Abstract
Six candidate overlapping genes have been detected in SARS-CoV-2, yet current methods struggle to detect overlapping genes that recently originated. However, such genes might encode proteins beneficial to the virus, and provide a model system to understand gene birth. To complement existing detection methods, I first demonstrated that selection pressure to avoid stop codons in alternative reading frames is a driving force in the origin and retention of overlapping genes. I then built a detection method, CodScr, based on this selection pressure. Finally, I combined CodScr with methods that detect other properties of overlapping genes, such as a biased nucleotide and amino acid composition. I detected two novel ORFs (ORF-Sh and ORF-Mh), overlapping the spike and membrane genes respectively, which are under selection pressure and may be beneficial to SARS-CoV-2. ORF-Sh and ORF-Mh are present, as ORF uninterrupted by stop codons, in 100% and 95% of the SARS-CoV-2 genomes, respectively.
Keywords: Codon usage, Membrane protein, Multivariate statistics, Overlapping reading frame, Selection pressure, Spike protein, Virus evolution
1. Introduction
Coronaviruses (subfamily Orthocoronavirinae, family Coronaviridae, order Nidovirales) are enveloped viruses with positive-sense single-stranded RNA genomes, which are unusually long (27–32 kb) if compared to those of other RNA viruses (reviewed by Gorbalenya et al., 2006). The 5′ terminal two-thirds of the genome encodes polyproteins pp1a and pp1ab, which are processed to yield 16 non-structural proteins (Snijder et al., 2003). The 3′ terminal one-third of the genome contains genes encoding the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as a variable number of ORFs encoding accessory proteins (reviewed by Liu et al., 2014; Cui et al., 2019). They are expressed from a nested 3′ coterminal set of subgenomic RNAs having at their 5’ end a common leader sequence (reviewed by Sola et al., 2015).
The 3’ genome region of Alpha, Beta, Gamma, and Deltacoronavirus (the four genera in the subfamily Orthocoronavirinae) shows conservation of structural genes and gain of new accessory ORFs by overprinting (Cui et al., 2019). Overprinting is the process by which a pre-existing gene that encodes only one protein undergoes nucleotide substitutions inducing the expression of a novel protein from an alternative reading frame (Miyata and Yasunaga, 1978; Keese and Gibbs, 1992). Overlapping genes are particularly abundant in viruses (Chirico et al., 2010; Schlub and Holmes, 2020), where they constitute a rich source of new proteins (Rancurel et al., 2009).
The gain of new accessory ORFs by overprinting was demonstrated in SARS-CoV, a virus of the species Severe acute respiratory syndrome-related coronavirus responsible for the 2002–2003 SARS outbreak in China. The genome of SARS-CoV, indeed, contains 2 overlapping ORFs whose expression in infected human tissues was validated experimentally (Chan et al., 2005). The first, called ORF3b and nested within the accessory gene ORF3a, encodes a 3b protein inducing apoptosis in transfected cells (Khan et al., 2006) and inhibiting the host interferon response (Kopecky-Bromberg et al., 2007). The other, called ORF9b and nested within the structural gene N, encodes a 9b virion-associated protein (Xu et al., 2009) acting as antagonist of the innate immune response to viral replication (Shi et al., 2014).
The finding that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease 2019 (Gorbalenya et al., 2020; Zhou et al., 2020), shows a nucleotide difference of 20% with respect to SARS-CoV has stimulated several studies. For example, Cagliani et al. (2020) found that SARS-CoV-2 has lost the ability to encode a full-length protein 3b, due to the appearance of premature stop codons in ORF3b. On the other hand, at least six ORFs, overlapping well-characterized SARS-CoV-2 genes (S, ORF3a and N) in alternative reading frames, have been hypothesized to encode functional proteins. Their names, in accordance to the consensus nomenclature by Jungreis et al. (2021a), are reported in Table 1 . Their location in the 3’ genome region of SARS-CoV-2 is shown in Fig. 1 : ORFs shaded in gray have been discovered experimentally or computationally, while dotted ORFs show evidence for translation or function but are undetectable by prediction methods (ORFs shaded in black have been discovered in the present study).
Table 1.
List of the six overlapping ORFs detected in SARS-CoV-2.
| ORF name | Type of experimental evidence for expression or function | Type of computational method for detection |
|---|---|---|
| ORF2b | Ribosome profiling (Finkel et al., 2021), immunopeptidomics (Weingarten-Gabbay et al., 2020) | – |
| ORF3b | Interferon antagonist, when expressed from a plasmid in Sendai-virus infected cells (Konno et al., 2020) | – |
| ORF3c | Ribosome profiling (Finkel et al., 2021) | Synplot2 (Firth, 2014, 2020), PhyloCSF (Lin et al., 2011; Jungreis et al., 2021b) |
| ORF3d | Ribosome profiling (Finkel et al., 2021), antibodies (Hachim et al., 2020) | Codon permutation method (Schlub et al., 2018; Nelson et al., 2020b), sequence-composition method (Pavesi, 2020) |
| ORF9b | Ribosome profiling (Finkel et al., 2021), immunopeptidomics (Weingarten-Gabbay et al., 2020), suppressor of interferon response (Jiang et al., 2020) | GOFIX (Michel et al., 2020), PhyloCSF (Lin et al., 2011; Jungreis et al., 2021b) |
| ORF9c | Suppressor of antiviral response, when expressed from a plasmid in transfected cells (Dominguez Andres et al., 2020) | GOFIX (Michel et al., 2020), sequence-composition method (Pavesi, 2020) |
Fig. 1.
Location of the eight overlapping ORFs detected in the 3′ genome region of SARS-CoV-2.
The aim of this study was the detection of candidate overlapping ORFs in the genome of SARS-CoV-2. The term “ORF” indicates a contiguous stretch of codons, beginning with the most upstream AUG codon, ending with the nearest downstream stop codon, and not interrupted by in-frame stop codons. Candidate ORF means an ORF which is under selection pressure and may be beneficial to the virus. Separate experimental evidence is needed to determine if the candidate ORF is indeed translated independently or in conjunction with another ORF and encodes a functional protein or its part during virus infection.
This study combines two statistical methods published previously into a unique prediction method. It includes the codon scrambling (CodScr) method, which is an extension of a codon usage test originally developed to predict a novel overlapping ORF in human hepatitis G virus (Pavesi, 2000). The method is also an extension of the sequence composition (SeqComp) method, which separated with high accuracy overlapping genes from non-overlapping genes in viruses using two prediction scores, both depending on a significantly different nucleotide and amino acid composition (Pavesi, 2020). The method is an extension because it adds two prediction scores to the two used previously.
