Abstract
Chemically induced dimerization of FKBP and FRB using rapamycin and rapamycin analogs has been utilized in a variety of biological applications. Formation of the FKBP-rapamycin-FRB ternary complex is typically used to activate a biological process and this interaction has proven to be essentially irreversible. In many cases, it would be beneficial to also have temporal control over deactivating a biological process once it has been initiated. Thus, we developed the first reactive oxygen species-generating rapamycin analog toward this goal. The BODIPY-rapamycin analog BORap is capable of dimerizing FKBP and FRB to form a ternary complex, and upon irradiation with 530 nm light, generates singlet oxygen to oxidize and inactivate proteins of interest fused to FKBP/FRB.
Utilization of a ROS-generating chromophore for the development of reversible control of rapamycin-induced protein dimerization via targeted oxidation.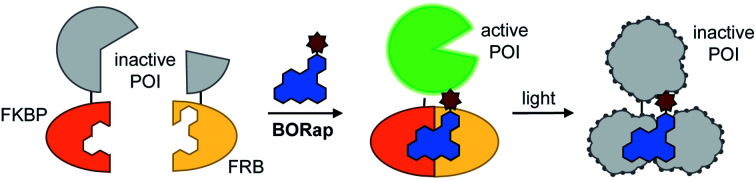
Introduction
Rapamycin is a natural product which functions as a chemical inducer of dimerization (CID) by binding FKBP (FK506 binding protein) and FRB (FKBP-rapamycin binding domain of mTOR) forming a ternary complex with a Kd of 12 nM.1 Chemically induced dimerization of proteins has allowed for conditional control over various biological processes.2 Rapamycin-induced dimerization of FKBP and FRB – and recently, trimerization using a split FRB3 – has been utilized in numerous applications, including the activation of split-proteins, such as tobacco etch virus protease,4 Abl kinase,5 and Cas9 nuclease,6 induction of translocation to the cell membrane7 or nucleus,8 and activation of G-protein coupled receptor signaling.9 The high binding affinity of the ternary complex, cell permeability of rapamycin, and small size of the FKBP/FRB proteins10 are all advantages of rapamycin-induced protein dimerization which have led to its extensive use. For this reason, several rapamycin analogs which selectively bind a mutant FRB, termed rapalogs, have also been developed.11 Furthermore, rapamycin (sirolimus) is an FDA approved drug and thus is safe and functional in higher organisms, including applications in rodents12,13 and non-human primates.14
Light provides unique opportunities for remote control of biological processes, including tunable activation and deactivation at precise time points and in precise locations.15,16 In order to gain spatiotemporal control over protein dimerization and the corresponding activation of biological processes, photocaged rapamycin analogs were developed.17–20 Proteins and processes that have been placed under light-activated control using photocaged rapamycin analogs include kinases, GTPases, proteases, Cre recombinase, and translation initiation.
While utilizing analogs of rapamycin as a way to spatiotemporally activate a biological process has proven to be a useful tool, it would be advantageous to have a method for turning these biological processes off at a defined time point. Reversibility has been achieved for some CIDs,21–23 but for rapamycin it has remained out of reach, due to the very high affinity FKBP-rapamycin-FRB complex. A completely different solution involves using a second CID to mislocalize the entire GAIs-YFP-FKBP-C2(LACT)-rapamycin-CFP-FRB-POI complex from the plasma membrane to the mitochondria.24 We aimed to expand the chemical biology toolbox to allow for reversible control of protein–protein interactions by addressing the lack of a proper “off-switch” for rapamycin-induced protein activation, while also taking advantage of the fast and precise perturbation light irradiation provides. In a recent example, we developed a photoswitchable arylazopyrazole-rapamycin analog which induced ternary complex formation upon irradiation with 365 nm light; however, the complex was unable to be disassembled upon irradiation with 530 nm light, again, due to its very strong affinity.25
In order to overcome the high binding affinity of FKBP-rapamycin-FRB, we now report a reactive oxygen species (ROS)-generating rapamycin analog as a tool to reverse the protein activity produced by rapamycin-induced dimerization via targeted protein oxidation.
Chromophore assisted light inactivation (CALI) is a method used for the targeted inactivation of proteins through application of a ROS-generating chromophore fused to a ligand of the protein of interest.26 There are many types of ROS, which can be categorized according to two main photosensitized oxidation pathways: type I oxidation – refers to oxidation by radicals or radical ions (via electron transfer) and type II oxidation – refers to oxidation by singlet oxygen (via energy transfer).27 Singlet oxygen has a lifetime of approximately 200 ns, during which it can diffuse 30 nm in the cell.28 When ROS, such as singlet oxygen, are generated in a cellular environment, amino acids such as methionine, cysteine, or tyrosine on nearby proteins can become oxidized and often lead to deactivation of those proteins.29 Genetically encoded ROS-generating proteins such as KillerRed30 and mini Singlet Oxygen Generator (miniSOG)31 have been used to induce oxidation of proteins of interest, but are mostly applied in cell ablation studies. Not surprisingly, these genetically encoded ROS generators result in a significant degree of cell death, as evident by the name.
Small molecule ROS-generators have also been used in various biological applications. Porphyrin dyes have been applied in photodynamic therapies to eradicate cancer cells through the use of a targeting group causing an accumulation of the chromophore in the tumor environment.32 Eosin33 and ruthenium(ii) tris-bipyridyl34 derivatives have been conjugated to HaloTag ligands to selectively inactivate proteins of interest. Fluorogen-activated proteins have been utilized to enhance fluorescence and oxidative properties of malachite green dyes.35 Boron dipyrromethene (BODIPY) chromophores have been shown to have tunable properties dependent on their core substituents – some of which are activatable with >500 nm light. Excitation of the chromophore in the visible light range is crucial to reducing cellular toxicity that can be caused by irradiation. The addition of halogen substituents to the BODIPY chromophore has shown to increase the ability of the chromophore to generate ROS through the heavy atom affect.36 Iodination of the specific chromophore used in this research has previously been determined to decease the fluorescence quantum yield from 0.50 to 0.04, while increasing the ROS-generating triplet-state quantum yield to 0.92.37 Because of the desirable photophysical properties of this BODIPY chromophore, we aimed to conjugate this chromophore to rapamycin to develop the first ROS-generating rapamycin analog, while maintaining its ability to induce protein dimerization, with the objective of gaining spatiotemporal control over oxidation of the FKBP-rapamycin-FRB ternary complex and associated proteins.
