Abstract
Bacterial species can adapt to significant changes in their environment by mutation followed by selection, a phenomenon known as “adaptive evolution.” With the development of bioinformatics and genetic engineering, research on adaptive evolution has progressed rapidly, as have applications of the process. In this review, we summarize various mechanisms of bacterial adaptive evolution, the technologies used for studying it, and successful applications of the method in research and industry. We particularly highlight the contributions of Dr. L. O. Ingram. Microbial adaptive evolution has significant impact on our society not only from its industrial applications, but also in the evolution, emergence, and control of various pathogens.
Keywords: Bacteria, Adaptive evolution mechanisms, Adaptive metabolic evolution
Introduction
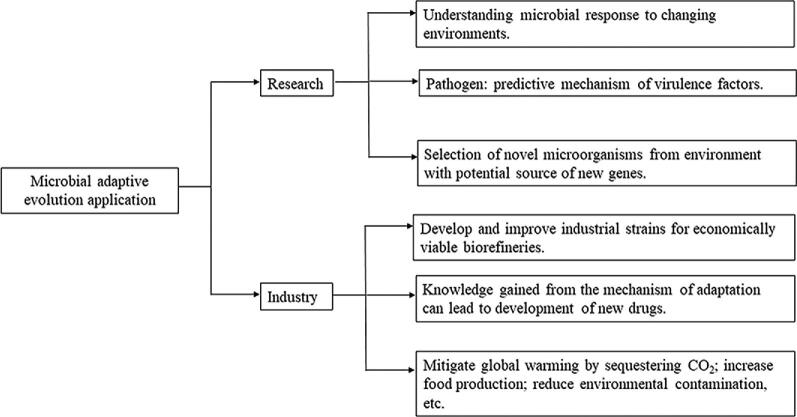
Adaptive evolution by natural selection, as proposed by Darwin and elaborated by Huxley (Darwin, 1882; Huxley, 1942), is readily observed in microorganisms, which can adapt to environmental changes, such as exposure to heavy metals, air pollution, insecticides, and herbicides (Horinouchi et al., 2015). In part, this is due to the large numbers of individual organisms on which selection can operate and the strong selective pressure to which these organisms are subjected. For example, Staphylococcus aureus adapts to its host by outgrowth of individuals with increased antibiotic resistance and decreased virulence (Goerke & Wolz, 2010; Kahl, 2010), while Pseudomonas aeruginosa does so by loss of factors essential for acute infection and activation of pathways related to chronic inflammation (Smith et al., 2006). The mutualistic Rhizobium–legume symbiosis has evolved for about 58 million years (Sprent, 2007) through acquisition of genes involved in nodulation (nod) and nitrogen fixation (nif/fix) (Remigi et al., 2016). Adaptive resistance to disparate toxins in Escherichia coli involves mutations of a set of genes associated with cell motility, transport, membrane structure, and metabolism, as well as genes that are not well characterized (Erickson et al., 2017). Adaptation has been reported for other species of bacteria, such as Salmonella enterica, Bacillus subtilis, and Clostridium sp., and for eukaryotic microorganisms such as Saccharomyces cerevisiae and Candida albicans (Foster, 2000). These cases show that microorganisms adapt to environmental changes through both temporary and permanent modifications. Temporary adaptation is realized by modulating the expression of genes related to phenotypic alterations (López-Maury et al., 2008), while adaptive evolution involves selection of mutations advantageous for survival in a particular new environment and represents permanent alteration in response to the changed environment. Generally, bacteria undergo adaptive evolution when the selection pressure is strong and persistent, while the temporary adaptation responds to short-term selection (Horinouchi & Furusawa, 2020). In this review, we summarize some of the reported mechanisms of bacterial adaptive evolution (Table 1), as well as the methods for bacterial adaptive evolution study and application of bacterial adaptive evolution in research and industrial development (Fig. 1).
Table 1.
The Process and Mechanism of Adaptive Evolution
| Process | Mechanisms | Reference |
|---|---|---|
| Horizontal gene transfer | Gene transfer between different species or different organisms. | Schönknecht et al. (2014) |
| Plasmids and phages as vectors for host evolution | Evolution-related genes transferred by plasmids or phages. | Davies et al. (2016), Robicsek et al. (2006) |
| Coevolution of bacteria in the same community | Bacterial adaptive mutations benefit the whole community. | Meyer & Kassen (2007) |
| Gene duplication and amplification (GDA) | Bacteria adapted to various environments by increasing the copy number of specific genes—can be flanked by insertion sequences (ISs). | Bergthorsson et al. (2007) |
| ISs | Mobilize specific genes within or among different microorganisms yielding desired characteristics. | Siguier et al. (2014) |
| Specific gene mutations promote adaptive evolution | Single or multiple mutations in the genome leads to the desired selectable phenotype. | Chauliac et al. (2020); Conrad et al. (2010); Kim et al. (2008) |
| Bacterial parallel evolution | The same mutation arises in all independent selections for the desired phenotype. | Bailey et al. (2017); Shi et al. (2020) |
Fig. 1.
Microbial adaptive evolution application.
The Process and Mechanisms of Adaptive Evolution in Bacteria
Several processes, such as genetic drift and gene conversion and mutation (Duret & Galtier, 2009), can alter genetic information of microorganisms and these processes can influence their phenotypes.
Horizontal Gene Transfer
Genetic information can be transferred either vertically or horizontally. Vertical gene transfer passes genetic information from parent to offspring, while horizontal gene transfer occurs between different organisms and often between different species (Schönknecht et al., 2014). Horizontal gene transfer has been reported to be extensive, with at least 5–10% of a typical bacterial genome deriving from divergent species (Ochman et al., 2000). Bacteria can express genes obtained from other species to allow adaptation to different environmental conditions, for instance, imparting novel metabolic capacities (Levin & Cornejo, 2009). For example, changes of metabolic networks in E. coli are mostly derived from apparent horizontal gene transfer in the past 100 million years. Metabolic networks grow by acquiring genes that can utilize the nutrients in the altered environment but without interrupting the central metabolic network. In addition, genes involved in one compound metabolic pathway are frequently transferred in an operon (Pal et al., 2005). Horizontal gene transfer was also reported in pathogens, such as Streptococcus pneumoniae and Haemophilus influenzae (Levin & Cornejo, 2009). In the laboratory, horizontal gene transfer can be realized by conjugation, transformation, and transduction, as a prelude to adaptive metabolic evolution.
