Abstract
Maintenance of a functional proteome is achieved through the mechanism of proteostasis that involves precise coordination between molecular machineries assisting a protein from its conception to demise. Although each organelle within a cell has its own set of proteostasis machinery, inter-organellar communication and cell non-autonomous signaling bring forth the multidimensional nature of the proteostasis network. Exposure to extrinsic and intrinsic stressors can challenge the proteostasis network, leading to the accumulation of aberrant proteins or a decline in the proteostasis components, as seen during aging and in several diseases. Here, we summarize recent advances in understanding the role of proteostasis and its regulation in aging and disease, including monogenetic and infectious diseases. We highlight some of the emerging as well as unresolved questions in proteostasis that need to be addressed to overcome pathologies associated with damaged proteins and to promote healthy aging.
Keywords: Proteostasis, protein folding, protein degradation, molecular chaperones, Unfolded Protein Response, aging, metabolism, autophagy, neurodegeneration
Defining the need to understand the role of proteostasis
Proteins need to be maintained in their native structures at required cellular concentrations and specific locations. Deviations lead to aberrant enzyme activity, binding, stoichiometry in complexes, or aggregation, all of which may impose a penalty on cellular fitness, finally leading to ailing pathophysiological states or cell death. Proteostasis is the homeostasis of maintaining the proteome by regulating protein synthesis, translocation (if required), post-translational modifications (if required for stabilizing folded states), folding, and degradation or conversion of toxic oligomers into benign and less toxic amyloid aggregates1. An effective interplay between components of the proteostasis network consisting of chaperones (protein and chemical), co-chaperones, degradation machinery, translation control machinery, and adaptor proteins take cares of the balance in proteostasis. Hence, a thorough understanding of proteostasis will allow us to not only tackle the protein folding problems at hand but also open gates to a healthy aging paradigm.
Proteostasis components, problems, and regulation
Components of the proteostasis network
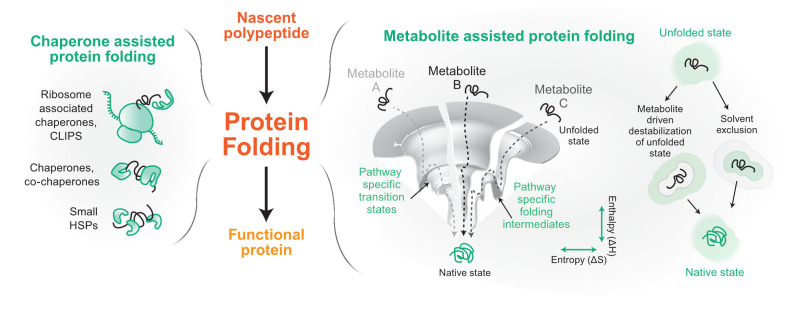
Proteins are translated on ribosomes that also act as scaffolds to assemble protein chaperones, which aid the nascent chains to reach their final functional structure. Protein chaperones recognize and bind nascent chains or folding intermediates that exhibit exposed hydrophobic patches2. Binding and unbinding of the substrate protein with its chaperone follow a cycle dictated by either the ATP-hydrolysis rate of the chaperone/co-chaperone system (like Hsp70, Hsp90, and Hsp60 chaperone systems)3,4 or the dissociation and association rate of the substrate on ATP-independent chaperones (small heat shock proteins)5 (Figure 1). Ribosome-associated chaperones are called the CLIPS (chaperones linked to protein synthesis), which act as guardians of the nascent chains and comprise the Hsp70, Hsp40, Hsp110, and Hsp60 group of chaperones6. However, the Hsp70 system can perform diverse functions, including disaggregation of amyloid fibrils owing to its binding to a myriad of Hsp40s and Hsp110s as co-chaperones7–9. After protein translation, translocation is mediated by cytosolic and compartment-specific chaperones. Many of the proteins destined for cytosol, nucleus, and peroxisomes are folded in the cytosol by a cycle of binding and unbinding to the protein chaperones10. For proteins destined to the endoplasmic reticulum (ER) and mitochondria, the nascent chains are maintained in an unfolded state at the site of translation, targeted to the appropriate compartment, and subsequently folded inside either post-translationally or co-translationally using the chaperone machinery present in the targeted organelle11.
Figure 1. Components of proteostasis helping in protein folding.
From its conception on the ribosome, a protein molecule folds to its native functional state with the help of two major proteostasis components. The left panel shows some of the representative components of a chaperone-based protein folding arm. In the right panel, we show metabolite-assisted folding of a protein. The folding landscape of a protein has been shown in the presence of three metabolites A, B, and C. On the extreme right, possible molecular mechanisms of metabolite-based protein folding are depicted. Gray represents solvent and green represents metabolite. CLIPS, chaperones linked to protein synthesis; HSP, heat shock protein.
Each organelle that handles non-native protein chains has its complement of protein chaperones to aid folding. ER, which handles most of the membrane and secretory proteome, contains specialized chaperones that help in N-glycosylation and disulphide bond formation along with ER-localized forms of the canonical Hsp70 and Hsp90 chaperone machineries12. The mitochondrial matrix (MM), on the other hand, handles a large number of metabolic enzymes and proteins involved in oxidative phosphorylation; it has chaperones that are homologous to the Hsp70, Hsp90, and Hsp60 class of chaperone systems. Mitochondrial intermembrane space (IMS) resembles ER in terms of oxidative folding and has disulphide-forming chaperones along with other small chaperones. This space is thought to be devoid of the canonical Hsp70, Hsp90, and Hsp60 chaperoning machineries13.
Protein chaperones have diverged and specialized for organelle-specific proteostasis. Although the protein chaperones in the different compartments play a central role in protein quality assurance, recent reports have shown that the cellular milieu comprising the different metabolites also plays an important role in assisting proteins to their native states14–17. The exact mechanism of metabolite assistance is unknown, but metabolites may change the solvent structure or interaction of the nascent chain with the solvent or also aid the process of folding directly or indirectly through protein chaperones18–20 (Figure 1). More importantly, these reports have opened new avenues by placing metabolites into the proteostasis network.
Problems in proteostasis
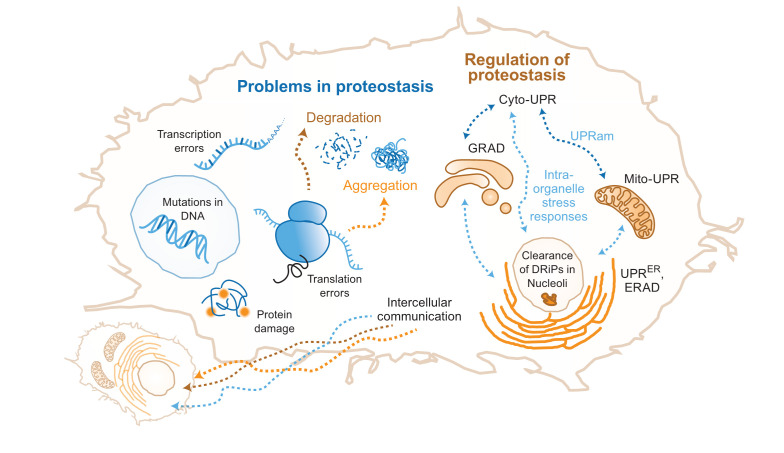
A hitch in any of the steps of proteostasis can lead to its disequilibrium. It may start when one or more proteins take more than their allocated time to reach its folded state. Although this timer mechanism is important to ensure that the proteins are given sufficient time to fold to their native structure before tagging it as a non-foldable protein, evidence for such a mechanism is prominently established only in the ER21. A protein that has a problem in folding can be degraded either by the collaboration of chaperones, C-terminal Hsp70-interacting protein (CHIP), BAG3, ubiquitin ligase, and proteasome or by the chaperone-dependent autophagy22,23. In some cases, when both are unable to take care of the misfolded protein, it may sequester essential chaperones and other proteins to form small soluble aggregates that confer cellular toxicity as seen for multiple neurodegenerative diseases24. Alternatively, the misfolded protein may be packaged into vesicles to form harmless molecular aggregates25,26. These small aggregates can also be detoxified by cellular processes that coagulate them to form larger seemingly harmless aggregates27.
Proteostasis problems can also start with an incompletely translated nascent chain resulting from a damaged mRNA or an mRNA with base misincorporation, faulty editing, or splicing (Figure 2). These nascent chains are actively degraded by the ribosome quality control (RQC) system that couples protein and damaged mRNA degradation28,29. Mistranslated proteins are ubiquitinated and transported to the nucleolus for degradation30. Mislocalization of the proteins can also precipitate problems; if ER proteins are mistargeted to mitochondria (or vice versa) during stress, incompatibility between the chaperone systems and the mistargeted proteins leads to the loss of proteostasis31,32.
Figure 2. Problems in proteostasis and regulation of proteostasis.
The left side of the cell shows the molecular origins of problems in protein folding. Errors at various steps in the life cycle of a protein leading to protein degradation and protein aggregation, resulting in imbalance of proteostasis. On the right, a few of the different organelle-specific cellular responses to the imbalance of proteostasis are shown. They include inter-organelle and inter-cellular responses. Cyto-UPR, cytosolic unfolded protein response; DRiP, defective ribosomal product; ERAD, endoplasmic reticulum–associated degradation; GRAD, Golgi apparatus–related degradation; Mito-UPR, mitochondrial unfolded protein response; UPRam, unfolded protein response activated by mistargeting of proteins; UPRER, endoplasmic reticulum–associated unfolded protein response.
