Abstract
The Coronavirus disease, 2019 (COVID-19) is caused by severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2), which poses a major threat to human life and health. Given its continued development, limiting the spread of COVID-19 in the population remains a challenging task. Currently, multiple therapies are being tried around the world to deal with SARS-CoV-2 infection, and a variety of studies have shown that natural products have a significant effect on COVID-19 patients. The combination of SARS-CoV-2 S protein with Angiotensin converting enzyme II(ACE2) of host cell to promote membrane fusion is an initial critical step for SARS-CoV-2 infection. Therefore, screening natural products that inhibit the binding of SARS-CoV-2 S protein and ACE2 also provides a feasible strategy for the treatment of COVID-19. Establishment of high throughput screening model is an important basis and key technology for screening S protein-ACE2 blockers. Based on this, the molecular structures of SARS-CoV-2 and ACE2 and their processes in the life cycle of SARS-CoV-2 and host cell infection were firstly reviewed in this paper, with emphasis on the methods and techniques of screening S protein-ACE2 blockers, including Virtual Screening (VS), Surface Plasmon Resonance (SPR), Biochromatography, Biotin-avidin with Enzyme-linked Immunosorbent assay and Gene Chip Technology. Furthermore, the technical principle, advantages and disadvantages and application scope were further elaborated. Combined with the application of the above screening technologies in S protein-ACE2 blockers, a variety of natural products, such as flavonoids, terpenoids, phenols, alkaloids, were summarized, which could be used as S protein-ACE2 blockers, in order to provide ideas for the efficient discovery of S protein-ACE2 blockers from natural sources and contribute to the development of broad-spectrum anti coronavirus drugs.
Keywords: COVID-19, ACE2, S protein, Drug screening technology, Natural products
Graphical abstract
This paper introduces the screening methods and technologies of screening S protein-ACE2 blockers, including Virtual Screening, Surface Plasmon Resonance, Biochromatography, Biotin-avidin with Enzyme-linked Immunosorbent assay and Gene Chip Technology. Combined with the application of the above screening technologies in S protein-ACE2 blockers, a variety of natural products, such as flavonoids, terpenoids, phenols, alkaloids, were summarized, which can be used as an alternative supplement for people at risk of COVID-19 infection.
1. Introduction
COVID-19 is the most widespread global pandemic to hit humans in the last century, with more than 165 million people infected worldwide as of May 21, 2021. Because of its high infectivity and mortality, it has been declared as an international public health emergency by the World Health Organization, which has brought a major threat to human life safety and health, and led to a huge economic and social burden to the world [1,2]. COVID-19 is caused by SARS-CoV-2, which is structurally related to two other coronaviruses that cause SARS and Middle East respiratory syndrome (MERS) [3]. One of the main ways to prevent SARS-CoV-2 infection is to develop vaccines. At present, major breakthroughs have been made in the development of safe and effective vaccines against SARS-CoV-2 to prevent further spread of COVID-19, with a total of 201 candidate vaccines under development worldwide, 45 of which are in clinical trials and 156 in preclinical studies [4]. However, mutations in the virus have resulted in changes in the infectivity and pathogenicity of SARS-CoV-2. The mutated Alpha strain, which first circulated in the United Kingdom, increased the transmissibility by about 56%. Subsequently, Beta strain circulating in South Africa also increased the transmissibility, and Gamma strain circulating in South America increased the transmissibility by 1.4–2.2 times [5]. Recently, the mutated new coronavirus with at least two key point mutations was found in India and spread to many countries and regions in the world. Virology experts widely believe that the mutation occurred in a key part of SARS-CoV-2, making it significantly more infectious and potentially making existing vaccines less effective [6]. Delta strain that is currently circulating around the world has the characteristics of rapid transmission, certain immune escape ability, and may aggravate the disease, so its threat has attracted the attention of the whole world. Delta strain has multiple mutations in the spike protein that give it both greater ACE2 receptor binding ability and immune escape ability. Delta strain grows more rapidly in the lung and throat of infected people and has a thousand-fold increase in viral load levels, most likely associated with the ACE2 receptor [7]. In view of its continued development, limiting the spread of COVID-19 in population remains a challenging task. Although several highly effective vaccines have recently been widely deployed around the world, widespread adoption and equitable distribution of vaccines face challenges in the context of global inequality in wealth and education. Therefore, we need to pay attention not only to the development of vaccines, but also to the application of therapeutic drugs. (see Table 1 )
Table 1.
Natural products inhibiting S protein to bind to ACE2.
| Compounds | Chemical class | Source | Effect | Screening method | Reference |
|---|---|---|---|---|---|
| Quercetin (1) | Flavonoids |
Bupleurum chinense DC. Cyathula officinalis Kuan |
Binds to ACE2, impairs the interaction between S protein and ACE2 | Net-work pharmacology, Molecular docking, SPR | [123] |
| Quercetin (1) | Flavonoids | In silico | Targets ACE2 expression and alters the expression of genes encoding protein targets of SARS-CoV-2 in human cells | GSEA, expression profiling experiments | [115] |
| Quercetin (1) | Flavonoids | In silico | Binds to ACE2 and MPro | Molecular docking, Net-work pharmacology | [62] |
| Quercetin (1) | Flavonoids | Forsythia suspensa (Thunb.) Vahl Lonicera japonica Thunb. Bupleurum chinense DC. | Binds to the RBD region of S protein and inactivate it and then prevent S protein binding to ACE2 of epithelial cell surface | Net-work pharmacology, SPR, Molecular docking | [86] |
| Quercetin (1) | Flavonoids | In silico | Decreases ACE2 expression via regulation of transcription factors or miRNAs of ACE2 | Net-work pharmacology | [155] |
| Isorhamnetin (2) | Flavonoids | In silico | Bind to ACE2 and MPro | Molecular docking, Net-work pharmacology | [62] |
| Isorhamnetin (2) | Flavonoids | Hippophae rhamnoides L. | Binds to S protein and ACE2, inhibites SARS-CoV-2 spike pseudotyped virus entering ACE2hcell | CMC, Molecular docking, SPR | [130] |
| Puerarin (3) | Flavonoids | Pueraria lobata (Willd.) Ohwi | Binds to ACE2 and S protein, impairs the interaction between S protein and ACE2 | Net-work pharmacology, Molecular docking, SPR | [123] |
| Puerarin (3) | Flavonoids | In silico | Binds to ACE2 | Molecular docking | [156] |
| Rutin (4) | Flavonoids | Carthamus tinctorius L. | Binds to S protein and ACE2 | Molecular docking | [157] |
| Rutin (4) | Flavonoids |
Bupleurum chinense DC. Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hsiao |
Binds to ACE2 | Net-work pharmacology, SPR, Molecular docking | [158] |
| Oroxylin A (5) | Flavonoids | Scutellaria baicalensis Georgi | Binds to ACE2, suppresses entrance of SARS-CoV-spiked pseudo typed virus into ACE2 cells, inhibits LC3-mediated autophagy of ACE2 cells | CMC, Molecular docking, SPR | [100] |
| Procyanidin (6) | Flavonoids | In silico | Binds to ACE2 and S protein | Molecular docking, Molecular dynamics simulations | [124] |
| Irisolidone (7) | Flavonoids | In silico | Binds to ACE2 | Molecular docking | [156] |
| Luteolin (8) | Flavonoids | In silico | Binds to ACE2 and MPro | Molecular docking, Network pharmacology | [62] |
| Hesperidin (9) | Flavonoids | Valeriana Jatamansi | Binds to S protein and ACE2, destabilize S protein binding to human host ACE2 receptor | Molecular docking, Molecular dynamics simulations | [125] |
| Chrysin (10) | Flavonoids | Oroxylum indtcum (L.)Vent. | Binds to S protein and ACE2, destabilize S-protein binding to human host ACE2 receptor | Molecular docking, Molecular dynamics simulations | [125] |
| Anhydrosafflor yellow B (11) | Flavonoids | Carthamus tinctorius L. | Binds to S protein and ACE2 | Molecular docking | [157] |
| Glabridin (12) | Flavonoids | In silico | Decreases ACE2 expression via regulation of transcription factors or miRNAs of ACE2 | Net-work pharmacology | [155] |
| Myricitrin (13) | Flavonoids | Myristica fragrans Houtt. | Strong binding affinity to ACE2 and RNA dependent RNA polymerase | Molecular docking | [158] |
| Euchrenone (14) | Flavonoids | Glycyrrhiza uralensis Fisch | Binds to ACE2, Mpro and RdRp | Net-work pharmacology, Molecular docking | [66] |
| Epigallocatechin-3- gallate (15) | Flavonoids | Camellia sinensis (L.) O. Kuntze | Binds to S protein to impair the interaction between S protein and ACE2 | Molecular docking, in vitro antiviral experiment | [128,129] |
| Glycyrrhizic acid (16) | Terpenoids | Glycyrrhiza uralensis Fisch | Binds to ACE2, Mpro and RdRp | Net-work pharmacology, Molecular docking | [66] |
| Glycyrrhizic acid (16) | Terpenoids | In silico | Binds to RBD domain of the SARS-CoV-2 S protein | Molecular docking, Molecular dynamics simulations | [139] |
| Glycyrrhizic acid (16) | Terpenoids | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [60] |
| Glycyrrhizic acid (16) | Terpenoids | Glycyrrhiza uralensis Fisch | Binds to S protein and ACE2, destroys the interaction between S-RBD and ACE2 | SPR, Molecular docking | [140] |
| Uncaric acid (17) | Terpenoids | Uncaria tomentosa | Binds to RBD/ACE-2 interface and the ACE2 binding site on SARS-CoV-2 RBD viral spike | Molecular docking, Molecular dynamics simulations | [159] |
| Ursolic acid (18) | Terpenoids | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [60] |
| Demethylzeylasteral (19) | Terpenoids | Tripterygium wilfordii Hook.f. | Binds to ACE2 and S protein, impairs the interaction between S protein and ACE2 | SPR, Molecular docking | [85] |
| Maslinic acid (20) | Terpenoids | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [60] |
| Obacunone (21) | Terpenoids | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [60] |
| Andrographolide (22) | Terpenoids | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [160] |
| Atractylenolide III(23) | Terpenoids | Atractylodes lancea (Tunb.) Dc. (Cangzhu) | Binds to ACE2 and has anti-inflammatory effects and antiviral effects in vitro | Molecular docking | [161] |
| Astragaloside IV(24) | Terpenoids | Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hsiao | Binds to ACE2 | Net-work pharmacology, SPR, Molecular docking | [86] |
| Taraxerol (25) Daturaolone (26) |
Terpenoids | Clerodendrum D. innoxia | Bind to ACE2, S protein and Mpro | Molecular docking, Molecular dynamics simulations | [77] |
| Limonin (27) | Terpenoids | In silico | Binds to ACE2 to impair the interaction between S protein and ACE2 | Molecular docking, Molecular dynamics simulations | [162] |
| Cucurbitacin G 2-glucoside (28) | Terpenoids | Cucurbita pepo L. | Binds to ACE2 | Molecular docking | [163] |
| Citronellol (29) | Terpenoids | Pelargonium graveolens L'Hér. | Downregulates ACE2 expression in epithelial cells | Spectrum-effect relationship analysis | [119] |
| Limonene (30) | Terpenoids | Lemon essential oils | Downregulates ACE2 expression in epithelial cells | Spectrum-effect relationship analysis | [119] |
| Salvianolic acid A (31), Salvianolic acid B (32), Salvianolic acid C (33) |
Phenols | Salvia miltiorrhiza Bge. | Bind to S protein and ACE2 | Molecular docking, SPR | [146] |
| Neochlorogenic acid (34) | Phenols | Lonicera japonica Thunb. | Binds to ACE2 and inhibits ACE2 activity, binds to the contact surface of ACE2 and spike complex | MBC, SPR, Molecular docking | [95] |
| Kobophenol A (35) | Phenols | Caragana sinica (Buc'hoz) Rehder | Inhibits SARS-CoV-2 binding to cells through blocking spike RBD to the host ACE2 receptor | Molecular docking, Molecular dynamics simulations, ELISA | [107] |
| Resveratrol (36) | Phenols | In silico | Binds to ACE2 | Molecular docking | [150] |
| Chlorogenic acid (37) | Phenols | Lonicera japonica Thunb. | Binds to ACE2 to impair the interaction between S-protein and ACE2 | Molecular docking, Network pharmacology | [132] |
| Gallic acid (38) | Phenols | In silico | Decreases ACE2 expression via regulation of transcription factors or miRNAs of ACE2 | Net-work pharmacology | [155] |
| Bis-demethoxycur-cumin (39) | Phenols | Curcuma longa L. | Binds to RBD domain of the SARS-CoV-2 S-protein | Molecular docking, Molecular dynamics simulations | [164] |
| δ-viniferin (40) | Phenols | Syzygium jambos (L.) Alston | Strong binding affinity to ACE-2 and RNA dependent RNA polymerase | Molecular docking | [158] |
| Pterostilbene (41) | Phenols | In silico | Binds to ACE2 | Molecular docking, Molecular dynamics simulations | [160] |
| Ephedrine (42), Pseudoephedrine (43), Methylephedrine (44) |
Alkaloids | Ephedra sinica Stapf | Bind to ACE2 and spike RBD, inhibit SARS-CoV-2 spike pseudovirus entering ACE2hcells | CMC-HPLC-IT-TOF-MS, SPR, Molecular docking |
[99] |
| Cryptospirolepine (45) Cryptoquindoline (46) |
Alkaloids | Cryptolepis sanguinolenta (Lindl.) Schlt (Periplocaceae) | Bind to ACE2 and S-protein, destroy the stability of ACE2-RBD complex | Molecular docking, Molecular dynamics simulations | [150] |
| Speciophylline (47), Uncarine F (48) |
Alkaloids | Uncaria rhynchophylla (Miq.) Miq. | Bind to RBD/ACE-2 interface and the ACE2 binding site on SARS-CoV-2 RBD viral spike | Molecular docking, Molecular dynamics simulations | [159] |
| Solanine (49) | Alkaloids | Solanum Sps. | Inhibits spike RBD and main protease | Molecular docking combined Molecular dynamics simulations |
[27] |
| Bicuculline (50) | Alkaloids | In silico | Binds to ACE2 | Molecular docking | [156] |
| Anisodamine(51) | Alkaloids | Anisodus tanguticus (Maxim.) Pascher. | Binds to ACE2 | Net-work pharmacology, Molecular docking | [165] |
| Thymoquinone (52) | Quinones | Nigella sativa L. | Covers active sites of ACE2 and inhibits the activity of ACE2 | Molecular docking | [166] |
| Emodin (53) | Quinones | Rheum palmatum L. | Binds to S-protein and ACE2, destabilizes S-protein binding to human host ACE2 receptor | Molecular docking, Molecular dynamics simulations | [125] |
| Rhein (54) | Quinones | Rheum palmatum L. | Binds to ACE2 and inhibits ACE2 activity, binds to the contact surface of ACE2 and spike complex | MBC, SPR, Molecular docking | [95] |
| Forsythoside A (55), Forsythoside I (56) |
Glycosides | Forsythia suspensa (Thunb.) Vahl | Bind to ACE2 and inhibits ACE2 activity, bind to the contact surface of ACE2 and spike complex | MBC, SPR, Molecular docking | [95] |
| Amygdalin (57) | Glycosides | Prunus armeniaca L. var. ansu Maxim. | Binds to ACE2, Mpro and RdRp | Net-work pharmacology, Molecular docking | [66] |
| Urso-deoxycholic acid (58) | Steroids | Ipomoea obscura (L.) | Binds to ACE2 and MPro | Molecular docking, Molecular dynamics simulations | [167] |
| Withanolide A (59) | Steroids | Withania somnifera | Binds to ACE2, S-protein and Mpro | Molecular docking, Molecular dynamics simulations | [77] |
| β-sitosterol (60) | Steroids | Urtica dioica | Binds to ACE2 | Molecular docking | [168] |
| Sesamin (61) Sesaminol (62) Sesamolin (63) |
Lignans | Sesamum indicum L. | Bind to ACE2 and S-protein | Molecular docking | [61] |
| Phillyrin (64) | Lignans | Forsythia suspense (Thunb.) Vahl | Binds to ACE2 to impair the interaction between S-protein and ACE2 | Molecular docking, Network pharmacology | [132] |
| Farnesiferol B (65) | Coumarins |
Ferula sinkiangensis K. M. Shen |
Binds to ACE2 and S-protein | Molecular docking | [61] |
Natural products, widely found in nature, have long been an important source of innovative drug discovery. According to statistics, more than 60% of the drugs on the market are obtained from natural products or natural products as lead compounds after structural optimization and derivation [8]. Natural products are widely used in the treatment of antiviral infection and enhancement of host immune response. In previous coronavirus infections including SARS-CoV or MERS-CoV, natural products have significant therapeutic effects. Natural products also play an important role in the control of COVID-19, as SARS-CoV-2 is similar to SARS-CoV or MERS-CoV in certain genomic sequences [9]. For patients with mild symptoms, early intervention of natural products can effectively prevent the deterioration of the disease. For patients who have developed to advanced stage, natural products can improve symptoms, reduce many complications and mortality [10]. In addition, some natural products with antiviral activity come from a variety of spices, fruits and vegetables, which can reduce the risk and severity of various viral infections by antiviral and enhancing immune response [11]. Therefore, natural products are not only expected to be used to develop effective and less toxic anti SARS-CoV-2 drugs, but also can be used as an alternative supplement for people at risk of COVID-19 infection.