Demonstrating that horizontal gene transfer can promote fitness, Power et al. co-cultured two B. subtilis strains that had 7% DNA sequence difference over multiple rounds of growth and stationary phase (Power et al., 2021). They observed the rapid emergence of hybrid organisms with gene transfer swaps of approximately 12% of the core genome in just 200 generations and 60% of core genes replaced in at least one population. Moreover, many of the individual hybrids exhibited increased fitness, particularly in survival at stationary phase (Power et al., 2021). Thus, horizontal gene transfer can promote adaptive evolution.
Plasmids and Phages as Vectors for Host Evolution
Plasmids and phages also serve as vectors for horizontal gene transfer. Toxins and antibiotic resistance genes in bacteria are often found on plasmids or temperate phages. As one of many examples, fluoroquinolone resistance of gram-negative pathogens is conferred by the Qnr proteins (quinolone resistance proteins) that protect DNA gyrase from quinolone inhibition. The qnr genes achieved global distribution through a variety of plasmid environments and bacterial genera (Robicsek et al., 2006).
DNA sequences from phages are more common in pathogens than nonpathogenic bacteria (Busby et al., 2013) and are linked to increased virulence (Figueroa-Bossi & Bossi, 1999). This is due in part to genes carried on the phages but also to their potential to mutate the host genome by insertion (Letarov & Kulikov, 2009). For example, P. aeruginosa evolved in the laboratory with or without the transposable phage ɸ4 showed that the phage ɸ4 could integrate into the bacterial genome randomly, especially into the motility-related genes and quorum-sensing-system regulators that were essential for virulence (Davies et al., 2016).
Coevolution of Bacteria in the Same Community
Bacteria that share the same environment can collectively engage in adaptive evolution in a way that mutually benefits the community as a whole. For the study of community composition effects on evolution, Lawrence et al. (Lawrence et al., 2012) prepared bacterial communities composed of either one species or a five-species mixture and cultured them on a beech tea medium. Their results showed that the complexity of the community influenced bacterial adaptive evolution and this impact could not be predicted with results obtained with a single species alone. Thus, complex communities can drive adaptive evolution through multiple pathways that are not available to single species (Meyer & Kassen, 2007).
Gene Duplication and Amplification
Gene duplications are pervasive (Bergthorsson et al., 2007) and provide opportunities for increasing expression of a gene and for introducing genomic variability, which in turn facilitates adaptation to various environments. For example, antibiotics resistance can be realized by increasing the gene copies encoding antibiotic hydrolytic enzymes, efflux pumps, or target enzymes (Sandegren & Andersson, 2009). The pathogen S. enterica subsp. enterica serovar Typhimurium LT2 exhibited tandem genetic duplications in all regions of the chromosome with sizes from a few kilobases to several megabases in response to adaptation to various environmental challenges (Andersson et al., 1998; Nilsson et al., 2006; Straus & Straus, 1976). Spirochetes of the Borrelia burgdorferi sensu lato (s.l.) species can cause Lyme disease (Centers for Disease Control and Prevention, 2007; Piesman & Gern, 2004). Their success as pathogens depends, in part, on the PFam54-related genes in the B. burgdorferi genome, which exist as a tandem array and whose diverse products can target different innate immune proteins (Wywial et al., 2009). Finally, it should be noted that the high ethanol productivity of an ethanologenic E. coli strain KO11 developed by Dr. Ingram and his co-workers is a result of a 20-fold duplication of a single PET operon (pdc and adhB genes of Zymomonas mobilis) (Ohta et al., 1991; Turner et al., 2012). These studies show that gene duplication is not uncommon during adaptive evolution and differs from more stable mutation-derived adaptation.
Insertion Sequences
Insertion sequences (ISs) are small pieces of DNA that transpose within or between genomes and can influence host genome evolution. IS elements are widespread and can occur in high copy numbers in bacterial genomes. Different IS elements have different functions and their effects on the host genome depend on their particular transposition mechanism (Siguier et al., 2014).
As an example of IS contribution in adaptive evolution, insertion of IS10 in yqhC that encodes a regulatory protein in the ethanologenic E. coli strain EMFR9 led to higher resistance to the inhibitor furfural (Miller et al., 2009; Turner et al., 2011). Another example is that transposon insertion in a Sinorhizobium strain that was incompatible with soybean led to effective derivatives that could generate N2-fixing nodules in soybean roots (Zhao et al., 2018).
Specific Gene Mutations Promote Adaptive Evolution
In some instances, adaptive evolution results from gain of function mutations in specific gene(s) that allow adaptation to a specific environment. A strain of Bacillus coagulans selected for growth under oxygen-limiting conditions carried mutations in a gene encoding a glycerol (polyol) dehydrogenase that established a new fermentation pathway to d-(−)-lactic acid (Chauliac et al., 2020; Wang, Ingram & Shanmugam, 2011). Similarly, a specific mutation in lpd-encoding lipoamide dehydrogenase, a component of the pyruvate dehydrogenase complex, supported anaerobic growth of a negative E. coli mutant during metabolic evolution (Kim et al., 2008). This mutation also introduced a homo-ethanol fermentation pathway in the adapted derivative that is not detectable in the wild-type E. coli. In addition, specific mutations in the RNA polymerase (RNAP) genes exhibited wide effects on bacterial phenotypes (Klein-Marcuschamer et al., 2009). Conrad et al. repeatedly found rpoC mutations (RNAP β′-subunit) during E. coli K-12 adaptive evolution for optimal growth in M9 minimal medium with various carbon sources (glycerol, glucose, and lactate) (Conrad et al., 2010). These studies show that specific but unexpected mutations allow bacteria to overcome a constraining physiological stress.
Bacterial Parallel Evolution
Given that specific mutations can allow adaptation to a specific environmental condition, it is not surprising that the same mutation or mutations may arise repeatedly in bacteria subjected to the same condition. For instance, Dr. Ingram's laboratory adapted two separate cultures of an ethanologenic E. coli strain LY180 for growth in lignocellulosic acid hydrolysate over hundreds of generations. After genome sequence analysis, six common mutations were found in two lineages, strains AQ15 and SL112, indicating parallel evolution (Shi et al., 2020). A second example is provided by Burkholderia dolosa, a pathogen that can cause disease in humans. Lieberman et al. compared the genetic adaptation of B. dolosa in patients during the epidemic spread and found that the bacteria acquired 17 overlapping nonsynonymous mutated genes in multiple subjects, suggesting parallel evolution (Lieberman et al., 2011). Another example of parallel evolution is Bacteroides fragilis, abundant in healthy people (Faith et al., 2013; Schloissnig et al., 2013). To evaluate the evolution within individual microbiomes, Zhao et al sequenced the genomes of 602 isolates of B. fragilis from 12 healthy subjects (Zhao et al., 2019). They found that 16 genes in B. fragilis had mutations within tested individuals, 5 of which were mutated in multiple individuals. These mutated genes were involved in polysaccharide utilization, cell-envelope biosynthesis, and other uncharacterized pathways. In summary, parallel evolution emerges after strong selection and suggests limited avenues for mutational adaptation to a specific environmental condition (Bailey et al., 2017).