Even folded proteins may initiate proteostasis disequilibrium due to environmental insults that result in protein damage and aggregation33 (Figure 2). These aggregates need to be disaggregated or cleared while damaged proteins need to be degraded to prevent aberrant interactions and signaling events.
Regulation of proteostasis machinery
Problems in proteostasis are sensed in the different compartments by specialized sensors. Heat shock factor 1 (HSF1) is the canonical eukaryotic sensor in the cytosol for proteostasis disequilibrium and is maintained in an inactive state by the Hsp70 and Hsp90 chaperones. When misfolded proteins accumulate, they titrate out the chaperones and free up HSF1, which then is free to translocate to the nucleus to signal cytosolic unfolded protein response (cyto-UPR)10,34. HSF1 activation upregulates chaperones, degradation machinery, and metabolic and stress response–related genes. More recently, heme-regulated kinase (HRI) was discovered as an additional cytosolic sensor of protein aggregation arising because of blocked protein degradation, which attenuates protein translation to decrease the intracellular protein concentration35.
In the ER, proteostasis disequilibrium is sensed by IRE1 (the most conserved sensor among all eukaryotes), ATF6, and PERK (the last two branches are currently known to be present only in metazoans). These sense proteostasis problems in the ER to elicit an integrated stress response that increases ER chaperones, components of ER-associated degradation (ERAD) (clears misfolded proteins of ER) or autophagy, and ER volume (to decrease protein concentration)36. This concerted response decreases global protein translation and alters metabolism among a host of other pathways that are still being investigated37. Although IRE1 and ATF6 seem to be redundant in the pathways that they regulate, PERK primarily prevents protein translation by phosphorylating eIF2α and inactivating it. Recent studies of these signaling processes have shed light on the nuances that govern this complex integration of stress response38,39. Activation of the ER sensors also seems to follow the same titration model as HSF1, where the ER-resident Hsp70 (Bip/GRP78) prevents activation of the sensors as long as it is not titrated away by the accumulation of the client proteins40.
Whereas regulation of proteostasis machinery in the ER is well studied, little is known about proteostasis regulation in the mitochondria. Mitochondria, being a double-membrane-bound organelle, creates additional complexity with different compartments vis-à-vis MM and IMS having different redox environments. Proteostasis stress due to mitochondrial DNA damage is sensed by ATFS-1, first discovered in Caenorhabditis elegans to upregulate mitochondrial chaperones in response41. Subsequently, ATF5 was found to play a similar role in mammalian cells42. However, mitochondria respond differently to misfolding inside the MM (ROX1)43 and proteostasis imbalance due to overburdened protein translocation machinery44. The MM along with the nucleolus45,46 plays an important role in clearing cytosolic misfolded proteins. Nucleoli and promyelocytic leukemia protein bodies act as clearance hubs for defective ribosomal products as well30. Currently, there is limited knowledge regarding proteostasis disequilibrium in the two sub-compartments and whether they can report the compartment-specific problems back to the nucleus. Given the importance of mitochondria in global misfolding stress, sensors and signaling mechanisms of the two sub-compartments are of immediate interest. In a recent study, Rao et al. showed sub-compartment-specific stress-response pathways in MM and IMS during proteotoxic stress-induced by misfolded proteins, where upregulation of TOM complex mediates the IMS response while Vms1 is important in MM stress response47.
As stress may disturb organellar membranes, reorganization of some membrane-bound organelles is also reported during stress. For example, Golgi apparatus–related degradation is aided by 26S proteasome in the cytosol and controls Golgi dispersal48. Another study reported the shuttling of mammalian ubiquitin ligase CHIP from chaperones to the membranes during acute stress. CHIP then acts on its organelle specific substrates leading to reorganization of that organelle49. Remarkably, the stress-response pathways of the different compartments seem to communicate intracellularly: ER-UPR activation can clear cytoplasmic aggregates50,51, and downregulation of cytosolic chaperones can upregulate ER stress response in specific cells52 (Figure 2). Similarly, mitochondrial proteostasis can take care of cytosolic misfolded proteins45, whereas ER can act as a hub for protein quality control when mitochondrial proteostasis is perturbed53. Also, mitochondrial precursor proteins, when accumulated in the cytosol, result in the activation of another type of stress response, referred to as UPRam (UPR activated by mistargeting of proteins), which was found to be beneficial for cells54. Another study discovered similar mitochondrial precursor over-accumulation stress (mPOS) in Saccharomyces cerevisiae disturbing cytosolic proteostasis55. Response to mPOS includes upregulation of ribosome-associated proteins, ultimately leading to cell survival55. Thus, the stress-response pathways in organelles are interconnected and this connection extends to inter-organelle contact sites as well. For example, contact sites of mitochondria with lysosomes and ER are quite important for mitochondrial quality control (reviewed elsewhere56). Organisms not only have intracellular stress sensors and response mechanisms but the response pathways are also regulated by cell non-autonomous signaling57. Intercellular communication (Figure 2) of this sort is seen in C. elegans, where the upregulation of mitochondrial unfolded protein response (Mito-UPR) in the brain seems to activate the HSF1-dependent pathway in the intestinal cells58,59. ER-UPR upregulation in the glial cells of C. elegans leads to the upregulation of ER-UPR in distal cells using neuropeptide signaling conferring ER stress tolerance and longevity60. In a striking discovery, a tyrosine phosphatase that negatively regulates HSF1 activity was found to be downregulated by endogenous small interfering RNA (endo-siRNA) in germline-less C. elegans61. This suggests that in metazoans multiple pathways upregulate the proteostasis network even when intracellular signals are missing, thus opening new avenues to modulate proteostasis using hormonal or neurotransmitter-based signaling in organisms62–64.
Although we have a large catalogue of proteostasis members and pathways that regulate it, the knowledge is by no means exhaustive. New members of the proteostasis network are being identified not only in complex metazoans but also in simple and well-studied model organisms like Escherichia coli and S. cerevisiae65,66. There are many unknowns in the way that proteostasis network members are regulated. For example, we have only started to understand whether different types of misfolded proteins in the same compartment would signal similar or different pathways and how the amplitude and the spectrum of response would depend on the number of misfolded molecules and their type67. The regulation of these members is being studied, and quantitative models have been developed to simulate the proteostasis network of different organisms68,69. The ever-increasing knowledge in this field contributes immensely to our understanding of the role of proteostasis in health and diseases.
Aging and proteostasis
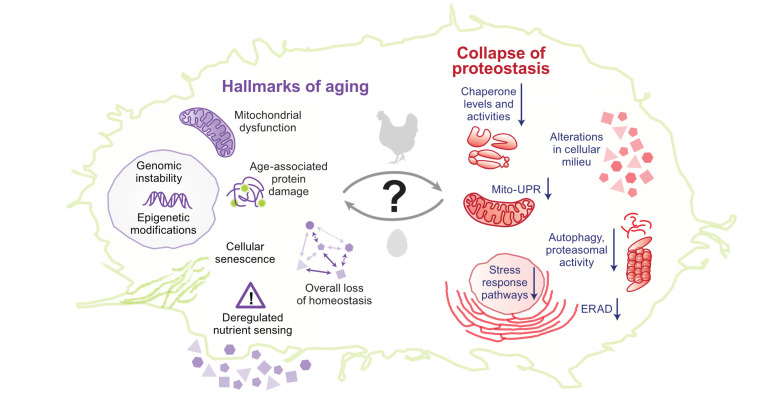
Multiple pieces of evidence suggest that aging leads to a progressive decline in proteostasis70–72. Unstable transgenic proteins are found to form punctate structures in aged C. elegans, and a large fraction of the endogenous proteome aggregates in an age-dependent manner73–75. In an interesting short-lived vertebrate model, Nothobranchius furzeri, protein aggregates comprising primarily ribosomal subunits increased with age, indicating a loss of ribosome stoichiometry, aggregation, and proteostasis imbalance76. Another comprehensive proteomics study reported that mice showed only minimal change in proteome with age77; however, the authors focused only on the soluble proteome and its alteration with age. Hence, it is possible that the aggregated proteome changes with age. Direct correlation of protein aggregation with age was shown by an aggregated protein fraction from the brain of young mice having enhanced ability to seed Aβ aggregation in aged mice, underlining their compromised proteostasis78. Supporting this, in humans, amyloid-type aggregates show an age-dependent increase in plasma79. Environmental and genetic conditions that lead to delayed aging also delay age-associated protein aggregation in C. elegans74. However, the connection between aging and proteostasis is not simple; rather, aging in metazoans is a complex process involving multiple cell types, and each cell type may have a different rate of molecular aging determining their dependence on proteostasis80. Although aging leads to a decline in proteostasis, a decline in proteostasis may also contribute to aging: expression of aggregation-prone proteins in worms and mouse decreases their life span and accelerates aging-associated phenotypes24,74. Other conditions that perturb proteostasis, like the mismatch between mitochondrial and nuclear genotype, can also accelerate aging81. Evidence from the “anti-aging” program in stem cells also suggests that aging is partially governed by the maintenance of proteostasis82–84. Proteostasis decline makes up for the primary hallmarks of aging along with genomic instability, epigenetic modifications, and telomere attrition. Secondary hallmarks or antagonistic hallmarks—deregulated nutrient sensing, cellular senescence, and mitochondrial dysfunction—arise after that, leading to tertiary hallmarks which are seen as phenotypes when homeostasis is not restored85. Since these hallmarks can occur simultaneously and continuously during aging, the connection between aging and proteostasis is two-way: proteostasis decline accelerates aging while aging accelerates the decline of proteostasis (Figure 3).