Studies have shown that ACE2 is the key target of SARS-CoV-2 entering host cells and the key point of drug intervention [12]. A trimer fusion protein S protein in SARS-CoV-2 structure is the main target of vaccine design [13], it is responsible for attaching the virus to the receptor of host cell surface, especially ACE2 [14]. ACE2 is widely expressed in the cell membranes of a variety of epithelial cells, including the lung, respiratory tract, heart, kidney, liver, pancreas, and intestine [15]. Receptor-binding domain (RBD) in S protein is a fragment of SARS-CoV-2 that binds to the ACE2 receptor of host cells and promotes the receptor-mediated endocytosis of the virus within host cells [16]. With the disclosure of ACE2 as the target of SARS-CoV-2 invasion into human body, blocking or competitively inhibiting the binding of SARS-CoV-2 and ACE2, so as to disrupt the cell process of virus infection, has become one of the therapeutic directions. Therefore, screening active ingredients from natural products that can inhibit the binding of SARS-CoV-2 and ACE2 also provides a feasible strategy for the treatment of COVID-19. It is worth noting that ACE2 located on the surface of host cells can also bind effectively with S protein of other coronavirus due to the genetic similarity between coronavirus [17]. Therefore, the active components which inhibit the binding of SARS-CoV-2 and ACE2 may also be effective for other types of coronaviruses, and can be used to fight other emerging coronavirus infection. Some of the natural products showed antiviral activity against different viruses in vitro and in vivo [18], so it is more conducive to the development of broad-spectrum anti-coronavirus drugs. At the same time, the establishment of a simple, rapid, sensitive, economic and efficient drug screening model is also an important basis and key technology for screening out active ingredients from natural products that block or competitively inhibit the binding of SARS-CoV-2 and ACE2. To this end, this paper firstly reviews the molecular structure of SARS-CoV-2, ACE2 and their roles in the life cycle of SARS-CoV-2 as well as the infection of host cells, and focuses on the research progress of screening methods and technologies for SARS-CoV-2 and ACE2 binding inhibitors. Finally, the active natural products that inhibit the binding of SARS-CoV-2 and ACE2 were summarized, in order to provide ideas for the efficient discovery of natural S protein-ACE2 blockers, and contribute to the development of broad-spectrum anti-coronavirus drugs.
2. The characteristics of SARS-CoV-2 and ACE2
2.1. SARS-CoV-2
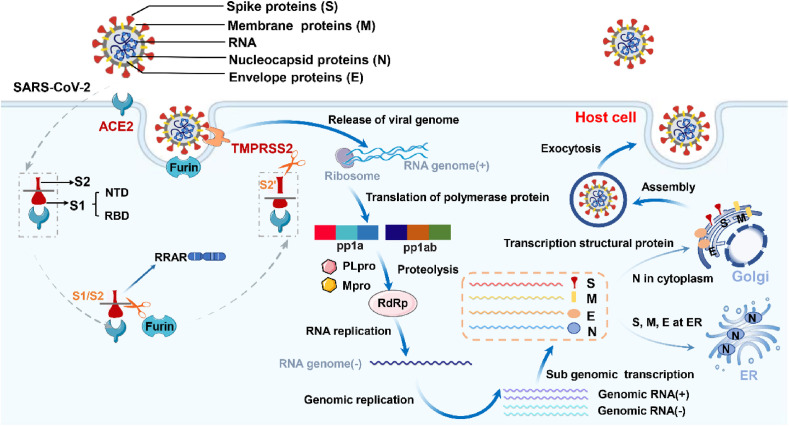
Coronavirus belongs to the genus Coronavirinae, which contains four genera, α, β, δ and γ. Its genome consists of a single-stranded position-sense RNA with a length of 27–32 kb, which is the largest RNA virus identified so far [19,20]. SARS-CoV-2 is the seventh coronavirus of the genus β that can be transmitted from person to person. Sequencing results showed that the genome of SARS-CoV-2 was composed of 29,903 nucleotide bases and 10 open reading frames (ORFs), with a size of about 29.9 kb [21]. ORF1ab encodes large replicase polyprotein PP1ab, which cleaved Papain Like protease (Plpro) and 3-chymotrypsin Like protease (3Clpro) into nonstructural proteins (NSP1-16), ORF2-10 encodes four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) [22]. Among these structural proteins, S protein plays the most critical role in viral attachment, fusion and entry into target cells [23].
When imaged by cryo-electron microscopy, SARS-CoV-2 appears as approximately spherical particles with a diameter of about 100 nm. The surface of SARS-CoV-2 was covered with dense viral substance by a prominent S protein trimer lipid bilayer [24]. S protein is a trimeric fusion protein of SARS-CoV-2, which exists in a metastable prefusion conformation after translation [25]. Each monomer contains two functional subunits, S1 and S2. S1 subunit is exposed to the surface of the envelope and is important site for recognizing and binding to the receptor on the surface of host cells, including an N-terminal domain and RBD. SARS-CoV-2 binds to ACE2 of host cells through RBD. S2 subunit is the key structure of virus invasion and fusion between virus envelope and host cell membrane [26,27], which is embedded in the envelope and contains an N-terminal hydrophobic fusion peptide, two heptad repeats (HR1 and HR2)、a transmembrane domain and a cytoplasmic tail [28]. Due to the mutation of the RBD amino acid sequence of SARS-CoV-2 S protein, the Q493 and P499 amino acid residues are more stable to ACE2 [28,29], and the fusion core structure formed by the HR1 and HR2 domains in the S2 subunit is highly stable [30]. Therefore, in the process of virus adsorption and entry, SARS-CoV-2 is more capable than SARS-CoV. At the same time, S protein has multiple N-glycosylation sites, glycosyl is linked to protein through covalent bonds to form glycoprotein, and the existence of a large number of glycosyl can change the spatial structure of protein molecules through glycosylation and block or destroy antigen epitopes, thereby inhibiting the immune response of the body and playing a protective role on the virus [31]. Therefore, SARS-CoV-2 spreads from person to person at a high rate of infection.
2.2. The key factor of SARS-CoV-2 entering host cells——ACE2
In 2000, ACE2 attracted wide attention as the first reported ACE homolog. ACE2 is also the anti-regulator of renin-angiotensin system, which plays a key role in the cardiovascular system [32]. Meanwhile, ACE2, as the receptor for coronavirus to enter host cells, plays a certain role in lung diseases. In 2003, it was confirmed that SARS-CoV entered host cells by binding to ACE2 through the S protein on its surface, so ACE2 was confirmed as the SARS-CoV receptor [33]. SARS-CoV has 79.6% sequence identity with SARS-CoV-2 [34], and several studies have confirmed that ACE2 is also a SARS-CoV-2 receptor [[34], [35], [36]].
ACE2 is widely expressed in the cell membranes of a variety of epithelial cells, including the lung, respiratory tract, heart, kidney, liver, pancreas and intestine, with the highest distribution in the small intestine. Some COVID-19 patients show digestive tract symptoms such as vomiting and diarrhea in the early stage, which may also be related to the distribution of ACE2 in intestinal tissues [15,37]. Analysis of RNA sequencing data of human lung cells revealed that 83% of ACE2 was expressed in the type II apical surface of epithelial cells (AT2) [38]. Therefore, intestinal tissue and AT2 may be the main targets of SARS-CoV-2 infection. ACE2 is a transmembrane protein, which is difficult to exist stably in vitro. Yan et al. [37] first found that ACE2 could form a stable complex with an amino acid transporter B0AT1 in the intestine by using cryo-electron microscopy. By analyzing the full-length protein structure of ACE2, it was found that ACE2 existed in the form of dimer and had two conformational changes of open and close. However, both conformations contain interfaces for mutual recognition with SARS-CoV-2. In terms of function and distribution, there are two forms of ACE2 (mACE2 and sACE2). mACE2 is located on the cell membrane and contains a transmembrane anchor and an extracellular domain. The extracellular domain has been shown to be a receptor site for SARS-CoV-2 [21]. sACE2 is a soluble form that lacks a membrane anchor and circulates in the blood in small amounts [39]. Some studies have shown that sACE2 can inhibit the binding between SARS-CoV-2 and mACE2, so high level of sACE2 may have a protective effect on the body [40]. However, a recent study confirmed that sACE2 can interact with S protein of SARS-CoV-2, and the formed sACE2-S protein complex can enter host cells through AT1 receptor mediated endocytosis [41]. Therefore, sACE2 also plays an important role in SARS-CoV-2 infection.
3. The process of S protein binding to ACE2
SARS-CoV-2 mainly enters cells through endocytosis. Since S protein belongs to type I virus fusion protein, it needs protease cleavage to activate its fusion potential [42,43]. There are two S protein hydrolysis sites: one located at the S1/S2 junction and the other located inside S2 (S2’). S protein hydrolyzed into S1 and S2 subunits and connected through non-covalent bonds to obtain the ability of membrane fusion [44], which is crucial for its mediated SARS-CoV-2 infection. Therefore, the activation of S protein is an important step for SARS-CoV-2 to enter host cells. Transmembrane protease serine 2 (TMPRSS2) is a type II transmembrane serine protease, which is widely expressed in the epithelial cells of the respiratory tract, gastrointestinal tract and urogenital tract. It cleans S protein mainly through the S2’ site and is the key enzyme to activate S protein [45,46]. In addition, the insertion of a “RRAR” sequence recognized by furin enzyme at S1/S2 is a unique structure of SARS-CoV-2 S protein. The experiment verified that the furin enzyme cleaves S protein through S1/S2 sites, inhibiting the furin enzyme, TMPRSS2 or simultaneously inhibiting the two proteases will affect the fusion activity of S protein. Therefore, TMPRSS2 and furin enzyme play an important role in the activation of S protein [47,48].
As mentioned before, S Protein exists in a metastable trimer conformation before activation. RBD of S1 subunit exists in two states, one is “up” conformation with poor stability, the other is stable “down” conformation, which exposes and hides receptor binding sites respectively [28]. The structural study of S protein showed that RBD in the “down” conformation was closer to the center of trimer, with fewer exposed sites, and it was not easy to recognize ACE2 [37]. Thus only “up” RBD can bind ACE2, while “down” RBD is in the receptor inactive binding conformation [28,37]. Studies have shown that ACE2 binds to SARS-CoV-2 S protein with an affinity of about 15 nmol/L, which is about 10–20 times higher than that of SARS-CoV S protein [49]. When RBD binds ACE2 of the host cell with a high affinity, the virus attaches to the cell surface and destroys the stability of the trimer before S protein fusion. S protein cleavage usually occurs sequentially, with furin enzyme cleavage at S1/S2 site first, and followed by TMPRSS2 cleavage at S2’ site [48,50]. Under protease cleavage, the S1 (RBD)-ACE2 complex was separated from the S2 subunit, and the S2 subunit changed dramatically to a stable fusion conformation. The interaction between HR-1 and HR-2 in the S2 subunit formed a stable six-helix bundle structure. These changes expose the FP of the S2 subunit and insert it into the host membrane to initiate the fusion process [51]. Once viral RNA is released into the host cell, ss-positive sense RNA acts as messenger RNA and is translated through the host cells, and the life cycle of SARS-CoV-2 is shown in Fig. 1 .
Fig. 1.
The life cycle of SARS-CoV-2. When SARS-CoV-2 S protein binds ACE2 of the host cell with a high affinity, the virus attaches to the cell surface and destroys the stability of the trimer before S protein fusion. S protein cleavage usually occurs sequentially, with furin enzyme cleavage at the S1/S2 site first, followed by TMPRSS2 cleavage at the S2’ site. Under protease cleavage, the S1 (RBD)–ACE2 complex was separated from the S2 subunit, and SARS-CoV-2 enters cells through endocytosis. SARS-CoV-2 releases the single chain positive RNA, then translates into pp1a and pp1ab with the help of ribosomes of host cells, and produces hydrolytic enzymes that can precisely cut polyprotein by means of self-shearing, namely main protease (Mpro) and papain-like protease (Plpro). Under the hydrolysis of Mpro and Plpro, RNA dependent RNA polymerase (RdRp) was formed. RdRp synthesized RNA genome (−) and then synthesized virus genome through genome replication. N protein binds to genomic RNA and replicates, transcribes and synthesizes in the cytoplasm. S, M and E proteins integrate into the membrane of the endoplasmic reticulum (ER). Virus are transported to the host cell membrane and released by exocytosis, and then infect other tissues and cells.
The binding of S protein to ACE2 is the first step of viral infection, in order to prevent SARS-CoV-2 from entering host cells, ACE2 has become one of the ideal targets for the development of novel broad-spectrum anti-coronavirus drugs. Firstly, the natural products that can bind to ACE2 were screened. On the one hand, their affinity for binding to other proteins was reduced biologically, and on the other hand, available sites were saturated to prevent viruses from binding to ACE2 and entering host cells. Secondly, similar to SARS-CoV, targeting SARS-CoV-2 S protein can also interfere with the virus. By binding to S protein through RBD and destroying S protein which directly binds to ACE2, the interaction between S protein and host cells can be prevented and infection can be prevented.
4. Screening methods for S protein-ACE2 blockers
4.1. Virtual screening
Virtual screening (VS) is a common strategy to obtain small molecule active compounds with high affinity for target proteins before biological activity test by preliminary screening multiple target molecules in the database of compounds with the help of computer in the early stage of drug development [52]. It is mainly divided into Structure-Based Virtual Screening (SBVS) and Ligand-Based Virtual Screening (LBVS) [53]. SBVS is primarily used to predict the optimal interaction mode or the formation of stable complexes between two molecules, and to evaluate the force of non-covalent interactions between ligands and molecular targets using the scoring function [54]. LBVS is based on the known structural information and physical and chemical properties of active compounds, and then a new active compound is selected according to the similarity principle [53]. At present, the research foundation of S protein-ACE2 blockers is weak, and there are few drugs with definite clinical efficacy and clinical trials. Therefore, the application of LBVS method is limited to a certain extent. Most studies on natural products as S protein-ACE2 blockers have focused on SBVS.