Results & discussion
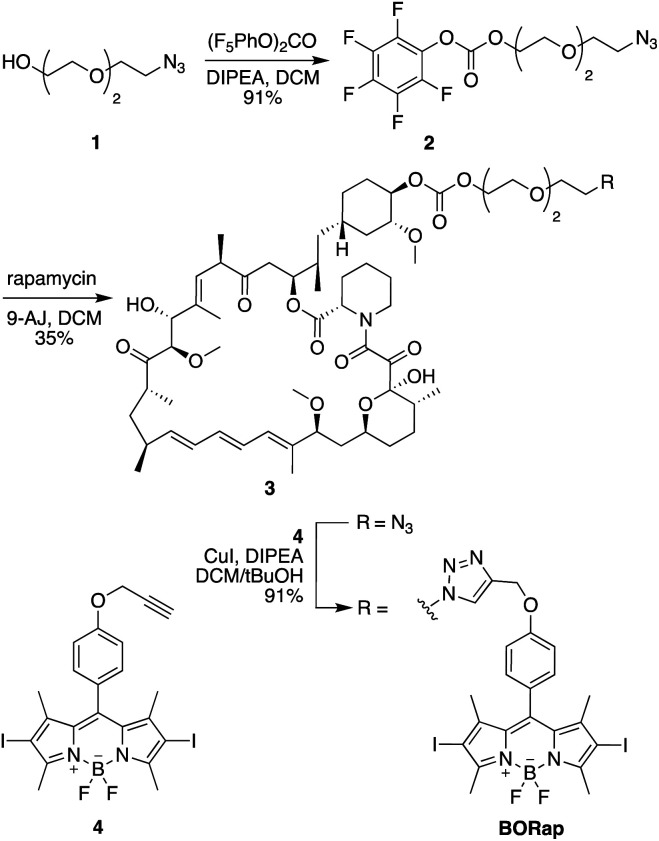
In initial studies, a propyl-linked BODIPY-rapamycin analog was synthesized but was found to be an inefficient dimerizer of FKBP and FRB (data not shown). In order to increase the distance between the chromophore and the binding site, as to not inhibit ternary complex formation, a PEG spacer was introduced. The synthesis of the PEG-linked BODIPY-rapamycin BORap (Fig. 1) began with the propargylation of 4-hydroxybenzaldehyde (6) to form the propargyl ether 7.1 The BODIPY chromophore 8 was formed through the condensation of dimethyl pyrrole in the presence of the benzaldehyde 7 and trifluoroacetic acid. Upon oxidation with DDQ, the resulting dipyrromethane was converted to dipyrromethene, which reacts with boron trifluoride in the presence of triethylamine to yield the BODIPY chromophore. This chromophore was iodinated using N-iodosuccinimide, generating 4, in order to increase the quantum yield for ROS generation. In a convergent synthesis, triethylene glycol was mono-tosylated, and the tosyl group was subsequently converted to an azide yielding 1. The alcohol 1 was activated using (bis)pentafluorocarbonate and selectively coupled to the C-40 position38 of rapamycin in the presence of 4-dimethylaminopyridine, yielding the azido-modified rapamycin 3. A Cu-catalyzed [3 + 2] cycloaddition reaction with the BODIPY alkyne 4 yielded the BODIPY-rapamycin BORap.
Fig. 1. Synthesis of the BODIPY-modified rapamycin analog BORap.
Prior to testing in a biological environment, we first characterized the stability of BORap in standard buffers. The stability of BORap was tested in phosphate buffered saline (pH 7.4) (ESI Fig. 1A†) or 10% fetal bovine serum (FBS) in phosphate buffered saline (pH 7.4) (ESI Fig. 1B†) over an 8 hour period at 37 °C to closely mimic physiological conditions. Monitoring the peak area by HPLC, we found BORap to be completely stable over this time period in both settings, suggesting this compound should be stable in cells over the time frame required for protein activation through dimerization followed by optical inactivation.
As BODIPY chromophores are also utilized as photocaging groups, one possible concern is potential cleavage from rapamycin upon irradiation. To address this, the stability of BORap to the irradiation and incubation conditions was monitored via HPLC, and the compound was found to be completely stable (ESI Fig. 3†).
In order to confirm BORap was capable of generating ROS, the singlet oxygen sensor, DPBF (1,3-diphenylisobenzofuran) was used. DPBF is a fluorescent molecule that reacts with singlet oxygen, generating an unstable peroxide intermediate, which, through the loss of a water molecule, forms a non-fluorescent product (ESI Fig. 5A†).39 Because the absorbance maximum of BORap was determined to be 542 nm (ESI Fig. 2†), we first tested the effects of irradiation using 530 nm light. A solution of DPBF with or without BORap was irradiated for 0, 10, 30, or 60 seconds at 530 nm using an LED light source. With increasing irradiation times, a concomitant decrease in fluorescence was observed in samples treated with BORap, thus demonstrating that BORap does generate ROS and, more specifically, singlet oxygen (ESI Fig. 5B†). To validate 530 nm as the most appropriate irradiation wavelength to use in subsequent experiments for the generation of singlet oxygen, various commonly utilized LED light sources were compared. The power level of all LEDs was adjusted to approximately 70 mW and solutions of DPBF with or without BORap were irradiated for 60 seconds with each LED. BORap-containing samples irradiated at 365, 405, 490, and 530 nm showed comparable reductions in fluorescence, with all other irradiation wavelengths being less efficient (ESI Fig. 5C†). In order to avoid any irradiation-induced cellular toxicity, the longest wavelength (530 nm) was selected for use in all future experiments.