The rationale behind this approach was to provide a highly selective method (CodScr + SeqComp) for the prediction of overlapping ORFs in viruses. Detection of a candidate overlapping ORF, indeed, depends on a match to five prediction criteria: one from the CodScr method and four from the SeqComp method. When applied to the 3’ genome region of SARS-CoV-2, CodsScr + SeqComp identified two overlooked overlapping ORFs that are under selection pressure. They were named ORF-Sh (h stands for hypothetical) and ORF-Mh, because they are nested within the spike and membrane genes respectively.
2. Materials and methods
2.1. Description of the codon scrambling (CodScr) method
The method is based on the assumption that in overlapping genes the use of synonymous codons in the ancestral frame is significantly biased, to avoid the appearance of premature stop codons in the novel frame. To validate this assumption, and also to evaluate the sensitivity of the method, it was essential to have a dataset of overlapping genes with known genealogy. The “genealogy” of the overlap is identifying which frame is ancestral and which one is de novo. This can be done by examining their phylogenetic distribution, under the assumption that the frame with the most restricted distribution is the de novo one (Rancurel et al., 2009). When this approach is not applicable, because the two frames have an identical phylogenetic distribution, the genealogy of the overlap can be inferred using the codon usage method. It assumes that the ancestral frame, which has co-evolved with the other viral genes over a long period of time, has a distribution of synonymous codons significantly closer to that of the viral genome than the de novo frame (Pavesi et al., 2013).
In a previous study, I could predict the genealogy of 46 viral overlapping genes using the phylogenetic and codon usage methods (Table S1 in Pavesi, 2020). In the same study, by extending the inferred genealogy to the respective homologs, I obtained a dataset of 194 overlapping genes with a known ancestral and novel frame: 126 overlaps with a novel frame shifted one nucleotide 3′ with respect to the ancestral one (+1 overlap) and 68 overlaps with a novel frame shifted two nucleotides 3’ (+2 overlap) (see Files S2 and S3 in Pavesi, 2020).
In the present study, I used this dataset as benchmark to evaluate the sensitivity of the CodScr method. The sensitivity, also called the true-positive rate, was the percent frequency of overlapping genes correctly classified as true positive. For each of the 194 ancestral frames, I first obtained by codon scrambling a total of 10,000 replicates, all encoding the same amino acid sequence but using different synonymous codons. The change of synonyms was proportional to the pattern of codon usage in the virus under examination. Consider, for example, an ancestral frame encoding a 100 amino acid (aa) long protein. In each replicate, the change of synonyms concerns all the amino acids with a six-fold, four-fold, three-fold, and two-fold codon degeneracy. Thus, the number of scrambled codons is 100 minus the number of codons for methionine and tryptophan, the two amino acids encoded by a single codon.
I then calculated the percent frequency of replicates in which the change of synonyms in the ancestral frame yielded an alternative frame interrupted by stop codons. A frequency higher than 95% supported the hypothesis that the use of synonyms in the ancestral frame is significantly biased (P <0.05), to avoid premature stop codons in the novel frame. The sensitivity of the method was the percent frequency of the overlapping genes examined (a total of 194) having a P-value <0.05, and thus correctly classified as true positives.
I evaluated the specificity, also called the true-negative rate, of the CodScr method by first accessing a large dataset of non-overlapping genes (1,723,968 nt) that I previously collected from 244 viral genome sequences (File S1 in Pavesi, 2020). By sequence analysis of the dataset, I assembled a smaller dataset composed of 3868 spurious overlapping ORFs with a minimum length of 90 nt (from AUG to stop codon). The great majority of them (86.4%) were shifted one nucleotide 3′ with respect to the protein-coding sequence, while the remaining ones (13.6%) were shifted two nucleotides 3’. I used this dataset as benchmark to assess the specificity of the method.
For each overlap, I first obtained by codon scrambling a total of 1000 replicates of the protein-coding sequence, all encoding the same amino acids but using different synonymous codons. The change of synonyms was proportional to the codon usage in the virus under examination and the number of scrambled codons was as detailed above. I then calculated the percent frequency of replicates in which the change of synonyms in the protein-coding sequence yielded an alternative frame interrupted by stop codons. In the case of a frequency lower than 95%, I classified the overlap as true negative. The specificity of the method was the percent frequency of the spurious overlapping ORFs examined (a total of 3868) correctly classified as true negatives.
2.2. Prediction of overlapping ORFs in the 3’ genome region of SARS-CoV-2
I analyzed the 3’ genome region of the reference sequence of SARS-CoV-2 (Ac. Number NC_045512.2) starting from the AUG initiation codon of gene S (nt 21,563) and ending to the stop codon of ORF10 (nt 29,674). In addition to S and ORF10, the region contains three structural genes (E, M and N) and six accessory ORFs (ORF3a, ORF6, ORF7a, ORF7b, and ORF8) (Fig. 1). Using as cut-off a minimum length of 90 nt, I selected a total of 30 ORFs overlapping genes S, ORF3a, M, N, and ORF8 (see Results). This cut-off is lower than the arbitrary cut-offs used in genome annotation (150 or 300 nt), because translation of small viral ORFs was proven by ribosome profiling and there exist proteins shorter than 50 aa with established roles in virus infection (reviewed by Finkel et al., 2018).
I analyzed each overlap using five prediction criteria. The first came from the CodScr method, which evaluated if the use of synonymous codons in the SARS-CoV-2 gene is significantly biased (P <0.05 from 10,000 replicates), to avoid the appearance of premature stop codons in the overlapping ORF.
The other criteria came from the SeqComp method. It analyzed the nucleotide and amino acid composition of the overlap, yielding four prediction scores: PLS-DA score, LDA-score, LDA-ancestral score, and LDA-novel score. The usefulness of PLS-DA score stems from the finding (Pavesi, 2020) that the partial least-squares discriminant analysis (PLS-DA) correctly classified 95% of overlapping genes (PLS-DA score <0) and 98% of non-overlapping genes (PLS-DA score >0) (Supplementary Fig. S1). The usefulness of LDA score depends on the finding (Pavesi, 2020) that the Fisher's linear discriminant analysis (LDA) correctly classified 96% of overlapping genes (LDA score below the cut-off −35.31) and 97% of non-overlapping genes (LDA score above the cut-off −35.31) (Supplementary Fig. S2).
