Abstract
Stable, long-term culture of primary B lymphocytes has many potential scientific and medical applications, but remains an elusive feat. A major obstacle to long-term culture is that in vitro mitogens quickly drive B cells to differentiate into short-lived plasmacytes (PCs). PC differentiation is governed by opposing teams of transcription factors: Pax5, Bach2, and Bcl6 suppress PC commitment, while IRF4 and Blimp1 promote it. To determine whether transcriptional programming could prolong B-cell culture by blocking PC commitment, we generated mouse primary B cells harboring gain- or loss-of-function in the key transcription factors, continuously stimulated these cells with CD154 and IL-21, and determined growth potential and phenotypes in vitro. We found that transgenic expression of Bach2 prohibits PC commitment and endows B cells with extraordinary growth potential in response to external proliferation and survival cues. Long-term Bach2-transgenic B cell lines have genetically stable BCRs (i.e., do not acquire V(D)J mutations), express high levels of MHCII and molecules for co-stimulation of T cells, and transduce intracellular signals when incubated with BCR ligands. Silencing the Bach2 transgene in an established transgenic cell line causes the cells to secrete large quantities of Ig. This system has potential applications in mAb production, BCR signaling studies, Ag presentation to T cells, and ex vivo clonal expansion for adoptive cell transfer. Additionally, our results provide insight into molecular control over activated B-cell fate, and suggest that forced Bach2 expression in vivo may augment germinal center B cell or memory B cell differentiation at the expense of PC commitment.
INTRODUCTION
Common techniques for primary B cell culture limit clonal expansion to a few weeks (1-3), precluding the isolation of long-term, monoclonal, Ag-specific cell lines that would have numerous potential applications, e.g., in mAb production, BCR signaling studies, in vitro screening of vaccine candidates (4), or expansion of specific B-cell clones for adoptive cell transfer experiments. A critical obstacle to long-term culture of primary cells is that the exogenous mitogens used to induce B-cell activation and proliferation also drive B cells to terminally differentiate into short-lived plasma cells (PCs) (1-3, 5). Short culture periods can be circumvented by infecting B cells with EBV (6) or hybridization with immortal myeloma cells (7), but these techniques drastically alter B-cell physiology, limiting the applications of the resultant cell lines. Moreover, EBV infection is restricted to primate cells (8).
Prior work showed that primary B cell culture can be prolonged by forcing B cells to express Bcl6, a transcriptional repressor of the PC master regulator Blimp1 (Prdm1) (9). In vitro, primary B cells transduced with retroviruses co-expressing Bcl6 and the anti-apoptotic Bcl-xL protein clonally expand rapidly for months when stimulated with CD154-expressing feeder cells and exogenous IL-21 (10, 11). Bcl6-/Bcl-xL- transgenic B cell cultures have some valuable applications, including identification and cloning of high-affinity protective Abs against important pathogens (10, 11), and serving as an abundant source of autologous APCs for human T-cell epitope-mapping studies (12).
However, programming B cell fate by forced expression of Bcl6 has some important shortcomings. First, the transduced B cells continuously acquire BCR mutations in culture (10), albeit at a mutation rate ≥10-fold lower than that estimated for germinal center (GC) B cells (13, 14). Although ongoing BCR mutation allows for in vitro selection of higher-affinity variants (10), it also confounds the derivation of genetically stable, monoclonal cell lines, and in turn, diminishes the usefulness of Bcl6-/Bcl-xL-transgenic B cells for BCR signaling assays or direct production of mAbs. Second, Bcl6-/Bcl-xL-transduced IgM+ memory B cells reportedly do not undergo class-switch recombination (CSR) to IgG (10, 11), despite the fact that this commonly occurs during in vitro culture of non-transduced cells (3, 5). The inability to switch to IgG complicates analysis of mAbs secreted from IgM+ input cells, since IgM and IgGs have different valences and distinct effector functions (15, 16).
Alternative strategies for disrupting PC differentiation might recapitulate the extraordinary growth potential of Bcl6-/Bcl-xL-transgenic cultures while avoiding their shortcomings. PC differentiation is regulated by a transcription factor network comprising five main players, divided into two teams: Pax5, Bach2, and Bcl6 oppose PC differentiation, while IRF4 and Blimp1 promote it (17). Pax5 and Bach2 are broadly expressed in the B-cell lineage, but are silenced in PCs (18-20). Pax5 enforces B-cell identity by activating expression of many B cell-critical signaling molecules and transcription factors (including Bach2) (21, 22), and by repressing numerous non-B lineage genes (21). Bach2 represses cell-cycle checkpoint genes to enable activated B cells to proliferate rapidly (23), and antagonizes PC differentiation by repressing Prdm1 (20, 24). Forced expression of Pax5 or Bach2 might prevent PC differentiation of cultured primary B cells, since 1) these transcription factors are silenced during PC differentiation (18, 20), 2) overexpression of Pax5 in immortalized B-cell lines diminishes their PC traits (25), and 3) Pax5- or Bach2-knockout accelerates PC differentiation in immortalized cell lines or in response to immunization (20, 26-28).
Conversely, preventing the expression of IRF4 or Blimp1 might block PC commitment and maintain the growth potential of cultured B cells. B cells express low to intermediate levels of IRF4 until they are strongly stimulated by Ag, costimulatory signals from follicular TH cells, or other activating receptors (29-31). Strong cumulative signaling increases the concentration of IRF4 (29), which then induces Prdm1 and represses Bcl6 (31, 32). Finally, Blimp1 solidifies PC commitment by repressing Pax5 (33), Myc (34), Bcl6, and other critical B-cell genes (35), thereby silencing B cell-identity and extinguishing growth potential. Blimp1 also activates a suite of genes critical for PC maturation and function (35-37). Neither Irf4−/− nor Prdm1−/− B cells can differentiate into mature PCs (32, 38, 39), and Prdm1−/− B cells are proposed to arrest PC differentiation at a pre-plasmablast stage in response to in vitro activation (40). The de-repression of some Pax5-repressed genes in this pre-plasmablast stage suggests inactivation of Pax5 and the B-cell identity (40); however, the extent to which this state retains fundamental B-cell identity and sustains continuous clonal expansion in response to mitogens has not been determined.
We set out to systematically test whether programming activated B cells by gain- or loss-of-function in Pax5, Bach2, IRF4, or Blimp1 could block PC differentiation and sustain continuous B-cell growth in response to exogenous proliferation and survival signals. To accomplish this, we cultured retrovirus-transduced or genetic knockout B cells in the 40LB culture system, which uses CD154- and BAFF-expressing feeder-cells supplemented by exogenous IL-4 and IL-21 to mimic T-cell help (1). We found that IRF4- or Blimp1-deficiency impairs PC differentiation, but does not support prolonged B cell culture. Transgenic Pax5 also fails to support continuous growth of activated B cells. In contrast, a Bach2 transgene prohibits PC commitment and endows B cells with extraordinary growth potential, allowing them to proliferate and accumulate for months when stimulated by CD154 and IL-21. Long-term Bach2-transgenic B cell lines express high levels of MHCII and molecules for co-stimulation of TH cells, express high levels of surface BCR, do not acquire V(D)J mutations during culture, and transduce intracellular signals when incubated with BCR ligands. Extinguishing the Bach2 transgene in an established cell line causes the cells to differentiate into PCs and secrete large quantities of IgG. Thus, forced expression of Bach2 may be an attractive option for long-term primary B cell culture, and might also be useful for programming activated B cell fate in vivo.
MATERIALS AND METHODS
Reagents
All cell culture reagents and buffers were from GIBCO, unless otherwise specified. Crude supernatant from the mouse anti-BACH2 hybridoma (PCRP-BACH2-5B11; IgG2a,κ), produced by the Protein Capture Reagents Program (Johns Hopkins University/CDI Laboratories), was acquired from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA).
Mice
C57BL/6J, B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ (Cd79aCre) (41), B6.129S1-Irf4tm1Rdf/J (Irf4frt-fl) (38), B6.Cg-Tg(ACTFLPe)9205Dym/J (42), and B6.129P2(C)-Ightm2Cgn/J (B1-8i+/+) (43) mice were from The Jackson Laboratory. Prdm1fl mice (39) were a gift from K. Calame, via D. Allman. Homozygous 2F5 heavy- and light-chain knockin (2F5 KI) mice (44) were a gift from B. Haynes. Irf4frt-fl mice were bred with flippase (FLP)-expressing deleter mice (42) to generate Irf4+/− mice, and then Irf4+/− and Irf4−/− mice were obtained by interbreeding. Irf4+/− and Irf4−/− littermates were co-housed and continuously supplied with drinking water containing sulfamethoxazole (1 mg/ml) and trimethoprim (0.025 mg/ml) to control the otherwise high incidence of rectal prolapse in Irf4−/− mice. All animals were on a C57BL/6 background. All experiments used age- and sex-matched, 8- to 12-week-old male or female mice. Mice were housed under specific-pathogen-free conditions at the Duke University Animal Care Facility. All experiments involving animals were approved by the Duke University Institutional Animal Care and Use Committee.
Cell lines
40LB cells (1) (a kind gift from D. Kitamura) were maintained in DMEM with 10% FBS (Cytiva), 100 units/ml penicillin, 100 μg/ml streptomycin, and MEM non-essential amino acids. Gryphon Eco cells (Allele Biotechnology) were maintained in DMEM with 10% FBS, penicillin, streptomycin, MEM non-essential amino acids, 10 mM HEPES, and 4 mM sodium pyruvate. J558L cells (45) were maintained in B cell medium (BCM: RPMI-1640 with 10% FBS, 5.5x10−5 M 2-mercaptoethanol, 10 mM HEPES, 1 mM sodium pyruvate, penicillin, streptomycin, and MEM nonessential amino acids). All cells were maintained in a humidified atmosphere at 37°C with 5% CO2.
