Abstract
Background
Unravelling autoimmune targets triggered by SARS‐CoV‐2 infection may provide crucial insights into the physiopathology of the disease and foster the development of potential therapeutic candidate targets and prognostic tools. We aimed at determining (a) the association between anti‐SARS‐CoV‐2 and anti‐apoA‐1 humoral response and (b) the degree of linear homology between SARS‐CoV‐2, apoA‐1 and Toll‐like receptor 2 (TLR2) epitopes.
Design
Bioinformatics modelling coupled with mimic peptides engineering and competition experiments were used to assess epitopes sequence homologies. Anti‐SARS‐CoV‐2 and anti‐apoA‐1 IgG as well as cytokines were assessed by immunoassays on a case‐control (n = 101), an intensive care unit (ICU; n = 126) and a general population cohort (n = 663) with available samples in the pre and post‐pandemic period.
Results
Using bioinformatics modelling, linear sequence homologies between apoA‐1, TLR2 and Spike epitopes were identified but without experimental evidence of cross‐reactivity. Overall, anti‐apoA‐1 IgG levels were higher in COVID‐19 patients or anti‐SARS‐CoV‐2 seropositive individuals than in healthy donors or anti‐SARS‐CoV‐2 seronegative individuals (P < .0001). Significant and similar associations were noted between anti‐apoA‐1, anti‐SARS‐CoV‐2 IgG, cytokines and lipid profile. In ICU patients, anti‐SARS‐CoV‐2 and anti‐apoA‐1 seroconversion rates displayed similar 7‐day kinetics, reaching 82% for anti‐apoA‐1 seropositivity. In the general population, SARS‐CoV‐2‐exposed individuals displayed higher anti‐apoA‐1 IgG seropositivity rates than nonexposed ones (34% vs 16.8%; P = .004).
Conclusion
COVID‐19 induces a marked humoral response against the major protein of high‐density lipoproteins. As a correlate of poorer prognosis in other clinical settings, such autoimmunity signatures may relate to long‐term COVID‐19 prognosis assessment and warrant further scrutiny in the current COVID‐19 pandemic.
Keywords: anti‐apolipoprotein A‐1 autoantibodies, COVID‐19, molecular mimicry, spike protein, toll‐like receptor 2
1. INTRODUCTION
Several lines of evidence point to a SARS‐CoV‐2‐triggered maladaptive immune response as an important determinant of coronavirus disease 2019 (COVID‐19) severity. 1 , 2 Relying on a complex interplay between pathogens and host factors, the B‐cell immune–mediated response is characterized by a polyclonal activation leading to the production of numerous antibodies which may cross‐react with self‐antigens when shared molecular homology between self and non‐self‐antigens occurs. 3 , 4 , 5 Because infection‐triggered autoantibodies have been shown to enhance tissue damage and the host inflammatory response, molecular mimicry between self and exogenous epitopes is considered to represent an important mechanism underlying the triad between infectious diseases, autoimmunity and poorer outcomes. 3 , 4 , 5
Furthermore, concurring emerging data demonstrate that humoral autoimmune mechanisms are frequent in COVID‐19, with several autoantibodies being detectable in up to 69% of COVID‐19 acute cases. 6 , 7 , 8 , 9 Sequence/structural homologies between SARS‐CoV‐2 immunodominant epitopes, the receptor‐binding domain (RBD) and numerous host antigens have been proposed to underlie such phenomenon. 10 , 11 , 12 Recently, we identified three epitopes from the Spike(S) sub‐domains S1 and S2, and the C terminus (c‐ter) of Spike as potential immunodominant epitopes of the Spike protein, 13 the c‐ter of Spike (amino acid region 1140‐1170) having been independently confirmed. 14 , 15 , 16 Previous unpublished observations indicated that this Spike c‐ter region shares sequence homology with the c‐ter of apolipoprotein A‐1 (apoA‐1), the major protein fraction component of high‐density lipoprotein (HDL), while a more proximal RBD region (amino acid region 455‐487), interacting with ACE2 receptor, 15 displayed homology with Toll‐like receptor 2 (TLR2). TLR2 engagement and subsequent activation have been shown to be required for autoantibodies against apoA‐1 (anti‐apoA‐1 IgG) to mediate their pro‐atherogenic effects. 17 , 18 , 19 , 20 Because anti‐apoA‐1 IgGs were shown to represent an independent cardiovascular (CV) risk factor associated with poor prognosis, 21 , 22 , 23 , 24 , 25 , 26 to be elevated after certain viral infections, 27 , 28 and to be preferentially oriented against the c‐ter part of apoA‐1, 29 , 30 we hypothesized that SARS‐CoV‐2 infection could elicit an anti‐apoA‐1 IgG response with substantial overlap with anti‐SARS‐CoV‐2 IgG serology.
Therefore, we used bioinformatics modelling coupled with mimetic engineered peptides and competition assay to validate linear sequence homologies between apoA‐1 and spike epitopes followed by the screening of three independent COVID‐19 adult cohorts for the presence of anti‐apoA‐1 IgG, including a case‐control (n = 101), a prospective intensive care unit (ICU) (n = 126) and a general population cohort (n = 663).
2. MATERIAL AND METHODS
2.1. Sequence homology analyses, Spike‐apoA‐1 and Spike‐TLR2 mimic peptides synthesis
Homologies between Spike and nucleocapsid epitopes, TLR2 or c‐ter apoA‐1 were assessed using Clustal Omega and BlastP sequence alignment. These methods have been extensively described in the Supplementary Material.
2.2. Study populations and sample collection
The case‐control cohort, the ICU cohort and the general population cohort have been extensively described in the Supplementary Material.
2.3. SARS‐CoV‐2 RT‐PCR analyses
As previously reported, 31 , 32 SARS‐CoV‐2 RT‐PCR was performed according to the manufacturers’ instructions on various platforms, including initially in house method using the BD SARS‐CoV‐2 reagent kit for BD Max system (Becton, Dickinson and Co, US) and Cobas 6800 SARS‐CoV‐2 RT‐PCR (Roche, Switzerland).
2.4. Anti‐SARS‐CoV‐2 against Spike 1 domain IgG assessment
We used the Euroimmun IgG enzyme‐linked immunosorbent assays (ELISA) (Euroimmun AG, Lübeck, Germany # EI 2606‐9601 G; CE‐marked) to assess SARS‐CoV‐2 IgG serology against the S1 domain of the Spike protein (anti‐S1 IgG) as explained in the Supplementary Material.
2.5. Assessment of total antibodies against N antigen of SARS‐CoV‐2
Total antibodies against the N antigen of SARS‐CoV‐2 were measured on a Cobas e801 analyser (Roche Diagnostics) according to the manufacturer's instructions. Results are reported as numeric values in form of a cut‐off index (signal sample/cut‐off or signal calibrator ratio) and are considered as positive when equal to or above 1. Inter‐assay variation was 14.3% at a ratio of 2.97 (n = 17).
2.6. Anti‐apoA‐1 IgG assessment
Anti‐apoA‐1 IgGs were measured as previously described. 21 , 22 , 25 , 26 , 27 , 28 , 33 Those methods are explained in the Supplementary Material.
2.7. Cytokines and anti‐pneumococcal IgG (P14 serotype) assessment
All cytokines and the anti‐pneumococcal IgG (P14 serotype) measurements were done using Meso Scale Discovery (MSD) platform on the SQ120 instrument. The methods are extensively described in the Supplementary Material.
2.8. Cross‐reactivity and competition experiments
To assess the degree of cross‐reactivity between anti‐SARS‐CoV‐2 and anti‐apoA‐1 IgG with their respective antigens and mimic peptides, two kinds of competition experiments were performed and are extensively reported in the Supplementary Material.
2.9. Statistics
Analyses were performed with Statistica software (version 13.5.0.17; TIBCO Software, Inc). Statistical methods are explained in the Supplementary Material.
3. RESULTS
3.1. Sequence homology assessment, corresponding Spike mimic peptides synthesis and competition experiments
Capitalizing on prior findings indicating that: (a) anti‐apoA‐1 IgG has to bind to TLR2 due to molecular mimicry in order to generate a pro‐inflammatory response by inducing the formation of a TLR2/TLR4/CD14 heterotrimer 18 , 19 and (b) anti‐apoA‐1 IgG are preferentially oriented against the c‐ter of apoA‐1 in humans 29 , 30 ; we searched for linear sequence similarities between the Spike protein epitopes, 13 , 14 , 15 , 16 apoA‐1 and the extracellular part of TLR2.