Figure 3. Association between hallmarks of aging and collapse of proteostasis.
Despite decades-long research, it is still difficult to pinpoint to a definite cause-and-effect relationship between hallmarks of aging and collapse of proteostasis. On the left, a few important cellular hallmarks of aging are shown. On the right, age-associated global changes to proteostasis are shown. ERAD, endoplasmic reticulum–associated degradation; Mito-UPR, mitochondrial unfolded protein response.
The reasons behind the age-dependent decline of proteostasis are not clear since many factors can dictate proteostasis collapse: (1) accumulation of age-dependent damage to proteins, (2) deregulation of protein synthesis causing aberrant accumulation of proteins involved in functional complex formation86, (3) the inability of aged cells to respond to altered proteostasis demands87,88, and (4) alterations in cellular milieu because of age-dependent changes in metabolism and hence impairment of the metabolite-dependent folding arm of proteostasis17,58,89. Any of these factors either alone or in combination could lead to the age-dependent collapse of proteostasis. The major focus has been on the age-associated accumulation of damaged proteins and the age-dependent loss of responsivity of stress-response pathways that maintain proteostasis (Figure 3).
Protein damage
Proteins accumulate damage during aging90. Modifications such as carbonylation, oxidation, glycation, and deamidation can change protein structure, function, and aggregation propensity of the folded proteins, thereby causing global protein aggregation91,92. Pathways nullifying this damage can delay aging, indicating that this damage may have a significant contribution in increasing the load on the proteostasis network during aging93. Interestingly, pathways that alleviate this damage are thought to play an important role in life-span extension when organisms are treated with sub-lethal stresses early in life (a phenomenon termed hormesis)51. Although damage to proteins may play a crucial role in determining the load on the proteostasis network, error in mRNA editing, mRNA splicing, and translation fidelity can also increase the load on the proteostasis network by producing faulty proteins that are unable to fold94–96.
Protein misfolding may escalate during aging if damaged proteins are not cleared efficiently. Supporting this, increasing protein degradation by enhancing autophagy97 or proteasomal pathway98 enhances life span99. Although chemically inducing autophagy by rapamycin increases life span100, it also represses protein translation, a well-known modifier of aging101. The specific role of autophagy was substantiated by upregulating the adaptor protein, p62, which increases autophagy and hence life span in both C. elegans102 and Drosophila melanogaster103. The link between life span and the capacity to clear the damaged proteins is further validated by the observations that long-lived mammals tend to have enhanced protein degradation pathways104. However, it is important to understand whether there is a threshold above which the increase of the degradation pathways, particularly autophagy, causes deleterious effects105. Given the benefit of upregulating these pathways and the threshold, these studies are really helpful for designing therapeutics wherever needed.
Age-dependent loss of proteostasis network–restoring pathways
Some of the stress-response pathways that regulate proteostasis exhibit an abrupt programmed decrease with aging, specifically during the reproductive phase of worms, partially explaining the age-dependent loss of proteostasis88,106,107. Experiments with mammals have shown conflicting results including decrease or no change in heat shock response (HSR) with aging108,109. Even while this is being investigated, the consensus is that proteostasis-restoring pathways decline with age. For example, protein turnover through the autophagic flux marginally decreases with age110, and in a vertebrate model, killifish, a decrease in expression of proteasome components could predict the life span of the organisms with high confidence76. Similarly, in humans, lysosomal function and particularly autophagic flux and proteostasis are compromised in T cells with aging111. Corroborating the loss of proteostasis with aging, upregulation of proteostasis restorative pathways has shown promising results in aging: an increase in the HSR pathway by upregulating the transcription factor itself increases life span in yeast (chronological aging)112 and C. elegans113. An increase in Mito-UPR in a tissue-specific manner also increases life span114. Activating the ERAD arm through IRE1 mitigates the age-dependent collapse of proteostasis51,115. This suggests that there is a loss of proteostasis restoration pathways with aging.
If an age-dependent decline in stress-response pathways decreases protein clearance and increases the concentration of damaged proteins, we expect to see a decrease in protein turnover during aging. Autophagic flux in C. elegans seems to follow this trend; it decreases with aging and its attenuation works as a mode of rescue in the long-lived mutants116. However, protein degradation seems to show a confusing trend: whereas some of the proteins show muted degradation, many others show an increase in degradation, arguing against a general loss of degradation capacity with age117. Decreased activity and dysfunction of the proteasome are associated with many late-onset disorders. Furthermore, enhancement in proteasome activity not only extends life span but also increases stress resistance. Thus, a decline in proteasome functionality is an important determinant of aging98. The chaperones may be unable to recognize and engage specifically with defective proteins in aged organisms because of post-translational modifications118–121. In summary, forced upregulation of quality control branches increases life span. Nevertheless, it remains to be seen whether aging is indeed associated with the muted clearance of defective proteins.
Thus, the role of proteostasis in governing aging and associated phenotypes is still an active field. Given the complex connection between stress-response pathways and the breadth of the response, it is hard to pinpoint a single reason for the age-associated decline in proteostasis and vice versa; most likely, it does not depend on only a single pathway. Because aging depends on multiple pathways, obtaining their quantitative contribution would require quantitative genetic tools that can measure the contribution of each pathway and their epistasis, as has been developed in yeast122. This needs to be supplemented with biochemical experiments to allow a holistic understanding of organismal proteostasis and its regulation with aging.
Role of proteostasis in diseases
Role of proteostasis in monogenetic diseases
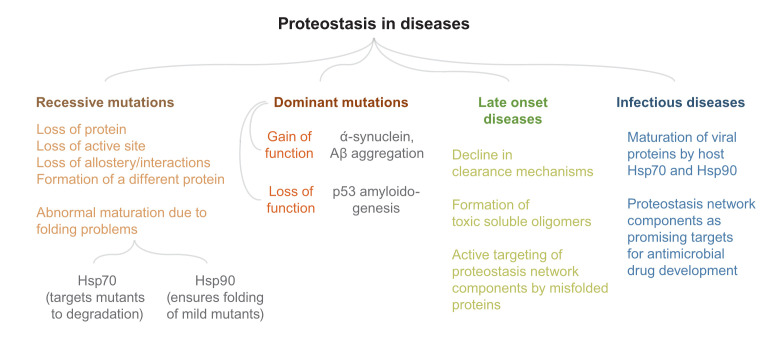
The role of proteostasis is well documented in neurodegenerative diseases123, but little is known about its role in monogenetic recessive diseases. Monogenetic recessive diseases linked to protein malfunction are caused by mutations that lead to loss of function of a protein. This can happen if the mutation (1) causes loss of the protein, (2) inactivates the protein by changing its active site, (3) disturbs the normal maturation of the protein, or (4) causes production of a different protein124. The majority of the recessive diseases that have been mapped to protein-coding regions do not occur in proteins’ active sites. Although these alterations may still affect protein function by altering allosteric sites or affecting macromolecular interactions125, many of them have the potential to affect protein folding and maturation126,127. Although its role in the first two cases is not as important, proteostasis may play a major role in the last two cases. HSR activation can cause a maladaptive response primarily for loss-of-function mutations and cause the degradation of mutant proteins128. On the same line, some of the disease-causing mutations in Fanconi anemia resulted in mutant proteins that had a stronger association with Hsp70 chaperone machinery than the wild-type (WT) proteins, causing degradation of mutant variants129. The work also predicts that mutations that predispose a patient to severe forms of recessive diseases may generally associate more with the Hsp70 chaperone system whereas mild mutations generally tend to associate strongly with the Hsp90 chaperone system129. This corroborates nicely with a recent finding which shows that Hsp70 prevents aggregation at the cost of preventing folding of mutant proteins but Hsp90 may relieve this brake and ensure proper maturation130. Similarly, cystic fibrosis causing mutations in the channel protein CFTR (cystic fibrosis transmembrane conductance regulator) shows the alleviation of phenotype when the mutant protein is dissociated from its cognate chaperone Aha1131,132. However, opposing trends where the upregulation of Hsp70 machinery helped protein maturation have also been observed. Arimoclomol, a proteostasis regulator that amplifies HSR, can decrease the lysosomal dysfunction in cells derived from Gaucher disease–affected patients133. Treatment with the drug, as expected, upregulated HSR but interestingly also enhanced the levels of Bip (an ER-resident Hsp70) and helped in the maturation of the mutant GCases (beta-glucosidases) that represented the different mutations found in the patients133. So it is difficult to predict the outcome of altering proteostasis on the function of mutant proteins: whereas some mutants may fold more efficiently and regain activity upon induction of proteostasis network members, other mutants may be rendered inactive by the degradation machinery. Nonetheless, proteostasis contributes significantly to the phenotypes of recessive diseases. Large-scale testing with deep mutational scanning of proteins has opened up avenues to understand the role of proteostasis in specific mutations17,20,134 and may help in designing tools to efficiently predict the effect of proteostasis modulators on mutants of proteins that cause loss-of-function diseases.
