Abstract
Recent studies have revealed that treatment-resistant cancer stem-like cells (CSCs)/cancer-initiating cells (CICs) can be targeted by cytotoxic T lymphocytes (CTLs). CTLs recognize antigenic peptides derived from tumor-associated antigens; thus, the identification of tumor-associated antigens expressed by CSCs/CICs is essential. Human leucocyte antigen (HLA) ligandome analysis using mass spectrometry enables the analysis of naturally expressed antigenic peptides; however, HLA ligandome analysis requires a large number of cells and is challenging for CSCs/CICs. In this study, we established a novel bladder CSC/CIC model from a bladder cancer cell line (UM-UC-3 cells) using an ALDEFLUOR assay. CSCs/CICs were isolated as aldehyde dehydrogenase (ALDH)-high cells and several ALDHhigh clone cells were established. ALDHhigh clone cells were enriched with CSCs/CICs by sphere formation and tumorigenicity in immunodeficient mice. HLA ligandome analysis and cap analysis of gene expression using ALDHhigh clone cells revealed a distinctive antigenic peptide repertoire in bladder CSCs/CICs, and we found that a glutamate receptor, ionotropic, kainite 2 (GRIK2)-derived antigenic peptide (LMYDAVHVV) was specifically expressed by CSCs/CICs. A GRIK2 peptide-specific CTL clone recognized GRIK2-overexpressing UM-UC-3 cells and ALDHhigh clone cells, indicating that GRIK2 peptide can be a novel target for bladder CSC/CIC-targeting immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-03025-z.
Keywords: Bladder cancer, Cancer stem cell, GRIK2, Antigen, Cytotoxic T lymphocytes
Introduction
Bladder cancer is the most common malignancy of the urinary system with 81,400 estimated new cases and 17,980 estimated deaths yearly in the USA. [1] Of the newly diagnosed bladder cancers, 15–20% are muscle-invasive bladder cancer (MIBC), and the prognosis of MIBC is unsatisfactory with a 5-year overall survival rate of 60% after cystectomy. [2] Cisplatin-based chemotherapy in neoadjuvant therapy and adjuvant therapy improves the overall survival of patients with MIBC [3–5]; however, treatment resistance directly affects prognosis. In the past few decades, cancer stem-like cells (CSCs)/cancer-initiating cells (CICs) have been revealed to play important roles in resistance to chemotherapy and radiotherapy, and novel approaches to target treatment-resistant bladder CSCs/CICs are expected to improve current bladder cancer therapy. [6]
CSCs/CICs are defined as a small subpopulation of cancer cells that are endowed with higher tumor-initiating ability, self-renewal capability, and differentiation ability. [6] CSCs/CICs are resistant to standard therapies including chemotherapy and radiotherapy owing to their high expression of transporter genes and anti-apoptotic genes and exist in a dormant state. [7] CSCs/CICs are less immunogenic than non-CSCs/CICs because of their lower expression of human leucocyte antigen (HLA) class 1 antigen-presenting machinery-related genes and higher expression of immune-suppressive cytokines including interleukin-4, interleukin-10, and tumor growth factor-β. [8] However, CSCs/CICs can be targeted by cytotoxic T lymphocytes (CTLs), and several CSC/CIC-related antigens have been described [9–13], indicating that cancer immunotherapy that can cause an anti-CSC/CIC-specific CTL reaction might be a promising approach for targeting CSCs/CICs. To identify antigenic peptides, reverse immune genetics based on gene expression profiles and HLA ligandome analysis using mass spectrometry are major approaches [14, 15]. HLA ligandome analysis is an ideal approach because the antigenic peptides presented by cancer cells can be analyzed directly; however, HLA ligandome analysis is challenging since the number of CSCs/CICs is limited. Previously, a stable CSC/CIC model enabled HLA ligandome analysis of human CSCs/CICs. [13, 16]
Bladder CSCs/CICs can be isolated by using several markers including CD44, CD90, CK5, aldehyde dehydrogenase (ALDH), and ABCG2. [17–19] We also identified bladder CSCs/CICs as ALDHhigh cells from human bladder cancer cell lines and found that glutamate receptor, ionotropic, kainite 2 (GRIK2) is expressed at higher levels in bladder CSCs/CICs than in non-CSCs/CICs. [20] However, no stable bladder CSC/CIC model that is applicable for HLA ligandome analysis has been generated yet.
In this study, we established a novel bladder CSC/CIC model from a human bladder cancer cell line (UM-UC-3 cells). Using this model, we identified antigenic peptides expressed on bladder CSCs/CICs. The antigenic peptide GRIK2 was found to be preferentially expressed on bladder CSCs/CICs. An antigenic peptide derived from GRIK2 may be a promising molecule for bladder CSC/CIC-targeting immunotherapy.
Materials and Methods
Ethics statement
The mice were maintained and experimented on in accordance with the guidelines of and after approval by the Committee of Sapporo Medical University School of Medicine, Animal Experimentation Center (permit number 08–006). Any animal found unhealthy or sick was promptly euthanized.
Cell lines and cell culture
The following human cell lines were used in this study: bladder carcinoma (UM-UC3), erythroleukemia (K562), and TAP-deficient (T2) cells. UM-UC3, K562, and T2 cells were purchased from the American Type Culture Collection (Rockville, MD). Cell lines were maintained in Dulbecco’s modified Eagle’s medium or RPMI (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (5 mg/mL penicillin, 5 mg/mL streptomycin; Thermo Fisher Scientific, Waltham, MA). Cells were cultured in an incubator at 37 °C with humidified air and 5% CO2.
ALDEFLUOR assay and establishment of ALDHhigh and ALDHlow clone cells
An ALDEFLUOR assay (STEMCELL Technologies, Vancouver, Canada) using UM-UC-3 cells was performed as described previously. [20] Briefly, 1.0 × 106 cells were suspended in 1 mL assay buffer containing 1.5 mM of the ALDH1 substrate BODIPY®-aminoacetaldehyde and incubated for 50 min at 37 °C. The ALDH1 inhibitor diethylamino-benzaldehyde (DEAB), at a tenfold molar excess, was used as a negative control. Flow cytometry and cell sorting were performed using a FACSAria II cell sorter (BD Biosciences, Bedford, MA). ALDHhigh and ALDHlow cells were single cell sorted using FACSAria II into 96-well plates. Growing cells were transferred to T25 culture flasks (Corning, Inc., Corning, NY) and cultured for more than 1 month. ALDHhigh clone cells (H-1, H-6, and H-10) and ALDHlow clone cells (L-1, L-3, and L-8) were examined by an ALDEFLUOR assay.
