Abstract
Ipomoea obscura, small white morning glory, is an ornamental plant belonging to the family Convolvulaceae, and cultivated worldwide. I. obscura generates white petals including a pale-yellow colored star-shaped center (flower vein). Its fully opened flowers were known to accumulate trace amounts of carotenoids such as β-carotene. In the present study, the embryogenic calli of I. obscura, were successfully produced through its immature embryo culture, and co-cultured with Agrobacterium tumefaciens carrying the β-carotene 4,4′-ketolase (crtW) and β-carotene 3,3′-hydroxylase (crtZ) genes for astaxanthin biosynthesis in addition to the isopentenyl diphosphate isomerase (idi) and hygromycin resistance genes. Transgenic plants, in which these four genes were introduced, were regenerated from the infected calli. They generated bronze (reddish green) leaves and novel petals that exhibited a color change from pale-yellow to pale-orange in the star-shaped center part. Especially, the color of their withered leaves changed drastically. HPLC-PDA-MS analysis showed that the expanded leaves of a transgenic line (T0) produced astaxanthin (5.2% of total carotenoids), adonirubin (3.9%), canthaxanthin (3.8%), and 3-hydroxyechinenone (3.6%), which indicated that these ketocarotenoids corresponded to 16.5% of the total carotenoids produced there (530 µg g−1 fresh weight). Furthermore, the altered traits of the transgenic plants were found to be inherited to their progenies by self-crossing.
Keywords: astaxanthin, embryogenic callus, Ipomoea obscura, transgenic plant
Introduction
For ornamental crops, color variations in their flowers and also in their leaves are attractive and important traits. Japanese morning glory (Ipomoea nil), which is a well-known and popular ornamental plant in Japan, retains such flower color traits (Iida et al. 2004; Yoneda 1990). Its petals produce flavonoids with pink, red, sky-blue, blue, or purple color, while carotenoids that are responsible for yellow, orange or red pigmentation are entirely absent there (Yamamizo et al. 2010). Small white morning glory, Ipomoea obscura, is another ornamental species belonging to the genus Ipomoea (the family Convolvulaceae), and cultivated worldwide. Unlike diversity of the I. nil flowers, I. obscura generates only white petals including a pale-yellow colored star-shaped center (flower vein), along with deep purple throats. Its fully opened flowers were reported to accumulate not flavonoids but carotenoids such as β-carotene with their trace amounts (Yamamizo et al. 2010). The I. obscura plant has been positioned as a minor ornamental crop because of very narrow color variations in its flowers and leaves.
Among ornamental Ipomea plant species, pathway engineering researches have been performed only in Japanese morning glory (I. nil). It was reported in I. nil that white flower was generated by targeting and knocking out the dihydroflavonol-4-reductase-B (InDFR-B) gene with the CRISPR/Cas9 system (Watanabe et al. 2017a). The CRISPR/Cas9-based knocking-out of the carotenoid cleavage dioxygenase (InCCD4) gene caused pale yellow flowers in the white-flowered cultivar I. nil cv. AK77 (Watanabe et al. 2018). Hoshino et al. (2019) generated yellow flowers by introducing the snapdragon (Antirrhinum majus) aurone biosynthetic genes into I. nil. As for the I. obscura plants, any pathway engineering research has not been carried out. The main purpose of the present study is to confer new flower and leaf colors on this plant species by engineering carotenoid biosynthetic pathway.
Astaxanthin is a red carotenoid (ketocarotenoid) pigment with stronger antioxidative activity (Miki 1991). Considerable numbers of reports have indicated that the introduction of astaxanthin biosynthesis genes, which contain the β-carotene 4,4′-ketolase and β-carotene 3,3′-hydroxylase genes, into higher plants lead to color changes in their flowers and leaves due to the accumulation of ketocarotenoids. The higher-plant species demonstrated above contained Petunia hybrida (Umemoto et al. 2006), Lotus japonicus (Suzuki et al. 2007), Arabidopsis thaliana (Zhong et al. 2011), Nicotiana glauca (Mortimer et al. 2017), Nicotiana tabacum (tobacco; Hasunuma et al. 2008; Hirschberg 2001), Solanum lycopersicum (tomato; Huang et al. 2013), Solanum tuberosum (potato; Mortimer et al. 2016), and Lactuca sativa (lettuce; Harada et al. 2014). Thus, we can expect that the introduction of the genes for astaxanthin biosynthesis causes attractive color alteration in the flowers and leaves of Ipomoea species.
Modern techniques in plant biotechnology, such as somatic hybrid formation and plant-host transformation followed by its regeneration, are expected to enable further breeding of ornamental Ipomoea species. As for Japanese morning glory (I. nil), many reports have been published on its tissue culture (Jia and Chua 1992; Otani and Shimada 1998; Shimizu et al. 2003, 2005; Yoneda and Nakamura 1987), on its transformation (Kikuchi et al. 2005; Ono et al. 2000; Shibuya et al. 2014; Takatori et al. 2015; Watanabe et al. 2017b) and on its genome editing (Watanabe et al. 2017a, 2018), whereas few reports have described embryogenic callus formation and plant regeneration in other ornamental Ipomoea species (Ishikuro et al. 2014).