Analytical Methods for Bacterial Adaptive Evolution
Adaptive Laboratory Evolution
Adaptive laboratory evolution (ALE) allows rapid evaluation of the adaptive potential of microorganisms. Such an evolution helps us understand fundamental evolutionary principles, including drug resistance in microbial pathogens (Donald & Van Helden, 2009), insect resistance toward pesticides (Renton, 2013), and acquisition of novel metabolic activities (Miller et al., 2009; Shi et al., 2016). For instance, long-term evolution in the laboratory of an ethanologenic E. coli strain LY195 in mineral salts medium with lignocellulosic acid hydrolysate containing growth inhibitors led to the strain MM160. Strain MM160 produced 29 g/l ethanol with a yield of 0.21 g ethanol/g bagasse dry weight during liquefaction plus simultaneous saccharification and cofermentation (L + SScF) (Geddes et al., 2011). Similarly, a strain of E.coli K-12 adapted for growth on l-1,2-propanediol (l-1,2-PDO) as the carbon source in M9 minimal medium was analyzed by Lee and Palsson (Lee & Palsson, 2010). Six mutations accumulated in the adapted strain during the process of evolution, and five of them were within the coding regions of fucO, ilvG1, rrlD, and ylbE1 (with two mutations). IS5 was found between two fuc regulons (fucAO and fucPIKUR operons). Two mutations (fucO and its promoter) involved in the l-1,2-PDO catabolic pathway occurred early during adaptive evolution and supported growth in this new medium, while the other later mutations apparently improved fitness (Lee & Palsson, 2010).
Flux Balance Analysis
Flux balance analysis (FBA) has been developed to analyze metabolic flux change in reconstructed metabolic networks (Varma & Palsson, 1994). More recently, researchers combined FBA with ecological interactions among multiple bacteria to perform metabolic network study of species in a specific environment (Louca & Doebeli, 2015; O'Brien et al., 2013; Zomorrodi & Maranas, 2012). To generate credible predictions between evolutionary and ecosystem-level outcomes, researchers integrated the FBA model with both evolutionary dynamics and cellular constraints. The combined framework is called evolution with flux balance analysis (evoFBA). Some studies found that the metabolic, physiological, and ecological properties of those coexisting species are consistent with evoFBA predictions (Grosskopf et al., 2016).
Different Omics Approaches
With the development of bioinformatics, different omics approaches have been used for understanding and predicting the molecular mechanisms of bacterial temporary and permanent responses to different environments. These omics are used independently to study bacterial adaptation to a specific environment, such as identifying genes related to tolerance to lignocellulosic acid hydrolysates by genome sequencing (Shi et al., 2020), and capturing expression patterns of growth-dependent genes in Salmonella and Pseudomonas by transcriptome analysis (Imdahl et al., 2020).
Often, researchers have combined two or more omics tools to obtain a better understanding of bacterial adaptive evolution. For example, O'Brien et al. analyzed a respiratory pathogen S. pneumoniae, which causes about 1 million deaths per year worldwide, by genome and transcriptome studies to predict the mechanism of antibiotic resistance as well as in vivo infection conditions (O’Brien et al., 2009). Proteomics combined with genomics was used to analyze different lineages of Mycobacterium tuberculosis (Yimer et al., 2020) and in a separate study (Felix et al., 2021) Francisella tularensis, a pathogen causing the acute human respiratory disease tularemia, to decipher the virulence factors and infection mechanisms in these pathogens. Similarly, Agostini et al. applied genomics, proteomics, and metabolomics in concert to identify the adaptive mechanism of E. coli to fluorinated indoles (Agostini et al., 2021). By analyzing E. coli strains before and after adaptation for growth with fluorinated amino acids, they found that, in addition to specific mutations, gene expression regulation networks as well as membrane integrity and protein folding were also changed in the adapted derivatives (Agostini et al., 2021). Inverse metabolic engineering (IME) is also a useful method to understand the mechanism of adaptive evolution (Hong & Nielsen, 2012). With the development of genome editing systems and sequencing technology, IME has been extensively used for modifying industrial bacteria to decrease side products and improve target product titer and yield (Jaffe et al., 2015; Zhu et al., 2014).
Adaptive Evolution Application in Research and Industry
After designing an appropriate selection, growth-based metabolic adaptive evolution is a powerful tool for selecting beneficial mutations. This is usually carried out in a continuous culture or by sequential transfers of the culture under selecting conditions (Wang et al., 2011). Dr. Ingram and his group included adaptive metabolic evolution as a critical step during the development of various industrially relevant microorganisms and some of these are detailed subsequently to highlight the work in this special volume dedicated to Dr. Ingram's contributions to industrial microbiology (Table 2).
Table 2.