Sphere-formation assay, mouse xenograft assay, and resistance to cisplatin and radiation
A sphere-formation assay and estimation of CSCs/CICs were performed as described previously. [21] A mouse xenograft assay was performed as described previously. [20] Briefly, 1.0 × 100, 101, 102, and 103 UM-UC-3 H-10 cells and 1.0 × 101, 102, 103, and 104 UM-UC-3 L-3 cells were transplanted into the subcutaneous space of 4–6-week-old female BALB/c-nu/nu mice. Tumor size was assessed weekly using a caliper and calculated using the following formula: tumor size (mm3) = (longest diameter × shortest diameter2)/2. To estimate the number of CSCs/CICs, the ELDA web site (http://bioinf.wehi.edu.au/software/elda/) was used. [22]
Resistance to cisplatin (CDDP) and radiation was assessed as described previously. [11, 20] Briefly, UM-UC-3 H-10 and L-3 cells were incubated with cisplatin at several concentrations for 72 h and cell viability was assessed using a WST-8 assay (Dojindo Molecular Technologies, Kumamoto, Japan). Radiation was delivered by using an M100W (SOFTEX, Inc., Austin, TX), and cell viability was assessed 72 h later by a WST-8 assay.
Antibody and HLA ligandome analysis
A hybridoma for anti-HLA-A2 (BB7.2) antibodies was cultured in Hybridoma-SFM (Thermo Fisher Scientific) supplemented with 1% penicillin/streptomycin in CELLine bioreactor flasks (Corning, Inc.). The produced monoclonal antibodies were condensed and collected through a semipermeable membrane during cell culture. HLA ligandome analysis using anti-HLA-A2 (BB7.2) antibodies was performed as described previously. [23]
CAGE analysis and quantitative RT-PCR
For comprehensive mRNA expression profiling, cap analysis of gene expression (CAGE) was performed. CAGE library preparation, sequencing, mapping, and gene expression and motif discovery analyses were performed by DNAFORM (Yokohama, Kanagawa, Japan). CAGE libraries were prepared from UM-UC-3 wild-type (WT), H-1, H-6, H-10, L-1, L-3, and L-8 cells. In brief, RNA quality was assessed by a Bioanalyzer (Agilent, Santa Clara, CA) to ensure that the RNA integrity number was > 7.0, and A260/280 and 260/230 ratios were > 1.7. First strand cDNAs were transcribed to the 5′-end of capped RNAs, attached to CAGE "bar code" tags, and the sequenced CAGE tags were mapped to the mouse mm9 genome using BWA software (v0.5.9) after discarding ribosomal or non-A/C/G/T base-containing RNAs.
For tag clustering, CAGE-tag 5′ coordinates were input for CAGEr clustering [24] using the Paraclu algorithm [25] with default parameters. Raw and processed CAGE data have been deposited in the NCBI GEO database (GSE166947).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed as described previously. [10]
CTL induction, ELISPOT assay, and establishment of a GRIK2 peptide-specific CTL clone
CTL induction and establishment of a CTL clone were performed as described previously. [14] Briefly, peripheral blood mononuclear cells (PBMCs) isolated from an HLA-A*02:01-positive donor were stimulated twice weekly with 10 μg/mL GRIK2 peptide (Cosmo Bio, Inc., Tokyo, Japan), and reactivity for GRIK2 peptide were assessed by an interferon-gamma (IFNγ) ELISPOT assay. GRIK2 peptide-reactive wells were stained with GRIK2-HLA-A2 tetramer (MBL, Nagoya, Japan), and tetramer-positive cells were single cell sorted using a FACSAria II (BD Biosciences) into 96-well plates (Corning, Inc.). Sorted CTLs were cultured with 100 Gy-irradiated PBMCs as feeder cells for 2 weeks, and growing cells were evaluated by staining with GRIK2-HLA-A2 tetramer.
Immune selection model
H-10 cells and L-3 cells were mixed at a ratio of 1:9 and evaluated by ALDEFLUOR assay. For CTL selection, CTL clone 9G23 were added at effector/target ratio of 5:1 and cultured over night. Then, the growing medium was changed and cultured for 2 days. The immune selected cells were evaluated by ALDEFLUOR assay, cisplatin sensitivity and radiation sensitivity.
Statistical analysis
Student’s t-test was used to compare two groups. P < 0.05 was considered to indicate a significant difference. Differences of the estimated frequencies of CSCs/CICs were analyzed by a Chi-square test.