Isolation and culture of primary B cells
Bone marrow isolated from the tibia and femurs of 2F5 KI mice was suspended in cold BCM and stained with propidium iodide and fluorochrome-conjugated mAbs against B220 (RA3-6B2, BioLegend), CD93 (AA4.1, BioLegend), IgM (II/41, eBioscience), IgD (11/26, BD Biosciences), CD43 (S7, BD Biosciences). Bulk immature B cells (B220intCD93+IgMloIgD−CD43−FSClo) were sorted on a BD FACSAria II (BD Biosciences). Naïve mature B cells were isolated from mouse spleens by negative selection: after tissue harvest and treatment with RBC lysis buffer (BioLegend), cells were blocked with anti-CD16/CD32 (2.4G2, BD Pharmingen) and then stained with biotinylated mAbs against CD4 (GK1.5), CD8α (53-6.7), F4/80 (BM8) – all from BioLegend; CD5 (53-7.3), CD11b (M1/70), CD90.2 (53-2.1), CD93 (AA4.1), Gr-1 (RB6-8C5), and TER-119 – all from eBioscience; CD43 (S7), CD95 (Jo2), GL7 – all from BD Pharmingen. Cells were washed in cold MACS buffer (Ca2+/Mg2+-free PBS, pH 7.4, 0.5% BSA, 2 mM EDTA) and incubated with Streptavidin MicroBeads (Miltenyi Biotech) before being passed through an LS column (Miltenyi Biotech). Unbound cells (≥98% B220+ B cells) were typically seeded at a density of 1.5×103 – 10×103 B cells per cm2 on a layer of 40LB feeder cells seeded the previous day at 1.05 × 104 cells per cm2. Cells were grown in BCM with recombinant mouse IL-4 (2 ng/ml, Peprotech) and recombinant mouse IL-21 (10 ng/ml, Peprotech). On culture days 2 and 3, IL-4 was diluted by replacing ¾ of the growth medium in each well with fresh BCM containing only 10 ng/ml IL-21. Thereafter, ¾ of the growth medium in each well was replaced every 1-2 days, with IL-21 maintained at 10 ng/ml. Every 3-5 days, B cells and feeder cells were harvested and washed in warm MACS buffer, resuspended in room-temperature Percoll PLUS (Cytiva, diluted to 40% [v/v] with Ca2+/Mg2+-free PBS, pH 7.4) and centrifuged at 610 × g, 10 min, 25°C. The pellet of feeder cell-depleted B cells (>95% purity) was washed once in BCM, then seeded (1.5×103 – 10×103 B cells per cm2) on fresh 40LB cells in BCM plus 10 ng/ml IL-21. Occasionally, aliquots of feeder cell-depleted B cells were lysed in TRIzol and stored at −80°C until RT-PCR analysis, while culture supernatants were harvested, spiked with 0.1% w/v NaN3, and stored at 4°C until ELISA.
B-cells expressing doxycycline-inducible Bach2 were cultured in the standard way and continuously treated with 300 ng/ml doxycycline.
Flow cytometry and cell sorting
Cultured B cells were harvested in MACS buffer (for sorting) or FACS buffer (MACS buffer plus 0.1% NaN3, for flow cytometry analysis) and blocked with anti-CD16/CD32 (2.4G2, BD Pharmingen) and rat serum IgG (10 μg/ml). Cells were then stained with fluorochrome-conjugated mAbs against CD19 (6D5), B220 (RA3-6B2), CD138 (281-2), CD38 (90), Igλ (RML42), I-A/I-E (M5/114.15.2), H-2Kd (SF1-1.1), CD86 (GL1) – all from BioLegend; Igλ (R26-46), Igκ (187.1), CD95 (Jo2), GL7 (GL7), CD43 (S7), IgG1 (A85-1), CD80 (16-10A1), hCD8α (G42-8) – all from BD Pharmingen; or IgM (II/41, eBioscience). Ag-specific B cells were labeled with NP14-PE (Biosearch Technologies) or MPER-streptavidin tetramers (46), followed by PE anti-Streptavidin (3A20.2, BioLegend). To label transcription factors, surface-stained cells were fixed and permeabilized with the Transcription Factor Buffer Set (BD Pharmingen), then incubated with fluorochrome-conjugated mAbs against Bcl6 (K112-91), Blimp1 (5E7), Pax5 (1H9) – all BD Pharmingen; IRF4 (3E4, eBioscience). Bach2 was labeled with anti-BACH2 mAb (5B11, IgG2aκ), followed by PE anti-IgG2a (RMG2a-62, BioLegend). Net mean fluorescence intensity values were calculated by subtracting the signal of isotype control-stained cells from the signal of transcription factor-specific staining. Dead cells were excluded from analysis based on propidium iodide (Sigma-Aldrich) or Live/Dead Fixable Near-IR (ThermoFisher) staining. Doublets were excluded based on FSC-A/FSC-H gating. Labeled cells were analyzed or sorted into culture plates with a FACSCanto II (BD Biosciences), LSR II (BD Biosciences), LSRFortessa X-20 (BD Biosciences), FACSAria sorter (BD Biosciences), or a FACSVantage sorter with DIVA option (BD Biosciences). Flow cytometric data were analyzed with FlowJo software (BD Biosciences).
Retroviral vector cloning
A plasmid encoding the open reading frame of mouse Bach2 (NM_001109661) was obtained from GenScript, and Bach2 was cloned by standard methods, using these primers (restriction enzyme sites are underlined): Bach2-5’S, 5′-ATTGGATCCGCCGCCACCATGTCTGTGGATGAGAAGCCTG-3′; Bach2-3’A, 5′-TATGATGCGGCCGCTCACTAGGCATAATCTTTCCTGGGC-3′. Pax5 and Bcl6 were cloned from C57BL/6J Peyer’s patch cell cDNA, using standard methods and these primers: Pax5-5’S, 5′-TAGTTAATTAAGCCGCCACCATGGATTTAGAGAAAAATTACCC-3′; Pax5-3’A, 5′-CTAGCGGCCGCTTATCAGTGACGGTCATAGGC-3′; Bcl6-5’S, 5′-ATTGGATCCAAACACAACATGGCCTCCCCG-3′; Bcl6-3’A, 5′-GATGCGGCCGCCTATCAGCAGGCTTTGGGGAGCTC-3′. PCR amplicands were subcloned into the pMX-IRES-GFP plasmid (47) to make pMX-Bach2-IRES-GFP, pMX-Pax5-IRES-GFP, pMX-Bcl6-IRES-GFP. Corresponding plasmids encoding human CD8α (48) instead of GFP were also constructed. pR-TetOne-Bach2-HA, a retroviral vector encoding constitutively expressed reverse tetracycline-controlled transactivator protein and a tetracycline responsive element-regulated Bach2 ORF, was generated by appending the coding sequence for a short linker (GSG) and a hemagglutinin epitope tag (YPYDVPDYA) to the C-terminus of Bach2, using PCR and these primers: BstZ17I-Bach2-5’S, 5′-GTATACGCCGCCACCATGTCTGTG-3′; BglII-HA-Bach2-3’A, 5′-agatcttatcaAGCGTAATCTGGCACGTCGTATGGGTAtccggatccggcataatctttcctgggctg-3′. Bach2-HA was subcloned into pRetroX-TetOne (Takara Bio) to make pR-TetOne-Bach2-HA. The DNA sequences of all PCR amplicands were verified by Sanger sequencing. All retroviral vectors were cloned using NEB Stable competent E. coli (New England BioLabs).
Retroviral transduction
Typically, freshly isolated or cryopreserved splenic B cells (1.6×105 per cm2) were cultured in BCM with IL-4 (10 ng/ml) and anti-CD40 (5 μg/ml, clone HM40-3, BioLegend) for 2 days prior to transduction. In some experiments, B cells were cultured with anti-CD40 and IL-4 for 3 or 4 days before transduction. Immature 2F5 KI B cells were cultured on 40LB cells plus IL-4 (2 ng/ml) for 2 days prior to transduction. Retrovirus was packaged in Gryphon Eco cells according to the manufacturer’s protocols. Virus-containing culture supernatant was harvested 48 hr post-transfection and was immediately mixed 1:1 with B cells in BCM containing 8 μg/ml Polybrene (MilliporeSigma), 10 ng/ml IL-21, and 3 μg/ml anti-CD40. Cells were spin-infected at 1258×g, 32°C, for 90 min, then incubated at 37°C. After 4.5 hr, 50% of the medium in each well was replaced with fresh BCM containing 10 ng/ml IL-21 and 3 μg/ml anti-CD40. The next day, B cells were washed in MACS buffer, and then plated on 40LB cells with 10 ng/ml IL-21.
Aicda RT-PCR
On culture day 29, B cells were depleted of feeder cells with Percoll (as described above), and 5×104 B cells were lysed in TRIzol and stored at −80°C. Expression of Aicda mRNA was determined by quantitative RT-PCR referenced to a synthetic standard, as described (49).
Measurement of intracellular free Ca2+
B cells were harvested and depleted of feeder cells as described above, then suspended at 107 cells ml−1 in loading buffer (HBSS [with 5.5 mM glucose, plus Ca2+ and Mg2+] plus 10 mM HEPES and 1% FBS). Indo-1-acetoxymethylester (50 μg, Thermo Fisher) was dissolved in 35 μl DMSO plus Pluronic F-127 (143 mg/ml, MilliporeSigma), then diluted with 103 μl loading buffer. An 8 μl aliquot of this solution was further diluted into every 1 ml of suspended cells, which were then incubated in the dark, 30 min, room temperature, with occasional mixing. Cells were washed twice in loading buffer and suspended in loading buffer at 107 cells ml−1. Stimulus-induced changes in intracellular Ca2+ concentration were determined with an LSRFortessa X-20 flow cytometer equipped with a 355 nm laser and 379/28 nm and 515/30 nm bandpass filters. The concentration of intracellular free Ca2+ was calculated as the 379/515 nm emission ratio (i.e., “Indo-1 ratio”). BCR ligation was triggered by addition of NP2-BSA, NP10-BSA (Biosearch Technologies), or F(ab′)2 fragment goat anti-mouse IgG(H+L) (Jackson ImmunoResearch).