We next aimed to confirm that BORap retains the ability to dimerize FKBP and FRB with the addition of the bulky BODIPY chromophore in vitro (ESI Fig. 6†). Ternary complex formation was assessed through gel shift assays with recombinantly expressed FKBP and FRB fusion proteins. Briefly, FKBP-YFP-His and FRB-CFP-His were expressed in BL21 (DE3) cells and purified using Ni-NTA resin. Purified FKBP-YFP-His and FRB-CFP-His were incubated with rapamycin or BORap (over a range of concentrations) for two hours, to allow for ternary complex formation. The samples were analyzed by native-PAGE and Coomassie stained to monitor the appearance of a higher molecular weight band for the ternary complex. Ternary complex formation is achieved using BORap; however, the small molecule does not have as high of an affinity as the rapamycin control. To determine the reason for the reduced affinity, we also tested 3 in the same assay and found that it functions as efficiently as rapamycin. Thus, it appears that the BODIPY chromophore is responsible for the decrease in ternary complex formation efficiency, requiring BORap use at higher concentrations.
After initially validating BORap functions as a ROS-generator and dimerizer of FKBP/FRB in vitro, we next aimed to demonstrate the utility of this compound in a cellular environment. As chemical inducers of dimerization, such as rapamycin, have been widely applied to the conditional activation of various split, inactivated proteins, we first demonstrated dimerization in response to BORap using a split luciferase enzyme in HEK293T cells. The split luciferase reporter consists of the C-terminus of a split firefly luciferase fused to FKBP and the N-terminus fused to FRB.40 Upon treatment with rapamycin or BORap, the two fragments of the inactive luciferase enzyme are brought together by the formation of the FKBP-rapamycin-FRB ternary complex to generate a reconstituted, active enzyme. In order to deactivate this enzyme, irradiation with 530 nm light can be employed to initiate ROS generation and subsequent oxidation of the proteins (Fig. 2A).
Fig. 2. (A) Schematic representation of chemically induced protein dimerization and activation of luciferase, followed by subsequent light exposure and resulting deactivation of the enzyme. (B) We found that 100 nM of rapamycin and 10 μM of BORap elicited optimal activity, and as such, were used in the subsequent experiment. (C) A decrease in luminescence was observed with increasing irradiation times for BORap-treated cells, while no light-induced effect was seen in the rapamycin control. Average signal intensity is plotted with error bars representing standard deviation. Statistical analysis of triplicate experiments was performed in Prism using a two-way ANOVA test to compare irradiated samples for each treatment condition to non-irradiated (0 s) samples. P values less than or equal to 0.05 are represented by *, less than or equal to 0.0001 are represented by ****. Comparisons of rapamycin-treated samples (10, 30, 60, or 90 s vs. 0 s) and comparison of BORap-treated samples (10 s vs. 0 s) are not significant.
To find the optimal compound concentration to induce dimerization for this reporter system, HEK293T cells transfected with the split luciferase reporter construct were treated with rapamycin or BORap at various concentrations and incubated for 2 hours. Luciferase substrates were added and recorded luminescence values were normalized to the DMSO control (Fig. 2B).41 The optimal rapamycin concentration was found to be 100 nM, while the optimal concentration of BORap was found to be 10 μM. These concentrations were used in subsequent experiments.
To demonstrate deactivation of the ternary complex in response to irradiation with 530 nm light, HEK293T cells expressing the split luciferase reporter were treated with rapamycin (100 nM) or BORap (10 μM) and incubated for 2 hours. This incubation period was followed by two 1 hour washes to remove any excess compound not engaged in a ternary complex. Cells were irradiated using 530 nm light for 0, 10, 30, 60, or 90 seconds, followed by a 2 hour incubation period. After luciferase substrates were added, luminescence values were recorded and normalized to a DMSO control (Fig. 2C). A decrease in normalized luminescence is observed in response to increasing irradiation times, suggesting ROS generated by BORap is capable of deactivating the luciferase enzyme. After using the split luciferase reporter to display the ability for BORap to dimerize FKBP/FRB and activate enzyme function, followed by light-induced inactivation of luciferase in cells, we next aimed to utilize this strategy to gain conditional control over the widely utilized tobacco etch virus (TEV) protease. TEV protease has found broad application in the engineering of biological function, including sensing of protein–protein interactions,42 regulation of transcription,43 and control of protein activity.44
Several proteases are naturally regulated through active site cysteine oxidation,45,46 and to mimic this regulatory mechanism we applied BORap to the on- and off-switching of TEV protease, which functions to cleave a specific peptide sequence with high catalytic activity.47 In order to reversibly control a rapamycin-inducible split-TEV protease system, the expression constructs pFKBP-TEVpCT and pFRB-TEVpNT were transfected into mammalian cells.4 To visualize TEV protease activity, we employed a GFP sensor which encodes for a GFP protein, a TEV cleavage site, and a GFP-quenching peptide sequence, PCS2-GFP-TEV. Upon ternary complex formation, the two halves of the TEV protease are reconstituted, forming the active TEV protease, allowing for cleavage of the GFP-quenching peptide from the sensor, and producing a fluorescent signal (Fig. 3A).48
Fig. 3. (A) Schematic representation of the split-TEV assay. (B) HEK293T cells expressing pFKBP-TEVpCT, pFRB-TEVpNT, and PCS2-GFP-TEV were treated with either rapamycin or BORap and irradiated to monitor change in fluorescence intensity. Cells were irradiated through a DsRed filter for 0 or 90 seconds, and post-irradiation images were acquired after 18 hours. Scale bar equals 50 μM. (C) Average fluorescence intensity of 20 cells for each condition was quantified and normalized to the DMSO negative control, with error bars representing standard deviations. Statistical analysis was performed in Prism using a one-way ANOVA test to compare each treatment condition to the DMSO negative control. P values less than or equal to 0.0001 are represented by ****, while the irradiated BORap sample shows no statistical difference from the DMSO negative control.
HEK293T cells co-expressing the split-TEVp reporter and GFP sensor were treated with DMSO, rapamycin, or BORap at 20 μM concentrations for six hours to allow time for cleavage of the sensor by the TEV protease. After this time, cells were washed to remove any excess dimerizer and were irradiated for 90 seconds to promote the formation of ROS and therefore deactivation of the TEV protease. Cells treated with rapamycin or with non-irradiated BORap showed an increase in fluorescent signal over the next 18 hours, corresponding to cleavage of the GFP sensor by TEV protease reconstituted through rapamycin-induced dimerization, while cells treated with BORap and irradiated for 90 seconds showed no increase in fluorescence over the DMSO negative control. This indicates that the enzymatic function of the TEV protease was inactivated upon irradiation and oxidation (Fig. 3B and C). Demonstrating the general use of BORap to both turn on and off TEV protease function, reversible control of the numerous processes that have been engineered to respond to protease cleavage can now be switched on/off with temporal precision.