In the present study, we developed a production system of embryogenic calli from I. obscura, regenerated transgenic plants after introducing the genes for astaxanthin biosynthesis into the callus, and altered their flower and leaf colors due to the accumulation of ketocarotenoids. We also proposed an astaxanthin biosynthetic pathway, which was generated in the leaves of the transgenic I. obscura plants. This is the first report concerning the alteration of flower and leaf colors in ornamental I. obscura using genes for carotenoid biosynthesis.
Materials and methods
Plant materials
Embryogenic calli were induced from immature embryos of Ipomoea obscura, according to the method for Ipomoea tricolor (Ishikuro et al. 2014). Immature embryos (2 to 10.9 mm in diameter) from I. obscura grown in a green-house were cultured on LS medium (Linsmaier and Skoog 1965) containing 1 mg l−1 of 4-fluorophenoxyacetic acid (4FA), 3% (w/v) sucrose, and 0.32% (w/v) gelrite (4F1 medium) (Otani and Shimada 1996) or 1 mg l−1 of 4FA, 6% (w/v) sucrose, and 0.32% (w/v) gelrite (4F1S60 medium) (Ishikuro et al. 2014) at 26°C in the dark. Induced embryogenic calli were sub-cultured on the same fresh media at 30-day intervals. Cultures were kept at 26°C in the dark. Thirty embryos were provided for each kind of media.
Construction of plasmids
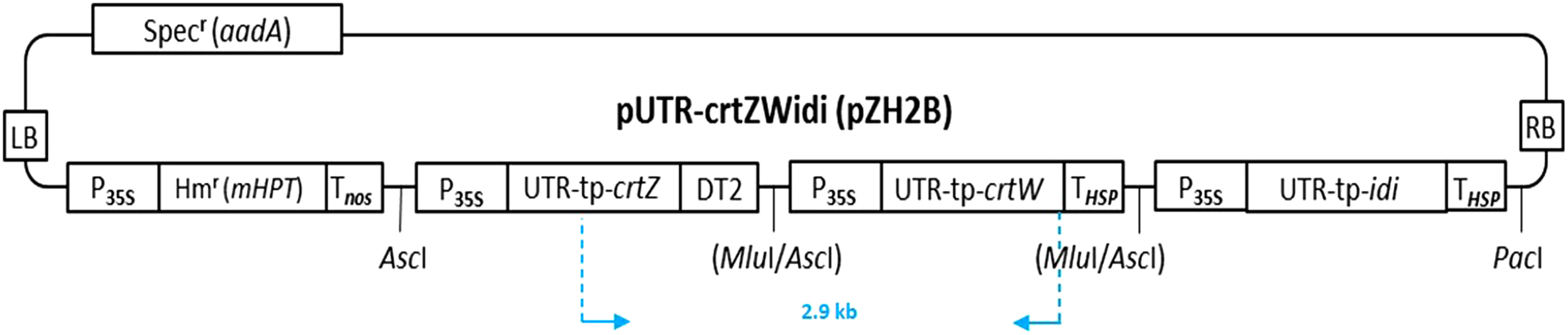
Plasmids for plant transformation, named pUTR-crtZWidi(pZH2B) (Figure 1), were constructed as described (Mortimer et al. 2017), in which the isopentenyl diphosphate (IPP) isomerase (idi) gene cassette flanked by the cauliflower mosaic virus 35S promoter and the HSP terminator was added to the construct, and binary vector pZH2B was used instead of pZK3B (Kuroda et al. 2010). The idi gene cassette was derived of the 5′-untranslated region (UTR) sequence of the tobacco alcohol dehydrogenase gene (Satoh et al. 2004) and the tp-idi gene as described (Fujisawa et al. 2009; Misawa et al. 1993).
Figure 1. Structure of the plasmid for plant transformation, pUTR-crtZWidi(pZH2B). Specr, the spectinomycin resistance gene; LB and RB, left and right borders; P35S, the cauliflower mosaic virus 35S promoter; Hmr, the hygromycin resistance gene; Tnos, the terminator of the nopaline synthase gene (nos); THSP, the terminator of the heat shock protein (HSP18.2) gene from Arabidopsis thaliana; DT2, double terminator that consists of THSP-Tnos; UTR, 5′-untranslated region of the alcohol dehydrogenase gene from tobacco (Nicotiana tabacum); tp, the transit peptide sequence from the pea RuBisCO small subunit gene; crtZ and crtW, the β-carotene 3,3′-hydroxylase and β-carotene 4,4′-ketolase genes, respectively, which code for the corresponding proteins from Brevundimonas sp. strain SD212; idi, the isopentenyl diphosphate (IPP) isomerase (type 2) gene that encodes the corresponding protein from Paracoccus sp. strain N81106.