Results
Establishment of novel stable cancer stem-like cell lines of human bladder cancer
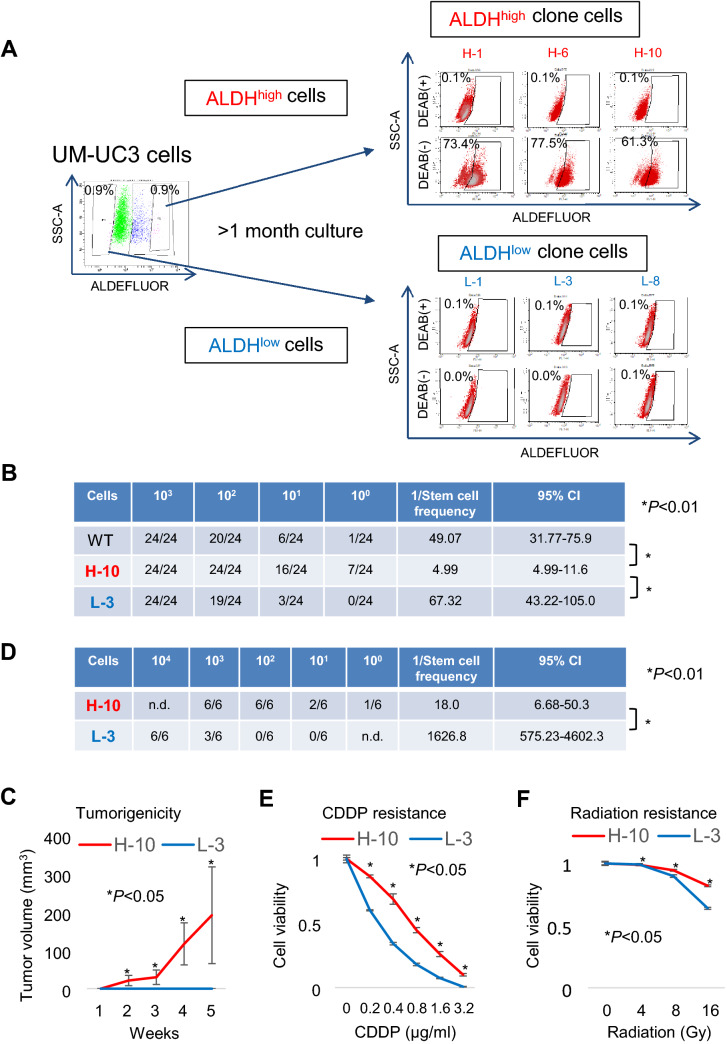
Previously, we isolated CSCs/CICs from the UM-UC-3 bladder cancer cell line as ALDHhigh cells by an ALDEFLUOR assay. [20] In this study, we aimed to identify novel immunological targets of ALDHhigh cells by HLA ligandome analysis. However, HLA ligandome analysis requires as many as 1.0 × 109 cells, and the collection of this number of ALDHhigh cells by the ALDEFLUOR assay is not realistic. Therefore, we established stable ALDHhigh cells from the UM-UC-3 human bladder cancer cell line. UM-UC-3 cells were analyzed by the ALDEFLUOR assay, and 0.9% of ALDHhigh cells were detected. ALDHhigh cells (0.9%) and ALDHlow cells (0.9%) were single cell sorted, and several clone cells were established. After culture for more than 1 month, we re-analyzed the clone cells. Clone cells derived from ALDHhigh cells showed high ALDHhigh ratios, whereas clone cells derived from ALDHlow cells showed low ALDHlow ratios (Fig. 1a), indicating that the ALDHhigh clone cells (H-1, H-6, and H-10) contain high ratios of ALDHhigh cells and that the ALDHlow clone cells (L-1, L-3, and L-8) contain extremely low ratios of ALDHlow cells.
Fig. 1.
Establishment of a bladder CSC/CIC model. A ALDEFLUOR assay of the UM-UC-3 human bladder cancer line cell. Left panel: UM-UC-3 cells were stained with BODIPY®-aminoacetaldehyde and analyzed by FACS. The ALDHhigh-positive gate was defined by a sample treated with the ALDH1 inhibitor DEAB. The top 0.9% (ALDHhigh) and lower 0.9% (ALDHlow) cells were single cell sorted and cultured for more than 1 month. Right panel: Clone cells established from ALDHhigh and ALDHlow cells were analyzed by an ALDEFLUOR assay. The ALDHhigh-positive gate was defined by a sample treated with DEAB. Percentages indicate the ALDHhigh-positive rates. Clone cells derived from ALDHhigh cells (H-1, H-6, and H-10) and ALDHlow cells (L-1, L-3, and L-8) were analyzed. B Sphere-formation assay of UM-UC-3 WT, H-10, and L-3 cells. Sphere-forming ability of WT UM-UC-3 cells, ALDHhigh cells (H-10), and ALDHlow cells (L-3) was assessed. The cells were seeded at 1.0 × 103, 102, 101, and 100 cells/well in an ultra-low attachment plate and cultured for 1 week. Sphere-forming wells were counted. Stem cell frequency was analyzed using the ELDA web site. CI: confidence interval. Differences of the estimated frequencies of CSCs/CICs were analyzed by a Chi-square test. C and D Tumor-forming ability of UM-UC-3 H-10 and L-3 cells. BALB/c-nu/nu mice were injected subcutaneously with 1.0 × 103, 102, 101, and 100 H-10 cells or 1.0 × 104, 103, 102, and 101 L-3 cells. The tumor growth curves of mice injected with 1.0 × 103 H-10 cells and 1.0 × 103 L-3 cells are shown in Fig. 1C. Data are shown as means ± standard deviation. Statistical analyses of the data were performed using a bilateral Student’s t-test. Tumor incidence and estimated CSC/CIC frequency are summarized in Fig. 1D. Stem cell frequency was analyzed using the ELDA web site. CI: confidence interval. Differences of the estimated frequencies of CSCs/CICs were analyzed by a Chi-square test. E and F. Resistance to cisplatin and radiation. H-10 and L-3 cells were treated with cisplatin (CDDP) or radiation at a serial dose, and cell viability was assessed by a WST-8 assay. Data are shown as means ± standard deviation. Statistical analyses for the data were performed using a bilateral Student’s t-test
To confirm that CSCs/CICs are enriched in ALDHhigh clone cells, we performed in vitro sphere-formation and in vivo tumor-formation assays. H-10 ALDHhigh clone cells showed greater sphere- and tumor-forming ability than L-3 ALDHlow clone cells (Fig. 1B, C, D), indicating that CSCs/CICs are enriched in H-10 cells. H-10 cells showed resistance to cisplatin (CDDP) and radiation compared to L-3 cells (Fig. 1E, F). These findings indicate that H-10 cells are a stable CSC/CIC model and L-3 cells are a stable non-CSC/CIC model.