The usefulness of LDA-ancestral score and LDA-novel score depends on the finding that LDA, when applied to overlapping genes with known genealogy, separated ancestral from novel frames with high accuracy (Pavesi, 2020). In the case of a novel frame shifted one nucleotide 3′ with respect to the ancestral one, LDA assigned a score above the cut-off 17.20 to 97% of ancestral frames and a score below 17.20 to 98% of novel frames (Supplementary Fig. S3A). In the case of a novel frame shifted two nucleotides 3’, LDA assigned a score above the cut-off −34.98 to 100% of ancestral frames and a score below −34.98 to 100% of novel frames (Supplementary Fig. S3B).
Calculation of the two latter scores requires a knowledge of the genealogy of the overlap. Here, I assumed that the structural (S, E, M, and N) and accessory genes (ORF3a, ORF6, ORF7a, ORF7b, and ORF8) of SARS-CoV-2 are pre-existing ancestral genes, because of their conservation across sarbecoviruses (Cui et al., 2019). Sarbecovirus is a subgenus of Betacoronavirus containing only the species Severe acute respiratory syndrome-related coronavirus, which includes many viruses of different hosts such as human, pangolin and bat. Further evidence for the ancestry of ORF3a and N was provided by a phylogenetic and codon usage analysis of the overlaps ORF3a/ORF3b and N/ORF9b in the closely related SARS-CoV (Pavesi, 2020).
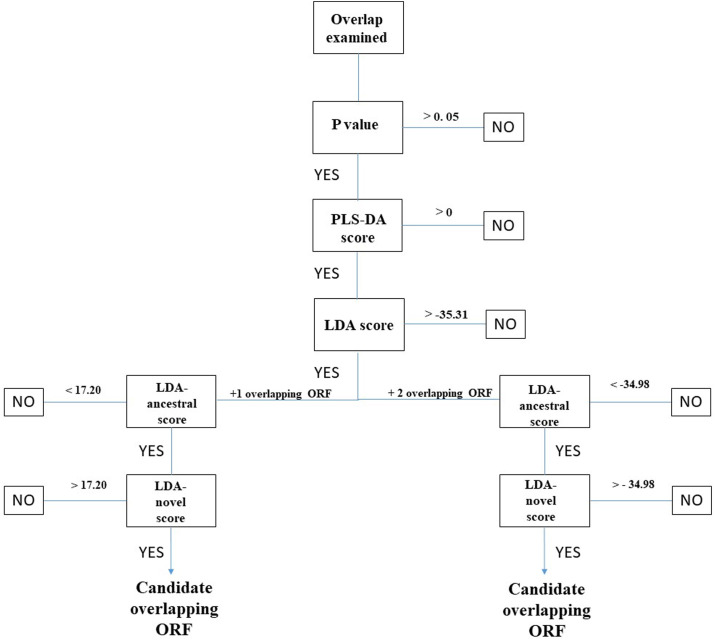
The workflow reported in Fig. 2 summarizes the five steps of the prediction method. In the analysis of the 30 ORFs overlapping genes S, ORF3a, M, N, and ORF8, I retained as candidates only those matching all five prediction criteria. Candidate means an ORF under selection pressure and potentially beneficial to the virus. A detailed example of calculation of the five prediction scores for an ORF nested within gene N with a shift of one nucleotide 3′, as well as for an ORF nested within gene M with a shift of two nucleotides 3’, is shown in Supplementary File S1.
Fig. 2.
Example workflow for CodScr + SeqComp analysis. As input data, CodScr + SeqComp requires the nucleotide sequence of a protein coding region (the ancestral reading frame) which contains an overlapping ORF shifted one nucleotide 3’ (+1 overlapping ORF) or an overlapping ORF shifted two nucleotides 3’ (+2 overlapping ORF). ORF indicates a contiguous stretch of codons, beginning with a start AUG codon, ending with a stop codon, not interrupted by premature stop codons, and having a length ≥ 90 nt. A detailed example of calculation of the five prediction scores (P-value, PLS-DA score, LDA score, LDA-ancestral score, and LDA-novel score) is shown in Supplementary File S1.
2.3. Comparative analysis of the candidate overlapping ORFs against the NCBI and GISAID databases
Using TBLASTN, I first compared the predicted protein of a candidate overlapping ORF against the nucleotide collection NCBI database translated in all reading frames, with the aim to assess if it is unique to SARS-CoV-2 or evolutionarily conserved across coronaviruses. Using BLASTN and TBLASTN, I then carried out a comparative analysis against the NCBI collection of SARS-CoV-2 genomes, updated to 28 May 2021 and containing over 500,000 complete genomes. By this analysis, I could determine the extent of conservation of a candidate overlapping ORF, after exclusion of the genome sequences in which the ORF is interrupted by premature stop codons. This analysis was extended using the sequence analysis pipeline included in the GISAID database (Elbe and Buckland-Merrett, 2017), updated to 28 May 2021 and containing over 1,000,000 genome sequences of SARS-CoV-2 (https://www.gisaid.org/). The search for transmembrane domains in the predicted protein of a candidate ORF was carried out using TMpred (https://embnet.vital-it.ch/software/TMPRED_form.htlm).
3. Results and discussion
3.1. Rationale of the study
To identify viral overlapping genes by sequence analysis, several groups have developed methods that detect the atypical pattern of nucleotide substitution induced by the overlap. For example, Synplot2 (Firth, 2014) analyzes alignments of protein-coding sequences to identify regions where there is a significantly reduced rate of synonymous substitution, a characteristic feature of overlapping functional elements such an overlapping gene or a conserved RNA structure. The same approach was followed by Sealfon et al. (2015), who developed a phylogenetic codon-model based method named FRESCo (Finding Regions of Excess Synonymous Constraints). Analogously, OLGenie (OLG means OverLapping Gene) is a method that estimates signs of strong purifying (negative) selection in aligned sequences, as hallmark of functional overlapping genes (Nelson et al., 2020a).
Although powerful, these methods are constrained by the requirement for multiple sequences of sufficient nucleotide diversity to predict overlapping genes. To overcome this drawback, Schlub et al. (2018) developed a codon permutation method that detects candidate overlapping genes in single viral sequences, by selecting ORFs that are significantly longer than expected by chance. Another method working on single sequences is GOFIX (Michel et al., 2020), which predicts overlapping ORFs on the basis of a significant enrichment in the X motif (a set of 20 codons over-represented in viral genes).