Measurement of phospho-ERK
Cultured B cells were depleted of feeder cells with Percoll (as described above), then rested in BCM for 4 hr, 37°C. Next, B cells were suspended at 107 cells ml−1 in RPMI-1640, 37°C. Aliquots of cells (95 μl) were transferred to 12 x 75 mm round-bottom polystyrene tubes, and then 5 μl of BCR ligand was spiked into each tube, immediately followed by pulse-vortexing and incubation in a 37°C water bath for 0, 1, 2, 3, or 4 min. Cells were fixed by adding an equal volume of pre-warmed (37°C) Fixation Buffer (BioLegend), and incubating 15 min, 37°C. Fixed cells were washed thrice with RPMI, permeabilized with 0.5 ml chilled (−20°C) True-Phos Perm Buffer (BioLegend), and incubated on ice, 30 min. Cells were washed twice with FACS buffer (MACS buffer plus 0.1% NaN3), stained with mAbs against phospho-ERK1/2 (4B11B69, BioLegend) and B220 (RA3-6B2), and analyzed by flow cytometry. NP2-BSA, NP10-BSA (Biosearch Technologies), or F(ab′)2 fragment goat anti-mouse IgG(H+L) (Jackson ImmunoResearch) was used as a BCR ligand.
ELISA
ELISA of Ig-containing culture supernatants was performed essentially as described (50). HRP-conjugated goat anti-mouse IgG, anti-mouse IgM, anti-mouse IgG3, anti-mouse IgG1, anti-mouse IgG2b, anti-mouse IgG2c, anti-mouse IgE, or anti-mouse IgA (all from Southern Biotech) was used to detect bound Ab. H33Lγ1 (mouse IgG1λ) standard Ab and anti-chicken gamma globulin (clone 16, mouse IgG1κ) standard Ab were produced in-house as described (51). Mouse IgMκ (MM-30, BioLegend), mouse IgG3κ (MG3-35, BioLegend), or mouse IgG2b (A-1), mouse IgG2c (G-3), mouse IgE (15.3), and mouse IgA (S107) – all from Southern Biotech – were also used as standard control Abs.
Amplification of V(D)J rearrangements from cultured B cells
Amplification and analysis of V(D)J rearrangements from cultured B cells was performed essentially as described (50).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software). Log-distributed data (i.e., cell numbers, geometric mean fluorescence intensities, and Ig concentrations) were log-transformed prior to analysis by one- or two-way ANOVA with Tukey’s post-test. Differences between groups were considered statistically significant at P < 0.05.
RESULTS
Transcription factor dynamics during PC differentiation in 40LB culture
Before attempting to transcriptionally re-program activated B cells, we sought to map the key events of B cell activation and PC differentiation onto the underlying expression dynamics of critical B-cell transcription factors. In the 40LB culture system, which uses exogenous IL-4, IL-21, and CD154- and BAFF-expressing feeder cells to mimic T-cell help (1), wildtype splenic B cells proliferate and accumulate logarithmically until culture day 8 (Fig. 1A). During this period, the B cells express Pax5 at a constant level comparable to that in resting mature B cells; Bach2 increases 4.5-fold by day 2, then gradually declines to its original level; and Bcl6 is consistently low (Fig. 1B-1C). Meanwhile, IRF4 is upregulated 20-fold by day 2, and thereafter remains high, whereas Blimp1 stays close to baseline (Fig. 1B, 1D). CSR occurs early in the culture period, with 80% of cells switching to IgG1 by day 4, and ~95% of cells expressing IgG1 at day 6 and thereafter (Fig. 1E-1F). Prior to day 8, PCs (CD43+CD138+) constitute ≤2% of the B-cell population; moreover, few cells singly express either CD43 or CD138, save for day 2, when ~50% of cells are CD138+CD43− (Fig. 1G-1H). The significance of this dramatic-but-transient increase in CD138 expression is unclear; however, because the average secretion rate for IgM and IgG is ≤3 molecules cell−1 sec−1 until at least day 6 (Fig. 1I), expression of CD138 at day 2 is not indicative of bona fide PC differentiation.
Fig. 1. Kinetics of wildtype B-cell clonal expansion, CSR, and PC differentiation in 40LB culture.

B6 splenic B cells co-cultured with 40LB feeder cells and IL-4 and IL-21 were periodically analyzed by flow cytometry, while culture supernatants were analyzed by ELISA. A, Growth kinetics of cultured B cells. B, Expression of key transcription factors (TFs) in cultured B cells and control B cells. Ex vivo B6 splenocytes were the positive control for Pax5 and Bach2 staining, ex vivo B6 Peyer’s patch GC B cells were the positive control for Bcl6 staining, and cultured J558L plasmacytoma cells were the positive control for IRF4 and Blimp1 staining. C and D, Dynamics of transcription factor expression in cultured B cells, normalized to control cells (as shown in B). E, Frequency of B cells expressing surface IgG1. F, Aggregate data for IgG1-CSR (gated as in E) at all time points. G, Frequencies of B cells expressing the PC markers CD43 or CD138 at various time points during 40LB culture. H, Aggregate data for expression of CD43 and CD138 (gated as in G) at all time points. I, The average rate of IgG and IgM secretion by 40LB-cultured B cells at various time points. GeoMFI, geometric mean fluorescence intensity. LOD, limit of detection. Flow cytometry analyses were pre-gated on viable H2-Kd−CD19+ cells (B-H). Error bars depict geometric mean ± 95% CI (A, I) or arithmetic mean ± 95% CI (C, D, F, H) of three replicates. Data are representative of one (B, C, D, I) or ≥3 independent experiments (A, E, F, G, H) with similar results.
Around day 8, a striking transition occurs in cultured B cells. Clonal expansion begins to plateau, Pax5 is extinguished, and Blimp1 increases (Fig. 1A-1D). Additionally, most B cells acquire a CD43+CD138+ PC phenotype or a presumably intermediate CD43+CD138− phenotype (Fig. 1G-1H), and the average IgG secretion rate increases 30-fold (Fig. 1I). By day 12, Bach2 expression declines to 50% of its initial value (or ~10% of its peak value) (Fig. 1C), while the concentrations of IRF4 and Blimp1 remain elevated (Fig 1D). At this point, >75% of B cells have become CD43+CD138+ PCs (Fig. 1G-1H), and the rate of IgG secretion increases to ~6x103 molecules cell−1 sec−1 (Fig. 1I), which is compatible with estimates of Ig secretion rates by PCs (52, 53).
These observations are largely congruent with the standard model (17) for B-cell activation and PC differentiation, and indicate that in 40LB culture, Pax5 and Bach2 may be the primary antagonists of PC differentiation, whereas Bcl6 plays little, if any, role. In contrast, high concentrations of IRF4 and especially Blimp1 silence the B cell identity and coordinate PC maturation.
IRF4-deficiency severely impairs B-cell clonal expansion
To determine whether IRF4 deficiency blocks PC differentiation and sustains continuous B-cell proliferation, we cultured Irf4+/+, Irf4+/−, and Irf4−/− B cells in the 40LB system. Numbers of both Irf4+/+ and Irf4+/− B cells increase ≥104-fold in 11 days (Fig. 2A). During this time, Irf4+/+ and Irf4+/− cells efficiently class-switch to IgG1 (Fig. 2B-2C), upregulate CD43 and CD138 (Fig. 2B, 2D), decrease expression of a B-cell marker, B220 (Fig. 2B, 2E), and increase Ig secretion (Fig. 2F). In stark contrast, numbers of Irf4−/− B cells increase at most 30-fold, peaking at culture day 7 before rapidly declining (Fig. 2A). This profound growth impairment occurs despite inhibition of PC differentiation in Irf4−/− B cells, evidenced by 1) limited downregulation of B220 (Fig. 2B, 2E), 2) dysregulated expression of CD43 and CD138 in day 7 cultures (Fig. 2B, 2D), and 3) a ~10-fold lower IgG secretion rate compared to IRF4-sufficient cells (Fig. 2F). The inability to secrete much IgG is not due to a failure in CSR, as ~50% of Irf4−/− B cells express surface IgG1 at culture day 7 (Fig. 2B-2C). Thus, rather than conferring increased growth potential by blocking PC differentiation, IRF4-deficiency imparts a severe growth defect to B cells stimulated with the T-cell help signals IL-4, IL-21, and CD154.
Fig. 2. IRF4-deficiency profoundly impairs both PC differentiation and B-cell clonal expansion.

Irf4+/+, Irf4+/−, and Irf4−/− splenic B cells were cultured and analyzed as in Fig. 1. A, Growth kinetics of cultured B cells. B, Flow cytometry plots showing expression of surface IgG1 vs. surface IgM (top) or CD43 vs. CD138 (middle) or B220 expression (bottom). C, Aggregate data for CSR (gated as in B) at culture day 7. D, Aggregate data for expression of PC markers (gated as in B) at culture days 4, 7, and 11. E, Aggregate data for expression of B220 (gated as in B) at culture day 7. F, The average rate of IgG secretion by 40LB-cultured B cells at various time points. GeoMFI, geometric mean fluorescence intensity. N.D., not detected. Flow cytometry analyses were pre-gated on viable H2-Kd−CD19+ cells (B-E). Error bars depict geometric mean ± 95% CI (A, F) or arithmetic mean ± 95% CI (C, D) of two replicates. Horizontal lines (E) depict geometric means. P values were determined by one- (E) or two-way (A, F) ANOVA, followed by Tukey’s test. *, P < 0.05; ****, P < 0.0001. Data are representative of three independent experiments with similar results.
Blimp1-deficiency blocks PC differentiation, but fails to sustain B-cell identity and growth potential
We next turned to knocking out Blimp1 as a potential strategy for blocking PC differentiation and preserving long-term growth potential. Although Blimp1 is required for PC differentiation (36, 37, 39), it has been proposed (36, 40) that Blimp1 is dispensable for the earliest steps of PC commitment, which instead may be driven by changes in Pax5 protein activity. Kallies et al. (40) reported that activated Prdm1−/− B cells arrest PC differentiation at a Pax5+ pre-plasmablast stage; however, it is unknown whether this state is stable and whether it permits long-term proliferation and survival.
Because global Blimp1 knockout is embryonic-lethal (54), we isolated B cells from Cd79awt/wtPrdm1fl/fl (Prdm1+/+), Cd79aCre/wtPrdm1wt/fl (Prdm1+/−), and Cd79aCre/wtPrdm1fl/fl (Prdm1−/−) mice, in which deletion of floxed Prdm1 exons is restricted to the B lineage (39, 41). When cultured on 40LB feeder cells, Prdm1+/+, Prdm1+/−, and Prdm1−/− B cells exhibit similar growth kinetics (Fig. 3A) and IgG1 CSR efficiency (Fig. 3B-3C). Strikingly, although Prdm1−/− cells do not differentiate into PCs – while Prdm1+/− and Prdm1+/+ B cells do by day 9 (Fig. 3D-3F) – B cells of all genotypes stop growing and die by culture day 15 (Fig. 3A). Therefore, Blimp1-deficient “pre-plasmablasts” are not permissive for continuous growth in response to T-cell help signals.