As shown in Figure 1, these analyses revealed that the amino acid (aa) sequence 1139‐1162 of the Spike protein shares sequence homology with the c‐ter part of apoA‐1 (amino acids 216‐243; Figure 1D) and that the aa region spanning 579‐587 of Spike protein had a good alignment with those of human TLR2 (aa 456‐464; 7 out of 9 amino acids; Figure 1D). Closer inspection of the position of the sequence showed that the secondary structure of the peptide was comparable for both sequence matches (Figure 1D). Specifically, the aa sequence 1139‐1162 in Spike is part of an alpha helical bundle with a portion of the peptide sequence unresolved, whereas the corresponding apoA‐1 sequence is also part of an alpha helical bundle with an unstructured portion. The Spike aa 579‐587 peptide is part of a beta turn and continues into a beta pleated sheet. There is remarkable structural homology with the structure of the corresponding peptide on TLR2, namely a beta turn followed by a beta pleated sheet segment (Figure 1D). The same analysis was performed to search for homology between TLR2, apoA‐1 and the N‐protein of SARS‐CoV‐2, using epitopes experimentally detected. 15 , 16 A good alignment between apoA‐1 (aa 131‐143) and N (aa 400‐412) was identified with 7 identical and 2 similar residues out of 11; however, the lack of structural data for the segment of the N‐protein precluded further analysis. Likewise, a good match was identified between TLR2 (aa 549‐557) and N (aa 217‐225) with 5 identical and 2 similar residues in a 8 amino acid stretch, but its structural homology could not be validated. In light of the lack of structural data for these N homology regions, further experimental validation of cross‐reactivity was not pursued. Peptide sequences and structures are presented in Figures S1 and S2
FIGURE 1.
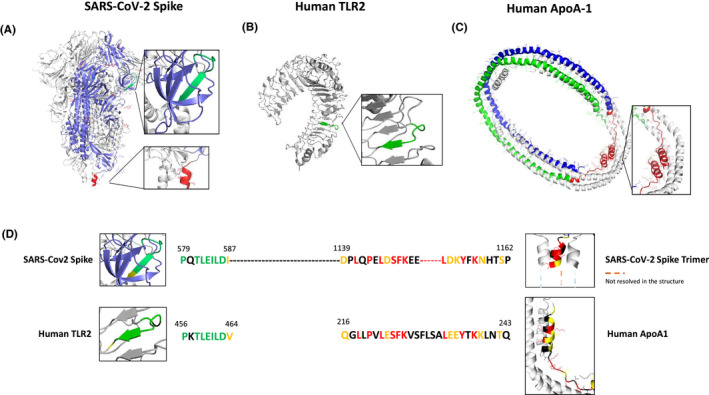
Localization of shared epitopes with apoA‐1 and TLR2 on the crystal structure of SARS‐CoV‐2 Spike protein. Panel (A) crystal structure of SARS‐CoV‐2 spike protein homotrimer (PDB ID 6VXX). Panel (B) human TLR2 crystal structure (PDB ID 6NIG). Panel (C) human ApoA‐1 tetramer crystal structure (PDB ID 1AV1). Epitope sequences conserved between Spike and TLR2 are highlighted in green (A and B) and conserved sequences between Spike and ApoA‐1 are represented in red (A and C). Panel (D) sequence alignment of SARS‐CoV‐2 Spike protein (QIV65088.1) with human TLR2 (H33756AA.1) using Clustal W. Conserved residues are indicated in green and the semiconserved one in yellow. BlastP sequence alignment of SARS‐CoV‐2 Spike sequence with human apoA‐1 (P02647.1). Conserved residues are shown in red and semiconserved (functional equivalent) ones in yellow
Following the bioinformatics identification of common epitopes between Spike and apoA‐1, we experimentally assessed the degree of cross‐reactivity between anti‐SARS‐CoV‐2 and anti‐apoA‐1 IgG with their respective antigens in our ELISA format. As shown in Figure 2A,B, we attempted to inhibit the anti‐apoA‐1 reactivity or the reactivity against the Spike protein incubating an anti‐SARS‐CoV‐2 and anti‐apoA‐1 IgG double‐positive sera with increasing concentration of Spike protein and C‐ter apoA‐1 peptides. As shown in Figure 2A,B, neither Spike protein nor c‐ter apoA‐1 competed for anti‐apoA‐1 IgG or anti‐SARS‐CoV‐2 IgG signal, respectively. However, when polyclonal anti‐Spike IgG pre‐incubation onto the plate was performed, we observed a weak but significant decrease of the anti‐apoA‐1 IgG signal derived from a pool of anti‐apoA‐1 IgG/anti‐SARS‐CoV‐2 IgG seropositive individuals (Figure 2C), while anti‐apoA‐1 IgG pre‐incubation did not affect the anti‐SARS‐CoV‐2 IgG signal of the pool of sera patients (Figure 2D).
FIGURE 2.
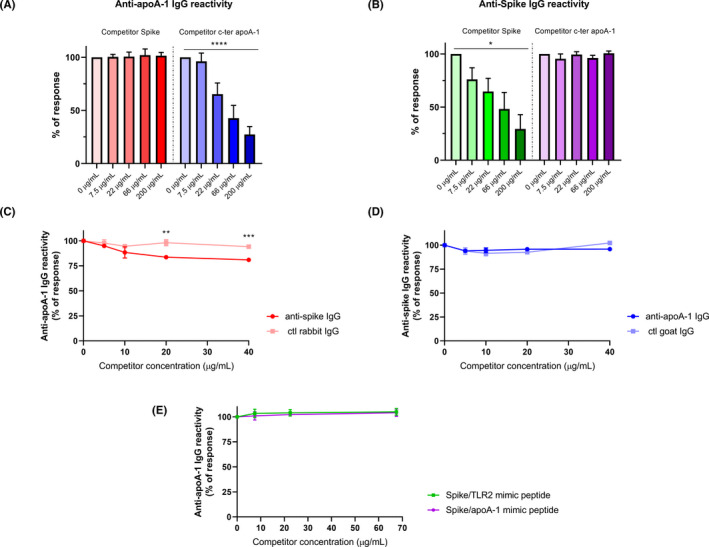
Absence of cross‐reactivity between anti‐apoA‐1 IgG and anti‐spike IgG. Panel (A and B). Four sera of COVID‐19 patients positive for both anti‐apoA‐1 IgG and anti‐spike IgG were pre‐incubated with or without Spike or c‐ter apoA‐1 peptides as competitors at the indicated concentrations prior to addition to assay well for anti‐apoA‐1 IgG or anti‐spike IgG measurements. Percentage of maximal ELISA signals were calculated as 100×{[signal in well]−[mean background signal (uncoated well)]}/{[mean maximal signal (no peptide)]−[mean background signal]}. Results are expressed as mean ± SD (n = 4). The statistical significance of the signal reduction was calculated by the one‐way ANOVA test: ****P < .0001 and *P = .012. Panel (C). Polyclonal anti‐spike antibodies and not the control rabbit IgGs slightly but significantly compete for apoA‐1 binding sites with anti‐apoA‐1 autoantibodies present in the pooled sera. Results are presented as the mean ± SD of three independent experiments (n = 3). Student's t test was used to determine the significant difference between the means of the two groups. **P = .002 and ***P = .0002. Panel (D). Anti‐apoA‐1 IgG did not compete for Spike protein. Results are presented as the mean ±SD of three independent experiments (n = 3). Panel (E) absence of anti‐apoA‐1 IgG signal inhibition when the pool of four anti‐apoA‐1 IgG/anti‐SARS‐CoV‐2 IgG seropositive individuals were pre‐incubated with or without Spike/TLR2 and Spike/apoA‐1 mimic peptides as competitors (n = 3)
To complete this set of experiment, Spike/ApoA‐1 and Spike/TLR2 mimic peptides were used in similar experimental competition procedures. As shown in Figure 2E, none of these mimic peptides competes for the anti‐apoA‐1 IgG signal derived from a pool of anti‐apoA‐1 IgG/anti‐SARS‐CoV‐2 IgG seropositive individuals.