Monogenetic dominant diseases can result from a mutation that either leads to dominant loss of function or forms toxic species. Among the dominant loss-of-function mutations, the most prevalent in cancer is on p53. p53 is stabilized upon DNA damage and ceases cell proliferation135. Cells harboring mutations in p53 proliferate and accumulate mutations in the genome. Since p53 forms a tetramer in vivo, the mutant proteins are thought to act dominantly by forming oligomers with WT copies of p53. Although many of the p53 mutations have problems binding to DNA muting its transcription factor activity, recent evidence shows that mutant p53 aggregates into amyloid-like fibrils, spreads to neighboring cells, and depletes WT p53 using a prion-like conformational switch mechanism136. Thus, if proteostasis alteration modulates protein quality control and aggregation, it is likely to play an important role in the process of p53 amyloidogenesis.
The latter class of monogenetic dominant diseases, that result from a toxic gain of function, includes Huntington’s disease and different classes of spinocerebellar ataxia. These share features of amyloid-like protein aggregation with complex neurological disorders like Alzheimer’s disease (AD) and Parkinson’s disease123. For the purpose of this commentary, we are treating both of them as one group comprising diseases showing evidence of amyloid-like protein aggregation. Nevertheless, there is no common pathophysiological background for these diseases; also, different HSPs have been associated with them137. For example, each of these diseases has a distinct barcode of HSPs which can rescue the aggregation. Furthermore, these age-related diseases are presented with extracellular or intracellular amyloid aggregates75. The aggregated proteins, except for polyglutamine (polyQ)-associated diseases, are divergent in sequence. Generally, repetitive sequences—for example, repeating sequences of amino acids like glycine-alanine dipeptide in amyotrophic lateral sclerosis138,139—are more prone to aggregation. Aggregation may also proceed in the absence of any repeat sequences, as in the case of α-synuclein and Aβ. Even for the canonical polyglutamine-containing protein Huntingtin (Htt), recent evidence suggests that toxicity can arise from the regions flanking the polyQ tract140 or from repeat-associated non-ATG (RAN) peptides generated by alternate translation frames of CAG repeats141. However, the transgenic mice model of Huntington’s disease exhibited toxicity only due to the aggregation of polyQ peptides and not due to RAN translated repeats142. Thus, identifying the toxic species in aggregation-associated diseases is still an active field. The target organs with different mutant proteins are different. For example, mutant Htt with polyQ extension in exon 1 aggregates in the cortex and striatum region of the brain while islet amyloid polypeptide (IAPP), an aggregate associated with type II diabetes, is found in the pancreas. For many of these diseases, the mutant protein is ubiquitously expressed in different regions of the body but the puzzling feature is the specificity of the site of aggregation. Although the late-onset nature of aggregation seems to be explained by the failing proteostasis, there is no consensus on the spatial specificity of aggregation72.
Many amyloid aggregates that are naturally found are not toxic143. Then why do some mutant protein aggregates show pathology associated with them? One possibility is that the aggregated proteins are not toxic but the soluble misfolded proteins, in either monomeric or oligomeric form, are toxic for the cells144. Misfolded proteins may sequester chaperones and make them unavailable for other essential substrates27,145. Mutant proteins like Htt form toxic soluble oligomers and seed further aggregation146–148. Thus, soluble oligomers extending to form large intracellular protein aggregates seem to be a recurrent feature of many late-onset aggregation-prone diseases. Although soluble aggregates play a major role in toxicity, both insoluble and soluble aggregates are toxic for the cells ex vivo in many of the canonical diseases75. Aggregates can also cause toxicity by permeabilizing membranes149 or sequestering other macromolecules150. Recent evidence also suggests that a large-scale metabolism change due to toxic misfolding in cells results from mitochondrial dysfunction151. Thus, toxicity seems to be multipronged and may depend partially on the aggregating protein, the spatial location of aggregates, and environmental stimuli.
Why do these toxic species accumulate? This may happen if these species are either overexpressed or hoarded by blocking the clearance pathway. Many of these aggregates are known to inhibit proteasomal activity, thereby inhibiting their clearance. For example, soluble aggregates of different pathological aggregation-prone proteins have similar structural features that enable them to target proteasome and block their activity with high affinity152. Some mutant proteins, like toxic oligomers of mutant Htt that bypass chaperone-mediated autophagy, may avoid clearance153. PolyQ expanded proteins may also block autophagy by decreasing Beclin-1 that is essential for autophagy. Beclin-1 (a key regulator of autophagy) interacts with the polyQ tracts of Ataxin-3 that prevents its degradation154. Expression of other proteins with polyQ tracts compete this interaction in a polyQ length-dependent manner, thereby causing Beclin-1 to be degraded by the proteasome. This leads to a decline in autophagic flux in cells harboring polyQ tracts in mice as well as human brain cells154. Proteotoxicity is resolved when autophagy block is relieved artificially, suggesting that the clearance arms of proteostasis are key to prevent the accumulation of these toxic proteins. As a corollary, conditions that compromise clearance mechanisms should precipitate the disease. Indeed, viral infections that block autophagy have been shown to precipitate α-synuclein aggregation155. However, the interdependence between autophagy and proteasomal degradation and between different branches of the autophagy pathways warrants more careful studies in unraveling the role of specific branches in the clearance of misfolded proteins, diffusely aggregated protein, insoluble protein aggregates, or organelles burdened terminally with proteotoxicity156–158.
Protein misfolding is known to induce an appropriate response by increasing the clearance capacity of the cells. Then why does it fail in these diseases? Failing response with age, as discussed above, could be the clue making these diseases late-onset diseases. However, these oligomers themselves have been found to perturb the proteostasis network and blunt cellular response, leading to a faster collapse of proteostasis with aging. Aggregates of α-synuclein found in patients with Parkinson’s disease can facilitate degradation of HSF1, thereby diminishing the sensory mechanisms that can mount a defensive response159,160. Aggregates of mutant Htt interact with the TIM23 translocase of mitochondria, thus perturbing mitochondrial proteostasis and possibly preventing its degradation through the MAGIC pathway45. Perturbed mitochondrial homeostasis may be a common crucial mediator in these diseases as even in sporadic cases of AD (without mutations in the known modulators), mito-UPR genes were found to be upregulated, attesting to the general role of mitochondrial proteostasis in AD161. These studies provide us with valuable knowledge of the underlying problems in these diseases, which have set forth new avenues that might lead to therapy.
Role of proteostasis in infectious diseases
Many intracellular pathogens use host cellular machinery for the efficient maturation of their proteins. This is particularly true for viruses that use the host chaperones to create their proteostasis network that aids viral replication. For example, ER-resident chaperones are important for the efficient maturation of viral surface proteins, and cytosolic chaperones are important for the folding of many of the non-structural proteins162,163. Large polyproteins made by viruses need to be processed and folded correctly. Mounting evidence shows that the maturation of large proteins is dependent upon the host chaperones like Hsp90162,164. The autophagy pathway is also crucial for generating replication sites for different viruses165. Interestingly, some of the viral protein aggregates are known to increase autophagy in the infected cells whereas others block the process of autophagy. Given the importance of the proteostasis machinery of the host in viral replication, the ER proteostasis inhibitor Castanospermine has shown pan-antiviral activity against many enveloped viruses166, and Hsp70 inhibitor has been shown to have strong anti-viral activity against the Zika virus163. Drug resistance in many viruses emerges fast as they can accumulate mutations rapidly. Many of the coding missense mutations can likely compromise protein folding, causing their evolution dependent upon the host proteostasis machinery. It has been shown that the mutations accumulated on the Influenza virus are regulated by the host proteostasis134 and inhibiting proteostasis prevents the emergence of drug-resistant Zika viruses163. Given the different networks present in different compartments, compartment-specific host proteostasis can show different effects on viral evolution167. Interestingly, a mutation (Pro283 nucleoprotein variant) fixed in the human Influenza virus strain, which provided immune tolerance, was found to be inactivated upon inhibition of HSF1 at febrile temperatures168. This proved that the proteostasis network may play an important role in buffering mildly deleterious mutations that have some functional advantage. However, HSF1, as mentioned earlier, controls many pathways, including the chaperone network. Thus, it remains to be seen whether this mutant takes the help of chaperones to fold in vivo. Intracellular bacteria communicate with the host cells through their secretory proteome. Deubiquitninases (DUBs) are among the prominent classes of enzymes secreted by bacteria that rewires the host proteostasis network to deubiquitinate proteins preventing the growth of intracellular pathogens169.
For extracellular pathogens like bacteria, proteostasis network members are major targets for the development of anti-microbial drugs. CLPP activators that constitutively activate CLPP protease and cause proteostasis imbalance are interesting candidates for antibiotics170. Bacteria can tolerate anti-microbials and evolve anti-microbial resistance (AMR). AMR can be affected by bacterial dormancy, which in turn results in persistence. In an intriguing study, Fan Bai’s group showed that protein aggregates determine bacterial dormancy and stress tolerance linking the role of proteostasis to stress-based cellular memory and possibly AMR171. Given that proteostasis members, chaperones (protein and chemical), can buffer mutations, it is tempting to speculate that alteration in the concentration of these members assists in the evolution of AMR. Intriguingly, metabolic alterations associated with osmotic shock have been shown to alter mutational buffering17. Thus, changes in metabolism either due to change in the niche of a microbe or due to the metabolic rewiring can potentiate genetic variations that may culminate in AMR. A summary of the above-mentioned diseases is shown in Figure 4.
