) |
| Shoot dry weight at maturity, % potassium (K), pollen fertility, % Sodium (Na), Na+/K+ ratio, panicle length, root dry weight, single-grain weight, sodium concentration at reproductive stage, Na+/K+ ratio at reproductive stage | Calapit-Palao et al. (2015), Hossain et al. (2015) |
Initial molecular studies were based on characterization and expression of salinity-induced tissue-specific proteins (e.g., Claes et al. 1990). Later, genetic studies based on populations derived from diverse parents differing for salt tolerance were utilized to locate the genomic regions associated with salt tolerance. With the development of an RFLP-based linkage map of rice (based on an F2 derived from O. sativa and O. longistaminata), the salT gene was linked to the RFLP marker RG 146B, localized on chromosome 1. This was the first gene reported to be associated with salinity tolerance (Causse et al. 1994). Later, genetic studies, mostly through biparental mapping populations, identified multiple genes/loci associated with salinity tolerance in rice along with their chromosomal locations and these findings helped in improvement of the trait (Gregorio and Senadhira 1993; Causse et al. 1994; Zhang et al. 1995; Ding et al. 1999; Quan et al. 2018). Numbers of QTLs were identified across the different chromosomes associated with seedling stage salinity tolerance (Gregorio et al. 1997; Ren et al. 2005; Thomson et al. 2010).
Saltol QTL and other genomic regions in seedling stage salinity tolerance
Saltol, a major QTL governing salinity tolerance, was mapped in F8 RILs of a cross between IR29 (salt sensitive) and Pokkali (salt tolerant) at the International Rice Research Institute (Gregorio 1997). The genomic region where this QTL was located contains a major gene found to possess three common QTLs for maintaining low Na+ uptake, high K+ uptake and Na+ /K+ homeostasis in shoots with 64.3–80.2% of total phenotypic variation (PV) conferring seedling stage salinity tolerance. Later, the Saltol region was precisely localized (Bonilla et al. 2002). Niones (2004) fine-mapped the common QTL region of Saltol in BC3F4 near isogenic lines (NILs) of IR 29/Pokkali. In addition to this major QTL (Saltol), 7 QTLs including three for Na+ uptake, two for K+ uptake and two for Na+ /K+ ratio were detected on chromosomes 3, 4, 10 and 12. One of the lines (IR 66946-3R-178-1-1, also known as FL478) was identified from a RIL population of the cross IR29/Pokkali that exhibited salt tolerance higher than or comparable to the tolerant parent, Pokkali. Using the same IR29/Pokkali-derived RIL population, Thomson et al. (2010) made a comprehensive study of the Saltol QTL and other major QTLs (other than Saltol) for shoot Na+ /K+ ratio, root K+ concentration, root Na+ /K+ ratio, seedling height, leaf chlorophyll content, initial SES tolerance score, final SES tolerance score and seedling survival across chromosomes 2, 3, 4, 6, 9 and 12. They (Thomson et al. 2010) found multiple Pokkali alleles introgressed into different RILs at different chromosomal regions including alleles at the Saltol locus, which is similar to that of QTL SKC 1 characterized from another highly salt-tolerant land race, Nona Bokra (Ren et al. 2005).
Saltol is a major QTL for salinity tolerance at the seedling stage; however, the contribution of Saltol to visual leaf injury at the seedling stage, as measured through the IRRI SES score (IRRI 2013), is not sufficient to provide a high degree of salt tolerance. Nine NILs, each with a single Pokkali introgression at the Saltol QTL, were evaluated in a saline field (stress conditions) in Iloilo, Philippines, and under controlled conditions (non-stress) at IRRI. The results showed that only two NILs exhibited a superior performance over the sensitive parent IR29 suggesting the need for combining seedling and reproductive stage tolerance while introgressing salinity tolerance into elite lines to address any yield penalty due to salt stress (Thomson et al. 2010).
Simultaneously, several research groups have explored different genetic resources to understand and dissect the genetic basis of salinity tolerance; this has led to identification of several QTLs spanning across the genome (Table 5; Online Resource 1). QTLs for percent seed germination, seedling root length, seedling dry matter and seedling vigor were reported (Prasad et al. 2000; Mardani et al. 2014). Several SSR and RFLP markers linked to QTL regions for shoot and root dry weight, Na+ and K+ absorption and Na+ /K+ ratio governing seedling salinity tolerance have also been reported (Lang et al. 2001a, b, 2008). Importantly, the net quantity of ions transported to shoots (Na+ uptake, K+ uptake and Na+ /K+ ratio) rather than their concentration is directly related to salinity tolerance with independent inheritance of Na+ and K+ uptake (with different pathways of apoplastic leakage and membrane transport respectively; Koyama et al. 2001). QTLs located away from Saltol or SalT regions were detected (Koyama et al. 2001). Although several QTLs were mapped for various traits associated with seedling tolerance, very few of them are being utilized in breeding because of difficulties in transferability of QTLs for physiological traits to an unrelated genetic population (Flowers et al. 2000).
Root QTLs for the total quantity of Na+ in the root (qRNTQ-1) and root K+ concentration (qRKC-4) underpinning salt tolerance were first reported in the cross Nona Bokra/Koshihikari (Lin et al. 2004). QTLs for root and shoot were reported to be located on different linkage groups suggesting that genes controlling transport of Na+ and K+ between shoots and roots may be different or induced uncoordinatedly by salt stress (Lin et al. 2004). QTLs for salinity tolerance rating (STR), weight of shoot dry matter and Na+ /K+ ratio at the seedling stage were also reported by Yao et al. (2005). Ren et al. (2005), fine-mapped qSKC-1, a major QTL localized within the Saltol locus, reported previously by Lin et al. (2004). The SKC1 gene (Os01g20160) controlling K+ /Na+ homeostasis encodes an OsHKT-type Na+ selective transporter and is preferentially expressed in parenchyma cells surrounding the xylem vessels. Thus, SKC1 affects K+ and Na+ translocation between roots and shoots and thereby regulates K+ /Na+ balance in the shoots. There are numerous studies on seedling stage salinity tolerance attributed to different underpinning traits spanning almost all the chromosomes of cultivated rice (Table 5). There are a few studies on salinity tolerance from wild rice accessions of Oryza rufipogon-derived introgression lines (ILs) associated with salt tolerance score (STS), relative root dry weight (RRW), relative shoot dry weight (RSW) and relative total dry weight (RTW), which identified a total of 15 QTLs for four traits (Tian et al. 2011). The detailed information on all the major reported QTLs for salinity tolerance is summarized in Table 5 and detailed in Online Resource 1.
Beyond seedling stage salinity tolerance; QTLs for reproductive stage tolerance of rice
Very few studies have attempted to dissect the genetic basis of tolerance at different growth stages, especially at tillering and flowering. Identification of main-effect QTLs governing salt tolerance at different growth stages will enable an understanding of the genetic nature of salt tolerance and hasten breeding for salt tolerance by facilitating pyramiding of component QTLs using molecular technologies. Takehisa et al. (2004) evaluated backcross inbred lines (BILs) derived from backcrossing Nipponbare/Kasalath//Nipponbare in BC1F9 to BC1F12 generations for four cropping seasons in a paddy field flooded with saline water and a separate non-saline paddy and detected 17 QTLs. Two QTLs for leaf bronzing with epistatic effects were later detected (Takehisa et al. 2006). Ammar et al. (2009) reported 25 QTLs for 17 traits including seedling-salt-injury score, Na+, K+, Cl– concentrations and Na+ /K+ ratio in leaf and stem at vegetative and reproductive stages using F2:F3 population derived from CSR27/MI48. Mohammadi et al. (2013) studied salinity tolerance at the reproductive stage and identified 35 QTLs for 11 traits, of which most were found to be novel for reproductive stage tolerance. However, the major issue in these studies was that the salinity treatment was given at the same time to all the genotypes irrespective of their growth stage at that time (reproductive stage or not).
The first report of QTL mapping for reproductive stage salinity tolerance in rice based on reproductive stage-specific phenotyping with selection pressure exerted exclusively at the time of flag leaf appearance (cf. Box 2) was carried out in a population derived from Cheriviruppu and Pusa Basmati 1 (Hossain et al. 2015). They (Hossain et al. 2015) identified 16 QTLs with LOD values ranging from 3.2 to 22.3 on chromosomes 1, 7 and 8 with the maximum number of QTL clusters for different component traits co-localized on the long arm of chromosomes 1 and 7. Pollen fertility, Na+ concentration and Na+/K+ ratio in the flag leaf were found as the most important mechanisms controlling salt tolerance at the reproductive stage in rice. Calapit-Palao et al. (2015) also carried out phenotyping specifically for the reproductive stage and identified QTLs for reproductive stage salinity tolerance using F2 population of the cross IR64/IR4630-22-2-5-1-3 for yield components, pollen fertility and physiological parameters under salt stress imposed at flag leaf emergence. Three significant (RM455, RM223, and RM271) marker loci on chromosomes 7, 8 and 10 were found to be significantly associated with Na+/K+ ratio. Two significant markers RM11 and RM455 for percent Na+ and K+ were co-localized on chromosome 7 and were responsible for 7.7% to 10.2% of the phenotypic variation. A few more studies conducted for reproductive stage salinity tolerance are reported in Table 5 (details in Online Resource 1), but their phenotyping was not carried out specifically at the reproductive growth stage (Box 2). These results have not been pursued for fine mapping and to develop closely linked markers, perhaps because of the low reliability of such studies.
