Abstract
Hepatitis B virus X (HBx) protein has been reported as a key protein regulating the pathogenesis of HBV-induced hepatocellular carcinoma (HCC). Recent evidence has shown that HBx is implicated in the activation of autophagy in hepatic cells. Nevertheless, the precise molecular and cellular mechanism by which HBx induces autophagy is still controversial. Herein, we investigated the molecular and cellular mechanism by which HBx is involved in the TRAF6-BECN1-Bcl-2 signaling for the regulation of autophagy in response to TLR4 stimulation, therefore influencing the HCC progression. HBx interacts with BECN1 (Beclin 1) and inhibits the association of the BECN1-Bcl-2 complex, which is known to prevent the assembly of the pre-autophagosomal structure. Furthermore, HBx enhances the interaction between VPS34 and TRAF6-BECN1 complex, increases the ubiquitination of BECN1, and subsequently enhances autophagy induction in response to LPS stimulation. To verify the functional role of HBx in liver cancer progression, we utilized different HCC cell lines, HepG2, SK-Hep-1, and SNU-761. HBx-expressing HepG2 cells exhibited enhanced cell migration, invasion, and cell mobility in response to LPS stimulation compared to those of control HepG2 cells. These results were consistently observed in HBx-expressed SK-Hep-1 and HBx-expressed SNU-761 cells. Taken together, our findings suggest that HBx positively regulates the induction of autophagy through the inhibition of the BECN1-Bcl-2 complex and enhancement of the TRAF6-BECN1-VPS34 complex, leading to enhance liver cancer migration and invasion.
Keywords: Hepatitis B virus, Autophagy, TNF receptor-associated factor 6, Beclin-1, Liver neoplasms
INTRODUCTION
Hepatitis B virus X (HBx) protein is functionally implicated in the pathogenesis of Hepatitis B virus (HBV)-induced hepatocellular carcinoma (HCC) (1,2). The HBx protein-induced HCC has been attributed to the epigenetic modification, including DNA hypermethylation, anti-apoptotic effects through the inhibition of p53-mediated apoptosis, telomerase activity by the increases of telomerase reverse transcriptase (TERT) expression, insulin-like growth factor II (IGF-II)-induced malignant transformation of hepatocytes and the regulation of the DNA repair by nucleotide excision repair (3). Accumulating evidence has recently suggested that autophagy is functionally implicated in the initiation and progression of HCC (4,5,6). Autophagy has multiple roles in the tumorigenesis and metastasis of HCC, and liver cellular contexts, such as liver homeostasis, genome stability in the liver cells, and the prevention of malignant transformation by removing harmful mitochondria and transformed liver cells (4,5,6,7). The Beclin1 (BECN1)–Vps34 complex and BECN1-Bcl-2 complex play pivotal roles in the biochemical and cellular mechanisms of autophagy induction in response to various cellular conditions, including nutrient deprivation and extra- or intracellular stress and signals (8,9,10,11).
TLR is well known to stimulate autophagy and functionally enhances host innate immune responses in the tumor environment (12,13). TLR signaling has been functionally implicated in the initiation, progression, and metastasis of HCC through autophagy induction (14,15,16,17). Recent evidence has suggested that TLR3/4-induced autophagy enhances cancer migration and invasion through the TRAF6-BECN1 signaling complex (18,19,20). BECN1 protein plays a central role in the induction of autophagy through the formation of TRAF6-BECN1 and BECN1-Vps34 complexes (18,19,20,21). In this case, TRAF6 interacts with BECN1 and induces the ubiquitination of BECN1, leading to the induction of autophagy (18,19,20). Similarly, BECN1-Vps34 proteins, along with Vps15, play an essential role in autophagy (21,22). On the contrast, BECN1 interacts with Bcl-2 to inhibit autophagy (22). It has been reported that mutations of either the BH3-only domain within the BECN1 or the BH3 receptor domain within the Bcl-2 or Bcl-XL, interrupt the formation of the BECN1–Bcl-2 complex and stimulate autophagy induction (22,23), suggesting that Bcl-2 is a negative regulator in the BECN1-dependent autophagy induction. Several reports have suggested that HBx is functionally involved in autophagy induction (24,25). During starvation-induced autophagy, HBx activates class III phosphatidylinositol 3-kinase (PtdIns3K) or induces the up-regulation of BECN1 expression, resulting in the enhancement of autophagy (24,25). Nevertheless, the precise molecular and cellular mechanism by which HBx regulates BECN1-Vps34 or BECN1-Bcl-2 complex formation remains elusive.
In this study, we investigated the functional role of HBx in the autophagy regulated by BECN1-Vps34 or BECN1-Bcl-2 complex. Our biochemical studies revealed that HBx enhances the interaction between Vps34 and TRAF6-BECN1 complex, therefore increase the ubiquitination of BECN1, and interrupt the formation of the BECN1-Bcl-2 complex, leading to enhanced autophagy induction. Notably, HBx-expressing human hepatocyte carcinoma HepG2, HBx-expressed human hepatic adenocarcinoma SK-Hep-1, and HBx-expressed human HCC SNU-761 cells markedly increased cancer migration, invasion, and cancer colony formation induced by TLR4 stimulation. These results suggest that HBx positively regulates the induction of autophagy through the enhancement of TRAF6-BECN1-VPS34 complex and inhibition of BECN1-Bcl-2 complex, thereby promoting liver cancer progression induced by TLR4 stimulation.
MATERIALS AND METHODS
Cells
Human embryonic kidney (HEK) 293T cells (ATCC, CRL-11268) and human hepatic adenocarcinoma SK-Hep-1 cells (ATCC, HTB-52) were cultured and maintained in DMEM (Thermo Fisher Scientific, 11965092) with 10% fetal bovine serum (FBS). Human HCC SNU-761 cells were purchased from the Korean Cell Line Bank (Seoul, Korea) and cultured according to the following recommendation (https://cellbank.snu.ac.kr). Control (Ctrl) HepG2 and HBx-HepG2 cells were obtained from Dr. K-H. Kim (Sungkyunkwan University School of Medicine, Suwon, Republic of Korea) and were cultured in DMEM containing 10% heat inactivated Fetal Bovine Serum (Capricorn; Ebsdorfergrund, Germany) and 1% of penicillin-streptomycin (Gibco; BRL, Grand Island, NY, USA) at 37°C in 5% CO2 (26).
Antibodies and reagents
Anti-Myc (2276), anti-GAPDH (2118), and anti-LC3A/B (4108) were purchased from Cell Signaling Technology (MA, USA). Anti-Flag (SAB4200071) was purchased from Sigma-Aldrich (St Louis, MO, USA). Anti-HA and anti-HBx antibodies were purchased from Abcam (Cambridge, MA). Lipopolysaccharide (LPS; serotype 0128:B12), chloroquine (CQ; C6628), dimethyl sulfoxide (DMSO; 472301), puromycin (P8833), paraformaldehyde (P6148), Triton X-100 (T8787), 3-methyladenine (3-MA;M9281), gentamicin (G1272), deoxycholate (D6750), and Dulbecco's phosphate-buffered saline (DPBS; D8537) were purchased from Sigma-Aldrich. Lipofectamine 2000 (11668019) was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Plasmid constructs
Flag-TRAF6 (21624), Flag-BECN1 (24388), HA-Vps34 (86749), and Flag-Bcl-2 (18003) were purchased from Addgene (Cambridge, MA, USA). HA-HBx and Flag-HBx were obtained from Dr. K-H. Kim (Sungkyunkwan University School of Medicine). HA-Ub plasmids were obtained from Dr. J. H. Shim (University of Massachusetts Medical School, USA). Using Flag-BECN1 plasmid, full-length Myc-BECN1 constructs were cloned into a pCMV-3Tag-7 (240202; Agilent technologies, Santa Clara, CA, USA). Using Flag-BECN1 wild type (WT) plasmid, Flag-BECN1 truncated mutants, Flag-BECN1 1-269 and Flag-BECN1 1-127, were generated by PCR (Polymerase Chain Reaction). HA-Vps34 truncated mutants, HA-Vps34 1-531 and HA-Vps34 1-260, were generated by PCR using HA-Vps34 WT plasmid.
Western blotting (WB) and immunoprecipitation (IP) assays
WB and IP assays were performed as previously described (19,20,27,28,29,30). Briefly, HEK-293T cells were seeded in 6-well plates, transfected, and treated, as described in the Results and Figures. After 38 to 48 hours, the cells were collected, and the cell lysates were immunoprecipitated with anti-Myc or anti-Flag antibodies. The IP complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 6–10%) and immune-probed with the different antibodies, as indicated in the text and Figures. For the ubiquitination assay, mock vector, Myc-BECN1, Flag-TRAF6, HA-Ub, HA-Vps34, and different concentrations of Flag-HBx were transfected into HEK-293T cells, as described in the text and Figures. Cell lysates were immunoprecipitated with anti-Myc Ab and probed with the different antibodies indicated in the text and Figures. Mock or Flag-HBx plasmid were transfected into SK-Hep-1 and SNU-761 cells, and the expression of Flag-HBx protein was confirmed through WB with anti-Flag Ab. Ctrl HepG2 and HBx-expressing HepG2 cells were treated with or without vehicle or CQ (10 μM) in the presence or absence of LPS (10 μg/mL) for 6 hr. The cell lysates were immunoblotted with anti-LC3A/B Ab and anti-GAPDH as a loading control.
Wound-healing migration assay
A wound-healing migration assay was performed following previous protocols (27,29). Briefly, Ctrl HepG2, HBx-expressing HepG2, Ctrl SK-Hep-1, HBx-expressed SK-Hep-1, Ctrl SNU-761, and HBx-expressed SNU-761 cells were seeded in 12-well plates and cultured to reach confluence. The cell monolayers were gently scratched and washed with culture medium. After the floating cells and debris were removed, the cells were treated with a vehicle (DMSO), 3-MA (5mM), and CQ (10 μM) in the presence or absence of LPS (10 μg/mL). Cell images were captured after culturing at different time periods as indicated in each experiment.
Transwell invasion assay
The Transwell invasion assay was performed following previous protocols (27,29). Briefly, Ctrl HepG2, HBx-expressing HepG2, Ctrl SK-Hep-1, HBx-expressed SK-Hep-1, Ctrl SNU-761, and HBx-expressed SNU-761 cells were suspended in culture medium (200 μL) without FBS, and cells were added to the upper compartments of a 24-well Transwell® chamber containing polycarbonate filters with 8-mm pores and coated with 60 mL of Matrigel (Sigma Aldrich, E1270; 1:9 dilution). Culture medium with 10% FBS was added to the lower chambers and incubated for 24 h. Cells in the upper compartments were removed, washed with PBS, and fixed. The invaded cells were stained with crystal violet (Sigma-Aldrich, C0775) and quantified by counting the number of fluorescent cells.
Single cell migration assay
Single cell migration assay was performed following the protocols provided by the Bio-protocol (www.bio-protocol.org/e3586). Briefly, Ctrl HepG2, HBx-expressing HepG2, Ctrl SK-Hep-1, and HBx-expressed SK-Hep-1 cells were seeded into 6-well culture plate, treated with vehicle (DMSO), and 3-MA (5mM) in the presence or absence of LPS (10 μg/mL), and allowed to attach overnight at 37°C and 5% CO2. 5-10 single cells were selected in multiple culture fields, and time-lapse imaging analysis was performed at different times using phase-contrast microscope (Olympus IX71 inverted microscope). Data analysis was performed following protocols provided by the Bio-protocol (www.bio-protocol.org/e3586).
Anchorage-independent soft agar colony formation assay
Anchorage-independent soft agar colony formation assay was performed following previous protocols (31). Briefly, Ctrl HepG2 and HBx-expressing HepG2 cells (1.0×104 cells per well) mixed with 0.3% Difco Noble Agar (BD) in complete medium were plated on the top of 0.5% agar layer in a 6-well plate with complete medium. Growth medium (1.5 mL) included with vehicle (DMSO), LPS (10 μg/mL), 3-MA (5mM), or CQ (10 μM) was added on the top of the layer and the cells were incubated at 37°C for 4 weeks.
Colony formation assay
The ability of a single cell to grow into a colony was passed through a colony formation assay as previously described (26,32). Ctrl HepG2, HBx-expressing HepG2, Ctrl SNU-761, and HBx-expressed SNU-761 cells were harvested with trypsin-EDTA and re-suspended in a singular form. The 1×103 cells (per well) were plated in a 6-well plate and treated with a vehicle (DMSO), 3-MA (5mM), and CQ (10 μM) in the presence or absence of LPS (10 μg/mL). After incubation for 18 days in HepG2 or for 12 days in SNU-761 cells, colonies were stained with 0.5% crystal violet (Sigma) for 30 min at room temperature.
Statistical analysis
The in vitro data were expressed as the mean±SEM of triplicate samples. Statistical significance was analyzed using ANOVA or Student's t-test in GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA).
RESULTS
HBx inhibits the interaction between BECN1 and Bcl-2
In the present study, we investigated whether HBx induces autophagy. We first examined the molecular role of HBx in the BECN1-Bcl-2 complex negative regulation of autophagy induction. Myc-BECN1 interacted with Flag-Bcl-2 (Fig. 1A, lane 3). Additionally, HA-HBx interacted with Flag-Bcl-2 (Fig. 1B, lane 3) and Flag-BECN1 (Fig. 1C, lane 3). Since Bcl-2 interacts with the N-terminal Bcl-2 binding domain of BECN1 (11), we sought to identify the interaction site of HBx on BECN1 using BECN1 truncated mutants (Fig. 1D). HA-HBx interacted with Flag-BECN1 WT, Flag-BECN1 1-269 mutant, and Flag-BECN1 1-127 mutant (Fig. 1E, lanes 6-8), indicating that HBx interacts with the N-terminal Bcl-2 binding domain of BECN1 (Fig. 1F). Given the results, we further investigated whether HBx affects the interaction between BECN1 and Bcl-2. Flag-Bcl-2 interacted with Myc-BECN1 in the absence of Flag-HBx (Fig. 1G, lane 1). Importantly, marked attenuations of the interaction between BECN1 and Bcl-2 were observed in the presence of Flag-HBx in a dose-dependent manner (Fig. 1G, lanes 2-4 in Flag-Bcl-2), whereas there was a significant increase in the interaction between BECN1 and HBx in a dose-dependent manner (Fig. 1G, lanes 2-4 in Flag-HBx). It has been reported that BECN1 interacts with Bcl-2 and inhibits autophagy formation (21,33,34) (Fig. 1H, left). Our results suggest that the interaction between HBx and BECN1 interrupts the association of Bcl-2 to BECN1 and may be positively involved in the autophagy induction (Fig. 1H, right).
Figure 1. HBx inhibits the interaction between BECN1 and Bcl-2 protein. (A) Mock, Myc-BECN1, and Flag-Bcl-2 plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-Myc Ab followed by IB using anti-Flag or anti-Myc Ab. (B) Mock, HA-HBx, and Flag-Bcl-2 plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-HA Ab. (C) Mock, Flag-BECN1, and HA-HBx plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-Flag Ab. (D) Truncated mutants of BECN1, as indicated, were generated as described in Materials and Methods. (E) Mock, Flag-BECN1 WT, Flag-BECN1 1-269, Flag-BECN1 1-127, and HA-HBx plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-Flag Ab, followed by IB using anti-Flag or anti-HA Ab. (F) A schematic model showing the interaction between BECN1 and HBx protein. (G) Mock, Myc-BECN1, Flag-Bcl-2, and different concentrations of Flag-HBx plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-Myc Ab followed by IB using anti-Flag or anti-Myc Ab. (H) A model showing how HBx inhibits the interaction between BECN1 and Bcl-2. BECN1 interacts with Bcl-2 therefore inhibiting the autophagy induction (left). In contrast, HBx interacts with BECN1 and inhibits the association of BECN1-Bcl-2 complex, leading to the autophagy induction (right).
HBx induces the formation of the BECN1-HBx-VPS34 complex
The BECN1-Vps34 (PI3KC3) complex with Atg14L protein is essential for autophagosome formation (18,19,20,21,22). The K63-linked ubiquitination of BECN1 by TRAF6 facilitates oligomerization of BECN1 and the activation of PI3KC3 (21). Next, we investigated whether HBx affects the formation of the BECN1-Vps34 complex, thereby regulates autophagy. HA-Vps34 interacted with Myc-BECN1 (Fig. 2A, lane 4) and Flag-HBx (Fig. 2B, lane 4). To identify the interaction site of Vps34 to BECN1 or BECN1 to Vps34, IP assay was performed with BECN1 (Fig. 1D) or Vps34 truncated mutants (Fig. 2C). HA-Vps34 interacted with Flag-BECN1 WT and Flag-BECN1 1-269 mutants (Fig. 2D, lanes 5 and 6), whereas no significant interaction was observed with Flag-BECN1 1-127 mutant (Fig. 2D, lane 7), indicating that Vps34 interacts with the coiled-coil domain of BECN1 (Fig. 2D, down). Additionally, Flag-BECN1 interacted with HA-Vps34 WT, HA-Vps34 1-531 mutant, and HA-Vps34 1-260 mutant (Fig. 2E, lanes 5-7), indicating that the C2 domain of Vps34 interacts with the coiled-coil domain of BECN1 (Fig. 2E, down). We next evaluated the interaction site of HBx to Vps34. Flag-HBx interacted with HA-Vps34 WT, HA-Vps34 1-531 mutant, and HA-Vps34 1-260 mutant (Fig. 2F, lanes 5-7), indicating that HBx interacts with the C2 domain of Vps34 (Fig. 2F, down).
Figure 2. HBx interacts with Vps34-BECN1 complex. (A) Mock, Myc-BECN1, and HA-Vps34 plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-Myc Ab followed by IB using anti-Myc or anti-HA Ab. (B) Mock, Flag-HBx, and HA-Vps34 plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-Flag Ab. (C) Truncated mutants of Vps34, were generated as described in Materials and Methods. (D) Mock, HA-Vps34, Flag-BECN1 WT, Flag-BECN1 1-269, and Flag-BECN1 1-127 plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-Flag Ab, and IB assay was performed with anti-Flag or anti-HA Ab. A schematic model for the interaction between BECN1 and Vps34 protein is shown (down). (E) Mock, Flag-BECN1, HA-Vps34 WT, HA-Vps34 1-531, and HA-Vps34 1-260 plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-Flag Ab, and IB assay was performed with anti-Flag or anti-HA Ab. A schematic model for the interaction between BECN1 and Vps34 protein is shown (down). (F) Mock, Flag-HBx, HA-Vps34 WT, HA-Vps34 1-531, and HA-Vps34 1-260 plasmids were transfected into HEK-293T cells, as indicated. IP assay was performed with anti-Flag Ab, and IB assay was performed with anti-Flag or anti-HA Ab. A schematic model for the interaction between HBx and Vps34 protein is shown (down).
HBx induces BECN1-VPS34 formation and the ubiquitination of BECN1, leading to enhanced autophagy induction in response to TLR4 stimulation
Given that HBx interacted with the Bcl-2 domain of BECN1 and the C2 domain of Vps34 (Fig. 3A), we evaluated if HBx is positively or negatively involved in the molecular association of BECN1-Vps34 complex. HA-Vps34 and Myc-BECN1 plasmids were transfected into HEK-293T cells with different concentrations of Flag-HBx, as shown in Fig. 3B, and IP assay was performed with an anti-HA Ab. As expected, HA-Vps34 interacted with Myc-BECN1 in the absence of Flag-HBx (Fig. 3B, lane 3). Interestingly, the interaction between HA-Vps34 and Myc-BECN1 significantly increased in the presence of Flag-HBx (Fig. 3B, lanes 4-6, Myc-BECN1). Moreover, the interaction between HA-Vps34 and Flag-HBx was markedly enhanced in a dose-dependent manner (Fig. 3B, lanes 4-6, Flag-HBx), suggesting that HBx facilitates the molecular association of BECN1-Vps34, as shown in Fig. 3A.
Figure 3. HBx enhances the ubiquitination of BECN1 and autophagy induction. (A) A schematic model showing the association among HBx, BECN1, and Vps34 protein. (B) Mock, HA-Vps34, Myc-BECN1, and different concentrations of Flag-HBx plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-HA Ab followed by IB using anti-HA, anti-Flag, or anti-Myc Ab. (C) Mock, Myc-BECN1, Flag-TRAF6, HA-Ub, HA-Vps34, and different concentrations of Flag-HBx plasmids were transfected into HEK-293T cells, as indicated. At 38 h post-transfection, the transfected cells were extracted and the cell lysates were subjected to IP with anti-Myc Ab followed by IB using anti-HA or anti-Myc Ab. (D, E) Ctrl HepG2 and HBx-HepG2 cells were treated with or without LPS (5 or 10 µg/mL) in the presence or absence of CQ (10 μM) for 6 hours, as indicated. The cells were lysed and subjected to SDS-PAGE followed by immunoblotting with LC3-I/-II, HBx, or GAPDH antibodies (D). Band intensity was quantified using Image J software (E) (*p<0.05, **p<0.01, ±SEM, n=3). (F) A schematic model showing how HBx facilitate the association of BECN1-Vps34 and the ubiquitination of BECN1, therefore enhancing autophagy induction.
Given that HBx enhanced the interaction between BECN1 and Vps34, we further evaluated whether HBx affects the ubiquitination of BECN1 associated with Vps34 using a ubiquitination assay. Myc-BECN1, Flag-TRAF6, HA-Ub, and HA-Vps34 plasmids were transfected into HEK-293T cells with different concentrations of Flag-HBx, as indicated (Fig. 3C). The ubiquitination of BECN1 was increased in the presence of Flag-TRAF6 and the absence of HA-Vps34 and Flag-HBx (Fig. 3C, lane 4). Specially, a significant increase in the ubiquitination of BECN1 was observed in the presence of HA-Vps34 (Fig. 3C, lane 5 vs. lane 4). Interestingly, the ubiquitination of BECN1 was significantly enhanced in the presence of HA-Vps34 and Flag-HBx (Fig. 3C, lane 6 and 7), strongly suggesting that HBx facilitates the ubiquitination of BECN1 through the enhancement of BECN1-Vps34 complex. TLR4 induces the autophagy through the ubiquitination of BECN1 by TRAF6, leading to increase cancer cells migration and invasion (19,20,27,28,29,30). To verify the functional role of HBx in autophagy induction stimulated by TLR4, Ctrl HepG2 and HBx-expressing HepG2 (HBx-HepG2) cells were treated with different concentrations of LPS (5 or 10 µg/ml) in the presence or absence of autophagy inhibitor CQ, as indicated in Fig. 3D, and the levels of LC3-II were evaluated. The basal level of LC3-II was slightly higher in HBx-HepG2 treated with vehicle than that of Ctrl HepG2 treated with vehicle (Fig. 3D and E, HBx-HepG2 with vehicle vs. Ctrl HepG2 with vehicle). Interestingly, upon LPS stimulation, the levels of LC3-II were markedly elevated in HBx-HepG2 in a dose-dependent manner compared with those of Ctrl HepG2 (Fig. 3D and E, HBx-HepG2 with LPS vs. Ctrl HepG2 with LPS). The co-treatment of CQ, which blocks the binding of autophagosomes to lysosomes by altering the acidic environment of lysosomes and induces the accumulation of LC3-II in cells, enhanced the levels of LC3-II in both cells (Fig. 3D and E, LPS + CQ vs. LPS in Ctrl HepG2 or HBx-HepG2). These results suggest that HBx facilitates the association of BECN1-Vps34-TRAF6 and induces the ubiquitination of BECN1, leading to the enhancement of autophagy induction, as shown in Fig. 3F.
HBx-expressing HepG2 cells enhance cell migration, invasion, and mobility in response to TLR4 stimulation
We next examined the functional role of HBx in cancer cell migration, invasion, and mobility. Ctrl HepG2 and HBx-HepG2 cells (Fig. 4A) were treated with vehicle, LPS, LPS plus 3-MA, and LPS plus CQ, as indicated in Fig. 4B, and a cancer cell invasion assay was performed using transwell invasion assay. The migration in HBx-HepG2 cells was significantly enhanced in HBx-HepG2 cells treated with vehicle, as compared to Ctrl HepG2 (Fig. 4B and C, HBx-HepG2 treated with vehicle vs. Ctrl HepG2 treated with vehicle). In addition, the migration was significantly higher in HBx-HepG2 cells treated with LPS than those of Ctrl HepG2 treated with LPS (Fig. 4b and 4c, HBx-HepG2 treated with LPS vs. Ctrl HepG2 treated with LPS). In contrast, marked inhibitions were detected in both cells co-treated with autophagy inhibitors, 3-MA or CQ (Fig. 4B and C, LPS + 3-MA and LPS + CQ). Consistent with these results, cancer migration was enhanced in HBx-HepG2 cells treated with vehicle, as compared to that of Ctrl HepG2 (Fig. 4d and 4e, HBx-HepG2 treated with vehicle vs. Ctrl HepG2 treated with vehicle), and a significant increase was observed in HBx-HepG2 treated with LPS (Fig. 4d and 4e, HBx-HepG2 treated with LPS vs. Ctrl HepG2 treated with LPS). To further verify the ability of single-cell migration, time-lapse image microscope analysis was performed. Upon LPS stimulation, the rate of single- cell migration in HBx-HepG2 was significantly faster than Ctrl HepG2 (Fig. 5A and B, HBx-HepG2 treated with LPS vs. Ctrl HepG2 treated with LPS). On the contrast, after the co-treatment of 3-MA, the ability of migration was markedly attenuated in both cells (Fig. 5A and B, LPS vs. LPS + 3-MA). Next, we examined HBx-induced cell proliferation using clonogenic assay (Fig. 5C and D) and anchorage-independent soft agar assay (Fig. 5E and F). The number of colonies in the clonogenic assay was significantly increased in the HBx-HepG2 treated with vehicle, as compared with those of Ctrl HepG2 treated with a vehicle (Fig. 5C and D, HBx-HepG2 treated with a vehicle vs. Ctrl HepG2 treated a vehicle). Moreover, upon LPS stimulation, the number of colonies was markedly higher in HBx-HepG2 than in Ctrl HepG2 (Fig. 5C and D, HBx-HepG2 treated with LPS vs. Ctrl HepG2 treated LPS), whereas a significant decrease was observed in the co-treatment of 3-MA or CQ (Fig. 5C and D, LPS vs. LPS + 3-MA or LPS + CQ in HBx-HepG2 and Ctrl HepG2). Notably, the anchorage-independent colonies were significantly increased in HBx-HepG2 treated with vehicle or LPS, as compared with those of Ctrl HepG2 cells (Fig. 5E and F, HBx-HepG2 vs. Ctrl HepG2 cells in vehicle or LPS). Consistently, an autophagy inhibitor, 3-MA or CQ, led to marked attenuation of the colony formation (Fig. 5E and F, LPS vs. LPS + 3-MA or LPS + CQ in HBx-HepG2 and Ctrl HepG2). These results suggest that HBx positively regulates the liver cancer progression induced by TLR4 stimulation.
Figure 4. HBx-HepG2 cells exhibited increased cell migration and invasion in response to TLR4 stimulation. (A) Ctrl HepG2 and HBx-HepG2 cells were extracted, and the cell lysates were subjected to WB with anti-HBx or anti-GAPDH Ab. (B, C) Ctrl HepG2 and HBx-HepG2 cells were suspended in culture medium including vehicle, LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL), and the invasive assay was performed as described in the Materials and Methods. Fixed cells were stained with crystal violet (B). The number of migrated cells was counted, and presented as the mean±SEM (C) (*p<0.05 and **p<0.01). (D, E) Ctrl HepG2 and HBx-HepG2 cells were seeded into 12-well cell culture plates, scraped with a sterile yellow Gilson-pipette tip, and treated with a vehicle (DMSO, <0.2% in culture medium), LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL) for different time periods, as indicated. A representative experiment is shown (D). The residual gap between the migrating cells from the opposing wound edge was expressed as a percentage of the initial scraped area (E) (±SEM, n=3; *p<0.05, **p<0.01, and ***p<0.001).
Figure 5. HBx-HepG2 cells exhibited increased cell mobility and colony formation in response to TLR4 stimulation. (A, B) Ctrl HepG2 and HBx-HepG2 cells were seeded into 6-well culture plate, treated with vehicle (DMSO), and 3-MA (5 mM) in the presence or absence of LPS (10 μg/mL), and time-lapse imaging analysis was performed for different times by using phase-contrast microscope (A), as described in Materials and Methods. Data analysis on the speed of cell mobility was performed following protocols provided by the Bio-protocol (B) (www.bio-protocol.org/e3586) (*p<0.05). (C, D) Ctrl HepG2 and HBx-HepG2 cells were harvested with trypsin-EDTA and re-suspended in a singular form. The 1×103 cells (per well) were plated in a 6-well plate and treated with a vehicle (DMSO), 3-MA (5 mM), and CQ (10 μM) in the presence or absence of LPS (10 μg/mL). After incubation for 18 days (C), colonies were stained with 0.5% crystal violet (Sigma) for 30 minutes at room temperature and counted (D) (±SEM, n=3; *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001). (E, F) Ctrl HepG2 and HBx-HepG2 cells (1.0×104 cells per well) mixed with 0.3% Difco Noble Agar in complete medium were plated on the top of 0.5% agar layer in a 6-well plate with complete medium. Growth medium (1.5 mL) with a vehicle (DMSO), LPS (10 μg/mL), 3-MA (5 mM), or CQ (10 μM) was added on top of the layer and the cells were incubated at 37°C for 4 weeks. For visualization, the foci were stained with 0.0005% crystal violet (E). The number of colonies were counted (F) (±SEM, n=3; *p<0.05 and **p<0.01).
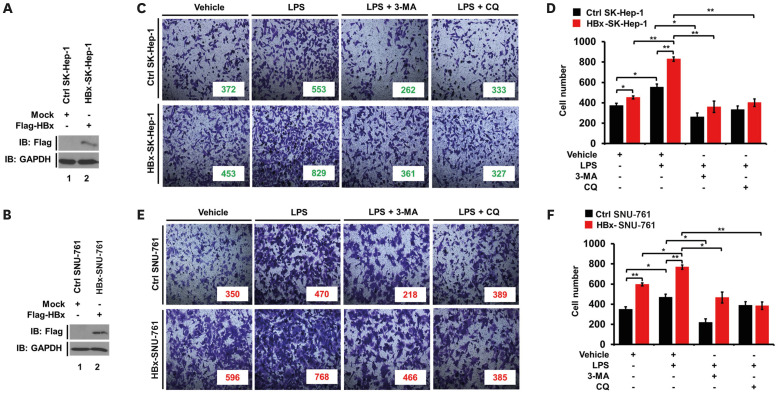
HBx-expressed SK-Hep-1 and SNU-761 cells exhibit enhanced cell migration, invasion, and mobility in response to TLR4 stimulation
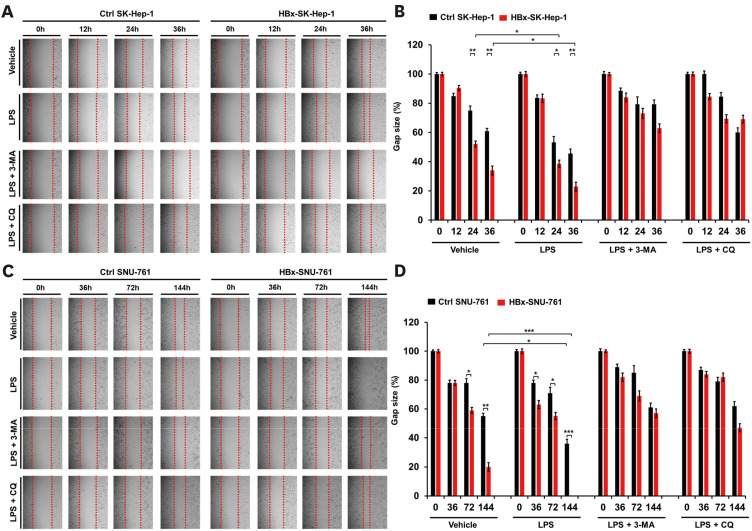
Given the results of the role of HBx in HBx-HepG2 cells, we further evaluated the role of HBx in SK-Hep-1 human hepatic adenocarcinoma and SNU-761 human HCC cell lines. Mock or Flag-HBx vector was transfected into SK-Hep-1 cells (Fig. 6A) or SNU-761 cells (Fig. 6B). Ctrl SK-Hep-1, HBx-expressed SK-Hep-1 (HBx-SK-Hep-1), Ctrl SNU-761, and HBx-SNU-761 cells were treated with vehicle, LPS, LPS plus 3-MA, and LPS plus CQ, as indicated. Cell invasion was significantly increased in the HBx-SK-Hep-1 treated with vehicle, and enhanced with the treatment of LPS, as compared to that of Ctrl SK-Hep-1 cells (Fig. 6C and D, HBx-SK-Hep-1 vs. Ctrl SK-Hep-1 in vehicle or LPS). In contrast, autophagy inhibitors 3-MA and CQ markedly abolished the LPS-induced invasion of Ctrl SK-Hep-1 cells and HBx-SK-Hep-1 cells (Fig. 6C and D, LPS vs. LPS + 3-MA or LPS + CQ in Ctrl SK-Hep-1 and HBx-SK-Hep-1). Similar results were observed in HBx-SNU-761 cells, as compared to those of Ctrl SNU-761 (Fig. 6E and F, HBx-SNU-761 vs. Ctrl SNU-761). Furthermore, upon LPS stimulation, cancer cell migration was markedly enhanced in HBx-SK-Hep-1 and HBx-SNU-761 cells (Fig. 7A and B, HBx-SK-Hep-1 treated LPS vs. Ctrl SK-Hep-1 treated LPS; Fig. 7C and B, HBx-SNU-761 treated LPS vs. Ctrl SNU-761 treated LPS), whereas there was marked attenuations with the co-treatment of 3-MA or CQ (Fig. 7A and B, HBx-SK-Hep-1 treated LPS vs. HBx-SK-Hep-1 treated LPS plus 3-MA or HBx-SK-Hep-1 treated LPS plus CQ; Fig. 7B and C, HBx-SNU-761 treated LPS vs. HBx-SNU-761 treated LPS plus 3-MA or HBx-SNU-761 treated LPS plus CQ), suggesting that HBx enhances cancer cell migration and invasion induced by TLR4 stimulation.
Figure 6. HBx-expressed SK-Hep-1 and HBx-expressed SNU-761 cells exhibited increased cell invasion in response to TLR4 stimulation. (A) SK-Hep-1 cells were transfected with mock (Ctrl) or Flag-HBx plasmid, extracted, and the cell lysates were subjected to WB with anti-Flag or anti-GAPDH Ab. (B) SNU-761 cells were transfected with mock (Ctrl) or Flag-HBx plasmid, extracted, and the cell lysates were subjected to WB with anti-Flag or anti-GAPDH Ab. (C, D) Ctrl SK-Hep-1 and HBx-SK-Hep-1 cells were suspended in culture medium including vehicle, LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL), and the invasive assay was performed as described in the Materials and Methods. Fixed cells were stained with crystal violet (C). The number of migrated cells was counted, and presented as the mean±SEM (D) (*p<0.05 and **p<0.01). (E, F) Ctrl SNU-761 and HBx-SNU-761 cells were suspended in culture medium with a vehicle, LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL), and the invasive assay was performed as described in the materials and methods. Fixed cells were stained with 4,6-diamidino-2-phenylindole (E). The number of migrated cells was counted, and presented as the mean±SEM (F) (*p<0.05 and **p<0.01).
Figure 7. HBx-expressed SK-Hep-1 and HBx-expressed SNU-761 cells exhibited increased cell migration in response to TLR4 stimulation. (A, B) Ctrl SK-Hep-1 and HBx-SK-Hep-1 cells were seeded into 12-well cell culture plates, scraped with a sterile yellow Gilson-pipette tip, and treated with vehicle (DMSO, <0.2% in culture medium), LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL) for different time periods, as indicated. A representative experiment is shown (A). The residual gap between the migrating cells from the opposing wound edge was expressed as a percentage of the initial scraped area (B) (±SEM, n=3; *p<0.05 and **p<0.01). (C, D) Ctrl SNU-761 and HBx-SNU-761 cells were seeded into 12-well cell culture plates, scraped with a sterile yellow Gilson-pipette tip, and treated with vehicle (DMSO, <0.2% in culture medium), LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL), and CQ (10 μM) plus LPS (10 μg/mL) for different time periods, as indicated. A representative experiment is shown (C). The residual gap between the migrating cells from the opposing wound edge was expressed as a percentage of the initial scraped area (D) (±SEM, n=3; *p<0.05, **p<0.01, and ***p<0.001).
We next analyzed the mobility of HBx-SK-Hep-1 cells using live time-lapse microscopy. The intrinsic mobility of HBx-SK-Hep-1 cells treated with vehicle was significantly higher than that of Ctrl SK-Hep-1 cells treated with vehicle (Fig. 8A and B, HBx-SK-Hep-1 cells treated with vehicle vs. Ctrl SK-Hep-1 cells treated with vehicle). Moreover, the mobility was markedly enhanced with the LPS treatment, whereas marked attenuation was observed in the co-treatment of 3-MA (Fig. 8A and B, LPS and LPS + 3-MA in HBx-SK-Hep-1 cells or Ctrl SK-Hep-1 cells). We further assessed the in vitro cancer formation of HBx-SNU-761 cells using a colony-forming assay. Consistent with the result observed in HBx-HepG2 cells, the number of colonies was increased in HBx-SNU-761 treated with vehicle as compared to that of Ctrl SNU-761 treated with vehicle, and a significant increase was observed with the treatment of LPS in HBx-SNU-761 (Fig. 8C and D, HBx-SNU-761 vs. Ctrl SNU-761 in vehicle or LPS). On the contrast, the co-treatment of autophagy inhibitors, 3-MA or CQ, resulted in a marked inhibition in both cells (Fig. 8C and D, LPS + 3-MA or LPS + CQ in HBx-SNU-761 and Ctrl SNU-761). Taken together, these results suggest that HBx functionally promotes the migration, invasion, and mobility of liver cancer cells induced by TLR4 stimulation.
Figure 8. HBx-expressed SK-Hep-1 and HBx-expressed SNU-761 cells exhibited increased cell mobility and colony formation in response to TLR4 stimulation. (A, B) Ctrl SK-Hep-1 and HBx-SK-Hep-1 cells were seeded into 6-well culture plate, treated with vehicle (DMSO), and 3-MA (5 mM) in the presence or absence of LPS (10 μg/mL), and time-lapse imaging analysis was performed for different times by using phase-contrast microscope (A), as described in Materials and Methods. Data analysis for measuring the speed of cell mobility was performed as following protocols provided by Bio-protocol (B) (www.bio-protocol.org/e3586) (*p<0.05). (C, D) Ctrl SNU-761 and HBx-SNU-761 cells were harvested with trypsin-EDTA and re-suspended in a singular form. The 1×103 cells (per well) were plated in a 6-well plate and treated with the vehicle (DMSO), 3-MA (5 mM), and CQ (10 μM) in the presence or absence of LPS (10 μg/mL). After incubation for 12 days (C), colonies were stained with 0.5% crystal violet (Sigma) for 30 minutes at room temperature and counted (D) (±SEM, n=3; *p<0.05 and **p<0.01).
DISCUSSION
HBx protein plays a pivotal role in the pathogenesis of HBV-related liver diseases through the functional regulation of various host proteins that regulate hepatocyte differentiation and proliferation (1,2,3,35). Previous reports have demonstrated that the interaction between HBx and different cellular proteins, including DNA repair proteins, damaged DNA-binding proteins, cell cycle-related proteins, and autophagy-related proteins is critically implicated in the pathogenesis of HCC following HBV infection (1,2,3,35,36). To elucidate the pathogenic mechanism by which HBx is involved in the development of liver carcinogenesis, it is essential to understand the molecular and cellular mechanisms involved in liver cancer progression in response to the cellular stimulus. Recent evidence has demonstrated that autophagy promotes carcinogenesis at the early stages, and tumor progression in HCC (37,38,39). Importantly, previous studies have demonstrated that HBx is functionally implicated in the autophagy processes or induction (25,36,40,41,42,43). HBx induced the formation of autophagosome, but inhibited autophagic degradation by impairing lysosomal maturation in Huh7 hepatoma cells (40). HBx has also been shown to induce autophagy in HepG2 and primary liver cancer cells through various cellular signals, thereby promotes liver cancer migration and invasion (41,42,43). These results suggest that HBx may have different roles in the regulation of autophagy in a context-dependent manner. Notably, TLR4 plays a vital role in the development and pathogenesis of HCC and the cancer progression by autophagy induction (14,20,21,22,23,24,25,26,27,28). Although several reports have suggested that HBx is functionally implicated in autophagy by regulating the mTOR pathway and the expression of BECN1 (25,36), the molecular and cellular mechanism by which HBx is involved in the TLR4-induced autophagy and thereby in liver cancer progression are still unclear. Herein, we investigated the regulatory mechanism of HBx in the autophagy induction and examined whether HBx is implicated in the progression of liver cancer induced by TLR4.
We proposed two regulatory mechanisms; i) HBx interacts with BECN1 and inhibits the association of Bcl-2, which is a negative regulator of the BECN1-induced autophagy (22,23), to BECN1, resulting in enhanced autophagy induction (Fig. 9, right), ii) HBx interacts with Vps34 and enhances the molecular association between Vps34 and BECN1 and the ubiquitination of BECN1, leading to the induction of autophagy (Fig. 9, left). It has been reported that the expression of Bcl-2 is increased in various human cancers and is associated with resistance against apoptosis or autophagy during tumorigenesis and chemotherapy (44,45). The molecular and cellular mechanism studies have revealed that the interaction of BECN1-Bcl-2 inhibits the BECN1-mediated autophagy (22,23). In the present study, we found that HBx interacted with the Bcl-2 binding domain of BECN1 and competitively interrupted the association of the BECN1-Bcl-2 complex. In addition, we found that HBx interacted with the C2 domain of Vps34 protein. The BECN1-Vps34 complex plays a critical role in autophagy induction (18,19,20,21,22). Notably, we found that HBx markedly facilitated the molecular association of BECN1-Vps34 and enhanced the ubiquitination of BECN1. In HBx-stable expressing HepG2 cells, the levels of LC3-II were significantly enhanced in response to LPS stimulation. These results strongly suggest that HBx positively regulates autophagy induction by TLR4 stimulation, presumably through the regulation of BECN1-Bcl-2 formation and BECN1-Vps34 complex. Given the molecular mechanism of HBx in the regulation of autophagy, we evaluated whether HBx is involved in liver cancer progression through autophagy induced by TLR4 stimulation. We utilized three different liver cancer cell lines, HBx-stable expressing HepG2, HBx-expressed SK-Hep-1, and HBx-expressed SNU-761 cells. Upon TLR4 stimulation, cancer cell migration, invasion, mobility, and colony formations were markedly enhanced in HBx-HepG2, HBx-SK-Hep-1, and HBx-SNU-761 cells, as compared to those of Ctrl HepG2, SK-Hep-1, and SNU-761 cells. In contrast, the co-treatment of autophagy inhibitors, 3-MA or CQ, resulted in the attenuation of these activities. Together, these results strongly suggest that HBx positively regulates liver cancer progression induced by TLR4 stimulation.
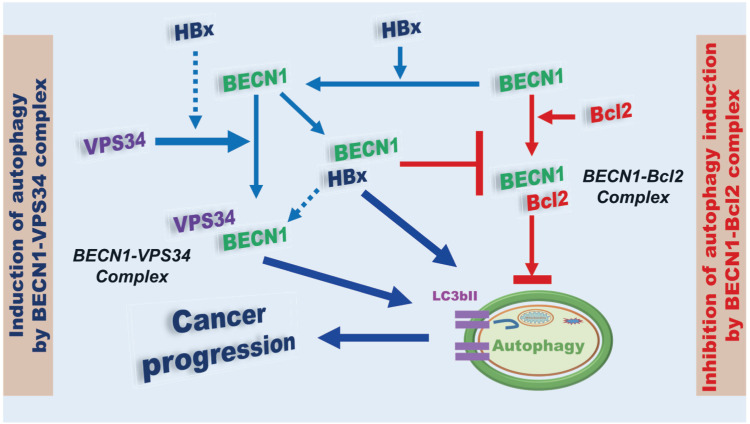
Figure 9. A model of how HBx is positively implicated in autophagy. BECN1 is either positively or negatively implicated in the autophagy through the formation of BECN1-Vps34 (left; induction of autophagy by BECN1-Vps34 complex) or BECN1-Bcl-2 complex (right; inhibition of autophagy induction by BECN1-Bcl-2 complex), respectively. HBx interacts with Vps34 and BECN1, and that facilitates the formation of BECN1-Vps34 complex for the induction of autophagy. Simultaneously, HBx interacts with BECN1 and that interrupts the formation of BECN1-Bcl-2 complex, resulting in the induction of autophagy. The HBx-mediated positive regulation of the formation of BECN1-Vps34 complex and negative regulation of the formation of BECN1-Bcl-2 complex eventually lead to cancer progression through the autophagy induction.
In summary, as illustrated in Fig. 9, BECN1 is either positively or negatively implicated in autophagy through the formation of BECN1-Vps34 (Fig. 9, left) or BECN1-Bcl-2 complex (Fig. 9, right), respectively. HBx interacts with Vps34 and BECN1 and enhances the formation of the BECN1-Vps34 complex (Fig. 9, left). Simultaneously, HBx interacts with BECN1 and inhibits the formation of the BECN1-Bcl-2 complex (Fig. 9, right). Eventually, HBx enhances the autophagy induction by facilitating BECN1-Vps34 complex and inhibiting the BECN1-Bcl-2 complex, resulting in enhanced liver cancer progression. Accumulating evidence has demonstrated that autophagy and TLR4 are functionally implicated in HCC progression and tumorigenicity (14,20,21,22,23,24,25,26,27,28). Although the functional role of HBx in the regulation of autophagy induced by TLR4 in vivo is further required, our results will provide insight into the pathogenesis of HBV-induced carcinogenesis and tumor progression, and the development of new therapeutic agents against HBV-induced liver diseases.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (NRF-2021R1F1A1049324, NRF-2021R1A2C1094478), Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (2020R1A6C101A191), Ministry of Science ICT and Future Planning (MSIP) funded by the Korean Government (NRF-2016R1A5A2945889).
Abbreviations
- BECN1
the Beclin1
- Ctrl
control
- HBV
hepatitis B virus
- HBx
hepatitis B virus X
- HCC
hepatocellular carcinoma
- HEK
human embryonic kidney
- IB
immunoblotting
- IGF-II
insulin-like growth factor II
- IP
immunoprecipitation
- TERT
telomerase reverse transcriptase
- WB
western blotting
- WT
wild type
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Son J, Kim MJ.
- Data curation: Son J, Kim MJ.
- Formal analysis: Son J, Kim MJ, Lee JS, Kim JY.
- Funding acquisition: Chun E, Lee KY.
- Investigation: Son J, Kim MJ, Lee JS, Kim JY.
- Methodology: Son J, Kim M-J.
- Resources: Son J, Kim MJ.
- Supervision: Chun E, Lee KY.
- Validation: Son J, Kim MJ.
- Writing-original draft: Chun E, Lee KY.
- Writing-review & editing: Chun E, Lee KY.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Marra M, Sordelli IM, Lombardi A, Lamberti M, Tarantino L, Giudice A, Stiuso P, Abbruzzese A, Sperlongano R, Accardo M, et al. Molecular targets and oxidative stress biomarkers in hepatocellular carcinoma: an overview. J Transl Med. 2011;9:171. doi: 10.1186/1479-5876-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26(Suppl 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 4.Yang S, Yang L, Li X, Li B, Li Y, Zhang X, Ma Y, Peng X, Jin H, Li H. New insights into autophagy in hepatocellular carcinoma: mechanisms and therapeutic strategies. Am J Cancer Res. 2019;9:1329–1353. [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Liao JZ, He XX, Li PY. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget. 2017;8:57707–57722. doi: 10.18632/oncotarget.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643–4651. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J, Shen HM, Lim LH. The role of autophagy in liver cancer: crosstalk in signaling pathways and potential therapeutic targets. Pharmaceuticals (Basel) 2020;13:432. doi: 10.3390/ph13120432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 10.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. 2012;2:214–221. [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang GM, Tan Y, Wang H, Peng L, Chen HT, Meng XJ, Li LL, Liu Y, Li WF, Shan H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol Cancer. 2019;18:17. doi: 10.1186/s12943-019-0944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janji B, Viry E, Moussay E, Paggetti J, Arakelian T, Mgrditchian T, Messai Y, Noman MZ, Van Moer K, Hasmim M, et al. The multifaceted role of autophagy in tumor evasion from immune surveillance. Oncotarget. 2016;7:17591–17607. doi: 10.18632/oncotarget.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Li M, Zheng QC. Emerging role of Toll-like receptor 4 in hepatocellular carcinoma. J Hepatocell Carcinoma. 2015;2:11–17. doi: 10.2147/JHC.S44515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung C, Seo W, Silwal P, Jo EK. Crosstalks between inflammasome and autophagy in cancer. J Hematol Oncol. 2020;13:100. doi: 10.1186/s13045-020-00936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Han W, Sui X, Fang Y, Pan H. Autophagy: a novel therapeutic target for hepatocarcinoma (Review) Oncol Lett. 2014;7:1345–1351. doi: 10.3892/ol.2014.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopes JA, Borges-Canha M, Pimentel-Nunes P. Innate immunity and hepatocarcinoma: can toll-like receptors open the door to oncogenesis? World J Hepatol. 2016;8:162–182. doi: 10.4254/wjh.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan Z, Xie X, Cao H, Zhou X, Zhang XD, Fan H, Liu Z. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy. 2014;10:257–268. doi: 10.4161/auto.27162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min Y, Kim MJ, Lee S, Chun E, Lee KY. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy. 2018;14:1347–1358. doi: 10.1080/15548627.2018.1474995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MJ, Min Y, Im JS, Son J, Lee JS, Lee KY. p62 is Negatively Implicated in the TRAF6-BECN1 signaling axis for autophagy activation and cancer progression by Toll-like receptor 4 (TLR4) Cells. 2020;9:1142. doi: 10.3390/cells9051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris DH, Yip CK, Shi Y, Chait BT, Wang QJ. Beclin 1-VPS34 complex architecture: understanding the nuts and bolts of therapeutic targets. Front Biol (Beijing) 2015;10:398–426. doi: 10.1007/s11515-015-1374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 26.Park S, Ha YN, Dezhbord M, Lee AR, Park ES, Park YK, Won J, Kim NY, Choo SY, Shin JJ, et al. Suppression of hepatocyte nuclear factor 4 α by long-term infection of hepatitis B virus contributes to tumor cell proliferation. Int J Mol Sci. 2020;21:948. doi: 10.3390/ijms21030948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MJ, Min Y, Son J, Kim JY, Lee JS, Kim DH, Lee KY. AMPKα1 regulates lung and breast cancer progression by regulating TLR4-mediated TRAF6-BECN1 signaling axis. Cancers (Basel) 2020;12:3289. doi: 10.3390/cancers12113289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MJ, Min Y, Kwon J, Son J, Im JS, Shin J, Lee KY. p62 negatively regulates TLR4 signaling via functional regulation of the TRAF6-ECSIT complex. Immune Netw. 2019;19:e16. doi: 10.4110/in.2019.19.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim MJ, Min Y, Shim JH, Chun E, Lee KY. CRBN is a negative regulator of bactericidal activity and autophagy activation through inhibiting the ubiquitination of ECSIT and BECN1. Front Immunol. 2019;10:2203. doi: 10.3389/fimmu.2019.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min Y, Wi SM, Shin D, Chun E, Lee KY. Peroxiredoxin-6 negatively regulates bactericidal activity and NF-κB activity by interrupting TRAF6-ECSIT complex. Front Cell Infect Microbiol. 2017;7:94. doi: 10.3389/fcimb.2017.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA. The soft agar colony formation assay. J Vis Exp. 2014;92:e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro . Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 33.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decuypere JP, Parys JB, Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells. 2012;1:284–312. doi: 10.3390/cells1030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gearhart TL, Bouchard MJ. The hepatitis B virus HBx protein modulates cell cycle regulatory proteins in cultured primary human hepatocytes. Virus Res. 2011;155:363–367. doi: 10.1016/j.virusres.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, Guo QS, Wang ZW, Qian HX. HBx induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway. Mol Cell Biochem. 2013;372:161–168. doi: 10.1007/s11010-012-1457-x. [DOI] [PubMed] [Google Scholar]
- 37.Kowalik MA, Perra A, Ledda-Columbano GM, Ippolito G, Piacentini M, Columbano A, Falasca L. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules. Oncotarget. 2016;7:5788–5799. doi: 10.18632/oncotarget.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huo X, Qi J, Huang K, Bu S, Yao W, Chen Y, Nie J. Identification of an autophagy-related gene signature that can improve prognosis of hepatocellular carcinoma patients. BMC Cancer. 2020;20:771. doi: 10.1186/s12885-020-07277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y, Song Z, Zheng Q, Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Li J, Wang S, Yang F, Zhou Y, Liu Y, Zhu W, Shi X. HBx-associated long non-coding RNA activated by TGF-β promotes cell invasion and migration by inducing autophagy in primary liver cancer. Int J Oncol. 2020;56:337–347. doi: 10.3892/ijo.2019.4908. [DOI] [PubMed] [Google Scholar]
- 42.Luo MX, Wong SH, Chan MT, Yu L, Yu SS, Wu F, Xiao Z, Wang X, Zhang L, Cheng AS, et al. Autophagy mediates HBx-induced nuclear factor-κB activation and release of IL-6, IL-8, and CXCL2 in hepatocytes. J Cell Physiol. 2015;230:2382–2389. doi: 10.1002/jcp.24967. [DOI] [PubMed] [Google Scholar]
- 43.Fu S, Wang J, Hu X, Zhou RR, Fu Y, Tang D, Kang R, Huang Y, Sun L, Li N, et al. Crosstalk between hepatitis B virus X and high-mobility group box 1 facilitates autophagy in hepatocytes. Mol Oncol. 2018;12:322–338. doi: 10.1002/1878-0261.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Aranda M, Pérez-Ruiz E, Redondo M. Bcl-2 inhibition to overcome resistance to chemo- and immunotherapy. Int J Mol Sci. 2018;19:3950. doi: 10.3390/ijms19123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas S, Quinn BA, Das SK, Dash R, Emdad L, Dasgupta S, Wang XY, Dent P, Reed JC, Pellecchia M, et al. Targeting the Bcl-2 family for cancer therapy. Expert Opin Ther Targets. 2013;17:61–75. doi: 10.1517/14728222.2013.733001. [DOI] [PMC free article] [PubMed] [Google Scholar]