Taken together, these results indicate that, despite the presence of common linear epitopes on Spike and apoA‐1 according to bioinformatics, in vitro cross‐reactivity between anti‐apoA‐1 IgG and S1 antigen seemed to be absent or modest at best if the results of Figure 5C would be predominantly considered.
3.2. Anti‐SARS‐CoV‐2 serologies and anti‐apolipoprotein A‐1 IgG associations
In order to validate our previous bioinformatics findings to humans, we explored the associations between anti‐apoA‐1 and anti‐SARS‐CoV‐2 serologies on three different cohorts, including a case‐control and the ICU cohort, as well as a general population cohort described in Figure S3 and S4. In the case‐control and ICU cohort, we aimed at replicating the previously reported correlations between anti‐apoA‐1 IgG with cytokine and lipid profile 20 , 34 as an additional orthogonal assessment of closely related serologies. The general population cohort was instrumental to generalize the results observed in acute settings at the population level and to determine whether the pre‐pandemic anti‐apoA‐1 serological status could modulate the anti‐SARS‐CoV‐2 response.
3.2.1. Results from the case‐control cohort
The case‐control cohort consisted 50 COVID‐19 RT‐PCR confirmed cases and 51 recruitment period‐matched healthy donors recruited over the same period (Figure S3 panel a). The baseline demographic characteristics of participants are summarized in Table 1. Briefly, RT‐PCR confirmed COVID‐19 patients were older with an over‐representation of male gender displaying a higher systemic pro‐inflammatory state when compared to the healthy blood donors (Table 1). Among COVID‐19 patients, the median delay between a positive SARS‐CoV‐2 RT‐PCR diagnostic test and current biomarker assessment was 10 days (IQR 5‐15 days). The proportion of patients within each days post‐diagnosis subgroup (delta between their molecular testing and serological testing) was 43.5% for 0‐6 days (n = 20), 30.4% for 7‐14 days (n = 14) and 26.1% for >14 days (n = 12).
TABLE 1.
Demographic and biological characteristics of the case‐control cohort
| The case‐control cohort | |||
|---|---|---|---|
| Patients clinical biological characteristics | COVID‐19 cases (n = 50) | Healthy controls (n = 51) | P‐value |
| Age | 70 (61‐76; 33‐85) | 47.0 (40‐62; 22‐62) | <.0001 |
| Male Gender; % (n) | 60.0 (30) | 19.6 (10) | .0001 |
| Cytokine | |||
| IFN‐γ, pg/mL | 26.6 (18.3‐63.2; 7.3‐9589) | 22.4 (16.3‐35.8; 9.6‐384.6) | .11 |
| IL‐6, pg/mL | 12.0 (5.8‐36; 3.02‐3831.4) | 1.1 (0.6‐2.0; 0.1‐22.3) | <.0001 |
| TNF‐α, pg/mL | 9.1 (6.5‐12.8; 2.2‐13.4) | 4.6 (3.7‐7.3; 1.5‐74.3) | <.0001 |
| MCP‐1, pg/mL | 1875 (1080‐2597; 404‐1292) | 1284.7 (949‐1629; 440‐3001) | .001 |
| IFN‐α2a, pg/mL | 2.8 (1.9‐4.4; 0.7‐39.4) | 1.3 (0.3‐1.9; 0.3‐24.9) | <.0001 |
| Serologies | |||
| Anti‐S1 IgG, ratio | 15.8 (1.4‐18; 0.2‐20.6) | 0.3 (0.3‐0.4; 2‐2.8) | <.0001 |
| Anti‐S1 IgG, seropositivity; %(n) | 74 (37) | 1.9 (1) | <.0001 |
| Anti‐N total ab, ratio | 29 (2.5‐73.3; 0.7‐143) | 0.07 (0.07‐0.08; 0.06‐53) | |
| Anti‐N total ab, seropositivity; %(n) | 74.0 (37) | 1.9 (1) | |
| Anti‐apoA‐1 IgG, OD450 | 1.58 (1.11‐1.9; 0.14‐2.32) | 0.7 (0.55‐0.93; 0.3‐1.30) | <.0001 |
| Anti‐pneumococcal (Pn14) IgG, mg/L | 0.44 (0.15‐1.36; 0.03‐81.1) | 0.76 (0.2‐2.11‐0.07‐102) | .40 |
| Splitting the derivation cohort according to anti‐S1 IgG seropositivity status | |||
|---|---|---|---|
| Anti‐S1 IgG seropositivity (n = 38) | Anti‐S1 IgG seronegativity (n = 63) | P‐value | |
| Age | 70 (61‐76; 50‐84) | 48 (42‐60; 22‐85) |
<.0001 <.0001 |
| Male Gender; % (n) | 78.9 (30) | 15.8 (10) | <.0001 |
| Cytokines | |||
| IFN‐γ, pg/mL | 25.8 (18.3‐63.2; 7.3‐664.9) | 22.6 (16.3‐41.1; 9.6‐9589) | .33 |
| IL‐6, pg/mL | 8.6 (4.9‐19.5; 1.6‐3831) | 1.6 (0.8‐6.7; 0.1‐1485) | <.0001 |
| TNF‐α, pg/mL | 9.5 (6.5‐12.8; 2.2‐131.4) | 5.5 (3.8‐8.5; 1.5‐74.3) | .0002 |
| MCP‐1, pg/mL | 1810 (1078‐2583; 538‐9465) | 1335 (958‐1795; 404‐12901) | .03 |
| IFN‐α2a, pg/mL | 2.7 (1.9‐3.8; 0.7‐10.9) | 1.5 (0.9‐2.5; 0.3‐39.4) | <.0001 |
| Serologies | |||
| Anti‐N total ab, ratio | 47.2 (17.4‐81.7; 0.26‐147) | 0.07 (0.07‐0.08; 0.06‐29) | <.0001 |
| Anti‐N total ab, seropositivity; % (n) | 92.1 (35) | 3.2 (2) | <.0001 |
| Anti‐apoA‐1 IgG, OD450 | 1.65 (1.35‐1.92; 0.14‐2.32) | 0.76 (0.57‐1.00; 0.17‐2.01) | <.0001 |
| Anti‐pneumococcal (Pn14) IgG, mg/L | 0.46 (0.15‐1.73; 0.03‐81.1) | 0.47 (0.17‐1.49; 0.07‐102) | .70 |
As shown in Table 1, COVID‐19 patients had higher median levels of all the pro‐inflammatory cytokines and serologies tested, with the exception of Pn14 pneumococcal IgG used as an unrelated serological control. The distribution of serological values between cases and controls is available in Figure S5. No difference between cases and controls was observed for circulating INF‐γ levels. Furthermore, when the cohort was split according to anti‐S1 IgG seropositivity status, identical differences were observed (Table 1, bottom panel).
These results were further corroborated by the significant and substantial correlations observed between anti‐apoA‐1 IgG and both (anti‐S1 and anti‐N) anti‐SARS‐CoV‐2 serologies (Table 2). On the other hand, none of the aforementioned antibodies correlated with anti‐pneumococcal IgG (Table 2). Furthermore, both anti‐SARS‐CoV‐2 serologies, anti‐apoA‐1 IgG, displayed similar strength of associations with most of the cytokines measured. Finally, anti‐pneumococcal IgG was not correlated to any of the cytokines tested (Table 2).
TABLE 2.
Spearman correlations between serologies and cytokines profile in the COVID‐19 cases of the case‐control cohort
| Anti‐S1 IgG r; P‐value |
Anti‐N total ab r; P‐value |
Anti‐apoA‐1 IgG r; P‐value |
Anti‐pneumococcal IgG r; P‐value |
|
|---|---|---|---|---|
| Serologies | ||||
| Anti‐S1 IgG; ratio | ND | 0.70;<.0001 | 0.60;<.0001 | −0.07; .50 |
| Anti‐N total ab; ratio | 0.70; <0.0001 | ND | 0.54;<.0001 | 0.07; .47 |
| Anti‐apoA‐1 IgG, OD450 | 0.60; <0.0001 | 0.54;<.0001 | ND | −0.03; .77 |
| Anti‐pneumococcal (Pn14) IgG, pg/mL | −0.07; 0.50 | 0.07; .47 | −0.03; .77 | ND |
| Cytokines | ||||
| IFN‐γ, pg/mL | 0.15; 0.14 | 0.12; .25 | −0.05; .63 | 0.01; .91 |
| IL‐6, pg/mL | 0.41; <0.0001 | 0.45; <.0001 | 0.36;<.0001 | −0.05; .59 |
| TNF‐α, pg/mL | 0.38; 0.0001 | 0.40; <.0001 | 0.26; .001 | −0.07; .47 |
| MCP‐1, pg/mL | 0.30; 0.003 | 0.14; .18 | −0.05; .73 | 0.09; .36 |
| IFN‐α2a, pg/mL | 0.43; <0.0001 | 0.37; .0001 | 0.26; .007 | −0.05; .62 |
Abbreviations: Anti‐N total ab, anti‐N antigen total antibodies; ND, not determined.
3.2.2. Results from the ICU cohort
To extend and validate these findings in severe COVID‐19 disease, we used a cohort of 126 consecutive patients admitted to the ICU for severe COVID‐19 disease who completed a follow‐up at 28 days, Figure S3 panel b. The baseline demographic and biological characteristics of ICU COVID‐19 patients are summarized in Table 3. Anti‐apoA‐1 IgG seropositivity upon ICU admission was found in 26.9% of the ICU patients (34/126), while anti‐S1 IgG and anti‐N seropositivity was 36.5% (46/126) and 42% (53/126), respectively. When split according to anti‐apoA‐1 IgG seropositivity status upon patient admission at the ICU, seropositive patients tended to have a more severe Simplified Acute Physiology Score II score, a higher number of DPSO at ICU admission, displayed higher median D‐dimers levels, but lower total cholesterol, LDL and triglycerides levels when compared to anti‐apoA‐1 seronegative individuals (Table 3). The proportion of anti‐S1 and anti‐N seroconversions were twofold higher in anti‐apoA‐1 IgG seropositive individuals compared to those tested negative for these autoantibodies (55.8% vs 28.2%, P = .006; and 64.7% vs 33.7%; P = .01, respectively). No other significant differences for the remaining parameters were identified between anti‐apoA‐1 IgG seropositive and seronegative individuals.
TABLE 3.
Baseline demographic and biological characteristics of ICU patients and according to anti‐apoA‐1 IgG serological status
| Demographic and biological characteristics | Overall (n = 126) | Anti‐apoA‐1 IgG seropositive patients (n = 34) | Anti‐apoA‐1 IgG seronegative patients (n = 92) | P‐value |
|---|---|---|---|---|
| Age, years | 63.5 (57‐73; 25‐86) | 65.5 (57‐73; 28‐86) | 62.5 (57‐72.5; 25‐83) | .48 |
| Female gender, % (n) | 22.2 (28) | 17.6 (6) | 23.4 (22) | .63 |
| BMI, kg/m2 | 28.1 (25.5‐32.1; 15.6‐52.4) | 28.3 (25.1‐32.8‐19.3‐52.4) | 28.1 (25.6‐31.8; 15.6‐50.8) | .92 |
| Current smoking, % (n) | 13.5 (17) | 14.7 (5) | 13.0 (12) | .78 |
| DPSO and ICU admission | 9.0 (7‐11; 0‐27) | 9.5 (7.5‐13.5; 3‐27) | 9.0 (7.0‐10; 0‐27) | .10 |
| Comorbidities | ||||
| Hypertension, % (n) | 47.6 (60) | 44.1 (15) | 48.9 (45) | .69 |
| Dyslipidaemia, % (n) | 58.3 (35) | 23.5 (8) | 29.3 (27) | .66 |
| Diabetes, % (n) | 26.7 (34) | 29.4 (10) | 26.0 (24) | .82 |
| Previous IC and or HF, % (n) | 23.8 (30) | 23.5 (8) | 23.9 (22) | 1 |
| Previous stroke, % (n) | 5.5 (7) | 0 (0) | 7.6 (7) | .19 |
| Known malignancy, % (n) | 7.9 (10) | 11.7 (4) | 6.5 (6) | .46 |
| Chronic kidney disease, % (n) | 7.1 (9) | 5.8 (2) | 7.6 (7) | 1 |
| Severity upon admission | ||||
| APACHE II score | 22 (14‐29; 3‐38) | 22 (14‐29; 3‐38) | 22 (13.5‐27.5; 4‐37) | .61 |
| SOFA score | 6 (4‐7; 1‐11) | 5 (4‐7; 2‐10) | 6 (4‐7; 1‐11) | .44 |
| SAPS II score | 53 (43‐65; 6‐82) | 58 (46‐69; 18‐78) | 52 (38.5‐61.5; 6‐82) | .09 |
| 28‐day mortality, %(n) | 16.7 (21) | 23.5 (8) | 14.1 (13) | .28 |
| Length of stay at ICU, days | 16 (10‐21; 1‐48) | 14 (10‐18; 1‐42) | 16 (10‐22; 1‐48) | .25 |
| Mechanical ventilation, % (n) | 96 (121) | 91.1 (31) | 97.8 (90) | .12 |
| Cytokines and inflammation | ||||
| CRP, mg/L | 154 (92‐205; 23.1‐402.8) | 162.7 (114.1‐219.5; 23.1‐402.8) | 144 (92.4‐201; 31‐311) | .35 |
| IFN‐γ, pg/mL | 411.1 (112‐988.1; 152.2‐37747) | 308.4 (112.1‐691.5; 5.2‐37747) | 426.7 (175.1‐1232.6; 175‐17778) | .22 |
| IL‐6, pg/mL | 155.0 (69.1‐324.3; 6.7‐7889.6) | 212.4 (61.4‐561.0; 6.7‐7689.8) | 140.8 (73.3‐284.6; 13.9‐2224.8) | .32 |
| TNF‐α, pg/mL | 6.7 (4.0‐16.6; 0.23‐164.5) | 9.3 (4.0‐16.9; 0.3‐71.2) | 6.4 (4.2‐14.8; 1.6‐164.5) | .33 |
| MCP‐1, pg/mL | 4199 (2509‐8488; 254‐36483) | 3620 (2314‐10281; 776‐36483) | 4300 (2634‐7609; 254‐30256) | .92 |
| ifn‐α2a, pg/mL | 7.1 (2.3‐22.6; 0.2‐468.5) | 6.3 (1.5‐19.1; 0.2‐92.3) | 8.9 (2.6‐30.9; 0.3‐468.5) | .13 |
| D‐dimers; ng/mL | 1531 (961‐2476; 220‐10001) | 1838 (1326‐3245; 439‐10001) | 1433 (863‐2174; 220‐9999) | .03 |
| Lipid profile | ||||
| Total cholesterol, mmol/L | 2.8 (2.2‐3.2; 1.1‐5.8) | 2.4 (1.9‐3.1; 1.1‐5.2) | 2.9 (2.4‐3.3; 1.1‐5.8) | .03 |
| HDL cholesterol, mmol/L | 0.63 (0.47‐0.76; 0.14‐1.95) | 0.52 (0.38‐0.61; 0.27‐0.88) | 0.66 (0.54‐0.79; 0.14‐1.95) | .0001 |
| LDL cholesterol, mmol/L | 1.29 (0.87‐1.76; 0.00‐3.70) | 1.00 (0.62‐1.57; 0.08‐3.70) | 1.43 (1.05‐1.79; 0.00‐3.58) | .02 |
| Triglycerides, mmol/L | 1.64 (1.18‐2.24; 0.79‐4.05) | 1.75 (1.38‐2.24; 0.81‐4.00) | 1.57 (1.15‐2.23; 0.79‐4.05) | .24 |
| Cardiac biomarkers | ||||
| Hs‐cTnT, ng/L | 16.0 (9.7‐34.9; 3.31‐971) | 13.0 (8.1‐40.5; 3.7‐665) | 17‐9 (9.7‐33.5; 3.3‐971) | .53 |
| NT‐proBNP, pg/mL | 308 (95.6‐1015; 15.1‐18772) | 363 (110‐1437; 22‐5928) | 278.5 (93.3‐909; 15.1‐18772) | .42 |
| Serologies | ||||
| Anti‐S1 IgG, ratio | 0.73 (0.4‐1.9; 0.3‐27.0) | 2.1 (0.5‐17.3; 0.4‐27.0) | 0.6 (0.4‐1.15; 0.3‐21.9) | .0009 |
| Anti‐S1 IgG, seropositivity; % (n) | 36.2 (46) | 55.8 (19) | 28.2 (26) | .006 |
| Anti‐N total ab, ratio | 0.46 (0.1‐4.70; 0.1‐36.3) | 2.71 (0.2‐11.3; 0.1‐36.3) | 0.31 (0.1‐1.5; 0.1‐29.1) | .0004 |
| Anti‐N total ab, seropositivity; % (n) | 42.0 (53) | 64.7 (22) | 33.7 (31) | .002 |
| Anti‐apoA‐1 IgG, OD450 | 0.43 (0.24‐0.70; 000‐2.60) | 0.99 (0.83‐1.70; 0.70‐2.60) | 0.33 (0.22‐0.44; 0.0‐0.67) | <.0001 |
| Renal function | ||||
| Creatinine; µmol/L | 81.0 (66.5‐105; 38‐769) | 81.5 (67.5‐110.5; 47‐173) | 80.5 (65‐98, 38‐769) | .36 |
All continuous variables are expressed as median (interquartile range; and range); *P‐value derived from the comparison between anti‐apoA‐1 IgG seropositive verse seronegative individuals.
Abbreviations: APACHE II, Acute Physiology And Chronic Health Evaluation II; DPSO, days post‐symptom onset; HF, heart failure; IC, ischaemic cardiopathy; SAPS, Simplified Acute Physiology Score; SOFA, Sequential Organ Failure Assessment.
When split according to anti‐S1 serological status, anti‐S1 IgG seropositive patients were less likely to be known to have chronic kidney disease, they tended to have a shorter length of ICU stay and they were less likely to require mechanical ventilation (Table S1). On the other hand, anti‐S1 IgG seropositive patients displayed higher number of DPSO before ICU admission, higher median levels of anti‐apoA‐1 IgG and D‐dimers, but lower median levels of INF‐γ and INF‐α2a. No significant differences were noted regarding the lipid profile and other biological parameters tested (Table S1). The proportion of anti‐apoA‐1 IgG seropositivity was increased by 2‐fold in anti‐S1 seropositive individuals compared to anti‐S1 IgG seronegative ones (41.3% vs 18.8%, P = .01; Table S1). As shown in Table 4, very similar correlations and strength of associations were found between anti‐apoA‐1 IgG, anti‐S1 and anti‐N serologies compared to those observed in the case‐control cohort. Furthermore, except for INF‐γ and INF‐α2a, similar observations were made regarding the associations between the different serologies and cytokines levels (Table 4).
TABLE 4.
Spearman correlations between serologies, cytokines and lipid profile in the ICU patients
| Anti‐S1 IgG r; P‐value | Anti‐N total ab r; P‐value | Anti‐apoA‐1 IgG r; P‐value | |
|---|---|---|---|
| Serologies | |||
| Anti‐S1 IgG; ratio | ND | 0.77; <.0001 | 0.43; <.0001 |
| Anti‐N total ab; ratio | 0.77; <.0001 | ND | 0.44; <.0001 |
| Anti‐apoA‐1 IgG, OD450 | 0.43; <.0001 | 0.44; <.0001 | ND |
| Cytokines | |||
| CRP; mg/L | 0.18; .05 | 0.16; .08 | 0.20; .02 |
| IFN‐γ, pg/mL | −0.25; .005 | −0.25; .005 | −0.19; .04 |
| IL‐6, pg/mL | −0.02; .81 | −0.07; .47 | 0.11; .24 |
| TNF‐α, pg/mL | −0.02; .83 | −0.09; .32 | 0.12; .19 |
| MCP‐1, pg/mL | 0.04; .69 | 0.006; .94 | 0.01; .93 |
| IFN‐α2a, pg/mL | −0.36; .0003 | −0.44; <.0001 | −0.30; .0005 |
| D‐dimers; ng/mL | 0.29; .004 | 0.11; .26 | 0.05; .63 |
| Lipid profile | |||
| Total cholesterol, mmol/L | 0.12; .21 | 0.04; .70 | −0.16; .06 |
| HDL cholesterol, mmol/L | −0.26; .004 | −0.27; .003 | −0.37; <.0001 |
| LDL cholesterol, mmol/L | 0.12; .21 | 0.09; .33 | −0.16; 0.08 |
| Triglycerides, mmol/L | 0.24; .008 | 0.10; .29 | 0.20; 0.03 |
3.2.3. One‐week serological kinetics in ICU patients
In the subgroup of 54 ICU patients for which additional serum samples were available at day 3 and day 7 of ICU admission, significant increases in median values of anti‐SARS‐CoV‐2 and anti‐apoA‐1 serologies were observed (Figure 3). Accordingly and as expected, anti‐SARS‐CoV‐2 seropositivity rates for both anti‐S1 and anti‐N serologies were high and reached values above 92% at day 7. Anti‐apoA‐1 IgG seropositivity evolution displayed a very similar temporal trend to anti‐SARS‐CoV‐2 serology, reaching 82.4% at day 7, indicating that anti‐apoA‐1 IgG serology kinetics closely follow the occurrence of anti‐SARS‐CoV‐2 antibodies over 7 days of severe COVID‐19 disease (Figure 3). No such kinetics were observed for anti‐Pn14 IgG.
FIGURE 3.
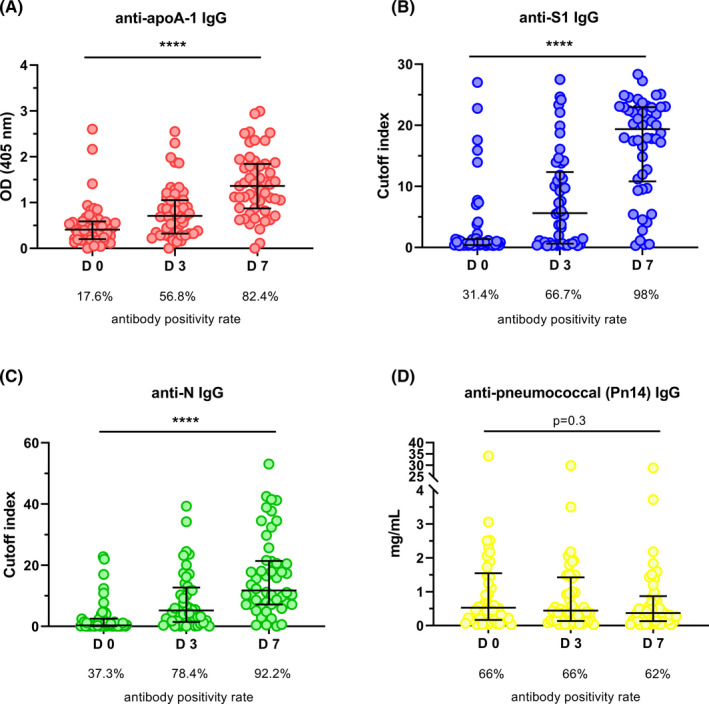
One‐week serological kinetics in ICU patients. In panels (A‐C), ICU patients showed a significant increase (P‐value of ****P < .0001 and P = .025) in antibody titre throughout seven days (days: 0, 3 and 7). In panel (D), the anti‐ pneumococcal (Pn14) antibody titre did not present any change over time (P = .3). Results are expressed as median with interquartile range and the Kruskal‐Wallis test was used to compare the three groups. Samples were analysed in duplicate
3.2.4. Results from the general population cohort
Because anti‐apoA‐1 IgG seropositivity has been shown to concern about one‐fifth of the general population and to be associated with a poorer prognosis over 5 years, 22 , 35 our results prompted us to investigate whether such SARS‐CoV‐2‐induced anti‐apoA‐1 IgG response in acute settings could be replicated in the general population. With this aim, we identified participants recruited in the ‘Bus Santé’ study between 2016 and 2018 and subsequently included in the SEROCoV‐POP study 32 during the COVID‐19 pandemic. Among these, we identified 663 individuals with available serum samples from both the pre‐ (2016‐2018) and post‐pandemic (2020) periods (Figure S4 and Table S2).