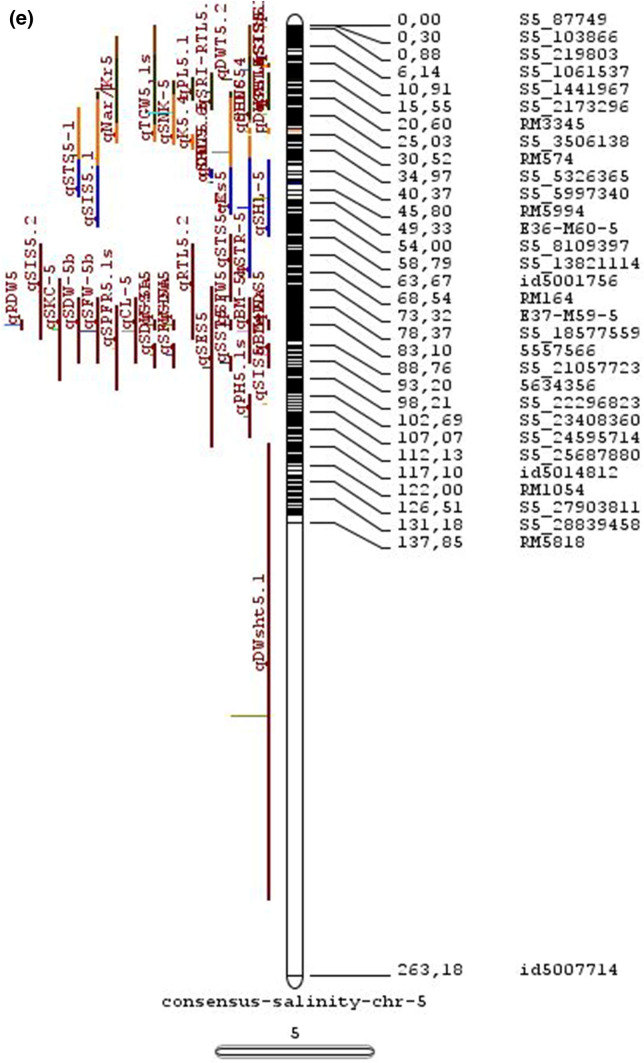
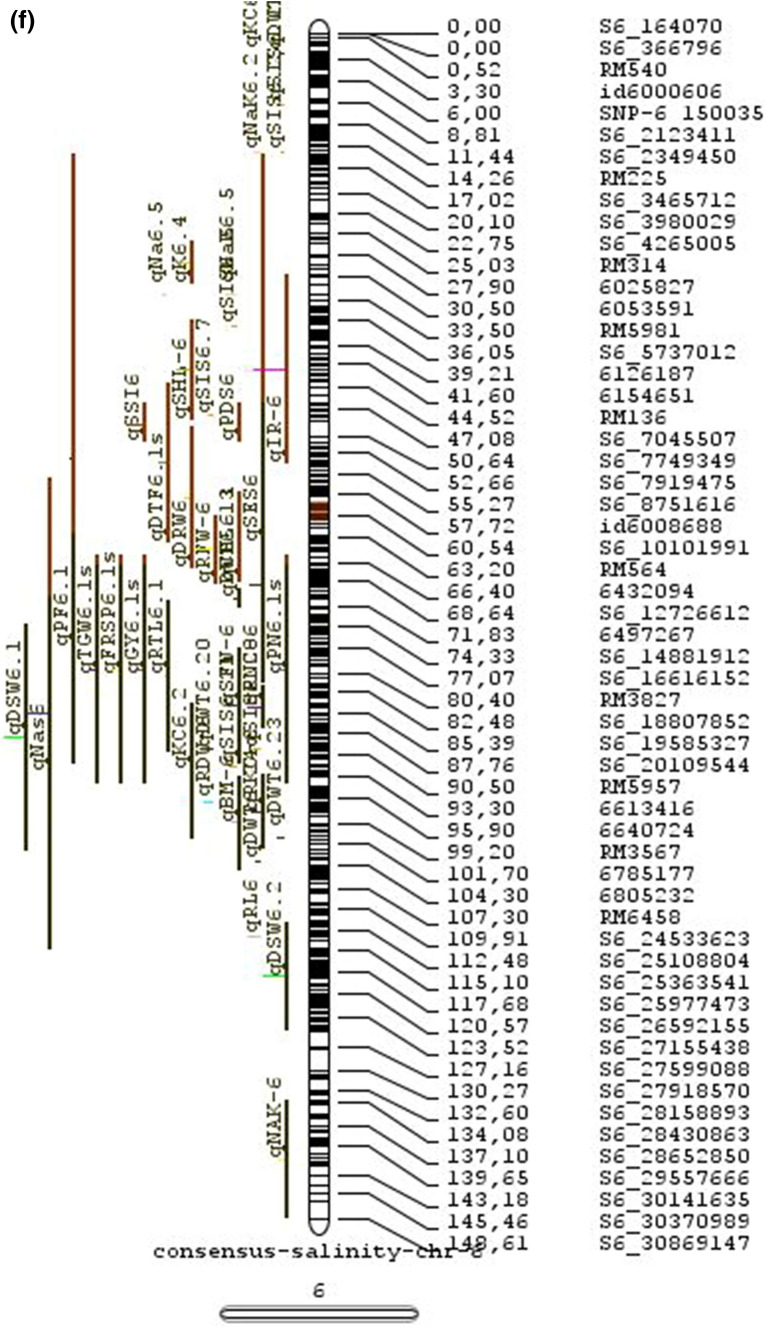
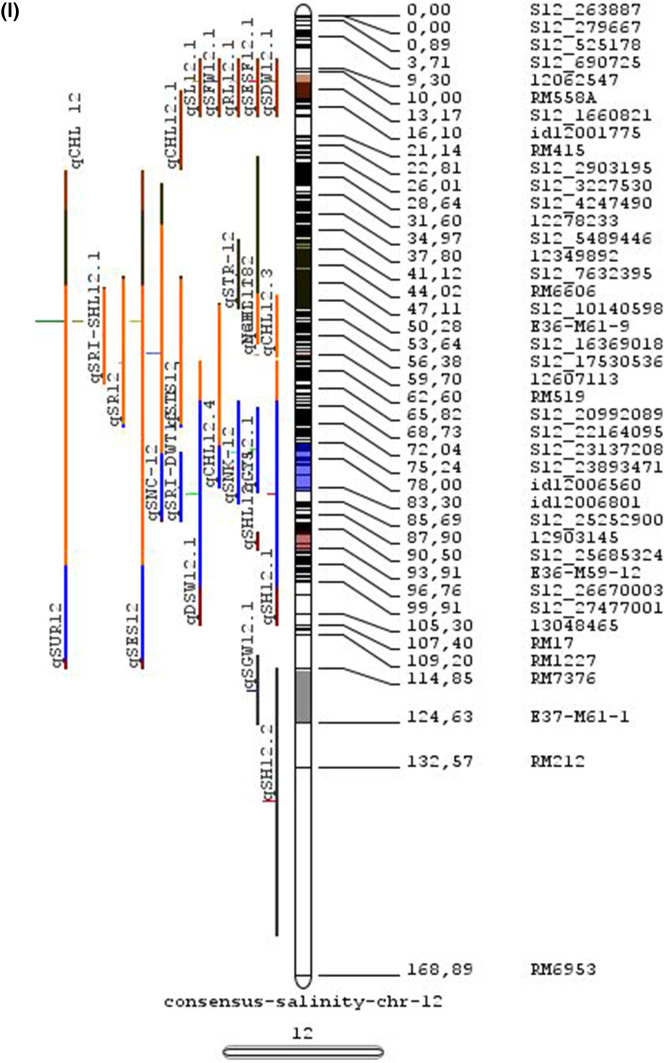
Meta-QTL analysis
Combining results from multiple studies allows greater statistical power for QTL detection and their potential use for genetics and breeding. We have carried out a meta-analysis of salinity tolerance QTLs to provide a reliable integration of information of multiple traits associated (MTA) and multiple QTLs located (MQL) in a single genomic region across various genetic backgrounds and various growth stages. The aim was to detect consistent QTLs that are promising for estimating the position of genes.
In the past two decades, many QTLs have been reported for different growth stages of salinity tolerance (Table 5 and Online Resource 1), but very few have been cloned to date, just, SKC1, qSE3 and OsHAK21 all related to K+ homeostasis (Ren et al. 2005; He et al. 2019). Among the reported 935 QTLs from 46 different QTL studies for salinity tolerance at both vegetative and reproductive stages in different genetic background of biparental mapping populations (detailed information including parents, type and size of mapping population and the reported QTLs are presented in Online Resource 1), only 567 QTLs (see below) were utilized in the meta-analysis (Fig. 2). The specific information on experimental conditions under which QTLs were detected, peak position and flanking markers of detected QTLs, logarithm of the odds (LOD) score, phenotypic variation explained by each QTL and the genetic map information of each study were collected from individual publications as well as from the Gramene database (http://gramene.org). Studies with missing information were excluded from the analysis. Chromosome-wise consensus genetic maps were developed for all chromosomes. The QTLs discovered were from 13 different genetic background mapping populations (BC1F9, BC2F5, BC2F8, BC3F2, BC3F4, BC3F5, BC4F4, BILs, DHs, F2, F2:F4, ILs and RILs) with a majority of the QTLs identified from RILs. The distribution of projected QTLs in the meta-analysis showed that chromosome 1 has highest number of QTLs followed by chromosomes 3, 2, 6 and 4. Most of the QTL mapping studies were based on genetic linkage maps which do not provide the exact physical position of the reported QTLs.
Fig. 2.
Chromosome locations of QTLs for salt tolerance in rice from mapping populations of different genetic backgrounds are distributed in all 12 chromosomes
The meta-analysis reported by Islam et al. (2019) revealed 11 meta-QTLs for three salinity tolerance traits with small confidence intervals that were localized on chromosomes 1 and 2. Our meta-analysis indicated 24 candidate genes in 15 meta-QTLs that spanned physical intervals < 0.2 Mb, including genes that have been cloned previously (e.g., EP3, LP, MIP1, HTD1, DSH1, and OsPNH1; Wu et al. 2016). A total of 63 meta-QTLs with CI of 95% were identified from 567 QTLs detected from different studies projected for salinity traits (Table 6 and Fig. 3). The meta-QTLs indicate the most important genomic regions that have the highest probability of success if specifically targeted for the introgression of salt tolerance in breeding materials through marker-assisted selection. A number of studies have used a meta-QTL region to develop reliable flanking markers for introgressions. Among them, the successful programs include the introgression of Saltol within first mQTL1.1: mQTL1.1 and mQTL1.2 include genes responsible for salinity tolerance, like OsCPK17, OsRMC, OsNHX1, OsHKT1;5 and SalT. The roles of these five genes have been substantiated by Negrao et al. (2012) who reported their allelic variants and their haplotypes associated with salinity tolerance. Eleven out of 32 SNPs identified from four of five tested genes were found to be significantly associated with salt tolerance. OsHKT1;5 (LOC_Os01g20160) for shoot K+ homeostasis was found to be the most diverse gene as evidenced from its 15 haplotypes in the germplasm based on 29 SNPs and two indels variants. NonaBokra, Koshihikari and Pokkali possessed the same haplotype, while other salt-tolerant genotypes like FL478, IR52724-2B-6-2B-1-1 or Hasawi exhibited different OsHKT1;5 haplotypes, although all of them are highly salt-tolerant at the seedling stage.
Table 6.
Meta-QTLs identified from the reported individual QTLs studies for seedling and reproductive stage salinity tolerance
| S.No | Chromosome | MetaQTL | Position (cM) | CI (95%) | Flanking markers |
|---|---|---|---|---|---|
| 1 | 1 | mQTL 1.1 | 48.81 | 0.24 | RM 577 -S1_7520107 |
| 2 | 1 | mQTL 1.2 | 64.39 | 0.9 | RM10745 -S1_9340031 |
| 3 | 1 | mQTL 1.3 | 92.41 | 1.59 | RM6880 -779469 |
| 4 | 1 | mQTL 1.4 | 114.78 | 5.34 | S1_24780749 -RM 486 |
| 5 | 1 | mQTL 1.5 | 121.02 | 0.28 | RM11542 -S1_26449563 |
| 6 | 1 | mQTL 1.6 | 145.61 | 3.83 | 1163456 -RM3422 |
| 7 | 1 | mQTL 1.7 | 172.71 | 0.02 | RM8049 -S1_38794029 |
| 8 | 2 | mQTL 2.1 | 47.93 | 1.25 | S2_7478506 -S2_8096678 |
| 9 | 2 | mQTL 2.2 | 103.85 | 2.6 | RM 6107 -2236772 |
| 10 | 3 | mQTL 3.1 | 35.42 | 0.59 | id3004633 -S3_6027506 |
| 11 | 3 | mQTL 3.2 | 43.87 | 0.84 | S3_7066823 -S3_7209963 |
| 12 | 3 | mQTL 3.3 | 49.29 | 0.6 | RG369 -S3_8357070 |
| 13 | 3 | mQTL 3.4 | 56.1 | 0.21 | RM282 -S3_9891061 |
| 14 | 3 | mQTL 3.5 | 81.25 | 3.61 | id3009433 -3096758 |
| 15 | 3 | mQTL 3.6 | 136.86 | 0.02 | RM2593 -S3_26845909 |
| 16 | 4 | mQTL 4.1 | 6.64 | 0.45 | S4_1966593 -S4_2036989 |
| 17 | 4 | mQTL 4.2 | 11.76 | 1.64 | S4_3570866 -S4_3881858 |
| 18 | 4 | mQTL 4.3 | 23.99 | 0.24 | S4_10625625 -3883297 |
| 19 | 4 | mQTL 4.4 | 23.99 | 0.4 | S4_10625625 -S4_10841800 |
| 20 | 4 | mQTL 4.5 | 43.47 | 7.1 | id4003259 -id4003690 |
| 21 | 4 | mQTL 4.6 | 64.87 | 1.32 | S4_20523929 -S4_20622937 |
| 22 | 4 | mQTL 4.7 | 83.2 | 2.68 | S4_23278361 -S4_24125704 |
| 23 | 4 | mQTL 4.8 | 92.96 | 7.49 | 4177005 -S4_27200682 |
| 24 | 4 | mQTL 4.9 | 106.6 | 2.6 | S4_28742183 -S4_29548991 |
| 25 | 4 | mQTL 4.10 | 121.54 | 2.3 | 4665219 -S4_31772822 |
| 26 | 5 | mQTL 5.1 | 11.76 | 0.27 | S5_1545025 -S5_1671090 |
| 27 | 5 | mQTL 5.2 | 15.3 | 0.59 | S5_2111966 -S5_2173849 |
| 28 | 5 | mQTL 5.3 | 29.95 | 1.21 | S5_4565557 -S5_4699921 |
| 29 | 5 | mQTL 5.4 | 43.84 | 0.56 | 5210158 -S5_644838 |
| 30 | 5 | mQTL 5.5 | 85.18 | 0.09 | S5_20342607 -S5_20461863 |
| 31 | 6 | mQTL 6.1 | 60.23 | 2.57 | ¡d6008704 -S6_10384890 |
| 32 | 6 | mQTL 6.2 | 74.04 | 0.27 | S6_13743866 -S6_14881912 |
| 33 | 7 | mQTL 7.1 | 27.53 | 2.44 | S7_3578352 -ud7000557 |
| 34 | 7 | mQTL 7.2 | 57.32 | 0.59 | S7_14598897 -RM1135 |
| 35 | 7 | mQTL 7.3 | 67.13 | 0.29 | S7_18588805–S7_19086057 |
| 36 | 8 | mQTL 8.1 | 3.69 | 4.29 | RM1959 -8024868 |
| 37 | 8 | mQTL 8.2 | 46.09 | 7.55 | S8_586637 -S8_10877789 |
| 38 | 8 | mQTL 8.3 | 59.13 | 0.2 | S8_19884635 -S8_20039575 |
| 39 | 8 | mQTL 8.4 | 67.51 | 5.51 | S8_21050940 -S8_21613952 |
| 40 | 8 | mQTL 8.5 | 94.3 | 4.79 | id8006485 -RM5485 |
| 41 | 8 | mQTL 8.6 | 101.95 | 2.43 | 8964581 -S8_25908509 |
| 42 | 8 | mQTL 8.7 | 108.37 | 0.02 | RM3571 -RM6019 |
| 43 | 9 | mQTL 9.1 | 12.52 | 1.54 | 9168923 -9186082 |
| 44 | 9 | mQTL 9.2 | 27.21 | 3.83 | 9302663 -9361710 |
| 45 | 9 | mQTL 9.3 | 38.96 | 0.3 | S9_14953982 -9466659 |
| 46 | 9 | mQTL 9.4 | 57.94 | 9.35 | S9_17109910 -9688613 |
| 47 | 9 | mQTL 9.5 | 70.75 | 7.58 | S9_20010716 -99776646 |
| 48 | 9 | mQTL 9.6 | 80.63 | 3.41 | 9805325 -S9_21917093 |
| 49 | 9 | mQTL 9.7 | 103.29 | 13.86 | RM6797 -RM225 |
| 50 | 9 | mQTL 9.8 | 136.37 | 2.22 | E12M55-4 -E12M48-2 |
| 51 | 9 | mQTL 9.9 | 145.32 | 0.6 | E12M31.1 -C506 |
| 52 | 10 | mQTL 10.1 | 3.05 | 0.93 | 9921967 -9941068 |
| 53 | 10 | mQTL 10.2 | 48.98 | 4.39 | S10_15613358 -S10_17272760 |
| 54 | 10 | mQTL 10.3 | 88.51 | 6.42 | S10_21407693 -S10_22060181 |
| 55 | 11 | mQTL 11.1 | 8.66 | 0.77 | S11_2167161 -S11_2379158 |
| 56 | 11 | mQTL 11.2 | 40.96 | 6.74 | S11_5945246 -S11_8343037 |
| 57 | 11 | mQTL 11.3 | 97.86 | 0.08 | S11_23429078 -11819865 |
| 58 | 12 | mQTL 12.1 | 12.68 | 4.42 | RM3483 -S12_2305577 |
| 59 | 12 | mQTL 12.2 | 45.85 | 12.49 | S12_7222741 -S12_15422550 |
| 60 | 12 | mQTL 12.3 | 60.06 | 0.31 | S12_18614318 -12617550 |
| 61 | 12 | mQTL 12.4 | 79.57 | 8.21 | S12_23893471 -S12_25142846 |
| 62 | 12 | mQTL 12.5 | 92.72 | 3.43 | S12_25696421 -S12_25927195 |
| 63 | 12 | mQTL 12.6 | 120.22 | 9.49 | RM7376 -RM212 |
Fig. 3.