Summary of the Work on Adaptive Metabolic Evolution by Dr. Ingram and His Group
| Product | Character | Strain | Titer (g/l) | Yield (g product/g sugar) | Cell density (g/l) | Productivity (g product/[l h]) | References |
|---|---|---|---|---|---|---|---|
| Ethanol | Ethanologenic Escherichia coli | KO3 | 10.4 | 0.13 | 0.03b | 0.4 | Ohta et al. (1991) |
| KO4 | 52.8 | 0.56 | 0.04 b | 1.5 | |||
| Ethanol tolerance | KO11 | 52.7 | 0.38 | 2.80 | Gonzalez et al. (2003), Yomano et al. (1998) | ||
| LY01 | 61 | 0.44 | 4.00 | ||||
| Fermentation in mineral salts medium | KO11LY168 | 26.945.5 | 0.300.51 | Jarboe et al. (2007) | |||
| Tolerance to inhibitors in lignocellulosic acid hydrolysate | LY180Various strainsa | ∼0.35∼42 | 0.063.60 | Geddes et al. (2014); Miller et al. (2009); Shi et al. (2016); Turner et al. (2011); Wang et al. (2012); Zheng et al. (2012) | |||
| Cellobiose metabolization in ethanologenic E. coli | KO11(pLOI1906)KO11(pLOI1910) | 145.4 | 0.010.51 | 0.303.10 | Moniruzzaman et al. (1997) | ||
| Succinate | E. coli C for succinate production under low-O2 condition | KJ012KJ060KJ122 | 0.7186.5680.89 | 0.130.920.96 | 0.302.202.10 | 0.040.90.84 | Jantama, Haupt et al., (2008); Jantama, Zhang et al. (2008); Zhang et al. (2009) |
| Succinate production with xylose | KJ122 | 37.49 | 0.81 | 1.80 | 0.31 | Sawisit et al. (2015) | |
| AS1600a | 84.26 | 0.88 | 2.14 | 0.96 | |||
| l-Alanine | l-Alanine production in mineral salts medium | XZ111XZ132 | 7.57114.04 | 0.060.94 | 2.03 | 4.04 | Zhang et al. (2007) |
| d-Lactate | d-(−)-Lactic acid production in mineral salts medium | W3110TG114 | 19.46117.10 | 0.460.97 | 0.602.31 | 0.212.88 | Grabar et al. (2006); Zhou et al. (2003) |
| Malate | Mineral salts medium fermentation | KJ060XZ658 | 33.41 | 01.04 | 1.902.50 | 0.47 | Zhang et al. (2011) |
Note. Rows in bold are after adaptation of the strain listed in the above row.
Here just shows the strain with highest ethanol production and cell yield among all the adapted strains.
g cell weight/g glucose.
Metabolic Engineering of E. coli to Produce Ethanol
E. coli KO3 is an ethanologenic strain carrying alcohol dehydrogenase II (adhB) and pyruvate decarboxylase (pdc) from Z. mobilis in the chromosome. Adaptive metabolic evolution of strain KO3 under appropriate conditions produced strain KO4, with increased ethanol yield from 0.13 to 0.56 (g ethanol/g glucose consumed) and an increase in volumetric productivity of ethanol from 0.4 (strain KO3) to 1.5 (strain KO4) (g ethanol/[l h]). The higher ethanol yield than the theoretical value of 0.51 is apparently a result of the rich medium containing unaccounted for sugars used in these experiments (Ohta et al., 1991). Specific genetic intervention by introducing casAB genes encoding Enzyme IIcellobiose and phospho-beta-glucosidase from Klebsiella oxytoca P2 followed by adaptive metabolic evolution expanded sugar utilization characteristics of the ethanologenic E. coli to include cellobiose (Moniruzzaman et al., 1997). Further adaptive metabolic evolution of the ethanologenic E. coli strains for growth in medium containing ethanol as high as 50 g/l led to strain LY01, which produced an ethanol titer of over 60 g/l at a theoretical yield of 0.44 (g/g glucose consumed) (Yomano et al., 1998). The mutations in strain LY01 that related to higher ethanol tolerance included a nonfunctional fnr gene and overexpression of gcv genes (involved in glycine metabolism), betIBA, and betT (increased betaine production) (Gonzalez et al., 2003). This group also used adaptive metabolic evolution to derive strains that could grow rapidly in mineral salts medium (to lower the cost of nutrients in an industrial process), in the presence of various inhibitors (for rapid fermentation of all the sugars in lignocellulosic biomass), etc. (Table 2).
Improved Succinate Production in E. coli
Succinic acid has been identified by the US Department of Energy as one of the top 12 building block chemicals that could be produced from renewable feedstock (Werpy et al., 2004). Microbial production of succinic acid from carbohydrates offers the opportunity to be “‘green,” since succinic acid is biosynthesized using CO2 as one of the substrates (Jantama, Haupt, et al., 2008). Dr. Ingram's laboratory engineered E. coli to produce succinic acid in M9 minimal medium using simple batch fermentations without introducing any foreign genes (strain KJ122) (Jantama, Zhang, et al., 2008). Metabolic evolution for high titer of succinate production led to overexpression of phosphoenolpyruvate carboxykinase (pck), GalP permease (galP), and glucokinase (glk), and inactivation of PtsI to support the energy requirements of an O2-limited production platform (Zhang et al., 2009). Further adaptive metabolic evolution of strain KJ122 in mineral salts medium enhanced the rate of growth and xylose fermentation to succinate by selecting for a unique mutation in the galactose permease that supported xylose transport (strain AS1600a) (Sawisit et al., 2015). This adaptive evolution increased succinate productivity from xylose threefold from 0.3 to 0.96 g/(l h), comparable to that of succinate productivity from glucose or a mixture of sugars (Table 2).
l-Alanine Production in E. coli With Metabolic Evolution and Genome Engineering
Metabolic engineering of E. coli to produce l-alanine was another success story from the lab of Dr. Ingram. After appropriate metabolic engineering of the required pathway in E. coli, adaptive metabolic evolution led to twofold increased specific productivity and about 20-fold increased volumetric productivity at a titer of 1.3 M (Zhang et al., 2007). The resulting strain and its derivatives are currently employed as microbial biocatalysts for commercial production of l-alanine (Table 2).
Metabolic Engineering and Metabolic Evolution of E. coli to Produce Lactate
Biopolymer based on polylactic acid (PLA) is a renewable, environmentally friendly polymer that is produced from an appropriate blend of the two isomers of lactic acid. Although l-(+)-lactic acid is produced by several bacteria, microorganisms that produce pure d-(−)-lactic acid as the sole fermentation product in mineral salts medium are uncommon. To address this problem, Dr. Ingram's group engineered E. coli to produce d-lactic acid in mineral salts medium (Zhou et al., 2003). Grabar et al. deleted the methylglyoxal synthase gene to realize d-lactate production with no detectable chiral impurity (Grabar et al., 2006) (Table 2).
In addition to the previous discussion, there are several additional microorganisms developed with the help of adaptive metabolic evolution, such as increasing ethanol tolerance (Sanchez et al., 1992) and realizing xylose metabolism (Diao et al., 2013; Runquist et al., 2009) in S. cerevisiae and improving lactate tolerance in Lactobacillus casei (Overbeck et al., 2017; Serrazanetti et al., 2009; Wu et al., 2012).