4.1.1. Molecular docking
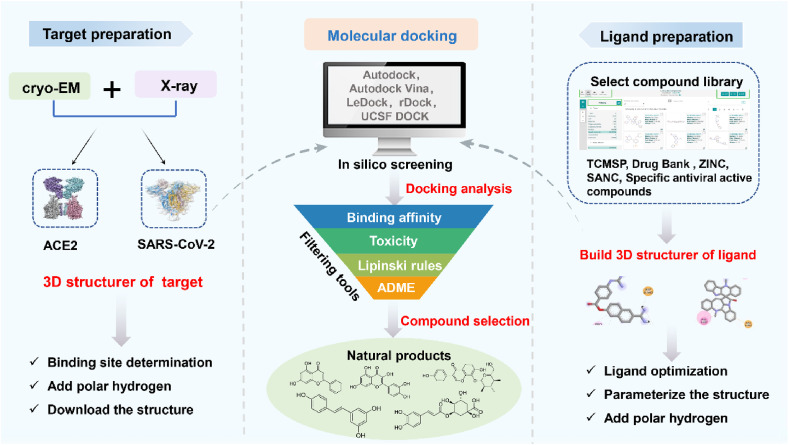
SBVS requires to know the 3D structure of the target protein, so that the interaction between the target and each compound can be predicted by computer [55]. Among the SBVS methods, molecular docking has attracted wide attention because of its low cost and good results. Molecular docking is a new drug design method based on the receptor characteristics and the interaction between receptor and drug molecules. As a new research method combining physical and chemical principles with scientific calculation algorithm, it provides a feasible strategy for exploring natural product as S protein-ACE2 blockers [56]. In the process of molecular docking for drug screening, the scoring function can be used to determine the binding site of ligand and the conformation between target and ligand to find out the allosteric site, and can also be used to predict the binding affinity between target and ligand [57]. According to the affinity of the evaluated ligands to the receptor sites, the ligands with some pharmacological activities can be identified more accurately. It is worth noting that even if the compound can specifically bind to the target, it does not mean that it can play a pharmacological role in vivo. Only when the compounds are absorbed by the body and have certain bioavailability to avoid being metabolized and excreted, can they play a good role in vivo. Therefore, in the process of molecular docking, the ADME (absorption, distribution, metabolism, and exception) properties of compounds should be evaluated on the computer with the help of mathematical algorithm [58], At the same time, the most classical Lipinski's rule can also be used to predict the bioavailability of compounds [59]. The process of molecular docking is shown in Fig. 2 .
Fig. 2.
Molecular docking process. Cryo-EM and X-ray reveal the structure of SARS-CoV-2 and ACE2. Downloading the structure and manually removing all binding ligands, ions and solvent molecules from the protein database for parameterization, and constructing 3D structure of targets. The ligands are mainly obtained from TCMSP, Drug Bank, ZINC, SANC and other databases. Then, Autodock tools is used to parameterize the structure to add complete hydrogen to the ligands. Finally, the ligands were subjected to molecular docking with SARS-CoV-2 and ACE2. The toxicity and pharmacokinetics of the compounds were analyzed. The ADME (Absorption, Distribution, Metabolism, and Excretion) properties of the compounds were evaluated by means of mathematical algorithm, and the bioavailability of compounds could be predicted by the most classic Lipinski rule to further obtain the potential drug-forming compounds with activity.
Vardhan [60]bases on molecular docking, ADME and drug-likeness prediction to screen potential inhibitors for ACE2 which is the therapeutic protein targets of SARS-CoV-2 from limonoids and triterpenoids. The results show that glycyrrhizic acid (16), ursolic acid (18), maslinic acid (20), and obacunone (21) were found binding at the catalytic site of ACE2 or the RBD site of ACE2. Natesh [61] uses molecular docking, ADME and Lipinski rules to analyze the binding efficiency of culinary spice bioactives to SARS-CoV-2 target protein. Among the 46 culinary spice bioactives, farnesiferol B (65), sesamin (61), sesaminol (62), and sesamolin (63) have good affinity with SARS-CoV-2 S protein and ACE2. Xu [62] screens the natural products acting on ACE2 from Traditional Chinese Medicine Systems Pharmacology Database (TCMSP). According to the binding affinity of natural products with ACE2, ADME and Lipinski rules, and the results showed that quercetin (1), luteolin (8), and isorhamnetin (2) have good binding ability with ACE2. Finally, the research focuses on the food rich in three active ingredients. Through large-scale literature search, it is concluded that the food rich in three active ingredients may have potential anti-COVID-19 bioactive are red wine, Chinese hawthorn, and blackberry.
Molecular docking is committed to identifying compounds with higher affinity for protein active sites from library or database [63]. However, for specific single traditional Chinese medicine (TCM) and compound preparations, it is usually necessary to use network pharmacology to find the material basis and molecular mechanism [64]. Therefore, the network pharmacology integrated the structure, pharmacological action and action pathway of TCM, and then combined with molecular docking technology to further clarify the material basis and action mechanism of TCM and compound preparation. Ephedra sinica Stapf-Prunus armeniaca L. is a common couplet medicine in anti-COVID-19 prescriptions. Gao [65] screed 47 potentially active ingredients from Ephedra sinica Stapf-Prunus armeniaca L. on the basis of the existing literature. They screened potential action targets of Ephedra sinica Stapf-Prunus armeniaca L. against COVID-19 from TCMSP database through network pharmacology, constructed herb-component-target (H–C-T) network and protein-protein interaction (PPI) network. Finally, they used ACE2 as the target to simulate the docking of 19 main compounds in Ephedra sinica Stapf-Prunus armeniaca L. with ACE2. The result showed that luteolin (8), β-sitosterol (60) had high binding activity to ACE2. Li [66] screened the active components and potential targets of Maxing Shigan Decoction through network pharmacology, and revealed its potential material basis and mechanism of action in antiviral and anti-inflammatory aspects. Through molecular docking, they further identified the components with potential binding ability to ACE2. Among them, amygdalin (57) of Prunus armeniaca L. had the strongest binding ability to ACE2.
In addition, using machine learning method in molecular docking can get more reasonable screening model from known data. In recent years, deep learning method based on neural network has shown great potential in the field of VS. SSnet is a deep neural network framework based on end-to-end, which uses protein structure and ligand information to predict protein ligand interaction probability [67]. SSnet is superior to Atomnet, 3D-CNN, Autodock Vina and other methods in identifying protein ligand complexes with high binding affinity [68]. At present, most of the studies focus on drug screening for S protein-ACE2 binding. This method only considers a single ACE2 structure and cannot identify potential allosteric inhibitors formed by ACE2-S protein [37]. Karki [68]first conducted drug screening for ACE2 in the open conformation and ACE2:S1 complex from the approved drug molecular library through SSnet, and verified the screened drugs through smina virtual docking. The results showed that natural products such as avonoids, flavonones, and polyketides have a strong binding ability with ACE2.
As an important method to assist new drug research and development, VS technology based on molecular docking plays a very important role in the early virtual screening of drug design and the search for new targets. Through the docking software, the screening time is effectively reduced and the screening efficiency is greatly improved. However, there are some common problems in molecular docking technology. First of all, the spatial conformation of the receptor structure is complex and changeable, so it may not be possible to obtain its complete and real three-dimensional structure. Secondly, during molecular docking, the molecular model of the receptor protein is almost impossible to be completely rigid and can undergo certain deformation. Therefore, whether the flexibility and solvation effect of receptor protein molecules in biological system can be truly simulated may directly affect the results of molecular docking.
4.1.2. Molecular dynamics simulations (MD)
Proteins are dynamic biomolecules, and their flexibility will affect the ligand recognition process. Molecular docking has limited ability to deal with the inherent flexibility of protein during docking, which greatly limits the expected applicability of the technology [69]. To solve this problem, several strategies have been tried, including flexible butting and local optimization of side chains, but these relatively simple approaches do not allow for extension and rearrangement of protein structures and significant conformational changes at the skeleton level [[70], [71], [72]]. Therefore, the flexibility of receptors remains a major challenge in ligand recognition. MD is a method based on the physical model of interatomic interaction to predict how each atom in a protein or other molecule changes with time. Based on virtual screening, the flexibility of protein is considered to obtain more reasonable ligand binding sites. MD can not only provide abundant dynamic structure information of biological macromolecules, but also provide a lot of energy information of protein ligand interaction, which provides a theoretical basis for understanding the nature of protein ligand interaction and guiding the process of drug design.
The recessive binding sites of protein ligand binding are invisible, which can only be seen in the crystal structure during the binding process. These sites can also be used as effective drug targets, but due to their concealment, it is difficult to find them through experimental screening. Mixing MD is one of the best methods to identify and characterize recessive binding sites [73]. It is a cosolvent simulation technique for identification of binding hots pots such as orthosteric, allosteric and cofactor binding sites as well as PPI sites [74]. Gopinath [75] used MixMD to detect potential inhibitor binding sites on S protein-ACE2 PPI interface. Drug-like organic probe molecules were added to the solvent to observe their localization in the simulation process, so as to detect the possible small molecule binding sites on the surface of S protein-ACE2 interface. Then, the natural products that bind to S protein and interfere with SARS-CoV-2 binding with host cells were screened through the database, and the compounds with strong and stable binding to RBD were further verified by MD.
MD can also be combined with molecular docking, which can be used to evaluate the structural stability of protein ligand complexes, and improve the accuracy of prediction of drug target binding ability, so as to obtain compounds with high activity [76]. Therefore, molecular docking and MD are highly complementary computational methods for drug screening. Mondal [77] selected 60 active components from 22 kinds of medicinal plants for molecular docking with S protein and ACE2 receptor proteins. The results showed that withanolide A (59) had a high affinity towards two targets of SARS-CoV-2 with the binding energies of −7.7 kcal/mol for S protein and −7.0 kcal/mol for ACE2, theoretically, it has stronger binding ability to S protein than ACE2. And the interaction between withanolide A (59) and ACE2 was almost stable after MD verification, indicating that withanolide A (59) could block the ACE2 receptor and make it difficult for SARS-CoV-2 to enter host cells.
Compared with molecular docking, MD takes full account of the flexibility of protein and can obtain a variety of target protein conformations. Combining with molecular docking is also a strategy to improve the enrichment factor of VS. In the study of S protein-ACE2 blockers, molecular docking is often used in combination with MD. MD can also simulate experimental conditions such as temperature, pressure, atomic number, ion concentration and solvent type used for solvating molecules [78]. All factors can be adjusted and controlled by the statistical mechanics integration under the MD, which is more conducive to drug screening and design.
At present, VS has become the main means for researchers to discover the lead compounds of enzyme inhibitors. VS greatly reduces the number of experimental screening compounds, shortens the research cycle, reduces the cost of drug research and development, and does not need the physical existence of compounds, so computational screening can be performed prior to synthesis. In addition, with the continuous improvement of virtual database, the continuous innovation of screening software, and the combination of the latest computer algorithm technology, the function of VS is more perfect and the results are more accurate. On the other hand, the flexibility of protein structure is a major limitation of VS. The binding sites of proteins are flexible, and there are many conformational states, which often involve the structural changes of rings and the secondary structural elements reconstruction caused by ligand binding [79,80], so false positive results will appear. Due to the complexity of receptor binding interaction, it is difficult to accurately predict the correct binding position of the complex, resulting in the best receptor-binding performance with some software, but poor performance with others [54]. Therefore, VS is usually combined with biological activity experiment.
4.2. Surface plasmon resonance (SPR)
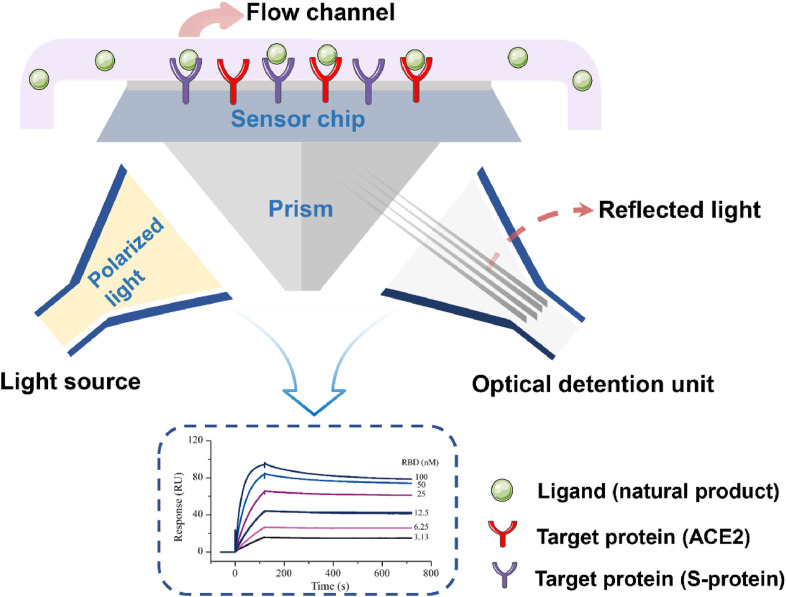
SPR is a kind of biosensor technology used to study the interaction between various biomolecules. As a new biochemical analysis and detection tool, SPR is widely used in drug screening with the characteristics of label free, high sensitivity and low consumption [81]. In the classical SPR screening method, the target protein is coupled to the chip surface as the receptor, and then the analyte molecular solution flows through the chip surface. The compounds in the analyte solution that can bind with the receptor are used as the ligands. The binding between the ligand and the receptor leads to the increase of the surface mass of the sensor and the change of the refractive index. Through the computer control system, it is transformed into the sensing signal graph of time and resonance signal. By analyzing the sensing signal graph, the binding affinity and kinetic parameters of ligand and protein can be obtained, the affinity of compounds can be evaluated, and the compounds with good affinity can be selected [82]. The process of SPR is shown in Fig. 3 . The detection principle of SPR is based on the change of the refractive index of the chip surface, and it does not need to label the small molecules to be tested. Therefore, it can eliminate the influence of modified labeling on the activity of compounds, and the detection results can more truly reflect the affinity between compounds and proteins [83,84].
Fig. 3.
Schematic diagram of SPR. The target protein ACE2 or S protein are coupled to the chip surface as the receptor, and then, the analyte molecular solution flows through the chip surface. The compounds in the analyte solution that can bind with the. The binding between the ligand and the receptor leads to the increase in the surface mass of the sensor. The computer control system is transformed into the sensing signal graph of time and the resonance signal. By analyzing the sensing signal graph, the binding affinity and kinetic parameters of the ligand and protein can be obtained. The affinity of the compounds was evaluated and the compounds with good affinity were screened.
SPR technology can be used to rapidly screen lead compounds by detecting the KD value of the interaction between the target protein and small molecules. Zhu [85]captured the ACE2-His protein as ligand, dissolved S-RBD-mFc in buffer solution as analyte to establish an SPR screening model of S-RBD and ACE2. When the compounds flowed through the chip surface fixed with ACE2 or S-RBD, the KD value was determined to screen the compounds that could bind to ACE2 or S-RBD. Among them, demethylzeylasteral (19) can bind to S-RBD-mFc in mobile buffer solution with the lowest dissociation rate constant and the longest residence time. Demethylzeylaster (18) was placed in the analytical solution containing RBD and continuously flowed over the chip surface of ACE2 to observe its signal response. The results showed that demethylzeylaster (18) could also block the binding of S-RBD to ACE2.
SPR also can be used as in vitro molecular authentication model combined with VS. For complex system screening of active compounds, some non-specific components can be eliminated through VS. Then SPR can monitor the whole process of the selected compounds binding to the target protein in real time to further verify the binding ability of the compounds obtained from VS. Ye [86]used network pharmacology, molecular docking and SPR technology to explore the potential compounds and interaction mechanism of Toujie Quwen granules in the treatment of COVID-19. Firstly, the H–C-T network was constructed by network pharmacology, and the key compounds were screened to dock with ACE2. The selected quercetin (1), astragaloside IV(24), rutin (4) were tested by SPR. S Protein and ACE2 are fixed in CM5 chip by covalent bond, and the screened compound solution flows through the chip respectively to test the binding energy with S Protein and ACE2. The results showed that quercetin (1) and astragaloside IV (24) bound to SARS-CoV-2 S protein and inactivated it, while rutin (4) bound to ACE2.
Compared with the traditional inhibitor screening based on enzyme activity, SPR technology does not depend on the enzyme reaction process and is more efficient. It does not need to label the small molecules to be detected, and it can monitor the whole process of the binding of compounds with protein targets in real time, so as to analyze the binding of compounds with protein and generate sensing images to evaluate the affinity and kinetic information of candidate compounds. In addition, SPR has strong specificity, and it does not require high matrix of samples. It can also detect colored solutions such as traditional Chinese medicine extracts, which further improves the screening efficiency.