Chromosome-wise consensus meta-QTLs based on 567 QTLs from 46 studies for salinity-related traits
QTL hotspots for introgression of salinity tolerance in rice
Meta-QTL analysis identified several genomic regions governing salinity tolerance across the rice genome spanning 12 chromosomes. Among the 63 meta-QTL regions encompassing 5970 genes within 567 initially identified QTLs for salinity tolerance, we propose 15 meta-QTL regions to be QTL hotspots underpinning major traits governing salinity tolerance in rice. These are (Online Resource 2):
On chromosome 1, mQTL 1.1, mQTL 1.2 and mQTL 1.6 with 26, 23 and 17 initial QTLs governing 20, 17 and 14 traits, respectively;
On chromosome 2, QTL 2.1 is the major QTL hotspot region with 63 initial QTLs for 37 traits under salinity;
Three meta-QTLs on chromosome 3, mQTL 3.1, mQTL 3.5 and mQTL 3.6 governing up to 15 traits;
Two meta-QTLs on chromosome 4, mQTL 4.1 and mQTL 4.9 with 12 and 10 traits;
On chromosome 5, one meta-QTL mQTL 5.5 governing 15 traits;
Two meta-QTLs on chromosome 6, meta-QTL 6.1 and meta-QTL6.2 with 17 and 14 traits, and
On chromosome 9, meta-QTL 9.6 with 12 and 15 traits.
In addition, these meta-QTL regions also possess candidate genes related to a wide range of functions including stress signaling and sensing pathways, genes coding integral membrane components, cell wall organization (wall-associated kinases), serine/threonine (Ser/Thr) kinases, pectinesterases, osmotic adjustment (chitinases, hydrolases), transcription factors regulating stress specific genes, ion homeostasis (Na+ and K+ transporters and vacuolar Na+/H+ exchangers) and other related genes. Some of the candidate genes present in these hotspot regions have been validated (Islam et al. 2019; Mirdar-Mansuri et al. 2020), while other needs to be validated for their tolerance in different genetic backgrounds. In addition to these QTL hotspots, there are several genomic regions with 5–10 traits associated with salinity tolerance.
Candidate genes associated with salinity tolerance
The meta-QTL regions in the present study were mined for potential candidate genes. Integrating differentially expressed genes (DEG’s) identified in microarray studies, RNA-Seq data and the reported candidate genes from 111 published papers resulted in about 60 candidate genes in roots, 4 in shoots, 98 in leaves and 28 in seedlings. Among them, 20 genes localized in the QTL hotspot regions for yield and ion homeostasis are promising potential candidates for enhancing salt tolerance in rice and are validated for differential gene expression using qRT-PCR. Our results are in broad agreement with those of Mirdar-Mansuri et al. (2020) for the families of candidate genes detected in meta-QTL. These potential candidate genes are listed in Online Resource 3 and include pectinesterase, peroxidase, oxidoreductase of the aldo/keto reductase family, inorganic phosphate transporter, transcription regulators and OsHKT1. Over expression of the transcription factor OsNAC45 improves salt and drought tolerance in rice through ABA signal responses and regulation of expression of two specific genes, OsPM1 and OsLEA3-1 (Zhang et al. 2020).
The role of halotolerant genes HAL1, HAL2, HAL3, HAL4 and HAL5 encoding proteins with physiological roles in salt stress of rice landraces has been elucidated in addition to those involved in ion homeostasis (Na+/H+, OsNHX antiporters), compatible organic solutes (glycinebetaine and proline), antioxidative genes (OsECS, OsVTE1, OsAPX and OsMSRA4.1), salt responsive regulatory elements and genes encoding protein kinases (MAPKs, SAPKs and STRKs) (Bhatt et al. 2020). Differential expression of genes related to calcium signaling and transport under salinity was observed in IR 64 colonized by an endophyte found in Pokkali (Ramaiah et al. 2020), while a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity-regulating ethylene production confers growth and salt tolerance in rice (Ji et al. 2020). In addition, genes related to ROS, Na+/K+ homeostasis, rice expansin 7 (OsEXPA7), encoding cell wall-loosening protein, response regulator 22 (OsRR22), a B-type response regulator protein involved in transcription factor regulating genes regulates salinity tolerance in rice (Qin et al. 2020).
Marker-assisted strategy for introgression of salinity tolerance in rice and rice varieties for salt-affected soils
Conventional breeding methodology involving hybridization followed by progeny screening under stress and recurrent selection led to the development of tolerant lines tested over multi-location trials before release for cultivation (summarized in Islam et al. 2008; Gregorio et al. 2013 for BRRI dhan 47). This involved a participatory approach involving farmers, which helped the adaptation of varieties suitable for specific locations. The Bangladesh Rice Research Institute (BRRI), in collaboration with IRRI, released BRRI dhan 47 (IR63307-4B-4-3) for saline-prone areas in Bangladesh through this participatory approach. Recently, considerable progress has been made in the development of varieties for salinity tolerance through combining traditional breeding and molecular-marker technology. Anther culture-derived dihaploid lines developed from the cross IR5657-33-2 between two indica breeding lines IR5657-33-2 × IR4630-22-2-5-1-3 evaluated for salinity tolerance and yield led to the release of a promising line IR51500-AC11-1 as PSBRc50 'Bicol' (Senadhira et al. 2002). However, in spite of more than 50 years of research on the effects of salinity on rice only a part of the knowledge gained has been utilized in applied research to develop of salt-tolerant varieties (Table 7).
Table 7.
Rice varieties released for salt tolerance globally
| Name of the variety | Year of release | Designation | Parentage/Cross combination | Breeding method | Released in Country/State/Provinces | Reference |
|---|---|---|---|---|---|---|
| Jhona 349 | 1933 | Jhona 349 | – | Local selection | India | GOI notification No.716 (E), dt.20/2/1970 |
| PVR 1 | 1968 | PVR 1 | SR 26-B/MTU 1 | Pedigree | India | INGER(https://www.irri.org/inger) |
| Shwewartun | 1974 | IR 5mutant | PETA/TANGKAI ROTAN | Mutation breeding | Myanmar | INGER(https://www.irri.org/inger) |
| Type 100 | 1978 | – | Selected from Bhanslot | Local selection | India | GOI notification No13(E), dt.9/12/1978 |
| Narendra 2 (IET 4555) | 1982 | IR 2071-625-1- 252 | IR 8 /Tadukan//TKM 6 /TN 1//IR 8 × IR 24 | Pedigree selection (Introduction from IRRI) | India | GOI notification No165 (E), dt.6/3/1987 |
| Vyttila 2 | 1982 | Culture 174 | Pure line sel. from Cheruviruppu | Pureline selection | India | GOI notification No.19 (E),dt 14/1/1982 |
| Vikas (IET 3116) | 1983 | RP 516-31-6 | IR 8/TKM 6 | Pedigree selection | India | GOI notification No499 (E),dt.8/7/1983 |
| Panvel 1 (IET 7337) | 1984 | PNL 5-30 | BR 4-10/IR 8 | Pedigree selection | India | GOI notification No540 (E),dt.24/7/1985 |
| Panvel 2 (IET 8118) | 1984 | PNL 32-10-1-1 | BR 4-10/IR 8 | Pedigree selection | India | GOI notification No.386, dt.15/5/1990 |
| Mohan (CSR 4) | 1984 | Mutant of IR 8 | Mutation breeding | India | INGER(https://www.irri.org/inger) | |
| Usar 1 | 1984 | – | Jaya/Getu | Bulk selection | India | GOI notification No540 (E) dt.24/7/1985 |
| ROK 5 | 1984 | SR 26/Wellington | Bulk selection | Sierra Leone | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| Sangankhan 4 | 1985 | – | Local selection | Myanmar | INGER(https://www.irri.org/inger) | |
| OM576 | 1986 | HUNGARI/OM 1630-108-2 | Pedigree selection | Vietnam | Singh et al. (2010) | |
| CSR 10 (IET 10349) | 1989 | 81-H21-2-4 | M 40-431-24-114/Jaya | Pedigree selection | India | GOI notification No. 915 (E), dt.6/11/1989 |
| CST 7-1 (IET 9341) | 1989 | CST 7-1 | Damodar/IR 24 | Pedigree selection | India | GOI notification No. 793 (E),dt.22/11/1991 |
| Vyttila 4 (IET 13418) | 1991 | KAU 906 | Chettivirippu/IR 4630-22-2-17 | Pedigree selection | India | http://agritech.tnau.ac.in/expert_system/paddy/KLvarieties.html |
| Lunishree (IET 10678) | 1991 | CRM 30 | Nonasal mutant | Mutation | India | GOI notification No. 814 (E),dt.4/11/1992 |
| WAR 1 | 1991 | IR 4595-4-1-5/Pafant 213 | Bulk Method | Sierra Leone | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| ITA 222 | 1992 | FARO 36 | Mahsuri/IET 1444 | Bulk Method | Gambia | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm |
| WAR 1 | 1992 | IR 4595-4-1-5/Pafant 213 | Bulk Method | Gambia | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 77-3-2-2 | 1992 | IR 4595-4-1-5/Pafant 213 | Pedigree selection | Gambia | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| Sagara | 1992 | – | Selection from Pureline Orumundakan | Pureline selection | India | GOI notification No. 599 (E),dt.25/4/2006 |
| WAR 1 | 1993 | IR 4595-4-1-5/Pafant 213 | Introduction from Sierra Leone | Guinea Bissau | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 77-3-2-2 | 1993 | IR 4595-4-1-5/Pafant 213 | Introduction from Sierra Leone | Guinea Bissau | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| Vyttila 5 (IET 14527) | 1993 | KAU 655 | Mutant of Mahsuri | Mutation breeding | India | http://agritech.tnau.ac.in/expert_system/paddy/KLvarieties.html |
| Giza 177 | 1994 | GZ 4120-205 | Giza 171/Yomji No.1//Pi No.4 | Recombination breeding | Egypt | Hassan et al. (2013) |
| Giza 178 | 1995 | GZ 4255-6-3 | Giza 175/Milyang 49 | Egypt | Hassan et al. (2013) | |
| Narendra Usar Dhan 2 (IET 13556) | 1995 | NDRK 5020 | IRRI Line F2 | Selection from introduction | India | GOI notification No. 401 (E),dt.15/5/1998 |
| B 38 D2 | 1995 | NA | Sierra Leone | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | ||
| WAR 73-1-M-1 | 1996 | Rice mill/I. Mahsuri | Recombination breeding | Guinea | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 77-3-2-2 | 1996 | IR 4595-4-1-5/Pafant 213 | Recombination breeding | Guinea | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 1 | 1997 | IR 4595-4-1-5/Pafant 213 | Recombination breeding | Senegal | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 77-3-2-2 | 1997 | IR 4595-4-1-5/Pafant 213 | Recombination breeding | Senegal | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 81-2-1-3-2 | 1997 | Miniku 33A/Bayerputih 462-10 | Recombination breeding | Senegal | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| WAR 1 | 1998 | IR 4595-4-1-5/Pafant 213 | Recombination breeding | Guinea | http://www.fao.org/docrep/006/Y4751E/y4751e09.htm | |