Conclusion and Future Perspective
Bacterial adaptive evolution requires designing appropriate selection conditions to identify desired mutations in the population. This process, both man-made and naturally occurring, has broad significance. Some of the evolved strains are beneficial, such as new or improved ability of industrial microorganisms to produce various desired products, while others are harmful, like pathogens’ resistance to antibiotics or evolving the ability to infect new hosts, including humans.
An understanding of the mechanisms leading to adaptive evolution in various environments is important to predict and control evolving diseases as well as to optimize industrial microorganisms. Recent developments in molecular biology and bioinformatics will likely contribute to future work on bacterial adaptive evolution mechanisms. For instance, recently developed single-cell sequencing has been applied to single human cells, such as normal immune cells (Tang et al., 2019) and acute myeloid leukemia tumor cells (Pellegrino et al., 2018). This method could be harnessed to observe the growth, expansion, and evolution of subpopulations within a community of cells in order to address the dynamic evolution behavior and its mechanisms. This technique has also been applied to perform transcriptome analysis of individual Salmonella and Pseudomonas bacteria cells (Imdahl et al., 2020), providing a new approach to explore bacterial evolution as well. Moreover, a large number of bacterial species cannot be engineered, because they cannot be cultured, as is the case of some human pathogens (Oliver, 2010). In the future, we can use single-cell sequencing to peer into the adaptive evolution mechanisms of these nonculturable bacteria.
Detailed analysis of the adapted derivatives has helped establish the physiological/biochemical functions of several genes that were annotated as unknown function, even in well-characterized organisms such as E. coli. In this context, it should be noted that the microbiome of the world has great untapped potential, in both metabolic pathways and secondary metabolites, and only a small fraction of this potential has been realized due to the lack of tools to engineer a vast array of these microorganisms. Genetic and omics tools combined with adaptive evolution are a promising approach to tap into this reservoir of metabolic versatility.
It should be mentioned that the current pandemic created by the SARS-CoV-2 is a result of adaptive evolution of the virus to human hosts (Rochman et al., 2021). Further adaptation of the virus to better suit the environment, in this case humans, is the delta variant of the virus (van Oosterhout et al., 2021). As the mechanisms of adaptive evolution are deciphered, the predictability of such evolution can be improved to better control emerging diseases.
As stated by Theodosius Dobzhansky, “Nothing in biology makes sense except in the light of evolution” (Ayala, 1977).
Acknowledgments
We thank Dr. Keelnatham T. Shanmugam for helping us go over the work done by Dr. Ingram and his lab members.
Contributor Information
Aiqin Shi, Institute for Personalized Medicine, Department of Biochemistry and Molecular Biology, Pennsylvania State University College of Medicine, Hershey, PA 17033, USA.
Feiyu Fan, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin, 300308, P.R. China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, 300308, P.R. China; National Innovation Center for Synthetic Biotechnology, Tianjin, 300308, P.R. China.
James R Broach, Institute for Personalized Medicine, Department of Biochemistry and Molecular Biology, Pennsylvania State University College of Medicine, Hershey, PA 17033, USA.
Funding
The work was financially supported by the National Key R&D Program of China (2019YFA0905300) and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-001).
Data Availability
The data that support the findings of this study are available from the related references.
Conflict of Interest
The authors declare no conflict of interest.
References
- Agostini F., Sinn L., Petras D., Schipp C. J., Kubyshkin V., Berger A. A., Dorrestein P. C., Rappsilber J., Budisa N., Koksch B. (2021). Multiomics analysis provides insight into the laboratory evolution of Escherichia coli toward the metabolic usage of fluorinated indoles. ACS Central Science, 7, 81–92. 10.1021/acscentsci.0c00679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., Slechta E. S., Roth J. R. (1998). Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science, 282, 1133–1135 [DOI] [PubMed] [Google Scholar]
- Ayala F. J. (1977). “Nothing in biology makes sense except in the light of evolution”: Theodosius Dobzhansky: 1900–1975. Journal of Heredity, 68, 3–10. 10.1093/oxfordjournals.jhered.a108767 [DOI] [PubMed] [Google Scholar]
- Bailey S. F., Blanquart F., Bataillon T., Kassen R. (2017). What drives parallel evolution? How population size and mutational variation contribute to repeated evolution. Bioessays, 39, 1–9. 10.1002/bies.201600176 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U., Andersson D. I., Roth J. R. (2007). Ohno's dilemma: Evolution of new genes under continuous selection. Proceedings of the National Academy of Sciences, 104, 17004–17009. 10.1073/pnas.0707158104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby B., Kristensen D. M., Koonin E. V. (2013). Contribution of phage-derived genomic islands to the virulence of facultative bacterial pathogens. Environmental Microbiology, 15, 307–312. 10.1111/j.1462-2920.2012.02886.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . (2007). Lyme disease—United States, 2003–2005. Morbidity and Mortality Weekly Report, 56, 573–576. https://www.ncbi.nlm.nih.gov/pubmed/17568368 [PubMed] [Google Scholar]
- Chauliac D., Wang Q., St John F. J., Jones G., Hurlbert J. C., Ingram L. O., Shanmugam K. T. (2020). Kinetic characterization and structure analysis of an altered polyol dehydrogenase with D-lactate dehydrogenase activity. Protein Science, 29, 2387–2397. 10.1002/pro.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T. M., Frazier M., Joyce A. R., Cho B.-K., Knight E. M., Lewis N. E., Landick R., Palsson B. Ø. (2010). RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proceedings of the National Academy of Sciences, 107, 20500–20505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1882). The origin of species by means of natural selection. John Murray [Google Scholar]
- Davies E. V., James C. E., Williams D., O'Brien S., Fothergill J. L., Haldenby S., Paterson S., Winstanley C., Brockhurst M. A. (2016). Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proceedings of the National Academy of Sciences, 113, 8266–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao L., Liu Y., Qian F., Yang J., Jiang Y., Yang S. (2013). Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnology, 13, 110. 10.1186/1472-6750-13-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald P. R., Van Helden P. D. (2009). The global burden of tuberculosis—Combating drug resistance in difficult times. New England Journal of Medicine, 360, 2393–2395 [DOI] [PubMed] [Google Scholar]