This study combines two previous methods (Pavesi, 2000, 2020) into a unique prediction method (CodScr + SeqComp). It can predict overlapping ORFs in single genome sequences as the Schlub's method, but has the advantage of greater sensitivity and specificity (see below paragraphs 3.3 and 3.6). As summarized in Fig. 2, the prediction of overlapping ORFs by CodScr + SeqComp does require a match to five criteria. They cover several features of known overlapping genes, such as a peculiar use of synonymous codons in the ancestral member of the pair and a peculiar nucleotide and amino acid composition.
3.2. Performance and limitations of the CodScr method in the prediction of overlapping genes
The sensitivity of the method was quite high (85.6%). With a cut-off P-value <0.05, CodScr correctly classified as true positive 166 out of 194 overlapping genes in the benchmark dataset. The sensitivity was 100% for the longest overlapping genes (57 overlaps with a length >600 nt). It decreased to 82.1% for the shortest overlapping genes (67 overlaps with a length from 144 to 300 nt) and to 77.1% for overlapping genes of intermediate length (70 overlaps with a length from 303 to 600 nt).
The sensitivity was remarkably high (98.5%) for overlapping genes with a novel frame shifted two nucleotides 3′ with respect to the ancestral one (67 overlaps correctly predicted out of 68). It decreased to 78.6% for overlapping genes with a novel frame shifted one nucleotide 3’ (99 overlaps correctly predicted out of 126).
These results validate the assumption that the shaping of codon usage in the ancestral frame, to avoid premature stop codons in the novel frame, is a relevant driving force in the stability and evolution of viral overlapping genes. However, a limitation of CodScr is that the sensitivity (85.6%) is lower than the threshold of statistical significance (95%). The most likely explanation is that the pattern of nucleotide substitution is affected by further constraints, such as the inherent adaptive conflict between two proteins encoded by the same gene (Peleg et al., 2004; Sabath et al., 2012; Simon-Loriere et al., 2013). Indeed, the evolution of overlapping genes is a complex process, which can be symmetric in some cases (similar selection pressures, strong or weak, on the two encoded proteins) or asymmetric in others (the novel protein shows a number of amino acid substitutions significantly higher than that of the ancestral protein) (Pavesi, 2019).
Another limitation of CodScr is that the specificity (65.8%) is largely below the threshold of statistical significance (95%). Indeed, CodScr correctly classified as true negative no more than two-thirds of the spurious overlapping ORFs examined (2543 out of 3868). The reason could depend on the presence of gene regions with a strong codon bias, which are paired purely by chance to short overlapping ORFs not interrupted by stop codons. In this case, codon bias can reflect other biological processes, such as slowdown of the translation elongation rate (Aragonés et al., 2010), regulation of gene expression (Shin et al., 2015) and stability of genomic RNA (Gumpper et al., 2019).
3.3. Comparison of the CodScr method with the Schlub's method
The method developed by Schlub et al. (2018) detects candidate overlapping genes in viruses by selecting overlapping ORFs that are significantly longer than expected by chance. It consists of a codon permutation test and a synonymous mutation test. In the first, the expected length of overlapping ORFs is estimated by randomly permuting codon positions in the original reading frame. In the other, the codon order is unchanged and random synonymous mutations are introduced in the original reading frame, before measuring ORF lengths in the other frames.
The sensitivity of the CodScr method was 85.6% with P <0.05 and 78% with P <0.01. As the minimum length of the overlapping genes I examined was 144 nt, I could compare the sensitivity of 78% with that reported in Schlub et al. (2018), as obtained from analysis of overlapping genes longer than 100 nt and with a chosen cut-off P-value <0.01. The sensitivity of CodScr (78%) was higher than that obtained both from the codon permutation test (65%) and the synonymous mutation test (71%). The specificity of CodScr, albeit low (65.8%), was considerably higher than that reported for both tests (around 40%, as deduced from Fig. 3 in Schlub et al., 2018).
Fig. 3.
Nucleotide and amino acid sequence of the two predicted overlapping ORFs in the 3′ genome region of SARS-CoV-2. (A) Overlapping ORF-Sh: the nucleotide sequence (from nt 24,050 to 24,172) encodes the region of protein S spanning residues 830–868, while the +1 overlapping ORF-Sh (from nt 24,051 to 24,170) encodes a predicted protein of 39 aa (underlined characters). Bold characters indicate a predicted transmembrane helix. (B) Overlapping ORF-Mh: the nucleotide sequence (from nt 26,691 to 26,873) encodes the region of protein M spanning residues 57–116, while the +2 overlapping ORF-Mh (from nt 26,693 to 26,872) encodes a predicted protein of 59 aa (underlined characters). Bold characters indicate two predicted transmembrane helices.
3.4. Prediction of eight overlapping ORFs in the 3’ genome region of SARS-CoV-2
In the 3′ genome region of the reference sequence of SARS-CoV-2 (NC_045512.2), I found a total of 30 overlapping ORFs with a length from 90 to 294 nt (Supplementary Table S1). The majority of them (25 out of 30) were shifted one nucleotide 3′ with respect to the protein-coding sequence, while the remaining ones were shifted two nucleotides 3’. In the case of multiple in-frame AUG initiation codons, I considered both the longest overlapping ORF and the shortest one(s).
I analyzed each overlap using the CodScr and SeqComp methods. CodScr yielded the P-value, while SeqComp yielded the PLS-DA, LDA, LDA-ancestral, and LDA-novel scores. The five prediction scores were compared to the respective cut-off values (Fig. 2). By this approach, I found eight overlaps meeting all five prediction criteria (Table 2 ) and thus containing a candidate overlapping ORF. Candidate means an ORF under selection pressure and potentially beneficial to the virus.
Table 2.