The median age of this cohort was 50‐year‐old (range 24‐78), 297 (44.7%) participants were male, while the baseline (pre‐pandemic) anti‐apoA‐1 IgG seropositivity rate was 25.0% (166/663) and was not associated with any factors commonly ascribed to autoantibodies such as age, gender or smoking (data not shown). The median time between the first anti‐apoA‐1 IgG assessment in the pre‐COVID‐19 period and the anti‐S1 IgG plus the second anti‐apoA‐1 IgG measurement during the post‐COVID‐19 period (after the first pandemic wave) was 3.2 years (IQR: 2.8‐3.6; range: 2.3‐4.3). In the post‐COVID‐19 period, 7.5% (50/663) were seropositive against SARS‐CoV‐2 according to anti‐S1 IgG levels. The rate of anti‐apoA‐1 IgG seropositivity, as well as median anti‐apoA‐1 IgG levels (data not shown) were significantly lower than in the pre‐COVID‐19 period, despite remaining of the same order of magnitude than previously reported in the general population 21 , 22 (Table S3). As shown in Table S3, in the post‐COVID‐19 samples, a modest but significant correlation was observed between anti‐apoA‐1 IgG and anti‐S1 IgG levels. Furthermore, in the post‐COVID‐19 period, anti‐S1 seropositive individuals displayed higher median anti‐apoA‐1 IgG levels (0.35 vs 0.57 OD; P = .0002) and higher median anti‐apoA‐1 seropositivity rates than anti‐S1 seronegative individuals (34%.0% vs 16.8%, P = .004) as shown in Table S3. Moreover, the strength of correlation between anti‐apoA‐1 IgG and anti‐S1 IgG in anti‐S1 seropositive individuals was of 0.31 (P = .03), whereas no association between these two serologies was found in anti‐S1 seronegative individuals (Table S3). Cox regression analyses indicated that the pre‐COVID‐19 anti‐apoA‐1 IgG status was a significant predictor of post‐COVID‐19 anti‐apoA‐1 IgG status (HR: 1.95; 1.52‐2.43; P <.0001), irrespective of age, gender and smoking status (HR: 1.57; 95%CI: 1.24‐1.99; P = .0001), but did not predict post‐COVID‐19 anti‐S1 seropositivity (HR: 1.30; 95%CI: 0.67‐2.54; P = .44). Finally, adjusted logistic regression analyses indicated that post‐pandemic anti‐SARS‐CoV‐2 seropositivity was significantly associated with a 3‐fold risk of post‐pandemic anti‐apoA‐1 IgG seropositivity, independently of age, gender and smoking status (OR: 2.46; 95%CI: 1.31‐4.60; P = .005).
4. DISCUSSION
The major findings of this study can be summarized by the fact that SARS‐CoV‐2 infection triggers a humoral response against native apoA‐1—the major HDL lipoprotein—in the vast majority infected individuals, displaying similar kinetics and marked correlations to anti‐SARS‐CoV‐2 responses. In this context, the bioinformatics identification of common linear epitopes between SARS‐CoV‐2 and those of apoA‐1 and TLR2 would have lend weight to the molecular mimicry hypothesis, especially as the known functions of identified epitopes would have been concordant with the correlations retrieved presently between anti‐apoA‐1 and anti‐SARS‐CoV2 responses, lipid profile and inflammation.
Indeed, the c‐ter sequence identified in Spike, aa 1139‐1162, 15 is known to be conserved in HCoV‐OC43 but not among other coronaviruses and was found to share linear homology with aa 216‐243 of apoA‐1. This c‐ter region of apoA‐1 structurally corresponds to an alpha helix bundle playing a key role in the cellular cholesterol efflux regulation by ATP‐binding cassette transporter A1 (ABCA1) and in HDL maturation 36 and was shown to be preferentially targeted by the polyclonal anti‐apoA‐1 IgG response. 29 , 30 Such regional targeting is therefore compatible with the previously reported inverse relationships between anti‐apoA‐1 IgG and HDL cholesterol, 21 , 33 , 34 the similar ones retrieved presently between HDL levels and anti‐SARS‐CoV‐2 serology titres in the ICU cohort. The second linear sequence homology identified concerned the aa 579‐587 of Spike, located in the S1 domain and close to the receptor‐binding domain, which has very homology to the aa 456‐464 sequence of TLR2. This sequence is part of the leucine‐rich repeats (LRR) ectodomain of TLR2, known to be key for proper pathogen‐associated or damage‐associated molecular pattern recognition and TLR2’s function. 37 The anti‐apoA‐1 IgGs engagement of this region due to sequence homology with apoA‐1 20 has been reported to mediate their pro‐inflammatory/pro‐atherogenic response, 18 , 19 , 20 , 34 , 38 and the present correlations retrieved between anti‐apoA‐1 IgG, anti‐SARS‐CoV‐2 serologies and pro‐inflammatory cytokines on the case‐control cohort would have lend further weight to the molecular mimicry hypothesis proposed to explain the occurrence of other cross‐reacting pathogenic antibodies in COVID‐19. 39 , 40 However, by failing to clearly demonstrate any cross‐reactivity between the different antigens and antibodies of interest, our experimental approach did not support the molecular mimicry hypothesis to explain the occurrence of COVID‐19‐induced anti‐apoA‐1 IgG response and its intricate relationship with anti‐SARS‐CoV‐2 serologies. Such discrepancy between bioinformatics modelling and competition experiments can be explained by the possible existence of common conformational epitope(s) that our epitope mapping systems could not detect, and/or by the likely existence of additional mechanisms, such as intermolecular epitope spreading, allowing the initial targeted humoral response to quickly broaden to antigens other than the inducing epitope. 41
From a physiopathological point of view, these results provide innovative perspectives. First of all, these results extend the coverage of virus‐mediated anti‐apoA‐1 IgG induction to SARS‐CoV‐2, as previously shown for HCV and HIV. 27 , 28 The inverse associations reported here between anti‐apoA‐1 IgGs and HDL levels are similar to what has been observed in HCV 27 and are reminiscent of the concept that HCV could hijack the scavenger receptor B‐1 (SR‐B1)–mediated HDL uptake to infect hepatocytes 27 , 42 which has recently been transposed to SARS‐CoV‐2 by the recent demonstration of Wei and colleagues who identified SR‐B1 as an additional receptor facilitating the SARS‐CoV‐2 entry into cells. 43 SR‐B1 being the canonical apoA‐1/HDL receptor involved in reverse cholesterol efflux and HDL maturation, our results suggest that the molecular mimicry‐based anti‐apoA‐1 IgG response in COVID‐19 may concur with other established inflammatory factors to explain the low HDL and apoA‐1 levels reported previously in COVID‐19. 44 , 45 Secondly and along the same line, our results lend further weight to the fact that host lipid metabolism may play an important role in COVID‐19 severity by modulating the intensity of the immune response. Lee et al 46 recently demonstrated that COVID‐19 activates regulatory element binding protein‐2 (SREBP‐2), a key transcription factory lying at the cross‐roads of inflammation modulation and cholesterol biosynthesis. Given the requirement of cholesterol biosynthesis for SARS‐CoV‐2 budding–driven exocytosis, any factor modulating SREBP‐2 pathway activation may influence the course of COVID‐19 disease. 46 In this respect, recent findings in human macrophages indicate that anti‐apoA‐1 IgGs increase the expression of SREBP‐2 in a TLR2/4‐dependent manner, culminating into enhanced foam cell formation, the hallmark of atherogenesis, 34 through anti‐apoA‐1 IgG‐dependent ACAT activation leading to the redirection of cellular cholesterol towards intracellular esterified cholesterol pools and decreased membrane free cholesterol content. 34 Because membrane free cholesterol is a key regulator of membrane ACE2R trafficking into dedicated lipid rafts for optimal SARS‐CoV‐1 endocytosis, 47 knowing whether anti‐apoA‐1 IgG could influence the course of COVID‐19 should be further investigated.
Although not designed to convey any actionable clinical implications, the results derived from this exploratory and hypothesis‐generating study may bare the following potential clinical implications. From a pragmatic analytical standpoint, our results indicate that despite the intimate relationship between anti‐apoA‐1 and anti‐S1 serology and the existence of linear sequence homology between defined apoA‐1 and S1 epitopes; our results indicate that the risk of potential analytical interference between these serologies can safely be ruled out. This point is important to make as 30%‐80% of COVID‐19 individuals will have high levels of both anti‐apoA‐1 IgG and anti‐SARS‐CoV‐2 antibodies, and the present associations would fuel such question. Importantly, although we could not assess the direct clinical implications of the COVID‐19‐induced anti‐apoA‐1 IgG response, our observations may well relate to patient prognosis for two reasons. Firstly, these pathogenic autoantibodies were shown to be active mediator of sterile inflammation 18 , 19 , 20 , 34 and independent predictors of overall morality and adverse CV events in numerous clinical settings, including general populations 21 , 22 , 23 , 24 , 25 , 26 ; these antibodies may well be of concern in COVID‐19 too. Secondly and along the same line, functional antibodies against G‐coupled receptors displaying identical biological activity to anti‐apoA‐1 IgGs were recently shown to be associated with prolonged symptoms persistence after COVID‐19 infection, 48 further supporting to the suspected clinical relevance of COVID‐19‐induced pathogenic autoantibodies for long‐term outcomes and potential enhanced patient risk stratification. 49 Until the formal demonstration of the harmlessness of such autoantibodies, such biological signature should be carefully evaluated, especially in the context of the long COVID syndrome.