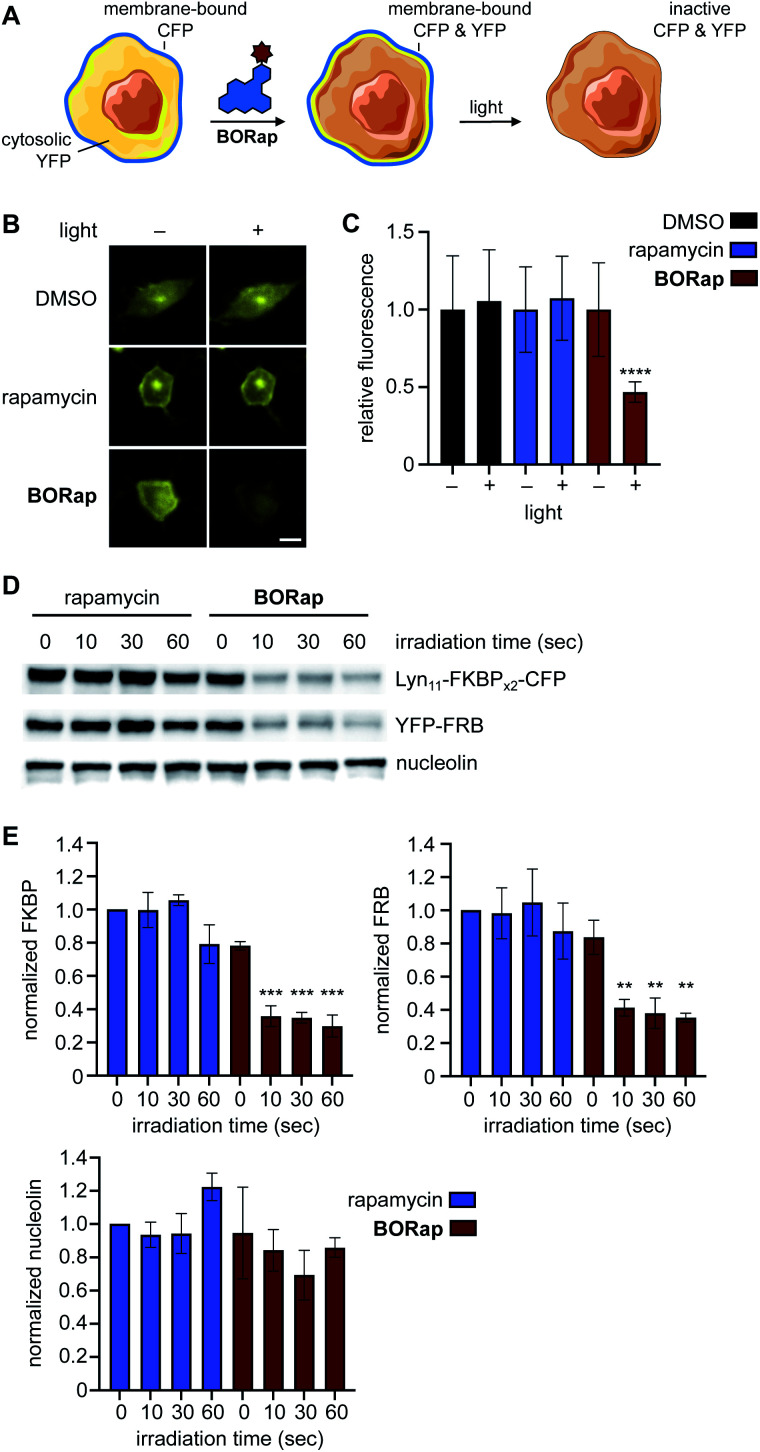
Another central application of chemically induced dimerization is translocation of proteins of interest to a certain compartment of the cell to, in turn, induce a distinct biological response. We next applied our targeted protein oxidation approach to protein translocation to the cell membrane, followed by optically triggered protein deactivation. Membrane reporter constructs Lyn11-FKBPx2-CFP and YFP-FRB were used to visualize dimerization of FKBP and FRB through monitoring co-localization of CFP and YFP at the plasma membrane using fluorescence microscopy (Fig. 4A).7 In HeLa cells expressing the reporter, YFP-FRB is diffuse through the cell and Lyn11-FKBPx2-CFP is localized to the plasma membrane. As expected, when cells were treated with rapamycin or BORap, YFP-FRB translocates to the membrane as a result of ternary complex formation (ESI Fig. 7†). Treatment with 20 μM of rapamycin or BORap resulted in the most efficient translocation to the membrane and this concentration was used in all subsequent experiments in which the membrane reporter constructs were used.
Fig. 4. (A) Schematic representation of the membrane reporter translocation assay. (B) Cells treated with either rapamycin or BORap and irradiated at 0 hours followed by YFP imaging at 2 hours. Scale bar equals 10 μM. (C) Quantification of the YFP signal intensity before and after irradiation (90 s), showed reduced fluorescence in BORap-treated cells, while all other conditions show little to no decrease in fluorescent signal. Average fluorescence intensity in 20 cells is shown normalized to pre-irradiated controls, with error bars representing standard deviations. Statistical analysis was performed in Prism using a two-way ANOVA test to compare irradiated samples for each treatment condition to pre-irradiated samples. P values less than or equal to 0.0001 are represented by ****, while all other comparisons are not significant. (D) HEK293T cells expressing the CFP and YFP constructs were treated with 20 μM of rapamycin or BORap prior to irradiating with 530 nm light. Western blot analysis using anti-GFP, anti-GAPDH, and anti-nucleolin antibodies showed a light-induced band decrease with BORap but not rapamycin treatment. (E) Triplicate experiments were performed and quantified using FIJI. Band intensity of FKBP, FRB, and nucleolin for each irradiation timepoint were normalized to the rapamycin-treated, non-irradiated sample and averaged. Statistical analysis was performed as above, comparing irradiated to non-irradiated samples within each treatment condition (10, 30, or 60 s vs. 0 s). P values less than or equal to 0.001 are represented by ***, less than or equal to 0.01 by **, while comparisons of rapamycin-treated samples (for FKBP, FRB, and nucleolin) and comparison of BORap-treated samples (for nucleolin) are not significant.