Figure 4. Proteostasis in diseases.
A summary of proteostasis-related diseases is presented. Many of the proteostasis-related diseases we know of have a genetic component. There has been considerable progress in deciphering the molecular mechanisms of many of these diseases. Still, a better understanding of eukaryotic stress-response pathways will take us a long way in the direction of treatment and cure.
Outlook
Long-term quests in the field have been to understand the role of proteostasis in regulating different physiological or pathological processes and to identify proteostasis-modulating strategies to alleviate problems associated with these processes. This would require the painstaking effort to catalogue the dependence of the different pathways on proteostasis. A specific problem that requires attention is the way that proteostasis is perturbed to investigate the dependence. Genetic deletion of chaperones or other members of the proteostasis network is often used to investigate the dependence, but their deletion most often upregulates other pathways of proteostasis that do not optimally complement the loss of the deleted pathway but rather increase the clearance processes172. The results obtained upon chaperone deletions in S. cerevisiae are hence convoluted; they could be an outcome of the loss of the pathway or a dominant effect of the upregulated pathways. So it is important to devise strategies to effectively downregulate functions of chaperones without complete deletion, a strategy that has already produced interesting results173.
To identify novel proteostasis modulators, we need to have comprehensive knowledge of pathways regulating proteostasis. There is a large gap of knowledge in our understanding of the pathways that regulate proteostasis in higher eukaryotic systems. For example, although we know that the deletion of some of these pathways (like certain branches of ER-UPR) can cause specific effects in certain tissues, we do not understand the reason that these pathways show tissue specificity although these are expressed ubiquitously in different tissues174,175. We do not know whether there is a threshold load of misfolded proteins that activate the different branches differentially. It is also possible that each of these pathways has a tissue-specific difference in their threshold of activation, a phenomenon that can change the way we target the pathways in different diseases. In fact, we understand little about how the response to proteostasis perturbations is guided in a cell type–specific manner.
However, the cumulative knowledge generated in this field has allowed the development of multiple therapeutic candidates that can be used to ameliorate human suffering176–179. As we unravel the role of proteostasis in different biological settings and close the gaps in knowledge, we hope that the excitement of discovery will be surpassed only by the usefulness of the discoveries in making our planet far more livable.
The peer reviewers who approve this article are:
Paula Ludovico, Life and Health Sciences Research Institute (ICVS), School of Health Sciences, University of Minho, Braga, Portugal; ICVS/3B's-PT Government Associate Laboratory, Braga/Guimarães, Portugal
Daniel N Hebert, Department of Biochemistry and Molecular Biology, University of Massachusetts, Amherst, United States; Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, United States
R Martin Vabulas, Charité - Universitätsmedizin Berlin, Institute of Biochemistry, Berlin, Germany
Funding Statement
The KC lab is supported by a Swarna Jayanthi Fellowship Grant from DST (DST/SJF/LSA-01/2015-16) and a National Bio-Science Award for career development from DBT. Infrastructure is provided by CSIR and CSIR-IGIB.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Douglas PM, Dillin A: Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010; 190(5): 719–29. 10.1083/jcb.201005144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balchin D, Hayer-Hartl M, Hartl FU: In vivo aspects of protein folding and quality control. Science. 2016; 353(6294): aac4354. 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 3.Clare DK, Saibil HR: ATP-driven molecular chaperone machines. Biopolymers. 2013; 99(11): 846–59. 10.1002/bip.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer MP, Gierasch LM: Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J Biol Chem. 2019; 294(6): 2085–97. 10.1074/jbc.REV118.002810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogk A, Ruger-Herreros C, Bukau B: Cellular Functions and Mechanisms of Action of Small Heat Shock Proteins. Annu Rev Microbiol. 2019; 73: 89–110. 10.1146/annurev-micro-020518-115515 [DOI] [PubMed] [Google Scholar]
- 6.Albanèse V, Reissmann S, Frydman J: A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol. 2010; 189(1): 69–81. 10.1083/jcb.201001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serlidaki D, van Waarde MAWH, Rohland L, et al. : Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J Biol Chem. 2020; 295(21): 7301–16. 10.1074/jbc.RA119.012449 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 8.Nillegoda NB, Kirstein J, Szlachcic A, et al. : Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015; 524(7564): 247–51. 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faust O, Abayev-Avraham M, Wentink AS, et al. : HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature. 2020; 587(7834): 489–94. 10.1038/s41586-020-2906-4 [DOI] [PubMed] [Google Scholar]
- 10.Hartl FU, Hayer-Hartl M: Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002; 295(5561): 1852–8. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 11.Ellgaard L, McCaul N, Chatsisvili A, et al. : Co- and Post-Translational Protein Folding in the ER. Traffic. 2016; 17(6): 615–38. 10.1111/tra.12392 [DOI] [PubMed] [Google Scholar]
- 12.Adams BM, Oster ME, Hebert DN: Protein Quality Control in the Endoplasmic Reticulum. Protein J. 2019; 38(3): 317–29. 10.1007/s10930-019-09831-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart RA, Cyr DM, Craig EA, et al. : Mitochondrial molecular chaperones: Their role in protein translocation. Trends Biochem Sci. 1994; 19(2): 87–92. 10.1016/0968-0004(94)90041-8 [DOI] [PubMed] [Google Scholar]
- 14.Bandyopadhyay A, Saxena K, Kasturia N, et al. : Chemical chaperones assist intracellular folding to buffer mutational variations. Nat Chem Biol. 2012; 8(3): 238–45. 10.1038/nchembio.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel A, Malinovska L, Saha S, et al. : ATP as a biological hydrotrope. Science. 2017; 356(6339): 753–6. 10.1126/science.aaf6846 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 16.Xie L, Jakob U: Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J Biol Chem. 2019; 294(6): 2180–90. 10.1074/jbc.REV118.002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma K, Saxena K, Donaka R, et al. : Distinct metabolic states of a cell guide alternate fates of mutational buffering through altered proteostasis. Nat Commun. 2020; 11(1): 2926. 10.1038/s41467-020-16804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu CY, Kokubo H, Lynch GC, et al. : Backbone additivity in the transfer model of protein solvation. Protein Sci. 2010; 19(5): 1011–22. 10.1002/pro.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao YT, Manson AC, DeLyser MR, et al. : Trimethylamine N-oxide stabilizes proteins via a distinct mechanism compared with betaine and glycine. Proc Natl Acad Sci U S A. 2017; 114(10): 2479–84. 10.1073/pnas.1614609114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dandage R, Pandey R, Jayaraj G, et al. : Differential strengths of molecular determinants guide environment specific mutational fates. PLoS Genet. 2018; 14(5): e1007419. 10.1371/journal.pgen.1007419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Xu C, Larrimore KE, et al. : Slp1-Emp65: A Guardian Factor that Protects Folding Polypeptides from Promiscuous Degradation. Cell. 2017; 171(2): 346–357.e12. 10.1016/j.cell.2017.08.036 [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Le WD: Autophagy and Ubiquitin-Proteasome System. Adv Exp Med Biol. 2019; 1206: 527–50. 10.1007/978-981-15-0602-4_25 [DOI] [PubMed] [Google Scholar]
- 23.Tekirdag K, Cuervo AM: Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J Biol Chem. 2018; 293(15): 5414–24. 10.1074/jbc.R117.818237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hipp MS, Kasturi P, Hartl FU: The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019; 20(7): 421–35. 10.1038/s41580-019-0101-y [DOI] [PubMed] [Google Scholar]
- 25.Arrasate M, Mitra S, Schweitzer ES, et al. : Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004; 431(7010): 805–10. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 26.Coleman BM, Hill AF: Extracellular vesicles--Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol. 2015; 40: 89–96. 10.1016/j.semcdb.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 27.Hill SM, Hanzén S, Nyström T, et al. : Restricted access: Spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 2017; 18(3): 377–91. 10.15252/embr.201643458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandman O, Hegde RS: Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016; 23(1): 7–15. 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guydosh NR, Kimmig P, Walter P, et al. : Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. eLife. 2017; 6: e29216. 10.7554/eLife.29216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mediani L, Guillén-Boixet J, Vinet J, et al. : Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J. 2019; 38(15): e101341. 10.15252/embj.2018101341 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 31.Costa EA, Subramanian K, Nunnari J, et al. : Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science. 2018; 359(6376): 689–92. 10.1126/science.aar3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamerdinger M, Hanebuth MA, Frickey T, et al. : The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science. 2015; 348(6231): 201–7. 10.1126/science.aaa5335 [DOI] [PubMed] [Google Scholar]