List of the eight overlapping ORFs in the 3’ genome region of SARS-CoV-2 meeting all five prediction criteria of the CodScr + SeqComp method.
| Overlapping ORFa | Genome positionb | Length (nt) | Within gene (genome position) | Shift of the overlapping ORF | P-value from the CodScr method | PLS-DA score | LDA score | LDA-ancestral score | LDA-novel score | Prediction criteria met |
|---|---|---|---|---|---|---|---|---|---|---|
| nORF1 | 24051–24170 | 120 | S (24050–24172) | +1 | 0.0001 | −0.45 | −37.03 | 37.07 | −1.54 | 5 |
| nORF2* | 24072–24170 | 99 | S (24071–24172) | +1 | 0.0001 | −0.13 | −35.41 | 37.08 | −8.74 | 5 |
| nORF3 | 25457–25582 | 126 | ORF3a (25456–25584) | +1 | 0.04 | −1.46 | −41.89 | 21.48 | 12.76 | 5 |
| nORF4 | 25524–25697 | 174 | ORF3a (25522–25698) | +2 | 0.02 | −1.32 | −40.44 | −27.80 | −59.59 | 5 |
| nORF5 | 26693–26872 | 180 | M(26691–26873) | +2 | 0.003 | −0.02 | −36.26 | −17.74 | −60.62 | 5 |
| nORF6 | 28284–28577 | 294 | N (28283–28579) | +1 | 0.0001 | −0.52 | −39.17 | 27.12 | 11.56 | 5 |
| nORF7* | 28305–28577 | 273 | N (28304–28579) | +1 | 0.0001 | −0.35 | −39.29 | 26.88 | 12.29 | 5 |
| nORF8* | 28359–28577 | 219 | N (28358–28579) | +1 | 0.0001 | −0.21 | −37.77 | 27.78 | 14.76 | 5 |
Term “n” stands for “new” and asterisk indicates an overlapping ORF starting with an AUG codon which is in frame with respect to the previous overlapping ORF.
The boundaries of the overlapping ORF are referred to the reference genome sequence of SARS-CoV-2 (NC_045512.2).
In the case of overlapping ORFs with multiple in-frame AUG initiation codons, Table 2 shows the prediction scores both for the longest overlapping ORF and the shortest one(s). For example, nested within gene N, I found three overlapping ORFs starting with different in-frame AUG codons (nORF6, nORF7* and nORF8*) and having a length of 294, 273 and 219 nt, respectively.
3.5. Identification of two novel overlapping ORFs in the 3’ genome region of SARS-CoV-2
I summarized the content of Table 2 by considering as candidate only the longest overlapping ORF. As shown in Table 3 , CodScr + SeqComp identified in the 3’ genome region of SARS-CoV-2 five candidate ORFs. The first three (ORF3c, ORF3d and ORF9b) were also predicted by other computational methods (Table 1). CodScr + SeqComp did not identify as candidate ORF9c, because of a P-value >0.05 (see ORF29 in Supplementary Table S1). Analogously, it did not identify as candidate ORF2b (three criteria met, see ORF1 in Supplementary Table S1), ORF3d-2 (a shorter isoform of ORF3d, four criteria met, see ORF19 in Supplementary Table S1), and ORF3b. The latter ORF (two criteria met, data not shown) was examined separately from the others, because of a length below the cut-off of 90 nt. The last two candidate overlapping ORFs in Table 3 were predicted only by CodScr + SeqComp. I called them ORF-Sh (h stands for hypothetical) and ORF-Mh, because they are nested within the spike and membrane genes respectively (see ORFs shaded in black in Fig. 1).
Table 3.
List of the five candidate overlapping ORFs in the 3’ genome region of SARS-CoV-2 (italic characters indicate ORFs predicted by other methods; bold characters indicate ORFs predicted only by the CodScr + SeqComp method).
| Candidate overlapping ORF (length) | Ancestral overlapping gene | Boundaries of candidate overlapping ORF | Shift vs. ancestral gene | P value from CodScr | PLS-DA score | LDA score | LDA-ancestral score | LDA-novel score | Prediction criteria met |
|---|---|---|---|---|---|---|---|---|---|
| ORF3c (126 nt) | ORF3a | 25457–25582 | +1 | 0.04 | −1.46 | −41.89 | 21.48 | 12.76 | 5 |
| ORF3d (174 nt) | ORF3a | 25524–25697 | +2 | 0.02 | −1.32 | −40.44 | −27.80 | −59.59 | 5 |
| ORF9b (294 nt) | N | 28284–28577 | +1 | 0.0001 | −0.52 | −39.17 | 27.12 | 11.56 | 5 |
| ORF-Sh (120 nt) | S | 24051–24170 | +1 | 0.0001 | −0.45 | −37.03 | 37.07 | −1.54 | 5 |
| ORF-Mh (180 nt) | M | 26693–26872 | +2 | 0.003 | −0.02 | −36.26 | −17.74 | −60.62 | 5 |
aBoundaries of the overlapping ORF are referred to the reference genome sequence of SARS-CoV-2 (NC_045512.2).
I found that the coding sequence of the putative ORF-Sh protein (39 aa) overlaps the coding sequence of the S protein from residue 830 to 868, and that it contains a predicted transmembrane helix (Fig. 3A). Using TBLASTN against the NCBI nucleotide collection, I found that the protein is potentially encoded by very few sarbecoviruses (90% of identity with the homologous predicted protein of pangolin coronavirus MP789, 85% with that of bat coronavirus RaTG13, and 76% with that of pangolin coronavirus PCoV_GX-P5L). In contrast, I found that the protein cannot be expressed by SARS-CoV, because the region homologous to ORF-Sh of SARS-CoV-2 contains 2 premature stop codons.
To see whether the sarbecoviruses having ORF-Sh are monophyletic or instead scattered over the phylogeny, I examined the two genomic trees reported respectively in Liu et al. (2020) and Lam et al. (2020). In the first, I found that SARS-CoV-2, Bat-CoV-RaTG13 and Pangolin-CoV-2020 (isolate MP789) cluster together into a clade which is significantly differentiated from that including Bat-CoV-ZC45 and Bat-CoV-ZXC21. This kind of phylogenetic relatedness was confirmed in the other tree, which contains also the pangolin coronavirus PCoV_GX-P5L. The finding that both Bat-CoV-ZC45 and Bat-CoV- ZXC21 lack ORF-Sh, because of one and two premature stop codons respectively, supports the hypothesis that ORF-Sh emerged in the ancestor of the clade including pangolin coronaviruses, Bat-CoV-RaTG13 and SARS-CoV-2, and that ORF-Sh has been kept because it is beneficial to the virus. The age of the clade was inferred in the middle seventeenth century (Boni et al., 2020).
I found that ORF-Sh is strongly conserved in SARS-CoV-2. Using TBLASTN and BLASTN against the NCBI collection of SARS-CoV-2 genomes (517,664 records), I detected only 65 sequences in which synonymous or non-synonymous substitutions in gene S caused the appearance of a premature stop codon in ORF-Sh. Using the sequence analysis pipeline from GISAID database (1,352,146 records covering the complete genome of SARS-CoV-2 or the nucleotide sequence of gene S), I detected only 132 ORF-Sh sequences interrupted by a premature stop codon. Both analyses demonstrated a conservation close to 100%.