Agrobacterium-mediated transformation
Transformation was performed according to the method of Otani et al. (1998) with some modifications. Around twenty-eight-day-old embryogenic calli of I. obscura were soaked in a bacterial suspension, which contained Agrobacterium tumefaciens strain EHA101 possessing plasmid pUTR-crtZWidi(pZH2B), for 2 min, and blotted with sterile double-layered filter paper to remove excess bacteria. Then, the calli were transferred onto a co-culture medium which was LS medium supplemented with 1 mg l−1 4FA, 10 mg l−1 acetosyringone (3′,5′-dimethoxy-4′-hydroxy-acetophenone; Sigma-Aldrich, St Louis, MO, USA), 2.3 g l−1 2-morpholinoethane-sulfonic acid (MES; Wako Pure Chemical Industries, Ltd., Osaka, Japan), 3% (w/v) sucrose, and 0.32% (w/v) gelrite. After 2 days of co-cultivation at 23°C in the dark, the infected embryogenic calli were washed five to six times with sterile distilled water supplemented with 500 mg l−1 carbenicillin and then transferred onto selection medium containing LS medium supplemented with 1 mg l−1 4FA, 25 mg l−1 hygromycin B, 500 mg l−1 carbenicillin, 3% (w/v) sucrose, and 0.32% (w/v) gelrite. The calli were sub-cultured onto fresh medium every 2 weeks. After 60 days of culture on the selection medium, the calli were transferred onto the somatic embryo formation medium containing LS medium supplemented with 4 mg l−1 abscisic acid (ABA), 1 mg l−1 gibberellic acid (GA3), 25 mg l−1 hygromycin B, 500 mg l−1 carbenicillin, 3% (w/v) sucrose, and 0.32% (w/v) gelrite, and cultured at 26°C under a 16-h photoperiod. Somatic embryos formed from hygromycin-resistant calli were transferred onto LS medium supplemented with 0.05 mg l−1 ABA, 25 mg l−1 hygromycin B, 500 mg l−1 carbenicillin, 3% (w/v) sucrose, and 0.32% (w/v) gelrite after 21 days of culture on the somatic embryo formation medium. Plantlets regenerated from hygromycin-resistant somatic embryos were cultured on LS PGR-free medium supplemented with 25 mg l−1 hygromycin B, 500 mg l−1 carbenicillin, 3% (w/v) sucrose, and 0.8% (w/v) agar.
Regenerated plants
Regenerated plants were cultured on LS PGR-free medium supplemented with 3% (w/v) sucrose and 0.8% (w/v) agar for further growth and good rooting. Then, the regenerated transgenic plants were planted into wagner pots (159 mm in diameter and 190 mm in height) containing culture soil [Engeibaido 1 (NIHON HIRYO CO., LTD. Tokyo, Japan)] in a biohazard green house.
PCR analysis
Genomic DNA was extracted according to the method of Liu et al. (1995). PCR was performed using Emerald Amp PCR Master Mix (TaKaRa, Ohtsu, Japan). The PCR conditions were as follows: 98°C for 10 s, 66°C for 20 s, 72°C for 3 min; 35 cycles. Primer pairs were designed at the start codon of crtZ (5′-ATG GCT TGG CTT ACT TGG ATC GCT CTT TTC CT-3′) and the stop codon of crtW (5′-TCA AGA CTC TCC TCT CCA AAG TCT CCA CCA AG-3′). PCR products were electrophoresed using a 1% (w/v) agarose gel.
Analysis of carotenoids in the transgenic plants
Analysis of carotenoids in the regenerated transgenic plants and the wild type plant was carried out based on the methods described (Maoka 2016, 2018; Maoka et al. 2020). Carotenoid pigments were extracted from the plant samples with acetone at room temperature, and then transferred to n-hexane: diethyl ether (Et2O) (1 : 1, v/v) by adding water. The n-hexane: Et2O phase was washed with water and dehydrated on anhydrous sodium sulphate. The total carotenoid amounts were calculated using the coefficient of E1%cm=2400 at λ max. Quantitative and qualitative analysis of the extracted carotenoids was carried out as follows: HPLC-PDA-MS analysis of carotenoids was carried out using a Waters Xevo G2S Q TOF (time-of-flight) mass spectrometer (Waters Corporation, Milford, CT, USA) equipped with an Acquity UPLC system. The electro-spray ionization (ESI) TOF MS spectra were acquired by scanning from m/z 100 to 1,500 with a capillary voltage of 3.2 kV, cone voltage of 20 eV, and source temperature of 120°C. Nitrogen was used as a nebulizing gas at a flow rate of 30 l h−1. UV-visible (VIS) absorption spectra were recorded from 200 to 600 nm by the PDA. An Acquity 1.7 µm BEH Shield RP18 (2.1 id X 100 mm) column (Waters Corporation, Milford, CT, USA) was used at 40°C, developed with acetonitrile (MeCN): H2O (85 : 15)—MeCN: methanol (MeOH) (65 : 35) (linear gradient 0 to 15 min) as a mobile phase, at a flow rate of 0.4 ml min−1 for the HPLC system. Carotenoids were identified by UV-VIS, and MS spectral data and the retention time in HPLC with a comparison of authentic samples that had been purified in T. Maoka’s Laboratory.