- Duret L., Galtier N. (2009). Biased gene conversion and the evolution of mammalian genomic landscapes. Annual Review of Genomics and Human Genetics, 10, 285–311. 10.1146/annurev-genom-082908-150001 [DOI] [PubMed] [Google Scholar]
- Erickson K. E., Otoupal P. B., Chatterjee A. (2017). Transcriptome-level signatures in gene expression and gene expression variability during bacterial adaptive evolution. Msphere, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J., Guruge J. L., Charbonneau M., Subramanian S., Seedorf H., Goodman A. L., Clemente J. C., Knight R., Heath A. C., Leibel R. L., Rosenbaum M., Gordon J. I. (2013). The long-term stability of the human gut microbiota. Science, 341, 1237439. https://doi: 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix J., Siebert C., Ducassou J. N., Nigou J., Garcia P. S., Fraudeau A., Huard K., Mas C., Brochier-Armanet C., Coute Y., Gutsche I., Renesto P. (2021). Structural and functional analysis of the Francisella lysine decarboxylase as a key actor in oxidative stress resistance. Scientific Reports, 11, 972. 10.1038/s41598-020-79611-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Bossi L. (1999). Inducible prophages contribute to Salmonella virulence in mice. Molecular Microbiology, 33, 167–176. 10.1046/j.1365-2958.1999.01461.x [DOI] [PubMed] [Google Scholar]
- Foster P. L. (2000). Adaptive mutation: Implications for evolution. Bioessays, 22, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes C. C., Mullinnix M. T., Nieves I. U., Peterson J. J., Hoffman R. W., York S. W., Yomano L. P., Miller E. N., Shanmugam K. T., Ingram L. O. (2011). Simplified process for ethanol production from sugarcane bagasse using hydrolysate-resistant Escherichia coli strain MM160. Bioresource Technology, 102, 2702–2711. 10.1016/j.biortech.2010.10.143 [DOI] [PubMed] [Google Scholar]
- Geddes R. D., Wang X., Yomano L. P., Miller E. N., Zheng H., Shanmugam K. T., Ingram L. O. (2014). Polyamine transporters and polyamines increase furfural tolerance during xylose fermentation with ethanologenic Escherichia coli strain LY180. Applied and Environmental Microbiology, 80, 5955–5964. 10.1128/AEM.01913-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Wolz C. (2010). Adaptation of Staphylococcus aureus to the cystic fibrosis lung. International Journal of Medical Microbiology, 300, 520–525 [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Tao H., Purvis J. E., York S. W., Shanmugam K. T., Ingram L. O. (2003). Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: Comparison of KO11 (parent) to LY01 (resistant mutant). Biotechnology Progress, 19, 612–623. 10.1021/bp025658q [DOI] [PubMed] [Google Scholar]
- Grabar T. B., Zhou S., Shanmugam K. T., Yomano L. P., Ingram L. O. (2006). Methylglyoxal bypass identified as source of chiral contamination in L(+) and D(-)-lactate fermentations by recombinant Escherichia coli. Biotechnology Letters, 28, 1527–1535. 10.1007/s10529-006-9122-7 [DOI] [PubMed] [Google Scholar]
- Grosskopf T., Consuegra J., Gaffe J., Willison J. C., Lenski R. E., Soyer O. S., Schneider D. (2016). Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evolutionary Biology, 16, 163. 10.1186/s12862-016-0733-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K.-K., Nielsen J. (2012). Recovery of phenotypes obtained by adaptive evolution through inverse metabolic engineering. Applied and Environmental Microbiology, 78, 7579–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T., Furusawa C. (2020). Understanding metabolic adaptation by using bacterial laboratory evolution and trans-omics analysis. Biophysical Reviews, 12, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T., Suzuki S., Hirasawa T., Ono N., Yomo T., Shimizu H., Furusawa C. (2015). Phenotypic convergence in bacterial adaptive evolution to ethanol stress. BMC Evolutionary Biology, 15, 180. 10.1186/s12862-015-0454-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley J. (1942). Evolution: The modern synthesis. London, UK: Allen & Unwin. [Google Scholar]
- Imdahl F., Vafadarnejad E., Homberger C., Saliba A. E., Vogel J. (2020). Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria. Nature Microbiology, 5, 1202–1206. 10.1038/s41564-020-0774-1 [DOI] [PubMed] [Google Scholar]
- Jaffe S. R., Strutton B., Pandhal J., Wright P. C. (2015). Inverse metabolic engineering for enhanced glycoprotein production in Escherichia coli. Methods in Molecular Biology, 1321, 17–35. 10.1007/978-1-4939-2760-9_2 [DOI] [PubMed] [Google Scholar]
- Jantama K., Haupt M. J., Svoronos S. A., Zhang X., Moore J. C., Shanmugam K. T., Ingram L. O. (2008). Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnology and Bioengineering, 99, 1140–1153. 10.1002/bit.21694 [DOI] [PubMed] [Google Scholar]
- Jantama K., Zhang X., Moore J. C., Shanmugam K. T., Svoronos S. A., Ingram L. O. (2008). Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnology and Bioengineering, 101, 881–893. 10.1002/bit.22005 [DOI] [PubMed] [Google Scholar]
- Jarboe L. R., Grabar T. B., Yomano L. P., Shanmugan K. T., Ingram L. O. (2007). Development of ethanologenic bacteria. Advances in Biochemical Engineering/Biotechnology, 108, 237–261. 10.1007/10_2007_068 [DOI] [PubMed] [Google Scholar]
- Kahl B. C. (2010). Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. International Journal of Medical Microbiology, 300, 514–519 [DOI] [PubMed] [Google Scholar]
- Kim Y., Ingram L. O., Shanmugam K. T. (2008). Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. Journal of Bacteriology, 190, 3851–3858. 10.1128/JB.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Marcuschamer D., Santos C. N. S., Yu H., Stephanopoulos G. (2009). Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Applied and Environmental Microbiology, 75, 2705–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D., Fiegna F., Behrends V., Bundy J. G., Phillimore A. B., Bell T., Barraclough T. G. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biology, 10, e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-H., Palsson B. Ø. (2010). Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, L-1, 2-propanediol. Applied and Environmental Microbiology, 76, 4158–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letarov A., Kulikov E. (2009). The bacteriophages in human- and animal body-associated microbial communities. Journal of Applied Microbiology, 107, 1–13. 10.1111/j.1365-2672.2009.04143.x [DOI] [PubMed] [Google Scholar]