We acknowledge several limitations of the present work. Firstly, we limited our analyses to autoantibodies directed against apoA‐1 and did not consider other autoantibodies of possible relevance in COVID‐19, such as anti‐phospholipid or anti‐platelet 4 autoantibodies. 50 , 51 Secondly, due to the fact that our hospital became a COVID‐19‐only hospital during the first pandemic wave could not identify a COVID‐19‐free control population matched for usual factors impacting anti‐SARS‐CoV‐2 serological response, such as age and gender during the recruitment period. However, because anti‐apoA‐1 IgG levels have been shown to be independent of most age, gender, smoking and most comorbidities (except CV ones), 20 , 21 , 22 , 23 , 24 , 25 , 26 and because of the kinetic observed on the ICU cohort together with the observations on the cohort population, we feel confident that using a recruitment‐matched case‐control cohort did induced a bias susceptible to blunt the conclusions of our present observations. Thirdly, in the context of the recent SARS‐COV‐2 variants unknown during the first epidemic wave, we could not assess the possible impact of such variants on the anti‐apoA‐1 IgG response. However, as the linear sequence homologies identified between apoA‐1 and Spike did not contain the characteristic epitope regions of the three main variants of concern (VOC) in Europe (United Kingdom, Brazilian and South‐African strains), it is unlikely that our results could be specific of a defined and currently existing SARS‐CoV‐2 strain. Fourth, if our results indicate that an acute exposure to SARS‐CoV‐2 rapidly increases the anti‐apoA‐1 IgG response, they do not allow inferring any conclusions about the possible longer term persistence of anti‐apoA‐1 IgG levels after COVID‐19 disease or the possible clinical relevance of phenomenon.
In conclusion, this report shows for the first time that in a substantial proportion of SARS‐CoV‐2‐exposed individuals, a marked humoral autoimmune response against the major lipoprotein of HDL occurs for reasons other than molecular mimicry despite the linear sequence homologies retrieved between Spike and apoA‐1 epitopes. Knowing whether the pre‐ or co‐existence of anti‐apoA‐1 IgG may modulate the course of COVID‐19 disease remains uncertain. However, as correlates of poorer prognosis in different settings, a better understanding of the possible clinical relevance of COVID‐19‐induced autoimmune biological signatures is warranted in the current COVID‐19 pandemic and ongoing vaccination programmes.
CONFLICT OF INTEREST
SP, OH and NV are named as co‐inventors on a patent related to c‐ter apoA‐1 mimetic peptides (‘Mimetic peptides for prognosis, diagnosis or treatment of a cardiovascular disease’, N° P1347EP00). N. Vuilleumier, S. Pagano and O. Hartley are named as co‐inventors of the patent related to cterA1, peptide (‘Mimetic peptides for prognosis, diagnosis or treatment of a cardiovascular disease’, N° P1347EP00) but have no other conflict of interest to disclose. The remaining authors have no conflict of interest to declare. Funding sources played no role in the design and conduct of the study, nor in the collection, analysis and interpretation of the data, nor in the preparation, review and approval of the article or decision to submit for publication.
Supporting information
Supplementary Material
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Prof. Cosson from the Geneva Antibody Facility for generous gift of reagents, Isabelle Arm‐Vernez from the virology laboratory of the laboratory medicine division for the assessment of routine SARS‐CoV‐2 IgG serology and Julien Virzi, Diego Andrey and Patrick Cohen from the laboratory medicine division. We are grateful to Dr Florence Pojer from the Protein Production and Structure Core Facility of the EPFL institute of technology of Lausanne who kindly provided purified Spike protein. Finally, we are indebted to Erik Boehm for his help in editing the manuscript and careful English revision. Open Access Funding provided by Universite de Geneve.
Pagano S, Yerly S, Meyer B, et al. SARS‐CoV‐2 infection as a trigger of humoral response against apolipoprotein A‐1. Eur J Clin Invest. 2021;51:e13661. 10.1111/eci.13661
Funding information
This study was funded by the Swiss Federal Office of Public Health, Swiss School of Public Health (Corona Immunitas research programme), the Fondation de Bienfaisance du Groupe Pictet, the Fondation Ancrage, the Fondation Privée des HUG, the Center for Emerging Viral Diseases, the De Reuter (grant No. 657) and the Schmidheiny Foundation
REFERENCES
- 1. Liu J, Li SM, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS‐CoV‐2 infected patients. Ebiomedicine. 2020;55:102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng FL, Linfeng L, Zeng J, et al. Can we predict the severity of coronavirus disease with a routine blood test? Pol Arch Intern Med. 2019;2020(130):400‐406. [DOI] [PubMed] [Google Scholar]
- 3. Carapetis JR, Beaton A, Cunningham MW, et al. Acute rheumatic fever and rheumatic heart disease. Nat Rev Dis Primers. 2016;2:1‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iseme RA, McEvoy M, Kelly B, et al. A role for autoantibodies in atherogenesis. Cardiovasc Res. 2017;113:1102‐1112. [DOI] [PubMed] [Google Scholar]
- 5. Stratton R, Slapak G, Mahungu T, Loes SKD. Autoimmunity and HIV. Curr Opin Infect Dis. 2009;22:49‐56. [DOI] [PubMed] [Google Scholar]
- 6. Gatto M, Perricone C, Tonello M, et al. Frequency and clinical correlates of antiphospholipid antibodies arising in patients with SARS‐CoV‐2 infection: findings from a multicentre study on 122 cases. Clin Exp Rheumatol. 2020;38:754‐759. [PubMed] [Google Scholar]
- 7. Guilmot A, Slootjes SM, Sellimi A, et al. Immune‐mediated neurological syndromes in SARS‐CoV‐2‐infected patients. J Neurol. 2021;268(3):751‐757. 10.1007/s00415-020-10108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schiaffino María Teresa, Di Natale Marisa, García‐Martínez Elena, et al. Immunoserologic detection and diagnostic relevance of cross‐reactive autoantibodies in coronavirus disease 2019 patients. J Infect Dis. 2020;222:1439‐1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vlachoyiannopoulos PGM, Magira Eleni, Alexopoulos Haris, et al. Autoantibodies related to systemic autoimmune rheumatic diseases in severely ill patients with COVID‐19. Ann Rheum Dis. 2020;79:1661‐1663. [DOI] [PubMed] [Google Scholar]
- 10. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe. 2020;27:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanduc D. From anti‐SARS‐CoV‐2 immune responses to COVID‐19 via molecular mimicry. Antibodies. 2020;9(3):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan M, Wu NC, Zhu XY, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS‐CoV‐2 and SARS‐CoV. Science. 2020;368:630‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farrera‐Soler L, Daguer JP, Barluenga S, et al. Identification of immunodominant linear epitopes from SARS‐CoV‐2 patient plasma. PLoS ONE. 2020;15:e0238089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poh CM, Carissimo G, Wang B, et al. Two linear epitopes on the SARS‐CoV‐2 spike protein that elicit neutralising antibodies in COVID‐19 patients. Nat Commun. 2020;11:2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shrock E, Fujimura E, Kula T, et al. Viral epitope profiling of COVID‐19 patients reveals cross‐reactivity and correlates of severity. Science. 2020;370(6520):eabd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H, Wu X, Zhang X, et al. SARS‐CoV‐2 proteome microarray for mapping COVID‐19 antibody interactions at amino acid resolution. ACS Central Sci. 2020;6(12):2238‐2249. 10.1021/acscentsci.0c00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mannic T, Satta N, Pagano S, et al. CD14 as a mediator of the mineralocorticoid receptor‐dependent anti‐apolipoprotein A‐1 IgG chronotropic effect on cardiomyocytes. Endocrinology. 2015;156:4707‐4719. [DOI] [PubMed] [Google Scholar]