Identification of antigenic peptides presented by HLA-A2 of bladder CSCs/CICs
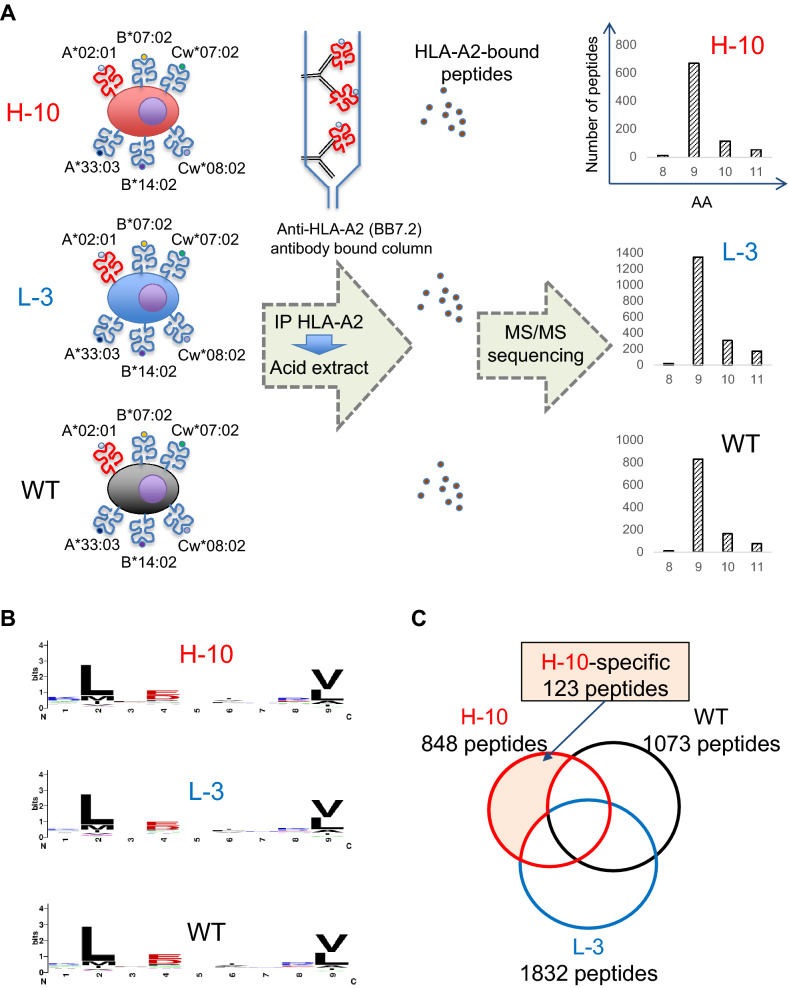
We previously established HLA ligandome analysis using HLA allele-specific antibodies that enable the comprehensive analysis of antigenic peptides that are presented by a specific HLA allele [13, 15]. Since UM-UC-3 cells are HLA-A*02:01-positive and HLA-A*02:01 is the most common HLA allele worldwide, we used an anti-HLA-A2 antibody (BB7.2) to purify antigenic peptides bound to HLA-A2 with 1.0 × 109 UM-UC-3 H-10 cells, L-3 cells, and WT cells. After immunoprecipitation and acid extraction, antigenic peptides were analyzed by tandem mass spectrometry (Fig. 2A). We identified 848 peptides from H-10 cells, 1,832 peptides from L-3 cells, and 1,073 peptides from WT cells (summarized in Tables S1, S2, and S3). The antigenic peptides were 8- to 11-mer in length, and 9-mer peptides were common in H-10, L-3, and WT cells (Fig. 2A). WebLogo analysis using 9-mer peptides revealed that hydrophobic amino acids (L and M) were common in the second position and V and L were common in the C-terminal ninth position, which are compatible with previously reported anchor motifs of HLA-A2. Of the 848 peptides derived from H-10 cells, 123 were H-10 cell-specific (Fig. 2C).
Fig. 2.
Isolation of HLA ligands expressed in bladder CSCs/CICs. A Schematic summary of HLA ligandome analysis. HLA-class 1 molecules were immunoprecipitated using an anti-HLA-A2 antibody (BB7.2), and HLA ligands were isolated by acid treatment. The HLA ligand landscape was analyzed by mass spectrometry. A summary of peptide length is shown in the right panel. UM-UC-3 H-10, L-3, and WT cells were used. B Summary of the antigenic peptides. WebLogo analysis using 9-mer peptides isolated from H-10, L-3, and WT cells is shown. L: leucine; V: valine. C Venn diagram of antigenic peptides. The diagram indicates the number of peptides isolated from H-10, L-3, and WT cells; 848, 1832, and 1073 peptides were isolated from H-10, L-3, and WT cells, respectively, with 123 H-10 cell-specific peptides
Gene expression analysis and identification of CSC/CIC-specific antigenic peptides
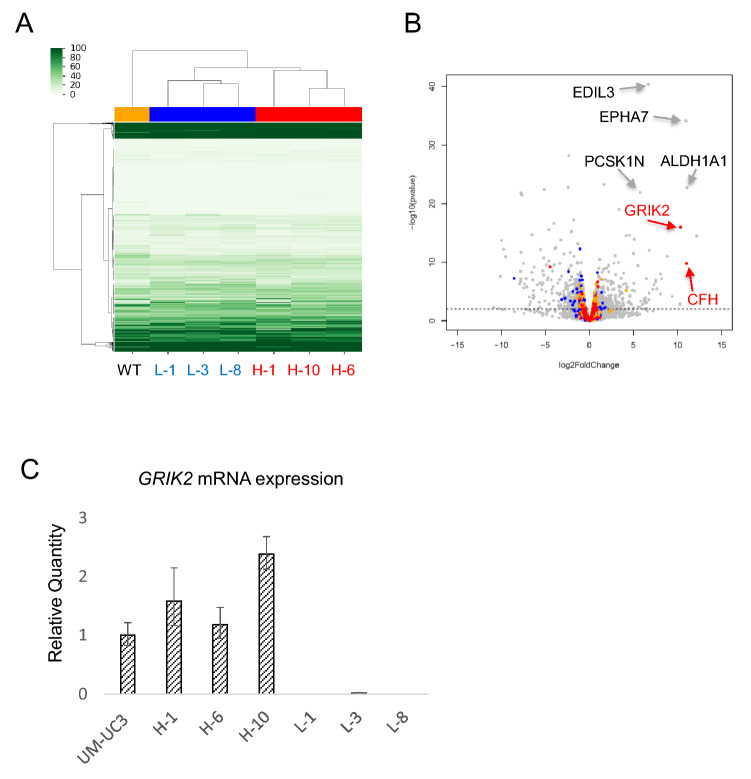
To reduce the number of candidate peptides, we added gene expression information. Comprehensive RNA expression was assessed by CAGE using WT, L-1, L-3, L-8, H-1, H-6, and H-10 cells. Cluster analysis revealed that L-1, L-3, and L-8 cells and H-1, H-6, and H-10 cells made clusters, suggesting similarities between those cells (Fig. 3A). Several genes were specifically expressed in ALDHhigh clone cells. Among them, only GRIK2 and complement factor H (CFH) were shared with the ALDHhigh clone cells assessed by HLA ligandome analysis (Fig. 3B). The expression of GRIK2 in ALDHhigh clone cells was confirmed by qRT-PCR (Fig. 3C). Furthermore, GRIK2 is expressed at higher level in ALDHhigh cells derived from J82 another human bladder cancer cell line compared with J82 wild-type cells (supplemental Figure S1) suggesting that the expression of GRIK2 might be common in bladder CSCs/CICs.
