Abstract
The Sigma 1 receptor (Sigmar1) is a ubiquitously expressed multifunctional inter-organelle signaling chaperone protein playing a diverse role in cellular survival. Recessive mutation in Sigmar1 have been identified as a causative gene for neuronal and neuromuscular disorder. Since the discovery over 40 years ago, Sigmar1 has been shown to contribute to numerous cellular functions, including ion channel regulation, protein quality control, endoplasmic reticulum-mitochondrial communication, lipid metabolism, mitochondrial function, autophagy activation, and involved in cellular survival. Alterations in Sigmar1’s subcellular localization, expression, and signaling has been implicated in the progression of a wide range of diseases, such as neurodegenerative diseases, ischemic brain injury, cardiovascular diseases, diabetic retinopathy, cancer, and drug addiction. The goal of this review is to summarize the current knowledge of Sigmar1 biology focusing the recent discoveries on Sigmar1’s molecular, cellular, pathophysiological, and biological functions.
Keywords: Sigmar1, molecular structure, physiological function, cellular function, biological function
Introduction
Sigma Receptor
Sigma receptors were first proposed to be a subclass of opioid receptors based on the observations of the psychotomimetic actions of (±)-SKF-10,047 (N-allylnormetazocine) and other racemic benzomorphans on behavior in dogs (Martin et al., 1976). The complex pharmacology of this racemic compound led to the naming of “Sigma opioid receptors” as a subtype of the opioid receptor family (Martin et al., 1976). Subsequent pharmacological and behavioral studies revealed (–)-SKF-10,047 binds to μ and κ opioid receptors, whereas the (+)-SKF-10,047 isomer binds with high affinity to the sigma receptor (Su, 1982). Therefore, the protein was named “Sigma receptor” by Su to distinguish it from opioid receptors (Su, 1982).
Two subtypes of Sigma receptors have been proposed based on their drug selectivity pattern and molecular mass: Sigma-1 receptor (Sigmar1) and Sigma-2 receptor (Sigmar2) (Su, 1982; Hellewell and Bowen, 1990). Sigmar1 is characterized by a higher affinity for dextrorotatory benzomorphans rather than its levorotatory isomers (Su, 1982). On the other hand, Sigmar2 exhibit an equal or greater affinity for the levorotatory benzomorphans isomers than their dextrorotatory counterparts (Hellewell and Bowen, 1990). Subsequent studies demonstrate that these two subtypes of Sigma receptors mediate different cellular and physiological functions. Though recent studies identified transmembrane protein 97 (TMEM97) as Sigmar2 (Alon et al., 2017), the literature contains conflicting evidence concerning the sequence, structure, and function of Sigmar2. Sigmar1 was successfully cloned in 1996 and has been more extensively examined in different research areas (Hanner et al., 1996). In this review article, we will focus on the recent discoveries concerning the molecular, cellular, pathophysiological, and biological functions of Sigmar1.
Molecular Characterization and Structure of Sigmar1
Sigmar1 is a multifunctional, ubiquitously expressed chaperone protein encoded by the SIGMAR1 gene. The Online Mendelian Inheritance in Man (OMIM) catalog entry describes SIGMAR1 to be located in the p arm of Chromosome 9 with the cytogenetic location of 9p13.3 and genomic coordinates of 9:34,634,721–34,637,825 [according to the National Center for Biological Information (NCBI)].
Structure
The molecular characterization of Sigmar1 began with the purification and cloning of Sigmar1-binding site from guinea pig liver using Sigmar1 specific probes such as benzomorphan (+) [3H] pentazocine and arylazide(−)[3H] azidopamil (Hanner et al., 1996). The molecular mass as determined by radiation inactivation of a pentazocine-labeled Sigmar1-binding site yielded a value of 24 ± 2 kDa. However, subsequent cloning of cDNA using degenerate oligonucleotides and cDNA library screening showed Sigmar1 protein isolated from guinea pig liver had 223 amino acids (aa) with a molecular mass of 25,314 Da (25.3 kDa) with at least one putative transmembrane segment. Human Sigmar1 (hSigmar1) cloned from human placental choriocarcinoma (JAR) cells cDNA library also predicted to have a protein of 223 amino acids with a single putative transmembrane domain (Kekuda et al., 1996). Sigmar1 mRNA (1.7 kb) was expressed in several human and guinea pig tissues, and the highest densities were found in liver, kidney, and steroid producing tissues such as placenta, ovary, and adrenal gland (Hanner et al., 1996; Kekuda et al., 1996). Subsequent cloning and functional characterization of mouse and rat Sigmar1 showed similar results having 223 aa (Seth et al., 1997, 1998; Mei and Pasternak, 2001). The sequence of murine Sigmar1 showed homology to guinea pigs (87% identity and 91% similarity), rats (92% identity and 96% similarity), and humans (90% identity and 93% similarity) (Seth et al., 1997). Rat Sigmar1 has an open reading frame of 672 base pairs (bp) flanked with non-coding regions of 30 bp at 5′ and 880 bp at 3′ of the coding region (Seth et al., 1998). Rat Sigmar1 has two transmembrane domains with 93.3% sequence homology with the mouse Sigmar1, 93.7% with that of guinea pig, and 96.0% with that from human (Mei and Pasternak, 2001).
Both the murine and human Sigmar1 gene (approximately 7 kb) is made up of 4 exons and 3 introns: exon 3 is the shortest one, and exon 4 of the protein is the longest one (Seth et al., 1997; Prasad et al., 1998; Figure 1A). Both mouse and rat Sigmar1 cDNA has a poly(A) tail, an upstream polyadenylation signal (AATAAA), and the protein has an amino acid sequence of MPWAVGRR at the N-terminal (believed to be ER retention signal) (Seth et al., 1997, 1998). Subsequent studies have identified the presence of a Phenyl-A region in Sigmar1 as a crucial structural feature in determining the substrate specificity for Sigmar1 ligands (Ablordeppey et al., 2002). Furthermore, the structural analysis showed that the two arginine motifs at the N-terminus of Sigmar1 are required for ER membrane targeting (Schutze et al., 1994). In addition, studies have shown that the presence of Ser99 to Leu106 residues in Sigmar1 protein located in the putative transmembrane domain play a crucial role in ligand binding and receptor-ligand interaction (Yamamoto et al., 1999). Investigation of Sigmar1 ligand binding sites in Jurkat human T lymphocyte suggested two spliced variants lacking exon 3 (deletion of 31 amino acids), which failed to bind with Sigmar1 ligands suggesting Exon 3 as the ligand-binding (Ganapathy et al., 1999). Subsequent studies identified six spliced variants in mouse Sigmar1 formed either by exon skipping or alternative 3′ and 5′ splicing to generate truncated proteins. Differential expressions of these Sigmar1 variants were observed across different organs (Pan et al., 2017).
FIGURE 1.
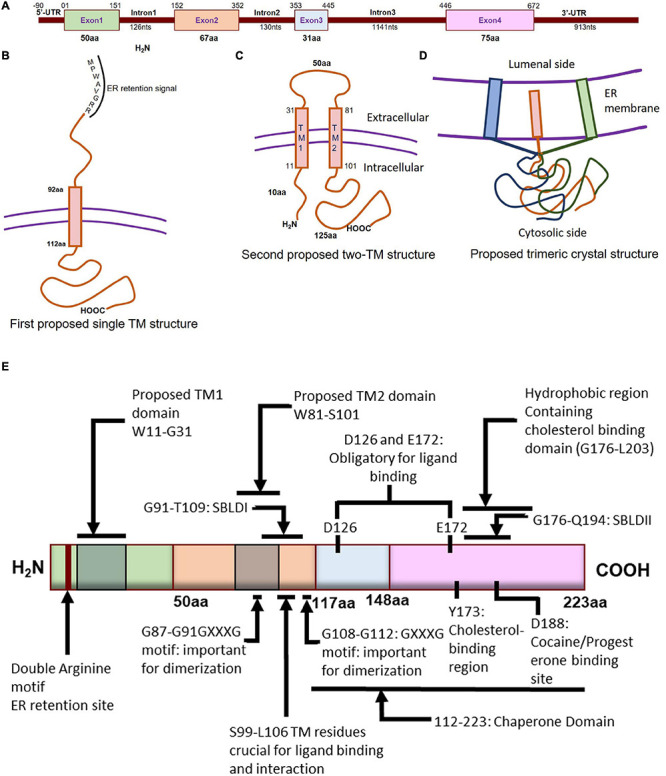
Molecular characterization of Sigmar1. (A) Schematic diagram representing the genetic structure of the SIGMAR1 gene, including the exon and, intron lengths in terms of nucleotides and amino acids. (B) Simplified schematic of full-length Sigmar1 topology showing the first predicted one transmembrane (TM) structure with a TM domain-containing amino acid (aa) residues from 92 to 112, MPWAVGRR as the ER retention site, and regions important for ligand binding (exon 3 and Ser99-Leu106). (C) Simplified schematic of the full-length Sigmar1 topology showing the second predicted two TM structures with two TM domains (TM1 containing aa residues 11 to 31 and TM2 containing 81 to 101) and an extracellular loop (containing 50aa). Both N- and C- terminal of the protein are on the same side. (D) Simplified schematic for the recent crystal structure of full-length Sigmar1 suggesting the trimeric structure of the protein with trimerization of three single transmembrane domains and each transmembrane domain being tightly associated with one promoter. The C-terminal is located on the cytosolic side. (E) Schematic diagram representing full protein structure of Sigmar1 with structural details for receptor dimerization, ligand binding, cholesterol-, cocaine-, progesterone- binding.
Transmembrane Topology
Guinea pig Sigmar1 showed substantial sequence homology with fungal sterol C8-C7 isomerase (ERG2: 30.3% identical and 66.4 similarity), and hydrophobicity plots predicted one putative transmembrane segment at the N terminus (Hanner et al., 1996; Figure 1B). However, unlike the fungal sterol C8-C7 isomerase (Hanner et al., 1996), the Sigmar1 lacks sterol isomerase activity and shares no sequence homology with any known mammalian proteins, including the mammalian C8-C7 sterol isomerase (Labit-Le Bouteiller et al., 1998). Further, studies from two independent groups (Aydar et al., 2002; Hayashi and Su, 2007) suggested Sigmar1 having two transmembrane domains where both N- and C- terminal of Sigmar1 resides on the same side of the membrane. However, the proposed model by Aydar et al. suggested that both the N- and C-termini being intracellular, whereas the proposed model by Hayashi et al. showed them extracellular. Aydar et al. further proposed transmembrane domain 1 with amino acid (aa) residues 9 to 28 and transmembrane domain 2 with residues 81–101 (Aydar et al., 2002). Studies also suggested that Sigmar1 has a short N-terminal (10 aa), an extracellular loop (50 aa), and a longer C-terminal (125 aa) with a sequence similar to sterol isomerase (Aydar et al., 2002). Moreover, Sigmar1’s both N- and C- terminal are hypothesized to reside intracellularly (Aydar et al., 2002; Figure 1C).
More recently, Sigmar1’s crystal structure determined by overexpressing the FLAG-tagged Sigmar1 in baculovirus, affinity purification and reconstitution of the protein into lipidic cubic phase, and crystallization by the hanging drop technique (Schmidt et al., 2016). The crystal structure of the Sigmar1 protein was suggested to possess a single TM domain with a short N-terminus facing the ER lumen, while most of the protein bulk was located on the cytosolic side of the ER membrane (Schmidt et al., 2016; Figure 1D). In contrast, Sigmar1 transmembrane topology determined by electron microscopic examination of ascorbate peroxidase 2 (APEX2)-tagged Sigmar1 protein in transfected ND7/23 cells suggested the N-terminus of Sigmar1 facing the cytosol and the C-terminus facing the ER lumen (Mavylutov et al., 2018). Though the Sigmar1’s topology identified in the GFP-APEX2-tagged Sigmar1 and the crystal structure study (Schmidt et al., 2016) showed similarity, they differ whether the facing of the N-terminus of Sigmar1 protein faces the cytosol or ER lumen.
Ligand Binding
Previous work using site-directed mutagenesis showed Asp126 and Glu172 are essential for high-affinity ligand binding, as mutation of either resulted in a profound loss of ligand-binding activity (Seth et al., 2001; Figure 1E). Study using the hydropathy plot suggested that a hydrophobic segment (aa 176 to 203) in Sigmar1 contains the cholesterol-binding domain with the conserved L/V-X1-5- Y-X1-5-K/R motif near the ligand-binding domain (Palmer et al., 2007). This study further suggested that amino acid residue Y173 is crucial for cholesterol binding (Palmer et al., 2007). Later studies involving photolabeling of the protein have suggested that Sigmar1 has two sterol binding domain-like motifs, namely SBLDI (aa 91–109) and SBLD II (aa 176–194). Further studies have demonstrated that these two domains are close enough and juxtaposed to form a ligand-binding site responsible for ligand binding and lipid raft remodeling (Fontanilla et al., 2008; Pal et al., 2008). 3D modeling of the Sigmar1 protein structure supports the above-described study, providing further evidence for the presence of β-strands in the C-terminal half of the protein (at residues 111–116, 133–135, 144–146, and 158–164) (Laurini et al., 2011). The crystal structure of Sigmar1 also suggested the Sigmar1 ligand binding through a charge-charge interaction with the highly conserved Glu172 and Asp126 (Schmidt et al., 2016).
Oligomerization
Several studies suggested the existence of the oliogomeric structure of Sigmar1 and ligand-dependent changes in Sigmar1 oligomerization as well as activity. Studies using photo-affinity labeling have demonstrated the dimeric structure or oligomer of dimeric structures of Sigmar1, which is associated with the presence of two GXXXG motifs at residues 87–91 (TM2) and residues 108–112 (C-terminal of SBLDII) (probable oligomerization motifs) (Chu et al., 2013). Fluorescence resonance energy transfer spectrometry analysis of heterologously expressed Sigmar1 in COS-7 cells showed the presence of multiple oligomeric forms. Treatment with Sigmar1 ligands altered these oligomer forms where Sigmar1 agonist [(+)-pentazocine] favored the monomers and dimers, and Sigmar1 antagonist (haloperidol) favored higher order Sigmar1 oligomers (Mishra et al., 2015). The crystal structure of Sigmar1 proposed a trimeric structure of Sigmar1 where Sigmar1 possesses a single transmembrane domain at N-terminus (Schmidt et al., 2016). Size-exclusion chromatography with multi-angle light scattering experiments as well as native polyacrylamide gel electrophoresis analysis suggested the presence of Sigmar1 oligomers ranging in size from hexamers to as large as 15-mers (Schmidt et al., 2016). All these studies suggested ligand mediated oligomerization as an important characteristics for Sigmar1 activity, but the molecular mechanism of Sigmar1 oligomerization and resultant changes in Sigmar1’s function remained elusive.
Tissue Distribution of Sigmar1
Extensive Northern blot assays carried out in animals and humans have demonstrated the ubiquitous expression of Sigmar1 throughout the body tissures, including heart, liver, brain, placenta, thymus, lung, kidney, stomach, skeletal muscle, and pancreas (Kekuda et al., 1996; Mei and Pasternak, 2001). The characterization of spliced variants in mice revealed the presence of full-length protein and all of the spliced variants of Sigmar1 across different organs, including lung, liver, heart, spleen, kidney, brain, and various regions (Pan et al., 2017). Sigmar1 has been shown to be expressed in the spleen in mice and guinea pigs (Su et al., 1988; Mei and Pasternak, 2001) and in peripheral blood leukocytes in humans (Wolfe et al., 1988). However, expression of Sigmar1 protein levels in different tissues varied, with the highest expression in the liver (Kekuda et al., 1996; Mei and Pasternak, 2001; Pan et al., 2017). The Human Protein Atlas (1 /ENSG00000147955 -SIGMAR1/tissue) summarizes the expression of Sigmar1 across different tissues of the human body and shows the highest level of Sigmar1 expression in the brain (cerebellum), liver and placenta; moderate levels in heart, skeletal muscle, different glands (parathyroid, adrenal, thyroid), pancreas, lungs, GI tract, kidneys, urinary bladder, and male and female reproductive organs; and low levels in soft tissue, with no report of expression in the bone marrow. Studies on Sigmar1 expression at the sub-tissue level have shown its presence in astrocytes, oligodendrocytes, gangliosides, and basal amygdala of the neuronal system (Palacios et al., 2003; Choi et al., 2016; Kasahara et al., 2017; Zhang et al., 2017a). Moreover, Sigmar1 also has been found in retinal tissue, bile duct, breast tissue, bone marrow-derived macrophages, endothelial cells (Amer et al., 2013; Barbieri et al., 2003; Xu et al., 2014; Mavlyutov et al., 2015a; Rosen et al., 2019). Despite its ubiquitous tissue distribution, studies to date have only attempted to explore the pathophysiological role of Sigmar1 in the neuronal, cardiovascular, kidney, and retinal systems.
Subcellular Localization
Extensive studies over the last 40 years have demonstrated that the subcellular localization of Sigmar1 is tissue-specific. Comprehensive studies have shown Sigmar1 localization at the mitochondrial-associated membranes (MAM) (co-localized with Mito-DsRed) in CHO Cells (Hayashi and Su, 2007) and the plasma membrane, where it interacts with ion channels (reviewed in Su et al., 2009). Studies have also shown localization of Sigmar1 at the endoplasmic reticulum (ER) and nuclear envelope in human immune cells (Dussossoy et al., 1999). Sigmar1 present in the cell membrane negatively regulating Kv1.4 potassium channel function (Aydar et al., 2002). Extensive immuno-electron microscopic (EM) data have shown that Sigmar1’s sub-cellular localization largely depends on cell and organ types (Mavlyutov and Ruoho, 2007; Mavlyutov et al., 2010, 2011, 2012, 2013, 2015a, 2016, 2017a; Mavlyutov and Guo, 2017; Yang et al., 2017). For example, Sigmar1 was localized to the nuclear envelope with no localization was observed in ER in the photoreceptor cells (Mavlyutov et al., 2015a), whereas Sigmar1 localization was observed in the nucleoplasmic reticulum and the nucleus in the NSC34 cell line (Mavlyutov et al., 2017; Figure 2 and Table 1). Moreover, immuno-EM examinations were unable to detect Sigmar1 at the plasma membrane (Mavlyutov and Ruoho, 2007; Mavlyutov et al., 2010, 2011, 2012, 2013, 2015a, 2016, 2017a; Mavlyutov and Guo, 2017; Yang et al., 2017). Mavlyutov group also showed that Sigmar1’s C-terminal resides inside ER-lumen and the N-terminus resides in the cytosol (Mavylutov et al., 2018), which is opposite to the recently derived crystal structure proposing that Sigmar1’s C-terminal reside on the cytosolic side of the ER (Schmidt et al., 2016). Sigmar1 was also detected on mitochondria of rat liver (depicted as Sigmar1-like receptor) using ligand-based studies and immunostaining (Klouz et al., 2002). Using ligand binding assays using (+) pentazocine and enzyme binding or activity assays (monoamine oxidases, cytochrome c oxidases) in the mitochondrial fraction from rat liver and brain, the group show Sigmar1 to reside in the outer mitochondrial membrane and this Sigmar1 in the liver to have a different binding site for ligands compared to that in the brain (Klouz et al., 2002). The existence of Sigmar1 on the mitochondria was confirmed by colocalization of Sigmar1 on mitochondria on liver tissue section when stained with Sigmar1 antibody and a mitochondrial marker (Klouz et al., 2002). Interestingly, subcellular fractionation of neural tissues from the mutant SOD1Tg mice showed Sigmar1 accumulation in mitochondrial fractions (Watanabe et al., 2016). The apparent discrepancies’ in Sigmar1’s subcellular localization results from the differences in cell types, methods of detection, and reagents used (i.e., antibody) (summarized in Table 1). All these studies to date suggest the organ- and tissue-specific localization and function of Sigmar1 (Mavlyutov and Ruoho, 2007; Mavlyutov et al., 2010, 2011, 2012, 2013, 2015a, 2016, 2017a; Mavlyutov and Guo, 2017; Yang et al., 2017).
FIGURE 2.
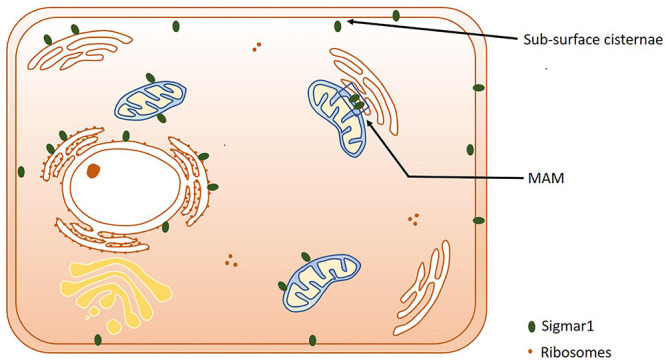
Subcellular localization of Sigmar1. Cartoon showing a summary of the sub-cellular localization of Sigmar1 as evidenced by several studies using various cell types. Overall, the presence of Sigmar1 has been detected on the mitochondria-associated ER membrane (MAM), plasma membrane, ER membrane, nuclear membrane, mitochondria-associated ER membrane, mitochondrial membrane, nucleoplasmic reticulum and sub-surface cisternae in different cell types including CHO cells, human B and T cells, photoreceptor cells, and neuronal cell lines including NSC34 cells and neuro2a cells.
TABLE 1.
Major studies on identifying Sigmar1’s subcellular organelle localization (in chronological order).
| Subcellular organelles | Experimental methods | Cells or Tissues | Antibody and antigen | Antibody validation | References |
| Mitochondria Microsomal Synaptosomal fractions | Western blotting on subcellular fractions | Rat brain | Anti-Sigmar1 Rabbit Polyclonal antibody raised against synthetic amino acid residues 143–163 of Sigmar1 (produced by authors) | Not listed | Yamamoto et al., 1999 |
| Plasma membrane Mitochondrial membranes Vesicles or elongated cisternae of ER | Immunoelectron microscopy | Adult rat brain hypothalamus and hippocampus sections | Anti-Sigmar1 Rabbit Polyclonal IgG raised against synthetic peptide of Sigmar1 amino acid sequence 143–162 (produced by authors) | Antibody specificity confirmed through absence of staining in brain tissue sections when co-incubated with Sigmar1 specific antigen | Alonso et al., 2000 |
| Perinuclear areas Plasmalemmal regions of cell-cell contact Growth cones of neurites | Fluorescence microscopy of co-immunocytohistochemistry and co-immunoprecipitation of endogenous Sigmar1 with Ankyrin B and IP3R3 receptors | Mouse neuroblastoma x rat glioma hybrid NG-108-15 cells | Anti-Sigmar1 Rabbit antibody raised against N-terminal residues of mouse Sigmar1 (produced by authors) | Not listed | Hayashi and Su, 2001 |
| Mitochondrial membranes | i. Radioisotope labeled Sigmar1 agonist [3H](+)-pentazocine binding assay ii. Immunofluorescence | i. Isolated rat liver mitochondria ii. Rat liver sections | Anti-Sigmar1 Rabbit Polyclonal IgG (Alonso et al., 2000) | Validated by original contributors at (Alonso et al., 2000) | Klouz et al., 2003 |
| Plasma membranes | i. Exogenous expression of N-terminal and C-terminal GFP tagged Sigmar1 constructs and fluorescence microscopy ii. Co-immunoprecipitation with anti-Kv1.4 antibody | i. Xenopus Oocytes ii. Triton X-100 solubilized membrane lysates of rat pituitary gland | Anti-Sigmar1 Rabbit Polyclonal IgG (Yamamoto et al., 1999) | Not listed | Aydar et al., 2002 |
| i. ER associated detergent insoluble lipid droplets/microdomains ii. Sigmar1 immunostained reticular structure was noted negative for endosomes (EEA-1), mitochondria (Mitotracker, bcl-2), lysosomes (Lyso Tracker, LAMP-1), synaptic vesicles (Synapsin II), plasma membrane (Fas and CTx-B), golgi (GM130), ER-associated proteins | Fluorescence microscopy; cells were specially treated with 0.02% SDS for 10 min | NG-108 cells (Hayashi and Su, 2003a, b), Rat hippocampal differentiated oligodendrocytes culture (Hayashi and Su, 2004) | i. Exogenously expressed C-terminal Enhanced Yellow Fluorescent Protein (EYFP)-tagged mouse Sigmar1 construct ii. Polyclonal rabbit anti-guinea pig Sigmar1 antibody raised against guinea pig Sigmar1 amino acid sequence 144–165 (Hayashi and Su, 2003b). | i. Exogenous overexpression ii. Anti-Sigmar1 staining pattern have been visually compared to expression pattern of exogenously expressed Sigmar1-EYFP plasmid construct (Hayashi and Su, 2003a, b) | Hayashi and Su, 2003a, b, 2004 |
| Mitochondria Mitochondria-associated ER membranes (MAM) Nuclear and microsomal fractions | i. Immunofluorescence staining of endogenous Sigmar1 in cells transfected with Mitochondria and ER fluorescent constructs, ii. Western blot analysis on ultracentrifugation based subcellular fractionations | Chinese hamster ovary (CHO) cells | Anti-Sigmar1 Rabbit antibodies raised against amino acid residues 52–69 and 143–165 of Rat Sigmar1 (produced by authors) | Western blot on CHO cell lysates and on different rat organs | Hayashi and Su, 2007 |
| Focal adhesion contacts (FACs) | Co-localization of Sigmar1 with FAC protein Talin in immunofluorescence staining | CHO-K1 cells | Anti-Sigmar1 Rabbit antibody raised against Maltose Binding Protein (MBP)-Guinea Pig full length Sigmar1 fusion protein (Ramachandran et al., 2007) (produced by authors) | Western blot analysis using guinea pig Sigmar1 overexpressing COS-7 cells as positive control (Ramachandran et al., 2007) | Mavlyutov and Ruoho, 2007 |
| i. Motor neurons of mouse brain Medulla and spinal cord ii. Cholinergic post-synaptic terminals co-localized to Kv2.1 potassium channels iii. Putative subsurface ER cisternae close to plasma membrane | i. Immunohistochemistry ii. Co-immunofluorescence with cholinergic neuron markers iii. Immuno-electron microscopy | Mouse brain and spinal cord sections | Anti-Sigmar1 Rabbit antibody raised against Maltose Binding Protein (MBP)-Guinea Pig full length Sigmar1 fusion protein (Ramachandran et al., 2007) | Sigmar1 knockout mice brain sections as negative control in immunohistochemistry | Mavlyutov et al., 2010, 2012 |
| Mitochondria ER | Fluorescence microscopy | Mouse neuroblastoma Neuro-2a cells | Exogenously expressed eGFP and mCherry tagged Sigmar1’s correlative, visual colocalization observation with erRFP and mtGFP labeled ER and mitochondria, respectively | Exogenous overexpression | Shioda et al., 2012 |
| Mitochondria and MAM fractions | Western blot analysis on subcellular fractions | Chinese hamster ovary (CHO) cells | Anti-Sigmar1 Rabbit antibodies raised against Rat Sigmar1 (Hayashi and Su, 2007) | Western blot on CHO cell lysates and on different rat organs (Hayashi and Su, 2007) | Mori et al., 2013 |
| ER and Nuclear Envelope | Confocal immunofluorescence staining | NG108 and Neuro-2a cells | Anti-Sigmar1 (Santa Cruz Biotechnology) | Commercial vendor | Tsai et al., 2015 |
| Predominantly in nuclear envelope, sparsely at ER cisternae located subsurface of plasma membranes | Immuno-electron microscopy | Mouse retinal photoreceptor cells | Anti-Sigmar1 Rabbit antibody raised against Maltose Binding Protein (MBP)-Guinea Pig full length Sigmar1 fusion protein (Ramachandran et al., 2007) | Reported at (Ramachandran et al., 2007) | Mavlyutov et al., 2015b |
| Plasma membrane ER Nuclear envelope | Immuno-electron microscopy | Mouse and rat dorsal root ganglion (DRG) cells | Anti-Sigmar1 Rabbit antibody raised against Maltose Binding Protein (MBP)-Guinea Pig full length Sigmar1 fusion protein (Ramachandran et al., 2007) | Validated through absence of immunofluorescence staining using Sigmar1 knockout mouse DRG tissue | Mavlyutov et al., 2016 |
| Nucleus Nucleoplasmic reticulum Plasma membrane subsurface ER cisternae | Immuno-electron microscopy | NSC34 cells (produced by fusion of motor neuron enriched, embryonic mouse spinal cord cells with mouse neuroblastoma), Sigmar1-null NSC34 cells | Endogenous Sigmar1 stained with antibody as reported in Ramachandran et al. (2007), Exogenously expressed full length Sigmar1-GFP-APEX2 (ascorbate peroxidase 2) fusion construct were enhanced using 3, 3′-diaminobenzidine (DAB) incubation | Reported at (Ramachandran et al., 2007) | Mavlyutov et al., 2017 |
| i. ER, Mitochondria associated ER membranes (MAM) ii. Cholesterol-containing giant unilamellar vesicles (GUVs) | Immuno-fluorescence microscopy | HEK293T cells | i. Exogenously expressed GFP-tagged human Sigmar1 colocalization assessed with mCherry tagged Sec16β protein and immunofluorescence staining of mitochondrial outer membrane protein Tom20 ii. GFP-tagged Sigmar1 fusion constructs | i. and ii. Exogenous overexpression of GFP-tagged fusion constructs | Zhemkov et al., 2021 |
Physiological and Pathological Role of Sigmar1
An enormous amount of studies in the current literature have attempted to elucidate Sigmar1’s molecular role under physiological and pathological conditions in different organs. Sigmar1 global knockout (Sigmar1–/–) mouse models were used to explore Sigmar1’s physiological functions’ in different organs. To date, two separate lines of Sigmar1–/– mice reported were generated by gene targeting (Oprs1tm1Lmon/Oprs1tm1Lmon) (Langa et al., 2003) and gene trapping [Oprs1Gt(IRESBetageo)33Lex/Oprs1Gt(IRESBetageo)33Lex]2. Both of these Sigmar1 homozygous knockout mouse lines were viable and fertile. They did not display any overt phenotype compared with their wild-type littermates. However, all these studies were limited by cursory observations of limited sample sizes from mouse strains with mixed genetic backgrounds (Langa et al., 2003). Extensive studies of this Sigmar1–/– mouse reported the development of multiple pathological phenotypes: locomotor defects (Langa et al., 2003), significant nerve denervation (Bernard-Marissal et al., 2015), loss of motor neurons (Bernard-Marissal et al., 2015), and age−dependent motor phenotype (Watanabe et al., 2016). Studies also demonstrated the development of a depressive-like phenotype (Sabino et al., 2009a) and a gender-related anxiety, depressive-like and memory related alterations in the Sigmar1–/– mouse (Chevallier et al., 2011). Extensive research using Sigmar1 ligands (agonists and antagonists) revealed Sigmar1’s roles in several pathological conditions in different organs summarized below:
Sigmar1 in Cardiovascular Pathophysiology
The presence of Sigmar1 in the heart was initially reported using ligand binding assays (Ela et al., 1994; Novakova et al., 1995), and since then, pharmacologic targeting has led to hypotheses concerning the potential importance of Sigmar1 protein in the heart (Tagashira et al., 2010, 2011; Bhuiyan and Fukunaga, 2011). All studies to date have been limited to pharmacologic approaches using less selective ligands for Sigmar1 due to the unavailability of genetic models to study the functionality of Sigmar1 in the heart (Tagashira et al., 2010, 2011; Bhuiyan and Fukunaga, 2011). Subsequent studies from our group using Western blot analysis of Sigmar1 protein levels in whole-cell extracts from the thoracic aorta, left ventricle, and right ventricle of rats has revealed ubiquitous expression of Sigmar1 in the major components of the cardiovascular system (Bhuiyan and Fukunaga, 2009, 2011). We recently reported the development of cardiac contractile dysfunction and cardiac fibrosis in Sigmar1 null mice with aging (Abdullah et al., 2018). Hearts of Sigmar1–/– mice developed significant accumulations of irregularly shaped mitochondria and defects in mitochondrial respiratory function. We demonstrated a potential molecular function of Sigmar1 in regulating the mitochondrial bioenergetics that are essential to maintain normal cardiac contractile function (Abdullah et al., 2018). Extensive research has explored the effects of Sigmar1 ligands (agonists and antagonists) on the cardiovascular system using different in vitro and in vivo cardiac injury models as summarized below:
Cardiac Contractility
Initial studies done to characterize Sigmar1 showed that cardiomyocytes exhibit sigma receptor ligand-binding sites, and that several of these Sigmar1 ligands may alter cardiac contractility. Among these Sigmar1 ligands, (+)−3-PPP, (+)-pentazocine, and haloperidol altered the contractility, calcium influx, and rhythmic activity of cultured cardiomyocytes (Ela et al., 1994, 1996; Novakova et al., 1995; Monassier et al., 2007). Several of these Sigmar1 ligands showed ionotropic action on isolated neonatal and adult cardiomyocytes (Novakova et al., 1995). A direct interaction has also demonstrated between the Sigmar1 and human Ether-à-go-go-Related Gene (hERG) that promotes hERG protein level in n myeloid leukemia and colorectal cancer cells (Crottes et al., 2016). Sigmar1 increased hERG current density via a regulation of channel subunit maturation and stability in a chronic myeloid cell line (K562), HEK-293 cells, and Xenopus oocytes (Crottes et al., 2011). The hERG channel is a voltage-dependent K+ channel that regulates cardiac repolarization (Trudeau et al., 1995), but the role of Sigmar1 or Sigmar1 ligands in hERG channel activity in cardiomyocytes has never been studied. It has also been shown that both Sigmar1 agonists (SKF-10047 and (+)-pentazocine) and antagonists (haloperidol and ditolylguanidine) reversibly inhibited Na(v)1.5 channels to varying degrees in HEK-293 cells and COS-7 cells (Johannessen et al., 2009). However, all these four Sigmar1 ligands four ligands also inhibited Na(+) current in neonatal mouse cardiac myocytes (Johannessen et al., 2009). Sigmar1 was also involved in the major Ca2+ influx pathway through inhibiting store-operated Ca2+ entry (SOCE) and reducing the Ca2+ content of the intracellular stores in HEK cells and Sigmar1 expressed HEK cells (Srivats et al., 2016). Stable expression of a Sigmar1 in HEK cells and treatment with Sigmar1 agonists [(+) SKF10047] in CHO cells inhibited SOCE (Srivats et al., 2016). In contrast, Sigmar1 siRNA knockdown and treatment with Sigmar1 antagonists in CHO cells enhanced SOCE (Srivats et al., 2016). Studies demonstrated that haloperidol treatment is frequently accompanied by cardiovascular side effects, including QT interval prolongation and the occurrence of even lethal arrhythmias. Haloperidol treatment in guinea pigs significantly decreased the relative heart rate and prolonged QT interval of the isolated hearts from the haloperidol-treated animals. These effects were associated with the increased expression of Sigmar1 and ITPR (type 1 and type 2) in the atria of haloperidol-treated animals (Stracina et al., 2015). However, Sigmar1 ligands (DTG, PB28, and (+) SKF10047) inhibited Na+ (Nav) channels activity in Sigmar1 siRNA knockout HEK-293 cells indicating Sigmar1 independent effect in the Na+ activity of these ligands (Johannessen et al., 2009, 2011). Similarly, Sigmar1-independent inhibition of the Kv2.1 channel was achieved by sigma ligands (both agonists and antagonists) using Kv2.1-overexpressing HEK-293 cells with and without CRISPR/Cas9 Sigmar1 knockout (Liu et al., 2017). Therefore, Sigmar1 ligands may affect various ion channels via Sigmar1 as well as through a direct action of the ligand on the ion channel function. Overall, ion channel modulation by different Sigmar1 ligands affecting in vitro cell contractility was inconsistent. The molecular mechanisms of Sigmar1 interactions and direct involvement with these ion channels remained unknown.
Cardiac Hypertrophy
Extensive studies have been focused on the effects of Sigmar1 ligands (both agonists and antagonists) in heart tissues to explore the pathophysiological role of Sigmar1. Temporal study performed to demonstrate the time-dependent changes in Sigmar1 protein levels in the heart showed a significant negative linear correlation with the development of cardiac dysfunction in pressure overload-induced (PO) or transverse aortic constriction (TAC)-induced cardiac hypertrophy (Bhuiyan et al., 2010; Tagashira et al., 2010). Moreover, cardiac hypertrophy in mice induced by aortic banding also exhibit reduced expression levels of Sigmar1 protein in the brain and depression-like behavior, along with the development of impaired cardiac function (Ito et al., 2011). Sigmar1 activation using agonists has been shown to elicit cardioprotection in these rodent models of cardiac hypertrophy and heart failure (Bhuiyan and Fukunaga, 2009, 2011; Bhuiyan et al., 2010, 2011a,b, 2013; [citeskum]BR294,BR293,BR295,BR297,BR291,BR292[citeekum]Tagashira et al., 2010, 2011, 2013b, 2013c, 2013a, 2014a,Tagashira and Kobori, 2013). A number of studies have shown that stimulation of Sigmar1 using its agonists (e.g., dehydroepiandrosterone and fluvoxamine) elicit protective effects against PO-induced cardiac hypertrophy in ovariectomized rats and TAC-induced cardiac hypertrophy in mice (Bhuiyan and Fukunaga, 2009; Bhuiyan et al., 2010; Tagashira et al., 2010). These studies demonstrated that Sigmar1 activation by the agonist ameliorates cardiac hypertrophy and contractile dysfunction by activating the Akt-eNOS signaling pathway (Bhuiyan et al., 2010; Tagashira et al., 2010). This protective effect of Sigmar1 activation (by the use of its agonists) was ablated using Sigmar1 antagonist (NE-100 and haloperidol) (Bhuiyan and Fukunaga, 2009; Bhuiyan et al., 2010; Tagashira et al., 2010). In association with the activation of Akt-eNOS signaling, Sigmar1 activation by agonist also restored TAC-induced alterations in mitochondrial calcium mobilization and ATP production (Tagashira et al., 2013a, c, 2014a). Studies also showed Sigmar1 agonists could restore TAC-mediated disrupted interaction of Sigmar1 with ITPR and negatively regulate ryanodine receptors (Tagashira et al., 2013a, 2014a). However, Sigma1 inhibition by treatment with antagonists aggravated cardiac pathology with aggravation of impaired mitochondrial calcium mobilization, decreased ATP production, increased autophagosome accumulation, and mitochondrial dysfunction with increased mitochondrial fragmentation (Shinoda et al., 2016).
Several studies have demonstrated that the neurosteroid dehydroepiandrosterone (DHEA) serves as an endogenous ligand for Sigmar1, and DHEA treatment ameliorated PO-induced cardiac hypertrophy in ovariectomized rats (Bhuiyan and Fukunaga, 2009; Bhuiyan et al., 2011a; Tagashira et al., 2011). Upregulation of the Sigmar1 protein levels following fluvoxamine and DHEA treatments has been suggested to be responsible for Sigmar1’s cardioprotective action. Several studies using different Sigmar1 ligands showed that these ligands have different effects on Sigmar1 expression. In rats, chronic treatment with the Sigmar1 ligand E-5842 increased Sigmar1 mRNA expression in the brain (Zamanillo et al., 2000), whereas chronic treatment with imipramine decreased levels of Sigmar1 binding sites in the brain (Shirayama et al., 1993). Similarly, chronic haloperidol (a Sigmar1 antagonist) treatment promoted a reduction of Sigmar1 binding sites (Inoue et al., 2000). On the other hand, treatment with the Sigmar1 antagonist NE-100 did not alter Sigmar1 expression in the heart in vivo (Tagashira et al., 2010). Moreover, a combination of NE-100 with fluvoxamine nullified fluvoxamine-mediated anti-hypertrophic effects without altering the protein levels of Sigmar1 in the heart (Tagashira et al., 2010). Apparently, these differences in the modulation Sigmar1 mRNA expression and protein level by ligands result from the different methodologies used to examine Sigmar1, including in vivo vs. in vitro tests and binding assays vs. immunodetection. However, the direct role of Sigmar1 using genetic models has never tested in these cardiac-injury models.
Myocardial Infarction
It has also been suggested that Sigmar1 ligands play a potential cardioprotective role in ischemia/reperfusion (I/R) injury. Treatment with Sigmar1 ligand afobazole prevented the development of pathologic remodeling of the myocardium, maintained its inotropic function, and decreased the plasma level of brain natriuretic peptide in a rat model of myocardial infarction. Interestingly, afobazole treatment down-regulated the mRNA expression of angiotensin, vasopressin, glucocorticoid receptor, and Epac2 protein level in the infarcted myocardium (Kryzhanovskii et al., 2018). Another study also showed delayed cardioprotective effects of afobazole, evaluated by using echocardiography in an experim ental myocardial infarction model (rat model of acute myocardial ischemia) (Kryzhanovskii et al., 2017). It has been proposed that the cardiotropic effects of the anxiolytic afobazole were associated with Sigmar1 agonistic effects in cardiomyocytes (Kryzhanovskii et al., 2017, 2018). However, a recent study showed that chronic Sigmar1 activation ameliorated ventricular remodeling and decreased susceptibility to ventricular arrhythmias after myocardial infarction in rats (Fo et al., 2020). Sigmar1 activation following treatment with fluvoxamine improved cardiac function through reduced susceptibility to ventricular arrhythmias, mitigated myocardial fibrosis, lightened sympathetic remodeling and electrical remodeling, and upregulated Sigmar1 protein levels (Fo et al., 2020). Fluvoxamine also significantly prolonged the ventricular effective refractory period, shortened action potential duration, and reduced susceptibility to ventricular arrhythmias after MI (Fo et al., 2020). Similarly, treatment with a Sigmar1 agonist (PRE-084) in rats with I/R injuries improved cardiac hemodynamic parameters, including LV pressure development and left ventricular systolic pressure (Gao et al., 2018). Mechanistically, the protective effect of PRE-084 was associated with the reduction of apoptotic cell death with increased Bcl-2 levels and decreased Bax levels in cardiomyocytes. Sigmar1 dependent activation of the PI3K/Akt/eNOS signaling pathways has been suggested to inhibit I/R injury-induced apoptotic cell death (Gao et al., 2018). A recent study in MI mice showed that a decreased brain Sigmar1 played a vital role in the coexistence of increased HF via sympathoexcitation and mental disorders, such as depression or cognitive impairment (Ito et al., 2013). Interestingly, intracerebroventricular infusion of PRE084 in MI mice improved both mental disorder and cardiac function with lowered sympathetic activity. These protective effects were associated with the PRE084 induced recovery of the Sigmar1 expression in both the hypothalamus and hippocampus (Ito et al., 2013).
Atrial Fibrillation
Stimulation of Sigmar1 has also been shown to exhibit cardioprotection in tachycardia, atrial fibrillation, and asphyxia cardiac arrest. Sigmar1 agonist-mediated activation of Sigmar1 decreased the duration of stress-induced tachycardia without altering the peak heart rate in rats (Delaunois et al., 2013). This protective effect of Sigmar1 agonists was abrogated by Sigmar1 antagonists (Delaunois et al., 2013). Similar to the effects of Sigmar1 in ventricles, inhibition of Sigmar1 by treatment with antagonists altered atrial electrophysiology, reducing effective refractory period, action potential duration, and leading to increased inducibility and time of atrial fibrillation (Ye et al., 2019). Furthermore, inhibition of Sigmar1 by antagonists resulted in increased atrial fibrosis and reduced the levels of connexin 40 (a gap junction protein) (Ye et al., 2019), leading to slow conduction of electrical impulses across atria (Ye et al., 2019). Treatment with a Sigmar1 agonist reversed these effects rescuing the effects of Sigmar1 inhibition (Ye et al., 2019). Similarly, Sigmar1 activation by selective Sigmar1 ligands also protected the depression-induced atrial fibrillation (Liu et al., 2018b, 2019).
Vascular Disease
Studies have also suggested that the presence of Sigmar1 in aortic vasculature and its involvement in vascular remodeling was induced by pressure overload. Decreased expression of Sigmar1 protein levels was observed in aortic cell lysate after the PO model of cardiac injury in rats and the TAC model of cardiac injury in mice. The decreased Sigmar1 protein levels were associated with inhibition of the Akt-eNOS signaling pathway in the aorta. Activation of Sigmar1 in these models of aortic injury activated the Akt-eNOS mediated signaling, rescued the aortic injury, and resulted in aortic relaxation (Tagashira et al., 2013b). In fact, Sigmar1 activation by DHEA and fluvoxamine restored Akt activity, ameliorated impaired eNOS expression, and eNOS phosphorylation in the thoracic aorta after cardiac injury (Bhuiyan et al., 2010, 2011a,b; Tagashira et al., 2010, 2011).
Recently, the role of Sigma receptor in angiogenesis was demonstrated by using (±)-haloperidol metabolite II valproate ester [(±)-MRJF22], which was a prodrug of haloperidol metabolite II (Sigmar1 antagonist/Sigmar2 agonist ligand) obtained by conjugation to valproic acid (histone deacetylase inhibitor) via an ester bond (Olivieri et al., 2016). (±)-Haloperidol metabolite II valproate ester [(±)-MRJF22] exhibited an antiangiogenic effect, significantly reduced cell viability, endothelial cell migration, and tube formation in vascular endothelial growth factor A (VEGF-A) stimulated human retinal endothelial cell cultures (Olivieri et al., 2016). However, the direct role of Sigmar1 in endothelial cell proliferation, migration, angiogenesis, and function remained elusive. Further studies are required to demonstrate the clinical efficacy of Sigmar1 ligands (agonists or antagonists) in endothelial cell pathologies in humans.
The lymphatic system is fundamentally important to several pathologies, including cardiovascular disease, edema, infection, Crohn’s disease, cancer, and obesity (Mortimer and Rockson, 2014). Sigmar1 mRNA and protein has been detected in lysates from isolated rat mesenteric collecting lymphatics, and Sigmar1 localization has been observed in the lymphatic endothelium using immunofluorescence confocal microscopy (Trujillo et al., 2017). Sigmar1 activation by the anxiolytic afobazole (an agonist of the Sigmar1) reduced lymphatic pump function elicited by an elevation in normalized end-systolic diameter, resulting in the decreased normalized amplitude of contraction, ejection fraction, and fractional pump flow (FPF) in isolated rat mesenteric lymphatics (Trujillo et al., 2017). Although simultaneous treatment with several Sigmar1 antagonists (BD 1047, BD 1063, and SM-21) reduced the effects of afobazole on lymphatic contraction, suggesting the involvement of Sigmar1, afobazole has been reported to be a mixed Sigmar1/Sigmar2 agonist (Katnik et al., 2016) that also has a high affinity for the melatonin MT1 receptor (Seredenin and Voronin, 2009). Afobazole-induced changes in lymphatic pump function were mediated via endothelial NO production in cultured lymphatic endothelial cells (Trujillo et al., 2017). However, it has also been suggested that Afobazole-induced NO-independent effects, as afobazole treatment in the presence of NOS inhibitor L-NAME led to a decrease in the normalized end-diastolic diameter of the isolated lymphatic vessel (Trujillo et al., 2017). Recently, Sigmar1’s role in lymphatic endothelial barrier function has been demonstrated by a study showing the contribution of Sigmar1 to basal lymphatic endothelial barrier function, potentially through the enhancement of glycolytic energy production in cultured adult human dermal lymphatic endothelial cells (Motawe et al., 2020). Despite all these pharmacologic data collected using non-selective ligands, the molecular role of Sigmar1 in the pathophysiology associated with the lymphatic system remains unknown. Therefore, future studies are required to determine the role of Sigmar1 in the lymphatic system, which could potentially be useful for individuals with lymphatic system disorders.
Drug-Induced Cardiomyopathy
We recently reported a potential protective role for Sigmar1 in methamphetamine-induced cardiomyopathy, where methamphetamine reduced Sigmar1 protein levels in mice, rats, and humans (Abdullah et al., 2020). Methamphetamine use in humans, rats (self-administered), mice (Binge-and-Crash model of injection) resulted in increased collagen and fibrosis, cardiac hypertrophy, mitochondrial dysfunction with altered morphology, dynamics, and reduced bioenergetics. Moreover, methamphetamine consumption reduced the levels of Sigmar1 correlated with methamphetamine-induced cardiac and mitochondrial dysfunction (Abdullah et al., 2018, 2020).
Maladaptive ER Stress
Recently, we also reported a cardioprotective role for Sigmar1 against maladaptive ER stress (Alam et al., 2017). An array of pathological stress responses that lead to cardiovascular disease results in ER stress characterized by the accumulation of unfolded and misfolded proteins. C/EBP-homologous protein (CHOP) is a ubiquitously expressed stress-inducible transcription factor whose expression is robustly induced by maladaptive endoplasmic reticulum (ER) stresses in a wide variety of cells. Sigmar1-siRNA knockdown in neonatal rat ventricular cardiomyocytes (NRCs) has been found to significantly increase the expression of CHOP and induced cellular toxicity by sustained activation of ER stress in cardiomyocytes. Conversely, adenovirus-mediated Sigmar1 overexpression decreased the expression of CHOP and significantly decreased cellular toxicity in cardiomyocytes. Sigmar1 overexpression significantly increased inositol requiring kinase 1α (IRE1α) phosphorylation and increased spliced X-box-binding proteins (XBP1s) expression as well as nuclear localization. In contrast, Sigmar1 knockdown significantly decreased IRE1α phosphorylation and decreased XBP1s expression as well as nuclear transport. Overall, Sigmar1-dependent activation of IRE1α-XBP1s ER-stress response pathways was associated with inhibition of CHOP expression and suppression of cellular toxicity. Therefore, Sigmar1 functions as an essential component of the adaptive ER-stress response pathways eliciting cellular protection in cardiomyocytes (Alam et al., 2017).
Despite the existence of knockout mice, all studies to date have been limited to pharmacologic approaches using less selective ligands for Sigmar1 (Bhuiyan et al., 2010; Bhuiyan and Fukunaga, 2011; Tagashira et al., 2011). The role of Sigmar1 in the heart has remained elusive, as all previously described Sigmar1 ligands [such as fluvoxamine (Omori et al., 2010), sertraline (Kim et al., 2016), (+) pentazocine (Hernandez and Appel, 1979), haloperidol (Chertkow et al., 2007), and cutamesine (SA4503) (Matsuno et al., 1996)] involve serotonin reuptake inhibitors (SSRIs) and also have a wide affinity for other receptors (Hayashi et al., 2011; Niitsu et al., 2012). Although approximately 35 publications have dealt with Sigmar1’s possible functions in cardiomyocytes, all current studies have been correlative, limited to pharmacologic approaches using less selective ligands (e.g., SSRIs), and the molecular mechanisms has not been unexplored (Fontanilla et al., 2009; Johannessen et al., 2009; Crottes et al., 2011; Amer et al., 2013). Sigmar1 has a significant therapeutic potential to treat the cardiovascular disease as reflected by two Sigmar1 ligands already in clinical trials: cutamesine (SA4503) for ischemic stroke (Phase II) (Urfer et al., 2014) and sertraline for depression in patients with heart failure (SADHART-CHF) (Serebruany et al., 2003, 2005; Swenson et al., 2003; Glassman et al., 2006; Jiang et al., 2008, 2011; O’Connor et al., 2010; Xiong et al., 2012, 2015). However, a direct role for cardiac Sigmar1 has not been defined. A major barrier to understanding the molecular functions of Sigmar1 is the lack of organ-specific genetic mouse models (either Tg or knockout) and selective ligands. Therefore, achieving an understanding of the molecular function of Sigmar1 would allow us to design selective Sigmar1 activators, which could be used to therapeutically to prevent cardiomyocytes loss and mitigate the clinical progression of heart failure in patients.
Neuromuscular Dysfunction
Neuromuscular disorder comprises a range of conditions that impair the functioning of the muscles, either directly due to pathologies of the voluntary muscle or indirectly due to pathologies of the peripheral nervous system or neuromuscular junctions. Progressive muscle weakness is the predominant condition associated with these disorders. Extensive studies have identified several recessive mutations in SIGMAR1 in association with a range of neuromuscular disorders, including amyotrophic lateral sclerosis (ALS) (Tagashira et al., 2014b; Fukunaga et al., 2015), ALS with or without frontotemporal lobar degeneration (Luty et al., 2010; Ullah et al., 2015), juvenile ALS (Al-Saif et al., 2011; Watanabe et al., 2016), distal hereditary motor neuropathy (dHMN) (Li et al., 2015; Gregianin et al., 2016; Horga et al., 2016; Lee et al., 2016; Almendra et al., 2018; Nandhagopal et al., 2018), frontotemporal lobe degeneration (FTLD) (Li et al., 2015; Gregianin et al., 2016; Horga et al., 2016; Lee et al., 2016; Almendra et al., 2018; Nandhagopal et al., 2018), and silver-like syndrome (Horga et al., 2016) as summarized in Table 2 and Figure 3.
TABLE 2.
Genetic and clinical features of patients with SIGMAR1 mutations.
| Genotype | Protein | Ethnic origin | Diagnosis | Muscle weakness onset age | Muscle weakness | Knee Jerks | Babinski Response | Brain MRI | Citations |
| c.505T > A/.622C > T | p.T169R/p.R208W | Japanese | ALS | 80y | + | Brisk | Present | Normal | Izumi et al., 2018 |
| c.283dupC | p.L95Pfs*29 | Hispanic | Juvenile ALS | 5y | + | Brisk | Present | Normal | Watanabe et al., 2016 |
| c.304G > C | p.E102Q | Saudi | Juvenile ALS | 1–2y | + | Brisk | N/A | Normal | Al-Saif et al., 2011 |
| *c.58T > C | 3′-UTR | Korean | Sporadic ALS | 55y | + | N/A | N/A | N/A | Kim et al., 2014 |
| *31A > G | 3’-UTR | Pakistani | Juvenile ALS | 30–36yrs | + | Brisk | N/A | Normal | Ullah et al., 2015 |
| c.412G > A | p.E138K | Italian | HMN | 10—18y | + | Brisk | N/A | Mild cerebral atrophy | Previtali et al., 2019 |
| c.247T > C/c.545T > C | p.F83L/p.L182P | German/French | HMN | 34y | + | Brisk | Present | Normal | Ma et al., 2020 |
| c.412G > C | p.E138Q | Italian | HMN | 9–12y | + | Brisk | Present | Normal | Gregianin et al., 2016) |
| c.448G > A | p.E150K | Italian | HMN | Infancy | + | Brisk | Present | Normal | Gregianin et al., 2016 |
| c.238C > T | p.Q80* | Omani | dHMN | 1–11y | + | Brisk | Present | Normal | Nandhagopal et al., 2018 |
| c.500A > T | p.N167I | Jordanian | dHMN | 6–10y | + | Brisk | Present | Normal | Christodoulou et al., 2000 |
| c.561_576del | p.D188Pfs*69 | Portuguese | dHMN | 4y | + | Absent | N/A | Normal | Almendra et al., 2018 |
| *51G > T | 3′-UTR | Australian | FTLD | No info | No info | No info | No info | No info | Luty et al., 2010; Belzil et al., 2013 |
| *26C > T | 3′-UTR | Australian | FTLD | No info | No info | No info | No info | No info | Luty et al., 2010; Belzil et al., 2013 |
| *47G > A | 3′-UTR | Polish | FTLD | No info | No info | No info | No info | No info | Luty et al., 2010; Belzil et al., 2013 |
| c.194T > A | p.L65Q | French/British | Silver-like syndrome | 3y | + | Brisk | Present | Normal | Horga et al., 2016 |
“*” means 3’-UTR.
FIGURE 3.
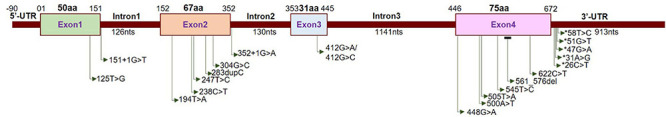
Localization of neuropathy-related mutations in the SIGMAR1 gene. Schematic diagram representing the genetic structure of the SIGMAR1 gene showing the locations of all the mutations related to skeletal muscle pathology.
Most of the genetic studies of Sigmar1 havedemonstrated an association between Sigmar1 mutations and ALS pathology. The clinical hallmarks of ALS pathology include progressive muscle wasting, speech and swallowing difficulties, fasciculation, altered reflexes, spasticity, and death due to respiratory complications (Loeffler et al., 2016). Juvenile cases of ALS have been associated with a missense mutation (c.304G > C, p.E102Q) (Al-Saif et al., 2011) and a frameshift mutation (c.283dupC, p.L95 fs) in Sigmar1 (Watanabe et al., 2016). Progressive development of skeletal muscle pathology was observed in E102Q mutations bearing patients, including weakness of the hand and forearm muscles (at the age of 9 to 10 years) leading to paralysis of forearm extensors and triceps. These patients had no respiratory or bulbar muscle weakness and demonstrated normal sphincteric, sensory, and cerebellar functions (Al-Saif et al., 2011). Similarly, the patient with the L95 fs mutations developed progressive muscle weakness with significant atrophy of distal muscles with development of pes cavus and wasting of the calf muscles and the intrinsic muscles of the hands (Watanabe et al., 2016). Interestingly, examination of a biopsy of vastus lateralis muscle showed severe type II fiber predominance with scattered angular esterase positive fibers, and also showed intense staining with nicotinamide adenine dinucleotide tetrazolium reductase (NADH−TR) (Watanabe et al., 2016). Patients bearing these mutations showed normal brain and spinal cord magnetic resonance imaging (MRI) (Al-Saif et al., 2011; Watanabe et al., 2016).
These clinical skeletal muscle phenotypes, all of which were observed in Sigmar1 mutation-bearing patients, have also been observed in patients with distal hereditary motor neuropathy (dHMN). In fact, several of the truncations/deletions or point mutations in Sigmar1 have also beenreported in association with the development of dHMN (Li et al., 2015; Gregianin et al., 2016; Horga et al., 2016; Lee et al., 2016; Almendra et al., 2018; Nandhagopal et al., 2018). The dHMN comprise a heterogeneous group of diseases having the common feature of slowly progressive, symmetrical, and distal-predominant neurogenic weakness and amyotrophy. All dHMN patients with the Sigmar1 mutations manifest identical clinical features: progressive muscle wasting/weakness in the lower and upper limbs without sensory loss (Li et al., 2015; Gregianin et al., 2016; Horga et al., 2016; Lee et al., 2016; Almendra et al., 2018; Nandhagopal et al., 2018) accompanied by normal brain and spine MRI (Gregianin et al., 2016).
Studies have also shown an association of Sigmar1 mutations in the 3′−untranslated region with the frontotemporal lobar degeneration (FTLD)-motor neuron disease (MND). Sigmar1 normally localizes to cytoplasmic membranes in healthy individuals, while in the c.672∗51G > T carriers showed intense Sigmar1 immunoreactivity in the nucleus dentate granule and CA1 pyramidal cells. However, the details of the clinical features in these patients remain unknown. Patients bearing a homozygous missense variant (c.194T > A, p.Leu65Gln) of Sigmar1 have been associated with autosomal recessive Silver-like syndrome (Horga et al., 2016). The clinical feature of this Sigmar1 mutation-bearing patient includes bilateral foot drop and frequent falls (at age 3 years), and development of progressive muscle weakness and atrophy in the lower limbs. This patient developed clawed hands with no fixed contractures, bilateral finger and foot drop, knee bobbing, marked muscle atrophy from mid-forearms and knees down, and weakness of wrist extension at the age of 17 years. However, the patient had normal intellect, no sensory symptoms, and no sphincter problems with normal brain and spinal cord MRIs.
Despite the evidence in these reports, proof of a direct association between mutations in Sigmar1 and human diseases remains elusive, as this association has only been identified in small, isolated families, with limited genetic and functional studies. Functional studies to determine molecular mechanism showed that ALS associated Sigmar1 mutations (p.E102Q and p.L95 fs) (Al-Saif et al., 2011; Watanabe et al., 2016) are uniformly unstable and non−functional when expressed in Neuro2a (N2a) cells, suggesting a role of Sigmar1’s loss of function in ALS (Al-Saif et al., 2011; Watanabe et al., 2016). Moreover, expression of the Sigmar1 E102Q carrying mutation in Drosophila (which lacks a Sigmar1 homolog) alters locomotor activity and eye development (Couly et al., 2020b). Whereas fu+nctional studies using two of dHMN associated mutations (p.E138Q and p.E150K) in several neuronal cell lines (two human neuroblastoma cell lines, SH-SY5Y and SK-N-BE, and the murine motor neuron-like NSC-34 line) suggested the pathogenicity of the mutations may involve the alterations in ER-mitochondria tethering, calcium homeostasis, and autophagy. The presence of the c.672∗26C > T, c.672∗47G > A, and c.672∗51G > T mutations within the 3′−UTR of SIGMAR1 affect transcript stability resulting in increased Sigmar1 transcript in human neuroblastoma SK−N−MC and HEK-293 cells (Luty et al., 2010). Though studies using Sigmar1 global knockout mice provided a molecular tool to understand the physiological function of Sigmar1 (Langa et al., 2003), these mice did not show any pathological phenotype associated with the human diseases observed in Sigmar1 mutation bearing patients. The neuronal dysfunction reported in Sigmar1–/– mice were locomotor defects (Mavlyutov et al., 2010), nerve denervation (Bernard-Marissal et al., 2015), loss of motor neurons (Bernard-Marissal et al., 2015), age−dependent motor dysfunction (Watanabe et al., 2016), and development of depressive-like behavior (Langa et al., 2003; Sabino et al., 2009a).
The most common clinical feature observed in patients with Sigmar1 mutation is muscle weakness caused possibly as a result of myofiber injury or by motor neuron injury resulting in denervation. However, the physiological function of Sigmar1 in skeletal muscle has never been studied and remains elusive.
The Involvement of Sigmar1 in Physiological and Pathological Conditions in the Brain
Since the discovery of Sigmar1, most studies have focused on elucidating the role of Sigmar1 under physiological and pathological conditions in the brain. Studies have demonstrated that the absence of Sigmar1 in Sigmar1–/– mice affected a wide range of brain functions (Couly et al., 2020a), including regulation of cognition and memory (Chevallier et al., 2011), motor activity (Bernard-Marissal et al., 2015), psychiatry-related behaviors (Chevallier et al., 2011; Di et al., 2017), sensory system and pain (Cendan et al., 2005). However, studies carried out to demonstrate the role of Sigmar1 in memory regulation using the Sigmar1 null mice resulted in inconsistent data. Behavioral studies using Sigmar1 null mice in different age groups (7–48 weeks) and of both sexes showed an array of effects ranging from no impact to a loss of long-term memory without alterations in short-term memory (reviewed in Couly et al., 2020a). Studies of the alteration in motor-related behaviors included movements from spontaneous locomotion to motor coordination and muscle strength. Sigmar1 null mice did not show any affects to spontaneous locomotion, as demonstrated by the open-field test and Y-maze test (Langa et al., 2003; Chevallier et al., 2011). However, the absence of Sigmar1 impairs motor coordination in an age-dependent manner, as shown by the lower motor coordination scores of older Sigmar1 null mice during the rotarod test. Similarly, Sigmar1 null mice showed reduced muscle strength compared to age-matched controls (Bernard-Marissal et al., 2015). The absence of Sigmar1 also resulted in an abnormal swimming patterns in Sigmar1 null mice without altering the swimming efficacy and speed (Chevallier et al., 2011; Di et al., 2017). Sigmar1 has a prominent effect on the psychiatric behaviors in mice, including depression and anxiety. The absence of Sigmar1 results in an increased depressive phenotype as shown by increased immobility in forced swimming test and tail suspension test (Chevallier et al., 2011; Sha et al., 2015). Additionally, a lack of Sigmar1 showed normal anxiety-like behavior as shown during the elevated plus maze test and light-dark transfer test (Sabino et al., 2009a).
Since the discovery of Sigmar1, alterations in the it’s function have been reported to associate with the development of neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD). Broadly, Sigmar1 activation using ligands elicit potent neuroprotective effects, promotes neuronal survival, and restores neuronal plasticity to slow disease progression, whereas dysfunction in Sigmar1 may worsen the progression of neurodegenerative diseases. Here, we summarize the different neurodegenerative diseases associated with the dysfunction of Sigmar1.
Role of Sigmar1 in the Pathogenesis of Alzheimer’s Disease (AD)
Alzheimer’s disease is a progressive neurodegenerative disease characterized by an accumulation of protein aggregates (e.g., Aβ-containing amyloid plaques and tau-derived neurofibrillary tangles), memory loss [both short and long term (late-stage)], cognitive deficits with impaired reading, writing, and learning abilities, behavioral changes (increased aggression, loss of empathy), loss of motor coordination and exhaustion (late-stage) (Förstl and Kurz, 1999). Earlier studies have demonstrated an association between Sigmar1’s polymorphism and the risk of developing AD (Feher et al., 2012). Further genetic studies have shown an association between Sigmar1’s genetic polymorphisms and apolipoprotein E (APOE), which influences the severity of AD across multiple ethnic populations (Huang et al., 2011). Additional studies using postmortem samples from AD patients revealed reduced Sigmar1 binding sites (Jansen et al., 1993). Notably, in vivo brain imaging using [11C]-SA4503 showed a reduced Sigmar1 density in the frontal, temporal, occipital lobes, cerebellum, and thalamus of early stage AD patients (Mishina et al., 2008; Toyohara et al., 2009). Consisted with these observations in humans, both knockout of Sigmar1 and pharmacological inactivation with the antagonist (NE-100) aggravated Aβ25–35-induced toxicity with the concurrent development of learning impairment, oxidative stress, and BDNF alteration in animal models of AD (Maurice et al., 2018). Extensive studies have shown that treatment with Sigmar1 agonists improved cognition in various preclinical animal models, including Aβ25–35 peptide-induced neurodegeneration (Maurice et al., 1998; Meunier et al., 2006b; Villard et al., 2009), cholinergic deficits (Matsuno et al., 1993; Senda et al., 1997; Maurice et al., 2001), aging-induced memory loss (Maurice et al., 1996; Phan et al., 2003), hypoxia, and toxin-induced neurodegeneration (Maurice et al., 1994b, 1999), as well as drug-induced glutamatergic, serotonergic, or calcium channel deficits models of neurodegeneration (Matsuno et al., 1994; Maurice et al., 1994a, 1995). The molecular mechanisms by which Sigmar1 agonists induced protective effects were mediated through the modulation of glutamate release, calcium homeostasis, functional modulation of ion channels, NMDA activity, neuroplasticity, reduction of oxidative stress, and modulation of mitochondrial function (Jansen et al., 1993; Maurice et al., 1998; Uchida et al., 2005; Mishina et al., 2008; Feher et al., 2012). Studies also demonstrated that Sigmar1 ligands (e.g., OZP002, donepezil, ANAVEX2-73) prevented toxicity and memory impairment in pharmacologic and genetic mouse models of AD (Maurice et al., 2019; Ryskamp et al., 2019). Although several studies suggest a protective roles of Sigmar1 ligands in AD pathology (Ryskamp et al., 2019), the direct role for Sigmar1 in AD pathobiology has never been studied using genetic mouse models (organ-specific knockout and transgenic mouse for Sigmar1).
Role of Sigmar1 in the Pathogenesis of Huntington’s Disease (HD)
Huntington’s disease is a progressive and inherited neurodegenerative disease exhibiting phenotypes such as motor defects, cognitive decline, and psychiatric symptoms. HD is characterized by the accumulation of huntingtin protein aggregates [caused by a mutation in the huntingtin (Htt) gene]. Huntingtin protein regulates multiple cellular functions, including cell division, vesicle recycling and trafficking, autophagy (aids in cargo recognition), cell survival, and several other functions. Mutation in huntingtin protein disrupts all these cellular functions leading to increased apoptosis, cellular degeneration, impaired autophagic clearance, and dysfunctional vesicle transport. Additionally, mutant huntingtin protein interacts with mitochondria causing mitochondrial dysfunction with altered metabolism and increased reactive oxygen species (ROS) (Zuccato et al., 2010; Saudou and Humbert, 2016). Additionally, mutant huntingtin protein is associated with decreased levels of NF-κB-p65 and activated calpastatin levels (leading to increased ROS levels) (Hyrskyluoto et al., 2013).
The evidence for the Sigmar1’s role in HD pathology was provided by the initial in vitro studies in which the expression of N-terminal huntingtin fragment proteins with 120 polyQ repeats or the full-length Htt protein with 75 repeats downregulated Sigmar1 level in neuronal PC6.3 cells. Treatment with Sigmar1 agonist (PRE-084) in this model increased cellular survival and prevented the deleterious effects of Htt (Hyrskyluoto et al., 2013). It has also beenreported that accumulation of Sigmar1 was common to neuronal nuclear inclusions in the brains of patients with five HD, dentatorubral-pallidoluysian atrophy, spinocerebellar ataxia types 1–3, and intranuclear inclusion body disease (Miki et al., 2014). In the cellular model of HD, silencing of Sigmar1 significantly increased the number of nuclear inclusions and caused the accumulation of high-molecular-mass GFP-labeled mutant huntingtin (Miki et al., 2015). Mechanistically, Sigmar1 knockdown studies showed Sigmar1 degrades aberrant mutant huntingtin proteins in the nucleus via activation of the ER-related degradation machinery (Miki et al., 2015). Moreover, a recent study showed that pridopidine (a therapeutic drug for HD) has an affinity for Sigmar1 and acts via Sigmar1 at a nanomolar level. A PET scan study of human HD patients also showed complete Sigmar1 occupancy by pridopidine (Grachev et al., 2020; Battista et al., 2021), and the neuroprotective effects of pridopidine were abolished in Sigmar1 knockout mice (Francardo et al., 2019). Activation of Sigmar1 by pridopidine rescued mitochondrial dysfunction induced by oxidative damage in YAC128 transgenic mice, human HD lymphoblasts, and human HD neural stem cells (NSCs) (Naia et al., 2021). Moreover, early pridopidine treatment was effective in delaying onset of HD-related motor symptoms in YAC128 HD mice (Naia et al., 2021).
Role of Sigmar1 in the Pathogenesis of Parkinson’s Disease (PD)
Parkinson’s disease is a slowly progressing brain disease characterized by abnormal locomotion (such as shaking, stiffness, and difficulty in walking, balance, and coordination), and the deposition of Lewy bodies, α-synuclein, ubiquitin, and neurofilaments. Patients with PD exhibit a low Sigmar1 ligand (SA4503) binding potential in the putamen of the brain visualized by PET scan compared to healthy controls (Mishina et al., 2005). Chronic treatment with Sigmar1 agonists (PRE-084 and pridopidine) elicits gradual improvement of parkinsonian-like motor deficits in PD model mice developed by intrastriatal 6-hydroxydopamine (6-OHDA) (Francardo et al., 2014, 2019). Similarly, studies have also shown that PRE-084 treatment in animals reduced neuroinflammation, increased density of dopaminergic fibers in the denervated striatal regions, increased the levels of neurotrophic factors [e.g., brain-derived neurotrophic factor (BDNF) and glial-derived neurotrophic factor (GDNF) in the striatum and nigra], and monoamines (dopamine, DA, and serotonin, 5-HT) (Francardo et al., 2014). In contrast to these studies, treatment with Sigmar1 antagonists (NE 100) and Sigmar1 null mice showed reduced 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced dopaminergic neurons death and parkinsonism by suppressing N-methyl-d-aspartate receptor (NMDAr) function and dopamine transporter (DAT) expression (Hong et al., 2015). Additionally, inhibition of Sigmar1 by treatment with antagonists (NE-100) prevented neurotoxin-induced neurodegeneration through facilitating TRPC1-mediated calcium influx in SH-SY5Y cells (Sun et al., 2020). As these studies have suggested, either pharmacologic activation/inhibition of Sigmar1 could be useful in slowing down the progression of PD. Robust preclinical studies are required using more clinically relevant mouse models of PD (such as transgenic alpha-synuclein overexpressing mice) to move forward the preclinical research to clinical trials.
Overall, Sigmar1 activation has shown protective effects in different neurodegenerative diseases (AD, HD, PD) through the involvement of different cellular pathway modulation, including mitochondrial function regulation, autophagy, calcium homeostasis regulation, and chaperone function. This makes Sigmar1 a possible target in treating pathologies where modulating Sigmar1 activity could be used as a therapeutic approach in the treatment of neurodegenerative diseases.
Ischemic Brain Injury
A major therapeutic goal during the subacute and chronic phases of stroke is the enhancement of functional recovery, as a significant number of patients suffer from persistent neurological deficits. Sigmar1 has also been implicated in brain injuries; the expression of Sigmar1 increased in the penumbral or peri-infarct region, making it useful as a molecular marker and therapeutic target in the treatment of acute ischemic stroke (Zhang et al., 2017c). Subsequent studies have shown that pharmacologic activation of Sigmar1 in endothelial cells reduces infarct size, protects blood-brain-barrier integrity, and protective against dementia and learning disabilities (Liu et al., 2018a). Studies have also shown that treatment with Sigmar1 agonists (such PRE-084) reduced infarct volume, neurological deficits, levels of pro-inflammatory cytokines, and enhanced the actions of anti-inflammatory cytokines after embolic stroke in rats (Allahtavakoli and Jarrott, 2011). Similarly, treatment with the Sigmar1 ligand 4-phenyl-1-(4-phenylbutyl)-piperidine (PPBP) decreased cortical infarction volume without altering neurobehavior after transient focal ischemia and prolonged reperfusion in the rat (Harukuni et al., 2000). A preclinical study using permanent middle cerebral artery occlusion in rats showed that chronic treatment with the Sigmar1 agonist cutamesine (SA4503) for a period of 28 days enhanced functional recovery after experimental stroke without affecting infarct size when treatment was initiated within 48 h (Ruscher et al., 2011). A Phase 2 clinical trial was conducted to explore the safety, tolerability, dose range, and functional effects of cutamesine in patients with ischemic stroke (Urfer et al., 2014). Post hoc analysis of moderately and severely affected patients (baseline National Institutes of Health Stroke Scale, ≥ 7 and ≥ 10) showed greater National Institutes of Health Stroke Scale improvements compared with placebo (Urfer et al., 2014). Although cutamesine was safe and well-tolerated at the tested dosage levels, the study did not show any significant effects on functional end points in the population as a whole (Urfer et al., 2014).
Drug Addiction
Extensive research has documented Sigmar1’s involvement in drug addiction pathobiology. In fact, selective Sigma1 ligands modulate monoaminergic systems, particularly dopaminergic as well as serotoninergic systems. Sigmar1 has strong affinities and interacts with several addictive drugs, such as (±)-3,4-methylenedioxymeth-amphetamine (MDMA; derivative of amphetamine), methamphetamine (METH), and cocaine, mediating their locomotor stimulatory and neurotoxic effects (Nguyen et al., 2005; Brammer et al., 2006).
Methamphetamine
Sigmar1 has been extensively studies as a possible therapeutic target for use in disrupting the methamphetamine-induced addictive process and toxicity (Sambo et al., 2017, 2018), as studies have reported that methamphetamine binds to Sigmar1 at physiologically relevant concentrations (Ki 2.16 ± 0.25 μM) (Nguyen et al., 2005). Although the molecular consequences of methamphetamine binding to Sigmar1 remain unknown, studies suggest that methamphetamine may exhibit antagonist activity (Hayashi and Su, 2007) and/or act as an inverse agonist for Sigmar1 (Yasui and Su, 2016). Pharmacologic activation of Sigmar1 by treatment with agonists have been shown to attenuate methamphetamine-induced behavioral responses, hyperthermia, and neurotoxicity. Pretreatment with the Sigmar1 agonist PRE-084 decreases methamphetamine-induced psychomotor responses, drug-seeking behavior, and enhancement of the brain reward function (Sambo et al., 2017, 2018). In contrast, studies have also shown that Sigmar1 antagonist, N-[2-(3,4-dichlorophenyl)ethyl]-N-methyl-2-2(dimethylamino)ethylamine (BD1047), exert a protective effect against MDMA-, and methamphetamine-induced locomotion stimulatory effects (Nguyen et al., 2005; Brammer et al., 2006). Despite the evidence from all these studies demonstrating Sigmar1’s role in methamphetamine-induced cellular dysfunction, it remains unknown whether and how Sigmar1 contributes to cellular protection.
Cocaine
Sigmar1 had been implicated in the cocaine-induced addictive process and toxicity, and studies have shown that cocaine binds to Sigmar1 (at Asp188) at physiologically reward-relevant concentrations (2–7 μM) (Sharkey et al., 1988; Kahoun and Ruoho, 1992; Matsumoto et al., 2002; Chen et al., 2007). Several studies reported that treatment with selective Sigmar1 antagonists mitigates cellular and behavioral toxicities induced by cocaine, including convulsion and death (Matsumoto et al., 2004, 2014; Robson et al., 2012). Mechanistically, the addictive and neurotoxic actions of cocaine were mediated through Sigmar1 activation, enhancing IP3-dependent Ca2+ signaling (Barr et al., 2015). Cocaine also transcriptionally suppresses the expression of monoamine oxidase B (MAOB) through Sigmar1-dependent recruitment of HDACs (Tsai et al., 2015). The Sigmar1 dependent addictive effect of cocaine was confirmed in studies using Sigmar1 null mice where the absence of Sigmar1 abrogated the suppression of MAOB expression (Tsai et al., 2015). Extensive studies have demonstrated that pharmacologic Sigmar1 antagonists (BD1063, BD1047, NE-100) elicit protection against cocaine-mediated addictive effects on locomotion and neurotoxicity (Maurice et al., 2002; Romieu et al., 2002). However, treatment with the Sigmar1 antagonist, BD1047, did not block cocaine self-administration; however, it did attenuate the cocaine reinstatement.
Alcohol
Extensive studies on alcohol use disorder have suggested Sigmar1 as a potential mediator of alcohol reward and reinforcement (Quadir et al., 2019). Several studies have shown that inhibition of Sigmar1 by treatment with Sigmar1 antagonists (BD1063, NE-100) reduced ethanol consumption and ethanol-induced rewarding effects such locomotion stimulation and taste aversion. These effects were reverted by treatment with Sigmar1 agonists in rats (voluntary consumption) and mice (intraperitoneal injection) (Maurice et al., 2003; Sabino et al., 2009b; Blasio et al., 2015). However, Sigmar1 null mice subjected to voluntary ethanol consumption showed increased alcohol drinking with increased taste aversion and hypothermia with no effects on locomotion (Valenza et al., 2016). The contrasting effects of Sigmar1 on ethanol consumption and ethanol-induced locomotor effects observed by different groups might be due to differences in animal models used in the study (mice vs. rats), and the protocol used for ethanol consumption [voluntary vs. intraperitoneal injection (used in Sigmar1 null mice)]. Sigmar1 is also involved in both the chronic effects and withdrawal effects of alcohol consumption, where modulation of Sigmar1 using a selective agonist reduced hyper-responsiveness and mitigate the effects of chronic alcohol consumption induced cognitive decline (Meunier et al., 2006a). Despite these contradictory findings concerning the role of Sigmar1 ligands in alcohol consumption and rewarding effects, all these studies suggested Sigmar1’s involvement in alcohol consumption (acute and chronic) mediated toxic effects. Further studies are required to demonstrate whether modulation of Sigmar1 using selective ligands can be used therapeutically to reverse alcohol consumption-mediated adverse effects.
Cancer
The role of Sigmar1 has been widely studied in different types of cancers, including prostate cancer, colorectal cancer, breast cancer, and hepatocarcinoma. Clinical studies have shown high levels of Sigmar1 in tumor tissues from breast cancer patients and has been proposed to be used as a clinical marker of breast cancer (Simony-Lafontaine et al., 2000). Similar results were seen in lung cancer patients’ samples where Sigmar1 was secreted by tumor cells and increased the viability of squamous lung cancer cells, and correlated with increased survival of the cancer cells (Mir et al., 2012). Elevated levels of Sigmar1 (mostly cytoplasmic) were also reported in hilar cholangiocarcinoma tissue samples, and correlated with poor prognosis of the patients and their decreased longevity (Xu et al., 2014). Patients with colorectal cancer also exhibit upregulated levels of Sigmar1 depending on the stage of the disease, especially in the upper colon (Skrzycki and Czeczot, 2013). Similar results were observed in experiments carried out in vitro and in vivo (rodent models), where Sigmar1 was essential for the growth of prostate cancer, breast cancer, and colorectal cancer cells. Upregulated levels of Sigmar1 in different cancer cell lines drive cell migration, invasiveness, and promote cell survival by increasing calcium entry in the cells and regulating membrane electrical activity (Crottes et al., 2016; Gueguinou et al., 2017). Inhibition of Sigmar1 by ligands limited the translocation of androgen receptor and mediated protective effects in prostate cancer cells (Thomas et al., 2017). Additionally, Sigmar1 inhibition altered calcium homeostasis increased apoptotic cell death, and inhibit cancer cell proliferation and migration in breast cancer and colorectal cancer cells (Azzariti et al., 2006; Crottes et al., 2016; Gueguinou et al., 2017). In contrast, hepatocarcinoma cells have been shown to have reduced Sigmar1 expression in clinical samples and human liver cancer cells (HepG2) concurrent with reduced apoptosis and increased NF-κB levels. Overexpression of Sigmar1 in vitro in HepG2 cells has demonstrated protective effects by reducing cell proliferation, increasing apoptosis, and decreasing NF-κB levels (Xu et al., 2018). However, inhibition of Sigmar1 in hepatocarcinoma also showed protective effects through the reduction of iron metabolism and ferroptosis (Bai et al., 2019). The discrepancy between study results may be related to the focused signaling pathway (ferroptosis vs. apoptosis, cell proliferation vs. mitochondrial ROS).
All these studies demonstrate a potential regulatory role for Sigmar1 in cancer biology, as Sigmar1 has been shown to be upregulated in all types of cancer and functions in driving cell migration, increasing membrane invasiveness, and further enhancing cell proliferation, contributing to disease progression. However, further studies are required to understand the exact mechanism and functions of Sigmar1 in different cancer cells.
Retinal Diseases
Sigmar1 expression in the various ocular tissues was first reported using pharmacological ligand binding assays (Senda et al., 1998) and biochemical experiments (Ola et al., 2001, 2002). Subsequent studies have shown Sigmar1 expression in multiple retinal cell types, including photoreceptor cells, ganglion cells, and Müller and pigment epithelial cells (Ola et al., 2002; Jiang et al., 2006). Sigmar1 null mice demonstrate normal retina development, but as they aged, these mice developed apoptosis in the optic nerve head, decreased ganglion cell function, and eventually loss of ganglion cells (Ha et al., 2011). Extensive research on the molecular function of Sigmar1 showed pharmacologic Sigmar1 activation by agonists attenuated ganglion cell death (Campana et al., 2002; Smith et al., 2008), mitigated retinal glial cell reactivity (Zhao et al., 2014; Wang et al., 2015; Vogler et al., 2016), and diminished light-induced photoreceptor cell loss (Shimazawa et al., 2015). Sigmar1’s role in diabetic retinopathy was evident from two murine models of diabetic retinopathy, the streptozotocin-induced model (Ola et al., 2002) and the Ins2Akita/+ mouse (Smith et al., 2008). Both of these diabetic models showed a similar level of Sigmar1 expression in the retinal tissues compared to control mice (Ola et al., 2002; Smith et al., 2008). Pharmacologic activation of Sigmar1 in the Ins2Akita/+ mouse conferred significant neuroprotection, reduced oxidative stress, and preserved retinal architecture (Smith et al., 2008). The protective effect of Sigmar1 in diabetes-induced retinal neurodegeneration has also been demonstrated using Sigmar1 null mice, where the absence of Sigmar1 aggravated retinal ganglionic cell dysfunction in streptozotocin-injected diabetic mice (Ha et al., 2012). Sigmar1 activation by treatment with agonists also showed neuroprotective effects associated with attenuated Müller cell gliosis, reduced microglial activation, and decreased oxidative stress in an inherited photoreceptor degeneration model, the Pde6brd10/J (rd10) mouse model (Wang et al., 2016). Extensive in vivo and in vitro mechanistic studies have been performed in isolated retinal cells, such as Müller glial cells, microglial cells, optic nerve head astrocytes, and retinal ganglion cells as well as in the intact retina to determine the molecular signaling pathways regulated by Sigmar1 (Smith et al., 2018). All these studies together demonstrate that Sigmar1 dependent retinal neuroprotection involved activation of functions associated with ion channel regulation, chaperone activity, oxidative stress modulation, and regulation of cellular calcium (Smith et al., 2018).
Kidney Injury
Expression of Sigmar1 has also been detected in kidneys using Northern blot and Western blot analysis, suggesting a potential role for Sigmar1 in kidney pathophysiology. Sigmar1 expression level were found to be significantly reduced during kidney injury in rats induced by pressure overload following bilateral ovariectomy (Bhuiyan and Fukunaga, 2010). Sigmar1 activation following dehydroepiandrosterone treatment was found to prevent kidney injury by activating the Akt-eNOS signaling pathways and restoring NO levels (Bhuiyan and Fukunaga, 2010). Similarly, Sigmar1 agonists (Fluvoxamine) have been shown to improve postischemic survival and renal function via activation of Akt-mediated nitric oxide signaling in the kidney in rats model of ischemia-reperfusion injury (Hosszu et al., 2017). A recent study has provided evidence for increased Sigmar1 expression in distal tubular kidney cells of young and streptozotocin (STZ) induced diabetic rats (Milardovic et al., 2020). However, the molecular role for Sigmar1 in the postnatal development of the rat kidneys and in distal tubular damage in the pathogenesis of diabetes requires further investigation.
COVID-19
Sigma ligands has recently been explored as a therapeutic target in COVID-19 repurposing therapy (Abate et al., 2020; Vela, 2020; Hashimoto, 2021). Both Sigmar1 agonist (fluvoxamine) (Lenze et al., 2020) and antagonist (haloperidol) (Hoertel et al., 2021) underwent clinical trial for possible therapy in patients with COVID-19. The interaction map for SARS-CoV-2 protein reveals Sigmar1 interaction with Nsp6 (SARS-CoV-2 viral protein) and proposed Sigmar1 ligands as a possible therapeutic target for COVID-19 (Gordon et al., 2020b). Validation of the interaction map showed Sigma ligands (both Sigmar1 and Sigmar2) inhibit viral activity (Gordon et al., 2020b). Further studies of the role of Sigmar1 in COVID-19 have suggested a functional host-dependency factor for SARS-CoV-2; the absence of Sigmar1 by knockdown reduced the replication of SARS-CoV-2 protein, delaying disease progression and presenting Sigmar1 as an attractive therapeutic target (Gordon et al., 2020a). Extensive future research are required to elucidate the molecular role of Sigmar1 in COVID-19 pathobiology.
Others
In addition to the association of Sigmar1 with the above mentioned pathologies (summarized in Figure 4), the protective role of Sigmar1 has also been explored in inflammation and sepsis. During sepsis, Sigmar1 has been shown to interact with IRE1, reducing the splicing of XBP1, resulting in reduced production of inflammatory cytokines and increasing the longevity of mice in sub-lethal models of sepsis (Rosen et al., 2019). These protective effects of Sigmar1 were confirmed using Sigmar1 null mice, where the absence of Sigmar1 further increased pro-inflammatory cytokines, XBP1 splicing, and reduced survival of mice (Rosen et al., 2019). Sigmar1 has also been implicated in liver ischemia where the use of Sigmar1 ligand BHDP (a sigmar1 ligand) has been shown to maintain membrane integrity, restore metabolic capacities of the liver, restore mitochondrial respiration and tissue integrity, reducing the deleterious effects caused by ischemia in the liver (Klouz et al., 2008). All these studies suggest strong involvement of Sigmar1 in different pathologies, and more studies are required to further explore it as a therapeutic target.
FIGURE 4.
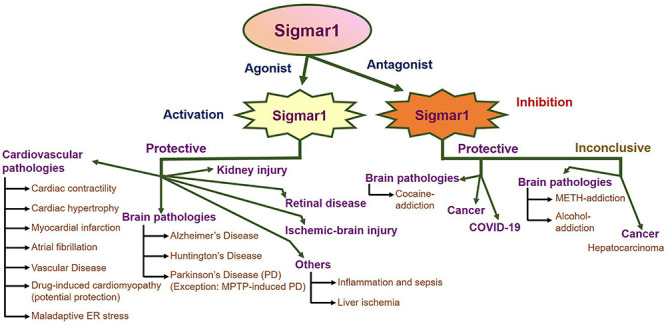
Summary of the pathophysiological functions of Sigmar1. Schematic showing an overall summary of the role of Sigmar1 in the pathophysiology related to different organs as present in the current literature. Briefly, agonist-mediated activation of Sigmar1 has protective effects in pathological conditions of several organs including heart (cardiac hypertrophy, myocardial infarction, atrial fibrillation, vascular disease, drug-induced cardiomyopathy and maladaptive ER stress), brain (neurodegenerative diseases including AD, HD and PD (with the exception of MPTP-induced PD) and ischemic brain injuries), kidneys, retina, liver, and the immune system. Inhibition of Sigmar1 using its antagonists is reported to be protective in several pathologies including cancer, cocaine addiction, and COVID-19. However, due to conflicting reports on whether activation or inhibition of Sigmar1 is protective, the field remains inconclusive about the effects of Sigmar1 on methamphetamine and alcohol addiction.
Biological Functions of Sigmar1
Despite extensive research on Sigmar1 over the last fifty years, we are still exploring and learning about the molecular role of Sigmar1 in different cells and organs. Research to date suggested that Sigmar1 mediates cellular signaling pathways by acting as a ligand-operated chaperone protein, and lack its intrinsic signaling machinery (Figure 5). Sigmar1 functions primarily via translocation and protein-protein interactions by ligands to modulate the activity of various ion channels and signaling molecules, including inositol phosphates, protein kinases, and calcium channels (Su et al., 2010). Most of these studies were performed in cell culture models using Sigmar1 ligands (agonist or antagonists), and most of these ligands possess affinities for other receptors or even elicit pleiotropic effects. Moreover, the molecular characteristics of Sigmar1’s interactions in each of these signaling pathways remain elusive.
FIGURE 5.
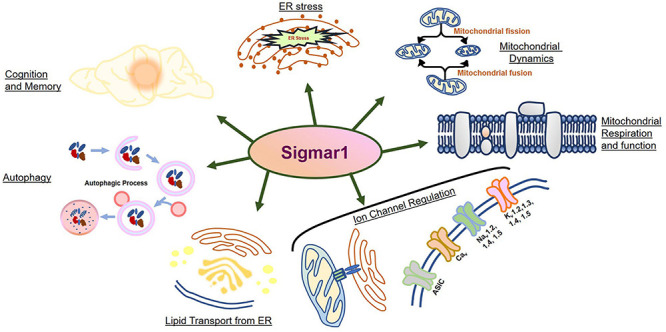
Biological functions of Sigmar1. Schematic diagram summarizing the functions of Sigmar1 including its involvement in cognition, memory, ER stress, mitochondrial dynamics, mitochondrial respiration and function, autophagy, lipid transport from ER, and ion channel regulation.
Ion Channel Regulation
Earlier studies on Sigmar1 have suggested that Sigmar1 dependent modulation of ion channel signaling pathways occurs via protein-protein interaction as summarized in Table 3. Extensive research has suggested that Sigmar1 may be involved in intracellular calcium signaling and inositol triphosphate (IP3) turnover as evident by Sigmar1 interaction with Inositol triphosphate receptors (ITPR) using co-immunoprecipitation (Co-IP). Sigmar1 interaction with all three types of ITPRs (ITPR1, ITPR2, and ITPR3) has been reported using in vitro experiments in different cells, and ligand-dependent activation of Sigmar1-ITPR interaction modulates the intracellular calcium levels (Hayashi and Su, 2007). Sigmar1 was reported to form a trimeric complex with ITPR3 and Ankyrin B regulating ER-mitochondrial communication through regulating Ca2+ efflux from the ER into the cytosol (Hayashi and Su, 2001; Wu and Bowen, 2008). Sigmar1 inhibition by antagonists also showed to decrease in calcium response in neuroblastoma cells (Gasparre et al., 2012). In hepatocytes, Sigmar1 stimulation decreased IP3R1 dependent calcium signaling by inhibiting the synthesis of IP3 receptors in PKC dependent manner (Abou-lovergne et al., 2011). In cardiomyocytes, Sigmar1 stimulation reduced upregulation of pressure-overload induced IP3R-2 (Tagashira et al., 2013a). This study further reported binding of Sigmar1 to ryanodine receptor (RyR), and Sigmar1 activation by ligands showed inhibition of ryanodine-induced calcium release from the sarcoplasmic reticulum (SR) in cardiomyocytes (Tagashira et al., 2013a). All these studies point toward a cell-type dependent effect of Sigmar1 stimulation by agonist on ITPR dependent calcium release.
TABLE 3.
Sigmar1 dependent regulation of ion channels through protein-protein interaction.
| Ion channel | Cell types | Method of interaction detection | Sigmar1 construct | Ion Channel construct | Expression | References |
| Inositol tri-phosphate receptor (IP3R) | NG-108, CHO, bovine brain mitochondria | Co-IP | Native Sigmar1 and Sigmar1-EGFP | Native IP3R3 | Native expression and transient expression | Hayashi and Su, 2001, 2007; Natsvlishvili et al., 2015 |
| NG-108 | Co-IP and PLA | Native Sigmar1 | Native IP3R1 | Native expression | Kubickova et al., 2018 | |
| Rat heart tissue | Co-IP | Native Sigmar1 | Native IP3R2 | Native expression | Tagashira et al., 2013a | |
| Ryanodine receptor | Rat heart tissue | Co-IP | Native Sigmar1 | Native RYR2 | Native expression | Tagashira et al., 2013a |
| L-ype Voltage-gated calcium channel (VGCC) | RGC-5 cells | Co-IP | Wild-type Sigmar1 | Native L-type VGCC | Stable and native expression | Tchedre et al., 2008 |
| Voltage-dependent N-type calcium channel (Cav2.2) | HEK293T | FRET and Co-IP | Sigmar1-dsred | EGFP-Cav2.2 | Transient overexpression | Zhang et al., 2017c |
| Calcium release-activated calcium channel | tSA-201 | Co-IP | Sigmar1-FLAG | ORAI-Myc | Transient overexpression | Srivats et al., 2016 |
| Voltage-dependent potassium channel | Mouse nucleus accumbens lysate, NG108-15 | Co-IP and Co-IP with cross-linking | Native Sigmar1 and Sigmar1-V5-His | Native Kv1.2, wild-type Kv1.2 | Native expression and transient expression | Kourrich et al., 2013 |
| HEK293 | Co-IP | Sigmar1-FLAG | Kv1.3-HA | Transient expression | Kinoshita et al., 2012 | |
| Rat posterior pituitary lysate | Co-IP | Native Sigmar1 | Native Kv1.4 | Native expression | Aydar et al., 2002 | |
| SK3 channel | SKmel28 cells, HEK293 | Co-IP and HTRF | Sigmar1-Myc, HALO-Sigmar1-Myc | Wild-type SK3, SK3-HA | Transient and stable overexpression | Gueguinou et al., 2017 |
| Nav1.5 Na+ channel | tSA201, MDA-MB-468, MDA-MB-231 | Anti-FLAG chromatography, PLA, Co-IP | Sigmar1-FLAG, Native Sigmar1 | Nav1.5-HA, Native Nav1.5 | Transient overexpression, native expression | Balasuriya et al., 2012 |
| Voltage-dependent anion channel 2 (VDAC2) | MA-10 | Co-IP | Native Sigmar1 | Native VDAC2 | Native expression | Marriott et al., 2012 |
| Acid sensing ion channel 1a (ASIC1a) | HEK293 | Ni affinity chromatography | Sigmar1-FLAG-His | ASIC1a-His | Stable overexpression (ASIC1a) and transient overexpression (Sigmar1) | Carnally et al., 2010 |
Co-IP, Co-immunoprecipitation; PLA, proximity ligation assay; FRET, fluorescence resonance energy transfer; HTRF, homogeneous time resolved fluorescence.
Apart from the ITPRs, Sigma ligands have also been shown to block all calcium channel subtypes (N-, L-, P/Q-, and R-type calcium channels) in neonatal rat intracardiac and superior cervical (SCG) ganglia (Zhang and Cuevas, 2002). Several studies showed that the Sigmar1 ligand could modulate calcium influx in the cells by regulating voltage-dependent calcium channel (VDCC) (Hayashi et al., 2000). Use of Sigmar1 ligands inhibited calcium re-entry resulting in decreased calcium current in the cells (Katnik et al., 2006; Amer et al., 2013; Pan et al., 2014). This antagonism by Sigmar1 on VDCC and calcium current were used to treat painful neuropathies, reduce the unwanted increase in vascular permeability and angiogenesis, target excitotoxicity-induced neurodegenerative disease, and confer neuroprotection (Katnik et al., 2006; Amer et al., 2013; Pan et al., 2014). Another mechanism of Sigmar1 dependent regulation of Ca2+ dynamics has been demonstrated through store-operated calcium entry by modulating Orai to STIM1 coupling (Brailoiu et al., 2015; Srivats et al., 2016). Studies have shown that Sigmar1 activation by agonists induced Sigmar1-STIM1 interactions and de-coupled it from the Orai complex, and thereby inhibiting calcium influx (Brailoiu et al., 2015; Srivats et al., 2016).
Several Sigmar1 ligands have been shown to regulate the activity of a different type of potassium channels (Kv1.2, Kv1.3, Kv1.4, and Kv1.5) in different cell types (NG108, and oocytes) (Mavlyutov et al., 2010; Kinoshita et al., 2012; Kourrich et al., 2013). Activation of Sigmar1 by agonists decreased the transient outward potassium current (Soriani et al., 1999a) and the sustained outward potassium current (Soriani et al., 1999b) in cultured frog pituitary melanotrope cells. Sigmar1 activation also inhibited Kv1.4 potassium channels in Xenopus oocytes by interaction with Kv1.4 (Aydar et al., 2002). Sigmar1 activation by ligands also reversibly blocked delayed outwardly rectifying potassium channels, large-conductance Ca2+-sensitive K+ channels, and the M-current by direct coupling to potassium channels in rat parasympathetic intracardiac neurons (Zhang and Cuevas, 2005). In addition, Sigmar1 has also been shown to bind to a voltage-dependent sodium channel (Nav1.5) and Sigmar1 ligands (haloperidol and pentazocine) altered Sigmar1: sodium channel interaction (Balasuriya et al., 2012; Aydar et al., 2016). Activation of Sigmar1 with agonists reduced voltage-gated Na+ channel (namely Nav1.5, Nav1.2/1.4) function in neonatal rat intracardiac ganglia, cardiomyocytes, and HEK-293 cells. These resulted in alterations in the action potential configuration with increased latency and reduced firing (Johannessen et al., 2009; Zhang et al., 2009; Gao et al., 2012). Sigmar1 was also reported to modulate acid-sensing ion channels (ASIC) (specifically, ASIC1a, a channel with higher permeability for ca2+ ions) through direct interaction (Herrera et al., 2008; Carnally et al., 2010). Activation of Sigmar1 using agonists inhibited ASIC1a function, decreased membrane currents, reduced acidosis induced ion flux, and increased calcium-induced current ([Ca2+]i) (Herrera et al., 2008).
Despite extensive studies that support the modulation of ion channel signaling by Sigmar1 ligands, more research is required to determine the direct involvement of Sigmar1 through Sigmar1-protein interactions. Studies have shown that several Sigmar1 ligands directly modulate ion channel function independently of the Sigmar1 (Gao et al., 2012; Amer et al., 2013; Brindley et al., 2017; Asano et al., 2019). Two Sigmar1 ligands (BD1047/BD1063 and 4−IBP) have been shown to inhibit chemically activated calcium entry channels [transient receptor potential (TRP)], acting relatively directly and independently of the Sigmar1 in human saphenous vein endothelial cells and HEK-293 cells (Amer et al., 2013). (+)-SKF 10047 inhibited NaV1.2 and NaV1.4 channels independently of sigma-1 receptor activation in HEK-293T cells and COS-7 cells (Gao et al., 2012). Similarly, some of the Sigmar1 ligands used have a very weak affinity for Sigmar1, requiring high concentrations (>10 μM) of the Sigmar1 ligands to allow observation of ion channel activity (Hayashi et al., 2000; Zhang and Cuevas, 2002; Johannessen et al., 2009; Zhang et al., 2017b). Overall, more research is required to understand the molecular mechanism responsible for Sigmar1 dependent modulation of ion channels.
Chaperone Function
It has been proposed that Sigmar1 functions as a ligand-operated chaperone following identifications of a large number of protein-protein interactions between Sigmar1 and other proteins. The chaperone activity shown by Sigmar1 has been demonstrated by using reconstituted biochemical experiments where a purified C-terminal fragment of the Sigmar1 (residues 116-223) minimized the aggregation of proteins in a light scattering assay (Ortega-Roldan et al., 2013; Gregianin et al., 2016). Additional indirect studies have also shown evidence for Sigmar1’s chaperone function as Sigmar1 overexpression increased the whole-cell or surface expression of various proteins (Crottes et al., 2011; Kourrich et al., 2013; Pabba et al., 2014). Similarly, Sigmar1 knockdown in vitro also decreased the expression of several proteins suggesting chaperone function (Hayashi and Su, 2007; Mori et al., 2013; Aydar et al., 2016). The increased stability of different proteins, including IP3R channels (at the ER-mitochondria interface) (Gregianin et al., 2016), ankyrin (Hayashi and Su, 2001), potassium channels (Aydar et al., 2002), opioid receptors (Kim et al., 2010; Hong et al., 2017), and dopamine receptors (D1 and D2) (Kim et al., 2010; Navarro et al., 2010, 2013), following ligand-dependent activation of Sigmar1 also supports Sigmar1’s chaperone activity. Additionally, it has been proposed that the chaperone function of Sigmar1 is part of ER-associated degradation (ERAD) machinery involving in degradation of sphingolipid enzymes (Hayashi et al., 2012). Studies have also reported that Sigmar1 modulates the ER stress response and subsequent unfolded protein response (UPR), influencing protein stability and localization (Mori et al., 2013; Rosen et al., 2019). In addition, Sigmar1’s chaperone activity has been reported to degrade intranuclear inclusions and provide neuroprotection in Huntington’s disease, Alzheimer’s disease, and Parkinson’s disease (Miki et al., 2014, 2015; Yamoah et al., 2020). More studies are required to demonstrate the molecular mechanism responsible for the chaperone function of Sigmar1 in vivo.
Regulation of Mitochondrial Morphology, Dynamics, and Function
Evidence for the molecular role played by Sigmar1 in mitochondrial morphology, dynamics, and function comes from studies involving the Sigmar1 null mouse and phenotypes of the disease-causing mutations found in Sigmar1. The involvement of Sigmar1 in mitochondrial morphology has been made evident by studies where the loss of Sigmar1, either by knockdown or by pharmacologic inactivation with the antagonist (using NE-100) resulted in increased mitochondrial length and development of mitochondrial dysfunction (Bernard-Marissal et al., 2015). In contrast to the existing notion stating Sigmar1 inhibition results in mitochondrial elongation, one study showed overexpression of full length and a spliced variant of Sigmar1 (with a deletion of 47 base pairs starting at amino acid 106 further resulting in a shorter form of Sigmar1) increased mitochondrial length (Shioda et al., 2012). This study involved in vitro experiments using Sigmar1 overexpressed (full length and spliced Sigmar1) Neuro-2a cells. However, further studies are required to demonstrate the role of Sigmar1 in regulating mitochondrial dynamics and define the molecular mechanisms thereof.
The molecular role played by Sigmar1 in regulating mitochondrial calcium signaling has been reported by studies showing that Sigmar1 interacts with IP3R3 to regulate ER-mitochondrial calcium levels under ER-stress conditions in CHO and neuroblastoma cells (Hayashi and Su, 2007). Expression of the mutant SIGMAR1 resulted in a non-functional Sigmar1 caused by mislocalization from MAM, impairing Sigmar1-IP3R3 interaction and thereby altering intracellular and mitochondrial calcium handling as in human neuroblastoma cell lines (SH-SY5Y and SK-N-BE), murine motor neuron-like NSC-34 cells, and N2a cells (Gregianin et al., 2016; Watanabe et al., 2016). Similarly, the absence of Sigmar1 in motor neuron cells from Sigmar1 null mice and inactivation by antagonist NE-100 (in motor neuron cells) showed impairment of ER-mitochondria contacts, deregulation of calcium homeostasis, and activation of ER stress pathways. Sigmar1 activation by agonist (PRE-084) showed protective effects in these cell lines restoring the Sigmar1-IP3R3 interaction and preserving the calcium homeostasis (Bernard-Marissal et al., 2015; Watanabe et al., 2016). Moreover, overexpression of mutant Sigmar1 in cells resulted in collapsing of mitochondrial-associated ER membrane leading to deregulated calcium signaling (Dussossoy et al., 1999; Gregianin et al., 2016). Similarly, Sigmar1 mutant overexpression in neuro2A cells resulted in mitochondrial dysfunction and reduced ATP production. Supplementation with methyl pyruvate (TCA cycle substrate) enhanced ATP production and rescued the mitochondrial dysfunction (Fukunaga et al., 2015). Moreover, Sigmar1 has been shown to regulate mitochondrial metabolism where Sigmar1 drives cholesterol influx to the mitochondria through interaction with VDAC2 (Marriott et al., 2012).
Expression of a spliced variant of Sigmar1 results in a shorter version of the Sigmar1 protein and subsequently reduced ATP production, increase ER stress, increase autophagosome formation, and increased apoptosis in Neuro2a cells (Shioda et al., 2012). Additionally, Sigmar1 has been shown to be associated with mitochondrial metabolic regulation regulating conversion of cholesterol to pregnenolone and mediate steroidogenesis in MA-10 cells (Leydig tumor cells) (Marriott et al., 2012). Knockdown of Sigmar1 by siRNA in MA-10 cells reduced pregnenolone synthesis by more than 75% (Marriott et al., 2012). Pharmacologic activation of Sigmar1 by agonists enhances mitochondrial complex I activity and increased mitochondrial ROS production at the physiological condition in the forebrain of mice (Goguadze et al., 2019). However, under pathological conditions, i.e., in a mouse model of Alzheimer disease (mice expressing the Aβ1-42 and, Aβ25–35 peptides), activation of Sigmar1 has been shown to reduce mitochondrial ROS production (Lahmy et al., 2015; Goguadze et al., 2019). Further studies have suggested that activation of Sigmar1 using agonists provided protection against Aβ25–35 peptide induced reduction in oxygen consumption during all states of mitochondrial respiration, mitochondrial complex IV activity, and mitochondrial damage (as assessed by increased cytochrome C release) (Lahmy et al., 2015).
Sigmar1’s molecular role in the regulation of mitochondrial function has also been reported in the retinal cells. Sigmar1 overexpression and pharmacologic activation protected and restored mitochondrial membrane potential and cytochrome C release in retinal ganglion cells isolated from rat pups exposed to hypoxia (Ellis et al., 2017). Additionally, the absence of Sigmar1 in retinal explants in Sigmar1 null mice and neuronal cell line showed reduced mitochondrial clearance upon mitophagy induction without the involvement of PINK/Parkin mitophagy pathway (Yang et al., 2019). The converse was also true when using dopaminergic neurons where Sigmar1 activation by agonists (PRE-084) rescued the defects in mitophagy clearance in Parkinson’s disease in a PINK/Parkin dependent pathway (Wang et al., 2020). The involvement of Sigmar1 in neuroprotection was confirmed when dopaminergic neuronal cells isolated from Sigmar1 null mice showed impaired mitochondrial clearance with reduced levels of PINK and Parkin during mitophagy induction (Wang et al., 2020).
Similar effects have been observed in cardiomyocytes where the activation of Sigmar1 by agonists (SA4503, fluvoxamine) restored mitochondrial calcium mobilization and ATP production in angiotensin-induced cardiomyocyte hypertrophy (Tagashira et al., 2013c, 2014a). Similarly, pressure-overload induced hypertrophy in mice showed impaired calcium uptake and reduced ATP production, which were restored upon treatment with Sigmar1 agonists (SA4503, fluvoxamine) (Tagashira et al., 2013c, 2014a). These results were supported by experiments using Sigmar1 antagonists (NE-100, haloperidol), which demonstrated aggravated cardiac pathology in transverse aortic constriction (TAC)-subjected mice and angiotensin treated in cardiomyocytes (Tagashira et al., 2013c; Shinoda et al., 2016).
All these studies together suggest a molecular role for Sigmar1 in mitochondrial dynamics and functions in different cell types (including neuronal, retinal, and cardiac systems), modulation of mitochondrial calcium mobilization, mitochondrial ATP production, and mitochondrial lipid metabolism. However, the molecular mechanism responsible for Sigmar1 dependent regulation of mitochondrial dynamics and functions still remains unknown.
Sigmar1 in Autophagy
Sigmar1’s molecular role in the autophagy pathway has been made evident by studies where Sigmar1 siRNA knockdown in HEK293 and NSC34 cells led to the accumulation of numerous autophagic vacuoles often filled with non-degraded autophagic substrates and deformities of ER ultrastructure (Prause et al., 2013; Vollrath et al., 2014). Subsequent studies using NIH-3T3 cells stably expressing RFP-GFP-LC3 showed that Sigmar1 siRNA knockdown impaired the fusion of endosomes or autophagosomes to lysosomes (Vollrath et al., 2014). TEM images also showed the accumulation of several double-membrane autophagosomes (AV) filled with cargos that failed to fuse with lysosomes in Sigmar1-deficient GFP–RFP-LC3 expressing NIH-3T3, NSC34, and HEK-293 cell lines (Vollrath et al., 2014). All these biochemical analysis suggested an impairment of endolysosomal pathways in Sigmar1-deficient cells indicated by the accumulation of various autophagic substrates and defects in endosomal trafficking.
In addition, Sigmar1 has been identified as a novel regulator of autophagosome expansion during starvation in a siRNA screen in RPE1 cells (MacVicar et al., 2015). The effect of Sigmar1 on autophagy has also been reported by studies showing Sigmar1 ligands modulated the autophagic process in a dose-dependent manner uveal melanoma cells (Schrock et al., 2013). Moreover, studies using cancer cell lines (e.g., breast cancer, hepato-carcinoma, pancreatic adenocarcinoma, prostate adenocarcinoma) have also shown Sigmar1 mediated regulation of autophagy where Sigmar1 inhibition by antagonists (IPAG [1-(4-iodophenyl)-3-(2-adamantyl) guanidine], haloperidol) increased autophagosome formation. Surprisingly, Sigmar1 activation by agonists [PRE-084, (+) SKF10047] in these cell lines did not show any effect onautophagy (Maher et al., 2018; Yang et al., 2019). Contrary to the results obtained from studies in cancer cells, knockout of Sigmar1 impaired mitochondrial clearance without altering the PINK1/Parkin signaling in mouse retinal explants and cultured cells (HEK-293, NSC34, and SH-SY5Ycell lines) (Yang et al., 2019). The study further showed that the absence of Sigmar1 partially impaired autophagosome and lysosome fusion in SNARE-dependent mechanism, with no effects on autophagosome closure or lysosome functional activity (Yang et al., 2019). Recently, a Sigmar1 agonist [tetrahydro-N,N-dimethyl-2,2-diphenyl-3-furanmethanamine hydrochloride (ANAVEX2-73)] has been shown to increase autophagy through ULK1 phosphorylation and to reduce proteotoxicity by decreasing protein aggregation in HeLa, HEK 293A cells, and C. elegans (Christ et al., 2019).
Despite the contradictory findings concerning the role of Sigmar1 in autophagy made by different groups (which may be due to the use of different cell types and the systems used under the study conditions), Sigmar1 has been showed to have a substantial role in the modulation of autophagy in maintaining cellular homeostasis. However, since all these studies were limited to in vitro cell culture models, which lacked proper monitoring of autophagy flux, the molecular mechanism responsible for Sigmar1 dependent regulations of autophagy (either activation or inhibition) in vivo remained elusive. Further studies are required to dissect the types of autophagy (macro-autophagy vs. mitophagy) regulated by Sigmar1, determine molecular mechanism responsible for Sigmar1-dependent regulation of macro-autophagy/mitophagy, and assess the effects of acute and chronic Sigmar1 dependent regulation of macro-autophagy/mitophagy under conditions of cellular pathobiology.
Sigmar1 in Lipid Metabolism
Sigmar1’s molecular function in lipid metabolism has been suggested by several studies. Sigmar1 is specifically targeted to lipid storage sites (lipid droplets) (Hayashi and Su, 2003b) and regulates the compartmentalization of ER-synthesized neutral lipids (triglycerides and cholesteryl esters) in NG 108 cells (Hayashi and Su, 2003a, b; Hayashi and Fujimoto, 2010). Sigmar1 also accumulates at lipids rafts by forming a complex with cholesterol and galactosylceramides (GalCer) regulating the GalCer (Hayashi and Su, 2004) and regulates cholesterol transport in NG108 cells (Hayashi and Su, 2005). Sigmar1 also causes the remodeling of lipid rafts by increasing the level of lipid raft-forming gangliosides in PC12 cells (Takebayashi et al., 2004). All these studies were correlative, limited to in vitro experiments in cell lines [NG-108 cells (Hayashi and Su, 2003a, b)] and pharmacologic approaches using less selective ligands [such as (+)-pentazocine (Hayashi and Su, 2003a, b)]. Despite the preponderance of evidence provided by these studies, the molecular mechanisms of Sigmar1-dependent regulation of lipid metabolism remain unknown.
Sigmar1 in ER Stress Response
The role of Sigmar1, as well as that of Sigmar1 ligands (agonists and antagonists), has been extensively studied under ER stress conditions in different cell systems and demonstrated a wide range of cellular effects depending on the cell type. The Sigmar1 agonist [(+)-Pentazocine] suppressed oxidative stress-induced cell death and suppressed the induction of the ER stress proteins BiP and EIF2α in the human lens cell line (Wang et al., 2012). In contrast, the Sigmar1 antagonist (NE-100) protected against the ER stress-induced cell death in murine hippocampal HT22 cells via CHOP expression by the upregulation of GRP78 through the ATF6 pathway, and these protective effects were independent of Sigmar1 antagonistic effect (Ono et al., 2013). However, Sigmar1 antagonist (NE-100) did not change the expression of phosphorylated eukaryotic initiation factor 2α (p-eIF2α) and splicing of X-box-binding protein 1 (XBP-1) in HT22 cells (Ono et al., 2013). Another study showed imipramine treatment in HT22 cell inhibited tunicamycin-induced cell death, which was abolished by treatment with NE-100 (Ono et al., 2012). ER-stress (induced by tunicamycin or thapsigargin) transcriptionally increased Sigmar1 protein levels via the PERK/eIF2α/ATF4 pathway and ameliorated cell death signaling under in HEK293 cells or mouse neuroblastoma (Neuro2a) cells (Mitsuda et al., 2011). In contrast, Sigmar1 agonist (fluvoxamine) induced Sigmar1 level by increasing ATF4 translation without the involvement of the whole PERK pathway in Neuro2a (Omi et al., 2014). Sigmar1 dependent regulation of the stress-inducible transcription factor, C/EBP-homologous protein (CHOP), was reported in primary cardiomyocytes in the tunicamycin-induced ER-stress model (Alam et al., 2017). Sigmar1-siRNA knockdown in neonatal rat ventricular cardiomyocytes (NRCs) could significantly increase the expression of CHOP and induced cellular toxicity by sustained activation of tunicamycin-induced ER stress. Conversely, adenovirus-mediated Sigmar1 overexpression decreased the expression of CHOP and significantly decreased cellular toxicity in cardiomyocytes. Mechanistically, Sigmar1-dependent activation of IRE1α-XBP1s ER-stress response pathways was associated with inhibition of CHOP expression and suppression of cellular toxicity in cardiomyocytes (Alam et al., 2017). All these studies together suggested that Sigmar1 functions as an essential component of the adaptive ER-stress response pathways but that the molecular ER-stress signaling and mechanisms varied depending on the ER-stress inducer and cell type. Therefore, more studies are required to explore the role of Sigmar1 in different pathophysiological conditions in vivo using Sigmar1 genetic mouse models.
Perspective
Despite extensive studies carried out over the last fifty years, we are still at the beginning of our understanding of the molecular functions and cellular signaling mediated by Sigmar1. The majority of the studies to date have been limited to pharmacologic Sigmar1 ligands using in vitro cell culture models; data from in vivo genetic mouse models are needed to validate the cellular pathways mediated by these ligands. Extensive pharmacologic studies of the therapeutic role played by Sigmar1 in disease models has resulted in conflicting and confusing data due to the non-selectivity of the pharmacologic ligands, most of which possess affinity for other receptors or pleiotropic effects. To date the various Sigmar1 ligands tested to demonstrate the molecular role of Sigmar1 ranges from agonist, antagonists, reverse agonists, as well as some are Sigmar2 ligand (reviewed in Chu and Ruoho, 2016). Moreover, some of these ligands may also serve as a positive and negative allosteric modulators for Sigmar1 (reviewed in Vavers et al., 2019). The apparent complexity of these pharmacologic ligands requires elaborative review (Chu and Ruoho, 2016; Vavers et al., 2019). We presented a table of the most common ligands cited in this review article (Table 4).
TABLE 4.
Most common Sigmar1 ligands cited in this manuscript with their respective affinities and selectivity.
| Ligand | Binding Affinity | Effect on Sigmar1 | Effect on Sigmar2 | Selectivity for others | Pleiotropic effects |
| PRE-084 | Sigmar1 (Ki 44–53 nM) > Sigmar2 (Ki 32.1 μM) (Su et al., 1991; Garcés-Ramírez et al., 2011) | Agonist (Maurice et al., 1994b; Hayashi and Su, 2007; Garcés-Ramírez et al., 2011; Hyrskyluoto et al., 2013; Gao et al., 2018) | − | Low binding affinity for dopamine D2, muscarinic acetylcholine, serotonin, and adrenergic receptors (Su et al., 1991) | − |
| SA4503 (cutamesine) | Sigmar1 (Ki 4.6 nM) > Sigmar2 (Ki 63.1 nM) (Matsuno et al., 1996) | Agonist (Kobayashi et al., 1996; Matsuno et al., 1996) | − | Little affinity for 36 other receptors, ion channels and second messenger systems (Matsuno et al., 1996) | − |
| (+) Pentazocine | Sigmar1 (Ki 3.9–23.3 nM) > Sigmar2 (Ki 1,542–6,611 nM) (Hellewell et al., 1994; John et al., 1999) | Agonist (Pan et al., 2014; Mishra et al., 2015) | Binds at low affinity (Hellewell and Bowen, 1990) | k-opioid receptor-partial agonist (analgesic), enhances acetylcholine release (Hiramatsu and Hoshino, 2005), low affinity partial agonist to mu receptor (Chien and Pasternak, 1994) | − |
| ANAVEX 2-73 (blarcamesine) | Moderate Sigmar1 (Ki 860 nM) (Villard et al., 2011) | Agonist (Vamvakidès, 2002; Villard et al., 2011; Christ et al., 2019) | − | Muscarinic acetylcholine receptor-inhibitor (Villard et al., 2011; Christ et al., 2019) | − |
| (+)-SKF-10047 | Sigmar1 (Ki 54–597 nM) > Sigmar2 (Ki 11.17–39.74 μM) (Hellewell et al., 1994; Vilner and Bowen, 2000) | Agonist (Hellewell et al., 1994; Vilner and Bowen, 2000) | − | NMDA receptor antagonist, enhances acetylcholine release (Hiramatsu and Hoshino, 2005) | Inhibits Nav channels in Sigmar1 knockout cells (Johannessen et al., 2009, 2011) |
| Fluvoxamine | Sigmar1 (Ki 36 nM) > Sigmar2 (Ki 8,439 nM) (Narita et al., 1996) | Agonist (Narita et al., 1996; Tagashira et al., 2010; Bhuiyan et al., 2011b) | − | Serotonin reuptake inhibitor (Fu et al., 2012; Narita et al., 1996) | − |
| DTG | Sigmar1 (Ki 38–203 nM) =Sigmar2 (Ki13–58 nM) (Koe et al., 1989; Hellewell et al., 1994; Vilner and Bowen, 2000) | Agonist (Walker et al., 1992; Marrazzo et al., 2011; Zampieri et al., 2016) | Agonist (Walker et al., 1992; Marrazzo et al., 2011; Zampieri et al., 2016) | − | Inhibits Nav channels in Sigmar1 knockout cells (Johannessen et al., 2009, 2011) |
| DHEA | Moderate Sigmar1 (Ki 2.96 μM) (Maurice et al., 1996) | Agonist (Monnet et al., 1995; Waterhouse et al., 2007) | − | Neurosteroid, NMDA receptors, GABA-A receptors, nuclear receptor (Wu et al., 1991; Paul and Purdy, 1992; Waterhouse et al., 2007) | − |
| Cocaine | Sigmar1 (Ki 2.5–19 μM) > Sigmar2 (Ki 31 μM) (McCann and Su, 1991; Matsumoto et al., 2002; Garcés-Ramírez et al., 2011) | Agonist (Hayashi and Su, 2001, 2007; Matsumoto et al., 2002) | − | Dopamine transporters, Neurotransmitter reuptake blocker (Matsumoto et al., 2002; Beggiato et al., 2017) | − |
| 4-IBP | Sigmar1 (Ki 1.70 nM) > Sigmar2 (Ki 25.2 nM) (John et al., 1995, 1998) | Agonist (Amer et al., 2013) Inverse agonist (Bermack and Debonnel, 2005; Schmidt et al., 2016) | Antagonist (John et al., 1995, 1998) | − | Inhibits Ca2+ entry and block TRPCs and TRPMs in the absence of Sigmar1 (Amer et al., 2013) |
| PPBP | Sigmar1 (Ki 0.8 nM) = Sigmar2 (Ki 1.14 nM) (Glennon et al., 1991; Hashimoto and London, 1993) | Agonist (Goyagi et al., 2001; Nishimura et al., 2008; Yang et al., 2010) | − | nNOS inhibitor (Goyagi et al., 2001; Yang et al., 2010) | − |
| Fabomotizole (Afobazole) | Moderate Sigmar1 (Ki 5.9 μM) (Seredenin and Voronin, 2009) | Agonist (Katnik et al., 2016) | Agonist (Katnik et al., 2016) | Melatonin receptors (MT1 and MT3), MAO-A receptive site (Kryzhanovskii et al., 2018) | − |
| Pridopidine | Sigmar1 (Ki 57 nM) > Sigmar2 (Ki 5,450 nM) (Johnston et al., 2019) | Agonist (Shenkman et al., 2021; Dyhring et al., 2010; Johnston et al., 2019) | Binds at comparatively lower affinity (Johnston et al., 2019) | adrenergic-α2C, Dopamine D2 and D3 receptors, serotoninergic-5HT1A (Dyhring et al., 2010; Johnston et al., 2019) | − |
| Haloperidol | Sigmar1 (Ki 1–40 nM) Sigmar2 (Ki 12–221 nM) (Su et al., 1988: Hellewell et al., 1994; Vilner and Bowen, 2000; Choi et al., 2001) | Antagonist (Hellewell and Bowen, 1990; Hellewell et al., 1994; Mishra et al., 2015) | Antagonist (Hellewell and Bowen, 1990; Hellewell et al., 1994; Choi et al., 2001) | Dopamine (D2 and D3) receptor antagonist (Fox et al., 1994) | − |
| Sertraline | Sigmar1 (Ki 57 nM) > Sigmar2 (Ki 5,297 nM) (Narita et al., 1996) | Antagonist (Ishima et al., 2014) Inverse Agonist (Matsushima et al., 2019) | − | Serotonin reuptake inhibitor (Ishima et al., 2014) | − |
| PB28 | Sigmar2 (Ki 0.28 nM) > Sigmar1 (Ki 10 nM) (Azzariti et al., 2006) | Antagonist (low affinity) (Azzariti et al., 2006) | Agonist (Azzariti et al., 2006) | − | Inhibits Nav channels, and Kv2.1 current in the absence of Sigmar1 (Johannessen et al., 2009, 2011; Liu et al., 2017) |
| NE-100 | Sigmar1 (Ki 1.5 nM) > Sigmar2 (Ki 84.6 nM) (Okuyama et al., 1993) | Antagonist (Tanaka et al., 1995; Ono et al., 2013) | In vivo regulation of serotonin 5-HT2a receptors (Hashimoto et al., 1997) | Inhibits Kv2.1 current in the absence of Sigmar1 (Liu et al., 2017) | |
| E-5842 | Sigmar1 (Ki 4 nM) > Sigmar2 (Ki 220 nM) (Guitart and Farré, 1998; Guitart et al., 1998; Romero et al., 2000) | Antagonist (Zamanillo et al., 2000) | − | Dopamine receptors, serotonin receptors, acetylcholine and muscarinic receptors, adrenergic receptors (Guitart and Farré, 1998) | Induces micronucleated polychromatic erythrocyte-dependent hypothermia in Sigmar1 null mice (Guzmán et al., 2008) |
| BD1047 | Sigmar1 (Ki 0.6–5.3 nM) > Sigmar2 (Ki 47 nM) (Matsumoto et al., 1995) | Antagonist (Matsumoto et al., 1995; Pan et al., 2014) | Binds with 10 times lower affinity (Matsumoto et al., 1995) | Beta adrenergic receptor ligand (Matsumoto et al., 1995) | Inhibits Ca2+ entry and block TRP channels, and Kv2.1 current in the absence of Sigmar1 (Amer et al., 2013; Liu et al., 2017) |
| BD1063 | Sigmar1 (Ki 47 nM) > Sigmar2 (Ki 449 nM) (Matsumoto et al., 1995) | Antagonist (Matsumoto et al., 1995; Pan et al., 2014) | − | − | Inhibits Ca2+ entry and block TRP channels in the absence of Sigmar1 (Amer et al., 2013) |
| SM21 | − | − | Antagonist (Ghelardini et al., 2000) | − | Inhibits Kv2.1 current in the absence of Sigmar1 (Liu et al., 2017) |
| Methamphetamine | − | Antagonist (Hayashi and Su, 2007) Inverse agonist (Yasui and Su, 2016) | − | Dopamine Transporters, Monoamine transporters (Siciliano et al., 2014; McFadden et al., 2015) | − |
| IPAG | Sigmar1 (Ki 3 nM) > Sigmar2 (Ki 500–8000 nM) (Brimson et al., 2011; Schrock et al., 2013) | Antagonist (Schrock et al., 2013; Maher et al., 2018) | − | − | − |
Overall, Sigmar1 may be involved in a wide range of vital cellular functions, including regulation of ion channel dynamics, modulation of protein stability via its chaperone activity, regulation of mitochondrial dynamics and function, and regulation of autophagy. This makes Sigmar1 an attractive through which to modulate the above mentioned cellular processes in different pathologies involving alteration or dysfunction of calcium homeostasis, protein aggregation, accumulation of dysfunctional mitochondria, and altered lipid content and metabolism. Though Sigmar1 is ubiquitously present in different organs, its molecular role and signaling mechanisms remain elusive in different cell types using genetic mouse models.
Author Contributions
RA and MSB conceptualized, designed, and wrote the manuscript. RA, CSA, MM, and NSR participated in the conceptualization and editing of the manuscript. All co-authors edited and proofread the manuscript and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Institutes of Health Grants: R00HL122354, R01HL145753, R01HL145753-01S1, and R01HL145753-03S1; LSUHSC-S CCDS Finish Line Award, COVID-19 Research Award, and LARC Research Award to MSB; LSUHSC-S Malcolm Feist Cardiovascular and AHA Postdoctoral Fellowship to CSA (20POST35210789); and LSUHSC-S Malcolm Feist Pre-doctoral Fellowship to RA.
References
- Abate C., Niso M., Abatematteo F. S., Contino M., Colabufo N. A., Berardi F. (2020). PB28, the Sigma-1 and Sigma-2 receptors modulator with potent Anti-SARS-CoV-2 activity: a review about its pharmacological properties and structure affinity relationships. Front. Pharmacol. 11:589810. 10.3389/fphar.2020.589810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah C. S., Aishwarya R., Alam S., Morshed M., Remex N. S., Nitu S., et al. (2020). Methamphetamine induces cardiomyopathy by Sigmar1 inhibition-dependent impairment of mitochondrial dynamics and function. Commun. Biol. 3:682. 10.1038/s42003-020-01408-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah C. S., Alam S., Aishwarya R., Miriyala S., Panchatcharam M., Bhuiyan M. A. N., et al. (2018). Cardiac dysfunction in the sigma 1 receptor knockout mouse associated with impaired mitochondrial dynamics and bioenergetics. J. Am. Heart Assoc. 7:e009775. 10.1161/JAHA.118.009775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablordeppey S. Y., Fischer J. B., Law H., Glennon R. A. (2002). Probing the proposed phenyl-A region of the sigma-1 receptor. Bioorg. Med. Chem. 10 2759–2765. 10.1016/S0968-0896(02)00096-2 [DOI] [PubMed] [Google Scholar]
- Abou-lovergne A., Collado-Hilly M., Monnet F. P., Koukoui O., Prigent S., Coquil J. F., et al. (2011). Investigation of the role of sigma1-receptors in inositol 1,4,5-trisphosphate dependent calcium signaling in hepatocytes. Cell Calcium 50 62–72. 10.1016/j.ceca.2011.05.008 [DOI] [PubMed] [Google Scholar]
- Alam S., Abdullah C. S., Aishwarya R., Orr A. W., Traylor J., Miriyala S., et al. (2017). Sigmar1 regulates endoplasmic reticulum stress-induced C/EBP-homologous protein expression in cardiomyocytes. Biosci. Rep. 37:BSR20170898. 10.1042/bsr20170898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahtavakoli M., Jarrott B. (2011). Sigma-1 receptor ligand PRE-084 reduced infarct volume, neurological deficits, pro-inflammatory cytokines and enhanced anti-inflammatory cytokines after embolic stroke in rats. Brain Res. Bull. 85 219–224. 10.1016/j.brainresbull.2011.03.019 [DOI] [PubMed] [Google Scholar]
- Almendra L., Laranjeira F., Fernandez-Marmiesse A., Negrao L. (2018). SIGMAR1 gene mutation causing distal hereditary motor neuropathy in a portuguese family. Acta Myol. 37 2–4. [PMC free article] [PubMed] [Google Scholar]
- Alon A., Schmidt H. R., Wood M. D., Sahn J. J., Martin S. F., Kruse A. C. (2017). Identification of the gene that codes for the sigma2 receptor. Proc. Natl. Acad. Sci. U.S.A 114 7160–7165. 10.1073/pnas.1705154114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso G., Phan V., Guillemain I., Saunier M., Legrand A., Anoal M., et al. (2000). Immunocytochemical localization of the sigma(1) receptor in the adult rat central nervous system. Neuroscience 97 155–170. 10.1016/s0306-4522(00)00014-2 [DOI] [PubMed] [Google Scholar]
- Al-Saif A., Al-Mohanna F., Bohlega S. (2011). A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Annal. Neurol. 70 913–919. 10.1002/ana.22534 [DOI] [PubMed] [Google Scholar]
- Amer M. S., McKeown L., Tumova S., Liu R., Seymour V. A. L., Wilson L. A., et al. (2013). Inhibition of endothelial cell Ca2+ entry and transient receptor potential channels by Sigma-1 receptor ligands. Br. J. Pharmacol. 168 1445–1455. 10.1111/bph.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M., Motoike S., Yokota C., Usuki N., Yamamoto H., Urabe T., et al. (2019). SKF-10047, a prototype Sigma-1 receptor agonist, augmented the membrane trafficking and uptake activity of the serotonin transporter and its C-terminus-deleted mutant via a Sigma-1 receptor-independent mechanism. J. Pharmacol. Sci. 139 29–36. 10.1016/j.jphs.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Aydar E., Palmer C. P., Klyachko V. A., Jackson M. B. (2002). The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron 34 399–410. 10.1016/s0896-6273(02)00677-3 [DOI] [PubMed] [Google Scholar]
- Aydar E., Stratton D., Fraser S. P., Djamgoz M. B. A., Palmer C. (2016). Sigma-1 receptors modulate neonatal Nav1.5 ion channels in breast cancer cell lines. Eur. Biophys. J. 45 671–683. 10.1007/s00249-016-1135-0 [DOI] [PubMed] [Google Scholar]
- Azzariti A., Colabufo N. A., Berardi F., Porcelli L., Niso M., Simone G. M., et al. (2006). Cyclohexylpiperazine derivative PB28, a σ2 agonist and σ1 antagonist receptor, inhibits cell growth, modulates P-glycoprotein, and synergizes with anthracyclines in breast cancer. Mol. Cancer Ther. 5 1807–1816. 10.1158/1535-7163.Mct-05-0402 [DOI] [PubMed] [Google Scholar]
- Bai T., Lei P., Zhou H., Liang R., Zhu R., Wang W., et al. (2019). Sigma-1 receptor protects against ferroptosis in hepatocellular carcinoma cells. J. Cell Mol. Med. 23 7349–7359. 10.1111/jcmm.14594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasuriya D., Stewart A. P., Crottès D., Borgese F., Soriani O., Edwardson J. M. (2012). The sigma-1 receptor binds to the nav1.5 voltage-gated Na+ channel with 4-fold symmetry. J. Biol. Chem. 287 37021–37029. 10.1074/jbc.M112.382077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri F., Sparatore A., Alama A., Novelli F., Bruzzo C., Sparatore F. (2003). Novel sigma binding site ligands as inhibitors of cell proliferation in breast cancer. Oncol. Res. 3 455–461. 10.3727/000000003108747974 [DOI] [PubMed] [Google Scholar]
- Barr J. L., Deliu E., Brailoiu G. C., Zhao P., Yan G., Abood M. E., et al. (2015). Mechanisms of activation of nucleus accumbens neurons by cocaine via sigma-1 receptor–inositol 1,4,5-trisphosphate–transient receptor potential canonical channel pathways. Cell Calcium 58 196–207. 10.1016/j.ceca.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista T., Pascarella G., Staid D. S., Colotti G., Rosati J., Fiorillo A., et al. (2021). Known drugs identified by structure-based virtual screening are able to bind sigma-1 receptor and increase growth of huntington disease patient-derived cells. Int. J. Mol. Sci. 22:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggiato S., Borelli A. C., Borroto-Escuela D., Corbucci I., Tomasini M. C., Marti M., et al. (2017). Cocaine modulates allosteric D2-σ1 receptor-receptor interactions on dopamine and glutamate nerve terminals from rat striatum. Cell Signal. 40 116–124. 10.1016/j.cellsig.2017.09.007 [DOI] [PubMed] [Google Scholar]
- Belzil V. V., Daoud H., Camu W., Strong M. J., Dion P. A., Rouleau G. A. (2013). Genetic analysis of SIGMAR1 as a cause of familial ALS with dementia. Eur. J. Hum. Genet. 21 237–239. 10.1038/ejhg.2012.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermack J. E., Debonnel G. (2005). Distinct modulatory roles of sigma receptor subtypes on glutamatergic responses in the dorsal hippocampus. Synapse 55 37–44. 10.1002/syn.20085 [DOI] [PubMed] [Google Scholar]
- Bernard-Marissal N., Médard J.-J., Azzedine H., Chrast R. (2015). Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain 138 875–890. 10.1093/brain/awv008 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Fukunaga K. (2009). Stimulation of Sigma-1 receptor signaling by dehydroepiandrosterone ameliorates pressure overload-induced hypertrophy and dysfunctions in ovariectomized rats. Expert Opin. Ther. Targets 13 1253–1265. 10.1517/14728220903264064 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Fukunaga K. (2011). Targeting sigma-1 receptor signaling by endogenous ligands for cardioprotection. Expert Opin. Ther. Targets 15 145–155. 10.1517/14728222.2011.546350 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Tagashira H., Fukunaga K. (2011a). “Dehydroepiandrosterone-mediated stimulation of sigma-1 receptor activates Akt-eNOS signaling in the thoracic aorta of ovariectomized rats with abdominal aortic banding. Cardiovasc. Ther. 29 219–230. 10.1111/j.1755-5922.2010.00196.x [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Tagashira H., Fukunaga K. (2011b). Sigma-1 receptor stimulation with fluvoxamine activates Akt-eNOS signaling in the thoracic aorta of ovariectomized rats with abdominal aortic banding. Eur. J. Pharmacol. 650 621–628. 10.1016/j.ejphar.2010.10.055 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Tagashira H., Fukunaga K. (2013). Crucial interactions between selective serotonin uptake inhibitors and sigma-1 receptor in heart failure. J. Pharmacol. Sci. 121 177–184. 10.1254/jphs.12r13cp [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S., Tagashira H., Shioda N., Fukunaga K. (2010). Targeting sigma-1 receptor with fluvoxamine ameliorates pressure-overload-induced hypertrophy and dysfunctions. Expert Opin. Ther. Targets 14 1009–1022. 10.1517/14728222.2010.509348 [DOI] [PubMed] [Google Scholar]
- Bhuiyan S., Fukunaga K. (2010). Stimulation of Sigma-1 receptor by dehydroepiandrosterone ameliorates hypertension-induced kidney hypertrophy in ovariectomized rats. Exp. Biol. Med. (Maywood) 235 356–364. 10.1258/ebm.2009.009177 [DOI] [PubMed] [Google Scholar]
- Blasio A., Valenza M., Iyer M. R., Rice K. C., Steardo L., Hayashi T., et al. (2015). Sigma-1 receptor mediates acquisition of alcohol drinking and seeking behavior in alcohol-preferring rats. Behav. Brain Res. 287 315–322. 10.1016/j.bbr.2015.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailoiu G. C., Deliu E., Console-Bram L. M., Soboloff J., Abood M. E., Unterwald E. M., et al. (2015). Cocaine inhibits store-operated Ca2+ entry in brain microvascular endothelial cells: critical role for sigma-1 receptors. Biochem. J. 473 1–5. 10.1042/bj20150934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammer M. K., Gilmore D. L., Matsumoto R. R. (2006). Interactions between 3,4-methylenedioxymethamphetamine and σ1 receptors. Eur. J. Pharmacol. 553 141–145. 10.1016/j.ejphar.2006.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimson J., Brown C., Safrany S. (2011). Antagonists show GTP-sensitive high-affinity binding to the sigma-1 receptor. Br. J. Pharmacol. 164 772–780. 10.1111/j.1476-5381.2011.01417.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley R. L., Bauer M. B., Hartley N. D., Horning K. J., Currie K. P. M. (2017). Sigma-1 receptor ligands inhibit catecholamine secretion from adrenal chromaffin cells due to block of nicotinic acetylcholine receptors. J. Neurochem. 143 171–182. 10.1111/jnc.14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana G., Bucolo C., Murari G., Spampinato S. (2002). Ocular hypotensive action of topical flunarizine in the rabbit: role of sigma 1 recognition sites. J. Pharmacol. Exp. Ther. 303 1086–1094. 10.1124/jpet.102.040584 [DOI] [PubMed] [Google Scholar]
- Carnally S. M., Johannessen M., Henderson R. M., Jackson M. B., Edwardson J. M. (2010). Demonstration of a direct interaction between σ-1 receptors and acid-sensing ion channels. Biophys. J. 98 1182–1191. 10.1016/j.bpj.2009.12.4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendan C. M., Pujalte J. M., Portillo-Salido E., Montoliu L., Baeyens J. M. (2005). Formalin-induced pain is reduced in sigma(1) receptor knockout mice. Eur. J. Pharmacol. 511 73–74. 10.1016/j.ejphar.2005.01.036 [DOI] [PubMed] [Google Scholar]
- Chen Y., Hajipour A. R., Sievert M. K., Arbabian M., Ruoho A. E. (2007). Characterization of the cocaine binding site on the sigma-1 receptor. Biochemistry 46 3532–3542. 10.1021/bi061727o [DOI] [PubMed] [Google Scholar]
- Chertkow Y., Weinreb O., Youdim M. B., Silver H. (2007). Dopamine and serotonin metabolism in response to chronic administration of fluvoxamine and haloperidol combined treatment. J. Neural. Transm. (Vienna) 114 1443–1454. 10.1007/s00702-007-0753-1 [DOI] [PubMed] [Google Scholar]
- Chevallier N., Keller E., Maurice T. (2011). Behavioural phenotyping of knockout mice for the sigma-1 chaperone protein revealed gender-related anxiety, depressive-like and memory alterations. J. Psychopharmacol. 25 960–975. 10.1177/0269881111400648 [DOI] [PubMed] [Google Scholar]
- Chien C. C., Pasternak G. W. (1994). Selective antagonism of opioid analgesia by a sigma system. J. Pharmacol. Exp. Ther. 271 1583–1590. [PubMed] [Google Scholar]
- Choi S.-R., Roh D.-H., Yoon S.-Y., Kwon S.-G., Choi H.-S., Han H.-J., et al. (2016). Astrocyte sigma-1 receptors modulate connexin 43 expression leading to the induction of below-level mechanical allodynia in spinal cord injured mice. Neuropharmacology 111 34–46. 10.1016/j.neuropharm.2016.08.027 [DOI] [PubMed] [Google Scholar]
- Choi S.-R., Yang B., Plössl K., Chumpradit S., Wey S.-P., Acton P. D., et al. (2001). Development of a Tc-99m labeled sigma-2 receptor-specific ligand as a potential breast tumor imaging agent. Nucl. Med. Biol. 28 657–666. 10.1016/S0969-8051(01)00234-7 [DOI] [PubMed] [Google Scholar]
- Christ M. G., Huesmann H., Nagel H., Kern A., Behl C. (2019). Sigma-1 receptor activation induces autophagy and increases proteostasis capacity in vitro and in vivo. Cells 8:211. 10.3390/cells8030211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou K., Zamba E., Tsingis M., Mubaidin A., Horani K., Abu-Sheik S., et al. (2000). A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1-p12. Ann. Neurol. 48 877–884. [DOI] [PubMed] [Google Scholar]
- Chu U. B., Ruoho A. E. (2016). Biochemical pharmacology of the sigma-1 receptor. Mol. Pharmacol. 89 142–153. 10.1124/mol.115.101170 [DOI] [PubMed] [Google Scholar]
- Chu U. B., Ramachandran S., Hajipour A. R., Ruoho A. E. (2013). Photoaffinity labeling of the sigma-1 receptor with N-[3-(4-Nitrophenyl)propyl]-N-dodecylamine: evidence of receptor dimers. Biochemistry 52 859–868. 10.1021/bi301517u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly S., Goguadze N., Yasui Y., Kimura Y., Wang S.-M., Sharikadze N., et al. (2020a). Knocking out sigma-1 receptors reveals diverse health problems. Cell Mol. Neurobiol. 10.1007/s10571-020-00983-3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly S., Khalil B., Viguier V., Roussel J., Maurice T., Lievens J. C. (2020b). Sigma-1 receptor is a key genetic modulator in amyotrophic lateral sclerosis. Hum. Mol. Genet. 29 529–540. 10.1093/hmg/ddz267 [DOI] [PubMed] [Google Scholar]
- Crottes D., Martial S., Rapetti-Mauss R., Pisani D. F., Loriol C., Pellissier B., et al. (2011). Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. J. Biol. Chem. 286 27947–27958. 10.1074/jbc.M111.226738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crottes D., Rapetti-Mauss R., Alcaraz-Perez F., Tichet M., Gariano G., Martial S., et al. (2016). SIGMAR1 regulates membrane electrical activity in response to extracellular matrix stimulation to drive cancer cell invasiveness. Cancer Res. 76 607–618. 10.1158/0008-5472.CAN-15-1465 [DOI] [PubMed] [Google Scholar]
- Delaunois A., De Ron P., Detrait E., Guyaux M. (2013). Inhibitory effects of sigma-1 ligands on handling-induced tachycardia in conscious tethered rats. Fundam. Clin. Pharmacol. 27 354–363. 10.1111/j.1472-8206.2012.01042.x [DOI] [PubMed] [Google Scholar]
- Di T., Zhang S., Hong J., Zhang T., Chen L. (2017). Hyperactivity of hypothalamic-pituitary-adrenal axis due to dysfunction of the hypothalamic glucocorticoid receptor in sigma-1 receptor knockout mice. Front. Mol. Neurosci. 10:287. 10.3389/fnmol.2017.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussossoy D., Carayon P., Belugou S., Feraut D., Bord A., Goubet C., et al. (1999). Colocalization of sterol isomerase and sigma(1) receptor at endoplasmic reticulum and nuclear envelope level. Eur. J. Biochem. 263 377–386. 10.1046/j.1432-1327.1999.00500.x [DOI] [PubMed] [Google Scholar]
- Dyhring T., Nielsen E. Ø, Sonesson C., Pettersson F., Karlsson J., Svensson P., et al. (2010). The dopaminergic stabilizers pridopidine (ACR16) and (−)−OSU6162 display dopamine D2 receptor antagonism and fast receptor dissociation properties. Eur. J. Pharmacol. 628 19–26. 10.1016/j.ejphar.2009.11.025 [DOI] [PubMed] [Google Scholar]
- Ela C., Barg J., Vogel Z., Hasin Y., Eilam Y. (1994). Sigma receptor ligands modulate contractility, Ca++ influx and beating rate in cultured cardiac myocytes. J. Pharmacol. Exp. Ther. 269 1300–1309. [PubMed] [Google Scholar]
- Ela C., Hasin Y., Eilam Y. (1996). Apparent desensitization of a sigma receptor sub-population in neonatal rat cardiac myocytes by pre-treatment with sigma receptor ligands. Eur. J. Pharmacol. 295 275–280. [DOI] [PubMed] [Google Scholar]
- Ellis D. Z., Li L., Park Y., He S., Mueller B., Yorio T. (2017). Sigma-1 receptor regulates mitochondrial function in glucose- and oxygen-deprived retinal ganglion cells. Invest Ophthalmol. Vis. Sci. 58 2755–2764. 10.1167/iovs.16-19199 [DOI] [PubMed] [Google Scholar]
- Feher A., Juhasz A., Laszlo A., Kalman J., Jr., Pakaski M., Kalman J., et al. (2012). Association between a variant of the sigma-1 receptor gene and Alzheimer’s disease. Neurosci. Lett. 517 136–139. 10.1016/j.neulet.2012.04.046 [DOI] [PubMed] [Google Scholar]
- Fo Y., Zhang C., Chen X., Liu X., Ye T., Guo Y., et al. (2020). Chronic sigma-1 receptor activation ameliorates ventricular remodeling and decreases susceptibility to ventricular arrhythmias after myocardial infarction in rats. Eur. J. Pharmacol. 889:173614. 10.1016/j.ejphar.2020.173614 [DOI] [PubMed] [Google Scholar]
- Fontanilla D., Hajipour A. R., Pal A., Chu U. B., Arbabian M., Ruoho A. E. (2008). Probing the steroid binding domain-like I (SBDLI) of the sigma-1 receptor binding site using N-substituted photoaffinity labels. Biochemistry 47 7205–7217. 10.1021/bi800564j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanilla D., Johannessen M., Hajipour A. R., Cozzi N. V., Jackson M. B., Ruoho A. E. (2009). The hallucinogen N, N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 323 934–937. 10.1126/science.1166127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstl H., Kurz A. (1999). Clinical features of Alzheimer’s disease. Eur. Arch. Psychiatry Clin. Neurosci. 249 288–290. 10.1007/s004060050101 [DOI] [PubMed] [Google Scholar]
- Fox C. A., Mansour A., Watson S. J. (1994). The effects of haloperidol on dopamine receptor gene expression. Exp. Neurol. 130 288–303. 10.1006/exnr.1994.1207 [DOI] [PubMed] [Google Scholar]
- Francardo V., Bez F., Wieloch T., Nissbrandt H., Ruscher K., Cenci M. A. (2014). Pharmacological stimulation of sigma-1 receptors has neurorestorative effects in experimental parkinsonism. Brain 137 1998–2014. 10.1093/brain/awu107 [DOI] [PubMed] [Google Scholar]
- Francardo V., Geva M., Bez F., Denis Q., Steiner L., Hayden M. R., et al. (2019). Pridopidine induces functional neurorestoration via the sigma-1 receptor in a mouse model of parkinson’s disease. Neurotherapeutics 16 465–479. 10.1007/s13311-018-00699-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Yu S., Guo X., Li X., Li T., Li H., et al. (2012). Fluvoxamine increased glutamate release by activating both 5-HT(3) and sigma-1 receptors in prelimbic cortex of chronic restraint stress C57BL/6 mice. Biochim. Biophys. Acta 1823 826–837. 10.1016/j.bbamcr.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Fukunaga K., Shinoda Y., Tagashira H. (2015). The role of SIGMAR1 gene mutation and mitochondrial dysfunction in amyotrophic lateral sclerosis. J. Pharmacol. Sci. 127 36–41. 10.1016/j.jphs.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Ganapathy M. E., Prasad P. D., Huang W., Seth P., Leibach F. H., Ganapathy V. (1999). Molecular and ligand-binding characterization of the σ-receptor in the jurkat human T lymphocyte cell line. J. Pharmacol. Exp. Ther. 289 251–260. [PubMed] [Google Scholar]
- Gao Q. J., Yang B., Chen J., Shi S. B., Yang H. J., Liu X. (2018). Sigma-1 receptor stimulation with PRE-084 Ameliorates myocardial ischemia-reperfusion injury in rats. Chin. Med. J. (Engl) 131 539–543. 10.4103/0366-6999.226076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. F., Yao J. J., He Y. L., Hu C., Mei Y. A. (2012). Sigma-1 receptor agonists directly inhibit Nav1.2/1.4 channels. PLoS One 7:e49384. 10.1371/journal.pone.0049384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcés-Ramírez L., Green J. L., Hiranita T., Kopajtic T. A., Mereu M., Thomas A. M., et al. (2011). Sigma receptor agonists: receptor binding and effects on mesolimbic dopamine neurotransmission assessed by microdialysis. Biol. Psychiatry 69 208–217. 10.1016/j.biopsych.2010.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparre G., Abate C., Berardi F., Cassano G. (2012). The sigma-1 receptor antagonist PB212 reduces the Ca2+-release through the inositol (1, 4, 5)-trisphosphate receptor in SK-N-SH cells. Eur J Pharmacol 684 59–63. 10.1016/j.ejphar.2012.03.021 [DOI] [PubMed] [Google Scholar]
- Ghelardini C., Galeotti N., Bartolini A. (2000). Pharmacological identification of SM-21, the novel σ2 antagonist. Pharmacol. Biochem. Behav. 67 659–662. 10.1016/S0091-3057(00)00405-6 [DOI] [PubMed] [Google Scholar]
- Glassman A. H., Bigger J. T., Gaffney M., Shapiro P. A., Swenson J. R. (2006). Onset of major depression associated with acute coronary syndromes: relationship of onset, major depressive disorder history, and episode severity to sertraline benefit. Arch. Gen. Psychiatry 63 283–288. 10.1001/archpsyc.63.3.283 [DOI] [PubMed] [Google Scholar]
- Glennon R. A., Yousif M. Y., Ismaiel A. M., el-Ashmawy M. B., Herndon J. L., Fischer J. B., et al. (1991). Novel 1-phenylpiperazine and 4-phenylpiperidine derivatives as high-affinity sigma ligands. J. Med. Chem. 34 3360–3365. 10.1021/jm00116a003 [DOI] [PubMed] [Google Scholar]
- Goguadze N., Zhuravliova E., Morin D., Mikeladze D., Maurice T. (2019). Sigma-1 receptor agonists induce oxidative stress in mitochondria and enhance complex i activity in physiological condition but protect against pathological oxidative stress. Neurotox Res. 35 1–18. 10.1007/s12640-017-9838-2 [DOI] [PubMed] [Google Scholar]
- Gordon D. E., Hiatt J., Bouhaddou M., Rezelj V. V., Ulferts S., Braberg H., et al. (2020a). Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370:eabe9403. 10.1126/science.abe9403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020b). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyagi T., Goto S., Bhardwaj A., Dawson V. L., Hurn P. D., Kirsch J. R. (2001). Neuroprotective effect of sigma (1)-Receptor Ligand 4-Phenyl-1-(4-Phenylbutyl) Piperidine (PPBP) is linked to reduced neuronal nitric oxide production. Stroke 32 1613–1620. 10.1161/01.STR.32.7.1613 [DOI] [PubMed] [Google Scholar]
- Grachev I. D., Meyer P. M., Becker G. A., Bronzel M., Marsteller D., Pastino G., et al. (2020). Sigma-1 and dopamine D2/D3 receptor occupancy of pridopidine in healthy volunteers and patients with Huntington disease: a [18F] fluspidine and [18F] fallypride PET study. Eur. J. Nucl. Med. Mol. Imaging 48 1103–1115. 10.1007/s00259-020-05030-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregianin E., Pallafacchina G., Zanin S., Crippa V., Rusmini P., Poletti A., et al. (2016). Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum. Mol. Genet. 25 3741–3753. 10.1093/hmg/ddw220 [DOI] [PubMed] [Google Scholar]
- Gueguinou M., Crottès D., Chantôme A., Rapetti-Mauss R., Potier-Cartereau M., Clarysse L., et al. (2017). The SigmaR1 chaperone drives breast and colorectal cancer cell migration by tuning SK3-dependent Ca 2+ homeostasis. Oncogene 36 3640–3647. 10.1038/onc.2016.501 [DOI] [PubMed] [Google Scholar]
- Guitart X., Codony X., Ballarín M., Dordal A., Farré A. J. (1998). E-5842: a new potent and preferential sigma ligand. Preclinical Pharmacological Profile. CNS Drug Rev. 4 201–224. 10.1111/j.1527-3458.1998.tb00065.x [DOI] [Google Scholar]
- Guitart X., Farré A. J. (1998). The effect of E-5842, a σ receptor ligand and potential atypical antipsychotic, on Fos expression in rat forebrain. Eur. J. Pharmacol. 363 127–130. 10.1016/S0014-2999(98)00812-7 [DOI] [PubMed] [Google Scholar]
- Guzmán A., Marín A.-P., García C., de Henestrosa A. R. F., Ruiz M. T., Tortajada A., et al. (2008). Induction of hypothermic conditions associated with increased micronuclei formation in sigma-1 receptor knockout mice after administration of the antipsychotic compound E-5842. Environ. Mol. Mutagen. 49 727–733. 10.1002/em.20428 [DOI] [PubMed] [Google Scholar]
- Ha Y., Saul A., Tawfik A., Williams C., Bollinger K., Smith R., et al. (2011). Late-onset inner retinal dysfunction in mice lacking sigma receptor 1 (sigmaR1). Invest Ophthalmol. Vis. Sci. 52 7749–7760. 10.1167/iovs.11-8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y., Saul A., Tawfik A., Zorrilla E. P., Ganapathy V., Smith S. B. (2012). Diabetes accelerates retinal ganglion cell dysfunction in mice lacking sigma receptor 1. Mol. Vis. 18 2860–2870. [PMC free article] [PubMed] [Google Scholar]
- Hanner M., Moebius F. F., Flandorfer A., Knaus H. G., Striessnig J., Kempner E., et al. (1996). Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc. Natl. Acad. Sci. U.S.A. 93 8072–8077. 10.1073/pnas.93.15.8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harukuni I., Bhardwaj A., Shaivitz A. B., DeVries A. C., London E. D., Hurn P. D., et al. (2000). sigma(1)-receptor ligand 4-phenyl-1-(4-phenylbutyl)-piperidine affords neuroprotection from focal ischemia with prolonged reperfusion. Stroke 31 976–982. 10.1161/01.str.31.4.976 [DOI] [PubMed] [Google Scholar]
- Hashimoto K. (2021). Repurposing of CNS drugs to treat COVID-19 infection: targeting the sigma-1 receptor. Eur. Arch. Psychiatry Clin. Neurosci. 271 249–258. 10.1007/s00406-020-01231-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., London E. D. (1993). Further characterization of [3H]ifenprodil binding to sigma receptors in rat brain. Eur. J. Pharmacol. 236 159–163. 10.1016/0014-2999(93)90241-9 [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Narita N., Tomitaka S.-I., Iyo M., Minabe Y. (1997). In vivo regulation of serotonin 5-HT2a receptors in rat brain by subchronic administration of σ receptor ligand NE-100. Life Sci. 60 2245–2254. 10.1016/S0024-3205(97)00239-7 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujimoto M. (2010). Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 77 517–528. 10.1124/mol.109.062539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2001). Regulating ankyrin dynamics: roles of sigma-1 receptors. Proc. Natl. Acad. Sci. U.S.A. 98 491–496. 10.1073/pnas.021413698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2003a). Intracellular dynamics of sigma-1 receptors (sigma(1) binding sites) in NG108-15 cells. J. Pharmacol. Exp. Ther. 306 726–733. 10.1124/jpet.103.051292 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2003b). Sigma-1 receptors (sigma(1) binding sites) form raft-like microdomains and target lipid droplets on the endoplasmic reticulum: roles in endoplasmic reticulum lipid compartmentalization and export. J. Pharmacol. Exp. Ther. 306 718–725. 10.1124/jpet.103.051284 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2004). Sigma-1 receptors at galactosylceramide-enriched lipid microdomains regulate oligodendrocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 101 14949–14954. 10.1073/pnas.0402890101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2005). The potential role of sigma-1 receptors in lipid transport and lipid raft reconstitution in the brain: implication for drug abuse. Life Sci. 77 1612–1624. 10.1016/j.lfs.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2007). Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131 596–610. 10.1016/j.cell.2007.08.036 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hayashi E., Fujimoto M., Sprong H., Su T.-P. (2012). The Lifetime of UDP-galactose:ceramide galactosyltransferase is controlled by a distinct endoplasmic reticulum-associated degradation (ERAD) regulated by sigma-1 receptor chaperones. J. Biol. Chem. 287 43156–43169. 10.1074/jbc.M112.380444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Maurice T., Su T.-P. (2000). Ca2+ Signaling via ς1-receptors: novel regulatory mechanism affecting intracellular Ca2+concentration. J. Pharmacol. Exp. Ther. 293 788–798. [PubMed] [Google Scholar]
- Hayashi T., Tsai S. Y., Mori T., Fujimoto M., Su T. P. (2011). Targeting ligand-operated chaperone sigma-1 receptors in the treatment of neuropsychiatric disorders. Expert Opin. Ther. Targets 15 557–577. 10.1517/14728222.2011.560837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellewell S. B., Bowen W. D. (1990). A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 527 244–253. 10.1016/0006-8993(90)91143-5 [DOI] [PubMed] [Google Scholar]
- Hellewell S. B., Bruce A., Feinstein G., Orringer J., Williams W., Bowen W. D. (1994). Rat liver and kidney contain high densities of σ1 and σ2 receptors: characterization by ligand binding and photoaffinity labeling. Eur. J. Pharmacol. 268 9–18. 10.1016/0922-4106(94)90115-5 [DOI] [PubMed] [Google Scholar]
- Hernandez L. L., Appel J. B. (1979). Dopaminergic involvement in the mechanism of action of pentazocine. Behav. Neural Biol. 26 384–400. 10.1016/s0163-1047(79)91392-x [DOI] [PubMed] [Google Scholar]
- Herrera Y., Katnik C., Rodriguez J. D., Hall A. A., Willing A., Pennypacker K. R., et al. (2008). σ-1 Receptor modulation of acid-sensing ion channel a (ASIC1a) and ASIC1a-Induced Ca2+ influx in rat cortical neurons. J. Pharmacol. Exp. Ther. 327 491–502. 10.1124/jpet.108.143974 [DOI] [PubMed] [Google Scholar]
- Hiramatsu M., Hoshino T. (2005). Improvement of memory impairment by (+)- and (−)−pentazocine via sigma, but not kappa opioid receptors. Brain Res. 1057 72–80. 10.1016/j.brainres.2005.07.028 [DOI] [PubMed] [Google Scholar]
- Hoertel N., Sanchez-Rico M., Vernet R., Jannot A. S., Neuraz A., Blanco C., et al. (2021). Observational study of haloperidol in hospitalized patients with COVID-19. PLoS One 16:e0247122. 10.1371/journal.pone.0247122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Sha S., Zhou L., Wang C., Yin J., Chen L. (2015). Sigma-1 receptor deficiency reduces MPTP-induced parkinsonism and death of dopaminergic neurons. Cell Death Dis. 6:e1832. 10.1038/cddis.2015.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. C., Yano H., Hiranita T., Chin F. T., McCurdy C. R., Su T.-P., et al. (2017). The sigma-1 receptor modulates dopamine transporter conformation and cocaine binding and may thereby potentiate cocaine self-administration in rats. J. Biol. Chem. 292 11250–11261. 10.1074/jbc.M116.774075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horga A., Tomaselli P. J., Gonzalez M. A., Laura M., Muntoni F., Manzur A. Y., et al. (2016). SIGMAR1 mutation associated with autosomal recessive Silver-like syndrome. Neurology 87 1607–1612. 10.1212/WNL.0000000000003212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu A., Antal Z., Lenart L., Hodrea J., Koszegi S., Balogh D. B., et al. (2017). σ1-Receptor Agonism protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 28 152–165. 10.1681/asn.2015070772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zheng L., Halliday G., Dobson-Stone C., Wang Y., Tang H. D., et al. (2011). Genetic polymorphisms in sigma-1 receptor and apolipoprotein E interact to influence the severity of Alzheimer’s disease. Curr. Alzheimer Res. 8 765–770. 10.2174/156720511797633232 [DOI] [PubMed] [Google Scholar]
- Hyrskyluoto A., Pulli I., Törnqvist K., Huu Ho T., Korhonen L., Lindholm D. (2013). Sigma-1 receptor agonist PRE084 is protective against mutant huntingtin-induced cell degeneration: involvement of calpastatin and the NF-κB pathway. Cell Death Dis. 4:e646. 10.1038/cddis.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Sugita S., Shoji H., Ichimoto H., Hide I., Nakata Y. (2000). Repeated haloperidol treatment decreases sigma(1) receptor binding but does not affect its mRNA levels in the guinea pig or rat brain. Eur. J. Pharmacol. 401 307–316. [DOI] [PubMed] [Google Scholar]
- Ishima T., Fujita Y., Hashimoto K. (2014). Interaction of new antidepressants with sigma-1 receptor chaperones and their potentiation of neurite outgrowth in PC12 cells. Eur. J. Pharmacol. 727 167–173. 10.1016/j.ejphar.2014.01.064 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirooka Y., Sunagawa K. (2013). Brain sigma-1 receptor stimulation improves mental disorder and cardiac function in mice with myocardial infarction. J. Cardiovasc. Pharmacol. 62 222–228. 10.1097/FJC.0b013e3182970b15 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirooka Y., Matsukawa R., Nakano M., Sunagawa K. (2011). Decreased brain sigma-1 receptor contributes to the relationship between heart failure and depression. Cardiovasc. Res. 93 33–40. 10.1093/cvr/cvr255 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Morino H., Miyamoto R., Matsuda Y., Ohsawa R., Kurashige T., et al. (2018). Compound heterozygote mutations in the SIGMAR1 gene in an oldest-old patient with amyotrophic lateral sclerosis. Geriatr. Gerontol. Int. 18 1519–1520. 10.1111/ggi.13506 [DOI] [PubMed] [Google Scholar]
- Jansen K. L., Faull R. L., Storey P., Leslie R. A. (1993). Loss of sigma binding sites in the CA1 area of the anterior hippocampus in Alzheimer’s disease correlates with CA1 pyramidal cell loss. Brain Res. 623 299–302. 10.1016/0006-8993(93)91441-t [DOI] [PubMed] [Google Scholar]
- Jiang G., Mysona B., Dun Y., Gnana-Prakasam J. P., Pabla N., Li W., et al. (2006). Expression, subcellular localization, and regulation of sigma receptor in retinal muller cells. Invest Ophthalmol. Vis. Sci. 47 5576–5582. 10.1167/iovs.06-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Krishnan R., Kuchibhatla M., Cuffe M. S., Martsberger C., Arias R. M., et al. (2011). Characteristics of depression remission and its relation with cardiovascular outcome among patients with chronic heart failure (from the SADHART-CHF Study). Am. J. Cardiol. 107 545–551. 10.1016/j.amjcard.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., O’Connor C., Silva S. G., Kuchibhatla M., Cuffe M. S., Callwood D. D., et al. (2008). Safety and efficacy of sertraline for depression in patients with CHF (SADHART-CHF): a randomized, double-blind, placebo-controlled trial of sertraline for major depression with congestive heart failure. Am. Heart J. 156 437–444. 10.1016/j.ahj.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Fontanilla D., Mavlyutov T., Ruoho A. E., Jackson M. B. (2011). Antagonist action of progesterone at sigma-receptors in the modulation of voltage-gated sodium channels. Am. J. Physiol. Cell Physiol. 300 C328–C337. 10.1152/ajpcell.00383.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Ramachandran S., Riemer L., Ramos-Serrano A., Ruoho A. E., Jackson M. B. (2009). Voltage-gated sodium channel modulation by sigma-receptors in cardiac myocytes and heterologous systems. Am. J. Physiol. Cell Physiol. 296 C1049–C1057. 10.1152/ajpcell.00431.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John C. S., Gulden M. E., Li J., Bowen W. D., McAfee J. G., Thakur M. L. (1998). Synthesis, In Vitro binding, and tissue distribution of radioiodinated 2-[125I]N-(N-Benzylpiperidin-4-yl)-2-Iodo Benzamide, 2-[125I]BP: a potential σ receptor marker for human prostate tumors. Nucl. Med. Biol. 25 189–194. 10.1016/S0969-8051(97)00168-6 [DOI] [PubMed] [Google Scholar]
- John C. S., Vilner B. J., Geyer B. C., Moody T., Bowen W. D. (1999). Targeting sigma receptor-binding benzamides as in vivo diagnostic and therapeutic agents for human prostate tumors. Cancer Res. 59 4578–4583. [PubMed] [Google Scholar]
- John C. S., Vilner B. J., Gulden M. E., Efange S. M. N., Langason R. B., Moody T. W., et al. (1995). Synthesis and pharmacological characterization of 4-[125I]- N-(N-Benzylpiperidin-4-yl)-4-iodobenzamide: a high affinity σ receptor ligand for potential imaging of breast cancer. Cancer Res. 55 3022–3027. [PubMed] [Google Scholar]
- Johnston T. H., Geva M., Steiner L., Orbach A., Papapetropoulos S., Savola J.-M., et al. (2019). Pridopidine, a clinic-ready compound, reduces 3,4-dihydroxyphenylalanine-induced dyskinesia in Parkinsonian macaques. Mov. Disord. 34 708–716. 10.1002/mds.27565 [DOI] [PubMed] [Google Scholar]
- Kahoun J. R., Ruoho A. E. (1992). (125I)iodoazidococaine, a photoaffinity label for the haloperidol-sensitive sigma receptor. Proc. Natl. Acad. Sci. U.S.A. 89 1393–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara R., Yamamoto N., Suzuki K., Sobue K. (2017). The σ1 receptor regulates accumulation of GM1 ganglioside-enriched autophagosomes in astrocytes. Neuroscience 340 176–187. 10.1016/j.neuroscience.2016.10.058 [DOI] [PubMed] [Google Scholar]
- Katnik C., Garcia A., Behensky A. A., Yasny I. E., Shuster A. M., Seredenin S. B., et al. (2016). Activation of sigma1 and sigma2 receptors by afobazole increases glial cell survival and prevents glial cell activation and nitrosative stress after ischemic stroke. J. Neurochem. 139 497–509. 10.1111/jnc.13756 [DOI] [PubMed] [Google Scholar]
- Katnik C., Guerrero W. R., Pennypacker K. R., Herrera Y., Cuevas J. (2006). Sigma-1 receptor activation prevents intracellular calcium dysregulation in cortical neurons during in vitro ischemia. J. Pharmacol. Exp. Ther. 319 1355–1365. 10.1124/jpet.106.107557 [DOI] [PubMed] [Google Scholar]
- Kekuda R., Prasad P. D., Fei Y.-J., Leibach F. H., Ganapathy V. (1996). Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem. Biophys. Res. Commun. 229 553–558. 10.1006/bbrc.1996.1842 [DOI] [PubMed] [Google Scholar]
- Kim F. J., Kovalyshyn I., Burgman M., Neilan C., Chien C.-C., Pasternak G. W. (2010). σ1 Receptor modulation of G-protein-coupled receptor signaling: potentiation of opioid transduction independent from receptor binding. Mol. Pharmacol. 77 695–703. 10.1124/mol.109.057083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J., Kwon M.-J., Choi W.-J., Oh K.-W., Oh S.-i., Ki C.-S., et al. (2014). Mutations in UBQLN2 and SIGMAR1 genes are rare in Korean patients with amyotrophic lateral sclerosis. Neurobiol. Aging 35 1957.e1957–1957.e1958. 10.1016/j.neurobiolaging.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Li H., Kim H. W., Shin S. E., Choi I. W., Firth A. L., et al. (2016). Selective serotonin reuptake inhibitor sertraline inhibits voltage-dependent K+ channels in rabbit coronary arterial smooth muscle cells. J. Biosci. 41 659–666. 10.1007/s12038-016-9645-6 [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Matsuoka Y., Suzuki T., Mirrielees J., Yang J. (2012). Sigma-1 receptor alters the kinetics of Kv1.3 voltage gated potassium channels but not the sensitivity to receptor ligands. Brain Res. 1452 1–9. 10.1016/j.brainres.2012.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klouz A., Saïd D. B., Ferchichi H., Kourda N., Ouanes L., Lakhal M., et al. (2008). Protection of cellular and mitochondrial functions against liver ischemia by N-benzyl-N’-(2-hydroxy-3,4-dimethoxybenzyl)-piperazine (BHDP), a sigma1 ligand. Eur. J. Pharmacol. 578 292–299. 10.1016/j.ejphar.2007.09.038 [DOI] [PubMed] [Google Scholar]
- Klouz A., Sapena R., Liu J., Maurice T., Tillement J.-P., Papadopoulos V., et al. (2002). Evidence for sigma-1-like receptors in isolated rat liver mitochondrial membranes. Br. J. Pharmacol. 135 1607–1615. 10.1038/sj.bjp.0704626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klouz A., Tillement J.-P., Boussard M.-F., Wierzbicki M., Berezowski V., Cecchelli R., et al. (2003). [3H]BHDP as a novel and selective ligand for σ1 receptors in liver mitochondria and brain synaptosomes of the rat. FEBS Lett. 553 157–162. 10.1016/S0014-5793(03)01011-1 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Matsuno K., Nakata K., Mita S. (1996). Enhancement of acetylcholine release by SA4503, a novel sigma 1 receptor agonist, in the rat brain. J. Pharmacol. Exp. Ther. 279 106–113. [PubMed] [Google Scholar]
- Koe B. K., Burkhart C. A., Lebel L. A. (1989). (+)-[3H]3-(3-hydroxyphenyl)-N-(1-propyl)-piperidine binding to sigma receptors in mouse brain in vivo. Eur. J. Pharmacol. 161 263–266. 10.1016/0014-2999(89)90857-1 [DOI] [PubMed] [Google Scholar]
- Kourrich S., Hayashi T., Chuang J.-Y., Tsai S.-Y., Su T.-P., Bonci A. (2013). Dynamic interaction between sigma-1 receptor and Kv1.2 shapes neuronal and behavioral responses to cocaine. Cell 152 236–247. 10.1016/j.cell.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryzhanovskii S. A., Kozhevnikova L. M., Tsorin I. B., Sukhanova I. F., Ionova E. O., Stolyaruk V. N., et al. (2018). On the mechanism of the cardioprotective action of sigma1 receptor agonist anxiolytic fabomotizole hydrochloride (Afobazole). Bull. Exp. Biol. Med. 165 660–664. 10.1007/s10517-018-4236-1 [DOI] [PubMed] [Google Scholar]
- Kryzhanovskii S. A., Tsorin I. B., Stolyaruk V. N., Ionova E. O., Vititnova M. B. (2017). Delayed results of experimental afobazole therapy in rats after acute myocardial infarction. Bull. Exp. Biol. Med. 163 180–183. 10.1007/s10517-017-3761-7 [DOI] [PubMed] [Google Scholar]
- Kubickova J., Lencesova L., Csaderova L., Stracina T., Hudecova S., Babula P., et al. (2018). Haloperidol affects plasticity of differentiated NG-108 cells through σ1R/IP3R1 Complex. Cell Mol. Neurobiol. 38 181–194. 10.1007/s10571-017-0524-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit-Le Bouteiller C., Jamme M. F., David M., Silve S., Lanau C., Dhers C., et al. (1998). Antiproliferative effects of SR31747A in animal cell lines are mediated by inhibition of cholesterol biosynthesis at the sterol isomerase step. Eur. J. Biochem. 256 342–349. 10.1046/j.1432-1327.1998.2560342.x [DOI] [PubMed] [Google Scholar]
- Lahmy V., Long R., Morin D., Villard V., Maurice T. (2015). Mitochondrial protection by the mixed muscarinic/σ1 ligand ANAVEX2-73, a tetrahydrofuran derivative, in Aβ25–35 peptide-injected mice, a nontransgenic Alzheimer’s disease model. Front. Cell Neurosci. 8:463. 10.3389/fncel.2014.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langa F., Codony X., Tovar V., Lavado A., Gimenez E., Cozar P., et al. (2003). Generation and phenotypic analysis of sigma receptor type I (sigma 1) knockout mice. Eur. J. Neurosci. 18 2188–2196. [DOI] [PubMed] [Google Scholar]
- Laurini E., Col V. D., Mamolo M. G., Zampieri D., Posocco P., Fermeglia M., et al. (2011). Homology model and docking-based virtual screening for ligands of the σ1 receptor. ACS Med. Chem. Lett. 2 834–839. 10.1021/ml2001505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J. Y., van Karnebeek C. D. M., Drogemoller B., Shyr C., Tarailo-Graovac M., Eydoux P., et al. (2016). Further Validation of the SIGMAR1 c.151+1G>T mutation as cause of distal hereditary motor neuropathy. Child Neurol. Open. 3:2329048X16669912. 10.1177/2329048X16669912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenze E. J., Mattar C., Zorumski C. F., Stevens A., Schweiger J., Nicol G. E., et al. (2020). Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID-19: a randomized clinical trial. JAMA 324 2292–2300. 10.1001/jama.2020.22760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hu Z., Liu L., Xie Y., Zhan Y., Zi X., et al. (2015). A SIGMAR1 splice-site mutation causes distal hereditary motor neuropathy. Neurology 84 2430–2437. 10.1212/wnl.0000000000001680 [DOI] [PubMed] [Google Scholar]
- Liu D.-Y., Chi T.-Y., Ji X.-F., Liu P., Qi X.-X., Zhu L., et al. (2018a). Sigma-1 receptor activation alleviates blood-brain barrier dysfunction in vascular dementia mice. Exp. Neurol. 308 90–99. 10.1016/j.expneurol.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Liu X., Fu Y., Yang H., Mavlyutov T., Li J., McCurdy C. R., et al. (2017). Potential independent action of sigma receptor ligands through inhibition of the Kv2.1 channel. Oncotarget 8 59345–59358. 10.18632/oncotarget.19581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Qu C., Shi S., Ye T., Wang L., Liu S., et al. (2019). The reversal effect of sigma-1 receptor (S1R) Agonist, SA4503, on atrial fibrillation after depression and its underlying mechanism. Front. Physiol. 10:1346. 10.3389/fphys.2019.01346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Qu C., Yang H., Shi S., Zhang C., Zhang Y., et al. (2018b). Chronic stimulation of the sigma-1 receptor ameliorates autonomic nerve dysfunction and atrial fibrillation susceptibility in a rat model of depression. Am. J. Physiol. Heart Circ. Physiol. 315 H1521–H1531. 10.1152/ajpheart.00607.2017 [DOI] [PubMed] [Google Scholar]
- Loeffler J. P., Picchiarelli G., Dupuis L., Gonzalez De Aguilar J. L. (2016). The role of skeletal muscle in amyotrophic lateral sclerosis. Brain Pathol. 26 227–236. 10.1111/bpa.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luty A. A., Kwok J. B., Dobson-Stone C., Loy C. T., Coupland K. G., Karlstrom H., et al. (2010). Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann. Neurol. 68 639–649. 10.1002/ana.22274 [DOI] [PubMed] [Google Scholar]
- Ma M. T., Chen D.-H., Raskind W. H., Bird T. D. (2020). Mutations in the SIGMAR1 gene cause a distal hereditary motor neuropathy phenotype mimicking ALS: report of two novel variants. Neuromuscul. Disord 30 572–575. 10.1016/j.nmd.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T. D., Mannack L. V., Lees R. M., Lane J. D. (2015). Targeted siRNA screens identify ER-to-mitochondrial calcium exchange in autophagy and mitophagy responses in RPE1 Cells. Int. J. Mol. Sci. 16 13356–13380. 10.3390/ijms160613356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C. M., Thomas J. D., Haas D. A., Longen C. G., Oyer H. M., Tong J. Y., et al. (2018). Small-molecule sigma1 modulator induces autophagic degradation of PD-L1. Mol. Cancer Res. 16 243–255. 10.1158/1541-7786.Mcr-17-0166 [DOI] [PubMed] [Google Scholar]
- Marrazzo A., Cobos E. J., Parenti C., Aricò G., Marrazzo G., Ronsisvalle S., et al. (2011). Novel potent and selective σ ligands: evaluation of their agonist and antagonist properties. J. Med. Chem. 54 3669–3673. 10.1021/jm200144j [DOI] [PubMed] [Google Scholar]
- Marriott K.-S. C., Prasad M., Thapliyal V., Bose H. S. (2012). σ-1 Receptor at the mitochondrial-associated endoplasmic reticulum membrane is responsible for mitochondrial metabolic regulation. J. Pharmacol. Exp. Ther. 343 578–586. 10.1124/jpet.112.198168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. R., Eades C. G., Thompson J. A., Huppler R. E., Gilbert P. E. (1976). The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J. Pharmacol. Exp. Ther. 197 517–532. [PubMed] [Google Scholar]
- Matsumoto R. R., Bowen W. D., Tom M. A., Vo V. N., Truong D. D., De Costa B. R. (1995). Characterization of two novel σ receptor ligands: antidystonic effects in rats suggest σ receptor antagonism. Eur. J. Pharmacol. 280 301–310. 10.1016/0014-2999(95)00208-3 [DOI] [PubMed] [Google Scholar]
- Matsumoto R. R., Gilmore D. L., Pouw B., Bowen W. D., Williams W., Kausar A., et al. (2004). Novel analogs of the sigma receptor ligand BD1008 attenuate cocaine-induced toxicity in mice. Eur. J. Pharmacol. 492 21–26. 10.1016/j.ejphar.2004.03.037 [DOI] [PubMed] [Google Scholar]
- Matsumoto R. R., McCracken K. A., Pouw B., Zhang Y., Bowen W. D. (2002). Involvement of sigma receptors in the behavioral effects of cocaine: evidence from novel ligands and antisense oligodeoxynucleotides. Neuropharmacology 42 1043–1055. 10.1016/S0028-3908(02)00056-4 [DOI] [PubMed] [Google Scholar]
- Matsumoto R. R., Nguyen L., Kaushal N., Robson M. J. (2014). Sigma (sigma) receptors as potential therapeutic targets to mitigate psychostimulant effects. Adv. Pharmacol. 69 323–386. 10.1016/B978-0-12-420118-7.00009-3 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Nakazawa M., Okamoto K., Kawashima Y., Mita S. (1996). Binding properties of SA4503, a novel and selective sigma 1 receptor agonist. Eur. J. Pharmacol. 306 271–279. 10.1016/0014-2999(96)00201-4 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Senda T., Matsunaga K., Mita S. (1994). Ameliorating effects of sigma receptor ligands on the impairment of passive avoidance tasks in mice: involvement in the central acetylcholinergic system. Eur. J. Pharmacol. 261 43–51. 10.1016/0014-2999(94)90298-4 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Senda T., Matsunaga K., Mita S., Kaneto H. (1993). Similar ameliorating effects of benzomorphans and 5-HT2 antagonists on drug-induced impairment of passive avoidance response in mice: comparison with acetylcholinesterase inhibitors. Psychopharmacology (Berl) 112 134–141. 10.1007/BF02247374 [DOI] [PubMed] [Google Scholar]
- Matsushima Y., Terada K., Kamei C., Sugimoto Y. (2019). Sertraline inhibits nerve growth factor-induced neurite outgrowth in PC12 cells via a mechanism involving the sigma-1 receptor. Eur. J. Pharmacol. 853 129–135. 10.1016/j.ejphar.2019.03.032 [DOI] [PubMed] [Google Scholar]
- Maurice T., Casalino M., Lacroix M., Romieu P. (2003). Involvement of the sigma 1 receptor in the motivational effects of ethanol in mice. Pharmacol. Biochem. Behav. 74 869–876. 10.1016/s0091-3057(03)00002-9 [DOI] [PubMed] [Google Scholar]
- Maurice T., Hiramatsu M., Itoh J., Kameyama T., Hasegawa T., Nabeshima T. (1994a). Behavioral evidence for a modulating role of sigma ligands in memory processes. I. Attenuation of dizocilpine (MK-801)-induced amnesia. Brain Res. 647 44–56. 10.1016/0006-8993(94)91397-8 [DOI] [PubMed] [Google Scholar]
- Maurice T., Martin-Fardon R., Romieu P., Matsumoto R. R. (2002). Sigma1 (σ1) receptor antagonists represent a new strategy against cocaine addiction and toxicity. Neurosci. Biobehav. Rev. 26 499–527. 10.1016/S0149-7634(02)00017-9 [DOI] [PubMed] [Google Scholar]
- Maurice T., Phan V. L., Privat A. (2001). The anti-amnesic effects of sigma1 (sigma1) receptor agonists confirmed by in vivo antisense strategy in the mouse. Brain Res. 898 113–121. 10.1016/s0006-8993(01)02152-7 [DOI] [PubMed] [Google Scholar]
- Maurice T., Phan V. L., Noda Y., Yamada K., Privat A., Nabeshima T. (1999). The attenuation of learning impairments induced after exposure to CO or trimethyltin in mice by sigma (sigma) receptor ligands involves both sigma1 and sigma2 sites. Br. J. Pharmacol. 127 335–342. 10.1038/sj.bjp.0702553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T., Roman F. J., Su T. P., Privat A. (1996). Beneficial effects of sigma agonists on the age-related learning impairment in the senescence-accelerated mouse (SAM). Brain Res. 733 219–230. 10.1016/0006-8993(96)00565-3 [DOI] [PubMed] [Google Scholar]
- Maurice T., Strehaiano M., Duhr F., Chevallier N. (2018). Amyloid toxicity is enhanced after pharmacological or genetic invalidation of the sigma1 receptor. Behav. Brain Res. 339 1–10. 10.1016/j.bbr.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Maurice T., Su T. P., Privat A. (1998). Sigma1 (sigma 1) receptor agonists and neurosteroids attenuate B25-35-amyloid peptide-induced amnesia in mice through a common mechanism. Neuroscience 83 413–428. 10.1016/s0306-4522(97)00405-3 [DOI] [PubMed] [Google Scholar]
- Maurice T., Su T. P., Parish D. W., Privat A. (1995). Prevention of nimodipine-induced impairment of learning by the selective sigma ligand PRE-084. J. Neural. Transm. Gen. Sect. 102 1–18. 10.1007/BF01276561 [DOI] [PubMed] [Google Scholar]
- Maurice T., Su T.-P., Parish D. W., Nabeshima T., Privat A. (1994b). PRE-084, a σ selective PCP derivative, attenuates MK-801-induced impairment of learning in mice. Pharmacol. Biochem. Behav. 49 859–869. 10.1016/0091-3057(94)90235-6 [DOI] [PubMed] [Google Scholar]
- Maurice T., Volle J. N., Strehaiano M., Crouzier L., Pereira C., Kaloyanov N., et al. (2019). Neuroprotection in non-transgenic and transgenic mouse models of Alzheimer’s disease by positive modulation of sigma1 receptors. Pharmacol. Res. 144 315–330. 10.1016/j.phrs.2019.04.026 [DOI] [PubMed] [Google Scholar]
- Mavlyutov T. A., Guo L. W. (2017). Peeking into sigma-1 receptor functions through the retina. Adv. Exp. Med. Biol. 964 285–297. 10.1007/978-3-319-50174-1_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Ruoho A. E. (2007). Ligand-dependent localization and intracellular stability of sigma-1 receptors in CHO-K1 cells. J. Mol. Signal. 2:8. 10.1186/1750-2187-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Baker E. M., Losenegger T. M., Kim J. R., Torres B., Epstein M. L., et al. (2017a). The sigma-1 receptor-a therapeutic target for the treatment of ALS? Adv. Exp. Med. Biol. 964 255–265. 10.1007/978-3-319-50174-1_17 [DOI] [PubMed] [Google Scholar]
- Mavlyutov T. A., Duellman T., Kim H. T., Epstein M. L., Leese C., Davletov B. A., et al. (2016). Sigma-1 receptor expression in the dorsal root ganglion: reexamination using a highly specific antibody. Neuroscience 331 148–157. 10.1016/j.neuroscience.2016.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Epstein M., Guo L. W. (2015a). Subcellular localization of the sigma-1 receptor in retinal neurons - an electron microscopy study. Sci. Rep. 5:10689. 10.1038/srep10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Epstein M. L., Andersen K. A., Ziskind-Conhaim L., Ruoho A. E. (2010). The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience 167 247–255. 10.1016/j.neuroscience.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Epstein M. L., Liu P., Verbny Y. I., Ziskind-Conhaim L., Ruoho A. E. (2012). Development of the sigma-1 receptor in C-terminals of motoneurons and colocalization with the N, N’-dimethyltryptamine forming enzyme, indole-N-methyl transferase. Neuroscience 206 60–68. 10.1016/j.neuroscience.2011.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Epstein M. L., Verbny Y. I., Huerta M. S., Zaitoun I., Ziskind-Conhaim L., et al. (2013). Lack of sigma-1 receptor exacerbates ALS progression in mice. Neuroscience 240 129–134. 10.1016/j.neuroscience.2013.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Guo L. W., Epstein M. L., Ruoho A. E. (2015b). Role of the Sigma-1 receptor in Amyotrophic Lateral Sclerosis (ALS). J. Pharmacol. Sci. 127 10–16. 10.1016/j.jphs.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Nickells R. W., Guo L. W. (2011). Accelerated retinal ganglion cell death in mice deficient in the Sigma-1 receptor. Mol. Vis. 17 1034–1043. [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Yang H., Epstein M. L., Ruoho A. E., Yang J., Guo L. W. (2017). APEX2-enhanced electron microscopy distinguishes sigma-1 receptor localization in the nucleoplasmic reticulum. Oncotarget 8 51317–51330. 10.18632/oncotarget.17906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavylutov T., Chen X., Guo L., Yang J. (2018). APEX2- tagging of Sigma 1-receptor indicates subcellular protein topology with cytosolic N-terminus and ER luminal C-terminus. Protein Cell. 9 733–737. 10.1007/s13238-017-0468-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann D. J., Su T. P. (1991). Solubilization and characterization of haloperidol-sensitive (+)-[3H]SKF-10,047 binding sites (sigma sites) from rat liver membranes. J. Pharmacol. Exp. Ther. 257 547–554. [PubMed] [Google Scholar]
- McFadden L. M., Vieira-Brock P. L., Hanson G. R., Fleckenstein A. E. (2015). Prior methamphetamine self-administration attenuates the dopaminergic deficits caused by a subsequent methamphetamine exposure. Neuropharmacology 93 146–154. 10.1016/j.neuropharm.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J., Pasternak G. W. (2001). Molecular cloning and pharmacological characterization of the rat sigma1 receptor. Biochem. Pharmacol. 62 349–355. 10.1016/s0006-2952(01)00666-9 [DOI] [PubMed] [Google Scholar]
- Meunier J., Demeilliers B., Célérier A., Maurice T. (2006a). Compensatory effect by sigma1 (σ1) receptor stimulation during alcohol withdrawal in mice performing an object recognition task. Behav. Brain Res. 166 166–176. 10.1016/j.bbr.2005.07.019 [DOI] [PubMed] [Google Scholar]
- Meunier J., Ieni J., Maurice T. (2006b). The anti-amnesic and neuroprotective effects of donepezil against amyloid beta25-35 peptide-induced toxicity in mice involve an interaction with the sigma1 receptor. Br. J. Pharmacol. 149 998–1012. 10.1038/sj.bjp.0706927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Mori F., Kon T., Tanji K., Toyoshima Y., Yoshida M., et al. (2014). Accumulation of the sigma-1 receptor is common to neuronal nuclear inclusions in various neurodegenerative diseases. Neuropathology 34 148–158. 10.1111/neup.12080 [DOI] [PubMed] [Google Scholar]
- Miki Y., Tanji K., Mori F., Wakabayashi K. (2015). Sigma-1 receptor is involved in degradation of intranuclear inclusions in a cellular model of Huntington’s disease. Neurobiol. Dis. 74 25–31. 10.1016/j.nbd.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Milardovic I., Vitlov Uljevic M., Vukojevic K., Kostic S., Filipovic N. (2020). Renal expression of sigma 1 receptors in diabetic rats. Acta Histochem. 122:151580. 10.1016/j.acthis.2020.151580 [DOI] [PubMed] [Google Scholar]
- Mir S. U. R., Ahmed I. S. A., Arnold S., Craven R. J. (2012). Elevated progesterone receptor membrane component 1/sigma-2 receptor levels in lung tumors and plasma from lung cancer patients. Int. J. Cancer 131 E1–E9. 10.1002/ijc.26432 [DOI] [PubMed] [Google Scholar]
- Mishina M., Ishiwata K., Ishii K., Kitamura S., Kimura Y., Kawamura K., et al. (2005). Function of sigma1 receptors in Parkinson’s disease. Acta Neurol. Scand. 112 103–107. 10.1111/j.1600-0404.2005.00432.x [DOI] [PubMed] [Google Scholar]
- Mishina M., Ohyama M., Ishii K., Kitamura S., Kimura Y., Oda K., et al. (2008). Low density of sigma1 receptors in early Alzheimer’s disease. Ann. Nucl. Med. 22 151–156. 10.1007/s12149-007-0094-z [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Mavlyutov T., Singh D. R., Biener G., Yang J., Oliver J. A., et al. (2015). The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands. Biochem. J. 466 263–271. 10.1042/BJ20141321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda T., Omi T., Tanimukai H., Sakagami Y., Tagami S., Okochi M., et al. (2011). Sigma-1Rs are upregulated via PERK/eIF2alpha/ATF4 pathway and execute protective function in ER stress. Biochem. Biophys. Res. Commun. 415 519–525. 10.1016/j.bbrc.2011.10.113 [DOI] [PubMed] [Google Scholar]
- Monassier L., Manoury B., Bellocq C., Weissenburger J., Greney H., Zimmermann D., et al. (2007). sigma(2)-receptor ligand-mediated inhibition of inwardly rectifying K(+) channels in the heart. J. Pharmacol. Exp. Ther. 322 341–350. 10.1124/jpet.107.122044 [DOI] [PubMed] [Google Scholar]
- Monnet F. P., Mahé V., Robel P., Baulieu E. E. (1995). Neurosteroids, via sigma receptors, modulate the [3H]norepinephrine release evoked by N-methyl-D-aspartate in the rat hippocampus. Proc. Natl. Acad. Sci. U.S.A. 92 3774–3778. 10.1073/pnas.92.9.3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Hayashi T., Hayashi E., Su T. P. (2013). Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8:e76941. 10.1371/journal.pone.0076941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer P. S., Rockson S. G. (2014). New developments in clinical aspects of lymphatic disease. J. Clin. Invest 124 915–921. 10.1172/JCI71608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motawe Z. Y., Farsaei F., Abdelmaboud S. S., Cuevas J., Breslin J. W. (2020). Sigma-1 receptor activation-induced glycolytic ATP production and endothelial barrier enhancement. Microcirculation 27:e12620. 10.1111/micc.12620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naia L., Ly P., Mota S. I., Lopes C., Maranga C., Coelho P., et al. (2021). The Sigma-1 receptor mediates pridopidine rescue of mitochondrial function in huntington disease models. Neurotherapeutics 10.1007/s13311-021-01022-9 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandhagopal R., Meftah D., Al-Kalbani S., Scott P. (2018). Recessive distal motor neuropathy with pyramidal signs in an Omani kindred: underlying novel mutation in the SIGMAR1 gene. Eur. J. Neurol. 25 395–403. 10.1111/ene.13519 [DOI] [PubMed] [Google Scholar]
- Narita N., Hashimoto K., Tomitaka S.-I., Minabe Y. (1996). Interactions of selective serotonin reuptake inhibitors with subtypes of σ receptors in rat brain. Eur. J. Pharmacol. 307 117–119. 10.1016/0014-2999(96)00254-3 [DOI] [PubMed] [Google Scholar]
- Natsvlishvili N., Goguadze N., Zhuravliova E., Mikeladze D. (2015). Sigma-1 receptor directly interacts with Rac1-GTPase in the brain mitochondria. BMC Biochem. 16:11. 10.1186/s12858-015-0040-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G., Moreno E., Aymerich M., Marcellino D., McCormick P. J., Mallol J., et al. (2010). Direct involvement of σ-1 receptors in the dopamine D1 receptor-mediated effects of cocaine. Proc. Natl. Acad. Sci. U.S.A. 107 18676–18681. 10.1073/pnas.1008911107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G., Moreno E., Bonaventura J., Brugarolas M., Farré D., Aguinaga D., et al. (2013). Cocaine inhibits dopamine D2 receptor signaling via sigma-1-D2 receptor heteromers. PLoS One 8:e61245. 10.1371/journal.pone.0061245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen E. C., McCracken K. A., Liu Y., Pouw B., Matsumoto R. R. (2005). Involvement of sigma (σ) receptors in the acute actions of methamphetamine: receptor binding and behavioral studies. Neuropharmacology 49 638–645. 10.1016/j.neuropharm.2005.04.016 [DOI] [PubMed] [Google Scholar]
- Niitsu T., Iyo M., Hashimoto K. (2012). Sigma-1 receptor agonists as therapeutic drugs for cognitive impairment in neuropsychiatric diseases. Curr. Pharm. Des. 18 875–883. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Ishima T., Iyo M., Hashimoto K. (2008). Potentiation of nerve growth factor-induced neurite outgrowth by fluvoxamine: role of sigma-1 receptors, IP3 receptors and cellular signaling pathways. PLoS One 3:e2558. 10.1371/journal.pone.0002558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova M., Ela C., Barg J., Vogel Z., Hasin Y., Eilam Y. (1995). Inotropic action of sigma receptor ligands in isolated cardiac myocytes from adult rats. Eur. J. Pharmacol. 286 19–30. [DOI] [PubMed] [Google Scholar]
- O’Connor C. M., Jiang W., Kuchibhatla M., Silva S. G., Cuffe M. S., Callwood D. D., et al. (2010). Safety and efficacy of sertraline for depression in patients with heart failure: results of the SADHART-CHF (Sertraline Against Depression and Heart Disease in Chronic Heart Failure) trial. J. Am. Coll. Cardiol. 56 692–699. 10.1016/j.jacc.2010.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama S., Imagawa Y., Ogawa S.-I., Araki H., Ajima A., Tanaka M., et al. (1993). NE-100, a novel sigma receptor ligand: In vivo tests. Life Sci. 53 L285–L290. 10.1016/0024-3205(93)90588-T [DOI] [PubMed] [Google Scholar]
- Ola M. S., Moore P., El-Sherbeny A., Roon P., Agarwal N., Sarthy V. P., et al. (2001). Expression pattern of sigma receptor 1 mRNA and protein in mammalian retina. Brain Res. Mol. Brain Res. 95 86–95. 10.1016/s0169-328x(01)00249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ola M. S., Moore P., Maddox D., El-Sherbeny A., Huang W., Roon P., et al. (2002). Analysis of sigma receptor (sigmaR1) expression in retinal ganglion cells cultured under hyperglycemic conditions and in diabetic mice. Brain Res. Mol. Brain Res. 107 97–107. 10.1016/s0169-328x(02)00444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri M., Amata E., Vinciguerra S., Fiorito J., Giurdanella G., Drago F., et al. (2016). Antiangiogenic Effect of (±)-haloperidol metabolite II valproate Ester [(±)-MRJF22] in human microvascular retinal endothelial cells. J. Med. Chem. 59 9960–9966. 10.1021/acs.jmedchem.6b01039 [DOI] [PubMed] [Google Scholar]
- Omi T., Tanimukai H., Kanayama D., Sakagami Y., Tagami S., Okochi M., et al. (2014). Fluvoxamine alleviates ER stress via induction of Sigma-1 receptor. Cell Death Dis. 5:e1332. 10.1038/cddis.2014.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori I. M., Watanabe N., Nakagawa A., Cipriani A., Barbui C., McGuire H., et al. (2010). Fluvoxamine versus other anti-depressive agents for depression. Cochrane Database Syst. Rev. 3:CD006114. 10.1002/14651858.CD006114.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Shimazawa M., Ishisaka M., Oyagi A., Tsuruma K., Hara H. (2012). Imipramine protects mouse hippocampus against tunicamycin-induced cell death. Eur. J. Pharmacol. 696 83–88. 10.1016/j.ejphar.2012.09.037 [DOI] [PubMed] [Google Scholar]
- Ono Y., Tanaka H., Tsuruma K., Shimazawa M., Hara H. (2013). A sigma-1 receptor antagonist (NE-100) prevents tunicamycin-induced cell death via GRP78 induction in hippocampal cells. Biochem. Biophys. Res. Commun. 434 904–909. 10.1016/j.bbrc.2013.04.055 [DOI] [PubMed] [Google Scholar]
- Ortega-Roldan J. L., Ossa F., Schnell J. R. (2013). Characterization of the human sigma-1 receptor chaperone domain structure and binding immunoglobulin protein (BiP) interactions. J. Biol. Chem. 288 21448–21457. 10.1074/jbc.M113.450379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabba M., Wong A. Y., Ahlskog N., Hristova E., Biscaro D., Nassrallah W., et al. (2014). NMDA receptors are upregulated and trafficked to the plasma membrane after sigma-1 receptor activation in the rat hippocampus. J. Neurosci. 34 11325–11338. 10.1523/JNEUROSCI.0458-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A., Chu U. B., Ramachandran S., Grawoig D., Guo L.-W., Hajipour A. R., et al. (2008). Juxtaposition of the steroid binding domain-like I and II regions constitutes a ligand binding site in the σ-1 receptor. J. Biol. Chem. 283 19646–19656. 10.1074/jbc.M802192200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Muro A., Vela J. M., Molina-Holgado E., Guitart X., Ovalle S., et al. (2003). Immunohistochemical localization of the σ1-receptor in oligodendrocytes in the rat central nervous system. Brain Res. 961 92–99. 10.1016/S0006-8993(02)03892-1 [DOI] [PubMed] [Google Scholar]
- Palmer C. P., Mahen R., Schnell E., Djamgoz M. B. A., Aydar E. (2007). Sigma-1 receptors bind cholesterol and remodel lipid rafts in breast cancer cell lines. Cancer Res. 67 11166–11175. 10.1158/0008-5472.Can-07-1771 [DOI] [PubMed] [Google Scholar]
- Pan B., Guo Y., Kwok W.-M., Hogan Q., Wu H.-E. (2014). Sigma-1 receptor antagonism restores injury-induced decrease of voltage-gated Ca2+ current in sensory neurons. J. Pharmacol. Exp. Ther. 350 290–300. 10.1124/jpet.114.214320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Pasternak D. A., Xu J., Xu M., Lu Z., Pasternak G. W., et al. (2017). Isolation and characterization of alternatively spliced variants of the mouse sigma1 receptor gene. Sigmar1. PLoS One 12:e0174694. 10.1371/journal.pone.0174694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. M., Purdy R. H. (1992). Neuroactive steroids. FASEB J. 6 2311–2322. [PubMed] [Google Scholar]
- Phan V. L., Urani A., Sandillon F., Privat A., Maurice T. (2003). Preserved sigma1 (sigma1) receptor expression and behavioral efficacy in the aged C57BL/6 mouse. Neurobiol. Aging 24 865–881. 10.1016/s0197-4580(02)00231-2 [DOI] [PubMed] [Google Scholar]
- Prasad P. D., Li H. W., Fei Y.-J., Ganapathy M. E., Fujita T., Plumley L. H., et al. (1998). Exon-intron structure, analysis of promoter region, and chromosomal localization of the human type 1 σ receptor gene. J. Neurochem. 70 443–451. 10.1046/j.1471-4159.1998.70020443.x [DOI] [PubMed] [Google Scholar]
- Prause J., Goswami A., Katona I., Roos A., Schnizler M., Bushuven E., et al. (2013). Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22 1581–1600. 10.1093/hmg/ddt008 [DOI] [PubMed] [Google Scholar]
- Previtali S. C., Zhao E., Lazarevic D., Pipitone G. B., Fabrizi G. M., Manganelli F., et al. (2019). Expanding the spectrum of genes responsible for hereditary motor neuropathies. J. Neurol. Neurosurg. Psychiatry. 90 1171–1179. 10.1136/jnnp-2019-320717 [DOI] [PubMed] [Google Scholar]
- Quadir S. G., Cottone P., Sabino V. (2019). Role of sigma receptors in alcohol addiction. Front. Pharmacol. 10:687. 10.3389/fphar.2019.00687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Lu H., Prabhu U., Ruoho A. E. (2007). Purification and characterization of the guinea pig sigma-1 receptor functionally expressed in Escherichia coli. Protein Expr. Purif. 51 283–292. 10.1016/j.pep.2006.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson M. J., Noorbakhsh B., Seminerio M. J., Matsumoto R. R. (2012). Sigma-1 receptors: potential targets for the treatment of substance abuse. Curr. Pharm. Des. 18 902–919. [DOI] [PubMed] [Google Scholar]
- Romero G., Pérez M. P., Carceller A., Monroy X., Farré A. J., Guitart X. (2000). Changes in phosphoinositide signalling activity and levels of the alpha subunit of Gq/11 protein in rat brain induced by E-5842, a sigma1 receptor ligand and potential atypical antipsychotic. Neurosci. Lett. 290 189–192. 10.1016/S0304-3940(00)01353-7 [DOI] [PubMed] [Google Scholar]
- Romieu P., Phan V.-L., Martin-Fardon R., Maurice T. (2002). Involvement of the sigma1 receptor in cocaine-induced conditioned place preference: possible dependence on dopamine uptake blockade. Neuropsychopharmacology 26 444–455. 10.1016/S0893-133X(01)00391-8 [DOI] [PubMed] [Google Scholar]
- Rosen D. A., Seki S. M., Fernández-Castañeda A., Beiter R. M., Eccles J. D., Woodfolk J. A., et al. (2019). Modulation of the sigma-1 receptor–IRE1 pathway is beneficial in preclinical models of inflammation and sepsis. Sci. Transl. Med. 11:eaau5266. 10.1126/scitranslmed.aau5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscher K., Shamloo M., Rickhag M., Ladunga I., Soriano L., Gisselsson L., et al. (2011). The sigma-1 receptor enhances brain plasticity and functional recovery after experimental stroke. Brain 134(Pt 3) 732–746. 10.1093/brain/awq367 [DOI] [PubMed] [Google Scholar]
- Ryskamp D. A., Korban S., Zhemkov V., Kraskovskaya N., Bezprozvanny I. (2019). Neuronal sigma-1 receptors: signaling functions and protective roles in neurodegenerative diseases. Front. Neurosci. 13:862. 10.3389/fnins.2019.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino V., Cottone P., Parylak S. L., Steardo L., Zorrilla E. P. (2009a). Sigma-1 receptor knockout mice display a depressive-like phenotype. Behav. Brain Res. 198 472–476. 10.1016/j.bbr.2008.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino V., Cottone P., Zhao Y., Iyer M. R., Steardo L., Steardo L., et al. (2009b). The σ-receptor antagonist BD-1063 decreases ethanol intake and reinforcement in animal models of excessive drinking. Neuropsychopharmacology 34 1482–1493. 10.1038/npp.2008.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambo D. O., Lebowitz J. J., Khoshbouei H. (2018). The sigma-1 receptor as a regulator of dopamine neurotransmission: a potential therapeutic target for methamphetamine addiction. Pharmacol. Ther. 186 152–167. 10.1016/j.pharmthera.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambo D. O., Lin M., Owens A., Lebowitz J. J., Richardson B., Jagnarine D. A., et al. (2017). The sigma-1 receptor modulates methamphetamine dysregulation of dopamine neurotransmission. Nat. Comm. 8:2228. 10.1038/s41467-017-02087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F., Humbert S. (2016). The biology of huntingtin. Neuron 89 910–926. 10.1016/j.neuron.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Schmidt H. R., Zheng S., Gurpinar E., Koehl A., Manglik A., Kruse A. C. (2016). Crystal structure of the human σ1 receptor. Nature 532 527–530. 10.1038/nature17391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrock J. M., Spino C. M., Longen C. G., Stabler S. M., Marino J. C., Pasternak G. W., et al. (2013). Sequential cytoprotective responses to sigma1 ligand–induced endoplasmic reticulum stress. Mol. Pharmacol. 84 751–762. 10.1124/mol.113.087809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze M. P., Peterson P. A., Jackson M. R. (1994). An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J 13 1696–1705. 10.1002/j.1460-2075.1994.tb06434.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda T., Matsuno K., Kobayashi T., Mita S. (1997). Reduction of the scopolamine-induced impairment of passive-avoidance performance by sigma receptor agonist in mice. Physiol. Behav. 61 257–264. 10.1016/s0031-9384(96)00447-7 [DOI] [PubMed] [Google Scholar]
- Senda T., Mita S., Kaneda K., Kikuchi M., Akaike A. (1998). Effect of SA4503, a novel sigma1 receptor agonist, against glutamate neurotoxicity in cultured rat retinal neurons. Eur. J. Pharmacol. 342 105–111. 10.1016/s0014-2999(97)01450-7 [DOI] [PubMed] [Google Scholar]
- Serebruany V. L., Glassman A. H., Malinin A. I., Nemeroff C. B., Musselman D. L., van Zyl L. T., et al. (2003). Platelet/endothelial biomarkers in depressed patients treated with the selective serotonin reuptake inhibitor sertraline after acute coronary events: the Sertraline AntiDepressant Heart Attack Randomized Trial (SADHART) Platelet Substudy. Circulation 108 939–944. 10.1161/01.CIR.0000085163.21752.0A [DOI] [PubMed] [Google Scholar]
- Serebruany V. L., Suckow R. F., Cooper T. B., O’Connor C. M., Malinin A. I., Krishnan K. R., et al. (2005). Relationship between release of platelet/endothelial biomarkers and plasma levels of sertraline and N-desmethylsertraline in acute coronary syndrome patients receiving SSRI treatment for depression. Am. J. Psychiatry 162 1165–1170. 10.1176/appi.ajp.162.6.1165 [DOI] [PubMed] [Google Scholar]
- Seredenin S. B., Voronin M. V. (2009). [Neuroreceptor mechanisms of the afobazole effect]. Eksp Klin. Farmakol. 72 3–11. [PubMed] [Google Scholar]
- Seth P., Fei Y.-J., Li H. W., Huang W., Leibach F. H., Ganapathy V. (1998). Cloning and functional characterization of a σ receptor from rat brain. J. Neurochem. 70 922–931. 10.1046/j.1471-4159.1998.70030922.x [DOI] [PubMed] [Google Scholar]
- Seth P., Ganapathy M. E., Conway S. J., Bridges C. D., Smith S. B., Casellas P., et al. (2001). Expression pattern of the type 1 sigma receptor in the brain and identity of critical anionic amino acid residues in the ligand-binding domain of the receptor. Biochim. Biophys. Acta 1540 59–67. 10.1016/S0167-4889(01)00117-3 [DOI] [PubMed] [Google Scholar]
- Seth P., Leibach F. H., Ganapathy V. (1997). Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem. Biophys. Res. Commun. 241 535–540. 10.1006/bbrc.1997.7840 [DOI] [PubMed] [Google Scholar]
- Sha S., Hong J., Qu W. J., Lu Z. H., Li L., Yu W. F., et al. (2015). Sex-related neurogenesis decrease in hippocampal dentate gyrus with depressive-like behaviors in sigma-1 receptor knockout mice. Eur. Neuropsychopharmacol. 25 1275–1286. 10.1016/j.euroneuro.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Sharkey J., Glen K. A., Wolfe S., Kuhar M. J. (1988). Cocaine binding at sigma receptors. Eur. J. Pharmacol. 149 171–174. [DOI] [PubMed] [Google Scholar]
- Shenkman M., Geva M., Gershoni-Emek N., Hayden M. R., Lederkremer G. Z. (2021). Pridopidine reduces mutant huntingtin-induced endoplasmic reticulum stress by modulation of the Sigma-1 receptor. J. Neurochem. 10.1111/jnc.15366 [DOI] [PubMed] [Google Scholar]
- Shimazawa M., Sugitani S., Inoue Y., Tsuruma K., Hara H. (2015). Effect of a sigma-1 receptor agonist, cutamesine dihydrochloride (SA4503), on photoreceptor cell death against light-induced damage. Exp. Eye Res. 132 64–72. 10.1016/j.exer.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Shinoda Y., Tagashira H., Bhuiyan M. S., Hasegawa H., Kanai H., Fukunaga K. (2016). Haloperidol aggravates transverse aortic constriction-induced heart failure via mitochondrial dysfunction. J. Pharmacol. Sci. 131 172–183. 10.1016/j.jphs.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Shioda N., Ishikawa K., Tagashira H., Ishizuka T., Yawo H., Fukunaga K. (2012). Expression of a truncated form of the endoplasmic reticulum chaperone protein, sigma1 receptor, promotes mitochondrial energy depletion and apoptosis. J. Biol. Chem. 287 23318–23331. 10.1074/jbc.M112.349142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama Y., Nishikawa T., Umino A., Takahashi K. (1993). p-chlorophenylalanine-reversible reduction of sigma binding sites by chronic imipramine treatment in rat brain. Eur. J. Pharmacol. 237 117–126. [DOI] [PubMed] [Google Scholar]
- Siciliano C. A., Calipari E. S., Ferris M. J., Jones S. R. (2014). Biphasic mechanisms of amphetamine action at the dopamine terminal. J. Neurosci. 34 5575–5582. 10.1523/jneurosci.4050-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simony-Lafontaine J., Esslimani M., Bribes E., Gourgou S., Lequeux N., Lavail R., et al. (2000). Immunocytochemical assessment of sigma-1 receptor and human sterol isomerase in breast cancer and their relationship with a series of prognostic factors. Br. J. Cancer 82 1958–1966. 10.1054/bjoc.2000.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzycki M., Czeczot H. (2013). Altered expression level of Sigma1 receptor gene in human colorectal cancer. J. Recept Signal. Transduct. Res. 33 313–318. 10.3109/10799893.2013.822891 [DOI] [PubMed] [Google Scholar]
- Smith S. B., Duplantier J., Dun Y., Mysona B., Roon P., Martin P. M., et al. (2008). In vivo protection against retinal neurodegeneration by sigma receptor 1 ligand (+)-pentazocine. Invest Ophthalmol. Vis. Sci. 49 4154–4161. 10.1167/iovs.08-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. B., Wang J., Cui X., Mysona B. A., Zhao J., Bollinger K. E. (2018). Sigma 1 receptor: a novel therapeutic target in retinal disease. Prog. Retin Eye Res. 67 130–149. 10.1016/j.preteyeres.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani O., Foll F. L., Roman F., Monnet F. P., Vaudry H., Cazin L. (1999a). A-Current down-modulated by sigma receptor in frog pituitary melanotrope cells through a G protein-dependent pathway. J. Pharmacol. Exp. Ther. 289 321–328. [PubMed] [Google Scholar]
- Soriani O., Le Foll F., Galas L., Roman F., Vaudry H., Cazin L. (1999b). The sigma-ligand (+)-pentazocine depresses M current and enhances calcium conductances in frog melanotrophs. Am. J. Physiol. 277 E73–E80. 10.1152/ajpendo.1999.277.1.E73 [DOI] [PubMed] [Google Scholar]
- Srivats S., Balasuriya D., Pasche M., Vistal G., Edwardson J. M., Taylor C. W., et al. (2016). Sigma1 receptors inhibit store-operated Ca2+ entry by attenuating coupling of STIM1 to Orai1. J. Cell Biol. 213 65–79. 10.1083/jcb.201506022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracina T., Slaninova I., Polanska H., Axmanova M., Olejnickova V., Konecny P., et al. (2015). Long-term haloperidol treatment prolongs QT interval and increases expression of sigma 1 and IP3 receptors in guinea pig hearts. Tohoku. J. Exp. Med. 236 199–207. 10.1620/tjem.236.199 [DOI] [PubMed] [Google Scholar]
- Su T. P. (1982). Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J. Pharmacol. Exp. Ther. 223 284–290. [PubMed] [Google Scholar]
- Su T. P., London E. D., Jaffe J. H. (1988). Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science 240 219–221. 10.1126/science.2832949 [DOI] [PubMed] [Google Scholar]
- Su T. P., Wu X. Z., Cone E. J., Shukla K., Gund T. M., Dodge A. L., et al. (1991). Sigma compounds derived from phencyclidine: identification of PRE-084, a new, selective sigma ligand. J. Pharmacol. Exp. Ther. 259 543–550. [PubMed] [Google Scholar]
- Su T. P., Hayashi T., Vaupel D. B. (2009). When the endogenous hallucinogenic trace amine N, N-dimethyltryptamine meets the sigma-1 receptor. Sci. Signal. 2:e12. 10.1126/scisignal.261pe12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. P., Hayashi T., Maurice T., Buch S., Ruoho A. E. (2010). The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 31 557–566. 10.1016/j.tips.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Sukumaran P., Singh B. B. (2020). Sigma1 receptor inhibits TRPC1-mediated Ca2+ entry that promotes dopaminergic cell death. Cell Mol. Neurobiol. 10.1007/s10571-020-00892-5 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson J. R., O’Connor C. M., Barton D., Van Zyl L. T., Swedberg K., Forman L. M., et al. (2003). Influence of depression and effect of treatment with sertraline on quality of life after hospitalization for acute coronary syndrome. Am. J. Cardiol. 92 1271–1276. [DOI] [PubMed] [Google Scholar]
- Tagashira H., Kobori T. (2013). [Development of experimental techniques and evaluation to support evidence-based medicine (EBM)]. Yakugaku Zasshi. 133:485. 10.1248/yakushi.12-00278-f [DOI] [PubMed] [Google Scholar]
- Tagashira H., Bhuiyan M. S., Fukunaga K. (2013a). Diverse regulation of IP3 and ryanodine receptors by pentazocine through σ1-receptor in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 305 H1201–H1212. 10.1152/ajpheart.00300.2013 [DOI] [PubMed] [Google Scholar]
- Tagashira H., Bhuiyan M. S., Shioda N., Fukunaga K. (2014a). Fluvoxamine rescues mitochondrial Ca2+ transport and ATP production through σ1-receptor in hypertrophic cardiomyocytes. Life Sci. 95 89–100. 10.1016/j.lfs.2013.12.019 [DOI] [PubMed] [Google Scholar]
- Tagashira H., Bhuiyan S., Shioda N., Fukunaga K. (2011). Distinct cardioprotective effects of 17beta-estradiol and dehydroepiandrosterone on pressure overload-induced hypertrophy in ovariectomized female rats. Menopause 18 1317–1326. 10.1097/gme.0b013e31821f915b [DOI] [PubMed] [Google Scholar]
- Tagashira H., Bhuiyan S., Shioda N., Hasegawa H., Kanai H., Fukunaga K. (2010). Sigma1-receptor stimulation with fluvoxamine ameliorates transverse aortic constriction-induced myocardial hypertrophy and dysfunction in mice. Am. J. Physiol. Heart Circ. Physiol. 299 H1535–H1545. 10.1152/ajpheart.00198.2010 [DOI] [PubMed] [Google Scholar]
- Tagashira H., Matsumoto T., Taguchi K., Zhang C., Han F., Ishida K., et al. (2013b). Vascular endothelial sigma1-receptor stimulation with SA4503 rescues aortic relaxation via Akt/eNOS signaling in ovariectomized rats with aortic banding. Circ. J. 77 2831–2840. [DOI] [PubMed] [Google Scholar]
- Tagashira H., Shinoda Y., Shioda N., Fukunaga K. (2014b). Methyl pyruvate rescues mitochondrial damage caused by SIGMAR1 mutation related to amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1840 3320–3334. 10.1016/j.bbagen.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Tagashira H., Zhang C., Lu Y.-M., Hasegawa H., Kanai H., Han F., et al. (2013c). Stimulation of σ1-receptor restores abnormal mitochondrial Ca2+ mobilization and ATP production following cardiac hypertrophy. Biochim. Biophys. Acta 1830 3082–3094. 10.1016/j.bbagen.2012.12.029 [DOI] [PubMed] [Google Scholar]
- Takebayashi M., Hayashi T., Su T. P. (2004). Sigma-1 receptors potentiate epidermal growth factor signaling towards neuritogenesis in PC12 cells: potential relation to lipid raft reconstitution. Synapse 53 90–103. 10.1002/syn.20041 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Shirasaki T., Kaku S., Muramatsu M., Otomo S. (1995). Characteristics of binding of [3H]NE-100, a novel sigma-receptor ligand, to guinea-pig brain membranes. Naunyn Schmiedeberg’s Arch. Pharmacol. 351 244–251. 10.1007/BF00233243 [DOI] [PubMed] [Google Scholar]
- Tchedre K. T., Huang R.-Q., Dibas A., Krishnamoorthy R. R., Dillon G. H., Yorio T. (2008). Sigma-1 receptor regulation of voltage-gated calcium channels involves a direct interaction. Invest. Ophthalmol. Vis. Sci. 49 4993–5002. 10.1167/iovs.08-1867 [DOI] [PubMed] [Google Scholar]
- Thomas J. D., Longen C. G., Oyer H. M., Chen N., Maher C. M., Salvino J. M., et al. (2017). Sigma1 targeting to suppress aberrant androgen receptor signaling in prostate cancer. Cancer Res. 77 2439–2452. 10.1158/0008-5472.Can-16-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyohara J., Sakata M., Ishiwata K. (2009). Imaging of sigma1 receptors in the human brain using PET and [11C]SA4503. Cent. Nerv. Syst. Agents Med. Chem. 9 190–196. [DOI] [PubMed] [Google Scholar]
- Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A. (1995). HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269 92–95. 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- Trujillo A. N., Katnik C., Cuevas J., Cha B. J., Taylor-Clark T. E., Breslin J. W. (2017). Modulation of mesenteric collecting lymphatic contractions by sigma1-receptor activation and nitric oxide production. Am. J. Physiol. Heart Circ. Physiol. 313 H839–H853. 10.1152/ajpheart.00702.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Chuang J. Y., Tsai M. S., Wang X. F., Xi Z. X., Hung J. J., et al. (2015). Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 112 E6562–E6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Ujike H., Tanaka Y., Sakai A., Yamamoto M., Fujisawa Y., et al. (2005). A variant of the sigma receptor type-1 gene is a protective factor for Alzheimer disease. Am. J. Geriatr. Psychiatry 13 1062–1066. 10.1176/appi.ajgp.13.12.1062 [DOI] [PubMed] [Google Scholar]
- Ullah M. I., Ahmad A., Raza S. I., Amar A., Ali A., Bhatti A., et al. (2015). In silico analysis of SIGMAR1 variant (rs4879809) segregating in a consanguineous Pakistani family showing amyotrophic lateral sclerosis without frontotemporal lobar dementia. Neurogenetics 16 299–306. 10.1007/s10048-015-0453-1 [DOI] [PubMed] [Google Scholar]
- Urfer R., Moebius H. J., Skoloudik D., Santamarina E., Sato W., Mita S., et al. (2014). Phase II trial of the Sigma-1 receptor agonist cutamesine (SA4503) for recovery enhancement after acute ischemic stroke. Stroke 45 3304–3310. 10.1161/STROKEAHA.114.005835 [DOI] [PubMed] [Google Scholar]
- Valenza M., DiLeo A., Steardo L., Cottone P., Sabino V. (2016). Ethanol-related behaviors in mice lacking the sigma-1 receptor. Behav. Brain Res. 297 196–203. 10.1016/j.bbr.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamvakidès A. (2002). [Anticonvulsant and forced swim anti-immobility effects of tetrahydro-N, N-dimethyl-2,2-diphenyl-3-furanemethanamine (AE37): common action mechanism?]. Ann. Pharm. Fr. 60 88–92. [PubMed] [Google Scholar]
- Vavers E., Zvejniece L., Maurice T., Dambrova M. (2019). Allosteric modulators of sigma-1 receptor: a review. Front. Pharmacol. 10:223. 10.3389/fphar.2019.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela J. M. (2020). Repurposing sigma-1 receptor ligands for COVID-19 therapy? Front. Pharmacol. 11:582310. 10.3389/fphar.2020.582310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villard V., Espallergues J., Keller E., Alkam T., Nitta A., Yamada K., et al. (2009). Antiamnesic and neuroprotective effects of the aminotetrahydrofuran derivative ANAVEX1-41 against amyloid beta(25-35)-induced toxicity in mice. Neuropsychopharmacology 34 1552–1566. 10.1038/npp.2008.212 [DOI] [PubMed] [Google Scholar]
- Villard V., Espallergues J., Keller E., Vamvakides A., Maurice T. (2011). Anti-amnesic and neuroprotective potentials of the mixed muscarinic receptor/sigma1 (σ1) ligand ANAVEX2-73, a novel aminotetrahydrofuran derivative. J. Psychopharmacol. 25 1101–1117. 10.1177/0269881110379286 [DOI] [PubMed] [Google Scholar]
- Vilner B. J., Bowen W. D. (2000). Modulation of cellular calcium by sigma-2 receptors: release from intracellular stores in human SK-N-SH neuroblastoma cells. J. Pharmacol. Exp. Ther. 292 900–911. [PubMed] [Google Scholar]
- Vogler S., Winters H., Pannicke T., Wiedemann P., Reichenbach A., Bringmann A. (2016). Sigma-1 receptor activation inhibits osmotic swelling of rat retinal glial (Muller) cells by transactivation of glutamatergic and purinergic receptors. Neurosci. Lett. 610 13–18. 10.1016/j.neulet.2015.10.042 [DOI] [PubMed] [Google Scholar]
- Vollrath J. T., Sechi A., Dreser A., Katona I., Wiemuth D., Vervoorts J., et al. (2014). Loss of function of the ALS protein SigR1 leads to ER pathology associated with defective autophagy and lipid raft disturbances. Cell Death Dis. 5:e1290. 10.1038/cddis.2014.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. M., Bowen W. D., Goldstein S. R., Roberts A. H., Patrick S. L., Hohmann A. G., et al. (1992). Autoradiographic distribution of [3H](+)-pentazocine and [3H]1,3-di-o-tolylguanidine (DTG) binding sites in guinea pig brain: a comparative study. Brain Res. 581 33–38. 10.1016/0006-8993(92)90340-F [DOI] [PubMed] [Google Scholar]
- Wang J., Saul A., Roon P., Smith S. B. (2016). Activation of the molecular chaperone, sigma 1 receptor, preserves cone function in a murine model of inherited retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 113 E3764–E3772. 10.1073/pnas.1521749113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shanmugam A., Markand S., Zorrilla E., Ganapathy V., Smith S. B. (2015). Sigma 1 receptor regulates the oxidative stress response in primary retinal Muller glial cells via NRF2 signaling and system xc(-), the Na(+)-independent glutamate-cystine exchanger. Free Radic. Biol. Med. 86 25–36. 10.1016/j.freeradbiomed.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eldred J. A., Sidaway P., Sanderson J., Smith A. J., Bowater R. P., et al. (2012). Sigma 1 receptor stimulation protects against oxidative damage through suppression of the ER stress responses in the human lens. Mech. Ageing Dev. 133 665–674. 10.1016/j.mad.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Wang M., Wan C., He T., Han C., Zhu K., Waddington J. L., et al. (2020). Sigma-1 receptor regulates mitophagy in dopaminergic neurons and contributes to dopaminergic protection. Neuropharmacology. 26:108360. 10.1016/j.neuropharm.2020.108360 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Ilieva H., Tamada H., Nomura H., Komine O., Endo F., et al. (2016). Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol. Med. 8 1421–1437. 10.15252/emmm.201606403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. N., Chang R. C., Atuehene N., Collier T. L. (2007). In vitro and in vivo binding of neuroactive steroids to the sigma-1 receptor as measured with the positron emission tomography radioligand [18F]FPS. Synapse 61 540–546. 10.1002/syn.20369 [DOI] [PubMed] [Google Scholar]
- Wolfe S. A., Kulsakdinun C., Battaglia G., Jaffe J. H., De Souza E. B. (1988). Initial identification and characterization of sigma receptors on human peripheral blood leukocytes. J. Pharmacol. Exp. Ther. 247 1114–1119. [PubMed] [Google Scholar]
- Wu F. S., Gibbs T. T., Farb D. H. (1991). Pregnenolone sulfate: a positive allosteric modulator at the N-methyl-D-aspartate receptor. Mol. Pharmacol. 40 333–336. [PubMed] [Google Scholar]
- Wu Z., Bowen W. D. (2008). Role of sigma-1 receptor C-terminal segment in inositol 1,4,5-trisphosphate receptor activation: constitutive enhancement of calcium signaling in MCF-7 tumor cells. J. Biol. Chem. 283 28198–28215. 10.1074/jbc.M802099200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G. L., Fiuzat M., Kuchibhatla M., Krishnan R., O’Connor C. M., Jiang W., et al. (2012). Health status and depression remission in patients with chronic heart failure: patient-reported outcomes from the SADHART-CHF trial. Circ. Heart Fail 5 688–692. 10.1161/CIRCHEARTFAILURE.112.967620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G. L., Prybol K., Boyle S. H., Hall R., Streilein R. D., Steffens D. C., et al. (2015). Inflammation markers and major depressive disorder in patients with chronic heart failure: results from the sertraline against depression and heart disease in chronic heart failure study. Psychosom. Med. 77 808–815. 10.1097/PSY.0000000000000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Yi W., Chen Y., Ma L., Wang J., Yu G. (2014). Overexpression of Sig1R is closely associated with tumor progression and poor outcome in patients with hilar cholangiocarcinoma. Med. Oncol. 31:261. 10.1007/s12032-014-0261-8 [DOI] [PubMed] [Google Scholar]
- Xu Q., Li L., Han C., Wei L., Kong L., Lin F. (2018). Sigma-1 receptor (σ1R) is downregulated in hepatic malignant tumors and regulates HepG2 cell proliferation, migration and apoptosis. Oncol. Rep. 39 1405–1413. 10.3892/or.2018.6226 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Miura R., Yamamoto T., Shinohara K., Watanabe M., Okuyama S., et al. (1999). Amino acid residues in the transmembrane domain of the type 1 sigma receptor critical for ligand binding. FEBS Lett. 445 19–22. 10.1016/s0014-5793(99)00084-8 [DOI] [PubMed] [Google Scholar]
- Yamoah A., Tripathi P., Sechi A., Köhler C., Guo H., Chandrasekar A., et al. (2020). Aggregates of RNA binding proteins and ER chaperones linked to exosomes in granulovacuolar degeneration of the Alzheimer’s Disease brain. J. Alzheimers Dis. 75 139–156. 10.3233/JAD-190722 [DOI] [PubMed] [Google Scholar]
- Yang H., Fu Y., Liu X., Shahi P. K., Mavlyutov T. A., Li J., et al. (2017). Role of the sigma-1 receptor chaperone in rod and cone photoreceptor degenerations in a mouse model of retinitis pigmentosa. Mol. Neurodegener. 12:68. 10.1186/s13024-017-0202-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Shen H., Li J., Guo L.-W. (2019). SIGMAR1/Sigma-1 receptor ablation impairs autophagosome clearance. Autophagy 15 1539–1557. 10.1080/15548627.2019.1586248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-J., Carter E. L., Torbey M. T., Martin L. J., Koehler R. C. (2010). Sigma receptor ligand 4-phenyl-1-(4-phenylbutyl)-piperidine modulates neuronal nitric oxide synthase/postsynaptic density-95 coupling mechanisms and protects against neonatal ischemic degeneration of striatal neurons. Exp. Neurol. 221 166–174. 10.1016/j.expneurol.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Su T. P. (2016). Potential molecular mechanisms on the role of the sigma-1 receptor in the action of cocaine and methamphetamine. J. Drug Alcohol. Res. 5:235970. 10.4303/jdar/235970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T., Liu X., Qu C., Zhang C., Fo Y., Guo Y., et al. (2019). Chronic inhibition of the sigma-1 receptor exacerbates atrial fibrillation susceptibility in rats by promoting atrial remodeling. Life Sci. 235:116837. 10.1016/j.lfs.2019.116837 [DOI] [PubMed] [Google Scholar]
- Zamanillo D., Andreu F., Ovalle S., Perez M. P., Romero G., Farre A. J., et al. (2000). Up-regulation of sigma(1) receptor mRNA in rat brain by a putative atypical antipsychotic and sigma receptor ligand. Neurosci. Lett. 282 169–172. 10.1016/s0304394000008843 [DOI] [PubMed] [Google Scholar]
- Zampieri D., Vio L., Fermeglia M., Pricl S., Wünsch B., Schepmann D., et al. (2016). Computer-assisted design, synthesis, binding and cytotoxicity assessments of new 1-(4-(aryl(methyl)amino)butyl)-heterocyclic sigma 1 ligands. Eur. J. Med. Chem. 121 712–726. 10.1016/j.ejmech.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Zhang B., Wang L., Chen T., Hong J., Sha S., Wang J., et al. (2017a). Sigma-1 receptor deficiency reduces GABAergic inhibition in the basolateral amygdala leading to LTD impairment and depressive-like behaviors. Neuropharmacology 116 387–398. 10.1016/j.neuropharm.2017.01.014 [DOI] [PubMed] [Google Scholar]
- Zhang H., Cuevas J. (2002). Sigma receptors inhibit high-voltage-activated calcium channels in rat sympathetic and parasympathetic neurons. J. Neurophysiol. 87 2867–2879. 10.1152/jn.2002.87.6.2867 [DOI] [PubMed] [Google Scholar]
- Zhang H., Cuevas J. (2005). σ Receptor activation blocks potassium channels and depresses neuroexcitability in rat intracardiac neurons. J. Pharmacol. Exp. Ther. 313 1387–1396. 10.1124/jpet.105.084152 [DOI] [PubMed] [Google Scholar]
- Zhang H., Katnik C., Cuevas J. (2009). Sigma receptor activation inhibits voltage-gated sodium channels in rat intracardiac ganglion neurons. Int. J. Physiol. Pathophysiol. Pharmacol. 2 1–11. [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zhao Z., Lan L., Wei X., Wang L., Liu X., et al. (2017b). Sigma-1 receptor plays a negative modulation on N-type calcium channel. Front. Pharmacol. 8:302. 10.3389/fphar.2017.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu F., Jiao Y., Tang T., Yang L., Lu C., et al. (2017c). An increase of sigma-1 receptor in the penumbra neuron after acute ischemic stroke. J. Stroke Cerebrovas Dis. 26 1981–1987. 10.1016/j.jstrokecerebrovasdis.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Zhao J., Ha Y., Liou G. I., Gonsalvez G. B., Smith S. B., Bollinger K. E. (2014). Sigma receptor ligand, (+)-pentazocine, suppresses inflammatory responses of retinal microglia. Invest Ophthalmol. Vis. Sci. 55 3375–3384. 10.1167/iovs.13-12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhemkov V., Ditlev J. A., Lee W. R., Wilson M., Liou J., Rosen M. K., et al. (2021). The role of sigma 1 receptor in organization of endoplasmic reticulum signaling microdomains. Elife 10:e65192. 10.7554/eLife.65192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C., Valenza M., Cattaneo E. (2010). Molecular mechanisms and potential therapeutical targets in huntington’s disease. Physiol. Rev. 90 905–981. 10.1152/physrev.00041.2009 [DOI] [PubMed] [Google Scholar]