- 18. Montecucco F, Braunersreuther V, Burger F, et al. Anti‐apoA‐1 auto‐antibodies increase mouse atherosclerotic plaque vulnerability, myocardial necrosis and mortality triggering TLR2 and TLR4. Thromb Haemost. 2015;114:410‐422. [DOI] [PubMed] [Google Scholar]
- 19. Pagano S, Carbone F, Burger F, et al. Anti‐apolipoprotein A‐1 auto‐antibodies as active modulators of atherothrombosis. Thromb Haemost. 2016;116:554‐564. [DOI] [PubMed] [Google Scholar]
- 20. Pagano S, Satta N, Werling D, et al. Anti‐apolipoprotein A‐1 IgG in patients with myocardial infarction promotes inflammation through TLR2/CD14 complex. J Intern Med. 2012;272:344‐357. [DOI] [PubMed] [Google Scholar]
- 21. Antiochos P, Marques‐Vidal P, Virzi J, et al. Association between anti‐apolipoprotein A‐1 antibodies and cardiovascular disease in the general population Results from the CoLaus study. Thromb Haemost. 2016;116:764‐771. [DOI] [PubMed] [Google Scholar]
- 22. Antiochos P, Marques‐Vidal P, Virzi J, et al. Impact of CD14 polymorphisms on anti‐apolipoprotein A‐1 IgG‐related coronary artery disease prediction in the general population. Arterioscler Thromb Vasc Biol. 2017;37:2342. [DOI] [PubMed] [Google Scholar]
- 23. Batuca J, Amaral M, Favas C, et al. Antibodies against hdlcomponents improve the diagnostic accuracy between ischemic stroke or coronary artery disease and healthy controls when added to traditional cardiovascular risk factors. Atherosclerosis. 2018;275:E88. [Google Scholar]
- 24. El‐Lebedy D, Rasheed E, Kafoury M, Abd‐El Haleem D, Awadallah E, Ashmawy I. Anti‐apolipoprotein A‐1 autoantibodies as risk biomarker for cardiovascular diseases in type 2 diabetes mellitus. J Diabetes Complications. 2016;30:580‐585. [DOI] [PubMed] [Google Scholar]
- 25. Vuilleumier N, Pagano S, Combescure C, et al. Non‐linear relationship between anti‐apolipoprotein A‐1 IgGs and cardiovascular outcomes in patients with acute coronary syndromes. J Clin Med. 2019;8(7):1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vuilleumier N, Rossier MF, Pagano S, et al. Anti‐apolipoprotein A‐1 IgG as an independent cardiovascular prognostic marker affecting basal heart rate in myocardial infarction. Eur Heart J. 2010;31:815‐823. [DOI] [PubMed] [Google Scholar]
- 27. Bridge SH, Pagano S, Jones M, et al. Autoantibody to apolipoprotein A‐1 in hepatitis C virus infection: a role in atherosclerosis? Hep Intl. 2018;12:17‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satta N, Pagano S, Montecucco F, et al. Anti‐apolipoprotein A‐1 autoantibodies are associated with immunodeficiency and systemic inflammation in HIV patients. J Infect. 2018;76:186‐195. [DOI] [PubMed] [Google Scholar]
- 29. Pagano S, Gaertner H, Cerini F, et al. The human autoantibody response to apolipoprotein A‐I Is focused on the C‐terminal helix: a new rationale for diagnosis and treatment of cardiovascular disease? PLoS ONE. 2015;10(7):e0132780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teixeira PC, Ducret A, Ferber P, et al. Definition of human apolipoprotein A‐I epitopes recognized by autoantibodies present in patients with cardiovascular diseases. J Biol Chem. 2014;289:28249‐28259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andrey DO, Cohen P, Meyer B, et al. Diagnostic accuracy of Augurix COVID‐19 IgG serology rapid test. Eur J Clin Invest. 2020;50(10):13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stringhini S, Wisniak A, Piumatti G, et al. Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Geneva, Switzerland (SEROCoV‐POP): a population‐based study. Lancet. 2020;396:313‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson JLC, Pagano S, Virzi J, et al. Autoantibodies to apolipoprotein A‐1 as independent predictors of cardiovascular mortality in renal transplant recipients. J Clin Med. 2019;8:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pagano S, Magenta A, D'Agostino M, et al. Anti‐ApoA‐1 IgGs in familial hypercholesterolemia display paradoxical associations with lipid profile and promote foam cell formation. J Clin Med. 2019;8(12):2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Antiochos P, Marques‐Vidal P, Virzi J, et al. Anti‐apolipoprotein A‐1 IgG predict all‐cause mortality and are associated with Fc receptor‐like 3 polymorphisms. Front Immunol. 2017;8:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chroni A, Liu T, Gorshkova I, et al. The central helices of ApoA‐I can promote ATP‐binding cassette transporter A1 (ABCA1)‐mediated lipid efflux ‐ Amino acid residues 220–231 of the wild‐type ApoA‐I are required for lipid efflux in vitro and high density lipoprotein formation in vivo. J Biol Chem. 2003;278:6719‐6730. [DOI] [PubMed] [Google Scholar]
- 37. Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1‐TLR2 heterodimer induced by binding of a tri‐acylated lipopeptide. Cell. 2007;130:1071‐1082. [DOI] [PubMed] [Google Scholar]
- 38. Vuilleumier N, Antiochos P, Marques‐Vidal P, et al. Prognostic and therapeutic considerations of antibodies against c‐ter apolipoprotein A‐1 in the general population. Clin Transl Immunol. 2020;9(12):1220. 10.1002/cti2.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crooke SN, Ovsyannikova IG, Kennedy RB, Poland GA. Immunoinformatic identification of B cell and T cell epitopes in the SARS‐CoV‐2 proteome. Sci Rep. 2020;10(1):14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woodruff MC, Ramonell RP, Nguyen DC, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID‐19. Nat Immunol. 2020;21(12):1506‐1516. 10.1038/s41590-020-00814-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smatti MK, Cyprian FS, Nasrallah GK, Al Thani AA, Almishal RO. Yassine HM. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11(8):762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamoto S, Fukuhara T, Ono C, et al. Lipoprotein receptors redundantly participate in entry of hepatitis C Virus. PLoS Pathog. 2016;12(5):e1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei C, Wan L, Yan Q, et al. HDL‐scavenger receptor B type 1 facilitates SARS‐CoV‐2 entry. Nat Metab. 2020;2(12):1391‐1400. 10.1038/s42255-020-00324-0 [DOI] [PubMed] [Google Scholar]
- 44. Tanaka S, De Tymowski C, Assadi M, et al. Lipoprotein concentrations over time in the intensive care unit COVID‐19 patients: Results from the ApoCOVID study. PLoS ONE. 2020;15:e0239573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang G, Zhang Q, Dong H, et al. Low high‐density lipoprotein level is correlated with the severity of COVID‐19 patients. Lipids Health Dis. 2020;19(1):204. 10.21203/rs.3.rs-34659/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee W, Ahn JH, Park HH, et al. COVID‐19‐activated SREBP2 disturbs cholesterol biosynthesis and leads to cytokine storm. Signal Transduct Target Ther. 2020;5(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu YN, Liu DX, Tam JP. Lipid rafts are involved in SARS‐CoV entry into Vero E6 cells. Biochem Biophys Res Comm. 2008;369:344‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wallukat G, Hohberger B, Wenzel K, et al. Functional autoantibodies against G‐protein coupled receptors in patients with persistent Long‐COVID‐19 symptoms. J Transl Autoimmun. 2021;4:100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khamsi R. Rogue antibodies could be driving severe COVID‐19. Nature. 2021;590:29‐31. [DOI] [PubMed] [Google Scholar]
- 50. Borghi MO, Beltagy A, Garrafa E, et al. Anti‐phospholipid antibodies in COVID‐19 are different from those detectable in the anti‐phospholipid syndrome. Front Immunol. 2020;11:584241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nazy I, Jevtic SD, Moore JC, et al. Platelet‐activating immune complexes identified in critically ill COVID‐19 patients suspected of heparin‐induced thrombocytopenia. J Thromb Haemost. 2021;19(5):1342‐1347. 10.1111/jth.15283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material