Fig. 3. Blimp1-deficiency fails to sustain B-cell clonal expansion, despite blocking PC differentiation.

Prdm1+/+, Prdm1+/−, and Prdm1−/− splenic B cells were cultured and analyzed as in Fig. 1. A, Growth kinetics of cultured B cells. B, Flow cytometry plots showing expression of surface IgG1 vs. surface IgM at culture days 2 and 5. C, Aggregate data for CSR (gated as in B). D, Flow cytometry plots showing expression of CD43 and CD138 at culture days 5 and 9. E, Aggregate data for expression of PC markers (gated as in D). F, The average rate of IgG secretion by 40LB-cultured B cells at culture days 5 and 9. N.D., not detected. Flow cytometry analyses were pre-gated on viable H2-Kd−CD19+ cells (B-E). Error bars depict geometric mean ± 95% CI (A), arithmetic mean ± 95% CI (C), arithmetic mean ± S.E.M. (E) of 2-4 replicates. Horizontal lines (F) depict geometric means. P values were determined by 2-way ANOVA, followed by Tukey’s test. ****, P < 0.0001. Data are representative of two independent experiments with similar results.
Forced expression of Bach2 or Bcl6 blocks PC commitment and confers long-term growth potential in response to external mitogens
Having failed to enable continuous B-cell proliferation by disrupting the master regulators of PC differentiation, we elected to focus instead on maintaining the B-cell identity program by forcibly expressing its principal coordinators. To this end, we infected B cells with retrovirus expressing Pax5- or Bach2-IRES-hCD8a, cultured the cells in the 40LB system, and tracked the fates of human CD8α+ (i.e., transduced) cells. B cells transduced with empty hCD8a retrovirus or Bcl6-IRES-hCD8a retrovirus respectively served as negative and positive controls for extended growth potential. To limit potential experimental variability that might arise from using B cells with a diverse BCR repertoire, the B cells in this experiment were derived from B1-8i heavy chain (HC) knockin (KI) mice, which express a specific rearrangement of VH1-72, JH2, and DH1-1 (43), that, when paired with Igλ light chain (LC), confers specificity for 4-hydroxy-3-nitrophenyl acetyl (NP) (55, 56). However, in principle, BCR specificity should not affect transcription factor function.
Transgenic Pax5 is ultimately ineffective at blocking growth arrest in 40LB-cultured B cells (Fig. 4A), despite impairing acquisition of PC markers (Fig. 4B-4D). After culture day 11, no additional accumulation of Pax5-transgenic cells occurs, although the cells survive longer than empty vector-transduced controls (Fig. 4A).
Fig. 4. Forced expression of Bcl6 or Bach2 prevents PC differentiation and enables long-term B-cell culture.

Splenic B1-8 HC KI B cells were transduced with retrovirus expressing Pax5-, Bcl6-, Bach2-IRES-hCD8a or empty IRES-hCD8a and then cultured on 40LB cells with exogenous IL-21. B cells were periodically enumerated, analyzed by flow cytometry, and transferred to fresh feeder cells for continued culture. A, Growth kinetics of cultured B cells. B, Flow cytometry plots showing expression of CD43 and CD138 on B cells at culture day 11. C, Kinetics of CD43 expression (gated as in B) among transduced (hCD8α+) B cells. D, Kinetics of CD138 expression (gated as in B) among transduced (hCD8α+) B cells. E, The average rate of IgG secretion by 40LB-cultured B cells at various time points. F, Growth kinetics for Bach2-transgenic B cell clone B1-8i.4 before (solid circles) and after (hollow circles) cryopreservation and thawing. Flow cytometry analyses were pre-gated on viable H2-Kd−CD19+ cells (B-D). Error bars depict geometric mean ± 95% CI (A) or arithmetic mean ± 95% CI (C, D) of two replicates. Data concerning Pax5 and Bcl6 transgenes represent one experiment; data concerning empty and Bach2 transgenes are representative of ≥6 independent experiments with similar results.
In contrast to the ineffectiveness of the Pax5 transgene, forced expression of Bach2 or Bcl6 is sufficient to block PC differentiation and avert growth arrest for as long as the cells are cultured (≥70 days) (Fig. 4A). By day 11, Bach2- or Bcl6-transgenic B cells are enriched for CD43−CD138− activated B cells (Fig. 4B-4D); thereafter, Bach2-transgenic cultures continue to enrich for non-PC activated B cells, and maintain an average rate of IgG secretion >50-fold lower than that of mature PCs from day 11 control cultures (Fig. 4E). Interestingly, Bcl6-transgenic cells behave somewhat differently, gradually increasing CD138 expression over the course of the culture period (Fig. 4D), while consistently suppressing CD43 expression (Fig. 4C) and secreting IgG at rates well below those typical of PCs (Fig. 4E). The significance of CD138 expression in Bcl6-transgenic cells (Fig. 4D) is unclear. Thus, forced expression of Bach2 – like Bcl6 – blocks PC differentiation and averts growth arrest in CD154- and IL-21-stimulated B-cell cultures.
Repeating the transduction experiments with either hCD8α- or GFP-marked retrovirus and B cells from B6 or various BCR KI mice, we obtained >50 independent Bach2-transgenic cell lines (Table I, Figure 5, and data not shown) that could be clonally expanded for as long as they were stimulated with 40LB cells and IL-21. In an attempt to determine the maximum number of cell divisions the cells could achieve before senescence, we cultured one Bach2-transgenic cell line continuously for 159 days (Fig. 4F). During this time, the population expanded >1040-fold (~133 population doublings), showing no evidence of replicative senescence. Additionally, all Bach2-transgenic cell lines tolerate cryopreservation and maintain their growth characteristics after thawing and subsequent culture (Fig. 4F and data not shown).
Table I.
V(D)J rearrangements recovered from Bcl6- or Bach2-transgenic B cell lines
| Parent cell line |
No. VHDJH recovered from subclonesa |
VH | D | JH | HCDR3 (aa) | No. ΔVH |
No. VLJL recovered from subclones |
VL | JL | LCDR3 (aa) | No. ΔVL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bcl6 #1b,c | 30 | VH1-72 | DH1-1 | JH2 | CARYDYYGSSYFDYW | 0 | 26 | Vκ17-121 | Jκ2 | CLQSDNLPLTF | 0 |
| 4 | Vλ1 | Jλ1 | CALWYSNHLVF | 0 | |||||||
| Bcl6 #2b | 7 | VH1-72 | DH1-1 | JH2 | CARYDYYGSSYFDYW | 0 | 6 | Vκ14-111 | Jκ5 | CLQYDEFPLTF | 0 |
| Bach2 #1b | 8 | VH1-72 | DH1-1 | JH2 | CARYDYYGSSYFDYW | 0 | 5 | Vκ11-125 | Jκ1 | CLQHSYLPTF | 0 |
| Bach2 #2b | 18 | VH1-72 | DH1-1 | JH2 | CARYDYYGSSYFDYW | 0 | 4 | Vκ17-121 | Jκ5 | CLQSDNLPLTF | 0 |
| 015.4 | N/Ad | VH1-54 | DH2-3 | JH3 | CARGVIYDGYSWFAYW | 0 | N/Ad | Vκ2-109 | Jκ1 | CAQNLELPRTF | 0 |
| 015.21 | N/Ad | VH5-4 | DH2-4 | JH2 | CARDQYDYALFDYW | 0 | N/Ad | Vκ13-84 | Jκ1 | CQQYWSTPWTF | 0 |
| 015.NB21 | N/Ad | VH1-55 | DH2-2 | JH3 | CARDQYDYALFDYW | 0 | N/Ad | Vκ6-17 | Jκ5 | CQQHYSTPLTF | 0 |
| 016.4 | N/Ad | VH14-1 | DH2-5 | JH4 | CYSNYDYYAMDYW | 0 | N/Ad | Vκ19-93 | Jκ1 | CLQYDNLWTF | 0 |
| 016.21 | N/Ad | VH1-18 | DH3-3 | JH2 | CARKGAAPFDYW | 0 | N/Ad | Vκ17-127 | Jκ2 | CLQSDNMPYTF | 0 |
| 2F5.1 | N/Ad | VH2-5e | DH3-3e | JH6e | CAHRRGPTTLFGVPIA-RGPVNAMDVW | 0f | N/Ad | Vκ1-13e | Jκ4e | CQQLHFYPHTF | 0f |
| B1-8.4b | N/Ad | VH1-72 | DH1-1 | JH2 | CARYDYYGSSYFDYW | 0 | N/Ad | Vλ1 | Jλ1 | CALWYSNHWVF | 0 |
Notes:
Single B cells were sorted after 67 days of culture, and then allowed to grow for 10 more days prior to V(D)J analysis.
These clonally independent cell lines were derived from B1-8 HC KI mice; thus, all express the B1-8 HC.
This bulk cell line includes two subpopulations of cells, bearing either Igκ or Igλ. During subcloning, Igκ+ cells and Igλ+ cells were sorted separately for outgrowth and V(D)J analysis.
V(D)J sequences were obtained from bulk (non-subcloned) cell lines.
Designations refer to human V(D)J gene segments.
Recovered sequences exactly matched the knockin 2F5 V(D)J sequences.
Fig. 5. 40LB-cultured Bach2-transgenic B cell lines have an activated B-cell phenotype.

Splenic B cells were stimulated for 2, 3, or 4 days with anti-CD40 and IL-4 before transduction with Bach2-expressing retrovirus. After an additional 30 days of culture, the established cell lines were analyzed by flow cytometry and ELISA. Ten independent lines were established for each activation condition. Each cell line is named as X.Y, where X represents the length (in days) of activation prior to transduction, and Y identifies the independent cell line within that cohort. A, Flow cytometry histograms showing expression of cell-surface markers on representative Bach2-transgenic cell lines and freshly isolated Peyer’s patch CD19+CD38hiGL7− quiescent B cells or CD19+CD38loGL7+ GC B cells. B-D, Geometric MFI of CD19 (B), GL7 (C), or CD95 (D) expression on Bach2-transgenic cell lines. E, Frequency of CD38lo cells (gated according to the dashed vertical line in A) within each cell line. F-H, Geometric MFI of MHCII (F), CD80 (G), or CD86 (H) expression on the cell lines. I, Flow cytometry histograms showing expression of B220 and cell-surface IgG1 on representative Bach2-transgenic cell lines or splenic CD19− non-B or CD19+ B cells. J,K, Frequencies of B220hi (J) or surface IgG1+ (K) cells in each cell line, gated according to the dashed vertical lines in I. L,M, Flow cytometry scatter plots showing surface immunoglobulin expression on representative Bach2-transgenic cell lines. N,O, Frequencies of IgG2b+ (N) or IgE+ (O) cells within each cell line. P, ELISA results showing the quantity of each Ig isotype in the culture supernatants of Bach2-transgenic cell lines. The left panel shows Igκ-containing Igs; the right panel shows Igλ. Flow cytometry analyses were pre-gated on viable CD19+ cells unless otherwise noted. Each symbol in B-H, J, K, and N-P represents an individual cell line. Solid and dashed long, horizontal lines in B-D and F-H respectively represent the geoMFI values for GC B cells or quiescent B cells, as depicted in A. Short horizontal lines (B-H, J, K, N, O) denote the median values.
Bach2-transgenic B cell cultures maintain an activated, non-GC B phenotype
The continuous accumulation of Bach2-transgenic B cells in response to signals mimicking T-cell help suggests that the Bach2 transgene blocks PC differentiation by locking in the B-cell identity. This conclusion is supported by flow cytometric analysis of the phenotypes of established Bach2-transgenic cell lines arising from B cells activated with anti-CD40 and IL-4 for 2, 3, or 4 days prior to transduction with Bach2-IRES-hCD8α retrovirus. Like GC B cells, Bach2-transgenic B cells exhibit an activated B cell phenotype, indicated by high expression of CD19 and upregulation of surface GL7 and CD95 (Fas) (Fig. 5A-5D). Numerous Bach2-transgenic cell lines also downregulate CD38 relative to resting B cells, although for unknown reasons the magnitude and frequency of downregulation varies considerably among cell lines (Fig. 5A, 5E). Bach2-transgenic B cell lines also express elevated surface MHCII and the costimulatory molecules CD80 and CD86 (Fig. 5A, 5F-5H), characteristic of activated B cells prepared to present Ag to TH cells. Importantly, Bach2-transgenic cells do not express detectable Bcl6 protein, distinguishing them from both GC B cells and Bcl6-transgenic B cell lines (Supp. Fig. 1).
We predicted that the duration of the initial cellular activation period prior to retroviral transduction might affect the phenotype of the resulting cell lines; however, this proved generally false for activation periods of 2-4 days (Fig. 5A-H). Only B220 and BCR expression are impacted by the length of the initial activation period (Fig. 5I-5O). Whereas cells activated for 2 days almost always produce cell lines with uniformly high B220 expression, extending the activation to 3 or 4 days frequently results in cell lines containing significant subpopulations of B220int/lo cells (Fig. 5I-5J). Additionally, cell lines arising from the 2-day activation condition typically express high levels of cell-surface IgG1, while 3 or 4 days of stimulation increases the frequency of surface IgG1− cells in the resultant cell lines (Fig. 5I, 5K). At least some of the IgG1− cells express substantial surface Ig LC (Fig. 5L) and surface IgE, or – less frequently – IgG2b (Fig. 5M-5O). Accordingly, ELISA analysis of culture supernatants showed that these cell lines secrete IgG2b and/or IgE in addition to (or instead of) IgG1 (Fig. 5P). We did not detect any other Ig isotypes in culture supernatants (save for a single culture with trace IgG2c; Fig. 5P) or on cell surfaces (as determined by flow cytometry; data not shown). Notably, the majority of the CD19+IgG1− cells express little or no surface Ig LC and have indeterminate surface Ig HC isotypes; however, these cells are not PCs, as they express neither CD43 nor CD138, and many are B220hi (data not shown). We tentatively conclude that these B cells express very little BCR, possibly because transgenic Bach2 represses the Igh 3′ enhancer (19) in a manner dependent on the initial activation conditions, although it is important to note that the expression levels of BCR did not correlate with those of the transgene reporter (i.e., hCD8α; data not shown). Whatever the underlying mechanism, it is clear that retroviral transduction after only two days of cellular activation minimizes the appearance of BCRlo cells, and moreover does not increase other phenotypic heterogeneity.
Bach2- and Bcl6-transgenic B cell cultures do not accumulate V(D)J mutations
Kwakkenbos et al. reported continuous V(D)J somatic hypermutation (SHM) in B cell lines established by co-transducing human B cells with BCL6- and BCL-XL-expressing retroviruses, followed by culture with CD154-expressing feeder cells and recombinant IL-21 (10). V(D)J SHM is driven by activation-induced deaminase (AID, Aicda) activity (57, 58). We determined the expression of Aicda in selected Bach2- or Bcl6-transgenic B cells at culture day 30 to be ~300-10,000 Aicda transcripts per thousand cells (Supp. Fig. 2), which is 2- to 100-fold lower than in GC B cells (49). The relatively high Aicda expression in some Bach2-transgenic cell lines suggested that they might also undergo AID-mediated SHM. To test this prediction, we sorted single-cell subclones from four clonally independent Bcl6- or Bach2-transgenic cell lines after 67 days of culture, allowed the subclones to clonally expand for 10 days, and then amplified V(D)J sequences from the expanded clones by RT-PCR. In total, we recovered HC rearrangements from 63 subclones (7-30 subclones per cell line), and light chain (LC) sequences from 45 subclones (4-26 subclones per cell line) (Table I). In these ~32,000 nucleotides of VH and VL regions, we did not detect any mutations. Thus, in our hands, neither Bcl6- nor Bach2-transgenic B cells undergo appreciable SHM, despite the long culture times and the latter’s relatively high expression of Aicda.
The isolation of subclones also afforded some insight into the clonality of transgenic B cell lines after extended culture. Since the parental cell lines for the subcloning experiment were all derived from B1-8 HC KI (43) B cells, it was unsurprising that each subclone expressed the B1-8 HC, comprising a specific rearrangement of VH1-72, JH2, and DH1-1 (Table I). However, the LCs from different parental cell lines were distinct from each other; moreover, for a given parental cell line, each subclone expressed the same LC rearrangement. The exception was Bcl6 #1, whose subclones expressed either a single VκJκ rearrangement or a single VλJλ arrangement, consistent with the observation that the Bcl6 #1 cell line comprises subpopulations expressing either Igκ or Igλ (data not shown). These data are consistent with the recovery of only one or two BCRs from bulk cell lines cultured for ~2 months (Table I), and imply that Bcl6- and Bach2-transgenic B cell lines either 1) grow out of rare founders selected during culture initiation, or 2) approach pauci- or monoclonality during extended culture as the fittest clones outcompete the others. Experiments are underway to test these hypotheses.
Generation of long-term cell lines with known Ag specificity
To determine whether the Bach2-transgene could be used to establish long-lived, Ag-specific B cell lines, we generated an Igκ−Igλ+ B1-8i HC KI B cell line, named B1-8.4. As expected, B1-8.4 binds NP-conjugated PE (Fig. 6A). We also derived a second Ag-specific Bach2-transgenic cell line from 2F5 KI IgM−/lo early immature B cells, which express a musinized HIV-1 broadly neutralizing Ab (2F5) specific for the membrane-proximal external region (MPER) of HIV-1 gp41 (44). This cell line, 2F5.1, does not bind NP-decorated PE (Fig. 6A), but does bind tetramers of MPER peptide containing the 2F5 linear epitope, ELDKWA (Fig. 6B) (46). Importantly, the 2F5 cell line cannot bind empty tetramers nor tetramers of MPER peptide containing an alanine substitution in the 2F5 epitope (ELAKWA) (46). As expected, B1-8.4 cells do not bind any of the tetramers (Fig. 6B).
Fig. 6. Cognate Ag binding by 2F5 KI and B1-8 HC KI cell lines.

Bach2-transgenic cell lines were established from 2F5 KI or Igλ+Igκ− B1-8i HC KI B cells. These cell lines were incubated with NP-conjugated PE (A) or PE-labeled tetramers (B) of peptides containing the 2F5 epitope (ELDKWA) or a mutated epitope (ELAKWA). Ag binding was measured by flow cytometry. Analyses were pre-gated on CD19+H2-Kd− cells.
BCR signaling in Bach2-transgenic B cell lines
Ag engagement by the BCR complex is signaled through multiple pathways, including intracellular calcium mobilization and phosphorylation of ERK (59). To determine whether Bach2-transgenic B cell lines are competent for BCR signal transduction, we stimulated several independent cell lines with BCR ligands, including anti-BCR Abs or cognate Ag, and measured changes in the concentrations of cytosolic free Ca2+ and phosphorylated ERK. In response to anti-BCR Abs, all cell lines tested exhibit time- and dose-dependent Ca2+-mobilization (5/5 cell lines) and/or increases in ERK phosphorylation (2/2 cell lines), with greater concentrations of ligand triggering proportionally greater magnitudes of signaling, up to a saturating limit (Fig. 7A-7D and data not shown). Not unexpectedly, cell lines differ from each other in the magnitude and kinetics of their responses to common stimuli (Fig. 7E), which may be attributable to differences in BCR expression levels (Fig. 7F). Importantly, incubation of the B1-8.4 cell line with NP2- or NP10-BSA elicits dose- and valence-dependent calcium mobilization and ERK phosphorylation (Fig. 7E-7F and data not shown). That Bach2-transgenic B cell lines generally are competent to transduce intracellular signals in response to BCR ligands indicates that these cells might be useful for studies of BCR signaling or for in vitro evaluation of potential immunogens.
Fig. 7. Bach2-transgenic B cell lines quantitatively transduce intracellular signals in response to BCR ligands.
A-D, 016.4 cells were stimulated with anti-IgG and the kinetics of calcium mobilization (A) or ERK1/2 phosphorylation (B-D) were determined. B shows the intensity of phospho-ERK1/2 staining after two minutes of stimulation. E, 2F5.1 and 016.4 cells were stimulated with anti-IgG and the kinetics of calcium mobilization were determined. F, Flow cytometry analysis of surface IgG1 and surface Igκ expression on 2F5.1 and 016.4 cells at the time the experiment depicted in E. G-H, B1-8.4 cells were stimulated with NP2-BSA (G) or NP10-BSA (H) and the kinetics of calcium mobilization were determined.
Inactivation of the Bach2 transgene enables rapid PC differentiation in cultured B cells
Constitutive expression of the Bach2 transgene prevents cells from secreting much Ig (Fig. 4E), thereby limiting their utility for mAb production. To solve this issue, we made use of a tetracycline-inducible gene expression system in which the activity of constitutively expressed reverse tetracycline-controlled transactivator protein (rtTA) is regulated by doxycycline (Dox), a tetracycline analog (60). On binding Dox, rtTA undergoes a conformational change that allows it to engage a tetracycline response element (TRE) to activate transcription of transgenic Bach2 (60). We transduced B6 splenic B cells with a retrovirus encoding rtTA and a TRE-regulated Bach2 transgene, and cultured the transduced cells in the 40LB system. In the presence of doxycycline, rtTA activated expression of the Bach2 transgene and enabled long-term (≥50 days) outgrowth of cultured B cells (Fig. 8A). In contrast, transduced B cells cultured without doxycycline differentiated into PCs and died by day 20, as expected (Fig. 8A).
Fig. 8. Deactivation of the Bach2 transgene in established cell lines results in rapid PC differentiation.
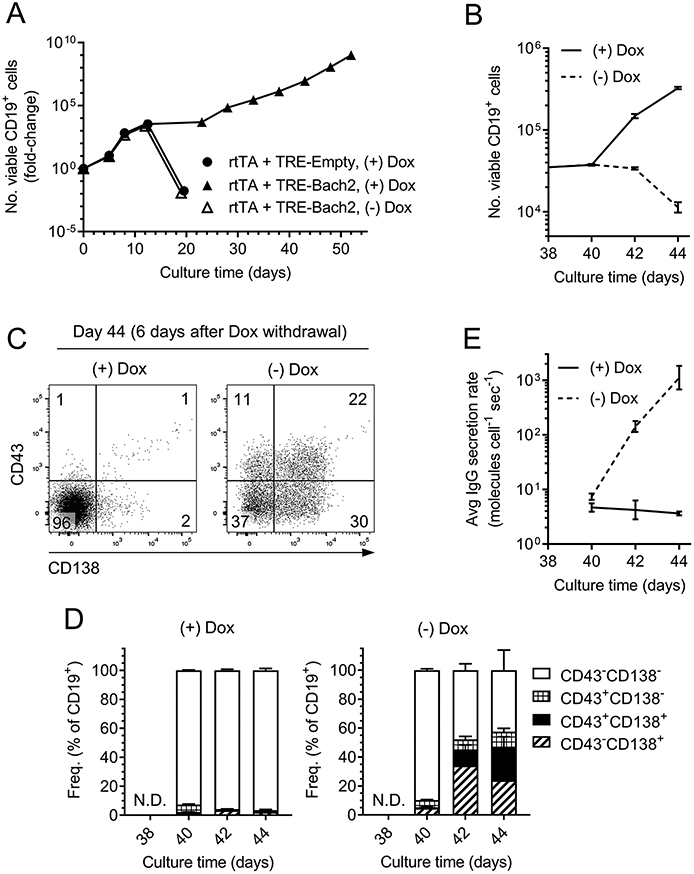
B6 splenic B cells were transduced with retrovirus encoding rtTA and a TRE-controlled Bach2 transgene. Transduced cells were cultured with 40LB feeder cells, and doxycycline (Dox) was added to some cultures to activate the Bach2 transgene. A, Growth kinetics of cultured B cells. B-E, The cell line expressing the doxycycline-inducible Bach2 transgene (solid triangles in A) was split at culture day 38 and cultured in parallel with (+) or without (−) Dox for 6 additional days. B, Growth kinetics. C, Flow cytometric analysis of PC marker expression. D, Kinetics of PC marker expression, gated as in C. E, Average rate of IgG secretion.
At culture day 38, when the Dox-dependent Bach2-transgenic cell line was established and growing robustly, we split the cells and continued growing one half of the population with Dox and one half without Dox. In the presence of Dox, the B cells continued proliferating; 6 days later, the cells had accumulated ~10-fold (Fig. 8B). Moreover, >90% of the B cells remained negative for the PC markers CD43 and CD138 (Fig. 8C-8D), and the average IgG secretion rate consistently hovered at a baseline value of 4 molecules cell−1 sec−1 (Fig. 8E). In contrast, the removal of Dox and the consequent inactivation of the Bach2 transgene caused the B cells to stop growing within 6 days (Fig. 8B). At that point, ~55% of the cells had acquired expression of CD43 and/or CD138 (Fig. 8C-8D), and the average IgG secretion rate increased ~100-fold relative to cells incubated with Dox (Fig. 8E), in which the Bach2 transgene was still active. Thus, deactivation of the Bach2 transgene in an established B-cell line allows the cells to differentiate into PCs and secrete substantial quantities of Ig.
DISCUSSION
A Bach2-transgene enables isolation of stable, long-term B cell lines with many applications
Commonly used techniques for primary B-cell culture are time-limited because in vitro exposure to external mitogens drives B cells to rapidly differentiate into short-lived PCs (1, 2, 61, 62). The short window for in vitro clonal expansion is an obstacle to isolating and maintaining monoclonal lines of primary B cells for ex vivo expansion of specific clones prior to adoptive transfer experiments, mAb production, in vitro evaluation of vaccine candidates (4), etc.
Prior work showed that enforced expression of Bcl6 and Bcl-xL in B cells enables the cells to proliferate for months when stimulated with CD154 and IL-21 (10, 11). These Bcl6-/Bcl-xL-transgenic B cell cultures have several important applications (10-12), but are reported to undergo significant AID-driven V(D)J SHM in vitro, precluding the isolation of stable, monoclonal cell lines (10). Additionally, Bcl6-/Bcl-xL-transgenic B cells were described as unable to class-switch from IgM to IgG (10, 11), preventing the conversion of IgM+ input cells into long-term, IgG+ cell lines for direct production of monoclonal IgG.
In light of these limitations, we sought to identify an alternative transcriptional programming regimen that enables long-term B cell culture without the drawbacks of Bcl6-/Bcl-xL-transgenic cultures. We discovered that constitutive expression of Bach2, like Bcl6, prevents PC differentiation and enables long-term growth of B cells stimulated with CD154 and IL-21. Importantly, Bach2-transgenic B cell lines readily switch from IgM to IgG1 in vitro, and do not accumulate V(D)J mutations during prolonged culture, enabling the isolation of truly monoclonal B-cell lines for mAb production or other applications. These monoclonal cell lines typically express high levels of BCR, enabling direct identification of antigen-specific cell lines. Bach2-transgenic B cell cultures also quantitatively transduce intracellular signals when incubated with BCR ligands, demonstrating their potential as a system for studying BCR signaling or for screening libraries of candidate immunogens (4). Additionally, high expression of MHCII and molecules for co-stimulation of T cells indicates that Bach2-transgenic B-cell lines may be useful as an unlimited source of potent, autologous APCs (3) for T-cell epitope-mapping studies in humans or non-human primates, for which it is typically challenging to obtain large numbers of autologous APCs (63), and where use of EBV-transformed B cells confounds assays by activating EBV-specific T cells (12). Finally, Bach2-transgenic cell lines are permissive for re-infection with a second retrovirus (data not shown), potentially opening additional avenues for investigating B-cell function.
Another obvious application of Bach2-transgenic cultures is the production of mAbs. We demonstrated that this works best with a chemically inducible transgene, which, when de-activated, permits rapid differentiation into PCs with specific productivity (~20 pg cell−1 day−1) comparable to that of commercially used hybridoma lines and recombinant Ab production systems (64, 65). Importantly, Bach2-transgenic B cells have some potential advantages over recombinant mAb expression systems and hybridomas. Recombinant mAb production requires cloning the paired HC and LC genes from B cells, which is labor intensive at large scales, and usually destroys the B cells (66, 67). In contrast, producing mAbs from Bach2-transgenic cultures precludes the need for genetic cloning. Hybridomas, the classical option for mAb production, are notoriously labor-intensive and inefficient to generate, but have an admitted advantage over Bach2-transgenic cultures in that the former grow rapidly in suspension culture without the need for feeder cells or cytokines. In principle, Bach2-transgenic cultures could be easily converted into hybridomas if desired, because the ability to pre-screen cultures for Ag specificity and produce large monoclonal populations for cellular fusion would alleviate the main difficulties of hybridoma generation (e.g., low efficiency of B-cell fusion with myeloma cells (68, 69), and the considerable effort required to screen hybridomas for clones of interest).
In future studies, it will be important to determine whether Bach2-transgenic B cells can be adoptively transferred back into hosts to participate in in vivo humoral responses, as can wildtype B cells briefly clonally expanded in 40LB culture (1). If so, the Bach2 culture system may be a valuable tool for ex vivo expansion and characterization of specific clones of interest prior to adoptive transfer experiments. Previously, producing large populations of monoclonal B cells for in vivo experiments required either generation of BCR transgenic mice or in vitro BCR editing (e.g., by CRISPR/Cas9 (70)). The former usually solves the problem of establishing an inexhaustible source of monoclonal B cells for in vivo or in vitro studies, but is expensive, time-consuming, and subject to complications from immunological tolerance controls (44, 50). In contrast, the latter is far faster and easier to implement, but still faces the problem of working with a limited supply of cells (70). Cultures of Bach2-transgenic B cells would strike a happy balance, establishing a practically unlimited supply of monoclonal cells for experiments, without the cost and effort involved in transgenic mouse generation. Moreover, Bach2-transgenic cultures could be paired with CRISPR/Cas9-based gene editing to quickly establish permanent, monoclonal cell lines bearing any desired BCR. An important consideration for these experiments will be whether Bach2-transgenic B cell lines retain genomic stability during prolonged in vitro culture. We have not yet ascertained this.
Finally, to increase culture throughput, we have generated KI mice expressing a doxycycline-inducible Bach2 transgene (Finney and Kelsoe, manuscript in preparation). By culturing individual GC B cells isolated from immunized KI animals, we have established long-lived, antigen-specific, monoclonal cell lines that have the capacity to secrete large quantities of Ig when doxycycline is withdrawn. We are currently optimizing conditions for high-throughput generation of Bach2-transgenic cultures from these mice.
Contradictions in the behavior of Bcl6-transgenic B cell cultures
Although our results with the Bcl6 transgene generally confirm those of earlier reports in which transgenic Bcl6 enabled long-term culture of primary B cells (10, 11), there are notable discrepancies, which likely can be attributed to differences in the culture systems or cultured B cell populations. Whereas the 40LB system used here employs CD154- and BAFF-expressing feeder cells and exogenous IL-4 and IL-21, Kwakkenbos et al. used only CD154-expressing feeder cells with exogenous IL-21 (10, 11). Thus, the authors’ observation (11) that Bcl6-transgenic B cell cultures grow very slowly unless they are co-transduced with an anti-apoptotic protein (e.g., Bcl-xL) may stem from their culture system’s lack of BAFF, a critical B-cell survival factor (71). Likewise, the inability of their system to induce IgM+ B cells to switch to IgG (10) is probably due to the absence of cytokines that stimulate IgG CSR (e.g., IL-4 or IL-10) (1, 3, 72-74). Harder to explain is Kwakkenbos et al.’s report that Bcl6-transgenic B cell cultures continuously undergo SHM (10, 11), in contrast to our finding that these cultures do not accumulate Ig mutations. Once again, this discrepancy might arise from differences in the culture systems, or, alternatively, differences in the starting cell population, since Kwakkenbos et al. typically cultured memory B cells (10), whereas our studies primarily used Ag-naïve, mature follicular B cells. The molecular signals sufficient for SHM in re-activated memory B cells may be insufficient for SHM in newly activated naïve B cells.
Biological insights into functions of key B-cell transcription factors
Our study provides important insights into the functions of transcription factors involved in activated B-cell fate specification. First, we observed that IRF4 is critical for B-cell proliferation and/or survival in response to T-cell help signals. Although our finding is at odds with an initial report (75) describing IRF4 as dispensable for the proliferation of B cells stimulated in vitro with anti-CD40 and IL-4, our results agree with several groups’ observations that IRF4-deficiency impairs B-cell clonal expansion in response T-dependent immunization, BCR crosslinking, or LPS exposure (31, 32, 75). Curiously, discussions (17, 31) of IRF4’s function in activated B cells tend to minimize its role in supporting clonal expansion, focusing instead on its role in activating the differentiation programs of PCs or GC B cells. We propose that in addition to these functions, IRF4 coordinates the proliferation and/or survival of activated, B cells, a function that may be physiologically relevant in pre-GC B cells and extrafollicular plasmablasts, although not in GC B cells, were Irf4 is silenced and dispensable for proliferation (38, 76). The mechanism by which IRF4 exerts its pro-growth function is unclear, but may include induction of Myc (c-Myc) (77), a transcription factor critical for activated B-cell proliferation (78). Additionally, analogous to its role in T cells, IRF4 may 1) coordinate metabolic adaptation as cells transition from quiescence to a metabolically demanding, proliferative state, and 2) repress genes involved in cell cycle arrest and apoptosis (79-81). That IRF4 may play a similar role in Ag-experienced B cells is supported by the recent discoveries that IRF4 regulates metabolism in PCs and is constitutively required for their survival (37, 82).
In regard to Blimp1, previous work (40) demonstrated that before inducing Blimp1, activated B cells enter a “pre-plasmablast” stage characterized by concurrent expression of abundant Pax5 protein and Pax5-repressed genes. Therefore, it was proposed that the initial step of PC commitment involves an undefined mechanism that interferes with Pax5 protein function, thereby de-activating the B-cell identity (40). However, prior studies did not determine whether the Blimp1-independent pre-plasmablast stage is capable of sustained self-renewal – a plausible outcome, since Myc expression might continue in the absence of Blimp1, a key repressor (83). We answered this question, showing that Blimp1-deficient and -sufficient B cells both enter growth arrest in 40LB culture by day 10. Additionally, our observation that PC commitment and growth arrest cannot be circumvented by overexpression of Pax5 supports Kallies et al.’s model (40) for PC commitment by inactivation of Pax5 protein activity, rather than silencing of Pax5.
An important corollary of this model is that transgenic Bcl6 or Bach2 cannot program B-cell fate solely by repressing Prdm1. Rather, Bcl6 and Bach2 probably also have important roles in preventing the inactivation of Pax5, or at the very least, in preventing growth arrest. The simplest model is that Bach2 and Bcl6 act through the same pathways, because Bach2 and Bcl6 can bind and/or regulate many of the same genes (24, 84). In particular, Bach2 and Bcl6 have been proposed to enable B-cell proliferation and survival by regulating several cell-cycle checkpoint genes (23, 24, 85), and these functions are likely important for conferring long-term growth potential in the transgenic cell lines.
Supplementary Material
KEY POINTS.
A Bach2 transgene enables unlimited growth of B cells cultured with CD154 and IL-21.
Bach2-transgenic cell lines have stable BCRs and transduce signals upon BCR ligation.
B cells secrete large quantities of Ig after the Bach2 transgene is silenced.
ACKNOLWEDGMENTS
We are grateful for the excellent technical support of D. Liao, X. Liang, W. Zhang, S. Langdon, B. Li, and S. Slater at Duke University. We thank Profs. Weiguo Zhang and Minghua Zhu for advice on BCR signaling experiments. This research was conducted in part using equipment and services provided by the Duke University DNA Analysis Facility, Duke Cancer Institute Flow Cytometry Shared Resource, and the Duke Human Vaccine Institute Research Flow Cytometry Facility.
Funding sources:
This research was supported in part by NIH grants AI 144372, AI 128832, and AI 131251 (to G.K.).
Abbreviations
- AID
activation-induced deaminase
- CSR
Ig class-switch recombination
- Dox
doxycycline
- GC
germinal center
- HC
BCR heavy chain
- KI
knockin
- LC
BCR light chain
- MPER
membrane proximal external region of HIV-1 gp41
- NP
4-hydroxy-3-nitrophenyl acetyl
- PC
plasma cell
- rtTA
reverse tetracycline-controlled transactivator protein
- SHM
V(D)J somatic hypermutation
- TRE
tetracycline response element
Footnotes
The sequences presented in this article have been submitted to GenBank (www.ncbi.nlm.nih.gov/GenBank) under accession numbers MZ450902-MZ450924.
REFERENCES
- 1.Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, Azuma T, and Kitamura D. 2011. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun 2: 465. [DOI] [PubMed] [Google Scholar]
- 2.Quintans J, and Lefkovits I. 1974. Clonal expansion of lipopolysaccharide-stimulated B lymphocytes. J Immunol 113: 1373–1376. [PubMed] [Google Scholar]
- 3.Su KY, Watanabe A, Yeh CH, Kelsoe G, and Kuraoka M. 2016. Efficient Culture of Human Naive and Memory B Cells for Use as APCs. J Immunol 197: 4163–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver GC, Villar RF, Kanekiyo M, Nabel GJ, Mascola JR, and Lingwood D. 2016. In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat Protoc 11: 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindner JM, Cornacchione V, Sathe A, Be C, Srinivas H, Riquet E, Leber XC, Hein A, Wrobel MB, Scharenberg M, Pietzonka T, Wiesmann C, Abend J, and Traggiai E. 2019. Human Memory B Cells Harbor Diverse Cross-Neutralizing Antibodies against BK and JC Polyomaviruses. Immunity 50: 668–676 e665. [DOI] [PubMed] [Google Scholar]
- 6.Henle W, Diehl V, Kohn G, Zur Hausen H, and Henle G. 1967. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science 157: 1064–1065. [DOI] [PubMed] [Google Scholar]
- 7.Kohler G, and Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256: 495–497. [DOI] [PubMed] [Google Scholar]
- 8.Munz C 2017. Humanized mouse models for Epstein Barr virus infection. Curr Opin Virol 25: 113–118. [DOI] [PubMed] [Google Scholar]
- 9.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, and Calame KL. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol 173: 1158–1165. [DOI] [PubMed] [Google Scholar]
- 10.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, and Beaumont T. 2010. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 16: 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwakkenbos MJ, van Helden PM, Beaumont T, and Spits H. 2016. Stable long-term cultures of self-renewing B cells and their applications. Immunol Rev 270: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, Visser M, Stratton MR, Haanen JB, Spits H, van der Burg SH, and Schumacher TN. 2015. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med 21: 81–85. [DOI] [PubMed] [Google Scholar]
- 13.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, and Weigert M. 1984. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci U S A 81: 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinstein SH, Louzoun Y, and Shlomchik MJ. 2003. Estimating hypermutation rates from clonal tree data. J Immunol 171: 4639–4649. [DOI] [PubMed] [Google Scholar]
- 15.Bournazos S, and Ravetch JV. 2017. Diversification of IgG effector functions. Int Immunol 29: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu LL, Suscovich TJ, Fortune SM, and Alter G. 2018. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 18: 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nutt SL, Hodgkin PD, Tarlinton DM, and Corcoran LM. 2015. The generation of antibody-secreting plasma cells. Nat Rev Immunol 15: 160–171. [DOI] [PubMed] [Google Scholar]
- 18.Barberis A, Widenhorn K, Vitelli L, and Busslinger M. 1990. A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev 4: 849–859. [DOI] [PubMed] [Google Scholar]
- 19.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, and Igarashi K. 1998. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3' enhancer. Embo J 17: 5734–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, and Igarashi K. 2006. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem 281: 38226–38234. [DOI] [PubMed] [Google Scholar]
- 21.Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, and Nutt SL. 2008. Identification of Pax5 target genes in early B cell differentiation. J Immunol 180: 1719–1728. [DOI] [PubMed] [Google Scholar]
- 22.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, and Busslinger M. 2007. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27: 49–63. [DOI] [PubMed] [Google Scholar]
- 23.Miura Y, Morooka M, Sax N, Roychoudhuri R, Itoh-Nakadai A, Brydun A, Funayama R, Nakayama K, Satomi S, Matsumoto M, Igarashi K, and Muto A. 2018. Bach2 Promotes B Cell Receptor-Induced Proliferation of B Lymphocytes and Represses Cyclin-Dependent Kinase Inhibitors. J Immunol 200: 2882–2893. [DOI] [PubMed] [Google Scholar]
- 24.Huang C, Geng H, Boss I, Wang L, and Melnick A. 2014. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood 123: 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usui T, Wakatsuki Y, Matsunaga Y, Kaneko S, Koseki H, and Kita T. 1997. Overexpression of B cell-specific activator protein (BSAP/Pax-5) in a late B cell is sufficient to suppress differentiation to an Ig high producer cell with plasma cell phenotype. J Immunol 158: 3197–3204. [PubMed] [Google Scholar]
- 26.Nera KP, Kohonen P, Narvi E, Peippo A, Mustonen L, Terho P, Koskela K, Buerstedde JM, and Lassila O. 2006. Loss of Pax5 promotes plasma cell differentiation. Immunity 24: 283–293. [DOI] [PubMed] [Google Scholar]
- 27.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, and Igarashi K. 2004. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429: 566–571. [DOI] [PubMed] [Google Scholar]
- 28.Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, and Kurosaki T. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol 17: 861–869. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Gadzinsky A, Gong L, Tong H, Calderon V, Li Y, Kitamura D, Klein U, Langdon WY, Hou F, Zou YR, and Gu H. 2018. Cbl Ubiquitin Ligases Control B Cell Exit from the Germinal-Center Reaction. Immunity 48: 530–541 e536. [DOI] [PubMed] [Google Scholar]
- 30.Zhang TT, Gonzalez DG, Cote CM, Kerfoot SM, Deng S, Cheng Y, Magari M, and Haberman AM. 2017. Germinal center B cell development has distinctly regulated stages completed by disengagement from T cell help. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, and Sciammas R. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38: 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, and Singh H. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25: 225–236. [DOI] [PubMed] [Google Scholar]
- 33.Lin KI, Angelin-Duclos C, Kuo TC, and Calame K. 2002. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol 22: 4771–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin KI, Lin Y, and Calame K. 2000. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol Cell Biol 20: 8684–8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, and Staudt LM. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17: 51–62. [DOI] [PubMed] [Google Scholar]
- 36.Minnich M, Tagoh H, Bonelt P, Axelsson E, Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M, and Busslinger M. 2016. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol 17: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK, Kallies A, Busslinger M, and Nutt SL. 2016. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol 17: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, and Dalla-Favera R. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7: 773–782. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, and Calame K. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19: 607–620. [DOI] [PubMed] [Google Scholar]
- 40.Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, and Nutt SL. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity 26: 555–566. [DOI] [PubMed] [Google Scholar]
- 41.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, and Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proceedings of the National Academy of Sciences of the United States of America 103: 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, and Dymecki SM. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140. [DOI] [PubMed] [Google Scholar]
- 43.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, and Rajewsky K. 1997. B cell development under the condition of allelic inclusion. Immunity 6: 225–233. [DOI] [PubMed] [Google Scholar]
- 44.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, Kelsoe G, and Haynes BF. 2011. Rescue of HIV-1 Broad Neutralizing Antibody-Expressing B Cells in 2F5 VH x VL Knockin Mice Reveals Multiple Tolerance Controls. J Immunol 187: 3785–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oi VT, Morrison SL, Herzenberg LA, and Berg P. 1983. Immunoglobulin gene expression in transformed lymphoid cells. Proc Natl Acad Sci U S A 80: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verkoczy L, Moody MA, Holl TM, Bouton-Verville H, Scearce RM, Hutchinson J, Alam SM, Kelsoe G, and Haynes BF. 2009. Functional, non-clonal IgMa-restricted B cell receptor interactions with the HIV-1 envelope gp41 membrane proximal external region. PLoS One 4: e7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura T 1998. New experimental approaches in retrovirus-mediated expression screening. Int J Hematol 67: 351–359. [DOI] [PubMed] [Google Scholar]
- 48.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, and Saito T. 2006. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol 7: 67–75. [DOI] [PubMed] [Google Scholar]
- 49.Kuraoka M, Holl TM, Liao D, Womble M, Cain DW, Reynolds AE, and Kelsoe G. 2011. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finney J, Yang G, Kuraoka M, Song S, Nojima T, Verkoczy L, Kitamura D, Haynes BF, and Kelsoe G. 2019. Cross-Reactivity to Kynureninase Tolerizes B Cells That Express the HIV-1 Broadly Neutralizing Antibody 2F5. J Immunol 203: 3268–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dal Porto JM, Haberman AM, Shlomchik MJ, and Kelsoe G. 1998. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol 161: 5373–5381. [PubMed] [Google Scholar]
- 52.Helmreich E, Kern M, and Eisen HN. 1961. The secretion of antibody by isolated lymph node cells. J Biol Chem 236: 464–473. [PubMed] [Google Scholar]
- 53.Hibi T, and Dosch HM. 1986. Limiting dilution analysis of the B cell compartment in human bone marrow. Eur J Immunol 16: 139–145. [DOI] [PubMed] [Google Scholar]
- 54.Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, and Nutt SL. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med 200: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bothwell AL, Paskind M, Reth M, Imanishi-Kari T, Rajewsky K, and Baltimore D. 1981. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell 24: 625–637. [DOI] [PubMed] [Google Scholar]
- 56.Reth M, Hammerling GJ, and Rajewsky K. 1978. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur J Immunol 8: 393–400. [DOI] [PubMed] [Google Scholar]
- 57.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, and Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563. [DOI] [PubMed] [Google Scholar]
- 58.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, and Durandy A. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102: 565–575. [DOI] [PubMed] [Google Scholar]
- 59.Kurosaki T, Shinohara H, and Baba Y. 2010. B cell signaling and fate decision. Annu Rev Immunol 28: 21–55. [DOI] [PubMed] [Google Scholar]
- 60.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, and Bujard H. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769. [DOI] [PubMed] [Google Scholar]
- 61.Andersson J, Sjoberg O, and Moller G. 1972. Mitogens as Probes for Immunocyte Activation and Cellular Cooperation. Transplant Rev-Denma 11: 131–177. [DOI] [PubMed] [Google Scholar]
- 62.Metcalf D, Warner NL, Nossal GJ, Miller JF, Shortman K, and Rabellino E. 1975. Growth of B lymphocyte colonies in vitro from mouse lymphoid organs. Nature 255: 630–632. [DOI] [PubMed] [Google Scholar]
- 63.Sette A, and Peters B. 2007. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Current Opinion in Immunology 19: 106–110. [DOI] [PubMed] [Google Scholar]
- 64.Kunert R, and Reinhart D. 2016. Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol 100: 3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li F, Vijayasankaran N, Shen AY, Kiss R, and Amanullah A. 2010. Cell culture processes for monoclonal antibody production. MAbs 2: 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of immunological methods 329: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tiller T, Busse CE, and Wardemann H. 2009. Cloning and expression of murine Ig genes from single B cells. Journal of immunological methods 350: 183–193. [DOI] [PubMed] [Google Scholar]
- 68.Listek M, Honow A, Gossen M, and Hanack K. 2020. A novel selection strategy for antibody producing hybridoma cells based on a new transgenic fusion cell line. Sci Rep 10: 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu X, McGraw PA, House FS, and Crowe JE Jr. 2008. An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. Journal of immunological methods 336: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartweger H, McGuire AT, Horning M, Taylor JJ, Dosenovic P, Yost D, Gazumyan A, Seaman MS, Stamatatos L, Jankovic M, and Nussenzweig MC. 2019. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J Exp Med 216: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mackay F, and Browning JL. 2002. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol 2: 465–475. [DOI] [PubMed] [Google Scholar]
- 72.Tangye SG, Ferguson A, Avery DT, Ma CS, and Hodgkin PD. 2002. Isotype switching by human B cells is division-associated and regulated by cytokines. J Immunol 169: 4298–4306. [DOI] [PubMed] [Google Scholar]
- 73.Pene J, Rousset F, Briere F, Chretien I, Bonnefoy JY, Spits H, Yokota T, Arai N, Arai K, Banchereau J, and et al. 1988. IgE production by normal human lymphocytes is induced by interleukin 4 and suppressed by interferons gamma and alpha and prostaglandin E2. Proc Natl Acad Sci U S A 85: 6880–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malisan F, Briere F, Bridon JM, Harindranath N, Mills FC, Max EE, Banchereau J, and Martinez-Valdez H. 1996. Interleukin-10 induces immunoglobulin G isotype switch recombination in human CD40-activated naive B lymphocytes. J Exp Med 183: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, and Mak TW. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275: 540–543. [DOI] [PubMed] [Google Scholar]
- 76.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, Smyth GK, Tarlinton DM, Belz GT, Corcoran LM, Kallies A, and Nutt SL. 2014. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol 192: 3200–3206. [DOI] [PubMed] [Google Scholar]
- 77.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, and Staudt LM. 2008. IRF4 addiction in multiple myeloma. Nature 454: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, and Alt FW. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55. [DOI] [PubMed] [Google Scholar]
- 79.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, and Kallies A. 2013. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 14: 1155–1165. [DOI] [PubMed] [Google Scholar]
- 80.Mahnke J, Schumacher V, Ahrens S, Kading N, Feldhoff LM, Huber M, Rupp J, Raczkowski F, and Mittrucker HW. 2016. Interferon Regulatory Factor 4 controls TH1 cell effector function and metabolism. Sci Rep 6: 35521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, and Sun J. 2013. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity 39: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Low MSY, Brodie EJ, Fedele PL, Liao Y, Grigoriadis G, Strasser A, Kallies A, Willis SN, Tellier J, Shi W, Gabriel S, O'Donnell K, Pitt C, Nutt SL, and Tarlinton D. 2019. IRF4 Activity Is Required in Established Plasma Cells to Regulate Gene Transcription and Mitochondrial Homeostasis. Cell Rep 29: 2634–2645 e2635. [DOI] [PubMed] [Google Scholar]
- 83.Lin Y, Wong K, and Calame K. 1997. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science 276: 596–599. [DOI] [PubMed] [Google Scholar]
- 84.Ochiai K, Muto A, Tanaka H, Takahashi S, and Igarashi K. 2008. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol 20: 453–460. [DOI] [PubMed] [Google Scholar]
- 85.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, Nowak D, Thoennissen GB, Rand V, Graeber TG, Koeffler HP, Carroll WL, Willman CL, Hall AG, Igarashi K, Melnick A, and Muschen M. 2013. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med 19: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