Results and discussion
Embryogenic callus formation
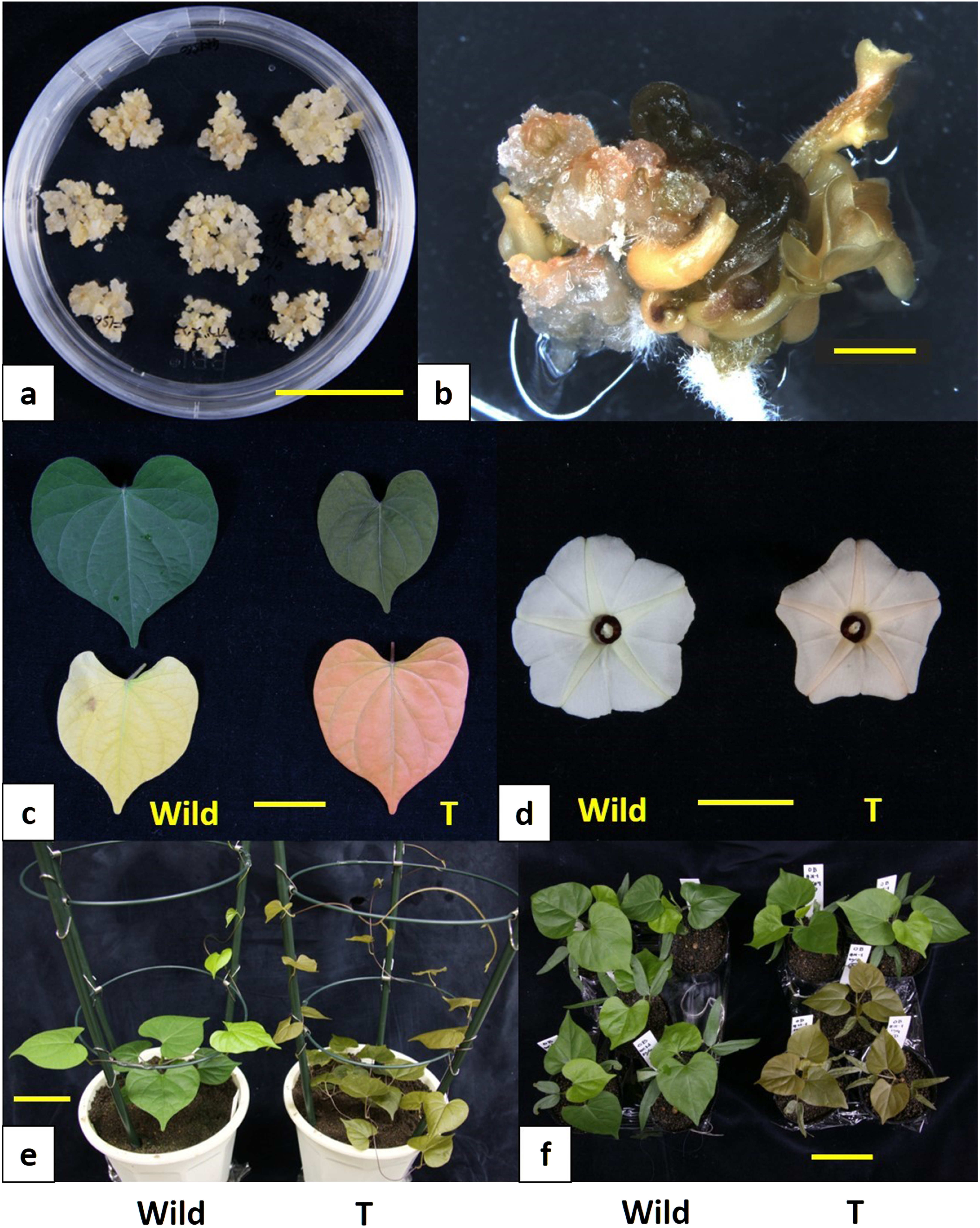
After three weeks of culture on both media (4F1 medium and 4F1S60 medium), the immature embryos of Ipomoea obscura produced embryogenic calli from their cotyledon parts. The highest frequency of embryogenic callus formation was 20% in the 4F1S60 medium (Figure 2a), while 10% of cultured immature embryos grew up to embryogenic calli on the 4F1 medium. This result indicated that the concentration of sucrose was an important factor for generating embryogenic calli from I. obscura. Similar results were also obtained in Ipomea purpurea and Ipomoea tricolor (Ishikuro et al. 2014). Thus, a higher concentration of sucrose in the callus induction medium is likely to be advantageous for producing embryogenic calli in the Ipomoea genus. In addition, supplementation with a synthetic auxin 4FA in the media was certainly considered effective in producing embryogenic calli from the immature embryos of I. obscura. Since we had also succeeded in forming embryogenic calli with 4FA in several Ipomoea genera including genotypes of sweetpotato (Ishikuro et al. 2014; Otani and Shimada 1996, 1998; Takahata et al. 2010), 4FA was strongly suggested to be effective for forming embryogenic calli in the Ipomoea genus.