- 33.Chernova TA, Chernoff YO, Wilkinson KD: Yeast Models for Amyloids and Prions: Environmental Modulation and Drug Discovery. Molecules. 2019; 24(18): 3388. 10.3390/molecules24183388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masser AE, Kang W, Roy J, et al. : Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. eLife. 2019; 8: e47791. 10.7554/eLife.47791 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 35.Alvarez-Castelao B, tom Dieck S, Fusco CM, et al. : The switch-like expression of heme-regulated kinase 1 mediates neuronal proteostasis following proteasome inhibition. Elife. 2020; 9: e52714. 10.7554/eLife.52714 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 36.Walter P, Ron D: The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011; 334(6059): 1081–6. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 37.Jaud M, Philippe C, Di Bella D, et al. : Translational Regulations in Response to Endoplasmic Reticulum Stress in Cancers. Cells. 2020; 9(3): 540. 10.3390/cells9030540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan BJ, van Hoef V, Jobava R, et al. : A Unique ISR Program Determines Cellular Responses to Chronic Stress. Mol Cell. 2017; 68(5): 885–900.e6. 10.1016/j.molcel.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cagnetta R, Wong HH, Frese CK, et al. : Noncanonical Modulation of the eIF2 Pathway Controls an Increase in Local Translation during Neural Wiring. Mol Cell. 2019; 73(3): 474–489.e5. 10.1016/j.molcel.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 40.Bakunts A, Orsi A, Vitale M, et al. : Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. eLife. 2017; 6: e27518. 10.7554/eLife.27518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nargund AM, Pellegrino MW, Fiorese CJ, et al. : Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012; 337(6094): 587–90. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 42.Fiorese CJ, Schulz AM, Lin YF, et al. : The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol. 2016; 26(15): 2037–43. 10.1016/j.cub.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 43.Poveda-Huertes D, Matic S, Marada A, et al. : An Early mtUPR: Redistribution of the Nuclear Transcription Factor Rox1 to Mitochondria Protects against Intramitochondrial Proteotoxic Aggregates. Mol Cell. 2020; 77(1): 180–188.e9. 10.1016/j.molcel.2019.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 44.Boos F, Krämer L, Groh C, et al. : Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat Cell Biol. 2019; 21(4): 442–51. 10.1038/s41556-019-0294-5 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 45.Ruan L, Zhou C, Jin E, et al. : Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017; 543(7645): 443–6. 10.1038/nature21695 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 46.Frottin F, Schueder F, Tiwary S, et al. : The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019; 365(6451): 342–7. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 47.Narayana Rao KB, Pandey P, Sarkar R, et al. : Localized Proteotoxic Stress in Mitochondrial Intermembrane Space and Matrix Elicits Sub-compartment Specific Response Pathways Governed by Unique Modulators. bioRxiv. 2020. 10.1101/2020.08.16.252734 [DOI] [Google Scholar]
- 48.Eisenberg-Lerner A, Benyair R, Hizkiahou N, et al. : Golgi organization is regulated by proteasomal degradation. Nat Commun. 2020; 11(1): 409. 10.1038/s41467-019-14038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 49.Kopp Y, Lang WH, Schuster TB, et al. : CHIP as a membrane-shuttling proteostasis sensor. eLife. 2017; 6: e29388. 10.7554/eLife.29388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duennwald ML, Lindquist S: Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008; 22(23): 3308–19. 10.1101/gad.1673408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matai L, Sarkar GC, Chamoli M, et al. : Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc Natl Acad Sci U S A. 2019; 116(35): 17383–92. 10.1073/pnas.1900055116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabnis AJ, Guerriero CJ, Olivas V, et al. : Combined chemical-genetic approach identifies cytosolic HSP70 dependence in rhabdomyosarcoma. Proc Natl Acad Sci U S A. 2016; 113(32): 9015–20. 10.1073/pnas.1603883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumoto S, Nakatsukasa K, Kakuta C, et al. : Msp1 Clears Mistargeted Proteins by Facilitating Their Transfer from Mitochondria to the ER. Mol Cell. 2019; 76(1): 191–205.e10. 10.1016/j.molcel.2019.07.006 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 54.Wrobel L, Topf U, Bragoszewski P, et al. : Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015; 524(7566): 485–8. 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Chen XJ: A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015; 524(7566): 481–4. 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picca A, Calvani R, Coelho-Junior HJ, et al. : Inter-Organelle Membrane Contact Sites and Mitochondrial Quality Control during Aging: A Geroscience View. Cells. 2020; 9(3): 598. 10.3390/cells9030598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morimoto RI: Cell-Nonautonomous Regulation of Proteostasis in Aging and Disease. Cold Spring Harb Perspect Biol. 2020; 12(4): a034074. 10.1101/cshperspect.a034074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labbadia J, Brielmann RM, Neto MF, et al. : Mitochondrial Stress Restores the Heat Shock Response and Prevents Proteostasis Collapse during Aging. Cell Rep. 2017; 21(6): 1481–94. 10.1016/j.celrep.2017.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 59.O'Brien D, van Oosten-Hawle P: Regulation of cell-non-autonomous proteostasis in metazoans. Essays Biochem. 2016; 60(2): 133–42. 10.1042/EBC20160006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frakes AE, Metcalf MG, Tronnes SU, et al. : Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science. 2020; 367(6476): 436–40. 10.1126/science.aaz6896 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 61.Cohen-Berkman M, Dudkevich R, Ben-Hamo S, et al. : Endogenous siRNAs promote proteostasis and longevity in germline-less Caenorhabditis elegans. Elife. 2020; 9: e50896. 10.7554/eLife.50896 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 62.Labbadia J, Morimoto RI: The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015; 84: 435–64. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finger F, Ottens F, Springhorn A, et al. : Olfaction regulates organismal proteostasis and longevity via microRNA-dependent signaling. Nat Metab. 2019; 1(3): 350–9. 10.1038/s42255-019-0033-z [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 64.Taylor RC, Berendzen KM, Dillin A: Systemic stress signalling: Understanding the cell non-autonomous control of proteostasis. Nat Rev Mol Cell Biol. 2014; 15(3): 211–7. 10.1038/nrm3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krzemień-Ojak Ł, Góral A, Joachimiak E, et al. : Interaction of a Novel Chaperone PhLP2A With the Heat Shock Protein Hsp90. J Cell Biochem. 2017; 118(2): 420–9. 10.1002/jcb.25669 [DOI] [PubMed] [Google Scholar]
- 66.Rössler I, Embacher J, Pillet B, et al. : Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res. 2019; 47(13): 6984–7002. 10.1093/nar/gkz317 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 67.Sui X, Pires DEV, Ormsby AR, et al. : Widespread remodeling of proteome solubility in response to different protein homeostasis stresses. Proc Natl Acad Sci U S A. 2020; 117(5): 2422–31. 10.1073/pnas.1912897117 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 68.Powers ET, Powers DL, Gierasch LM: FoldEco: A model for proteostasis in E. coli. Cell Rep. 2012; 1(3): 265–76. 10.1016/j.celrep.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agozzino L, Balázsi G, Wang J, et al. : How Do Cells Adapt? Stories Told in Landscapes. Annu Rev Chem Biomol Eng. 2020; 11: 155–82. 10.1146/annurev-chembioeng-011720-103410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klaips CL, Jayaraj GG, Hartl FU: Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018; 217(1): 51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santra M, Dill KA, de Graff AMR: Proteostasis collapse is a driver of cell aging and death. Proc Natl Acad Sci U S A. 2019; 116(44): 22173–8. 10.1073/pnas.1906592116 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 72.Taylor RC, Dillin A: Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011; 3(5): a004440. 10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Della David C, Ollikainen N, Trinidad JC, et al. : Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010; 8(8): e1000450. 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 74.Walther DM, Kasturi P, Zheng M, et al. : Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell. 2015; 161(4): 919–32. 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang C, Wagner-Valladolid S, Stephens AD, et al. : Intrinsically aggregation-prone proteins form amyloid-like aggregates and contribute to tissue aging in Caenorhabditis elegans. Elife. 2019; 8: e43059. 10.7554/eLife.43059 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 76.Kelmer Sacramento E, Kirkpatrick JM, Mazzetto M, et al. : Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol Syst Biol. 2020; 16(6): e9596. 10.15252/msb.20209596 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 77.Walther DM, Mann M: Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol Cell Proteomics. 2011; 10(2): M110.004523. 10.1074/mcp.M110.004523 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 78.Groh N, Bühler A, Huang C, et al. : Age-Dependent Protein Aggregation Initiates Amyloid-β Aggregation. Front Aging Neurosci. 2017; 9: 138. 10.3389/fnagi.2017.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 79.Xia K, Trasatti H, Wymer JP, et al. : Increased levels of hyper-stable protein aggregates in plasma of older adults. Age (Dordr). 2016; 38(3): 56. 10.1007/s11357-016-9919-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tain LS, Sehlke R, Jain C, et al. : A proteomic atlas of insulin signalling reveals tissue-specific mechanisms of longevity assurance. Mol Syst Biol. 2017; 13(9): 939. 10.15252/msb.20177663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, et al. : Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016; 535(7613): 561–5. 10.1038/nature18618 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 82.Noormohammadi A, Calculli G, Gutierrez-Garcia R, et al. : Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells. Cell Mol Life Sci. 2018; 75(2): 275–90. 