4.3. Biochromatography
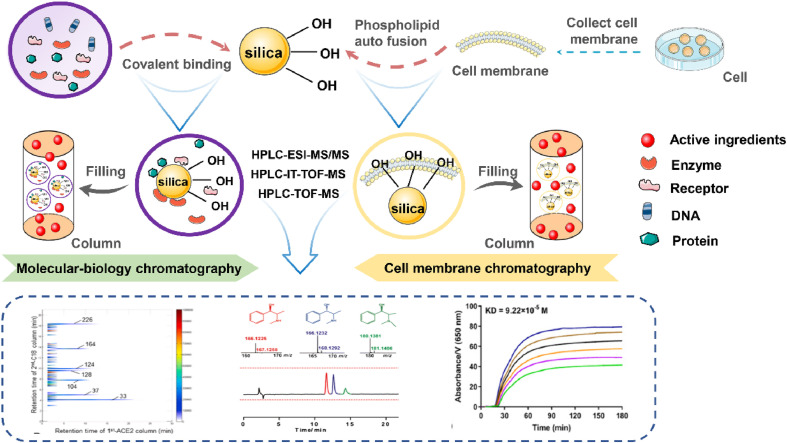
Biochromatography is a new chromatography technology developed by the combination of life science and chromatography in the 1980s. It has been widely used in the screening of active components of drugs and the study of action mechanism [87]. It uses bioactive materials (such as active macromolecules, active cell membranes, living cells, etc.) fixed on chromatographic carriers as stationary phases to simulate the interactions between drugs and biological macromolecules, targets or cells, or to simulate some key steps of biological activity expression of drugs in vivo under physiological and pathological conditions, avoiding the tedious and chemical separation process of the system [88]. The retention of a drug in a biochromatographic column is directly related to its activity, and it can exhibit certain pharmacological effects when it binds to biological macromolecules, targets or cells [89,90]. According to the different stationary phases, biochromatography can be divided into: Molecular-Biology Chromatography (MBC), Cell Membrane Chromatography (CMC), biomembrane chromatography, or artificial biomembrane chromatography [91]. Among them, MBC and CMC are widely used in the study of S protein-ACE2 blockers. The schematic diagram of MBC and CMC is shown in Fig. 4 .
Fig. 4.
Schematic diagram of Molecular-Biology Chromatography(MBC) and Cell Membrane Chromatography(CMC). MBC uses enzymes, receptors, DNA, transporters in plasma, and other biomacromolecules with essential physiological functions as stationary phases to separate and purify active compounds. CMC adsorbs the cell membrane containing specific receptors on the surface of SiO2. The self-fusion characteristics of living cell phospholipids and the strong binding characteristics of silicon hydroxyl (Si–OH) on SiO2 form the cell membrane stationary phase, which can identify the specific targets in complex samples that produce the biological effects of ligands.
4.3.1. Molecular-Biology Chromatography (MBC)
MBC is a new technology which can screen, separate and identify the active components in complex system [92]. When drugs are distributed in the body, they can interact specifically with biological macromolecules such as antibodies, enzymes and receptors. Based on the specific interaction of biomacromolecules, MBC uses enzymes, receptors, DNA, transporters in plasma and other biomacromolecules with important physiological functions as stationary phases to separate and purify active compounds and determine their biochemical parameters [93,94]. The ability to identify the chemical structure of bioactive compounds can be enhanced by establishing an integrated system with MBC as the core and combining with NMR, MS and other methods which can provide structural information.
Chen [95] first used UPLC-HRMS to analyze the prototype and metabolite components in plasma and urine after taking Lianhuaqingwen capsule (LHQW), covalently fixed ACE2 protein on silica gel stationary phase, and then used comprehensive 2D ACE2 column/C18 column/TOFMS system to screen potential ACE2 binding components in enriched urine extract. After preliminary screening, 8 active ingredients that may bind ACE2 were obtained. Through SPR, it was further confirmed that the active ingredients screened by biological chromatography comprehensive system had good binding force with ACE2, thus revealing the high efficiency of screening target compounds through this system.
The composition of natural products is complex, and the conventional chromatographic separation has impurity interference. MBC is simple, time-consuming, with high selectivity and good repeatability, which can eliminate most of the impurity interference. In addition, MBC can be directly related to some pharmacological parameters such as plasma protein binding rate or strength, which has certain pharmacological significance [88]. Therefore, MBC is widely used to screen, isolate and identify active components. However, there are still some problems in the research of MBC in drug active ingredient screening. As an in vitro analysis method of active components, MBC cannot fully represent the process of drug active components combined with biological macromolecules in vivo, and the preparation process of biological chromatographic column is relatively complex and its life is short, which hinders its application [91].
4.3.2. Cell membrane chromatography (CMC)
CMC is a biochromatographic method based on the selective interaction between membrane receptors and their ligands. The cell membrane containing specific receptors is adsorbed on the surface of SiO2. The self-fusion characteristics of living cell phospholipids and the strong binding characteristics of silicon hydroxyl (Si–OH) on SiO2 form the cell membrane stationary phase [96]. The binding characteristics between ligands and membrane receptors can effectively identify the specific target components in complex samples that produce ligand cell biological effects [88]. With the rapid development of molecular and cell biology and Bioengineering, CMC can screen active ingredients from complex samples such as natural plants, and use the specific affinity between drugs and membrane receptors to transform the drug interaction process in vivo into chromatographic process, so as to conduct biomimetic research on drug interaction in vitro [97]. Compared with the traditional methods, CMC has the dual functions of chromatography separation and activity characterization, and has a unique advantage in high-throughput screening of target components in complex samples. By combining CMC with HPLC and MS, the specificity, sensitivity and selectivity of target components identified from complex systems are improved, and the screening and identification efficiency of active components are greatly improved [98].
Lv [99] collected ACE2 h cells, separated cell membrane, mixed with pre activated SiO2 to make cell membrane stationary phase, and then loaded it into the standard column to prepare ACE2/CMC column. An ACE2/CMC bioaffinity chromatography model combined with HPLC-IT-TOF-MS system was established to screen three active components——Ephedrine (42), pseudoephedrine (43), and methylephedrine (44) from Ephedra sinica Stapf, which can act on ACE2. Gao [100] used ACE2-HEK293T cell membrane as stationary phase to prepare CMC column to screen potential ACE2 binding inhibitors in Scutellaria baicalensis Georgi. The result showed that oroxylin A (5) had the strongest binding ability to ACE2. At the same time, molecular docking and SPR verified that oroxylin A (5) bonded well to ACE2 from different perspectives. Further studies on autophagy and apoptosis of oroxylin A (5) showed that it could effectively reduce autophagy of ACE2 cells, and had no effect on apoptosis induced by ACE2 cells.
CMC, as a new receptor pharmacology research method, is simple and reliable, with the characteristics of specificity, applicability, saturation, multi-target, etc. It can combine the separation and screening of active ingredients, avoid the complex process of studying drug metabolism reaction in vivo, drug resistance difference between species and individuals, and eliminate the interference of impurity components, which can be used for specific, selective screening of active components in complex systems [101]. CMC has been widely used since its inception, but there are still some problems in the process of experiment and application. First of all, the membrane receptors attached to the silica gel of the cell membrane chromatography column are easy to fall off or lose activity, so the service life is short, and it is difficult to promote and apply in a wide range. There are many kinds of active protein receptors on the membrane, which may be interfered by non-specific binding in the process of CMC research. Therefore, blank carrier column should be used as the control group to eliminate the interference of non-specific binding on the phospholipid bimolecular membrane. In addition, as an in vitro analysis method of active ingredients, CMC cannot completely simulate the influence of complex environment and other systems in vivo on drug efficacy [98,102]. Therefore, it is necessary to carry out sufficient pharmacological evaluation experiments for further verification.
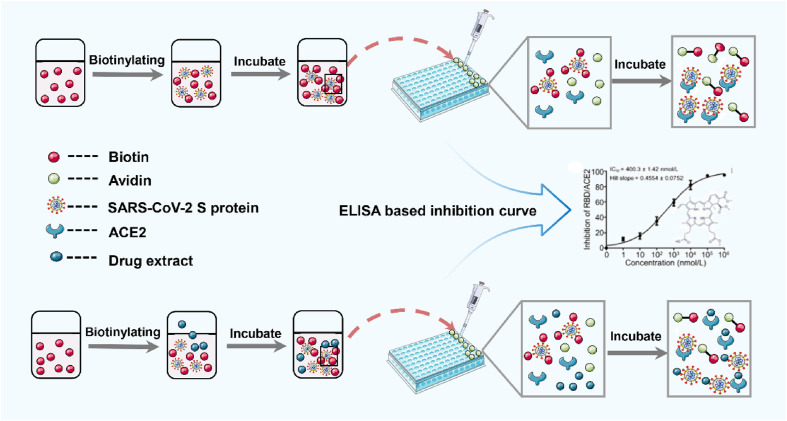
4.4. Biotin-avidin with enzyme-linked immunosorbent assay
Enzyme linked immunosorbent assay (ELISA) is a qualitative and quantitative method for immune reaction by adsorbing soluble antigen or antibody onto solid carrier such as polystyrene. It combines the specificity of antigen-antibody binding with the high efficiency of enzyme reaction. It can detect very low content of antigen such as protein, peptide, hormone or antibody in liquid samples, making the detection signal amplified. It has the advantages of high efficiency, high sensitivity and strong specificity, and has been widely used in various fields of life science [103]. According to the principle, it can be divided into many types, and competitive ELISA is the most widely used in the high-throughput screening of small molecule inhibitors. In order to improve the detection sensitivity, the conventional ELISA detection method has been improved, and a new method combining ELISA with other technologies has been developed. Biotin-Avidin with ELISA is one of the most widely used methods.
Biotin-Avidin System is a new type of amplification system of biological reaction, which can be combined with ELISA organically. Through the high affinity and multi-site binding between biotin and avidin, the majority of biological reaction signals can be amplified, and the specificity and sensitivity can be further improved [104]. In addition, the specific binding ability of avidin and biotin is 1 million times of antigen-antibody, and is not affected by the concentration of reaction reagent. The schematic diagram of biotin-avidin with ELISA is shown in Fig. 5 . Once the binding is extremely stable. Biotinylated ELISA has been proved to be able to evaluate the binding ability of SARS-CoV S proteins with ACE2. Ho [105] covalently linked biotin to SARS-CoV S protein for biotinylation and established competitive ELISA. The biotinylated S protein was mixed with different concentrations of drug extracts, and incubated at 37 °C for 2 h. Then the mixture was added to the adsorption plate coated with ACE2 for OD value determination. The results showed that emodin (53), the active component of Rheum palmatum L., blocked the binding of SARS-CoV S protein to ACE2 in a dose-dependent manner. Some scholars [106] selected 5 drugs from 958 FDA approved drug databases that could destroy the interaction between SARS-CoV-2 RBD and ACE2. Biotinylated RBD and 10-fold diluted drugs were added to 96 well plates coated with ACE2 respectively, and the RBD bound to ACE2 was detected by avidin HRP and TMB substrate. The results of ELISA-based high-throughput showed that RBD/ACE2 interaction could be inhibited by the 5 drugs at selected low and high concentrations. At present, the reports about screening drugs with anti-SARS-CoV-2 activity are all based on authentic virus, which limits the unqualified laboratories to carry out drug screening. The high-throughput ELISA based methodology can be better applied to the screening of anti-SARS-CoV-2 activity drugs.
Fig. 5.
Schematic diagram of biotin-avidin with ELISA. Biotin was added into S protein, and after the S protein was incubated with biotin, the biotinylated S protein was added to the pores of 96 plate coated with ACE2, and avidin was added to each hole. The binding ability of S protein and ACE2 was evaluated by ELISA. For competitive ELISA, biotinylated S protein was mixed with drug extract and incubated. After incubation, the mixture was added into the pores of 96 plate coated with ACE2. After adding avidin, the absorbance was measured by ELISA.
In addition, ELISA can also be used to quantitatively evaluate the inhibitory activity of compounds in vitro. Gangadevi [107] found that kobophenol A (35) and its three metabolites have strong binding ability in the ACE2/spike interface and the hydrophobic pocket of the ACE2 domain through molecular docking. Therefore, kobophenol A (35) is the primary target of ACE2/spike RBD in vitro. The recombinant 2019-nCoV S1-RBD was added ACE2 receptor protein and a certain concentration of kobophenol A (35) and verified by ELISA in vitro. It was found that kobophenol A (35) inhibited the binding of ACE2 to SARS-CoV-2 S1-RBD, indicating that kobophenol A (35) may inhibit the entry of S SARS-CoV-2 into host cells.
ELISA screening method has high sample handling capacity, high sensitivity and low false positive rate for detecting ligand-receptor interaction complexes on cell surface, and is especially suitable for high-throughput screening [108]. However, due to its high selectivity to reagents, ELISA is difficult to analyze multiple components at the same time, and has a certain degree of cross-reaction to compounds with similar structures. In the screening process, antibodies need to be incubated and washed for many times, which leads to high screening cost. Despite these limitations, ELISA has the potential to rapidly explore ligand-receptor interactions.
4.5. Gene Chip Technology
Gene chip Technology, also known as DNA microarray, is a kind of biological Chip [109]. It refers to the technology that high-density DNA fragments are attached to the surface of silicon, glass and other solid phases in a certain order or arrangement by microarray technology, DNA probes are labeled with isotope or fluorescence, and a large number of gene expression and monitoring are carried out by virtue of the principle of base complementary hybridization [110,111]. Among them, the expression profile chip technology is one of the most widely used technology, it can not only be used to analyze gene function and explore the pathogenesis of diseases, but also be used in high-throughput screening of drugs. Through the analysis of the parallel expression profile chip, the normal human cells or abnormal cells compared with pathological changes to detect differentially expressed genes in different specimens, so as to identify drug targets. It greatly improves the traditional experimental method of observing the expression of single or multiple genes, so as to speed up the identification of differentially expressed genes and the construction of differential expression profile [112]. It can also directly analyze the changes of gene expression profiles of different tissues and organs before and after medication, construct gene expression profiles, and screen new drugs or lead compounds efficiently by analyzing gene expression profiles, pathology and physiology [113,114].
Glinsky [115]first identified human genes that regulate the expression and function of ACE2 and FURIN genes through Gene Set Enrichment Analyses (GSEA) to establish a genomic regulatory interaction model that may influence SARS-CoV-2 infection. The expression profiling experiments (EPEs) revealed that ACE2 and FURIN were ubiquitous in human tissues. Then the transcription factor binding sites affecting the expression of ACE2 and FURIN were identified by Gene Expression Omnibus (GEO). The results showed that HMGA2, INSIG1, RUNX1, HNF-4α, JNK1/c-FOS activated the expression of ACE2 and FURIN. The GSEA of the drug perturbations from GEO database focused on downregulated genes identified estradiol and quercetin (1) among the top significantly enriched records, in which quercetin (1) targets the expression of ACE2, which may be an effective inhibitor of SARS-CoV-2 infection.
Compared with traditional biotechnology, gene chip can be used to analyze thousands of genes in the same test process, which can be used to track the changes of thousands of genes in cells after drug action [116]. It has the characteristics of high throughput and high sensitivity, which changes the limitation of traditional methods that can only study one or several genes. However, at present, this technology has high cost and lack of repeatability, which directly affects the evaluation results of differential genes and reduces the credibility of differential gene screening. In addition, gene screening experiments are mostly limited to the cell level, so we should pay more attention to the animal level and clinical level experiments.
4.6. Others
“Spectrum-effect relationship analysis” is a potential method to determine the effective components in complex systems. On the basis of modern chromatographic separation and analysis technology, we can obtain the most useful chemical information, evaluate the pharmacodynamic activity of the corresponding components, and finally connect the chemical information of each component with the pharmacodynamic results, so as to determine the compounds related to the pharmacodynamics [117]. At present, the spectrum-effect relationship has been introduced into the screening of enzyme inhibitors [118]. Through the determination method of enzyme catalyzed substrate reaction combined with the separation and identification of components with enzyme inhibitory activity, the target natural products can be quickly determined, so as to screen the lead compounds more effectively. Senthil Kumar [119]confirmed that geranium and lemon oils could significantly inhibit the expression of ACE2 in epithelial cells by Western blotting and qPCR analysis, and identified the main components of the two essential oils by GC-MS analysis. Finally, the inhibitory effect of each component on ACE2 was determined, citronellol (29) and limonene (30) in geranium and lemon oils were screened out to have strong inhibitory effect on ACE2. Amplified luminescent proximity homogeneous assay (AlphaLISA) is a kind of new homogeneous detection technology based on photoluminescence of nano-beads. It mainly uses the principle of intermolecular interaction, such as antigen-antibody specific interaction, and takes silica microspheres as the carrier. When the interaction between molecules makes the microspheres close to each other, the interaction between molecules can be detected by exciting fluorescence [120]. Hanson [121]has developed a method based on AlphaLISA to detect the binding ability of SARS-CoV-2 RBD to ACE2. A simplified Spike RBD-ACE2 interaction model was established by using recombinant SARS-CoV-2 RBD fused with Fc fragment and recombinant human ACE2 fused with biotinylated fragment. Using this assay platform and TruHits counterassay, compounds with inhibitory activity against ACE2-RBD were screened from 3384 small molecule drugs.
5. Natural products—S protein-ACE2 blockers
5.1. Flavonoids
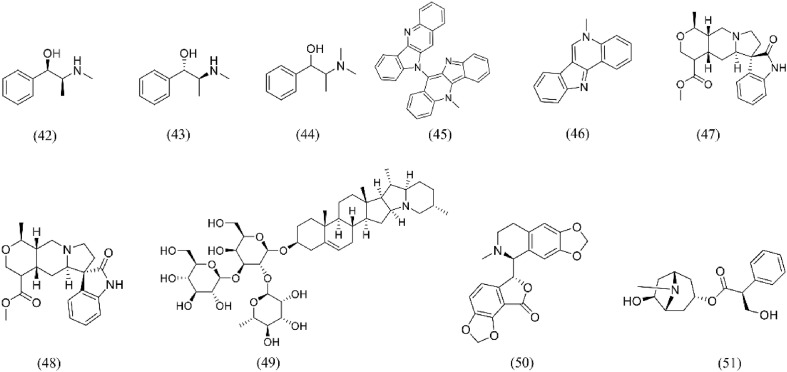
Flavonoids are natural products found in many plants. Modern pharmacological studies have shown that they have antiviral, anti-inflammatory, antioxidant, immunomodulatory and antitumor effects. As a potential antiviral agent, flavonoids can be traced back to the 1950s. In 1990, the antiviral effect of flavonoids on coronavirus was first found [122]. At present, a large number of drug screening studies have shown that flavonoids can block the combination of SARS-CoV-2 S protein and ACE2, thus inhibiting SARS-CoV-2 from entering the host cells. The chemical structure is shown in Fig. 6 . Many VS methods, such as molecular docketing, MD and net-work pharmacology, screened out quercetin (1), isorhamnetin (2), purearin (3), rutin (4), oroxylin A (5), procyanidin (6), irisulidone (7), luteolin (8), and hesperidin (9) as S-protein-ACE2 blockers [62,86,100,[123], [124], [125]]. Anamika [125] selected hesperidin (9), emodin (53), rhein (54) and chrysin (10) to dock with SARS-CoV2 S protein and its human receptor ACE2 molecule. The results showed that when S protein did not bind to ACE2, these natural products could form stable complexes with ACE2. Hesperidin (9) also binds ACE2 noncompetitively with S protein and the binding sites of S protein and hesperidin (9) are located at different sites in ACE2. Due to the existence of hesperidin (9), the binding structure of ACE2 and S protein becomes unstable. It is further confirmed that hesperidin (9) has antiviral activity as a noncompetitive regulator of S protein binding to its host receptor. The dihydroflavone moiety of hesperidin was predicted to be parallel to the β-6 RBD S protein sheet, while the sugar moiety fits into a shallow hole in the direction away from ACE2 [126]. Some of the natural products of flavonoids were tested for their activity in vitro after VS. Quercetin (1) and astragaloside IV(24) have been proved to bind and inactivate SARS-CoV-2 S protein by SPR in vitro, and then prevent S protein from binding to ACE2, while rutin (4) binds to ACE2, thus weakening the ability of SARS-CoV-2 to infect the host to a certain extent [86]. The antiviral studies in vitro have confirmed that isorhamnetin (2) and oxylin A (5) inhibit SARS-CoV-2 spike pseudotyped virus from entering ACE2h cells [100]. Epigallocatechin-3-gallate (EGCG,15) the most abundant component of catechins in tea (Camellia sinensis (L.) O. Kuntze), plays a role against viruses through inhibiting virus invasiveness, restraining gene expression and replication [127]. Molecular docking experiments revealed that EGCG had a higher atomic contact energy value, binding energy, ligand efficiency and surface area than hydroxy-chloroquine (HCQ) during binding with the S protein [128]. In vitro antiviral experiment results showed that EGCG inhibited SARS-CoV-2 virus replication and interfered directly with SARS-CoV-2 receptor binding [129]. Therefore, as a small compound with broad antiviral activity, EGCG could potentially be used as a lead structure to further develop highly effective antiviral drugs. In addition, there are also a variety of screening methods have been used to screen flavonoids of S protein-ACE2 blockers.
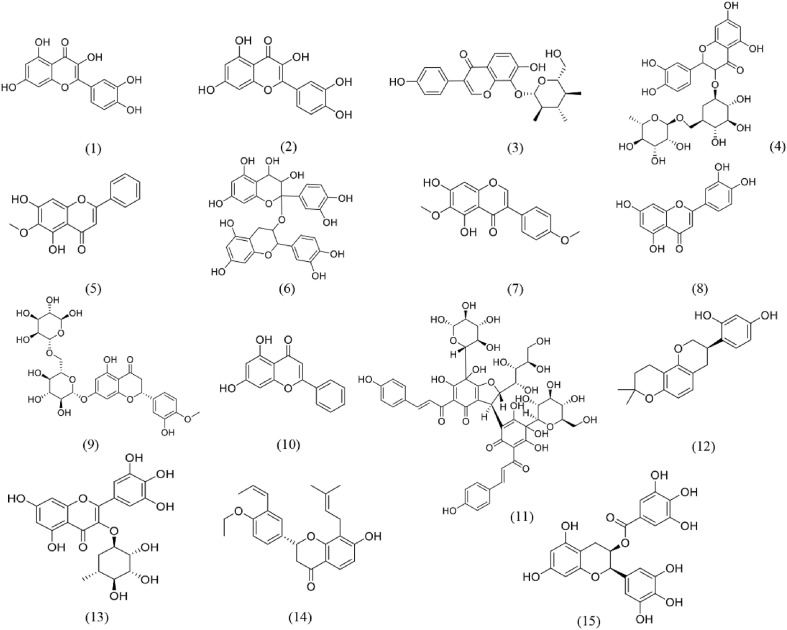
Fig. 6.
Structures of flavonoids for S protein-ACE2 repressors. (1) Quercetin; (2) Isorhamnetin; (3) Puerarin; (4) Rutin; (5) Oroxylin A; (6) Procyanidin; (7) Irisolidone; (8) Luteolin; (9) Hesperidin; (10) Chrysin; (11) Anhydrosafflor yellow B; (12) Glabridin; (13) Myricitrin; (14) Euchrenone; (15) Epigallocatechin-3- gallate.
Glinsky [115] found that quercetin (1) targeted ACE2 expression through GSEA and EPEs, which could inhibit the expression of c-FOS in human and rat cells, the expression of RUNX1 in rat cells and HNF-4α in human cells (RUNX1, HNF-4α and c-FOS all can activate the expression of ACE2). After quercetin (1) administration during the differentiation of human intestinal cells, the gene expression of ACE2 was significantly decreased. Furthermore, GSEA analysis of 332 human genes encoding SARS-CoV-2 showed that quercetin (1) changed the expression of 98 genes, which may interfere with 23 functions of SARS-CoV-2 protein in human cells. Therefore, quercetin (1) may be an effective inhibitor of SARS-CoV-2 infection. Zhan [130] found that quercetin (1) and isorhamnetin (2) in Hippophae rhamnoides L. had strong binding effect on ACE2-overexpressed HEK293 (ACE2h) cells through CMC analysis. It was confirmed by SPR that isorhamnetin (2) had a good binding effect with ACE2 and further verification in vitro showed that isorhamnetin (2) inhibited the entry of SARS-CoV-2 spike pseudotyped virus into ACE2 h cells. Therefore, isorhamnetin (2) may be a potential therapeutic candidate for COVID-19.
Flavonoids contain a flavan core with a 15-carbon skeleton. There are two benzene rings, connected by a heterocyclic pyran ring. Studies have shown that heterocyclic pyran ring comprises a C–C double bond and carbonyl groups that play an important role in the biological activities. The hydroxyl groups of two benzene rings are known to be responsible for the radical scavenging activity of flavonoids. Quantitative structure–activity relationship modeling was conducted, and the lack of the heterocyclic pyran ring in the flavonoid skeleton was shown to reduce the inhibitory activity of ACE by up to 91%. The absence of carbonyl groups in the heterocyclic pyran ring also reduced the inhibitory activity of ACE by 74% [131]. The resorcinol moiety might play a role in ACE2 inhibition, as this group might disrupt hydrogen bonds between Glu329/Gln325 of ACE2 and Arg426 of the S protein of SARS-CoV-2, which form a salt bridge to stabilize their interaction [132]. The functional groups of flavonoids, such as the pyran moiety in the heterocyclic pyran ring and hydroxyl groups of the two benzene rings, may play an important role in their ACE2 inhibition. Therefore, the application of flavonoid-based scaffolds in the design of new S protein-ACE2 blockers could be a good approach.
Among flavonoids, quercetin (1) is the most widely studied S protein-ACE2 blocker which has potential antiviral effects. In a randomized, double-blind, placebo-controlled trial, 1002 subjects took 500 or 1000 mg quercetin (1) or placebo daily for 12 days, in which 1000 mg/day quercetin (1) significantly reduced the symptoms of upper respiratory tract infection in middle-aged and elderly people [133]. A clinical study conducted in a hospital in Wuhan showed that 54 patients with COVID-19 were treated with TCM including high content of quercetin (1), which improved the symptoms of patients with COVID-19 and shortened the length of hospital stay of patients [134]. Therefore, quercetin (1) is expected to be used as a new anti- SARS-CoV-2 drug in clinical treatment. However, quercetin (1) has low bioavailability owing to biotransformation during digestion, absorption and metabolism, plants store quercetin (1) attached to sugars which substantially interfere with protein interactions but are released in the human digestive tract [135]. Therefore, other routes of delivery could be more effective in disrupting SARS-CoV-2 S protein/ACE2 interactions. For example, nasal spray containing quercetin (1) in a suitable form could deliver the appropriate concentration directly in the active molecular form, free unconjugated quercetin (1) [136].
5.2. Terpenoids
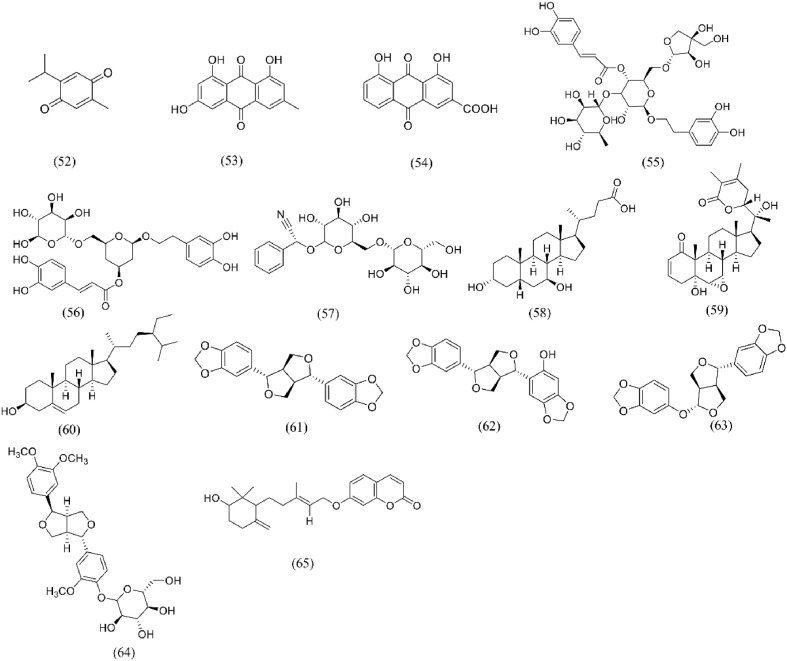
Terpenoids is a volatile substance distributed in essential oils from plant. It is mainly composed of monoterpenes, sesquiterpenes, diterpenes and triterpenes. It has been widely concerned about many pharmacological activities such as anti-tumor, antiviral, antibacterial, anti-inflammatory and immunomodulatory [137]. In recent years, many triterpenoids and their derivatives have been proved to have significant broad-spectrum antiviral activities. Cinatl [138] reported that glycyrrhizic acid (16) and its derivatives have anti-SARS-CoV activity, which can not only inhibit the virus replication process, but also inhibit the virus adsorption and penetration at the beginning of the virus life cycle. Several VS methods such as molecular docking, MD and Net-work pharmacology screened glycyrrhizic acid (16) as S protein-ACE2 blocker [60,66,139]. Yu [140] took S protein RBD of SARS-CoV-2 as the target, verified by SPR that the binding of glycyrrhizic acid (16) with S1 subunit of SARS-CoV-2 destroyed the binding between RBD and ACE2, and evaluated the cytotoxicity of glycyrrhizic acid (16) by MTT. The results showed that glycyrrhizic acid (16) was an effective anti-coronavirus active ingredient in vitro with low toxicity, which might be a potential candidate drug for anti-coronavirus. Zhu [85] established the SPR screening model of S-RBD and ACE2, the results showed that demethylzeylasteral (19) could bind to S-RBD and block the binding of S-RBD and ACE2. In addition, there are other triterpenoids such as ursolic acid (18), Maslinic Acid (20), obacunone (21), atractylenolide III (23), and astragaloside IV (24), which were confirmed that they could bind to ACE2 or S protein as S-protein-ACE2 blockers through a variety of VS methods, and the chemical structure is shown in Fig. 7 . Some diterpenoids such as citronellol (29) and limonene (30) were also proved to have ACE2 inhibitory activity. Senthil Kumar [119]found that citronellol (29) and limonene (30) in geranium and lemon oils significantly down regulated the expression of ACE2 in epithelial cells by spectrum-effect relationship. The cytotoxicity of citronellol (29) and limonene (30) was determined by MTT assay. The results showed that the HT-29 cells treated with two diterpenoids showed no cytotoxicity within 48 h at the concentration of 100 μM. Therefore, geranium and lemon oils and their derivatives citronellol (29) and limonene (30) may have potential antiviral effects.
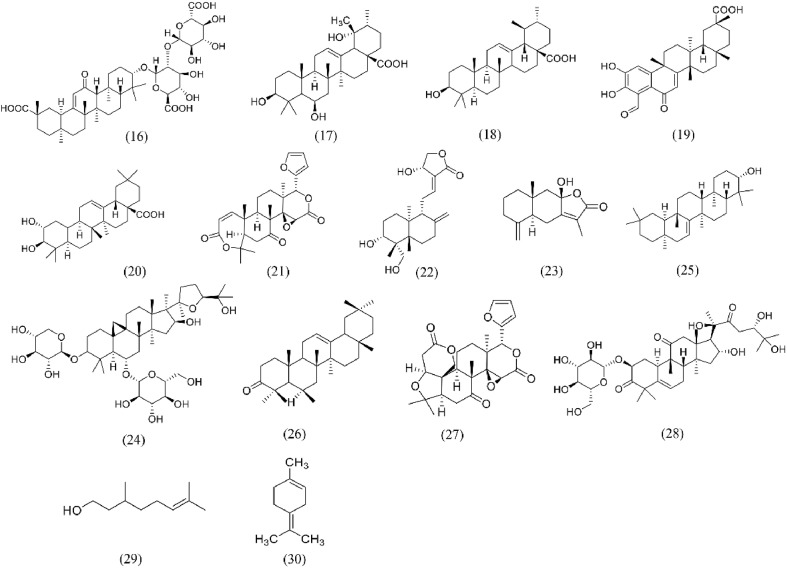
Fig. 7.
Structures of Terpenoids for S protein-ACE2 repressors. (16) Glycyrrhizic acid; (17) Uncaric acid; (18) Ursolic acid; (19) Demethylzeylasteral; (20) Maslinic acid; (21) Obacunone; (22) Andrographolide; (23) Atractylenolide III; (24) Astragaloside IV; (25) Taraxerol; (26) Daturaolone; (27) Limonin; (28) Cucurbitacin G 2-glucoside; (29) Citronellol; (30) Limonene.
Terpenoids play an important role in antiviral drugs, but they have a bottleneck as candidate drugs, such as low solubility, poor intestinal permeability and oral bioavailability. New methods should be introduced to improve their biopharmaceutical properties, especially drug delivery properties [141]. Currently, several delivery systems have been developed successfully, such as nanoemulsions, mesoporous silica nanoparticles, solid lipid nanoparticles, and liposomes, niosomal gels, and solid dispersions. Continuous progress will change the pharmacokinetics of terpenoids and increase their solubility, bioavailability, and even oral bioavailability. Glycyrrhizic acid (16) has an antiviral effect against coronavirus in vitro with low toxicity. It is considered as one potential candidate drug for anti-coronavirus to fight off SARS-CoV-2 by testing alone or combining with other drugs [142]. However, its cytotoxicity and poor solubility in water and biological fluids limit its further clinical application, as it may result in low bioavailability [143]. Therefore, to protect against COVID-19, highly biocompatible glycyrrhizic acid (16) nanoparticles (GANPs) based on the active component of glycyrrhizic acid (16) were synthesized. The results showed that GANPs had significant antiviral, anti-inflammatory, and antioxidant effects in vitro and in vivo. GANPs could target areas of severe inflammation through the EPR effect in the surrogate mouse model of COVID-19, which appeared to improve the accumulation of GANPs in the lungs and livers, further increasing the effectiveness of the treatment. Such a novel approach may provide an effective therapeutic solution for the pandemic, as well as treatment of hyperinflammation of other diseases [144].
5.3. Phenols
Phenols are the most abundant secondary metabolites in plants, which are widely used in food and medicine fields because of their antioxidant, anti-tumor, liver protection and other functions [145]. Salvianolic acid A (SAA,31), Salvianolic acid B(SAB,32), Salvianolic acid C(SAC,33) are phenolic carboxylic acids obtained from Salvia miltiorrhiza Bge. Hu [146] confirmed that the toxicity of the three compounds in ACE2h cells was low at a given concentration, and evaluated the binding ability of the three compounds with RBD and ACE2 by molecular docking. They further confirmed that SAA, SAB and SAC had good binding ability with RBD and ACE2 by SPR, and the binding ability of SAB was stronger than SAA and SAC. Finally, in vitro antiviral experiments shown that SAA, SAB and SAC can effectively inhibit the entry of 2019-nCoV pseudovirus into cells, and the inhibitory effect of SAB is greater than SAA and SAC, indicating that SAB may inhibit the binding of 2019-nCoV pseudovirus to ACE2. Chen [95] used the comprehensive 2D ACE2 column/C18 column/TOFMS system to screen the active components in LHQW that may bind to ACE2, and further confirmed that the selected active components have good binding force with ACE2 through SPR. In addition, the ACE2 Inhibitor Screening Kit was used to test the inhibitory activity of active components to ACE2 in vitro, and molecular docking technology was used to simulate the binding sites of the active components. The results showed that neochlorogenic acid (34), one of the phenolic acids, was highly exposed to human body and inhibited ACE2 in vivo. Gangadevi [107] found that kobophenol A (35) had a strong binding ability with the ACE2/spike interface and the hydrophobic pocket of the ACE2 domain through molecular docking, and it was verified by ELISA that kobophenol A (35) inhibited the binding of ACE2 to SARS-CoV-2 S1-RBD. The anti-SARS-CoV2 assay of kobophenol A (35) in Veroe6-EGFP cells further suggests that kobophenol A (35) may inhibit virus entry into the host to cause infection without cytotoxicity. In addition, resveratrol (36), chlorogenic acid (37), gallic acid (38), bis-demethoxycurcumin (39), δ-viniferin (40), pterostilbene (41) and other phenols that can be bind to ACE2 and SARS-CoV-2 spike were screened by a variety of VS methods. And the chemical structure is shown in Fig. 8 .
Fig. 8.
Structures of Phenols for S protein-ACE2 repressors. (31) Salvianolic acid A; (32) Salvianolic acid B; (33) Salvianolic acid C; (34) Neochlorogenic acid; (35) Kobophenol A; (36) Resveratrol; (37) Chlorogenic acid; (38) Gallic acid; (39) Bis-demethoxycur-cumin; (40) δ-viniferin; (41) Pterostilbene.
Recent studies have shown that the antiviral activity of these compounds is affected by the number of hydroxyl groups, substituents and the relative position of functional groups. Compounds with more hydroxyl groups showed higher antiviral activity. The location of hydroxyl groups in the benzene ring at the meta position (resorcinol structure) in the flavan ring which may disrupt hydrogen bonds between amino acid residues between ACE2 and S protein [132]. In addition, it appears that compounds with a methyl group have a stronger antiviral activity than compounds with a methoxy group, the position of methoxy group on benzene ring also affects antiviral activity [147]. Therefore, optimizing the structure of phenols compounds may be a new strategy for designing antiviral drugs. In addition, metal ions or metal-containing compounds show antiviral activity, and the presence of metal ions significantly affects the affinity of compounds to SARS-CoV-2 [148]. Complexation of phenols compounds with metals favorably changes their lipophilicity, water-solubility, bioavailability, stability and also allows the introduction of important macro-and microelements into the compound [149]. Therefore, metal complexation of phenols compounds may be one of the drug design strategies for SARS-CoV-2 to enhance the antiviral properties of phenols compounds.
5.4. Alkaloids
Alkaloids are a class of nitrogen-containing organic compounds with various structures, which have good antiviral activity and are an important source of antiviral drugs. Lv [99] established ACE2/CMC-HPLC-IT-TOF-MS system and screened three active components EP (42), PEP (43) and MEP (44) in Ephedra sinica Stapf to act on ACE2. Molecular docking and SPR were used to evaluate the binding ability of the three active compounds to ACE2 and SARS-CoV-2 RBD, and CCK-8 staining was used to evaluate the cytotoxicity of the selected active compounds to ACE2h cells. The results showed that the three active ingredients had double binding effect on ACE2 and SARS-CoV-2 RBD, and had little cytotoxicity to ACE2h cells at a certain concentration. In vitro simulation of SARS-CoV-2 spike pseudovirus entry into ACE2h cells showed that the selected compounds, especially EP (41), could inhibit the entry of SARS-CoV-2 spike pseudovirus into ACE2h cells. Gyebi [150] screened the alkaloids from African plants by molecular docking and MD simulations to block the receptor function of ACE2 and destroy the fusion of SARS-CoV-2 and host cells. The results showed that cryptospirolepine (45) and cryptoquindoline (46) have high affinity with ACE2, TMPRSS2 and SARS-CoV-2 S protein. It can bind to the complex formed by S protein RBD and ACE2 (RBD-ACE2), even when S1 subunit of S protein binds to ACE2, cryptospirolepine (45) and cryptoquindoline (46) may destroy the cell entry of SARS-CoV-2 and the stability of RBD-ACE2 complex. In addition, there are many alkaloids, such as speciophylline (47), uncarine F (48), solanine (49), bicuculline (50), anisodamine (51), which can bind to ACE2 and S-protein. The chemical structure is shown in Fig. 9 .
Fig. 9.
Structures of Alkaloids for S protein-ACE2 repressors. (42) Ephedrine; (43) Pseudoephedrine; (44) Methylephedrine; (45) Cryptospirolepine; (46) Cryptoquindoline; (47) Speciophylline; (48) Uncarine F; (49) Solanine; (50) Bicuculline; (51) Anisodamine.
Bis-benzylisoquinoline alkaloids can effectively inhibit S protein-ACE2 mediated cell membrane fusion by targeting host calcium channels, and the fusion rate is reduced by about 90%.These compounds block host calcium channels, thereby inhibiting Ca2+-mediated fusion and inhibiting virus entry. Although less research has been done on screening S protein-ACE2 blockers for bis-benzylisoquinoline alkaloids, these compounds also have potential as lead agents for the development of SARS-CoV-2 therapies, especially given the effectiveness of calcium channel blockers in controlling hypertension. Bis-benzylisoquinoline alkaloids should be considered for the treatment of SARS-COV-2 infection in hypertensive patients [151]. Due to the structural diversity and the screening methods of the alkaloids, it is not easy to establish a relationship between chemical structure and anti-SARS-CoV-2 activity of the alkaloids. Interestingly, the alkaloids are highly oxygenated due to the presence of several functional groups, including methoxyls, hydroxyls, methylene dioxide, and carbonyls of ketones, esters, and amides. Also, the nitrogens are heterocyclic atoms and are sometimes in ring fusion [152]. These may be the mechanism and influencing factors of alkaloids to anti-SARS-CoV-2.
5.5. Others
In addition to the natural products mentioned above, there are also many compounds that can block the binding of S-protein-ACE2, including quinones, glycosides, steroids and lignans, with the structural formula as shown in Fig. 10 . For example, the components of LHQW that may bind to ACE2 and have ACE2 inhibitory activity in vitro were screened by comprehensive 2D ACE2 column/C18 column/TOFMS system, including rhein (54), forsythoside A (55) and forsythoside I (56) [95]. Interestingly, the compounds in cooking spices including sesamin (61), sesaminol (62), sesamolin (63), and farnesifero B (65) have been shown to bind ACE2 and S proteins through molecular docking [61]. Because of its antioxidant, anti-inflammatory, immunomodulatory and antiviral effects, some scholars suggest that health products based on spices can be developed to prevent and treat COVID-19.
Fig. 10.
Structures of others S protein-ACE2 repressors. Quinones: (52) Thymoquinone; (53) Emodin; (54) Rhein; Glycosides: (55) Forsythoside A; (56) Forsythoside I; (57) Amygdalin; Steroids: (58) Urso-deoxycholic acid; (59) Withanolide A; (60) β-sitosterol; Lignans: (61) Sesamin; (62) Sesaminol; (63) Sesamolin; (64) Phillyrin; (65) Farnesiferol B.
6. Conclusion and perspectives
In recent years, with the combination of pharmaceutical analysis, molecular biology, bioinformatics and artificial intelligence, new vitality has been injected into the development of drug screening strategies and technologies, which greatly improves the efficiency of drug research and development from natural products. At present, S protein-ACE2 blocker screening methods mainly include molecular docking, MD and other VS methods. Except VS, other screening methods such as SPR, MBC, CMC and biotin avidin with ELISA are based on biochemical experiments. VS effectively reduces the screening time and improves the screening efficiency. However, due to the complex and changeable conformation of receptor structure, the binding sites of receptor protein are flexible and there are many conformational states, so false positive results will appear. Therefore, in order to improve the accuracy of VS results, it is usually combined with biological activity experiments. In addition to the above screening methods that have been applied to S protein-ACE2 blockers, a variety of screening methods used to detect protein-protein interactions can also be considered for the screening of S protein-ACE2 blockers. Affinity mass spectrometry, for example, is an effective method to efficiently find small molecular ligands with strong affinity to target proteins. It can efficiently find ligands with strong binding to target proteins from complex mixtures [153]. Fluorescence polarization assay is mainly based on the principle of positive correlation between the intensity of polarized fluorescence and the size of fluorescent molecules, which is widely used in high-throughput drug screening and drug analysis [154]. It should be noted that each of the above screening techniques has its own scope of application, and a single technique often has certain limitations in accuracy or efficiency. Therefore, in practical research, it is necessary to carry out multi-technology combination to screen and verify the accuracy of the results. For example, molecular docking combined with MD, on the one hand, can accurately locate the binding sites and recessive binding sites in the dynamic process of drug binding to the target, so as to more accurately explore the potential active compounds. On the other hand, after molecular docking is used to determine the active compounds with good binding ability to the target, MD can be used to further verify the stability of the active compounds and target complexes, and the results of molecular docking can be further verified. For complex systems, SPR can be combined with VS, and some non-specific components can be eliminated by VS. Then the whole process of the selected compounds binding to the target protein can be monitored by SPR in real time to further verify the binding ability of the compounds obtained by VS to the target protein.
As one of the important sources of new drug research and development, natural products have attracted the attention of researchers at home and abroad for a long time. The use of natural products to prevent or treat COVID-19 is largely inspired by the treatment of SARS. Many studies have proved the role of natural products in the treatment or prevention of viral diseases. A variety of natural products, such as flavonoids, terpenoids, phenols, alkaloids, were obtained by modern screening technology, which can be used as S protein-ACE2 blockers. Although a number of studies have proved that natural products have a significant effect on patients with COVID-19, the above natural products are lack of in vitro and clinical verification. Therefore, it is still necessary to increase the number of samples for randomized clinical trials to evaluate the efficacy and safety of natural products on patients with COVID-19.
Based on the history of drug development, the combination of natural products and chemical synthesis can produce more effective drugs. As for the active lead compounds screened for a variety of S Protein-ACE2 blockers, some of them may not have good ADMET parameters, so we can further modify their structures to obtain good ADMET parameters to enhance their biological activity, so as to develop as candidate drugs. Phenols and flavonoids are considered to have broad potential for preventing viral infections and modulating immune responses in nutritional and functional foods. Based on in vitro analysis and molecular docking models, it will be possible to design and chemically synthesize more efficient compounds based on phenols and flavonoid structures. Both natural and synthetic compounds require functional validation and are not limited to in vitro assays based on recombinant viral proteins. In addition, for the key problem of low bioavailability of candidate compounds, they can be administered through different routes from oral (e.g., inhalation, intravenous), which protects the drug molecule from intestinal metabolism and adsorption and improves its pharmacokinetic and pharmacodynamic parameters. Most of the current research work is based on theoretical calculations and in vitro bioassays, and there is still a lot of work to be done in the real sense of drug design and development. Nevertheless, these studies also laid the foundation for follow-up drug discovery and provided guidance for further in-depth in vivo research.
Author contributions
Jun-zhi Lin and Run-chun Xu put forward the idea; Le-le Ma wrote the manuscript and made the figures; Hui-min Liu, Xue-mei Liu, Xiao-yu Yuan, Chao Xu, and Fang Wang collected the literatures; Ding-kun Zhang contributed to the revisions.
Compliance with ethics guidelines
The authors declare that they have no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to the support of Outstanding Young Science and Technology Talents Project of the Science and Technology Department of Sichuan Province (2019JDJQ0007) and Open Project of State key Laboratory of Innovation Medicine and High Efficiency and Energy Saving Pharmaceutical Equipment in Jiangxi University of Traditional Chinese Medicine (GZSYS202003).
References
- 1.Ma L.L., Liu H.M., Luo C.H., He Y.N., Wang F., Huang H.Z., Han L., Yang M., Xu R.C., Zhang D.K. Fever and antipyretic supported by traditional Chinese medicine: a multi-pathway regulation. Front. Pharmacol. 2021;12:583279–583294. doi: 10.3389/fphar.2021.583279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wollina U. Challenges of COVID-19 pandemic for dermatology. Dermatol. Ther. 2020;33:e13430–13435. doi: 10.1111/dth.13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawat K., Kumari P., Saha L. COVID-19 vaccine: a recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharmacol. 2021;892:173751–173762. doi: 10.1016/j.ejphar.2020.173751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matta S., Rajpal S., Chopra K.K., Arora V.K. Covid-19 vaccines and new mutant strains impacting the pandemic. Indian J. Tubercul. 2021;68:171–173. doi: 10.1016/j.ijtb.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021 doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 7.Scudellari M. How the coronavirus infects cells - and why Delta is so dangerous. Natur. 2021;595:640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 8.Beutler J.A. Natural products as a foundation for drug discovery. Curr. Protoc. Pharmacol. 2019;86:e67–e88. doi: 10.1002/cpph.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J. Pharmacol. Exp. Therapeut. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boozari M., Hosseinzadeh H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytother Res. 2021;35:864–876. doi: 10.1002/ptr.6873. [DOI] [PubMed] [Google Scholar]
- 11.Mrityunjaya M., Pavithra V., Neelam R., Janhavi P., Halami P.M., Ravindra P.V. Immune-boosting, antioxidant and anti-inflammatory food supplements targeting pathogenesis of COVID-19. Front. Immunol. 2020;11:570122–570131. doi: 10.3389/fimmu.2020.570122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry F., Lavandero S., Xie X., Sabharwal B., Zheng Y.Y., Correa A., Narula J., Levy P. Manipulation of ACE2 expression in COVID-19. Open Heart. 2020;7:e001424–e001436. doi: 10.1136/openhrt-2020-001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254–266. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/ap-200220-0772. [DOI] [PubMed] [Google Scholar]
- 15.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/circresaha.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson A.M., Wysocki J., Batlle D. Interaction of SARS-CoV-2 and other coronavirus with ACE (Angiotensin-Converting enzyme)-2 as their main receptor: therapeutic implications. Hypertension. 2020;76:1339–1349. doi: 10.1161/hypertensionaha.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020;43:648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omrani M., Keshavarz M., Nejad Ebrahimi S., Mehrabi M., McGaw L.J., Ali Abdalla M., Mehrbod P. Potential natural products against respiratory viruses: a perspective to develop anti-COVID-19 medicines. Front. Pharmacol. 2020;11:586993–587005. doi: 10.3389/fphar.2020.586993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269–587283. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faheem, Kumar B.K., Sekhar K., Kunjiappan S., Jamalis J., Balaña-Fouce R., Tekwani B.L., Sankaranarayanan M. Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19. Bioorg. Chem. 2020;104:104269–104290. doi: 10.1016/j.bioorg.2020.104269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., Lu J.M., Peukes J., Xiong X., Kräusslich H.G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Natur. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 2020;288:198141–198153. doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296–306. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teli D.M., Shah M.B., Chhabria M.T. In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: targets for COVID-19. Front Mol Biosci. 2020;7:599079–599102. doi: 10.3389/fmolb.2020.599079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Othman H., Bouslama Z., Brandenburg J.T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W., Wang M., Yu D., Zhang X. Variations in SARS-CoV-2 spike protein cell epitopes and glycosylation profiles during global transmission course of COVID-19. Front. Immunol. 2020;11:565278–565289. doi: 10.3389/fimmu.2020.565278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina-Enríquez M.M., Lopez-León S., Carlos-Escalante J.A., Aponte-Torres Z., Cuapio A., Wegman-Ostrosky T. ACE2: the molecular doorway to SARS-CoV-2. Cell Biosci. 2020;10:148–159. doi: 10.1186/s13578-020-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Natur. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D., Xu Y., Cao Z., Gao Z. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24:422–430. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wysocki J., Ye M., Rodriguez E., González-Pacheco F.R., Barrios C., Evora K., Schuster M., Loibner H., Brosnihan K.B., Ferrario C.M., Penninger J.M., Batlle D. Targeting the degradation of angiotensin II with recombinant angiotensin-converting enzyme 2: prevention of angiotensin II-dependent hypertension. Hypertension. 2010;55:90–98. doi: 10.1161/hypertensionaha.109.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. (Lond.). 2020;134:543–545. doi: 10.1042/cs20200163. [DOI] [PubMed] [Google Scholar]
- 41.Yeung M.L., Teng J.L.L., Jia L., Zhang C., Huang C., Cai J.P., Zhou R., Chan K.H., Zhao H., Zhu L., Siu K.L., Fung S.Y., Yung S., Chan T.M., To K.K., Chan J.F., Cai Z., Lau S.K.P., Chen Z., Jin D.Y., Woo P.C.Y., Yuen K.Y. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184:2212–2228. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varghese P.M., Tsolaki A.G., Yasmin H., Shastri A., Ferluga J., Vatish M., Madan T., Kishore U. Host-pathogen interaction in COVID-19: pathogenesis, potential therapeutics and vaccination strategies. Immunobiology. 2020;225:152008–152029. doi: 10.1016/j.imbio.2020.152008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114–e105127. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3:e202000786–e202000796. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742–104748. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life science alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haga J.H., Ichikawa K., Date S. Virtual screening techniques and current computational infrastructures. Curr. Pharmaceut. Des. 2016;22:3576–3584. doi: 10.2174/1381612822666160414142530. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z., Sun H., Shen C., Hu X., Gao J., Li D., Cao D., Hou T. Combined strategies in structure-based virtual screening. Phys. Chem. Chem. Phys. 2020;22:3149–3159. doi: 10.1039/c9cp06303j. [DOI] [PubMed] [Google Scholar]
- 54.Lionta E., Spyrou G., Vassilatis D.K., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr. Top. Med. Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S., Alnammi M., Ericksen S.S., Voter A.F., Ananiev G.E., Keck J.L., Hoffmann F.M., Wildman S.A., Gitter A. Practical model selection for prospective virtual screening. J. Chem. Inf. Model. 2019;59:282–293. doi: 10.1021/acs.jcim.8b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirayama N. Docking simulations between drugs and HLA molecules associated with idiosyncratic drug toxicity. Drug Metabol. Pharmacokinet. 2017;32:31–39. doi: 10.1016/j.dmpk.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Li G.B., Yang L.L., Wang W.J., Li L.L., Yang S.Y. ID-Score: a new empirical scoring function based on a comprehensive set of descriptors related to protein-ligand interactions. J. Chem. Inf. Model. 2013;53:592–600. doi: 10.1021/ci300493w. [DOI] [PubMed] [Google Scholar]
- 58.Gimeno A., Ojeda-Montes M.J., Tomás-Hernández S., Cereto-Massagué A., Beltrán-Debón R., Mulero M., Pujadas G., Garcia-Vallvé S. The light and dark sides of virtual screening: what is there to know? Int. J. Mol. Sci. 2019;20:1375–1395. doi: 10.3390/ijms20061375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 60.Vardhan S., Sahoo S.K. In silIco ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020;124:103936–103948. doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natesh J., Mondal P., Penta D., Abdul Salam A.A., Meeran S.M. Culinary spice bioactives as potential therapeutics against SARS-CoV-2: computational investigation. Comput. Biol. Med. 2021;128:104102–104116. doi: 10.1016/j.compbiomed.2020.104102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J., Gao L., Liang H., Chen S.D. In Silico screening of potential anti-COVID-19 bioactive natural constituents from food sources by molecular docking. Nutrition. 2021;82:111049–111057. doi: 10.1016/j.nut.2020.111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glaab E. Building a virtual ligand screening pipeline using free software: a survey, Brief. Bioinformation. 2016;17:352–366. doi: 10.1093/bib/bbv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi Q., Li R., Li H.Y., Cao Y.B., Bai M., Fan X.J., Wang S.Y., Zhang B., Li S. Identification of the anti-tumor activity and mechanisms of nuciferine through a network pharmacology approach. Acta Pharmacol. Sin. 2016;37:963–972. doi: 10.1038/aps.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao K., Song Y.P., Song A. Exploring active ingredients and function mechanisms of Ephedra-bitter almond for prevention and treatment of Corona virus disease 2019 (COVID-19) based on network pharmacology. BioData Min. 2020;13:19–39. doi: 10.1186/s13040-020-00229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., Chu F., Li P., Johnson N., Li T., Wang Y., An R., Wu D., Chen J., Su Z., Gu X., Ding X. Potential effect of Maxing Shigan decoction against coronavirus disease 2019 (COVID-19) revealed by network pharmacology and experimental verification. J. Ethnopharmacol. 2021;271:113854–113867. doi: 10.1016/j.jep.2021.113854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verma N., Qu X., Trozzi F., Tao Y., Kraka E. SSnet: a deep learning approach for protein-ligand interaction prediction. Int. J. Mol. Sci. 2020;22:1392–1419. doi: 10.3390/ijms22031392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karki N., Verma N., Trozzi F., Tao P., Kraka E., Zoltowski B. Predicting potential SARS-COV-2 drugs-in depth drug database screening using deep neural network framework SSnet, classical virtual screening and docking. Int. J. Mol. Sci. 2021;22:1573–1590. doi: 10.3390/ijms22041573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okimoto N., Futatsugi N., Fuji H., Suenaga A., Morimoto G., Yanai R., Ohno Y., Narumi T., Taiji M. High-performance drug discovery: computational screening by combining docking and molecular dynamics simulations. PLoS Comput. Biol. 2009;5:e1000528–e1000541. doi: 10.1371/journal.pcbi.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 71.Huang S.Y., Zou X. Advances and challenges in protein-ligand docking. Int. J. Mol. Sci. 2010;11:3016–3034. doi: 10.3390/ijms11083016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Vivo M., Masetti M., Bottegoni G., Cavalli A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016;59:4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 73.Kuzmanic A., Bowman G.R., Juarez-Jimenez J., Michel J., Gervasio F.L. Investigating cryptic binding sites by molecular dynamics simulations. Acc. Chem. Res. 2020;53:654–661. doi: 10.1021/acs.accounts.9b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghanakota P., Carlson H.A. Moving beyond active-site detection: MixMD applied to allosteric systems. J. Phys. Chem. B. 2016;120:8685–8695. doi: 10.1021/acs.jpcb.6b03515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopinath K., Jokinen E.M., Kurkinen S.T., Pentikäinen O.T. Screening of natural products targeting SARS-CoV-2-ACE2 receptor interface - a MixMD based HTVS pipeline. Front Chem. 2020;8:589769–589782. doi: 10.3389/fchem.2020.589769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alonso H., Bliznyuk A.A., Gready J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 77.Mondal P., Natesh J., Abdul Salam A.A., Thiyagarajan S., Meeran S.M. Traditional medicinal plants against replication, maturation and transmission targets of SARS-CoV-2: computational investigation. J. Biomol. Struct. Dyn. 2020;5:1–18. doi: 10.1080/07391102.2020.1842246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ganesan A., Coote M.L., Barakat K. Molecular dynamics-driven drug discovery: leaping forward with confidence. Drug Discov. Today. 2017;22:249–269. doi: 10.1016/j.drudis.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 79.Luque F.J., Barril X., Bidonchanal A., Spyrakis F. Protein flexibility and ligand recognition: challenges for molecular modeling. Curr. Top. Med. Chem. 2011;11:192–210. doi: 10.2174/156802611794863571. [DOI] [PubMed] [Google Scholar]
- 80.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9:923–939. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patching S.G. Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta. 2014;1838:43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 82.Prabowo B.A., Purwidyantri A., Liu K.C. Surface plasmon resonance optical sensor: a review on light source technology. Biosensors. 2018;8:80–107. doi: 10.3390/bios8030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rich R.L., Myszka D.G. Biacore J: a new platform for routine biomolecular interaction analysis. J. Mol. Recogn. 2001;14:223–228. doi: 10.1002/jmr.535. [DOI] [PubMed] [Google Scholar]
- 84.Piliarik M., Homola J. Surface plasmon resonance (SPR) sensors: approaching their limits? Opt Express. 2009;17:16505–16517. doi: 10.1364/oe.17.016505. [DOI] [PubMed] [Google Scholar]
- 85.Zhu Z.L., Qiu X.D., Wu S., Liu Y.T., Zhao T., Sun Z.H., Li Z.R., Shan G.Z. Blocking effect of demethylzeylasteral on the interaction between human ACE2 protein and SARS-CoV-2 RBD protein discovered using SPR technology. Molecules. 2020;26:57–69. doi: 10.3390/molecules26010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ye M., Luo G., Ye D., She M., Sun N., Lu Y.J., Zheng J. Network pharmacology, molecular docking integrated surface plasmon resonance technology reveals the mechanism of Toujie Quwen Granules against coronavirus disease 2019 pneumonia. Phytomedicine. 2021;85:153401–1534014. doi: 10.1016/j.phymed.2020.153401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao H.Y., Jiang J.G. Application of chromatography technology in the separation of active components from nature derived drugs. Mini Rev. Med. Chem. 2010;10:1223–1234. doi: 10.2174/13895575110091223. [DOI] [PubMed] [Google Scholar]
- 88.Chen C., Yang F.Q., Zuo H.L., Song Y.L., Xia Z.N., Xiao W. Applications of biochromatography in the screening of bioactive natural products. J. Chromatogr. Sci. 2013;51:780–790. doi: 10.1093/chromsci/bmt002. [DOI] [PubMed] [Google Scholar]
- 89.Beigi F., Lundahl P. Immobilized biomembrane chromatography of highly lipophilic drugs. J. Chromatogr. A. 1999;852:313–327. doi: 10.1016/s0021-9673(99)00025-4. [DOI] [PubMed] [Google Scholar]
- 90.Lagerquist C., Beigi F., Karlén A., Lennernäs H., Lundahl P. Effects of cholesterol and model transmembrane proteins on drug partitioning into lipid bilayers as analysed by immobilized-liposome chromatography. J. Pharm. Pharmacol. 2001;53:1477–1487. doi: 10.1211/0022357011778016. [DOI] [PubMed] [Google Scholar]
- 91.Sun X., Liu H., Li J. Advances in the application of molecular biological chromatography in the study of traditional Chinese medicine. Her Med. 2010;29:1186–1189. [Google Scholar]
- 92.Kim H.S., Wainer I.W. Rapid analysis of the interactions between drugs and human serum albumin (HSA) using high-performance affinity chromatography (HPAC) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;870:22–26. doi: 10.1016/j.jchromb.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., Zou H., Kong L., Ni J. Analysis of bioactive components in traditional Chinese medicines by molecular biochromatography with alpha1-acid glycoprotein stationary phase. J. Basic Clin. Physiol. Pharmacol. 2000;11:155–172. doi: 10.1515/jbcpp.2000.11.2.155. [DOI] [PubMed] [Google Scholar]
- 94.Yoo M.J., Smith Q.R., Hage D.S. Studies of imipramine binding to human serum albumin by high-performance affinity chromatography. Journal of Chromatography B Analytical Technologies in the Biomedical & Life Sciences. 2009;877:1149–1154. doi: 10.1016/j.jchromb.2009.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., Chen J., Zhang L., Lv L., Zhang G., Yuan Y., Chai Y., Zhu M., Wu C. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm. Sin. B. 2021;11:222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He L., Wang S., Geng X. Coating and fusing cell membranes onto a silica surface and their chromatographic characteristics. Chromatographia. 2001;54:71–76. doi: 10.1007/BF02491836. [DOI] [Google Scholar]
- 97.Hou X., Zhou M., Jiang Q., Wang S., He L. A vascular smooth muscle/cell membrane chromatography-offline-gas chromatography/mass spectrometry method for recognition, separation and identification of active components from traditional Chinese medicines. J. Chromatogr. A. 2009;1216:7081–7097. doi: 10.1016/j.chroma.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 98.Zhang C., Chen G., Liu J. Advances in the application of cell membrane chromatography in the study of traditional Chinese medicine. Chin J Chin Pharmacol. 2013;38:1881–1886. [PubMed] [Google Scholar]
- 99.Lv Y., Wang S., Liang P., Wang Y., Zhang X., Jia Q., Fu J., Han S., He L. Screening and evaluation of anti-SARS-CoV-2 components from Ephedra sinica by ACE2/CMC-HPLC-IT-TOF-MS approach. Anal. Bioanal. Chem. 2021;413:2995–3004. doi: 10.1007/s00216-021-03233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao J., Ding Y., Wang Y., Liang P., Zhang L., Liu R. Oroxylin A is a severe acute respiratory syndrome coronavirus 2-spiked pseudotyped virus blocker obtained from Radix Scutellariae using angiotensin-converting enzyme II/cell membrane chromatography. Phytother Res. 2021;35:3194–3204. doi: 10.1002/ptr.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y., Guo J. Advances in application of cell membrane chromatography in screening of active ingredients in traditional Chinese medicine. Chin. Tradit. Herb. Drugs. 2012;43:383–387. doi: 10.11656/j.issn.1673-9043.2017.01.18. [DOI] [Google Scholar]
- 102.Hou X., Wang S., Zhang T., Ma J., Zhang J., Zhang Y., Lu W., He H., He L. Recent advances in cell membrane chromatography for traditional Chinese medicines analysis. J. Pharmaceut. Biomed. Anal. 2014;101:141–150. doi: 10.1016/j.jpba.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 103.Gan S.D., Patel K.R. Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Invest. Dermatol. 2013;133:e12–e15. doi: 10.1038/jid.2013.287. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y., Zhao Y., Lu S., Zhu M., He Y., Ruan C. A high-throughput biotin-avidin-ELISA for studying expression of platelet membrane glycoproteins and its clinical application. Tohoku J. Exp. Med. 2010;222:83–88. doi: 10.1620/tjem.222.83. [DOI] [PubMed] [Google Scholar]
- 105.Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fu W., Chen Y., Wang K., Hettinghouse A., Hu W., Wang J.Q., Lei Z.N., Chen Z.S., Stapleford K.A., Liu C.J. Repurposing FDA-approved drugs for SARS-CoV-2 through an ELISA-based screening for the inhibition of RBD/ACE2 interaction. Protein Cell. 2020:1–6. doi: 10.1007/s13238-020-00803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gangadevi S., Badavath V.N., Thakur A., Yin N., De Jonghe S., Acevedo O., Jochmans D., Leyssen P., Wang K., Neyts J., Yujie T., Blum G. Kobophenol A inhibits binding of host ACE2 receptor with spike RBD domain of SARS-CoV-2, a lead compound for blocking COVID-19. J. Phys. Chem. Lett. 2021;12:1793–1802. doi: 10.1021/acs.jpclett.0c03119. [DOI] [PubMed] [Google Scholar]
- 108.Ozgul S., von Daake S., Kakehi S., Sereni D., Denissova N., Hanlon C., Huang Y.J., Everett J.K., Yin C., Montelione G.T., Comoletti D. An ELISA-based screening platform for ligand-receptor discovery. Methods Enzymol. 2019;615:453–475. doi: 10.1016/bs.mie.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 109.McGall G., Labadie J., Brock P., Wallraff G., Nguyen T., Hinsberg W. Light-directed synthesis of high-density oligonucleotide arrays using semiconductor photoresists. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13555–13560. doi: 10.1073/pnas.93.24.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huo J., Miao Y., Zeng Y. Gene chip technology and its application. Let Biotech. 2007;2:329–332. [Google Scholar]
- 111.Zhu T. Global analysis of gene expression using GeneChip microarrays. Curr. Opin. Plant Biol. 2003;6:418–425. doi: 10.1016/s1369-5266(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 112.Wang X.W., Gao H.J., Fang D.C. Advances in gene chip technique in Barrett's metaplasia and adenocarcinoma. J. Dig. Dis. 2008;9:68–71. doi: 10.1111/j.1751-2980.2008.00324.x. [DOI] [PubMed] [Google Scholar]
- 113.Scherf U., Ross D.T., Waltham M., Smith L.H., Lee J.K., Tanabe L., Kohn K.W., Reinhold W.C., Myers T.G., Andrews D.T., Scudiero D.A., Eisen M.B., Sausville E.A., Pommier Y., Botstein D., Brown P.O., Weinstein J.N. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 114.Song B., Liu J., Han J. Application of gene chip technology in tumor diagnosis and treatment. Chi J Oncol Prevent Treat. 2003;17:349–351. [Google Scholar]
- 115.Glinsky G.V. Tripartite combination of candidate pandemic mitigation agents: vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID-19 pandemic defined by genomics-guided tracing of SARS-CoV-2 targets in human cells. Biomedicines. 2020;8:129–155. doi: 10.3390/biomedicines8050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Su N., Yang L., Liu J., Zhou H. Research progress in domestic application of gene chip technology. Let Biotech. 2016;27:289–292. doi: 10.3969/j.issn.1009-0002.2016.02.033. [DOI] [Google Scholar]
- 117.Zhu C.S., Lin Z.J., Xiao M.L., Niu H.J., Zhang B. The spectrum-effect relationship-a rational approach to screening effective compounds, reflecting the internal quality of Chinese herbal medicine. Chin. J. Nat. Med. 2016;14:177–184. doi: 10.1016/s1875-5364(16)30014-0. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y.L., Hu G., Zhang Q., Yang Y.X., Li Q.Q., Hu Y.J., Chen H., Yang F.Q. Screening and characterizing tyrosinase inhibitors from Salvia miltiorrhiza and Carthamus tinctorius by spectrum-effect relationship analysis and molecular docking. J Anal Methods Chem.2018. 2018:2141389–2141399. doi: 10.1155/2018/2141389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Senthil Kumar K.J., Gokila Vani M., Wang C.S., Chen C.C., Chen Y.C., Lu L.P., Huang C.H., Lai C.S., Wang S.Y. Geranium and lemon essential oils and their active compounds downregulate angiotensin-converting enzyme 2 (ACE2), a SARS-CoV-2 spike receptor-binding domain, in epithelial cells. Plants. 2020;9:770–782. doi: 10.3390/plants9060770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prabhu L., Chen L., Wei H., Demir Ö., Safa A., Zeng L., Amaro R.E., O'Neil B.H., Zhang Z.Y., Lu T. Development of an AlphaLISA high throughput technique to screen for small molecule inhibitors targeting protein arginine methyltransferases. Mol. Biosyst. 2017;13:2509–2520. doi: 10.1039/c7mb00391a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hanson Q.M., Wilson K.M., Shen M., Itkin Z., Eastman R.T., Shinn P., Hall M.D. Targeting ACE2-RBD interaction as a platform for COVID-19 therapeutics: development and drug-repurposing screen of an AlphaLISA proximity assay. ACS Pharmacol Transl Sci. 2020;3:1352–1360. doi: 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020;328:109211–109225. doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pan B., Fang S., Zhang J., Pan Y., Liu H., Wang Y., Li M., Liu L. Chinese herbal compounds against SARS-CoV-2: Puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Comput. Struct. Biotechnol. J. 2020;18:3518–3527. doi: 10.1016/j.csbj.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maroli N., Bhasuran B., Natarajan J., Kolandaivel P. The potential role of procyanidin as a therapeutic agent against SARS-CoV-2: a text mining, molecular docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020;22:1–16. doi: 10.1080/07391102.2020.1823887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Basu A., Sarkar A., Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020;10:17699–17714. doi: 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y.Q., Li Q.S., Zheng X.Q., Lu J.L., Liang Y.R. Antiviral effects of green tea EGCG and its potential application against COVID-19, molecules. 2021. Basel, Switzerland. 26. [DOI] [PMC free article] [PubMed]
- 128.Gowrishankar S., Muthumanickam S., Kamaladevi A., Karthika C., Jothi R., Boomi P., Maniazhagu D., Pandian S.K. Promising phytochemicals of traditional Indian herbal steam inhalation therapy to combat COVID-19 - an in silico study. Food Chem. Toxicol. : an international journal published for the British Industrial Biological Research Association. 2021;148:111966. doi: 10.1016/j.fct.2020.111966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Henss L., Auste A., Schürmann C., Schmidt C., von Rhein C., Mühlebach M.D., Schnierle B.S. The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection. J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhan Y., Ta W., Tang W., Hua R., Wang J., Wang C., Lu W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021;13:1–7. doi: 10.1002/ddr.21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guerrero L., Castillo J., Quiñones M., Garcia-Vallvé S., Arola L., Pujadas G., Muguerza B. Inhibition of angiotensin-converting enzyme activity by flavonoids: structure-activity relationship studies. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu J.W., Wang L., Bao L.D. Exploring the active compounds of traditional Mongolian medicine in intervention of novel coronavirus (COVID-19) based on molecular docking method. Journal of functional foods. 2020;71:104016. doi: 10.1016/j.jff.2020.104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heinz S.A., Henson D.A., Austin M.D., Jin F., Nieman D.C. Quercetin supplementation and upper respiratory tract infection: a randomized community clinical trial. Pharmacol. Res. 2010;62:237–242. doi: 10.1016/j.phrs.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo E., Zhang D., Luo H., Liu B., Zhao K., Zhao Y., Bian Y., Wang Y. Treatment efficacy analysis of traditional Chinese medicine for novel coronavirus pneumonia (COVID-19): an empirical study from Wuhan, Hubei Province, China. Chin. Med. 2020;15:34–47. doi: 10.1186/s13020-020-00317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rodriguez-Mateos A., Vauzour D., Krueger C.G., Shanmuganayagam D., Reed J., Calani L., Mena P., Del Rio D., Crozier A. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: an update. Arch. Toxicol. 2014;88:1803–1853. doi: 10.1007/s00204-014-1330-7. [DOI] [PubMed] [Google Scholar]
- 136.Williamson G., Kerimi A. Testing of natural products in clinical trials targeting the SARS-CoV-2 (Covid-19) viral spike protein-angiotensin converting enzyme-2 (ACE2) interaction. Biochem. Pharmacol. 2020;178:114123. doi: 10.1016/j.bcp.2020.114123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jaeger R., Cuny E. Terpenoids with special pharmacological significance: a review. Nat. Prod. Commun. 2016;11:1373–1390. [PubMed] [Google Scholar]
- 138.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/s0140-6736(03)13615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Br B., Damle H., Ganju S., Damle L. In silico screening of known small molecules to bind ACE2 specific RBD on Spike glycoprotein of SARS-CoV-2 for repurposing against COVID-19. F1000Res. 2020;9:663–686. doi: 10.12688/f1000research.24143.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu S., Zhu Y., Xu J., Yao G., Zhang P., Wang M., Zhao Y., Lin G., Chen H., Chen L., Zhang J. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine. 2021;85:153364–153375. doi: 10.1016/j.phymed.2020.153364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jinhua W. Ursolic acid: pharmacokinetics process in vitro and in vivo, a mini review. Arch. Pharmazie. 2019;352 doi: 10.1002/ardp.201800222. [DOI] [PubMed] [Google Scholar]
- 142.Torres T., Puig L. Managing cutaneous immune-mediated diseases during the COVID-19 pandemic. Am. J. Clin. Dermatol. 2020;21:307–311. doi: 10.1007/s40257-020-00514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A., Doerr H.W., Cinatl J., Jr. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 144.Zhao Z., Xiao Y., Xu L., Liu Y., Jiang G., Wang W., Li B., Zhu T., Tan Q., Tang L., Zhou H., Huang X., Shan H. Glycyrrhizic acid nanoparticles as antiviral and anti-inflammatory agents for COVID-19 treatment. ACS Appl. Mater. Interfaces. 2021;13:20995–21006. doi: 10.1021/acsami.1c02755. [DOI] [PubMed] [Google Scholar]
- 145.Dai J., Mumper R.J. Plant phenolics: extraction, analysis and their antioxidant and anticancer properties. Molecules. 2010;15:7313–7352. doi: 10.3390/molecules15107313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu S., Wang J., Zhang Y., Bai H., Wang C., Wang N., He L. Three salvianolic acids inhibit 2019-nCoV spike pseudovirus viropexis by binding to both its RBD and receptor ACE2. J. Med. Virol. 2021;93:3143–3151. doi: 10.1002/jmv.26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li R., Narita R., Nishimura H., Marumoto S., Yamamoto S.P., Ouda R., Yatagai M., Fujita T., Watanabe T. Antiviral activity of phenolic derivatives in pyroligneous acid from hardwood, softwood, and bamboo. ACS Sustain. Chem. Eng. 2017 acssuschemeng.7b01265. [Google Scholar]
- 148.Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Świderski G., Jabłońska-Trypuć A., Kalinowska M., Świsłocka R., Karpowicz D., Magnuszewska M., Lewandowski W. Spectroscopic, theoretical and antioxidant study of 3d-transition metals (Co (II), Ni(II), Cu(II), Zn(II) complexes with cichoric acid. Materials. 2020;13 doi: 10.3390/ma13143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gyebi G.A., Adegunloye A.P., Ibrahim I.M., Ogunyemi O.M., Afolabi S.O., Ogunro O.B. Prevention of SARS-CoV-2 cell entry: insight from in silico interaction of drug-like alkaloids with spike glycoprotein, human ACE2, and TMPRSS2. J. Biomol. Struct. Dyn. 2020;22:1–25. doi: 10.1080/07391102.2020.1835726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.He C.L., Huang L.Y., Wang K., Gu C.J., Hu J., Zhang G.J., Xu W., Xie Y.H., Tang N., Huang A.L. Identification of bis-benzylisoquinoline alkaloids as SARS-CoV-2 entry inhibitors from a library of natural products. Signal transduction and targeted therapy. 2021;6:131. doi: 10.1038/s41392-021-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fielding B.C., da Silva Maia Bezerra Filho C., Ismail N.S.M., Sousa D.P. vol. 25. 2020. (Alkaloids: Therapeutic Potential against Human Coronaviruses, Molecules). Basel, Switzerland. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Petre B.A. Affinity-mass spectrometry approaches for elucidating structures and interactions of protein-ligand complexes. Adv. Exp. Med. Biol. 2014;806:129–151. doi: 10.1007/978-3-319-06068-2_7. [DOI] [PubMed] [Google Scholar]
- 154.Chen Y., Fu Z., Li D., Yue Y., Liu X. Optimizations of a novel fluorescence polarization-based high-throughput screening assay for β-catenin/LEF1 interaction inhibitors. Anal. Biochem. 2021;612:113966–113976. doi: 10.1016/j.ab.2020.113966. [DOI] [PubMed] [Google Scholar]
- 155.Niu W., Wu F., Cui H., Cao W., Chao Y., Wu Z., Fan M., Liang C. Network pharmacology analysis to identify phytochemicals in traditional Chinese medicines that may regulate ACE2 for the treatment of COVID-19, evid. Based complement. Alternat. Med.2020. 2020:7493281–7493295. doi: 10.1155/2020/7493281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gao L.Q., Xu J., Chen S.D. In silico screening of potential Chinese herbal medicine against COVID-19 by targeting SARS-CoV-2 3CLpro and angiotensin converting enzyme II using molecular docking. Chin. J. Integr. Med. 2020;26:527–532. doi: 10.1007/s11655-020-3476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xing Y., Hua Y.R., Shang J., Ge W.H., Liao J. Traditional Chinese medicine network pharmacology study on exploring the mechanism of Xuebijing Injection in the treatment of coronavirus disease 2019. Chin. J. Nat. Med. 2020;18:941–951. doi: 10.1016/s1875-5364(20)60038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020;39:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yepes-Pérez A.F., Herrera-Calderon O., Quintero-Saumeth J. Uncaria tomentosa (cat's claw): a promising herbal medicine against SARS-CoV-2/ACE-2 junction and SARS-CoV-2 spike protein based on molecular modeling. J. Biomol. Struct. Dyn. 2020;29:1–17. doi: 10.1080/07391102.2020.1837676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alazmi M., Motwalli O. Molecular basis for drug repurposing to study the interface of the S protein in SARS-CoV-2 and human ACE2 through docking, characterization, and molecular dynamics for natural drug candidates. J. Mol. Model. 2020;26:338–348. doi: 10.1007/s00894-020-04599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sun Y., Yang A.W.H., Hung A., Lenon G.B. Screening for a potential therapeutic agent from the herbal formula in the 4(th) edition of the Chinese national guidelines for the initial-stage management of COVID-19 via molecular docking, evid. Based complement. Alternat. Med.2020. 2020:3219840–3219857. doi: 10.1155/2020/3219840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alazmi M., Motwalli O. In silico virtual screening, characterization, docking and molecular dynamics studies of crucial SARS-CoV-2 proteins. J. Biomol. Struct. Dyn. 2020;7:1–11. doi: 10.1080/07391102.2020.1803965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kapoor N., Ghorai S.M., Kushwaha P.K., Shukla R., Aggarwal C., Bandichhor R. Plausible mechanisms explaining the role of cucurbitacins as potential therapeutic drugs against coronavirus 2019. Inform Med Unlocked. 2020;21:100484–100486. doi: 10.1016/j.imu.2020.100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patel A., Rajendran M., Shah A., Patel H., Pakala S.B., Karyala P. Virtual screening of curcumin and its analogs against the spike surface glycoprotein of SARS-CoV-2 and SARS-CoV. J. Biomol. Struct. Dyn. 2021;5:1–9. doi: 10.1080/07391102.2020.1868338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Su J., Liu Z., Liu C., Li X., Wang Y., Zhao J., Wu Q., Zheng S., Zhang Y. Network pharmacology integrated molecular docking reveals the mechanism of anisodamine hydrobromide injection against novel coronavirus pneumonia. Evid. Based Complement. Alternat. Med.2020. 2020:5818107–5818118. doi: 10.1155/2020/5818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mehmood A., Khan S., Khan S., Ahmed S., Ali A., Xue M., Ali L., Hamza M., Munir A., Ur Rehman S., Mehmood Khan A., Hussain Shah A., Bai Q. In silico analysis of quranic and prophetic medicinals plants for the treatment of infectious viral diseases including corona virus. Saudi J. Biol. Sci. 2021;28:3137–3151. doi: 10.1016/j.sjbs.2021.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Poochi S.P., Easwaran M., Balasubramanian B., Anbuselvam M., Meyyazhagan A., Park S., Bhotla H.K., Anbuselvam J., Arumugam V.A., Keshavarao S., Kanniyappan G.V., Pappusamy M., Kaul T. Employing bioactive compounds derived from Ipomoea obscura (L.) to evaluate potential inhibitor for SARS-CoV-2 main protease and ACE2 protein. Food Front. 2020:1–12. doi: 10.1002/fft2.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Upreti S., Prusty J.S., Pandey S.C., Kumar A., Samant M. Identification of novel inhibitors of angiotensin-converting enzyme 2 (ACE-2) receptor from Urtica dioica to combat coronavirus disease 2019 (COVID-19) Mol. Divers. 2021;4:1–15. doi: 10.1007/s11030-020-10159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]