Figure 2. Embryogenic callus formation and transgenic plant regeneration in I. obscura. (a) Embryogenic callus derived from an immature embryo. Bar represents 30 mm; (b) hygromycin-resistant somatic embryo formation from an Agrobacterium-infected embryogenic callus. Bar represents 2 mm; (c) raw expanded leaves of non-transgenic (left, Wild) and transgenic (right, T) plants (upper side). Aged leaves of non-transgenic (left, Wild) and transgenic (right, T) plants (lower side). Bar represents 20 mm; (d) flowers of non-transgenic (left, Wild) and transgenic line #21-1 (right, T) plants. Bar represents 20 mm.; (e) appearance of non-transgenic (left, Wild) and transgenic line #21-1 (right, T) plants. Bar represents 50 mm; (f) self progenies of non-transgenic (left, Wild) and transgenic line #21-1 (right, T) plants. Bar represents 50 mm.
Transformation
Totally, 165 calli were infected with Agrobacterium tumefaciens possessing plasmid pUTR-crtZWidi (pZH2B). Consequently, 62 hygromycin-resistant calli or somatic embryos (37.6%) were generated from the infected embryogenic calli within 60 days after the bacterial-infection treatment. The color of those resistant calli and somatic embryos varied from green to orange (Figure 2b). Although deep orange colored cultures failed to regenerate plantlets, complete plantlets were regenerated from green or pale orange colored calli or somatic embryos in four months after the bacterial-infection treatment. More than ten independent hygromycin-resistant plants were regenerated. All the transgenic plants were grown for five months in a biohazard greenhouse. The transgenic plants could be classified into three groups showing bronze, pale bronze and green in terms of leaf color. PCR analysis was performed on arbitrarily selecting one transgenic plant lines from individual leaf color groups (#21-1 for bronze group, #21-3 for pale bronze group and #21-4 for green group).
Genomic DNA was extracted from expanded leaves (50 mg) of the non-transgenic plant and regenerated hygromycin-resistant plants according to the methods of Liu et al. (1995). Using these extracts as templates, PCR was performed to detect the crtZ and crtW genes. Fragments, whose size is anticipated to be 2.9 kb (Figure 1), corresponding to the length from the crtZ start codon to the crtW stop codon, were detected from the representative transgenic plants (Figure 3). A shorter fragment of non-specific amplification was observed in the wild type plant.
Figure 3. PCR analysis for detection of the crtW and crtZ genes in three independent transgenic plant lines of Ipomoea obscura. Lane WT non-transgenic plant, lanes 1, 3, and 4 independent transgenic plants #21-1, -3 and -4, lane plasmid plasmid DNA.
Morphological phenotypes of the transgenic plants
The morphological phenotypes of the transgenic plant lines #21-1, #21-3 and #21-4 were drastically different from those of the wild-type plants, especially the colors of the leaves and flowers changed remarkably. The leaf, petiole, stem, and petal colors of all the transformed plant lines were altered, while that of the wild type was green (Figure 2c–e). The leaf, petiole, and stem color of all the transformed plant lines were bronze (reddish). When chlorophyll contents became less, the aged leaf color of the transformed plants became bright salmon pink, which indicated accumulation of ketocarotenoids such as astaxanthin (Figure 2c). These transgenic plants may be used as autumn leaves plants, which is an important horticultural trait in Japan. The flower color of the transgenic plants was pale orange (changed from pale yellow) in the star-shaped center (flower vein) part (Figure 2d). It is very likely that the flower vein part produced ketocarotenoids. This altered flower color can be an attractive trait in this plant species.
HPLC analysis of transgenic plants
Carotenoid pigments were extracted from the expanded leaves of transgenic I. obscura plant #21-1 (T0) which showed bronze leaf color. HPLC-PDA-MS analysis of the extract showed the presence not only of astaxanthin but also of adonirubin, canthaxanthin, and 3-hydroxyechinenone (Figure 4), i.e., it was found to produce astaxanthin (5.2% of total carotenoids), adonirubin (3.9%), canthaxanthin (3.8%), and 3-hydroxyechinenone (3.6%) (Table 1). This result also indicated that the total ketocarotenoids that were newly generated in the leaves of the transgenic I. obscura plants corresponded to 16.5% of the total carotenoids produced there (530 µg g−1 fresh weight) (Table 1). The biosynthetic pathway of these ketocarotenoids was proposed as shown in Figure 5.