- Levin B. R., Cornejo O. E. (2009). The population and evolutionary dynamics of homologous gene recombination in bacteria. PLoS Genetics, 5, e1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman T. D., Michel J.-B., Aingaran M., Potter-Bynoe G., Roux D., Davis M. R., Skurnik D., Leiby N., LiPuma J. J., Goldberg J. B. (2011). Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nature Genetics, 43, 1275–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L., Marguerat S., Bähler J. (2008). Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nature Reviews Genetics, 9, 583–593 [DOI] [PubMed] [Google Scholar]
- Louca S., Doebeli M. (2015). Calibration and analysis of genome-based models for microbial ecology. Elife, 4, e08208. 10.7554/eLife.08208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. R., Kassen R. (2007). The effects of competition and predation on diversification in a model adaptive radiation. Nature, 446, 432–435. 10.1038/nature05599 [DOI] [PubMed] [Google Scholar]
- Miller E. N., Jarboe L. R., Turner P. C., Pharkya P., Yomano L. P., York S. W., Nunn D., Shanmugam K. T., Ingram L. O. (2009). Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Applied and Environmental Microbiology, 75, 6132–6141. 10.1128/AEM.01187-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M., Lai X., York S. W., Ingram L. O. (1997). Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Applied and Environmental Microbiology, 63, 4633–4637. 10.1128/aem.63.12.4633-4637.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A. I., Zorzet A., Kanth A., Dahlstrom S., Berg O. G., Andersson D. I. (2006). Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proceedings of the National Academy of Sciences, 103, 6976–6981. 10.1073/pnas.0602171103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien E. J., Lerman J. A., Chang R. L., Hyduke D. R., Palsson B. O. (2013). Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Molecular Systems Biology, 9, 693. 10.1038/msb.2013.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K. L., Wolfson L. J., Watt J. P., Henkle E., Deloria-Knoll M., McCall N., Lee E., Mulholland K., Levine O. S., Cherian T., Hib & Pneumococcal Global Burden of Disease Study Team . (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. The Lancet, 374, 893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature, 405, 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. (1991). Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Applied and Environmental Microbiology, 57, 893–900. 10.1128/aem.57.4.893-900.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiology Reviews, 34, 415–425. 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- Overbeck T. J., Welker D. L., Hughes J. E., Steele J. L., Broadbent J. R. (2017). Transient MutS-based hypermutation system for adaptive evolution of Lactobacillus casei to Low pH. Applied and Environmental Microbiology, 83, 20. 10.1128/AEM.01120-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Papp B., Lercher M. J. (2005). Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nature Genetics, 37, 1372–1375. 10.1038/ng1686 [DOI] [PubMed] [Google Scholar]
- Pellegrino M., Sciambi A., Treusch S., Durruthy-Durruthy R., Gokhale K., Jacob J., Chen T. X., Geis J. A., Oldham W., Matthews J., Kantarjian H., Futreal P. A., Patel K., Jones K. W., Takahashi K., Eastburn D. J. (2018). High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Research, 28, 1345–1352. 10.1101/gr.232272.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J., Gern L. (2004). Lyme borreliosis in Europe and North America. Parasitology, 129, S191. [DOI] [PubMed] [Google Scholar]
- Power J. J., Pinheiro F., Pompei S., Kovacova V., Yuksel M., Rathmann I., Forster M., Lassig M., Maier B. (2021). Adaptive evolution of hybrid bacteria by horizontal gene transfer. Proceedings of the National Academy of Sciences, 118, e2007873118. 10.1073/pnas.2007873118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi P., Zhu J., Young J. P. W., Masson-Boivin C. (2016). Symbiosis within symbiosis: Evolving nitrogen-fixing legume symbionts. Trends in Microbiology, 24, 63–75 [DOI] [PubMed] [Google Scholar]
- Renton M. (2013). Shifting focus from the population to the individual as a way forward in understanding, predicting and managing the complexities of evolution of resistance to pesticides. Pest Management Science, 69, 171–175. 10.1002/ps.3341 [DOI] [PubMed] [Google Scholar]
- Robicsek A., Jacoby G. A., Hooper D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. The Lancet Infectious Diseases, 6, 629–640 [DOI] [PubMed] [Google Scholar]
- Rochman N. D., Wolf Y. I., Faure G., Mutz P., Zhang F., Koonin E. V. (2021). Ongoing global and regional adaptive evolution of SARS-CoV-2. Proceedings of the National Academy of Sciences, 118, e2104241118. 10.1073/pnas.2104241118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runquist D., Fonseca C., Radstrom P., Spencer-Martins I., Hahn-Hagerdal B. (2009). Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Applied Microbiology and Biotechnology, 82, 123–130. 10.1007/s00253-008-1773-y [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Taulien J., Borkovich K. A., Lindquist S. (1992). Hsp104 is required for tolerance to many forms of stress. The EMBO Journal, 11, 2357–2364. https://www.ncbi.nlm.nih.gov/pubmed/1600951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L., Andersson D. I. (2009). Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nature Reviews Microbiology, 7, 578–588. 10.1038/nrmicro2174 [DOI] [PubMed] [Google Scholar]
- Sawisit A., Jantama K., Zheng H., Yomano L. P., York S. W., Shanmugam K. T., Ingram L. O. (2015). Mutation in galP improved fermentation of mixed sugars to succinate using engineered Escherichia coli AS1600a and AM1 mineral salts medium. Bioresource Technology, 193, 433–441. 10.1016/j.biortech.2015.06.108 [DOI] [PubMed] [Google Scholar]
- Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A., Waller A., Mende D. R., Kultima J. R., Martin J. (2013). Genomic variation landscape of the human gut microbiome. Nature, 493, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G., Weber A. P., Lercher M. J. (2014). Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays, 36, 9–20 [DOI] [PubMed] [Google Scholar]
- Serrazanetti D. I., Guerzoni M. E., Corsetti A., Vogel R. (2009). Metabolic impact and potential exploitation of the stress reactions in Lactobacilli. Food Microbiology, 26, 700–711. 10.1016/j.fm.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Shi A., Yomano L. P., York S. W., Zheng H., Shanmugam K. T., Ingram L. O. (2020). Chromosomal mutations in Escherichia coli that improve tolerance to nonvolatile side-products from dilute acid treatment of sugarcane bagasse. Biotechnology and Bioengineering, 117, 85–95 [DOI] [PubMed] [Google Scholar]