| CSR 13 (IET 10348) | 1998 | 80-H-3-13 | CSR I/Basmati 370//CSR 5 | Pedigree selection | India | GOI notification No. 425 (E),dt.8/6/1999 |
| Sakha 104 | 1999 | GZ 4096-8-1/GZ 4100-9-1 | Recombination breeding | Egypt | Zayed et al. (2014) | |
| CSR 27 (IET 13765) | 1999 | CSR-88IR-6 | Nona Bokra/IR 5657-33-2 | Recombination breeding | India | GOI notification No. 1050 (E),dt.26/10/1999 |
| Narendra Usar Dhan 3 (IET 14659) | 1999 | IR 46330 (NDRK 14659) | Lung Y AI 148/IR 9125-209-2-2-2-1//IR1872-27-3-1 | Selection from introduction | India | GOI notification No. 92 (E),dt.2/2/2001 |
| Panvel 3 (IET 15368) | 2000 | PNL 18-5-H 7-2 | Damodar/Pankaj | Pedigree selection | India | https://aicrp.icar.gov.in/sasusw/wp-content/uploads/2017/07/14.-Coastal-Saline-Soils-of-Maharashtra-Panvel-Centre.pdf |
| TRY 1 (IET 16643) | 2000 | TRY 1 | BR 153-2B-10-1-3 | Introduction from Bangladesh | India | GOI notification No. 92 (E),dt.2/2/2001 |
| TRY (R) 2 (IET 12863) | 2000 | RP 2597-14-250 | IET 6238/IR 36 | Recombination breeding | India | GOI notification No. 1134 (E),dt.15/11/2001 |
| OM2717 | 2000 | OM 1738/TN 128 | Recombination breeding | Vietnam | http://www.knowledgebank.irri.org/ricebreedingcourse/Breeding_for_salt_tolerance.htm | |
| Dandi (IET 14906) | 2001 | PNL 2-58-1-1 | PNL 2/IET 8320 | Pedigree selection | India | GOI notification No. 283 (E),dt.12/3/2003 |
| Basmati CSR 30 (Yamini: IET 14720) | 2001 | 88-H5-1-1-2 | Bhura Rata 4-10/Pak. Basmati | Pedigree selection | India | GOI notification No.1134 (E),dt 15/11/2001 |
| CSR 23 (IET 13769) | 2002 | CSR-89IR-5 | IR 64//IR 4630-22-2-5-1-3/IR 9764-45-2-2 | Recombination breeding | India | GOI notification No. 161 (E),dt.4/2/2004 |
| OM2517 | 2002 | OM 1352/OMCS 94 | Recombination breeding | Vietnam | http://www.knowledgebank.irri.org/ricebreedingcourse/Breeding_for_salt_tolerance.htm | |
| AS996 | 2002 | IR 64/O RUFIPOGON | Wide hybridization | Vietnam | INGER(https://www.irri.org/inger) | |
| BRRI dhan40 | 2003 | BR5331-93-2-8 | IR4595-4-1-15/BR10 | Recombination breeding | Bangladesh | INGER(https://www.irri.org/inger) |
| BRRI dhan41 | 2003 | BR5828-11-1-4 | BR23/BR1185-2B-16-1 | Recombination breeding | Bangladesh | GOI notification No. 161 (E),dt.4/2/2004 |
| Sumati (IET 13428) | 2003 | CSRC(S)2-1-7 | Pankaj/NC 678 | Pedigree selection | India | GOI notification No. 1566 (E),dt.5/11/2005 |
| Jarava (IET 15420) | 2003 | B 90-15 | B 32 Sel.4/O.rufipogon//B 29-6 | Wide hybridization | India | GOI notification No. 1566 (E),dt.5/11/2005 |
| Naina (CSR 36: IET 17340) | 2005 | CSR 36 | CSR 13/Panvel 2//IR 36 | Recombination breeding | India | GOI notification No. 1566 (E),dt.5/11/2005 |
| Narendra Usar Sankar Dhan 3 (IET 16651) | 2005 | NDURH 3 | IR 58025 A/NDRK 5026-1 R | Recombination breeding | India | |
| Vyttila 6 | 2005 | – | Chiriviruppu/IR 5//Jaya | Recombination breeding | India | GOI notification No. 1566 (E),dt.5/11/2005 |
| Bhutnath (IET 12855) | 2005 | CSRC(S) 5-2-2-5 | SR 26B/Pankaj | Pedigree selection | India | GOI notification No. 1572 (E),dt.20/9/2006 |
| Amal Mana (IET 18250) | 2006 | CSRC(S) 7-1-4 | Pankaj/SR 26 B | Pedigree selection | India | GOI notification No. 454 (E),dt.11/2/2009 |
| BRRI Dhan 47 | 2007 | IR63307-4B-4-3 | IR 51511-B-B-34-B/TCCP 266-2-49-B-B-3 | Introduction from IRRI | Bangladesh | STRASA (IRRI) |
| CSR 22 (IET 15942) | 2008 | CSR 22 | IR 64/IR 4630-22-2-5-1-3/IR 7969-45-2-2 | Recombination breeding | India | GOI notification No. 2187 (E),dt.27/8/2009 |
| DRR Dhan 39 (IET 19487) | 2008 | RP 4631-46-6-5-1-1-1 | CSR 3/Kasturi | Pedigree selection | India | GOI notification No. 211 (E),dt.29/1/2010 |
| Narendra Usar Dhan 2008(IET 18699) | 2009 | NDRK 5088 | TCCP 266-249-B-B-3/IR 262-43-8-1 | Selection from Introduction | India | GOI notification No. 2187 (E),dt.27/8/2009 |
| CR Dhan 402(Luna Sampad:IET 19470) | 2009 | CR 2095-181-1 | Mahsuri/Chakrakonda | Recombination breeding | India | https://krishi.icar.gov.in/jspui/bitstream/123456789/31960/1/1.7.pdf |
| NSIC Rc182 (Salinas 1) | 2009 | IR63307-4B-4-3 | IR 51511-B-B-34-B/TCCP 266-2-49-B-B-3 | Recombination breeding | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc184 (Salinas 2) | 2009 | PR26016-16-B-B-B | IR 8234-0T-9-2-4-2/GIZA 171 | Bulk method | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc186 (Salinas 3) | 2009 | PR30244-AC-V2 | WAGWAG (AC DERIVED) | Anther culture | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc188 (Salinas 4) | 2009 | PR28524-AC97-55 | TCCP 266-1-3B-10-2-1/PSB RC 10 | Recombination breeding | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc190 (Salinas 5) | 2009 | PR25997-B-B-B | IR 9764-45-2-2/IR 81491-AC-5-1 | Bulk method | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| BINA Dhan 8 | 2010 | IR66946-3R-149-1-1 | IR 29/POKKALI | Introduction from IRRI | Bangladesh | https://strasa.irri.org/ |
| BRRI Dhan 53 | 2010 | BR5778-156-1-3-HR14 | BR 10 (BR 51-46-5)//BR 23/BR 847-76-1-1 | Recombination breeding | Bangladesh | https://strasa.irri.org/ |
| BRRI Dhan 54 | 2010 | BR5999-82-3-2-HR1 | BR 1185-2B-16-1/BR 548-128-1-1-3 | Recombination breeding | Bangladesh | https://strasa.irri.org/ |
| CR Dhan 403 (Luna Suvarna:IET 18697) | 2010 | CR 2096-71-2 | Mahsuri/Ormundakan | Recombination breeding | India | https://krishi.icar.gov.in/jspui/bitstream/123456789/31960/1/1.7.pdf |
| Vytilla 8 | 2010 | – | IR 47310-94-4-3-1/CSR 10 | Recombination breeding | India | GOI notification No. 733 (E) dt.1/4/2010 |
| CR Dhan 406 (Luna Barial) | 2010 | CR 2092-158-3 | Jaya /Lunishree | Recombination breeding | India | https://krishi.icar.gov.in/jspui/bitstream/123456789/31960/1/1.7.pdf |
| Jagjeevan (IET 19487) | 2010 | RP 4631-46-6-5-1-1-1 | CSR 3/ Kasturi | Recombination breeding | India | GOI notification S.O. 211 (E), dt 1/29/2010 |
| BRRI Dhan 55 | 2011 | IR73678-6-9-B (AS996) | IR 73382-121/IR 64 | Wide hybridization (Introduction from IRRI) | Bangladesh | https://strasa.irri.org/ |
| Pyi Myanmar Sein (IR10T107) | 2011 | IR 83412-6-B-3-1-1(NSIC 110) | IRRI 126/IR 71606-1-1-4-2-3-1-2 | Recombination breeding (Introduction from IRRI) | Myanmar | http://cure.irri.org/events/myanmarpartnersproducehigh-yieldingsalinity-tolerantricevarieties |
| Shew ASEAN (CSR36) | 2011 | CSR 36 | CSR 13/Panvel 2//IR 36 | Recombination breeding (Introduction from India) | Myanmar | http://cure.irri.org/events/myanmarpartnersproducehigh-yieldingsalinity-tolerantricevarieties |
| NSIC Rc290 (Salinas 6) | 2011 | PR28377-AC97-54 | IRRI 113/PSB RC 10 (IR 50404-57-2-2-3) | Anther culture | Philippines | https://strasa.irri.org/ |
| NSIC Rc292 (Salinas 7) | 2011 | PR30244-AC-V19 | WAGWAG (AC DERIVED) | Anther culture | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc294 (Salinas 8) | 2011 | PR28378-AC96-36 | IRRI 113/IR 64 | Anther culture | Philippines | https://strasa.irri.org/ |
| NSIC Rc296 (Salinas 9) | 2011 | IR71896-3R-8-3-1 | IR 55182-3B-14-3-2/IR 65195-3B-13-2-3 (PSB RC 86) | Pedigree method | Philippines | https://strasa.irri.org/ |
| BINA Dhan 10 | 2012 | IR64197-3B-14-2 | IR 42598-B-B-B-B-12/NONA BOKRA | Recombination breeding (Introduction from IRRI) | Bangladesh | https://strasa.irri.org/ |
| CR Dhan 405 (Luna Sankhi) | 2012 | IR 72046-B-R-8-3-1-3 | IR 31406-333-1/2*IR 31142-14-1-1-3-2 | Recombination breeding (Introduction from IRRI) | India | https://krishi.icar.gov.in/jspui/bitstream/123456789/31960/1/1.7.pdf |
| CSR 43 (IET 18259) | 2012 | CSR-89IR-8 | KDML 105/IR 4630-22-2-5-1-3//IR 20925-33-3-1-28 | Recombination breeding | India | GOI notification S.O. 244 (E) dt.1/24/2014 |
| Gosaba 5 (IET 23403) | 2012 | Chinsurah Nona 1 (IR 55179-3B-11-3) | IR 4630-22-2-5-1-3/Nonabokra | Recombination breeding | India | GOI notification No. S.O.3540(E) dt.22/11/2016 |
| BRRI Dhan 61 | 2013 | BR7105-4R-2 | IR64419-3B-4-3 | Selection from Introduction | Bangladesh | https://strasa.irri.org/ |
| Sangankhan Sinthwelatt | 2013 | Yn 3220 MAS 62-2-4 | IR 53936-60-3-2-1/ Pokkali | Recombination breeding | Myanmar | https://strasa.irri.org/ |
| Saltol Sin thew Latt | 2013 | SarNganKhan Sin Thwe Latt | IR53936-60-3-2-3-1/Pokkali | MAS product | Myanmar | https://strasa.irri.org/ |
| NSIC Rc324 (Salinas 10) | 2013 | PR31607-2-B-B-B-B | IR 65185-3B-8-3-2 (PSB RC 84)/ASOMINORI | Bulk method | Philippines | https://strasa.irri.org/ |
| NSIC Rc326 (Salinas 11) | 2013 | IR84084-B-B-1-1 | IR 66946-3R-178-1-1/2*IR 64680-81-2-2-1-3 | Back crossing | Philippines | https://strasa.irri.org/ |
| NSIC Rc328 (Salinas 12) | 2013 | IR62700-2B-9-2-3 | IR 8192-200-3-3-1-1//BG 367-4/SUAKOKO 8 | Recombination breeding | Philippines | https://strasa.irri.org/ |
| NSIC Rc330 (Salinas 13) | 2013 | PR37435-30-1 | PSB RC 90/PR 29264-AC10//IR 64-1-1-4/IR 70030-7-2-2-1-2 | Recombination breeding | Philippines | https://strasa.irri.org/ |
| NSIC Rc332 (Salinas 14) | 2013 | PR38566-WAGWAG V9-3- 2-15-2 | Selection | Philippines | https://strasa.irri.org/ | |
| NSIC Rc334 (Salinas 15) | 2013 | IR83410-6-B-4-1-1-2 | IRRI 126/IR 64680-81-2-2-1-3 | Pedigree method | Philippines | https://strasa.irri.org/ |
| NSIC Rc336 (Salinas 16) | 2013 | IR84095-AJY3-8-SD01-B | IR 68144-2B-2-2-3-1/IR 66946-3R-78-1-1//IR 77080-B-4-2-2 | Shuttle breeding | Philippines | https://strasa.irri.org/ |
| NSIC Rc338 (Salinas 17) | 2013 | PR30665-1B-1-B-B-Cg | IR 52717-B-B-4-B-B-1-3//IR 9884-54-3/NONA BOKRA///POKKALI | Recombination breeding | Philippines | https://strasa.irri.org/ |
| NSIC Rc340 (Salinas 18) | 2013 | IR84096-AJY 4-2-SDO- 4-B | IR 72593-B-3-2-3-3/IR 72875-94-3-3-2//IR 66946-3R-156-1-1 | Shuttle breeding | Philippines | https://strasa.irri.org/ |
| BRRI Dhan 65 | 2014 | OM1490 | OM606/IR44592-62-1-1-3 | Recombination breeding (Introduction from Vietnam) | Bangladesh | https://strasa.irri.org/ |
| BRRI Dhan 69 | 2014 | BR 7100-R-6-6 | IR61247-3B-8-2-1/ BRRI dhan36 | Recombination breeding | Bangladesh | https://strasa.irri.org/ |
| ITA 212 | 2014 | FARO 35 | BG 90-2*4/TETEP | Back Cross | Gambia | https://strasa.irri.org/ |
| ARICA 11 | 2014 | IR 63275-B-1-1-1-3-3-2 | IR 68/TCCP 266-2-49-B-B-3 | Pedigree method | Gambia | https://strasa.irri.org/ |
| CSR 46 | 2014 | IR72/CSR23 | Recombination breeding | India | S.O. No. 6318(E) dt.26/12 2018 | |
| Inpari 34 Salin Agritan ((NSIC RC 106) | 2014 | IR78788-B-B-10-1-2-4-AJY1 | BR 41/IR 61920-3B-22-2-1 | Modified bulk pedigree | Indonesia | ASEAN%20Regional%20Guidelines%20on%20Promoting%20CSA%20Practices-endorsed%2037th%20AMAF.pdf |