Figure 4. HPLC chromatograms (at 450 nm) of extracts from the leaves of transgenic Ipomoea obscura #21-1 (T0) plant (a) and I. obscura wild type plant (b). Chl, Chlorophylls; Neo, Neoxanthin; Ast, Astaxanthin; Vio, Violaxanthin; Lut, Lutein; Ado, Adonirubin; OH-Echi, 3-Hydroxyechinenone; cis-Lut, cis-Lutein; Can, Canthaxanthin; b-Car, β-Carotene.

Table 1. Carotenoid content and composition in the leaves of non-transgenic wild type and transgenic plant #21-1 (T0) of Ipomoea obscura.
| Wild type | Transgenic #21-1 | |
|---|---|---|
| Total carotenoid content (µg g−1 FW) | 790 | 530 |
| Carotenoid composition (%) | ||
| β-Carotene | 25.2 | 22.3 |
| Canthaxanthin | 3.8 | |
| cis-Lutein | 8.2 | 3.9 |
| 3-Hydroxyechinenone | 3.6 | |
| Adonirubin | 3.9 | |
| Lutein | 60.4 | 48.9 |
| Astaxanthin | 5.2 | |
| Violaxanthin | 1.9 | 1.3 |
| Neoxanthin | 4.3 | 7.1 |
| Total ketocarotenoids (%) | 0 | 16.5 |
Figure 5. Proposed carotenoid biosynthetic pathway in the leaves of transgenic ornamental Ipomoea obscura plants. Heavy blue arrows and narrow black arrows represent metabolic routes mediated by the introduced foreign genes and the endogenous genes, respectively.

Progeny analysis
To investigate the stability of the introduced genes in the genome, transgenic plant line #21-1 (T0) which showed bronze leaf color were self-pollinated, and the seeds (T1) were harvested. The T1 seedlings were examined for the presence of the introduced astaxanthin biosynthesis genes, and subjected to segregation analysis. The T1 seedlings showed either green or bronze tops, while all the seedlings of the self-pollinated untransformed plants showed green tops (Figure 2f). PCR analysis revealed that bronze colored seedlings possessed the introduced foreign genes, while green colored seedlings had no introduced genes (Figure 6). The segregation ratio in the T1 generation was 6.5 : 1 for bronze to green. This result suggested that the astaxanthin biosynthesis genes were stably inherited to the progeny. However this segregation ratio did not reveal the number of inserted gene loci, while two to three copies of the foreign genes are likely to be present there. A more detailed study would be needed by increasing the number of seedlings.
Figure 6. PCR analysis for the detection of the crtW and crtZ genes in self cross progenies of transgenic plant line 1-1 of Ipomoea obscura. Lane WT non-transgenic plant, lanes 1, 2, 3, 5, 7 to 10 self-pollinated progenies, lane P plasmid DNA, lane M molecular size marker.

There are a few reports on tissue culture in Ipomoea obscura (Mungole et al. 2009; Sinha and Bandyopadhyay 2011), while definite plant regeneration from cultured tissues or calli has not yet been achieved in this plant species. In the present study, we succeeded in producing embryogenic calli from the immature embryos of I. obscura. Such a production system of embryogenic calli can provide a strong tool for plant biotechnology in this plant species.
Transgenic plants were regenerated after introducing the β-carotene 4,4′-ketolase (crtW) and β-carotene 3,3′-hydroxylase (crtZ) genes for astaxanthin biosynthesis in addition to the isopentenyl diphosphate (IPP) isomerase (idi) gene into the callus. They were found to generate bronze (reddish green) leaves that were changed from green, as well as novel petals that exhibited a color change from pale-yellow to pale-orange in the star-shaped center (flower vein) part, due to the production of ketocarotenoids. This is the first report concerning the alteration of organ colors in Ipomoea obscura by techniques of modern biotechnology. It was also confirmed that the changed colors have been maintained for more than six years in vitro and inherited to the progenies. Therefore, altered colors of the transgenic I. obscura plants are considered to be a new trait of I. obscura to provide an additional advantage for ornamental uses.
On the other hand, more drastic color changes may be required to raise product value. Hasunuma et al. (2008) and Harada et al. (2014) achieved direct transformation of the chloroplasts in tobacco (N. tabacum) and lettuce (Lactuca sativa) plants, respectively, using the same crtW and crtZ genes, and regenerated the corresponding transplastomic plants. They possessed much more reddish leaves, due to almost complete conversion from β-carotene to ketocarotenoids including astaxanthin, and further the transplastomic lettuce generated flowers with a vibrant salmon pink color. Its leaf and flower colors should be very attractive traits for ornamental plants. Thus, we want to try to perform chloroplast transformation of I. obscura plants for acquiring more changed colors.
It has additionally been reported that I. obscura is used not only for horticultural purposes but also as a medicinal plant (Londhe et al. 2017; Mungole et al. 2009; Sinha and Bandyopadhyay 2011). Therefore, systems that were established for tissue culture and genetic transformation in this plant species are expected to provide a great advantage to promote its cell agriculture and molecular breeding for medicinal ingredients.
Acknowledgments
We thank Dr. Atsushi Hoshino (National Institute for Basic Biology, Aichi, Japan) for useful advice.
Abbreviations
- HPLC-PDA-MS
high performance liquid chromatography-photodiode array detection-mass spectrometry; PGR, plant growth regulator
Disclosure of potential conflicts of interest
The authors declare that they have no conflicts of interest.
References
- Fujisawa M, Takita E, Harada H, Sakurai N, Suzuki H, Ohyama K, Shibata D, Misawa N (2009) Pathway engineering of Brassica napus seeds using multiple key-enzyme genes involved in ketocarotenoid formation. J Exp Bot 60: 1319–1332 [DOI] [PubMed] [Google Scholar]
- Harada H, Maoka T, Osawa A, Hattan J, Kanamoto H, Shindo K, Otomatsu T, Misawa N (2014) Construction of transplastomic lettuce (Lactuca sativa) dominantly producing astaxanthin fatty acid esters and detailed chemical analysis of generated carotenoids. Transgenic Res 23: 303–315 [DOI] [PubMed] [Google Scholar]
- Hasunuma T, Miyazawa SI, Yoshimura S, Shinzaki Y, Tomizawa KI, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55: 857–868 [DOI] [PubMed] [Google Scholar]
- Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Hoshino A, Mizuno T, Shimizu K, Mori S, Fukada-Tanaka S, Furukawa K, Ishiguro K, Tanaka Y, Iida S (2019) Generation of yellow flowers of the Japanese morning glory by engineering its flavonoid biosynthetic pathway toward aurones. Plant Cell Physiol 60: 1871–1879 [DOI] [PubMed] [Google Scholar]
- Huang JC, Zhong YJ, Liu J, Sandmann G, Chen F (2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17: 59–67 [DOI] [PubMed] [Google Scholar]
- Iida S, Morita Y, Choi JD, Park KI, Hoshino A (2004) Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Adv Biophys 39: 141–159 [PubMed] [Google Scholar]
- Ishikuro H, Nitasaka E, Otani M (2014) An efficient plant regeneration through embryogenic callus formation and direct somatic embryogenesis via immature embryo culture in Ipomoea purpurea and I. tricolor. Plant Biotechnol 31: 179–183 [Google Scholar]
- Jia SR, Chua NH (1992) Somatic embryogenesis and plant regeneration from immature embryo culture of Pharbitis nil. Plant Sci 87: 215–223 [Google Scholar]
- Kikuchi R, Sage-Ono K, Kamada H, Ono M (2005) Efficient transformation mediated by Agrobacterium tumefaciens with a ternary plasmid in Pharbitis nil. Plant Biotechnol 22: 285–302 [Google Scholar]
- Kuroda M, Kimizu M, Mikami C (2010) A simple set of plasmids for the production of transgenic plants. Biosci Biotechnol Biochem 74: 2348–2351 [DOI] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F (1965) Organic growth factor requirement of tobacco tissue culture. Physiol Plant 18: 100–127 [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Londhe DK, Neel RS, Bhuktar AS (2017) Ethno-medicinal uses of some species of genus Ipomoea L. from Maharashtra state. Internat J Appl Res 3: 82–84 [Google Scholar]
- Maoka T (2016) Structural studies of carotenoids in plants, animals, and food products. In: Kaczor A, Baranska A (eds) Carotenoids Nutrition, Analysis and Technology. Wiley Blackwell, UK, pp 103–129
- Maoka T (2018) New acetylenic carotenoid 6′-epimomadoxanthin from the rosary goby Gymnogobius castaneus. J Oleo Sci 97: 1259–1263 [DOI] [PubMed] [Google Scholar]
- Maoka T, Kawase N, Ueda T, Nishida R (2020) Carotenoids of doragonflies, from the perspective of comparative biochemical and chemical ecological studies. Biochem Syst Ecol 89: 104001 [Google Scholar]
- Miki W (1991) Biological functions and activities of animal carotenoids. Pure Appl Chem 63: 141–146 [Google Scholar]
- Misawa N, Yamano S, Linden H, De Felipe MR, Lucas M, Ikenaga H, Sandmann G (1993) Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtI in transgenic plants showing an increase of β-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J 4: 833–840 [DOI] [PubMed] [Google Scholar]
- Mortimer CL, Misawa N, Ducreux L, Campbell R, Taylor M, Bramley PM, Fraser PD (2016) Product stability and sequestration mechanisms in Solanum tuberosum engineered to biosynthesize high value ketocarotenoids. Plant Biotechnol J 14: 140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer CL, Misawa N, Perez-Fons L, Robertson FP, Harada H, Bramley PM, Fraser PD (2017) The formation and sequestration of non-endogenous ketocarotenoids in transgenic Nicotiana glauca. Plant Physiol 173: 1617–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungole A, Awati R, Dey S, Chaturvedi A, Zanwa P (2009) In-vitro callus induction and shoot regeneration in Ipomoea obscura (L.): Potent Indian medicinal plant. Indian J Sci Technol 2: 24–26 [Google Scholar]