Fig. 3.
Gene expression analysis of bladder CSCs/CICs. A Heatmap of CAGE. A summary of gene expression using CAGE is shown. WT, L-1, L-3, L-8, H-1, H-6, and H-10 cells were used. B Volcano plot. Gene expression of ALDHhigh clone cells (H-1, H-6, and H-10) and ALDHlow clone cells (L-1, L-3, and L-8) was analyzed by a Volcano plot. Several genes were specifically expressed in ALDHhigh clone cells, and only GRIK2 and CFH (indicated in red) were shared with the ALDHhigh-specific HLA ligandome data. C. Quantitative real-time PCR. Relative quantities of GRIK2 mRNAs in UM-UC3 cells, H-1, H-6, H-10, L-1, L-3 and L-8 cells were analyzed by qRT-PCR. Each value is the mean relative quantity ± SD
Antigenic peptide encoded by GRIK2 is a novel immunological target of bladder CSCs/CICs
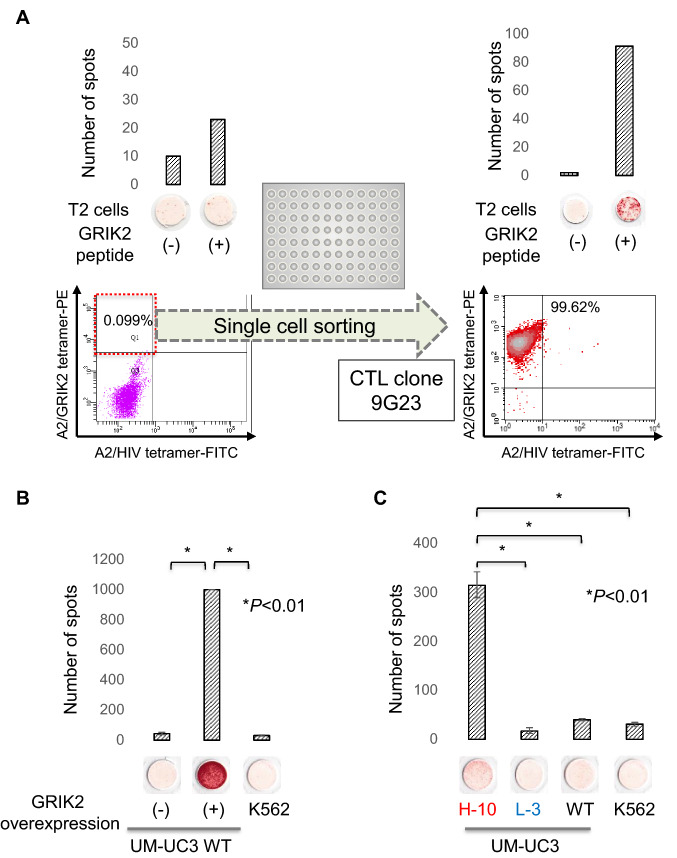
We previously reported that GRIK2 is overexpressed in bladder CSCs/CICs and has a role in the maintenance of these cells [20]. CFH is expressed in liver at high level and we hypothesized that CFH cannot be a target of immunotherapy. We therefore focused on GRIK2 peptide. GRIK2-derived peptide is LMYDAVHVV encoded at 322–330 amino acid residue of GRIK2 protein. In the present study, we thus assessed the immunogenicity of the antigenic peptide encoded by GRIK2. Human PBMCs derived from an HLA-A*02:01-positive donor were stimulated with GRIK2 peptide, and reactivity to GRIK2 peptide was analyzed by an IFNγ ELISPOT assay. The PBMCs showed relative higher IFNγ secretion to GRIK2 peptide-pulsed T2 cells compared to peptide-negative T2 cells (Fig. 4A), indicating that GRIK2 peptide might be immunogenic. For further analysis, a GRIK2 peptide-specific CTL clone was established. We stained the CTLs with a GRIK2 peptide/HLA-A2 tetramer, and the tetramer-positive rate was 0.9%. The tetramer-positive cells were single cell sorted (Fig. 4A). One of the growing wells showed GRIK2 peptide-specific IFNγ secretion by an ELISPOT assay and a rate as high as 99.62% to GRIK2/HLA-A2 tetramer was detected, indicating the CTL clone was GRIK2 peptide-specific.
Fig. 4.
Immunogenicity of a GRIK2-derived antigenic peptide. A Establishment of a GRIK2 peptide-specific CTL clone. PBMCs derived from an HLA-A2-positive donor were stimulated several times with GRIK2 peptide and analyzed by IFNγ ELISPOT and tetramer assays (left panel). GRIK2 peptide-pulsed T2 cells were used for the IFNγ ELISPOT assay. Peptide non-pulsed T2 cells were used as a negative control. For the tetramer assay, PBMCs were stained with phycoerythrin-labeled GRIK2 peptide-HLA-A2 tetramer and analyzed by FACS. The tetramer-positive cells were single cell sorted. Established CTL clones were evaluated by IFNγ ELISPOT and tetramer assays (right panel) for specificity. B GRIK2 peptide-specific CTL clone recognizes endogenously expressed GRIK2. GRIK2 peptide-specific CTL clones were evaluated for reactivity to GRIK2-overexpressing UM-UC-3 cells by an IFNγ ELISPOT assay. K562 cells were used as a negative control. Data are shown as means ± standard deviation. Statistical analyses for the data were performed using a bilateral Student’s t-test. c GRIK2 peptide-specific CTL clone recognizes H-10 clone cells. GRIK2 peptide-specific CTL clones were evaluated for reactivity to H-10, L-3, and WT cells by an IFNγ ELISPOT assay. K562 cells were used as a negative control. Data are shown as means ± standard deviation. Statistical analyses for the data were performed using a bilateral Student’s t-test
To confirm that the CTL clone 9G23 can recognize not only GRIK2 peptide-pulsed target cells but also endogenously presented GRIK2 peptide, we performed an ELISPOT assay using GRIK2-overexpressing UM-UC-3 WT cells. The CTL clone showed significantly higher IFNγ secretion to GRIK2-overexpressing WT cells compared with WT cells or negative control K562 cells (Fig. 4B), indicating that the CTL clone 9G23 can recognize endogenously expressed GRIK2 peptide. Finally, the reactivity of the CTL clone was examined using H-10, L-3, and WT cells. The CTL clone showed significantly higher IFNγ secretion to H-10 cells compared with L-3, WT, and negative control K562 cells (Fig. 4C), indicating that GRIK2 peptide-specific CTLs can recognize bladder CSCs/CICs.
Immune selection by CTL clone 9G23 resulted in chemo- and radio-sensitive non-CSCs/CICs
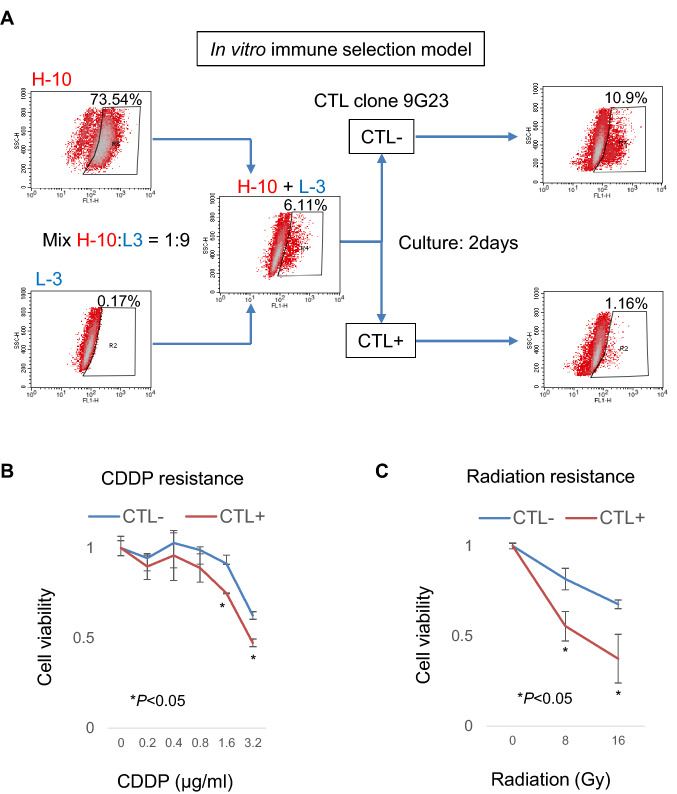
To evaluate targeting GRIK2 peptide in pre-clinical setting, we performed immune selection model in vitro. H-10 clone cells and L-3 clone cells were mixed at a ratio of 1:9. Mixed cell showed 6.11% ALDHhigh cells. Then, the mixed cells were selected by CTL clone 9G23 for over night and cultured 2 days. Immune-selection by CTL clone 9G23 decreased ALDHhigh cells (Fig. 5A). Furthermore, immune-selection increased chemo-sensitivity and radio-sensitivity (Fig. 5B, C).
Fig. 5.
In vitro immune selection model. A Summary of immune selection model. H-10 clone cells and L-3 clone cells were mixed at a ratio of 1:9. Mixture cells were cultured with (CTL +) or without CTL clone 9G23 (CTL-) for overnight at a effector/target ratio = 5:1 and then cultured for 2 days. Percentages indicate the ALDHhigh-positive rates. B and C Resistance to cisplatin and radiation. CTL + cells and CTL—cells were treated with cisplatin (CDDP) or radiation at a serial dose, and cell viability was assessed by a WST-8 assay. Data are shown as means ± standard deviation. Statistical analyses for the data were performed using a bilateral Student’s t-test
Discussion
In this study, we established ALDHhigh clone cells as a novel bladder CSC/CIC model for the first time. CSCs/CICs are defined as a subpopulation of cancer cells that are endowed with the ability to undergo self-renewal and differentiate, and ALDHhigh cells can differentiate into ALDHlow cells. The ALDHhigh clone cells established in this study were able to sustain the ALDHhigh phenotype during in vitro culture for several months. In a previous study, we described the establishment of side population clone cells as a novel colon CSC/CIC model that was also stable during in vitro culture for more than 2 months. These findings indicate that a small population of CSCs have high self-renewal ability and the stemness phenotype is sustainable during in vitro culture. In addition, the stable CSC model is a powerful tool to analyze CSCs in assays that require a large number of cells.
It has been discussed whether CSCs/CICs can be targeted by immunotherapy, because CSCs/CICs utilize several molecular mechanisms to escape from immunity. [26] CSCs/CICs isolated from mouse 4T1 cancer cells as ALDHhigh cells showed lower expression of antigen-presenting machinery-related genes, including Tap1, Tap2, and Tapbp, compared with ALDHlow cells by DNA methylation. [27] CSCs/CICs isolated as sphere cells from glioblastoma also showed lower expression of HLA class 1 molecules compared with non-CSCs/CICs. [28] In a mouse model study, CSCs/CICs that were induced by sphere-culture showed lower expression of MHC class 1 molecules and showed lower reactivity to CTLs than control cells. [29] In the present study, ALDHhigh clone cells expressed HLA class 1 molecules, but at lower level than ALDHlow cells (data not shown). However, antigenic peptides could be isolated from H-10 clone cells at comparative numbers as with WT cells, indicating that ALDHhigh cells derived from UM-UC-3 cells express sufficient levels of HLA class 1 molecules for antigen presentation and CTL recognition.
In our current CSC/CIC-targeting immune selection model (Fig. 5), immune selection using GRIK2-peptide-specific CTL clone resulted in higher ALDHlow population rates compared with CTL selection-negative cells indicating that GRIK2-specific CTL clone selectively recognized ALDHhigh cells. CSC/CIC-specific immunotherapy remains non-CSCs/CICs as shown in the immune selection model; however, non-CSCs/CICs are sensitive to chemotherapy and radiotherapy. These results strongly suggest the potency and rationale for combination therapy of CSC/CIC-targeting immunotherapy and chemotherapy or radiotherapy.
CSCs/CICs are related to treatment resistance; therefore, the development of novel therapies targeting CSCs/CICs is important. [7] Bladder CSCs/CICs express several distinctive molecules and signal transduction signatures [30]; however, most of them are shared with normal cells, and there are a very limited number of bladder CSC/CIC-specific targets. CD47 is expressed by bladder CSCs/CICs and works as a novel innate immune checkpoint molecule. [17, 31] CD47-targeting immunotherapy using an anti-CD47 antibody has been performed, and some promising results have been reported. [32, 33] To target bladder CSCs/CICs, a CD47-targeted near-infrared photoimmunotherapy model has been generated, suggesting that CD47 is a promising target for bladder CSC/CIC-targeting therapy. [34] However, we did not detect CD47-derived antigenic peptides by HLA ligandome analysis, and CTL-target antigenic peptides are limited.
GRIK2, also known as GluR6, is expressed in some neurons of the cerebral and cerebellar cortices [35], and GRIK2 has a role in autosomal recessive mental retardation. [36] These findings suggest possible adverse effects of GRIK2-targeting immunotherapy in the central nervous system, although neurons are considered to be immune privileged because they do not express HLA class 1 molecules. However, a recent study revealed that some neurons express MHC class 1 molecules. [37] Furthermore, neurons derived from induced pluripotent stem cells express MHC class 1 molecules following stimulation with IFNγ [38]. These findings indicate that neurons might also be targeted by CTLs, as suggested by a neurotropic viral infection model. [39] However, in the present study, we isolated GRIK2 peptide-specific CTLs from an HLA-A2+ healthy donor, suggesting that GRIK2 peptide-specific CTLs are not pathogenic. As GRIK2 and MHC class 1 molecules are expressed in some types of neurons, careful interpretation is needed for the further application and design of GRIK2-targeting immunotherapy.
Immune checkpoint blockade has also been approved for the treatment of bladder cancer. [40] In previous reports, CSCs/CICs have been described to express higher levels of PD-L1 as one mechanism for CSC immune escape in gliomas and head and neck cancers. [28, 41] In this bladder CSC/CIC model, H-10 cells expressed PD-L1 at an equivalent level as L-3 cells according to CAGE (data not shown) and the 9G23 CTL clone recognized H-10 clone cells. Thus, the bladder CSCs/CICs isolated in this study are immune competent and GRIK2-targeted immunotherapy combined with PD-L1/PD-1 axis blockade might be a powerful approach.
In summary, we established a novel human bladder CSC/CIC model derived from the UM-UC-3 human bladder cancer cell line. This bladder CSC/CIC model was stable during in vitro culture and was a useful tool to analyze bladder CSCs/CICs. We screened the antigenic peptides expressed by bladder CSCs/CICs and found that a GRIK2-derived antigenic peptide was expressed by these cells. GRIK2 peptide-specific CTLs specifically recognized bladder CSCs/CICs. These findings provide evidence that a GRIK2 peptide could be a target for novel bladder CSC/CIC-targeting immunotherapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to DNAFORM for performing CAGE. This work was supported by the Japan Society for the Promotion of Science, KAKENHI for T. Torigoe (17H01540) and Y. Hirohashi (20H03460). This work was also supported by the Japan Agency for Medical Research and Development, the Project for Cancer Research and Therapeutic Evolution (P-CREATE) for T. Torigoe (16770510) and T. Kanaseki (20cm0106352h0002), and the Japan Science and Technology Agency, CREST (JPMJCR15G3) for S. Hashimoto.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by HM, YH, SY, JY, AM, SH, ST, KH, TA, TK, TT, TK, NS, and TT. The first draft of the manuscript was written by HM, YH, NS, and TT, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Declarations
Conflicts of interest
The authors have no financial conflicts of interest to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoshihiko Hirohashi, Email: hirohash@sapmed.ac.jp.
Toshihiko Torigoe, Email: torigoe@sapmed.ac.jp.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 3.Advanced Bladder Cancer (ABC) Meta-analysis Collaboration (2005) Adjuvant chemotherapy in invasive bladder cancer: a systematic review and meta-analysis of individual patient data Advanced Bladder Cancer (ABC) Meta-analysis Collaboration. Eur Urol 48:189–199. Discussion 199–201 [DOI] [PubMed]
- 4.International Collaboration of Trialists, Medical Research Council Advanced Bladder Cancer Working Party (now the National Cancer Research Institute Bladder Cancer Clinical Studies Group), European Organisation for Research and Treatment of Cancer Genito-Urinary Tract Cancer Group et al. (2011) International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J Clin Oncol 29:2171–2177 [DOI] [PMC free article] [PubMed]
- 5.Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–423. doi: 10.3322/caac.21631. [DOI] [PubMed] [Google Scholar]
- 6.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 7.Park CY, Tseng D, Weissman IL. Cancer stem cell-directed therapies: recent data from the laboratory and clinic. Mol Ther. 2009;17:219–230. doi: 10.1038/mt.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirohashi Y, Torigoe T, Tsukahara T, et al. Immune responses to human cancer stem-like cells/cancer-initiating cells. Cancer Sci. 2016;107:12–17. doi: 10.1111/cas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishizawa S, Hirohashi Y, Torigoe T, et al. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem-like cells. Cancer Res. 2012;72:2844–2854. doi: 10.1158/0008-5472.CAN-11-3062. [DOI] [PubMed] [Google Scholar]
- 10.Morita R, Hirohashi Y, Torigoe T, et al. Olfactory receptor family 7 subfamily C member 1 is a novel marker of colon cancer-initiating cells and is a potent target of immunotherapy. Clin Cancer Res. 2016;22:3298–3309. doi: 10.1158/1078-0432.CCR-15-1709. [DOI] [PubMed] [Google Scholar]
- 11.Asano T, Hirohashi Y, Torigoe T, et al. Brother of the regulator of the imprinted site (BORIS) variant subfamily 6 is involved in cervical cancer stemness and can be a target of immunotherapy. Oncotarget. 2016;7:11223–11237. doi: 10.18632/oncotarget.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horibe R, Hirohashi Y, Asano T et al. (2017) Brother of the regulator of the imprinted site (BORIS) variant subfamily 6 is a novel target of lung cancer stem-like cell immunotherapy. PLoS One 12:e0171460 [DOI] [PMC free article] [PubMed]
- 13.Miyamoto S, Kochin V, Kanaseki T, et al. The antigen ASB4 on cancer stem cells serves as a target for CTL immunotherapy of colorectal cancer. Cancer Immunol Res. 2018;6:358–369. doi: 10.1158/2326-6066.CIR-17-0518. [DOI] [PubMed] [Google Scholar]
- 14.Morita R, Hirohashi Y, Nakatsugawa M, et al. Production of multiple CTL epitopes from multiple tumor-associated antigens. Methods Mol Biol. 2014;1139:345–355. doi: 10.1007/978-1-4939-0345-0_28. [DOI] [PubMed] [Google Scholar]
- 15.Kochin V, Kanaseki T, Tokita S et al (2017) HLA-A24 ligandome analysis of colon and lung cancer cells identifies a novel cancer-testis antigen and a neoantigen that elicits specific and strong CTL responses. Oncoimmunology 6:e1293214 [DOI] [PMC free article] [PubMed]
- 16.Takaya A, Hirohashi Y, Murai A et al (2016) Establishment and Analysis of Cancer Stem-Like and Non-Cancer Stem-Like Clone Cells from the Human Colon Cancer Cell Line SW480. PLoS One 11:e0158903 [DOI] [PMC free article] [PubMed]
- 17.Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y, Qiu Q, Zhang X, et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:327–337. doi: 10.1158/1055-9965.EPI-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volkmer J-P, Sahoo D, Chin RK, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci USA. 2012;109:2078–2083. doi: 10.1073/pnas.1120605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue R, Hirohashi Y, Kitamura H, et al. GRIK2 has a role in the maintenance of urothelial carcinoma stem-like cells, and its expression is associated with poorer prognosis. Oncotarget. 2017;8:28826–28839. doi: 10.18632/oncotarget.16259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasuda K, Hirohashi Y, Mariya T, et al. Phosphorylation of HSF1 at serine 326 residue is related to the maintenance of gynecologic cancer stem cells through expression of HSP27. Oncotarget. 2017;8:31540–31553. doi: 10.18632/oncotarget.16361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Shinkawa T, Tokita S, Nakatsugawa M, et al. Characterization of CD8+ T-cell responses to non-anchor-type HLA class I neoantigens with single amino-acid substitutions. Oncoimmunology. 2021;10:1870062. doi: 10.1080/2162402X.2020.1870062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haberle V, Forrest ARR, Hayashizaki Y et al. (2015) CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res 43:e51 [DOI] [PMC free article] [PubMed]
- 25.Frith MC, Valen E, Krogh A, et al. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maccalli C, Rasul KI, Elawad M, Ferrone S. The role of cancer stem cells in the modulation of anti-tumor immune responses. Semin Cancer Biol. 2018;53:189–200. doi: 10.1016/j.semcancer.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sultan M, Vidovic D, Paine AS, et al. Epigenetic silencing of TAP1 in aldefluor+ breast cancer stem cells contributes to their enhanced immune evasion. Stem Cells. 2018;36:641–654. doi: 10.1002/stem.2780. [DOI] [PubMed] [Google Scholar]
- 28.Di Tomaso T, Mazzoleni S, Wang E, et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16:800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison BJ, Steel JC, Morris JC. Reduction of MHC-I expression limits T-lymphocyte-mediated killing of cancer-initiating cells. BMC Cancer. 2018;18:469. doi: 10.1186/s12885-018-4389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abugomaa A, Elbadawy M, Yamawaki H, et al. emerging roles of cancer stem cells in bladder cancer progression, tumorigenesis, and resistance to chemotherapy: a potential therapeutic target for bladder cancer. Cells. 2020;9:235. doi: 10.3390/cells9010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logtenberg MEW, Scheeren FA, Schumacher TN. The CD47-SIRPα immune checkpoint. Immunity. 2020;52:742–752. doi: 10.1016/j.immuni.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Advani R, Flinn I, Popplewell L, et al. CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N Engl J Med. 2018;379:1711–1721. doi: 10.1056/NEJMoa1807315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikic BI, Lakhani N, Patnaik A, et al. First-in-human, first-in-class phase i trial of the anti-CD47 antibody Hu5F9-G4 in patients with advanced cancers. J Clin Oncol. 2019;37:946–953. doi: 10.1200/JCO.18.02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss B, van den Berg NS, Ertsey R, et al. CD47-targeted near-infrared photoimmunotherapy for human bladder cancer. Clin Cancer Res. 2019;25:3561–3571. doi: 10.1158/1078-0432.CCR-18-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paschen W, Blackstone CD, Huganir RL, Ross CA. Human GluR6 kainate receptor (GRIK2): molecular cloning, expression, polymorphism, and chromosomal assignment. Genomics. 1994;20:435–440. doi: 10.1006/geno.1994.1198. [DOI] [PubMed] [Google Scholar]
- 36.Motazacker MM, Rost BR, Hucho T, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang A, Yu H, He Y, et al. Developmental expression and localization of MHC class I molecules in the human central nervous system. Exp Brain Res. 2015;233:2733–2743. doi: 10.1007/s00221-015-4345-2. [DOI] [PubMed] [Google Scholar]
- 38.Clarkson BDS, Patel MS, LaFrance-Corey RG, Howe CL. Retrograde interferon-gamma signaling induces major histocompatibility class I expression in human-induced pluripotent stem cell-derived neurons. Ann Clin Transl Neurol. 2018;5:172–185. doi: 10.1002/acn3.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chevalier G, Suberbielle E, Monnet C et al (2011) Neurons are MHC class I-dependent targets for CD8 T cells upon neurotropic viral infection. PLoS Pathog 7:e1002393 [DOI] [PMC free article] [PubMed]
- 40.Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15:92–111. doi: 10.1038/nrurol.2017.179. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Shin JH, Longmire M, et al. CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin Cancer Res. 2016;22:3571–3581. doi: 10.1158/1078-0432.CCR-15-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.