| Inpari 35 Salin Agritan | 2014 | CSR-90IR-2 | IR 10206-29-2-1/SUAKOKO (SEL) | Recombination breeding | Indonesia | ASEAN%20Regional%20Guidelines%20on%20Promoting%20CSA%20Practices-endorsed%2037th%20AMAF.pdf |
| NSIC Rc390 (Salinas 19) | 2014 | IR 83140-B-28-B (IRRI 184) | IR 82869-11/IR 82870-11 | Modified bulk pedigree | Philippines | https://strasa.irri.org/ |
| NSIC Rc392 (Salinas 20) | 2014 | IR 84675-58-4-1-B-B (IRRI 185) | IR 64*3/MADHUKAR//IR 64*3/BINAM | Modified bulk pedigree | Philippines | https://strasa.irri.org/ |
| Rohyb 183-B-5-B-1 | 2014 | WAB0006141 | Recombination breeding | Sierra Leone | http://www.fao.org/3/y4347e/y4347e1l.htm | |
| Rohyb 162- B-1 | 2014 | Recombination breeding | Sierra Leone | http://www.fao.org/3/y4347e/y4347e1l.htm | ||
| ROK 37 (WAR77-3-2-2) | 2014 | WAR77-3-2-2 | IR 4595-4-1-5/PA FANT 213 | Pedigree selection | Sierra Leone | http://www.fao.org/3/y4347e/y4347e1l.htm |
| NERICA-L 20 | 2014 | WAS 122-IDSA-1-WAS | WAB 1291/3*IR 64 | Backcross | Sierra Leone | http://www.fao.org/3/y4347e/y4347e1l.htm |
| BR 78 (BRRI Dhan 78) | 2016 | IR 127-80-1-10/PANBIRA//IR 297-9-1-3-2-2-2 | Recombination breeding with help of MAS | Bangladesh | https://strasa.irri.org/ | |
| CSR 49 | 2016 | CSR-2 K-242 | Recombination breeding | India | GOI notification No. S.O. 3220(E) dt5/09/2019 | |
| NSIC Rc462 (Salinas 21) | 2016 | PR30025-99AC-WSAL-1086 | PR30025-99AC-WSAL-1086 | Anther culture | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc464 (Salinas 22) | 2016 | IR86385-38-1-1-B | IRRI 149/IRRI 128 | Recombination breeding with help of MAS | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc466 (Salinas 23) | 2016 | IR84089-7-3-AJY1-B | IR66946-312-178-1-1/IR72875-94-3-3-2//IR72875-94-3-3-2 | Pedigree Selection | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc468 (Salinas 24) | 2016 | IR06M139 | IR72158-16-3-3/IR74646-96-2-3-3 | Recombination breeding | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| NSIC Rc470 (Salinas 25) | 2016 | PR30245-IR640-ID-18-1-4 | IR64/AC97WP (Anther culture) | Anther culture | Philippines | https://nseedcouncil.bpinsicpvpo.com.ph/seed%20catalogue.php |
| CSR 52 | 2017 | CSR 23/CSR 27 | Recombination breeding | India | GOI notification No. S.O. 3220(E) dt5/09/2019 | |
| ROK 35 (No. 1 B.P 148) | 2017 | Selection | Sierra Leone | http://www.fao.org/3/y4347e/y4347e1l.htm | ||
| Vyttila 3 | – | Vyttila 1/TN 1 | Recombination breeding | India | ||
Pooling of physiological traits was suggested (Flowers and Yeo 1995) and proved successful in generating tolerant lines of rice for salinity tolerance (Gregorio et al. 2002; Thomson et al. 2010; Singh et al. 2021) and other abiotic stresses (Ali et al. 2017). Initially, screening of available germplasm resources to explore the natural variability for salt tolerance led to identification of promising genotypes which were further utilized in breeding for further improvement. Generation advance through conventional methods requires several years to stabilize the variability created before a cultivar is released for farmer cultivation. Recently, a technique called ‘Speed Breeding’ or Rapid Generation Advance (RGA) has been proposed that produces 4–5 generations per year. While there is much recent work in this area (Tanaka et al. 2016; Collard et al. 2017), it is out of the scope of this review, which is focused on reproductive stage phenotyping for salinity stress. Physiological parameters linked to salinity tolerance formed the basis of grouping the genotypes (Asch et al. 2000; Zeng et al. (2003), while other studies considered both physiological traits and yield as well as agronomic characteristics (Li and Xu 2007; Natarajan et al. 2005; Zeng and Shannon, 2000a,b; Zeng et al. 2002; Zeng et al. (2003). The complexity of breeding for salinity tolerance lies with the varied levels of tolerance during crop growth, differences between genotypes that are poorly correlated, with some reports indicating independent inheritance. Hence pyramiding of genetic components controlling tolerance at different growth stages could be the best approach.
Bohnert and Jensen (1996) suggested that successful releases of tolerant varieties of crop plants require large-scale metabolic engineering that include transfer of many genes. Despite the difficulties of dealing with physiological complexity of traits that are determined by sub-components each with a different set of genes in number and quantum effect (Flowers et al. 2000), MAS and marker-assisted backcrossing (MABC) have been vital tools in the transfer of tolerance-related genes/QTLs for the effects of drought, salinity and submergence to elite lines of rice. Several physiological mechanisms and their underlying genomic regions (genes/QTLs) have been tagged. Flanking markers have been found to differ in their reliability and stability for specific QTLs: for example, RM8094 and RM 3412 were found reliable and diagnostic for Saltol, being more closely flanking than others markers like RM 493 (Islam et al. 2012; Al-Amin et al. 2013; Babu et al. 2014). The successful utilization of MAS for salinity tolerance is illustrated by introgression of Saltol into several elite varieties of different countries like PB1121 and PB6 in India (Singh et al. 2011), AS996 BT7, Bac Thom 7 and Q5BD in Vietnam (Huyen et al. 2012, 2013; Linh et al. 2012; Vu et al. 2012; Luu et al. 2012), BRRI dhan 49 in Bangladesh (Hoque et al. 2015) and Novator in Russia (Usatov et al. 2015). Singh et al. 2016 reported the introgression of Saltol into seven popular varieties (ADT45, CR1009, Gayatri, MTU1010, PR114, Pusa 44 and Sarjoo 52) through a multi-institutional network project 'From QTL to variety: marker assisted breeding of abiotic stress tolerant rice varieties with major QTLs for drought, submergence and salt tolerance' a collaborative project of India and IRRI, Philippines. Saltol introgressed QTLs NILs in the genetic background of Pusa 44 and Sarjoo 52, the high yielding mega varieties of India, exhibited improved salinity tolerance at the seedling stage salinity (Krishnamurthy et al. 2020).
Coastal areas, especially in the wet season, need rice varieties not only with salinity tolerance but also submergence tolerance due to frequent inundation of the crop either by tides or estuarine river outflow. IRRI developed a number of salinity and submergence tolerance lines (like IR84649-81-4-B-B and IR84645-311-22-1-B) through introgression of Saltol from ‘Pokkali’ a saline-tolerant landrace and Sub 1 from Swarna-Sub1, a submergence-tolerant variety, through marker-assisted backcrossing (Gregorio et al. 2013). Recently, two dual tolerant two-in-one IRRI-derived rice varieties, BRRI dhan 78 in Bangladesh and Salinas 22 (IR86385-38-1-1-B) in Philippines were released for commercial cultivation (Table 7).
Conclusion
Recent developments in understanding the responses rice to salt stress need to be integrated to supplement conventional rice breeding and harness the maximum genetic improvement for salinity tolerance. A simple, reliable efficient phenotyping method, availability of adequate genetic variability, knowledge of the genetic control and physiological mechanisms governing salinity stress are tools a breeder can use to improve salt tolerance. For rice, a species whose tolerance to salinity varies over its life, understanding the mechanisms involved at different growth stages is of the utmost importance for generating genotypes that are salt-tolerant throughout the crop growing period. Various phenotyping methods, focused on screening at the seedling stage, have been proposed and utilized to develop varieties with enhanced salt tolerance. However, screening at the reproductive stage, which is more complex than screening seedlings but particularly important as tolerance at this later stage translates into grain yield, has yet to be utilized. A novel approach to phenotyping for reproductive stage screening, developed at IRRI since 2013 (Calapit-Palao et al 2013), has been described in detail. An inventory of salt-tolerant donors was made that can be used in breeding programs for generating breeding material with broad genetic base. A comprehensive compilation of previously reported QTLs for salinity tolerance has also been made. However, QTL confidence intervals are often too large to be utilized in marker-assisted introgression. Hence, a meta-QTL analysis was conducted to integrate the genetic linkage maps of different studies utilized for individual QTL mapping into a single consensus linkage map. Meta-analysis has redefined the confidence interval of QTL to a smaller physical and genetic interval that facilitates the identification of candidate genes for salinity tolerance. Validation of these meta-QTLs and the candidate genes would further facilitate their utilization and introgression through breeding programs. Conventional breeding combined with molecular and genomic approaches has supported the development of salt-tolerant rice varieties. However, the association between seedling and reproductive stage tolerance is known to be poor. Furthermore, none of the salt-tolerant donors possess all the desirable alleles for all salt-tolerant mechanisms. A recombination breeding strategy that involves combining all the favorable and complementary traits into a genetic background without any yield penalty would pave the way for the development of a variety with an outstanding performance in farmers’ fields throughout the growing period.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors acknowledge the support of Global Rice Science Partnership (GRISP) at the International Rice Research Institute (IRRI) Philippines that allowed to carry out the research through staff support. Support of Dr. M. Swamy, Sr. Scientist, IRRI, is greatly acknowledged for meta-QTL analytical tool.
Author contribution statement
RKS conceived the idea on novel phenotyping. SK did the literature search and conducted the meta-QTL study on reported QTLs. TJF and RKS contributed ideas for data analysis, design and presentation of the outputs. SK prepared the manuscript. TJF and RKS read and critically revised the manuscript.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest to declare that are relevant to the content of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rakesh Kumar Singh, Timothy Flowers and Suneetha Kota have contributed equally to this work.
References
- Abdullah Z, Khan MA, Flowers TJ. Causes of sterility in seed set of rice under salinity stress. J Agron Crop Sci. 2001;187:25–32. doi: 10.1046/j.1439-037X.2001.00500.x. [DOI] [Google Scholar]
- Abou-Khalifa AAB, Misra AN, Salem AEAKM. Effect of leaf cutting on physiological traits and yield of two rice cultivars. Afr J Plant Sci. 2008;2:147–150. doi: 10.5897/AJPS.9000084. [DOI] [Google Scholar]
- Adorada D, Ocampo RD, Mendoza R, Singh RK, Gregorio GB (2005) Identification of alternate sources of salinity tolerance for rice breeding program. Plant Breeding, Genetics and Biotechnology (PBGB) Division Biennial Report 2004–2005. Manila, Philippines: IRRI, p 22
- Agnihotri RK, Palni LMS, Pandey DK. Screening of landraces of rice under cultivation in Kumaun Himalaya for salinity stress during germination and early seedling growth. Indian J Plant Physiol. 2006;11:266–272. [Google Scholar]
- Ahmad P, Prasad MNV (2011) Abiotic stress responses in plants: metabolism, productivity and sustainability. Springer science and Business Media, New York. ISBN 978-1-4614-0633-4 e-ISBN 978-1-4614-0634-1. 10.1007/978-1-4614-0634-1
- Ahmadi J, Fotokian MH. Identification and mapping of quantitative trait loci associated with salinity tolerance in rice (Oryza sativa L.) using SSR markers. Iran J Biotechnol. 2011;9:21–30. [Google Scholar]
- Ahmadizadeh M, Vispo NA, Calapit-Palao CD, Pangaan ID, Vina CD, Singh RK. Reproductive stage salinity tolerance in rice: a complex trait to phenotype. Indian J Plant Physiol. 2016;12:528–536. doi: 10.1007/s40502-016-0268-6. [DOI] [Google Scholar]
- Alam MR, Rahman S, Seraj ZI, Thomson MJ, Ismail AM, Tumimbang-Raiz E, Gregorio GB. Investigation of seedling-stage salinity tolerance QTLs using backcross lines derived from Oryza sativa L. Pokkali Plant Breed. 2011;130:430–437. doi: 10.1111/j.1439-0523.2010.01837.x. [DOI] [Google Scholar]
- Al-Amin M, Islam MM, Begum SN, Alam MS, Moniruzzaman M, Patwary MAK. Evaluation of rice germplasm under salt stress at the seedling stage through SSR markers. Int J Agric Res Innov Technol. 2013;3:52–59. doi: 10.22004/ag.econ.305319. [DOI] [Google Scholar]
- Ali J, Xu JL, Gao YM, Ma XF, Meng LJ, Wang Y, Pang YL, Guan YS, Xu MR, Revilleza JE, Franje NJ, Zhou SC, Li ZK. Harnessing the hidden genetic diversity for improving multiple abiotic stress tolerance in rice (Oryza sativa L.) PLoS ONE. 2017;12:e0172515. doi: 10.1371/journal.pone.0172515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida DM, Almadanim MC, Lourenço T, Abreu IA, Saibo NJM, Oliveira MM (2016) Screening for abiotic stress tolerance in rice: salt, cold, and drought. In: Duque P (ed) Environmental responses in plants. Methods in molecular biology, vol 1398. Humana Press, New York, NY. 10.1007/978-1-4939-3356-3_14 [DOI] [PubMed]
- Amaranatha RM, Francies RM, Rasool Sk N, Prakash RVR. Breeding for tolerance to stress triggered by salinity in rice. Int J Appl Biol Pharm. 2014;5:167–176. [Google Scholar]
- Ammar MHM, Singh RK, Singh AK, Mohapatra T, Sharma TR, Singh NK. Mapping QTLs for salinity tolerance at seedling stage in rice (Oryza sativa L) African Crop Science Conference Proceedings. 2007;8:617–620. [Google Scholar]
- Ammar MHM, Pandit A, Singh RK, Sameen S, Chauhan M, Chauhan MS, Singh AK, Sharma PC, Sharma TR, Mohapatra T, Singh NK. Mapping of QTLs controlling Na+, K+ and CI- ion concentrations in salt tolerant indica rice variety CSR27. J Plant Biochem Biot. 2009;18:139–150. doi: 10.1007/BF03263312. [DOI] [Google Scholar]
- Asch F, Wopereis MCS. Responses of field-grown irrigated rice cultivars to varying levels of floodwater salinity in a semi-arid environment. Field Crops Res. 2001;70:127–137. doi: 10.1016/S0378-4290(01)00128-9. [DOI] [Google Scholar]
- Asch F, Dorffling K, Dingkuhn M. Response of rice varieties to soil salinity and air humidity: a possible involvement of root-borne ABA. Plant Soil. 1995;177:11–19. doi: 10.1007/BF00010333. [DOI] [Google Scholar]
- Asch F, Dingkuhn M, Dorffling K. Physiological stresses of irrigated rice caused by soil salinity in the Sahel. In: Miezan KM, Wopereis MCS, Dingkuhn M, Deckers J, Randolph TF, editors. Irrigated rice in the Sahel: prospects for sustainable development. BP: West Africa Rice Development Association; 1997. pp. 247–273. [Google Scholar]
- Asch F, Dingkuhn M, Dorffling K. Effects of transpiration on sodium and potassium distribution in salt-stressed irrigated rice. J Exp Bot. 1997;48:39. [Google Scholar]
- Asch F, Dingkuhn M, Dorffling K, Miezan K. Leaf K/Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica. 2000;113:109–118. doi: 10.1023/A:1003981313160. [DOI] [Google Scholar]
- Babu NN, Vinod KK, Gopala Krishnan S, Bhowmick PK, Vanaja T, Krishnamurthy SL, Ngarajan M, Singh NK, Prabhu KV, Singh AK. Marker based haplotype diversity of Saltol QTL in relation to seedling stage salinity tolerance in selected genotypes of rice. Indian J Genet Plant Breed. 2014;74:16–25. doi: 10.5958/j.0975-6906.74.1.003. [DOI] [Google Scholar]
- Bhatt T, Sharma A, Puri S, Minhas AP. Salt tolerance mechanisms and approaches: future scope of halotolerant genes and rice landraces. Rice Sci. 2020;27(5):368–383. doi: 10.1016/j.rsci.2020.03.002. [DOI] [Google Scholar]
- Bizimana JB, Luzi-Kihupi A, Murori RW, Singh RK. Identification of quantitative trait loci for salinity tolerance in rice (Oryza sativa L.) using IR29/ Hasawi mapping population. J Genet. 2017;96:571–582. doi: 10.1007/s12041-017-0803-x. [DOI] [PubMed] [Google Scholar]
- Bockheim JG, Gennadiyev AN. The role of soil-forming processes in the definition of taxa in Soil Taxonomy and the World Soil Reference Base. Geoderma. 2000;95:53–72. doi: 10.1016/S0016-7061(99)00083-X. [DOI] [Google Scholar]
- Bohnert HJ, Jensen RG. Metabolic engineering for increased salt tolerance -the next step. Aust J Plant Physiol. 1996;23:661–667. [Google Scholar]
- Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G. RFLP and SSL mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp Agric Sci. 2002;85:68–76. [Google Scholar]
- Calapit-Palao CD (2010) Identification of QTL for salinity tolerance at reproductive stage in rice (Oryza sativa L.). MS Thesis. University of the Philippines Los Baños, p 116
- Calapit-Palao CD, Vina CB, Gregorio GB, Singh RK. A new phenotyping technique for salinity tolerance at the reproductive stage in rice. Oryza. 2013;50:199–207. [Google Scholar]
- Calapit-Palao CD, Vina CB, Thomson MJ, Singh RK (2015) QTL identification for reproductive-stage salinity tolerance in rice (Oryza sativa L.). In: Proceedings of SABRAO 13th Congress and international conference, September 14–16, Bogor, Indonesia
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu KS, Xiao JH, Yu ZH, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HT, Jang H, Xue DW, Guo LB, Zeng DL, Zhan GH, Qian Q. Mapping of QTLs underlying tolerance to alkali at germination and early seedling stages in rice. Zuo Wu Xue Bao (acta Agronomica Sinica) 2008;34:1719–1727. [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, van den Bulcke M, Bauw G, van Montagu M. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell. 1990;2:19–27. doi: 10.1105/tpc.2.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont-Dauphin C, Suwannang N, Grunberger O, Claude H, Maeght JI. Yield of rice under water and soil salinity risks in farmers' fields in northeast Thailand. Field Crops Res. 2010;118:289–296. doi: 10.1016/j.fcr.2010.06.009. [DOI] [Google Scholar]
- Collard BCY, Beredo JC, Lenaerts B, Mendoza R, Santelices R, Lopena V, Verdeprado H, Raghavan C, Gregorio GB, Vial L, Demont M, Biswas PS, Iftekharuddaula KM, Rahman MA, Cobb JN, Islam MR. Revisiting rice breeding methods—evaluating the use of rapid generation advance (RGA) for routine rice breeding. Plant Prod Sci. 2017;20(4):337–352. doi: 10.1080/1343943X.2017.1391705. [DOI] [Google Scholar]
- Collins Terry and Associates (2014) http://www.terrycollinsassociates.com/2014/world-losing-2000-hectares-of-farm-soil-daily-to-salt-damage-un-university/ Accessed 14 Oct 2020
- Dahanayaka BA, Gimhani DR, Kottearachchi NS, Samarasighe WLG. QTL Mapping for salinity tolerance using an elite rice (Oryza sativa) breeding population. SABRAO J Breed Genet. 2017;49:123–134. [Google Scholar]
- De Leon TB, Linscombe S, Gregorio G, Subudhi PK. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front Plant Sci. 2015;6:374. doi: 10.3389/fpls.2015.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon TB, Linscombe S, Subudhi PK. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice. 2016;9:52. doi: 10.1186/s12284-016-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon TB, Linscombe S, Subudhi PK. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace 'Pokkali'. PLoS ONE. 2017;12:e0175361. doi: 10.1371/journal.pone.0175361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagnosis and improvement of saline and alkali soils. USSL Staff (1954) Richards LA (eds) US Salinity Laboratory Staff, US Department of Agriculture, Washington DC, p 160
- Ding H, Zhang G, Guo Y, Chen S, Chen S (1999) RAPD tagging of a salt tolerance gene in rice. Acad Sin, pp 63–66
- Enyi BAC. The contribution of different organs to grain weight in upland and swamp rice. Ann Bot. 1962;26:529–531. doi: 10.1093/oxfordjournals.aob.a083812. [DOI] [Google Scholar]
- Eynard A, Lal R, Wiebe K. Crop response in salt-affected soils. J Sustain Agric. 2005;27:5–50. doi: 10.1300/J064v27n01_03. [DOI] [Google Scholar]
- Fayed AM, Farid MA. Mapping of quantitative trait loci (QTL) for Na+ and K+ uptake controlling rice salt tolerance (Oryza sativa L.) Int J Curr Microbiol Appl Sci. 2017;6:462–471. doi: 10.20546/ijcmas.2017.601.054. [DOI] [Google Scholar]
- Flowers TJ, Yeo AR. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981;81:363–373. doi: 10.1111/j.1469-8137.1981.tb01731.x. [DOI] [Google Scholar]
- Flowers TJ, Yeo AR. Breeding for salinity resistance in crop plants—where next. Aust J Plant Physiol. 1995;22:875–884. [Google Scholar]
- Flowers TJ, Koyama ML, Flowers SA, Sudhakar C, Singh KP, Yeo AR. QTL: their place in engineering tolerance of rice to salinity. J Exp Bot. 2000;51:99–106. doi: 10.1093/jexbot/51.342.99. [DOI] [PubMed] [Google Scholar]
- Folkard A, Wopereis MCS. Responses of field-grown irrigated rice cultivars to varying levels of flood water salinity in a semi-arid environment. Field Crops Res. 2001;70:127–137. doi: 10.1016/S0378-4290(01)00128-9. [DOI] [Google Scholar]
- Ghassemi F, Jakeman AJ, Nix HA. Salinisation of land and water resources: human causes, extent, management and case studies. Wallingford: CABI Publishing; 1995. p. 526. [Google Scholar]
- Ghomi K, Rabiei B, Sabouri H, Sabouri A. Mapping QTLs for traits related to salinity tolerance at seedling stage of rice (Oryza sativa L.): an Agrigenomics study of an Iranian rice population. OMICS A J Integr Biol. 2013;17:242–251. doi: 10.1089/omi.2012.0097. [DOI] [PubMed] [Google Scholar]
- Gimhani DR, Gregorio GB, Kottearachchi NS, Samarasinghe WLG. SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa) Mol Genet Genom. 2016;291:2081–2099. doi: 10.1007/s00438-016-1241-9. [DOI] [PubMed] [Google Scholar]
- Glick BR, Cheng Z, Czarny J, Duan J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur J Plant Pathol. 2007;119:329–339. doi: 10.1007/s10658-007-9162-4. [DOI] [Google Scholar]
- Gregorio GB (1997) Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). University of the Philippines, Los Baños, the Philippines, p 118
- Gregorio GB, Senadhira D. Genetic analysis of salinity tolerance in rice (Oryza sativa L.) Theor Appl Genet. 1993;86:333–338. doi: 10.1007/BF00222098. [DOI] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. IRRI Disc Paper Series 22, International Rice Research Institute, Manila, Philippines. p 30
- Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002;76:91–101. doi: 10.1016/s0378-4290(02)00031-x. [DOI] [Google Scholar]
- Gregorio GB, Islam MR, Vergara GV, Thirumeni S. Recent advances in rice science to design salinity and other abiotic stress tolerant rice varieties. SABRAO J Breed Genet. 2013;45:31–41. [Google Scholar]
- Hakim MA, Juraimi AS, Begum M, Hanafi MM, Mohd R, Selamat A. Effect of salt stress on germination and early seedling growth of rice (Oryza sativa L.) Afr J Biotechnol. 2010;9:911–1918. doi: 10.5897/AJB09.1526. [DOI] [Google Scholar]
- Hassan HM, El-Khoby WM, El-Hissewy AA (2013) performance of some rice genotypes under both salinity and water stress conditions in Egypt. J Plant Prod 4(8):1235–1257. 10.21608/jpp.2013.74084
- He Y, Yang B, He Y, Zhan C, Cheng Y, Zhang J, Zhang H, Cheng J, Wang Z. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019;97(6):1089–1104. doi: 10.1111/tpj.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien DT, Jacobs M, Angenon G, Hermans C, Thu TT, Son LV, Roosens NH. Proline accumulation and D1-pyrroline-5-carboxylate synthetase gene properties in three rice cultivars differing in salinity and drought tolerance. Plant Sci. 2003;165:1059–1068. doi: 10.1016/S0168-9452(03)00301-7. [DOI] [Google Scholar]
- Hopmans JW, Qureshi AS, Kisekka I, Munns R, Grattan SR, Rengasamy P, Ben-Gal A, Assouline S, Javaux M, Minhas PS, Raats PAC, Skaggs TH, Wang G, De Jong Q, van Lier H, Jiao RS, Lavado N, Lazarovitch BL, Taleisnik E. Critical knowledge gaps and research priorities in global soil salinity. Adv Agron. 2021 doi: 10.1016/bs.agron.2021.03.001. [DOI] [Google Scholar]
- Hoque ABMZ, Haque MA, Sarker MRA, Rahman MA. Marker-assisted introgression of saltol locus into genetic background of BRRI Dhan-49. Int J Biosci. 2015;6:71–80. doi: 10.12692/ijb/6.12.71-80. [DOI] [Google Scholar]
- Hossain MS (2014) Effect of salinity on germination, growth, biochemical attributes and yield of rice mutants. M.S. Thesis. Bangladesh Agricultural University, Mymensingh
- Hossain H, Rahman MA, Alam MS, Singh RK. Mapping of quantitative trait loci associated with reproductive stage salt tolerance in rice. J Agron Crop Sci. 2015;201:17–31. doi: 10.1111/jac.12086. [DOI] [Google Scholar]
- Hu S, Tao H, Qian Q, Guo L. Genetics and molecular breeding for salt-tolerance in rice. Rice Genomics and Genetics. 2012;3:39–49. doi: 10.5376/rgg.2012.03.0007. [DOI] [Google Scholar]
- Huyen LTN, Cuc LM, Ismail AM, Ham LH. Introgression the salinity tolerance QTLs saltol into AS996, the elite rice variety of Vietnam. Am J Plant Sci. 2012;3:981–987. doi: 10.4236/ajps.2012.37116. [DOI] [Google Scholar]
- Huyen LTN, Cuc LM, Ham LH, Khanh TD. Introgression the Saltol QTL into Q5BD, the elite variety of Vietnam using marker assisted selection (MAS) Am J Plant Sci. 2013;1:80–84. doi: 10.11648/j.ajbio.20130104.15. [DOI] [Google Scholar]
- IRRI (2013) Standard evaluation system for rice (SES). 5th edition, International Rice Research Institute, Los Banos, Philippines, p 55
- Islam MR, Singh RK, Salam MA, Hassan L, Gregorio GB. Molecular diversity of stress tolerant rice genotypes using SSR markers. SABRAO J Breed Genet. 2008;40:127–139. [Google Scholar]
- Islam MR, Salam MA, Hassan L, Collard BCY, Singh RK and Gregorio GB (2011) QTL mapping for salinity tolerance at seedling stage in rice. Emir J Food Agric 23: 137–146. https://hdl.handle.net/10568/17296
- Islam MR, Gregorio GB, Salam MA, Collard BCY, Singh RK, Hassan L. Validation of SalTol linked markers and haplotype diversity on chromosome 1 of rice. Mol Plant Breed. 2012;3:103–114. doi: 10.5376/mpb.2012.03.0010. [DOI] [Google Scholar]
- Islam MS, Ontoy J, Subudhi PK. Meta-analysis of quantitative trait loci associated with seedling-stage salt tolerance in rice (Oryza sativa L.) Plants. 2019;8:33. doi: 10.3390/plants8020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed MA, Huyop FZ, Wagiran A, Salleh FM. Identification of QTLs for morph-physiological traits related to salinity tolerance at seedling stage in indica rice. Procedia Environ Sci. 2011;8:389–395. doi: 10.1016/j.proenv.2011.10.061. [DOI] [Google Scholar]
- Jena KK, Mackill DJ. Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 2008;48:1266–1276. doi: 10.2135/cropsci2008.02.0082. [DOI] [Google Scholar]
- Ji J, Yuan D, Jin C, Wang G, Li X, Guan C. Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol Plant. 2020;42:42. doi: 10.1007/s11738-020-3034-3. [DOI] [Google Scholar]
- Khan MSA, Hamid A, Karim MA. Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.) J Agron Crop Sci. 1997;179:163–169. doi: 10.1111/j.1439-037X.1997.tb00512.x. [DOI] [Google Scholar]
- Khan S, Javed MA, Jahan N, Fazilah AM. A short review on the development of salt tolerant cultivars in rice. Int J Public Health Sci. 2016;5:201–212. doi: 10.11591/ijphs.v5i2.4786. [DOI] [Google Scholar]
- Khatun S, Flowers TJ. Effects of salinity on seed set in rice. Plant Cell Environ. 1995;18:61–67. doi: 10.1111/j.1365-3040.1995.tb00544.x. [DOI] [Google Scholar]
- Kiani AR, Homaee M and Mirlatifi M (2006) Evaluating yield reduction functions under salinity and water stress conditions. Iran J Soil Water Sci. Spring. 20:73–83. https://www.sid.ir/en/journal/ViewPaper.aspx?id=53335
- Kim DM, Ju HG, Kwon TR, Oh CS, Ahn SN. Mapping QTLs for salt tolerance in an introgression line population between japonica cultivars in rice. J Crop Sci Biotechnol. 2009;12:121–128. doi: 10.1007/s12892-009-0108-6. [DOI] [Google Scholar]
- Koyama ML, Levesley A, Koebner RMD, Flowers TJ, Yeo AR. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 2001;125:406–422. doi: 10.1104/pp.125.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranto S, Chankaew S, Monkham T, Theerakulpisut P, Sanitchon J. Evaluation for salt tolerance in rice using multiple screening methods. J Agric Sci Technol. 2016;18:1921–1931. [Google Scholar]
- Krishnamurthy SL, Pundir P, Warraich AS, Rathor S, Lokeshkumar BM, Singh NK, Sharma PC. Introgressed Saltol QTL lines improve the salinity tolerance in rice at seedling stage. Front Plant Sci. 2020;11:833. doi: 10.3389/fpls.2020.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang NT, Yanagihara S, Buu BC. QTL analysis of salt tolerance in rice (Oryza sativa L.) SABRAO J Breed Genet. 2001;33:11–20. [Google Scholar]
- Lang NT, Li Z, Buu BC. Microsatellite markers linked to salt tolerance in rice. OmonRice. 2001;9:9–21. [Google Scholar]
- Lang NT, Buu BC, Ismail A. Molecular mapping and marker-assisted selection for salt tolerance in rice (Oryza sativa L.) OmonRice. 2008;16:50–56. [Google Scholar]
- Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, Lee KS, Eun MY. Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol Cells. 2006;21:192–196. [PubMed] [Google Scholar]
- Li ZK and Xu JL (2007) Breeding for drought and salt tolerant rice (Oryza sativa L.): progress and perspective. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerance crops. Springer, Dordrecht, The Netherlands, 2007, pp 531–564
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet. 2004;108:253–260. doi: 10.1007/s00122-003-1421-y. [DOI] [PubMed] [Google Scholar]
- Linh HL, Linh HT, Xuan DT, Ham HL, Ismail AM, Khanh DT. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the red river delta of Vietnam. Int J Plant Genomics. 2012 doi: 10.1155/2012/949038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu TNH, Luu MC, Ismail AM, Le HH. Introgression the salinity tolerance QTLs Saltol into AS996, the elite rice variety of Vietnam. Am J Plant Sci. 2012;3:981–987. doi: 10.4236/ajps.2012.37116. [DOI] [Google Scholar]
- Maas EV, Hoffmann GJ. Crop salt tolerance-current assessment. J Irrig Drain Div Am Soc Civ Enh. 1977;103:115–134. doi: 10.1061/JRCEA4.0001137. [DOI] [Google Scholar]
- Machado RMA, Serralheiro RP. Soil salinity: effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae. 2017;3:30. doi: 10.3390/horticulturae3020030. [DOI] [Google Scholar]
- Mardani Z, Rabiei B, Sabouri H, Sabouri A. Identification of molecular markers linked to salt-tolerant genes at germination stage of rice. Plant Breed. 2014;133:196–202. doi: 10.1111/pbr.12136. [DOI] [Google Scholar]
- Mirdar-Mansuri R, Shobbar ZS, Jelodar BN, Ghaffari M, Mohammadi SM, Daryani P. Salt tolerance involved candidate genes in rice: an integrative meta-analysis approach. BMC Plant Biol. 2020;20:452. doi: 10.1186/s12870-020-02679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B (1996) Highlights of research on crops and varieties for salt affected soils. Karnal, India: CSSRI
- Mohammadi-Nejad G, Arzani A, Singh RK, Gregorio GB. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL. Afr J Biotechnol. 2008;7(6):730–736. doi: 10.5897/AJB07.914. [DOI] [Google Scholar]
- Mohammadi R, Mendioro MS, Diaz GQ, Gregorio GB, Singh RK. Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice (Oryza sativa L.) J Genet. 2013;92:433–443. doi: 10.1007/s12041-013-0285-4. [DOI] [PubMed] [Google Scholar]
- Mohammadi R, Mendioro MS, Diaz GQ, Gregorio GB, Singh RK. Genetic analysis of salt tolerance at seedling and reproductive stages in rice (Oryza sativa) Plant Breed. 2014;133:548–559. doi: 10.1111/pbr.12210. [DOI] [Google Scholar]
- Mojakkir AM, Tareq MZ, Mottalib MA, Hoque ABMZ, Hossain MA. Effect of salinity at reproductive stage in rice. Int J Bus Social Sci. 2015;3:7–12. [Google Scholar]
- Moradi F, Ismail AM. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot. 2007;99:1161–1173. doi: 10.1093/aob/mcm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi F, Ismail AM, Gregorio GB, Egdane JA. Salinity tolerance of rice during reproductive development and association with tolerance at the seedling stage. Indian J Plant Physiol. 2003;8:105–116. [Google Scholar]
- Motamed MK, Asadi R, Rezaei M, Amiri E. Response of high yielding rice varieties to NaCl salinity in greenhouse circumstances. Afr J Biotechnol. 2008;7:3866–3873. [Google Scholar]
- Munns R, James RA, Lauchli A. Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot. 2006;57:1025–1043. doi: 10.1093/jxb/erj100. [DOI] [PubMed] [Google Scholar]
- Natarajan SK, Ganapathy M, Krishnakumar S, Dhanalakshmi R, Saliha BB. Grouping of rice genotypes for salinity tolerance based upon grain yield and Na: K ratio under coastal environment. Res J Agric Biol Sci. 2005;1:162–165. [Google Scholar]
- Negrao S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM. Recent updates on salinity stress in rice: from physiological to molecular responses. Crit Rev Plant Sci. 2011;30:329–377. doi: 10.1080/07352689.2011.587725. [DOI] [Google Scholar]
- Negrao S, Almadanim MC, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McKnally KL, Oliviera MM. New allelic variants found in key rice salt-tolerant genes: an association study. Plant Biotechnol. 2013;J11:87–100. doi: 10.1111/pbi.12010. [DOI] [PubMed] [Google Scholar]
- Niones JM (2004) Fine mapping of the salinity tolerance gene on chromosome 1 of rice (Oryza sativa L.), using near isogenic lines. MS dissertation. College, Laguna, Philippines: University of the Philippines Los Baños, Laguna
- Ologundudu AF, Adelusi AA, Akinwale RO. Effect of salt stress on germination and growth parameters of rice (Oryza sativa L.) Notulae Scientia Bioligicae. 2014;6:237–243. doi: 10.15835/nsb629163. [DOI] [Google Scholar]
- Pandit A, Rai V, Bal S, Sinha S, Kumar V, Chauhan M, Gautam RK, Singh R, Sharma PC, Singh AK, Gaikwad K, Sharma TR, Mohapatra T, Singh NK. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol Genet Genom. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- Prasad SR, Bagali P, Hittalmani S and Shashidhar HE (2000) Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr Sci 78: 162–164. https://www.jstor.org/stable/24103768
- Prusty MR, Kim SR, Vinarao R, Entila F, Egdane J, Diaz MGQ, Jena KK (2018) Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front Plant Sci 9:417. 10.3389/fpls.2018.00417 [DOI] [PMC free article] [PubMed]
- Puram VRR, Ontoy J, Linscombe S, Subudhi PK. Genetic dissection of seedling stage salinity tolerance in rice using introgression lines of a salt tolerant landrace Nona Bokra. J Hered. 2017;108:658–670. doi: 10.1093/jhered/esx067. [DOI] [PubMed] [Google Scholar]
- Puram VRR, Ontoy J, Subudhi PK. Identification of QTLs for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol Biol Rep. 2018;36:695–709. doi: 10.1007/s11105-018-1110-2. [DOI] [Google Scholar]
- Qin H, Li Y, Huang R. Advances and challenges in the breeding of salt-tolerant rice. Int J Mol Sci. 2020;21(21):8385. doi: 10.3390/ijms21218385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Yuan Z, Liu H, Xiang X, Yang L, He W, Du B, Ye G, Xu J, Xing D. Identification of salt tolerance-improving quantitative trait loci alleles from a salt-susceptible rice breeding line by introgression breeding. Plant Breed. 2015;134:653–660. doi: 10.1111/pbr.12321. [DOI] [Google Scholar]