- Ono M, Sage-Ono K, Kawakami M, Hasebe M, Ueda K, Masuda K, Inoue M, Kamada H (2000) Agrobacterium-mediates transformation and regeneration of Pharbitis nil. Plant Biotechnol 17: 211–216 [Google Scholar]
- Otani M, Shimada T (1996) Efficient embryogenic callus formation in sweet potato (Ipomoea batatas (L.) Lam.). Breed Sci 46: 275–280 [Google Scholar]
- Otani M, Shimada T (1998) Efficient embryogenic callus formation from immature embryo of Japanese morning glory (Pharbitis nil Choisy). Plant Biotechnol 15: 127–129 [Google Scholar]
- Otani M, Shimada T, Kimura T, Saito A (1998) Transgenic plant production from embryogenic callus of sweet potato (Ipomoea batatas (L.) Lam.) using Agrobacterium tumefaciens. Plant Biotechnol 15: 11–16 [Google Scholar]
- Satoh J, Kato K, Shinmyo A (2004) The 5′-untranslated region of the tobacco alcohol dehydrogenase gene functions as an effective translational enhancer in plant. J Biosci Bioeng 98: 1–8 [DOI] [PubMed] [Google Scholar]
- Shibuya K, Shimizu K, Niki T, Ichimura K (2014) Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J 79: 1044–1051 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Hashimoto M, Hashimoto F, Sakata Y (2003) Plant regeneration from suspension cultures in Japanese morning glory (Ipomoea nil (L.) Roth.). J Jpn Soc Hortic Sci 72: 409–414 [Google Scholar]
- Shimizu K, Tokumura T, Hashimoto F, Sakata Y (2005) In vitro propagation of sterile mutant strains in Japanese morning glory by sub-culturing embryoids derived from an immature embryo. J Jpn Soc Hortic Sci 74: 311–317 [Google Scholar]
- Sinha T, Bandyopadhyay A (2011) Induction of callogenesis in Ipomoea obscura (L.) Ker- Gawl, a little known medicinal plant. Afr J Biotechnol 10: 19161–19166 [Google Scholar]
- Suzuki S, Nishihara M, Nakatsuka T, Misawa N, Ogiwara I, Yamamura S (2007) Flower color alteration in Lotus japonicus by modification of the carotenoid biosynthetic pathway. Plant Cell Rep 26: 951–959 [DOI] [PubMed] [Google Scholar]
- Takahata Y, Tanaka M, Otani M, Katayama K, Kitahara K, Nakayachi O, Nakayama H, Yoshinaga M (2010) Inhibition of the expression of the starch synthase II gene leads to lower pasting temperature in sweetpotato starch. Plant Cell Rep 29: 535–543 [DOI] [PubMed] [Google Scholar]
- Takatori Y, Shimizu K, Ogata J, Endo H, Ishimaru K, Okamoto S, Hashimoto F (2015) Cloning of the flavonoid 3′-Hydroxylase gene of Eustoma grandiflorum (Raf.) shinn. (EgF3′H) and complementation of an F3′H-deficient mutant of Ipomoea nil (L.) Roth. by heterologous expression of EgF3′H. Hort J 84: 131–139 [Google Scholar]
- Umemoto N, Takano M, Shimada H, Mamiya K, Toguri T (2006) Flower color modification by xanthophyll biosynthetic genes in petunia. Plant Cell Physiol 47: Suppl S110 [Google Scholar]
- Watanabe K, Kobayashi A, Endo M, Sage-Ono K, Toki S, Ono M (2017a) CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci Rep 7: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Oda-Yamamizo C, Sage-Ono K, Ohmiya A, Ono M (2017b) Overexpression of carotenogenic genes in the Japanese morning glory Ipomoea (Pharbitis) nil. Plant Biotechnol 34: 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Oda-Yamamizo C, Sage-Ono K, Ohmiya A, Ono M (2018) Alteration of flower colour in Ipomoea nil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4. Transgenic Res 27: 25–38 [DOI] [PubMed] [Google Scholar]
- Yamamizo C, Kishimoto S, Ohmiya A (2010) Carotenoid composition and carotenogenic gene expression during Ipomoea petal development. J Exp Bot 61: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y (1990) Japanese morning glory. In: Ammirato PV, Evans DA, Sharp WR, Bajaj YPS (eds) Handbook of Plant Cell Culture, vol. 5: Ornamental Species. McGraw-Hill Publ, New York, pp 509–533
- Yoneda Y, Nakamura N (1987) Embryoid formation and plantlet regeneration from cultured immature embryos in three strains of Pharbitis nil. Rep Lib Arts. Shizuoka Univ (Sciences) 23: 11–20 [Google Scholar]
- Zhong YJ, Huang JC, Liu J, Li Y, Jiang Y, Xu ZF, Sandmann G, Chen F (2011) Functional characterization of various algal carotenoid ketolases reveals that ketolating zeaxanthin efficiently is essential for high production of astaxanthin in transgenic Arabidopsis. J Exp Bot 62: 3659–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]