- Shi A., Zheng H., Yomano L. P., York S. W., Shanmugam K. T., Ingram L. O. (2016). Plasmidic expression of nemA and yafC* increased resistance of ethanologenic Escherichia coli LY180 to nonvolatile side products from dilute acid treatment of sugarcane bagasse and artificial hydrolysate. Applied and Environmental Microbiology, 82, 2137–2145. 10.1128/AEM.03488-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Gourbeyre E., Chandler M. (2014). Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiology Reviews, 38, 865–891. 10.1111/1574-6976.12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. E., Buckley D. G., Wu Z., Saenphimmachak C., Hoffman L. R., D'Argenio D. A., Miller S. I., Ramsey B. W., Speert D. P., Moskowitz S. M. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proceedings of the National Academy of Sciences, 103, 8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J. I. (2007). Evolving ideas of legume evolution and diversity: A taxonomic perspective on the occurrence of nodulation. New Phytologist, 174, 11–25. https://doi: 10.1111/j.1469-8137.2007.02015.x [DOI] [PubMed] [Google Scholar]
- Straus D. S., Straus L. D. A. (1976). Large overlapping tandem genetic duplications in Salmonella typhimurium. Journal of Molecular Biology, 103, 143–153 [DOI] [PubMed] [Google Scholar]
- Tang X., Huang Y., Lei J., Luo H., Zhu X. (2019). The single-cell sequencing: New developments and medical applications. Cell & Bioscience, 9, 53. 10.1186/s13578-019-0314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Miller E. N., Jarboe L. R., Baggett C. L., Shanmugam K. T., Ingram L. O. (2011). YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. Journal of Industrial Microbiology & Biotechnology, 38, 431–439. 10.1007/s10295-010-0787-5 [DOI] [PubMed] [Google Scholar]
- Turner P. C., Yomano L. P., Jarboe L. R., York S. W., Baggett C. L., Moritz B. E., Zentz E. B., Shanmugam K. T., Ingram L. O. (2012). Optical mapping and sequencing of the Escherichia coli KO11 genome reveal extensive chromosomal rearrangements, and multiple tandem copies of the Zymomonas mobilis pdc and adhB genes. Journal of Industrial Microbiology and Biotechnology, 39, 629–639. 10.1007/s10295-011-1052-2 [DOI] [PubMed] [Google Scholar]
- van Oosterhout C., Stephenson J. F., Weimer B., Ly H., Hall N., Tyler K. M. (2021). COVID-19 adaptive evolution during the pandemic—Implications of new SARS-CoV-2 variants on public health policies. Virulence, 12, 2013–2016. 10.1080/21505594.2021.1960109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A., Palsson B. O. (1994). Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Applied and Environmental Microbiology, 60, 3724–3731. 10.1128/AEM.60.10.3724-3731.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ingram L. O., Shanmugam K. T. (2011). Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proceedings of the National Academy of Sciences, 108, 18920–18925. 10.1073/pnas.1111085108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Miller E. N., Yomano L. P., Shanmugam K. T., Ingram L. O. (2012). Increased furan tolerance in Escherichia coli due to a cryptic ucpA gene. Applied and Environmental Microbiology, 78, 2452–2455. 10.1128/AEM.07783-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werpy T, Petersen G, Aden A, Bozell J, Holladay J, White J, Manheim A, Eliot D, Lasure L, Jones S. (2004). Top value added chemicals from biomass. Results of screening for potential candidates from sugars and synthesis gas (Vol. I). Technical report. US Department of Energy [Google Scholar]
- Wu C., Zhang J., Chen W., Wang M., Du G., Chen J. (2012). A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Applied Microbiology and Biotechnology, 93, 707–722. 10.1007/s00253-011-3757-6 [DOI] [PubMed] [Google Scholar]
- Wywial E., Haven J., Casjens S. R., Hernandez Y. A., Singh S., Mongodin E. F., Fraser-Liggett C. M., Luft B. J., Schutzer S. E., Qiu W.-G. (2009). Fast, adaptive evolution at a bacterial host-resistance locus: The PFam54 gene array in Borrelia burgdorferi. Gene, 445, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer S. A., Kalayou S., Homberset H., Birhanu A. G., Riaz T., Zegeye E. D., Lutter T., Abebe M., Holm-Hansen C., Aseffa A., Tonjum T. (2020). Lineage-specific proteomic signatures in the Mycobacterium tuberculosis complex reveal differential abundance of proteins involved in virulence, DNA repair, CRISPR-Cas, bioenergetics and lipid metabolism. Frontiers in Microbiology, 11, 550760. 10.3389/fmicb.2020.550760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomano L. P., York S. W., Ingram L. O. (1998). Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. Journal of Industrial Microbiology and Biotechnology, 20, 132–138. 10.1038/sj.jim.2900496 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jantama K., Moore J. C., Jarboe L. R., Shanmugam K. T., Ingram L. O. (2009). Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proceedings of the National Academy of Sciences, 106, 20180–20185. 10.1073/pnas.0905396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jantama K., Moore J. C., Shanmugam K. T., Ingram L. O. (2007). Production of L-alanine by metabolically engineered Escherichia coli. Applied Microbiology and Biotechnology, 77, 355–366. 10.1007/s00253-007-1170-y [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang X., Shanmugam K. T., Ingram L. O. (2011). L-malate production by metabolically engineered Escherichia coli. Applied and Environmental Microbiology, 77, 427–434. 10.1128/AEM.01971-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Liu L. X., Zhang Y. Z., Jiao J., Cui W. J., Zhang B., Wang X. L., Li M. L., Chen Y., Xiong Z. Q. (2018). Adaptive evolution of rhizobial symbiotic compatibility mediated by co-evolved insertion sequences. The ISME journal, 12, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Lieberman T. D., Poyet M., Kauffman K. M., Gibbons S. M., Groussin M., Xavier R. J., Alm E. J. (2019). Adaptive evolution within gut microbiomes of healthy people. Cell Host & Microbe, 25, 656–667.e8. 10.1016/j.chom.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Wang X., Yomano L. P., Shanmugam K. T., Ingram L. O. (2012). Increase in furfural tolerance in ethanologenic Escherichia coli LY180 by plasmid-based expression of thyA. Applied and Environmental Microbiology, 78, 4346–4352. 10.1128/AEM.00356-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Causey T. B., Hasona A., Shanmugam K. T., Ingram L. O. (2003). Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Applied and Environmental Microbiology, 69, 399–407. 10.1128/AEM.69.1.399-407.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Tan Z., Xu H., Chen J., Tang J., Zhang X. (2014). Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metabolic Engineering, 24, 87–96. 10.1016/j.ymben.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Zomorrodi A. R., Maranas C. D. (2012). OptCom: A multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Computational Biology, 8, e1002363. 10.1371/journal.pcbi.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the related references.