To demonstrate deactivation of the membrane reporter using fluorescence microscopy, HeLa cells expressing Lyn11-FKBPx2-CFP and YFP-FRB were treated with 20 μM of rapamycin or BORap, followed by a 2 hour incubation period to allow for ternary complex formation and two 1 hour washes to remove unbound dimerizer. Irradiation was performed with a DsRed filter (550/25 nm) on the microscope and images of YFP were acquired (470/40 nm) for two hours (Fig. 4B and ESI Fig. 8†). To monitor oxidation and inactivation at the membrane, YFP images were analyzed over time. Average signal intensity after two hours was normalized to the level immediately prior to irradiation for each treatment condition. A ∼50% decrease in YFP signal intensity was observed for cells treated with BORap after a brief irradiation for 90 seconds. No decrease in signal intensity was observed for cells treated with DMSO, rapamycin, or non-irradiated BORap-treated cells (Fig. 4C). These results validate that compound treatment with BORap can induce ternary complex formation at a subcellular location, followed by optically triggered protein deactivation through irradiation with 530 nm light.
To further investigate the effects of BORap, these proteins were next analyzed via western blot. HEK293T cells expressing the membrane reporter system were treated with 20 μM of rapamycin or BORap, followed by a 2 hour incubation period and two 1 hour washes (media change followed by incubation). Cells were irradiated with 530 nm light and incubated for an additional 2 hours. Following cell lysis and protein denaturation, western blot analysis was performed. To visualize the levels of both Lyn11-FKBPx2-CFP and YFP-FRB, a GFP antibody was utilized. In addition, two housekeeping proteins, GAPDH and nucleolin, were monitored using their corresponding antibodies. Both Lyn11-FKBPx2-CFP and YFP-FRB signals decreased in response to increasing irradiation time when treated with BORap, but not rapamycin (Fig. 4D, E and ESI Fig. 9†). In contrast, signal intensity for both housekeeping proteins, GAPDH and nucleolin, was completely retained, suggesting ROS generation is not leading to any off-target oxidation but instead is targeted to proteins in close proximity to BORap. These results confirm that ROS generated through the irradiation of BORap is causing selective oxidation of the proteins of interest, presumably because of the short lifetime (200 ns) of singlet oxygen in a cellular environment and the resulting minimal diffusion radius (30 nm).28
To determine the cause of the band intensity decrease observed in the western blot, and as validation of the decreased fluorescence observed in the micrographs, a similar experiment as just described was performed; however, the samples were not boiled in order to avoid denaturing the fluorescent proteins. The samples were analyzed by SDS-PAGE and imaged using a ChemiDoc equipped with a GFP filter (560/50 nm) to visualize in-gel fluorescence. If non-denatured samples retained fluorescence, the decrease in signal could be credited to a lack of protein recognition by the antibody rather than deactivation of protein function. For both Lyn11-FKBPx2-CFP and FRB-YFP the same trend applies in which an increase in irradiation time corresponds to a decrease in fluorescent protein signal (ESI Fig. 10†). Thus, these results confirm that both the antibody recognition and the function of the target protein(s) is being inactivated upon irradiation.
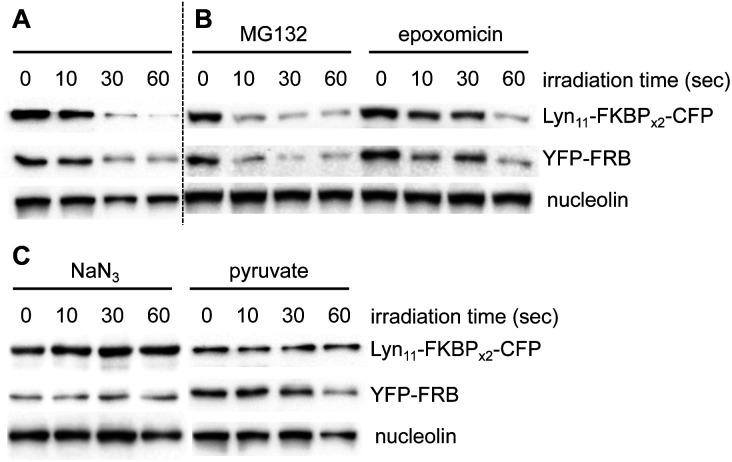
In addition to illustrating the general utility of this system as an optical off-switch, we also elucidated the mechanism through which deactivation occurs. We hypothesized that ROS generated by the chromophore might oxidize the proteins and subsequently signal for degradation through the proteasome.49 To investigate this hypothesis, two known proteasome inhibitors, epoxomicin50 and MG132,51 were used concurrently with BORap. If protein degradation was occurring via an active proteasome, no decrease in signal would be observed in the presence of a proteasome inhibitor. HEK293T cells expressing the membrane reporter were treated with 20 μM of BORap for two hours, then washed twice for one hour each time (in the presence or absence of the proteasome inhibitor at 10 μM), then irradiated as done previously. The same sample preparation was followed as discussed earlier and protein levels were detected via Western blot. A decrease in signal is still observed for the proteins of interest whether or not either proteasome inhibitor is present (Fig. 5A and B). This result suggests that an active proteasome is not involved in the mechanism of protein deactivation.
Fig. 5. (A) A western blot was performed as above with BORap-treated cells used as a control to show inactivation of the membrane reporter. (B) Treatment with MG132 or epoxomicin shows no change, suggesting that reduced band intensity is not the result of proteasomal protein degradation. (C) Sodium azide functions to block loss of signal, while sodium pyruvate has less or no effect, thus supporting that BORap-induced signal loss requires the generation of singlet oxygen.
After showing degradation does not occur through the proteasome, we hypothesized that the proteins are simply being rendered inactive through oxidation, and as a result, are not recognized efficiently by antibodies. Because oxidation could be occurring via singlet oxygen generation or by superoxide radical formation, we tested whether protein detection would be retained in the presence of two different types of ROS scavengers. Sodium azide and sodium pyruvate act as quenchers of singlet oxygen52 and hydrogen peroxide,53 respectively.
HEK293T cells expressing the membrane reporter were treated with 20 μM of BORap for two hours, followed by two 1 hour washes (with and without 10 mM of scavenger). Cells were irradiated using a 530 nm LED, followed by a 2 hour incubation. Western blot analysis was performed in the same manner as before. When treated with BORap in the presence of the hydrogen peroxide quencher sodium pyruvate, a decrease in GFP signal is still observed, suggesting hydrogen peroxide is not the cause of observed protein oxidation; however, in the presence of the singlet oxygen quencher sodium azide, GFP signal does not decrease upon irradiation. This suggests a type II energy pathway – generation of singlet oxygen – is responsible for the protein oxidation observed upon irradiation of BORap (Fig. 5C).
An XTT cell viability assay was performed and ROS generation showed no effect on general cell viability up to 72 hours. Non-transfected HEK293T cells and HEK293T cells expressing the membrane reporter were treated with 20 μM of rapamycin or BORap, followed by a 2 hour incubation period and two 1 hour washes. Cells were irradiated using a 530 nm LED, followed by a 72 hour incubation period, then an XTT assay was performed (ESI Fig. 11†).54 Absorbance values were normalized such that non-irradiated BORap-treated cells were equal to 100%. No decrease in normalized absorbance is observed in cells treated with BORap even upon irradiation and oxidation. A decrease in rapamycin cell viability was observed for non-transfected cells, but no effect due to irradiation was observed. This is consistent with literature reports of rapamycin-induced cell toxicity in a variety of different cell lines.55,56
In summary, we developed the BODIPY-modified rapamycin analog BORap, which works as both a dimerizer of FKBP/FRB – thereby activating protein function upon addition – and a ROS generator – thereby deactivating protein function upon irradiation. We have utilized this singlet oxygen-generating small molecule to inactivate FKBP-FRB induced protein–protein interactions and the resulting activities through exposure to 530 nm light, thereby offering a unique solution for achieving reversibility of biological processes placed under rapamycin control. We applied BORap to the reversible control of a luciferase enzyme, a cysteine protease, and membrane translocation in mammalian cells. While formation of the FKBP-rapamycin-FRB ternary complex has proven to be a useful tool for activating split-proteins, inducing protein–protein interactions, and controlling protein localization this has up until this point been an irreversible process. Studies into the mode-of-action of BORap confirmed singlet oxygen-generation of the BODIPY chromophore and resulting non-proteasomal inactivation of proteins of interest. Future work will lead to an expansion of this approach to different chromophores, additional protein targets, controlling function in multicellular model organisms, and investigating the precise amino acid oxidation mechanisms that occur.57,58 We hope that BORap will find widespread application in the targeted oxidation of proteins given the extensive number of biological processes that have been placed under control of rapamycin as a CID. Beyond being a highly functional off-switch, it also holds promise in the mapping of protein–protein interactions, using the BORap-FKBP module as a template for localizing a ROS chromophore to any protein of interest.
Data availability
Detailed experimental protocols, compound characterization data, NMR spectra, primer sequences, and supplementary figures are available in the ESI.†
Author contributions
T. M. C. and C. P. H. designed and performed the experiments. T. J. H. synthesized BORap. A. D. designed the project and oversaw the experiments. T. M. C., C. P. H., and A. D. wrote the manuscript.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
We thank the National Science Foundation (CHE-1904972) for support of this research. T. M. C. was supported by a National Science Foundation Graduate Research Fellowship. Cluc-FKBP (Addgene plasmid 31184) and FRB-NLuc (Addgene plasmid 31181) were gifts from David Piwnica-Worms. Lyn11-FKBP-FKBP-CFP (Addgene plasmid 20149) and YFP-FRB (Addgene plasmid 20148) were gifts from Tobias Meyer. Split TEV protease plasmids were gifts from Stephen Ikeda.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc04464h
References
- Wang P. Gao Z. Yuan M. Zhu J. Wang F. Mechanically linked poly[2]rotaxanes constructed from the benzo-21-crown-7/secondary ammonium salt recognition motif. Polym. Chem. 2016;7(22):3664–3668. doi: 10.1039/C6PY00494F. [DOI] [Google Scholar]
- Voß S. Klewer L. Wu Y. W. Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr. Opin. Chem. Biol. 2015;28:194–201. doi: 10.1016/j.cbpa.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Wu H. D. Kikuchi M. Dagliyan O. Aragaki A. K. Nakamura H. Dokholyan N. V. Umehara T. Inoue T. Rational design and implementation of a chemically inducible heterotrimerization system. Nat. Methods. 2020;17(9):928–936. doi: 10.1038/s41592-020-0913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. J. Puhl III H. L. Ikeda S. R. Rapid Modification of Proteins Using a Rapamycin-Inducible Tobacco Etch Virus Protease System. PLoS One. 2009;4(10):e7474. doi: 10.1371/journal.pone.0007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J. E. Morgan C. W. Minogue C. E. Hebert A. S. Coon J. J. Wells J. A. A Split-Abl Kinase for Direct Activation in Cells. Cell Chem. Biol. 2017;24(10):1250–1258. doi: 10.1016/j.chembiol.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B. Volz S. E. Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33(2):139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T. Heo W. D. Grimley J. S. Wandless T. J. Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2005;2(6):415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T. Johnson C. A. Gestwicki J. E. Kumar A. Conditionally controlling nuclear trafficking in yeast by chemical-induced protein dimerization. Nat. Protoc. 2010;5(11):1831–1843. doi: 10.1038/nprot.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putyrski M. Schultz C. Switching Heterotrimeric G Protein Subunits with a Chemical Dimerizer. Chem. Biol. 2011;18(9):1126–1133. doi: 10.1016/j.chembiol.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Putyrski M. Schultz C. Protein translocation as a tool: the current rapamycin story. FEBS Lett. 2012;586(15):2097–2105. doi: 10.1016/j.febslet.2012.04.061. [DOI] [PubMed] [Google Scholar]
- Bayle J. H. Grimley J. S. Stankunas K. Gestwicki J. E. Wandless T. J. Crabtree G. R. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 2006;13(1):99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Liu K. J. Arron J. R. Stankunas K. Crabtree G. R. Longaker M. T. Chemical rescue of cleft palate and midline defects in conditional GSK-3β mice. Nature. 2007;446(7131):79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- Rivera V. M. Clackson T. Natesan S. Pollock R. Amara J. F. Keenan T. Magari S. R. Phillips T. Courage N. L. Cerasoli F. Holt D. A. Gilman M. A humanized system for pharmacologic control of gene expression. Nat. Med. 1996;2(9):1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- Rivera V. M. Gao G.-p. Grant R. L. Schnell M. A. Zoltick P. W. Rozamus L. W. Clackson T. Wilson J. M. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105(4):1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- Bardhan A. Deiters A. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol. 2019;57:164–175. doi: 10.1016/j.sbi.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klewer L. Wu Y. W. Light-Induced Dimerization Approaches to Control Cellular Processes. Chemistry. 2019;25(54):12452–12463. doi: 10.1002/chem.201900562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov A. V. Zou Y. Shirvanyants D. Kota P. Dokholyan N. V. Young D. D. Hahn K. M. Deiters A. Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. J. Am. Chem. Soc. 2011;133(3):420–423. doi: 10.1021/ja109630v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N. Ueno T. Pohlmeyer C. Nagano T. Inoue T. A Photocleavable Rapamycin Conjugate for Spatiotemporal Control of Small GTPase Activity. J. Am. Chem. Soc. 2011;133(1):12–14. doi: 10.1021/ja108258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A. Zou Y. Shirvanyants D. Zhang J. Samanta S. Mantravadi P. K. Dokholyan N. V. Deiters A. Light-cleavable rapamycin dimer as an optical trigger for protein dimerization. Chem. Commun. 2015;51(26):5702–5705. doi: 10.1039/C4CC09442E. [DOI] [PubMed] [Google Scholar]
- Sadovski O. Jaikaran A. S. I. Samanta S. Fabian M. R. Dowling R. J. O. Sonenberg N. Woolley G. A. A collection of caged compounds for probing roles of local translation in neurobiology. Bioorg. Med. Chem. 2010;18(22):7746–7752. doi: 10.1016/j.bmc.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S. Laketa V. Stein F. Rutkowska A. MacNamara A. Depner S. Klingmüller U. Saez-Rodriguez J. Schultz C. A rapidly reversible chemical dimerizer system to study lipid signaling in living cells. Angew. Chem., Int. Ed. Engl. 2014;53(26):6720–6723. doi: 10.1002/anie.201402294. [DOI] [PubMed] [Google Scholar]
- Aonbangkhen C. Zhang H. Wu D. Z. Lampson M. A. Chenoweth D. M. Reversible Control of Protein Localization in Living Cells Using a Photocaged-Photocleavable Chemical Dimerizer. J. Am. Chem. Soc. 2018;140(38):11926–11930. doi: 10.1021/jacs.8b07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifferer M. Feng S. Stein F. Tischer C. Schultz C. Reversible chemical dimerizer-induced recovery of PIP2 levels moves clathrin to the plasma membrane. Bioorg. Med. Chem. 2015;23(12):2862–2867. doi: 10.1016/j.bmc.2015.03.048. [DOI] [PubMed] [Google Scholar]
- Lin Y. C. Nihongaki Y. Liu T. Y. Razavi S. Sato M. Inoue T. Rapidly reversible manipulation of molecular activity with dual chemical dimerizers. Angew. Chem., Int. Ed. Engl. 2013;52(25):6450–6454. doi: 10.1002/anie.201301219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney T. M. Horst T. J. Hankinson C. P. Deiters A. Synthesis and application of light-switchable arylazopyrazole rapamycin analogs. Org. Biomol. Chem. 2019;17(36):8348–8353. doi: 10.1039/c9ob01719d. [DOI] [PubMed] [Google Scholar]
- Jay D. G. Selective destruction of protein function by chromophore-assisted laser inactivation. Proc. Natl. Acad. Sci. U. S. A. 1988;85(15):5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista M. S. Cadet J. Di Mascio P. Ghogare A. A. Greer A. Hamblin M. R. Lorente C. Nunez S. C. Ribeiro M. S. Thomas A. H. Vignoni M. Yoshimura T. M. Type I and Type II Photosensitized Oxidation Reactions: Guidelines and Mechanistic Pathways. Photochem. Photobiol. 2017;93(4):912–919. doi: 10.1111/php.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. Rajfur Z. Vitriol E. Hahn K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 2008;18(9):443–450. doi: 10.1016/j.tcb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett B. S. Stadtman E. R. Protein Oxidation in Aging, Disease, and Oxidative Stress. J. Biol. Chem. 1997;272(33):20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Bulina M. E. Chudakov D. M. Britanova O. V. Yanushevich Y. G. Staroverov D. B. Chepurnykh T. V. Merzlyak E. M. Shkrob M. A. Lukyanov S. Lukyanov K. A. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24(1):95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- Qi Y. B. Garren E. J. Shu X. Tsien R. Y. Jin Y. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc. Natl. Acad. Sci. U. S. A. 2012;109(19):7499–7504. doi: 10.1073/pnas.1204096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Y. Tanaka M. Vecchio D. Garcia-Diaz M. Chang J. Morimoto Y. Hamblin M. R. Photodynamic therapy induces an immune response against a bacterial pathogen. Expert Rev. Clin. Immunol. 2012;8(5):479–494. doi: 10.1586/eci.12.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K. Matsuda T. McDougall M. Klaubert D. H. Hasegawa A. Los G. V. Wood K. V. Miyawaki A. Nagai T. Chromophore-Assisted Light Inactivation of HaloTag Fusion Proteins Labeled with Eosin in Living Cells. ACS Chem. Biol. 2011;6(5):401–406. doi: 10.1021/cb100431e. [DOI] [PubMed] [Google Scholar]
- Lee J. Yu P. Xiao X. Kodadek T. A general system for evaluating the efficiency of chromophore-assisted light inactivation (CALI) of proteins reveals Ru(II) tris-bipyridyl as an unusually efficient “warhead”. Mol. BioSyst. 2008;4(1):59–65. doi: 10.1039/b712307h. [DOI] [PubMed] [Google Scholar]
- He J. Wang Y. Missinato M. A. Onuoha E. Perkins L. A. Watkins S. C. St Croix C. M. Tsang M. Bruchez M. P. A genetically targetable near-infrared photosensitizer. Nat. Methods. 2016;13(3):263–268. doi: 10.1038/nmeth.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo T. Urano Y. Ishitsuka Y. Maniwa F. Nagano T. Highly Efficient and Photostable Photosensitizer Based on BODIPY Chromophore. J. Am. Chem. Soc. 2005;127(35):12162–12163. doi: 10.1021/ja0528533. [DOI] [PubMed] [Google Scholar]
- Banfi S. Nasini G. Zaza S. Caruso E. Synthesis and photo-physical properties of a series of BODIPY dyes. Tetrahedron. 2013;69(24):4845–4856. [Google Scholar]
- Wagner R. Mollison K. W. Liu L. Henry C. L. Rosenberg T. A. Bamaung N. Tu N. Wiedeman P. E. Or Y. Luly J. R. Lane B. C. Trevillyan J. Chen Y. W. Fey T. Hsieh G. Marsh K. Nuss M. Jacobson P. B. Wilcox D. Carlson R. P. Carter G. W. Djuric S. W. Rapamycin analogs with reduced systemic exposure. Bioorg. Med. Chem. Lett. 2005;15(23):5340–5343. doi: 10.1016/j.bmcl.2005.06.106. [DOI] [PubMed] [Google Scholar]
- Carloni P. Damiani E. Greci L. Stipa P. Tanfani F. Tartaglini E. Wozniak M. On the use of 1,3-diphenylisobenzofuran (DPBF). Reactions with carbon and oxygen centered radicals in model and natural systems. Res. Chem. Intermed. 1993;19(5):395–405. [Google Scholar]
- Luker K. E. Smith M. C. P. Luker G. D. Gammon S. T. Piwnica-Worms H. Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U. S. A. 2004;101(33):12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S. K. White S. Orth A. P. Reisdorph R. Miraglia L. Thomas R. S. DeJesus P. Mason D. E. Huang Q. Vega R. Yu D.-H. Nelson C. G. Smith B. M. Terry R. Linford A. S. Yu Y. Chirn G.-w. Song C. Labow M. A. Cohen D. King F. J. Peters E. C. Schultz P. G. Vogt P. K. Hogenesch J. B. Caldwell J. S. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2003;100(21):12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M. C. Rossner M. J. Split protein biosensor assays in molecular pharmacological studies. Drug Discovery Today. 2016;21(3):415–429. doi: 10.1016/j.drudis.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Morgan C. W. Diaz J. E. Zeitlin S. G. Gray D. C. Wells J. A. Engineered cellular gene-replacement platform for selective and inducible proteolytic profiling. Proc. Natl. Acad. Sci. U. S. A. 2015;112(27):8344–8349. doi: 10.1073/pnas.1504141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. C. Mahrus S. Wells J. A. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142(4):637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole L. B. Nelson K. J. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr. Opin. Chem. Biol. 2008;12(1):18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held J. M. Gibson B. W. Regulatory Control or Oxidative Damage? Proteomic Approaches to Interrogate the Role of Cysteine Oxidation Status in Biological Processes. Mol. Cell. Proteomics. 2012;11(4):R111.013037. doi: 10.1074/mcp.R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. K. Lin M. Z. On the cutting edge: protease-based methods for sensing and controlling cell biology. Nat. Methods. 2020;17(9):885–896. doi: 10.1038/s41592-020-0891-z. [DOI] [PubMed] [Google Scholar]
- Nicholls S. B. Chu J. Abbruzzese G. Tremblay K. D. Hardy J. A. Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J. Biol. Chem. 2011;286(28):24977–24986. doi: 10.1074/jbc.M111.221648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T. Höhn A. Grune T. The proteasome and the degradation of oxidized proteins: part II – protein oxidation and proteasomal degradation. Redox Biol. 2014;2:99–104. doi: 10.1016/j.redox.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. B. Myung J. Sin N. Crews C. M. Proteasome inhibition by the natural products epoxomicin and dihydroeponemycin: insights into specificity and potency. Bioorg. Med. Chem. Lett. 1999;9(23):3335–3340. doi: 10.1016/s0960-894x(99)00612-5. [DOI] [PubMed] [Google Scholar]
- Xia H. Redman C. M. Differential Degradation of the Three Fibrinogen Chains by Proteasomes: Involvement of Sec61p and Cytosolic Hsp70. Arch. Biochem. Biophys. 2001;390(1):137–145. doi: 10.1006/abbi.2001.2374. [DOI] [PubMed] [Google Scholar]
- Bancirova M. Sodium azide as a specific quencher of singlet oxygen during chemiluminescent detection by luminol and Cypridina luciferin analogues. Luminescence. 2011;26(6):685–688. doi: 10.1002/bio.1296. [DOI] [PubMed] [Google Scholar]
- Wang X. Perez E. Liu R. Yan L.-J. Mallet R. T. Yang S.-H. Pyruvate protects mitochondria from oxidative stress in human neuroblastoma SK-N-SH cells. Brain Res. 2007;1132:1–9. doi: 10.1016/j.brainres.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney T. M. Deiters A. Optical control of protein phosphatase function. Nat. Commun. 2019;10(1):4384. doi: 10.1038/s41467-019-12260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar Z. Millward A. B. Tse W. Demaine A. G. p21(WAF1/CIP1) Expression is Differentially Regulated by Metformin and Rapamycin. Int. J. Chronic Dis. 2014;2014:327640. doi: 10.1155/2014/327640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisching G. R. Loos B. Botha M. H. Engelbrecht A. M. The role of mTOR during cisplatin treatment in an in vitro and ex vivo model of cervical cancer. Toxicology. 2015;335:72–78. doi: 10.1016/j.tox.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Davies M. J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003;305(3):761–770. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- Pimenta F. M. Chiappetta G. Le Saux T. Vinh J. Jullien L. Gautier A. Chromophore Renewal and Fluorogen-Binding Tags: A Match Made to Last. Sci. Rep. 2017;7(1):12316. doi: 10.1038/s41598-017-12400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed experimental protocols, compound characterization data, NMR spectra, primer sequences, and supplementary figures are available in the ESI.†