10.1007/s00018-017-2602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vilchez D, Simic MS, Dillin A: Proteostasis and aging of stem cells. Trends Cell Biol. 2014; 24(3): 161–70. 10.1016/j.tcb.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 84.Vonk WIM, Rainbolt TK, Dolan PT, et al. : Differentiation Drives Widespread Rewiring of the Neural Stem Cell Chaperone Network. Mol Cell. 2020; 78(2): 329–345.e9. 10.1016/j.molcel.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 85.López-Otín C, Blasco MA, Partridge L, et al. : The hallmarks of aging. Cell. 2013; 153(6): 1194–217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 86.Anisimova AS, Meerson MB, Gerashchenko MV, et al. : Multifaceted deregulation of gene expression and protein synthesis with age. Proc Natl Acad Sci U S A. 2020; 117(27): 15581–90. 10.1073/pnas.2001788117 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 87.Liang V, Ullrich M, Lam H, et al. : Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell Mol Life Sci. 2014; 71(17): 3339–61. 10.1007/s00018-014-1558-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Labbadia J, Morimoto RI: Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol Cell. 2015; 59: 639–50. 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zečić A, Braeckman BP: DAF-16/FoxO in Caenorhabditis elegans and Its Role in Metabolic Remodeling. Cells. 2020; 9(1): 109. 10.3390/cells9010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baldensperger T, Eggen M, Kappen J, et al. : Comprehensive analysis of posttranslational protein modifications in aging of subcellular compartments. Sci Rep. 2020; 10(1): 7596. 10.1038/s41598-020-64265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Semba RD, Nicklett EJ, Ferrucci L: Does accumulation of advanced glycation end products contribute to the aging phenotype? J Gerontol A Biol Sci Med Sci. 2010; 65(9): 963–75. 10.1093/gerona/glq074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Höhn A, König J, Grune T: Protein oxidation in aging and the removal of oxidized proteins. J Proteomics. 2013; 92: 132–59. 10.1016/j.jprot.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 93.Golegaonkar S, Tabrez SS, Pandit A, et al. : Rifampicin reduces advanced glycation end products and activates DAF-16 to increase lifespan in Caenorhabditis elegans. Aging Cell. 2015; 14: 463–73. 10.1111/acel.12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kapur M, Ackerman SL: mRNA Translation Gone Awry: Translation Fidelity and Neurological Disease. Trends Genet. 2018; 34(3): 218–31. 10.1016/j.tig.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 95.Francisco S, Ferreira M, Moura G, et al. : Does proteostasis get lost in translation? Implications for protein aggregation across the lifespan. Ageing Res Rev. 2020; 62: 101119. 10.1016/j.arr.2020.101119 [DOI] [PubMed] [Google Scholar]
- 96.Vermulst M, Denney AS, Lang MJ, et al. : Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun. 2015; 6: 8065. 10.1038/ncomms9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rubinsztein DC, Mariño G, Kroemer G: Autophagy and aging. Cell. 2011; 146(5): 682–95. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- 98.Saez I, Vilchez D: The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases. Curr Genomics. 2014; 15(1): 38–51. 10.2174/138920291501140306113344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vilchez D, Saez I, Dillin A: The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 2014; 5: 5659. 10.1038/ncomms6659 [DOI] [PubMed] [Google Scholar]
- 100.Papadopoli D, Boulay K, Kazak L, et al. : mTOR as a central regulator of lifespan and aging [version 1; peer review: 3 approved]. F1000Res. 2019; 8: F1000 Faculty Rev-998. 10.12688/f1000research.17196.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nandagopal N, Roux PP: Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation (Austin). 2015; 3(1): e983402. 10.4161/21690731.2014.983402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kumsta C, Chang JT, Lee R, et al. : The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy. Nat Commun. 2019; 10(1): 5648. 10.1038/s41467-019-13540-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 103.Aparicio R, Rana A, Walker DW: Upregulation of the Autophagy Adaptor p62/SQSTM1 Prolongs Health and Lifespan in Middle-Aged Drosophila. Cell Rep. 2019; 28(4): 1029–1040.e5. 10.1016/j.celrep.2019.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodriguez KA, Valentine JM, Kramer DA, et al. : Determinants of rodent longevity in the chaperone-protein degradation network. Cell Stress Chaperones. 2016; 21(3): 453–66. 10.1007/s12192-016-0672-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thorburn A: Autophagy and its effects: Making sense of double-edged swords. PLoS Biol. 2014; 12(10): e1001967. 10.1371/journal.pbio.1001967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ben-Zvi A, Miller EA, Morimoto RI: Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009; 106(35): 14914–9. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 107.Taylor RC, Dillin A: XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013; 153(7): 1435–47. 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 108.Jurivich DA, Manocha GD, Trivedi R, et al. : Multifactorial Attenuation of the Murine Heat Shock Response With Age. J Gerontol A Biol Sci Med Sci. 2020; 75(10): 1846–52. 10.1093/gerona/glz204 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 109.Carnemolla A, Labbadia JP, Lazell H, et al. : Contesting the dogma of an age-related heat shock response impairment: Implications for cardiac-specific age-related disorders. Hum Mol Genet. 2014; 23(14): 3641–56. 10.1093/hmg/ddu073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Andrews B, Maslen S, Almeida-Souza L, et al. : Multidimensional dynamics of the proteome in the neurodegenerative and ageing mammalian brain. bioRxiv. 2018. 10.1101/472118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jin J, Li X, Hu B, et al. : FOXO1 deficiency impairs proteostasis in aged T cells. Sci Adv. 2020; 6(17): eaba1808. 10.1126/sciadv.aba1808 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 112.Ohtsuka H, Azuma K, Murakami H, et al. : hsf1+ extends chronological lifespan through Ecl1 family genes in fission yeast. Mol Genet Genomics. 2011; 285(1): 67–77. 10.1007/s00438-010-0588-6 [DOI] [PubMed] [Google Scholar]
- 113.Hsu AL, Murphy CT, Kenyon C: Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003; 300(5622): 1142–5. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 114.Jensen MB, Jasper H: Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014; 20(2): 214–25. 10.1016/j.cmet.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Narayan V, Ly T, Pourkarimi E, et al. : Deep Proteome Analysis Identifies Age-Related Processes in C. elegans. Cell Syst. 2016; 3(2): 144–59. 10.1016/j.cels.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 116.Chang JT, Kumsta C, Hellman AB, et al. : Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife. 2017; 6: e18459. 10.7554/eLife.18459 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 117.Dhondt I, Petyuk VA, Bauer S, et al. : Changes of Protein Turnover in Aging Caenorhabditis elegans. Mol Cell Proteomics. 2017; 16(9): 1621–33. 10.1074/mcp.RA117.000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Truttmann MC, Pincus D, Ploegh HL: Chaperone AMPylation modulates aggregation and toxicity of neurodegenerative disease-associated polypeptides. Proc Natl Acad Sci U S A. 2018; 115(22): E5008–E5017. 10.1073/pnas.1801989115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sieber SA, Cappello S, Kielkowski P: From Young to Old: AMPylation Hits the Brain. Cell Chem Biol. 2020; 27(7): 773–9. 10.1016/j.chembiol.2020.05.009 [DOI] [PubMed] [Google Scholar]
- 120.Nitika, Porter CM, Truman AW, et al. : Post-translational modifications of Hsp70 family proteins: Expanding the chaperone code. J Biol Chem. 2020; 295(31): 10689–708. 10.1074/jbc.REV120.011666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cho-Park PF, Steller H: Proteasome regulation by ADP-ribosylation. Cell. 2013; 153(3): 614–27. 10.1016/j.cell.2013.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 122.Roguev A, Wiren M, Weissman JS, et al. : High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007; 4(10): 861–6. 10.1038/nmeth1098 [DOI] [PubMed] [Google Scholar]
- 123.Chiti F, Dobson CM: Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev Biochem. 2017; 86: 27–68. 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 124.Eriksson M, Brown WT, Gordon LB, et al. : Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003; 423(6937): 293–8. 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 125.Sahni N, Yi S, Taipale M, et al. : Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015; 161(3): 647–60. 10.1016/j.cell.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Redler RL, Das J, Diaz JR, et al. : Protein Destabilization as a Common Factor in Diverse Inherited Disorders. J Mol Evol. 2016; 82(1): 11–6. 10.1007/s00239-015-9717-5 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 127.Taipale M: Disruption of protein function by pathogenic mutations: Common and uncommon mechanisms 1. Biochem Cell Biol. 2019; 97(1): 46–57. 10.1139/bcb-2018-0007 [DOI] [PubMed] [Google Scholar]