- Quan R, Wang J, Hui J, Bai H, Lyu X, Zhu Y, Zhang H, Zhang Z, Li S, Huang R. Improvement of salt tolerance using wild rice genes. Front Plant Sci. 2018;8:2269. doi: 10.3389/fpls.2017.02269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes D, Deckers J, Diallo M. Water requirements for salt control in rice schemes in the Senegal river delta and valley. Irrig Drain Syst. 1995;9:129–141. doi: 10.1007/BF00881672. [DOI] [Google Scholar]
- Raghavendra P, Dushyantha Kumar BM, Sachin Kumar HM, Madhuri R, Gangaprasad S, Krishnamurthy SL, Dhananjaya BC, Halingali BI, Hittalmani S. Exploration of genetic diversity in traditional landraces of rice for yield and its attributing traits under saline stress condition. Int J Curr Microbiol Appl Sci. 2018;7:3359–3366. doi: 10.20546/ijcmas.2018.706.394. [DOI] [Google Scholar]
- Rahman MA, Bimpong IK, Bizimana JB, Pascual ED, Arceta M, Swamy BPM, Diaw F, Rahman MS, Singh RK. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice. 2017;10:47. doi: 10.1186/s12284-017-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah MHS, Jagadheesh DP, Jambagi S, Kumari MMV, Oelmuller R, Nataraja KN, Ravishankar KV, Ravikanth G, Uma Shaanker R. An endophyte from salt-adapted Pokkali rice confers salt-tolerance to a salt-sensitive rice variety and targets a unique pattern of genes in its new host. Sci Rep. 2020;10:3237. doi: 10.1038/s41598-020-59998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PS, Mishra B, Gupta SR, Rathore A. Reproductive stage tolerance to salinity and alkalinity stresses in rice genotypes. Plant Breed. 2008;127:256–261. doi: 10.1111/j.1439-0523.2007.01455.x. [DOI] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Sabouri H, Biabani A. Toward the mapping of agronomic characters on a rice genetic map: Quantitative trait loci analysis under saline condition. Biotechnol. 2009;8:144–149. doi: 10.3923/biotech.2009.144.149. [DOI] [Google Scholar]
- Sabouri H, Sabouri A. New evidence of QTLs attributed to salinity tolerance in rice. Afr J Biotechnol. 2008;7:4376–4383. [Google Scholar]
- Sabouri H, Reizai AM, Mouomeni A, Kavoussi AM, Sabouri A. QTLs mapping of physiological traits related to salt tolerance in young rice seedlings. Biol Plant. 2009;53:657–662. doi: 10.1007/s10535-009-0119-7. [DOI] [Google Scholar]
- Safeena MIS, Sumanasinghe VA, Bandara DC. Identification of RAPD markers for salt tolerance in rice. Trop Agric Res. 2003;15:39–50. [Google Scholar]
- Sajid H, Jun-hua Z, Chu Z, Lian-feng Z, Xiao-chuang C, Sheng-miao Y, James AB, Ji-jie H, Qian-yu J. Effects of salt stress on rice growth, development characteristics, and the regulating ways: a review. J Integr Agric. 2017;16:2357–2374. doi: 10.1016/S2095-3119(16)61608-8. [DOI] [Google Scholar]
- Sarhadi E, Bazargani MM, Sajise AG, Abdolahi S, Vispo NA, Arceta M, Nejad GM, Singh RK, Salekdeh GH. Proteomic analysis of rice anthers under salt stress. Plant Physiol Biochem. 2012;58:280–287. doi: 10.1016/j.plaphy.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Senadhira D, Zapata-Arias FJ, Gregorio GB, Alejar MS, de la Cruz HC, Padolina TF, Galvez AM. Development of the first salt-tolerant rice cultivar through indica/indica anther culture. Field Crops Res. 2002;76:103–110. doi: 10.1016/S0378-4290(02)00032-1. [DOI] [Google Scholar]
- Shahbaz M, Ashraf M. Improving salinity tolerance in cereals. Crit Rev Plant Sci. 2013;32:237–249. doi: 10.1080/07352689.2013.758544. [DOI] [Google Scholar]
- Shereen A, Mumtaz S, Raza S, Khan MA, Solangi S. Salinity effects on seedling growth and yield components of different inbred rice lines. Pak J Bot. 2005;37:131–139. [Google Scholar]
- Singh RK, Flowers TJ (2010) The physiology and molecular biology of the effects of salinity on rice. In: Pessarakli M (ed) Handbook of plant and crop stress, 3rd edn. Publisher: Taylor and Francis, Florida, USA, pp 899–939
- Singh RK, Mishra B. Role of central soil salinity research institute in genetic improvement of rice in India. In: Sharma SD, Prasad Rao U, editors. Genetic improvement of rice varieties of India. New Delhi, India: Today and Tomorrow Printers & Publishers; 2004. pp. 189–242. [Google Scholar]
- Singh NK, Bracker CA, Hasegawa PM, Hand AK, Bucke S, Hermodso MA, Pfankoch E, Regnier FE, Bressan RA. Characterization of Osmotin: a Thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol. 1987;85:529–536. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, LaRosa PC, Handa AK, Hasegawa PM, Bressan RA. Hormonal regulation of protein synthesis associated with salt tolerance in plant cells. Proc Natl Acad Sci. 1987;84:739–743. doi: 10.1073/pnas.84.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Singh KN, Mishra B, Sharma SK, Tyagi NK (2004) Harnessing plant salt tolerance for overcoming sodicity constraints: an Indian experience. Advances in sodic land reclamation. In: Concept Paper for the International Conference on “Sustainable Management of Sodic Soils”. International Conference on “Sustainable Management of Sodic Soils”. Lucknow, India: UP Land Development Corporation, pp 81–120
- Singh RK, Adorada DL, Magsino C, Roque Z, Tamayo N, Gregorio GB (2005) Effect of relative humidity and temperature on salinity tolerance of rice. In: Plant breeding, genetics and biotechnology (PBGB) Division Biennial report 2004–2005. Manila, Philippines: IRRI, pp 19–21
- Singh RK, Gregorio G, Ismail A. Breeding rice varieties with tolerance to salt stress. J Indian Society of Coastal Agric Res. 2008;26:16–21. [Google Scholar]
- Singh RK, Redona ED, Refuerzo L (2010) Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice. In: Pareek A, Sopory SK, Bohnert HJ (eds) Abiotic stress adaptation in plants: physiological, molecular and genomic foundation Netherland, Springer, pp 387–415
- Singh AK, Gopalakrishnan S, Singh VP, Prabhu KV, Mohapatra T, Singh NK, Sharma TR, Nagarajan M, Vinod KK, Singh D, Singh UD, Chander S, Atwal SS, Seth R, Singh VK, Ranjith KE, Singh A, Anand D, Khanna A, Yadav S, Goel N, Singh A, Shikari AB, Singh A, Marathi B. Marker assisted selection: a paradigm shifts in basmati breeding. Indian J Genet Plant Breed. 2011;71:120–128. [Google Scholar]
- Singh R, Singh Y, Xalaxo S, et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016;242:278–287. doi: 10.1016/j.plantsci.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Singh RK, Krishnamurthy SL, Gautam RK (2021) Breeding approaches to develop rice varieties for salt-affected soils. In: Minhas PS, Yadav RK, Sharma PC (Eds), Managing salt-affected soils for sustainable agriculture. ICAR-DKMA, New Delhi, pp 227–251
- Solis CA, Yong MT, Vinarao R, Jena K, Holford P, Shabala L, Zhou M, Shabala S, Chen ZH. Back to the Wild: on a quest for donors toward salinity tolerant rice. Front Plant Sci. 2020;11:323. doi: 10.3389/fpls.2020.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa H, Shimodate T, Fukuta Y, Ueda T, Yano M, Yamaya T, Kameya T, Sato T. Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res. 2004;89:85–95. doi: 10.1016/j.fcr.2004.01.026. [DOI] [Google Scholar]
- Takehisa H, Ueda T, Fukuta Y, Obara M, Abe T, Yano M, Yamaya T, Kameya T, Higashitani A, Sato T. Epistatic interaction of QTLs controlling leaf bronzing in rice (Oryza sativa L.) grown in a saline paddy field. Breed Sci. 2006;56:287–293. doi: 10.1270/jsbbs.56.287. [DOI] [Google Scholar]
- Tanaka J, Hayashi T, Iwata H. A practical, rapid generation-advancement system for rice breeding using simplified biotron breeding system. Breed Sci. 2016;66:542–551. doi: 10.1270/jsbbs.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerakulpisut P, Bunnag S, Kongngern K. Genetic diversity, salinity tolerance and physiological responses of NaCl of six rice (Oryza sativa. L) cultivars. Asian J Plant Sci. 2005;4:562–573. doi: 10.3923/ajps.2005.562.573. [DOI] [Google Scholar]
- Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Tian L, Tan L, Liu F, Cai H, C and Sun C, Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J Genet Genom. 2011;38:593e601. doi: 10.1016/j.jgg.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Usatov AV, Alabushev AV, Kostylev PI, Azarin KV, Makarenko MS, Usatova OA. Introgression the Saltol QTL into the elite rice variety of Russia by marker-assisted selection. Am J Agric Biol Sci. 2015;10:165–169. doi: 10.3844/ajabssp.2015.165.169. [DOI] [Google Scholar]
- Vu HTT, Le DD, Ismail AM, Le HH. Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (Oryza sativa L.) to cope with climate change in Vietnam. Aus J Crop Sci. 2012;6:1649–1654. [Google Scholar]
- Wang Z, Wang J, Bao Y, Wu Y, Zhang H. Quantitative trait loci controlling rice seed germination under salt stress. Euphytica. 2011;178:297–307. doi: 10.1007/s10681-010-0287-8. [DOI] [Google Scholar]
- Wang Z, Cheng J, Chen Z, Huang J, Bao Y, Wang J, Zhang H. Identification of QTLs with main, epistatic and QTL x environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor Appl Genet. 2012;125:807–815. doi: 10.1007/s00122-012-1873-z. [DOI] [PubMed] [Google Scholar]
- Wang S, Cao M, Ma X, Chen W, Zhao J, Sun C, Tan L, Liu F. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front Plant Sci. 2017;8:1427. doi: 10.3389/fpls.2017.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Huang M, Tao X, Guo T, Chen Z, Xiao W. Quantitative trait loci identification and meta-analysis for rice panicle-related traits. Mol Genet Genomics. 2016;291:1927–1940. doi: 10.1007/s00438-016-1227-7. [DOI] [PubMed] [Google Scholar]
- Xie JH, Zapata-Arias FJ, Shen M, Afza R. Salinity tolerant performance and genetic diversity of four rice varieties. Euphytica. 2000;116:105–110. doi: 10.1023/A:1004041900101. [DOI] [Google Scholar]
- Yao M, Wang J, Chen H, Zhai H, Zhang H. Inheritance and QTL mapping of salt tolerance in rice. Rice Sci. 2005;12:25–32. [Google Scholar]
- Yeo AR, Flowers TJ. Accumulation and localization of sodium ions within the shoots of rice (Oryza sativa) varieties differing in salinity resistance. Physiol Plant. 1982;56:343–348. doi: 10.1111/j.1399-3054.1982.tb00350.x. [DOI] [Google Scholar]
- Yoshida S (1981) Fundamental of rice crop science. International Rice Research Institute, Los Baños, Laguna, Philippines, 269
- Yoshida S, Forno DA, Cock JK, Gomez KA (1976) Routine procedure for growing rice plants in culture solution. In: Laboratory manual for physiological studies of rice. 3rd edn. The International Rice Research Institute, Los Banos, Laguna, Philippines, pp 61–65
- Zang J, Sun Y, Wang Y. Dissection of genetic overlap of salt tolerance QTLs at the seedling and tillering stages using backcross introgression lines in rice. Sci China Series C- Life Sci. 2008;51:583–591. doi: 10.1007/s11427-008-0081-1. [DOI] [PubMed] [Google Scholar]
- Zayed BA, Abd El-azeem KS, Osama AMA (2014) Physiological characterization of Egyptian salt tolerantrice varieties under different salinity levels. Life Sci 11(10):1264–1272
- Zeng LH, Shannon MC. Salinity effects on seedling growth and yield components of rice. Crop Sci. 2000;40:996–1003. doi: 10.2135/cropsci2000.404996x. [DOI] [Google Scholar]
- Zeng LH, Shannon MC. Effects of salinity on grain yield and yield components of rice at different seeding densities. Agron J. 2000;92:418–423. doi: 10.2134/agronj2000.923418x. [DOI] [Google Scholar]
- Zeng LH, Shannon MC, Lesch SM. Timing of salinity stress affects rice growth and yield components. Agric Water Manag. 2001;48:191–206. doi: 10.1016/S0378-3774(00)00146-3. [DOI] [Google Scholar]
- Zeng L, Shannon MC, Grieve CM. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica. 2002;127:235–245. doi: 10.1023/A:1020262932277. [DOI] [Google Scholar]
- Zeng L, Poss JA, Wilson C, Draz ASE, Gregorio GB, Grieve CM. Evaluation of salt tolerance in rice genotypes by physiological characters. Euphytica. 2003;129:281–292. doi: 10.1023/A:1022248522536. [DOI] [Google Scholar]
- Zhang GY, Guo Y, Chen SL, Chen SY. RFLP tagging of a salt tolerance gene in rice. Plant Sci. 1995;110:227–234. doi: 10.1016/0168-9452(95)04219-K. [DOI] [Google Scholar]
- Zhang X, Long Y, Huang J, Xia J. OsNAC45 is involved in ABA response and salt tolerance in rice. Rice (NY) 2020;13(1):79. doi: 10.1186/s12284-020-00440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhao H, Liu H, Wang J, Zou D. QTL analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice. Euphytica. 2015;201:109–121. doi: 10.1007/s10681-014-1192-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.