No evidence for translation of ORF-Sh was provided by ribosome profiling studies (Noam Stern-Ginossar, personal communication). However, there is evidence for non-canonical subgenomic RNAs whose transcription starts immediately upstream the AUG initiation codon of ORF-Sh, located at nucleotide position 24051 (see Fig. 4A in Parker et al., 2021). As ORF-Sh is far from the 5’ end of the subgenomic RNA for S (Fig. 1), a mechanism of expression could be a non-canonical subgenomic RNA that allows access to ORF-Sh. A possible transcription regulatory sequence (TRS) ACAAAG, similar to the canonical TRS ACGAAC, occurs 18 nt upstream the AUG start codon of ORF-Sh.
The other new overlapping ORF detected by CodScr + SeqComp was ORF-Mh. The putative encoded protein (59 aa) overlaps protein M from residue 57 to 116, shows a high content (13.6%) of cysteine residues, and contains two predicted transmembrane helices (Fig. 3B). Using TBLASTN, I found that ORF-Mh is unique to SARS-CoV-2. Indeed, the region homologous to ORF-Mh in SARS-CoV and related sarbecoviruses contains 2 premature stop codons.
Although less conserved than ORF-Sh, ORF-Mh shows a conservation around 95%. Using TBLASTN and BLASTN against the NCBI collection of SARS-CoV-2 genomes (517,664 records), I detected a total of 7675 sequences in which a synonymous substitution in gene M caused a premature stop codon in ORF-Mh. The substitution was a third-base U to C transition at the following nucleotide positions: 26,735 (4982 sequences), 26,801 (1670 sequences), 26,822 (573 sequences), 26,858 (426 sequences), and 26,864 (24 sequences). Using the sequence analysis pipeline from GISAID database (1,030,210 records of the complete SARS-CoV-2 genome), I detected a total of 48,509 ORF-Mh sequences interrupted by a premature stop codon. Therefore, conservation of ORF-Mh ranged from 98.5% in NCBI database to 95.3% in GISAID database. The lack of ORF-Mh in 5% of SARS-CoV-2 sequences suggests that ORF-Mh, being a recently born overlapping ORF, is under still a low negative selection pressure.
Expression of the ORF encoding protein M is under the control of a transcription regulatory sequence (ACGAAC, immediately upstream the AUG start codon), which drives transcription of a canonical subgenomic RNA. In accordance to the consensus sequence for initiation of translation (GCCA/GCCAUGG) (Kozak, 1987), ORF-Mh has a favorable initiation context. Its AUG start codon, indeed, is preceded by G at −3 and −6, by C at −5, and it is followed by G at +4 (GCUGUUAUGG), suggesting that there is potential for efficient initiation of translation. However, no experimental evidence for translation of ORF-Mh was provided by ribosome profiling studies (Noam Stern-Ginossar, personal communication).
3.6. The CodScr + SeqComp method has the advantage of a good specificity (85.2%)
I used the dataset of 3868 spurious overlapping ORFs as benchmark to evaluate the specificity of CodScr + SeqComp. As reported in paragraph 3.2, the CodScr method has the advantage of a good sensitivity (85.6%) but the limitation of a poor specificity (65.8%). In a previous study (Pavesi, 2020), I found that the sensitivity of the PLS-DA and LDA scores used jointly is notably high, as they correctly classified 94.2% of overlapping genes and 97.1% of non-overlapping genes (Supplementary Fig. S4). In this study, however, I found that this approach has a specificity remarkably low (43.1%), because PLS-DA and LDA scores correctly classified as true negatives only 1667 out of 3868 spurious overlaps. The inclusion in SeqComp of two other prediction scores (LDA-ancestral and LDA-novel scores) increased the specificity to 61.8%, as 2392 out of 3868 spurious overlaps were correctly classified as true negatives. By combining CodScr with SeqComp, under the rule of a match to all five prediction criteria, I obtained an even higher specificity (85.2%). I found, indeed, that 3295 out of 3868 spurious overlaps were correctly classified as true negatives.
A further validation of CodScr + SeqComp came from analysis of the overlapping ORFs nested in the 5′ genome region of SARS-CoV-2. As these ORFs tend to be less accessible for translation, I used them as an additional negative control. I analyzed the 5′ genome region of the reference sequence of SARS-CoV-2 (Ac. Number NC_045512.2) starting from the AUG initiation codon of gene ORF1a (nt 266) and ending to the stop codon of gene ORF1ab (nt 21,555). I detected a total of 57 overlapping ORFs (38 nested within ORF1a and 19 within ORF1ab) having a length from 93 to 249 nt. As shown in Supplementary Table S2, the great majority of them (55 out of 57) were predicted by CodScr + SeqComp as spurious overlaps, because they did not match the five prediction criteria of the method. The number of the candidate overlapping ORFs found in the 5′ genome region of SARS-CoV-2 (2 out of 57 ORFs, see bold characters in Supplementary Table S2) was significantly lower than that found in the 3’ genome region (8 out of 30 ORFs, see Table 2) (chi-square = 8.21; P <0.005).
Overall, these features make CodScr + SeqComp a useful tool for predicting overlapping ORFs in viruses. Unlike Synplot2 (Firth, 2014), FRESCo (Sealfon et al., 2015) and OLGenie (Nelson et al., 2020a), it can examine single genome sequences, without the need for multiple protein-coding sequences with a substantial degree of nucleotide diversity. CodScr + SeqComp could be particularly effective in the prediction of overlapping ORFs born recently and thus showing a restricted phylogenetic distribution. Indeed, the two novel ORFs discovered in this study have both a narrow phylogenetic range: ORF-Sh occurs only in a clade including SARS-CoV-2 and sarbecoviruses infecting Manis javanica (pangolin) and Rhinolophus affinis (bat), while ORF-Mh is unique to SARS-CoV-2.
Analysis of the 3’ genome region of SARS-CoV-2 by the present method can be compared to that performed by Jungreis et al. (2021b) using PhyloCSF, a computational tool to detect evolutionary signatures of protein-coding regions (Lin et al., 2011). By analysis of 44 Sarbecovirus genomes, Jungreis et al. (2021b) found strong protein-coding signatures for the overlapping ORFs ORF3c and ORF9b, but not for the overlapping ORFs ORF2b, ORF3b, ORF3d and its isoform ORF3d-2, and ORF9c. CodScr + SeqComp differed from PhyloCSF because it predicted as candidate also ORF3d, in addition to ORF3c and ORF9b (Table 3), and because it detected ORF-Sh and ORF-Mh.
3.7. Limitations of the study
A first limitation of the study is that the detection of two novel candidate overlapping ORFs in SARS-CoV-2 depends on a match to prediction scores (PLS-DA, LDA, LDA-ancestral, and LDA-novel scores) yielded by an analysis of overlapping genes that belong to viruses infecting plant and animal hosts over a long period of time (Pavesi, 2020). For example, the dataset of overlapping genes contains twenty homologs, showing a nucleotide diversity from 28 to 50%, of the overlap replicase/movement protein. This overlap belongs to tymoviruses infecting as many as twenty different plant species. The GISAID database I examined is, instead, a massive collection of SARS-CoV-2 genome sequences from a virus evolving over a short period of time during the current pandemic of COVID-19, with a human host having little or no previous immunity (Sette and Crotty, 2020).
Another limitation of the study is that the codon bias in the regions of genes S and M overlapping respectively ORF-Sh and ORF-Mh could reflect the propensity to form stable RNA secondary structures. Tavares et al. (2020) developed an in silico pipeline to predict regions of high-base-pair content across the SARS-CoV-2 genome. They found a remarkable enrichment of structured regions in the 3’ terminal one-third of the genome. In detail, they also predicted an appreciable propensity to form stable RNA base-pairings both for the region of S overlapping ORF-Sh (from nt 24,050 to 24,172) and the region of M overlapping ORF-Mh (from nt 26,691 to 26,873) (see Fig. 4C in Tavares et al., 2020).
A last limitation concerns some aspects of the prediction method. As reported in the previous paragraph, the addition of LDA-ancestral and LDA-novel scores to PLS-DA and LDA scores has the advantage to increase the specificity of SeqComp from 43% to 62%. However, the two additional scores were obtained from a previous study (Pavesi, 2020) on a sample set of overlapping genes rather small (126 homologous overlaps with a known +1 de novo frame and 68 homologous overlaps with a known +2 de novo frame). To increase the sample size, further studies on the genealogy of overlapping genes are needed. The use of five prediction scores by CodScr + SeqComp is justified by the finding that the combined method has an even higher specificity (85%), under the rule of a matching to all five criteria. However, a limitation of CodScr + SeqComp is that it is not possible, unlike Synplot2 (Firth, 2014), to give a unified P-value combining the five prediction criteria.
4. Conclusions
The present study has evolutionary implications, because it demonstrated that the composition in synonymous codons of the ancestral reading frame, to avoid premature stop codons in the de novo frame, is a relevant driving force in the origin and retention of overlapping genes in viruses. I used this feature, as well as those concerning the peculiar nucleotide and amino acid composition of viral overlapping genes, to develop a multi-step statistical method (CodScr + SeqComp) for predicting overlapping ORFs in viruses. When applied to the reference genome sequence of SARS-CoV-2, it predicted two novel overlapping ORFs (ORF-Sh and ORF-Mh), both showing a high degree of conservation in SARS-CoV-2. The good sensitivity and specificity of the method, combined with the ability to examine single genome sequences or sequences with low genetic diversity, extends its field of application to large datasets of viral genome sequences. The method should be particularly effective to detect candidate overlapping ORFs with a restricted phylogenetic distribution, such as newborn or recently born overlapping ORFs.
Declaration of competing interest
None.
Acknowledgements
The author expresses a deep gratitude to the anonymous referees and to the Editor Alexander E. Gorbalenya for their valuable feedback. The author is grateful to Noam Stern-Ginossar (Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel) for helpful comments. Special thanks to Dr. Elisa Ronchei and Dr. Francesco Marchesini for suggestions and technical support. The author would like to extend his thanks to all who have contributed sequences to the NCBI and GISAID databases (http://www.ncbi.nlm.niv.gov/; https://www.gisaid.org/). This work has benefited from the equipment and framework of the COMP-HUB initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.07.011.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aragonés L., Guix S., Ribes E., Bosch A., Pintó R.M. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000797. https://doi:10.1371/journal.ppat.1000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. https://doi:10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. https://doi:10.1016/j.meegid.2020.104353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N., Vianelli A., Belshaw R. Why genes overlap in viruses. Proc. Biol. Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. https://doi:10.1098/rspb.2010.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. https://doi:10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez Andres A., Feng Y., Campos A.R., Yin J., Yang C.C., James B., Murad R., Kim H., Deshpande A.J., Gordon D.E., Krogan N., Pippa R., Ronai Z.A. SARS-CoV-2 ORF9c is a membrane-associated protein that suppresses antiviral responses in cells. bioRxiv. 2020 https://doi:10.1101/2020.08.18.256776. Preprint [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Ginossar N.S., Schwartz M. Viral short ORFs and their possible functions. Proteomics. 2018;18 doi: 10.1002/pmic.201700255. https://doi:10.1002/pmic.201700255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. https://doi:10.1038/s41586-020-2739-1 [DOI] [PubMed] [Google Scholar]
- Firth A.E. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 2014;42:12425–12439. doi: 10.1093/nar/gku981. https://doi:10.1093/nar/gku981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E. A putative new SARS-CoV protein, 3c, encoded in an ORF overlapping ORF3a. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001469. https://doi:10.1099/jgv.0.001469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. https://doi:10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. https://doi:10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumpper R.H., Li W., Luo M.J. Constraints of viral RNA synthesis on codon usage of negative strand RNA virus. J. Virol. 2019;93 doi: 10.1128/JVI.01775-18. https://doi:10.1128/JVI.01775-18 e01775-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., Malik Peiris J.S., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. https://doi:10.1038/s41590-020-0773-7 [DOI] [PubMed] [Google Scholar]
- Jiang H.W., Zhang H.N., Meng Q.F., Xie J., Li Y., Chen H., Zheng Y.X., Wang X.N., Qi H., Zhang J., Wang P.H., Han Z.G., Tao S.C. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. https://doi:10.1038/s41423-020-0514-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Nelson C.W., Ardern Z., Finkel Y., Krogan N.J., Sato K., Ziebuhr J., Stern-Ginossar N., Pavesi A., Firth A.E., Gorbalenya A., Kellis M. Conflicting and ambiguous names of overlapping ORFs in SARS-CoV-2: a homology-based resolution. Virology. 2021;558:145–151. doi: 10.1016/j.virol.2021.02.013. https://doi:10.20944/preprints202012.0048.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:2642. doi: 10.1038/s41467-021-22905-7. https://doi:10.1038/s41467-021-22905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese P.K., Gibbs A. Origin of genes: “big bang” or continuous creation? Proc. Natl. Acad. Sci. U.S.A. 1992;89:9489–9493. doi: 10.10173/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Fielding B.C., Tan T.H., Chou C.F., Shen S., Lim S.G., Hong W., Tan Y.J. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. https://doi:10.1016/j.virusres.2006.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Kovanagi Y., Sauter D., Gifford R.J., USFQ-COVID19 Consortium. Nakagawa S., Sato K. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. https://doi:10.1016/j.celrep.2020.108185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. https://doi:10.1128/JVI.01782-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. https://doi:10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. https://doi:10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. https://doi:10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. https://doi:10.1016/j.antiviral.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jiang J.-Z., Wan X.-F., Hua Y., Li L., Zhou J., Wang X., Hou F., Chen J., Zou J., Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008421. https://doi:10.1371/journal.ppat.1008421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17:131. doi: 10.1186/s12985-020-01402-1. https://doi:1186/s12985-020-01402-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T. Evolution of overlapping genes. Nature. 1978;272:532–535. doi: 10.1038/272532a0. https://doi:10.1038/272532a0 [DOI] [PubMed] [Google Scholar]
- Nelson C.W., Ardern Z., Wei X. OLGenie: estimating Natural Selection to predict functional overlapping genes. Mol. Biol. Evol. 2020;37:2440–2449. doi: 10.1093/molbev/msaa087. https://doi:10.1093/mollbev/msaa087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.W., Ardern Z., Goldberg T.L., Meng C., Kuo C.H., Ludwig C., Kolokotronis S.O., Wei X. Dinamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. Elife. 2020;9 doi: 10.7554/eLife.59633. https://doi:10.7554/eLife.59633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.D., Lindsey B.B., Leary S., Gaudieri S., Chopra A., Wyles M., Angyal A., Green L.R., Parsons P., Tucker R.M., Brown R., Groves D., Johnson K., Carrilero L., Heffer J., Partridge D.G., Evans C., Raza M., Keeley A.J., Smith N., Filipe A.D.S., Shepherd J.G., Davis C., Bennett S., Sreenu V.B., Kohl A., Aranday-Cortes E., Tong L., Nichols J., Thomson E.C., COVID-19 Genomics UK (COG-UK) Consortium. Wang D., Mallal S., de Silva T.I. Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data. Genome Res. 2021;31:645–658. doi: 10.1101/gr.268110.120. https://doi:10.1101/gr.268110.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. Detection of signature sequences in overlapping genes and prediction of a novel overlapping gene in hepatitis G virus. J. Mol. Evol. 2000;50:284–295. doi: 10.1007/s002399910033. https://doi:10.1007/s002399910033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation. Virology. 2019;532:39–47. doi: 10.1016/j.virol.2019.03.017. https://doi:10.1016/j.virol.2019.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology. 2020;546:51–66. doi: 10.1016/j.virol.2020.03.007. https://doi:10.1016/j.virol.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., Magiorkinis G., Karlin D.G. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of deltaretroviruses. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003162. https://doi:10.1371/journal.pcbi.1003162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg O., Kirzhner V., Trifonov E., Bolshoy A. Overlapping messages and survivability. J. Mol. Evol. 2004;59:520–527. doi: 10.1007/s00239-004-2644-5. https://doi:10.1007/s00239-004-2644-5 [DOI] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker A.K., Romero P.R., Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. https://doi:10.1128/JVI.0059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Wagner A., Karlin D. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 2012;29:3767–3780. doi: 10.1093/molbev/mss179. https://doi:10.1093/molbev/mss179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlub T.E., Holmes E.C. Properties and abundance of overlapping genes in viruses. Virus Evol. 2020;6 doi: 10.1093/ve/veaa009. https://doi:10.1093/ve/veaa009 veaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlub T.E., Buchmann J.P., Holmes E.C. A simple method to detect candidate overlapping genes in viruses using single genome sequences. Mol. Biol. Evol. 2018;35:2572–2581. doi: 10.1093/molbev/msy155. https://doi:10.1093/molbev/msy155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealfon R.S., Lin M.F., Jungreis I., Wolf M.Y., Kellis M., Sabeti P.C. FRESCo: finding regions of excess synonymous constraint in diverse viruses. Genome Biol. 2015;16:38. doi: 10.1186/s13059-015-0603-7. https://doi:10.1186/s13059-015-0603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. https://doi:10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. https://doi:10.4049/jimmunol.1303196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.C., Bischof G.F., Lauer W.A., Desrosiers R.C. Importance of codon usage for temporal regulation of viral gene expression. Proc. Natl. Acad. Sci. U.S.A. 2015;112:14030–14035. doi: 10.10173/pnas.1515387112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E.C., Pagán I. The effect of gene overlapping on the rate of RNA evolution. Mol. Biol. Evol. 2013;30:1916–1928. doi: 10.1093/molbev/mst094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. https://doi:10.1016/s0022-2836(03)00865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. https://doi:10.1146/annurevvirology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R.C.A., Mahadeshwar G., Wan H., Huston N.C., Pyle A.M. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. J. Virol. 2020;95 doi: 10.1128/JVI.02190-20. https://doi:10.1128/JVI.02190-020 e02190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S., Klaeger S., Sarkizova S., Pearlman L.R., Chen D.Y., Bauer M.R., Taylor H.B., Conway H.L., Tomkins-Tinch C.H., Finkel Y., Nachshon A., Gentili M., Rivera K.D., Keskin D.B., Rice C.M., Clauser K.R., Hacohen N., Carr S.A., Abelin J.G., Saeed M., Sabeti P.C. SARS-CoV-2 infected cells present HLA-I peptides from canonical and out-of-frame ORFs. bioRxiv. 2020 https://doi:10.1101/2020.10.02.324145. Preprint [Google Scholar]
- Xu K., Zheng B.J., Zeng R., Lu W., Lin Y.P., Xue L., Li L., Yang L.L., Xu C., Dai J., Wang F., Li Q., Dong Q.X., Yang R.F., Wu J.R., Sun B. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. J. Virol. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. https://doi:10.1016/j.virol.2009.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2012-7. https://doi:10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.