- 128.Roth DM, Hutt DM, Tong J, et al. : Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 2014; 12(11): e1001998. 10.1371/journal.pbio.1001998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karras GI, Yi S, Sahni N, et al. : HSP90 Shapes the Consequences of Human Genetic Variation. Cell. 2017; 168(5): 856–866.e12. 10.1016/j.cell.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 130.Morán Luengo T, Kityk R, Mayer MP, et al. : Hsp90 Breaks the Deadlock of the Hsp70 Chaperone System. Mol Cell. 2018; 70(3): 545–552.e9. 10.1016/j.molcel.2018.03.028 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 131.Wang X, Venable J, LaPointe P, et al. : Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006; 127(4): 803–15. 10.1016/j.cell.2006.09.043 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 132.Powers ET, Morimoto RI, Dillin A, et al. : Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009; 78: 959–91. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- 133.Fog CK, Zago P, Malini E, et al. : The heat shock protein amplifier arimoclomol improves refolding, maturation and lysosomal activity of glucocerebrosidase. EBioMedicine. 2018; 38: 142–53. 10.1016/j.ebiom.2018.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Phillips AM, Gonzalez LO, Nekongo EE, et al. : Host proteostasis modulates influenza evolution. eLife. 2017; 6: e28652. 10.7554/eLife.28652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Williams AB, Schumacher B: p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med. 2016; 6(5): a026070. 10.1101/cshperspect.a026070 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 136.Ghosh S, Salot S, Sengupta S, et al. : p53 amyloid formation leading to its loss of function: Implications in cancer pathogenesis. Cell Death Differ. 2017; 24(10): 1784–98. 10.1038/cdd.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kakkar V, Meister-Broekema M, Minoia M, et al. : Barcoding heat shock proteins to human diseases: Looking beyond the heat shock response. Dis Model Mech. 2014; 7(4): 421–34. 10.1242/dmm.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang YJ, Jeng US, Chiang YL, et al. : The Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-Cell Transmission Properties. J Biol Chem. 2016; 291(10): 4903–11. 10.1074/jbc.M115.694273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Freibaum BD, Taylor JP: The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD. Front Mol Neurosci. 2017; 10: 35. 10.3389/fnmol.2017.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shen K, Calamini B, Fauerbach JA, et al. : Control of the structural landscape and neuronal proteotoxicity of mutant Huntingtin by domains flanking the polyQ tract. eLife. 2016; 5: e18065. 10.7554/eLife.18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bañez-Coronel M, Ayhan F, Tarabochia AD, et al. : RAN Translation in Huntington Disease. Neuron. 2015; 88(4): 667–77. 10.1016/j.neuron.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang S, Yang H, Huang L, et al. : Lack of RAN-mediated toxicity in Huntington's disease knock-in mice. Proc Natl Acad Sci U S A. 2020; 117(8): 4411–7. 10.1073/pnas.1919197117 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 143.Jackson MP, Hewitt EW: Why are Functional Amyloids Non-Toxic in Humans? Biomolecules. 2017; 7(4): 71. 10.3390/biom7040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wallace EW, Kear-Scott JL, Pilipenko EV, et al. : Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 2015; 162(6): 1286–98. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park SH, Kukushkin Y, Gupta R, et al. : PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013; 154(1): 134–45. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 146.Ast A, Buntru A, Schindler F, et al. : mHTT Seeding Activity: A Marker of Disease Progression and Neurotoxicity in Models of Huntington's Disease. Mol Cell. 2018; 71(5): 675–688.e6. 10.1016/j.molcel.2018.07.032 [DOI] [PubMed] [Google Scholar]
- 147.Kayed R, Head E, Thompson JL, et al. : Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003; 300(5618): 486–9. 10.1126/science.1079469 [DOI] [PubMed] [Google Scholar]
- 148.Ghag G, Bhatt N, Cantu DV, et al. : Soluble tau aggregates, not large fibrils, are the toxic species that display seeding and cross-seeding behavior. Protein Sci. 2018; 27(11): 1901–9. 10.1002/pro.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ambadi Thody S, Mathew MK, Udgaonkar JB: Mechanism of aggregation and membrane interactions of mammalian prion protein. Biochim Biophys Acta Biomembr. 2018; 1860(9): 1927–35. 10.1016/j.bbamem.2018.02.031 [DOI] [PubMed] [Google Scholar]
- 150.Falsone A, Falsone SF: Legal but lethal: Functional protein aggregation at the verge of toxicity. Front Cell Neurosci. 2015; 9: 45. 10.3389/fncel.2015.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Münch C: The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018; 16(1): 81. 10.1186/s12915-018-0548-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thibaudeau TA, Anderson RT, Smith DM: A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun. 2018; 9(1): 1097. 10.1038/s41467-018-03509-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fu Y, Wu P, Pan Y, et al. : A toxic mutant huntingtin species is resistant to selective autophagy. Nat Chem Biol. 2017; 13(11): 1152–4. 10.1038/nchembio.2461 [DOI] [PubMed] [Google Scholar]
- 154.Ashkenazi A, Bento CF, Ricketts T, et al. : Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature. 2017; 545(7652): 108–11. 10.1038/nature22078 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 155.Marreiros R, Müller-Schiffmann A, Trossbach SV, et al. : Disruption of cellular proteostasis by H1N1 influenza A virus causes α-synuclein aggregation. Proc Natl Acad Sci U S A. 2020; 117(12): 6741–51. 10.1073/pnas.1906466117 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 156.Morel E, Mehrpour M, Botti J, et al. : Autophagy: A Druggable Process. Annu Rev Pharmacol Toxicol. 2017; 57: 375–98. 10.1146/annurev-pharmtox-010716-104936 [DOI] [PubMed] [Google Scholar]
- 157.Yu L: A special review collection on autophagy. Cell Res. 2020; 30(7): 553. 10.1038/s41422-020-0361-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khandia R, Dadar M, Munjal A, et al. : A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells. 2019; 8(7): 674. 10.3390/cells8070674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim E, Wang B, Sastry N, et al. : NEDD4-mediated HSF1 degradation underlies α-synucleinopathy. Hum Mol Genet. 2016; 25(2): 211–22. 10.1093/hmg/ddv445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kilpatrick K, Novoa JA, Hancock T, et al. : Chemical induction of Hsp70 reduces α-synuclein aggregation in neuroglioma cells. ACS Chem Biol. 2013; 8(7): 1460–8. 10.1021/cb400017h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Beck JS, Mufson EJ, Counts SE: Evidence for Mitochondrial UPR Gene Activation in Familial and Sporadic Alzheimer's Disease. Curr Alzheimer Res. 2016; 13(6): 610–4. 10.2174/1567205013666151221145445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Aviner R, Frydman J: Proteostasis in Viral Infection: Unfolding the Complex Virus-Chaperone Interplay. Cold Spring Harb Perspect Biol. 2020; 12(3): a034090. 10.1101/cshperspect.a034090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Taguwa S, Yeh MT, Rainbolt TK, et al. : Zika Virus Dependence on Host Hsp70 Provides a Protective Strategy against Infection and Disease. Cell Rep. 2019; 26(4): 906–920.e3. 10.1016/j.celrep.2018.12.095 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 164.Sakata M, Katoh H, Otsuki N, et al. : Heat Shock Protein 90 Ensures the Integrity of Rubella Virus p150 Protein and Supports Viral Replication. J Virol. 2019; 93(22): e01142–19. 10.1128/JVI.01142-19 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 165.Wileman T: Aggresomes and autophagy generate sites for virus replication. Science. 2006; 312(5775): 875–8. 10.1126/science.1126766 [DOI] [PubMed] [Google Scholar]
- 166.Heaton NS, Moshkina N, Fenouil R, et al. : Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity. 2016; 44(1): 46–58. 10.1016/j.immuni.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 167.Phillips AM, Doud MB, Gonzalez LO, et al. : Enhanced ER proteostasis and temperature differentially impact the mutational tolerance of influenza hemagglutinin. Elife. 2018; 7: e38795. 10.7554/eLife.38795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Phillips AM, Ponomarenko AI, Chen K, et al. : Destabilized adaptive influenza variants critical for innate immune system escape are potentiated by host chaperones. PLoS Biol. 2018; 16(9): e3000008. 10.1371/journal.pbio.3000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hermanns T, Hofmann K: Bacterial DUBs: Deubiquitination beyond the seven classes. Biochem Soc Trans. 2019; 47(6): 1857–66. 10.1042/BST20190526 [DOI] [PubMed] [Google Scholar]
- 170.Griffith EC, Zhao Y, Singh AP, et al. : Ureadepsipeptides as ClpP Activators. ACS Infect Dis. 2019; 5(11): 1915–25. 10.1021/acsinfecdis.9b00245 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 171.Pu Y, Li Y, Jin X, et al. : ATP-Dependent Dynamic Protein Aggregation Regulates Bacterial Dormancy Depth Critical for Antibiotic Tolerance. Mol Cell. 2019; 73(1): 143–156.e4. 10.1016/j.molcel.2018.10.022 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 172.Ghosh A, Gangadharan A, Verma M, et al. : Cellular responses to proteostasis perturbations reveal non-optimal feedback in chaperone networks. Cell Mol Life Sci. 2019; 76(8): 1605–21. 10.1007/s00018-019-03013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Patury S, Miyata Y, Gestwicki JE: Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem. 2009; 9(15): 1337–51. 10.2174/156802609789895674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reimold AM, Iwakoshi NN, Manis J, et al. : Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001; 412(6844): 300–7. 10.1038/35085509 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 175.Clauss IM, Gravallese EM, Darling JM, et al. : In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev Dyn. 1993; 197(2): 146–56. 10.1002/aja.1001970207 [DOI] [PubMed] [Google Scholar]
- 176.Rabouw HH, Langereis MA, Anand AA, et al. : Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc Natl Acad Sci U S A. 2019; 116(6): 2097–102. 10.1073/pnas.1815767116 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 177.Kelly JW: Pharmacologic Approaches for Adapting Proteostasis in the Secretory Pathway to Ameliorate Protein Conformational Diseases. Cold Spring Harb Perspect Biol. 2020; 12(5): a034108. 10.1101/cshperspect.a034108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stiegler SC, Rübbelke M, Korotkov VS, et al. : A chemical compound inhibiting the Aha1-Hsp90 chaperone complex. J Biol Chem. 2017; 292(41): 17073–83. 10.1074/jbc.M117.797829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Grandjean JMD, Madhavan A, Cech L, et al. : Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat Chem Biol. 2020; 16(10): 1052–61. 10.1038/s41589-020-0584-z [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation