Abstract
Malignant melanoma is one of the most highly ranked cancers in terms of years of life lost. Hereditary melanoma with its increased familial susceptibility is thought to affect up to 12% of all melanoma patients. In the past, only a few high-penetrance genes associated with familial melanoma, such as CDKN2A and CDK4, have been clinically tested. However, findings now indicate that melanoma is a cancer most likely to develop not only due to high-penetrance variants but also due to polygenic inheritance patterns, leaving no clear division between the hereditary and sporadic development of malignant melanoma. Various pathogenic low-penetrance variants were recently discovered through genome-wide association studies, and are now translated into polygenic risk scores. These can show superior sensitivity rates for the prediction of melanoma susceptibility and related mixed cancer syndromes than risk scores based on phenotypic traits of the patients, with odds ratios of up to 5.7 for patients in risk groups. In addition to describing genetic findings, we also review the first results of epigenetic research showing constitutional methylation changes that alter the susceptibility to cutaneous melanoma and its risk factors.
Keywords: Melanoma susceptibility, hereditary melanoma, familial melanoma, melanoma genetics, germline mutation, epigenetics, polygenic risk score
Introduction
Malignant melanoma is a type of skin cancer derived from neoplastic melanocytes. Once a rare cancer, incidence numbers have been growing worldwide for decades and are estimated to rise further [1]. Presently, malignant melanoma is one of the most frequent cancers with an estimated lifetime risk of 2% in Western populations. In the United States, it is the fifth most frequent type of cancer in men, and the sixth most frequent in women. Between 1973 and now, the annual number of cases has risen by more than 270%; partially explained by an aging society, imprudent tanning behavior, and loss of the ozone layer [2,3]. In Europe, incidence rates follow a gradient from the southern Mediterranean with lower, to northern Scandinavian countries with higher malignant melanoma rates [2]. Europe accounts for 45% of all malignant melanoma deaths worldwide [1]. Depending on the geographic region, melanoma can rank as high as third place in a population based on the number of years of life lost (YLL) [4,5].
However, malignant melanoma is not the most common type of skin cancer, but is the most lethal. This is due to early dissemination and metastasis formation, as well as high resistance to treatment [6]. Currently, the only curative treatment is early detection, followed by tumor excision [7].
Certain somatic mutations driving tumorigenesis have been identified in sporadic cases of malignant melanoma. In contrast, 5-12% of malignant melanoma cases are thought to develop due to genetic germline alterations, referred to as hereditary melanoma [8,9]. Several genes with pathogenic variants predisposing for a higher risk of developing malignant melanoma have been identified, with CDKN2A (cyclin-dependent kinase inhibitor 2A) being the most commonly altered gene, which accounts for an estimated 20-45% of hereditary melanoma cases [8-10]. Genetic alterations can be inherited, as well as constitutional epigenetic modifications. Studies have shown that certain DNA methylation changes increase the susceptibility to melanoma risk factors, such as dysplastic nevi, or for cutaneous melanoma itself [8,11,12].
As most hereditary tumors, also hereditary melanoma presents itself with an earlier age of cancer development onset, as well as familial clustering of specific cancer types [13].
As the available evidence on inherited melanoma susceptibility has advanced rapidly in recent years, the aim of this review was to provide a comprehensive and updated overview of the topic. This includes recent clinical and genome-wide association study (GWAS) findings, as well as a summary of the results from studies investigating the epigenetic factors of malignant melanoma.
Methods
Sources for this review were identified on Medline, UpToDate, and Cochrane Central Register with scientific publications spanning at least a five-year period up until May 17, 2021. Search terms included “melanoma”, “hereditary melanoma” and “familial melanoma”. Peer-reviewed sources were included in this article if they contained specific results on germline mutations and/or constitutional epigenetic alterations. Sources also included a medical textbook and a published doctoral dissertation. We did not include non-English language sources or articles on somatic melanoma driver mutations (exclusion criteria).
Genes with high-penetrance variants
Genes containing pathogenic variants associated with malignant melanoma are usually grouped by penetrance of the variant(s), biological pathomechanism or cumulative number of single nucleotide variants (SNVs) [9,10].
Penetrance for genes or variants, defined as the ratio of gene expression or susceptibility, can be high, intermediate, or low with respective odds ratios of over 5, 2 to 5, or lower than 2, although slightly varying in the literature. For intermediate-and low-penetrance variants, predicting melanoma occurrence is still very imprecise [9].
CDKN2A (Cyclin-dependent kinase inhibitor 2A)
CDKN2A is a gene located on chromosome 9p21. Germline alterations in this gene show an autosomal dominant inheritance pattern, and multiple single-nucleotide variants (SNVs) are associated with a higher risk of malignant melanoma [1,8,9].
It is the most common gene with pathogenic variants in hereditary melanoma, with variants varying by geographic region [9,14-16]. One of the most frequent of these so-called founder mutations is p.G101W, which is mostly detected in French, Spanish and Italian populations [9,17].
CDKN2A itself consists of four exons containing the genetic code for two unrelated proteins, p16 inhibitor of cyclin-dependent kinase 4 (p16INK4A) and p14 alternate reading frame (p14ARF) [9,18]. Both protein products act as tumor suppressors. P16INK4A is an inhibitor of CDK4 (cyclin-dependent kinase 4) and CDK6 (cyclin-dependent kinase 6), preventing the cell from entering the S-phase of the cell cycle, whereas p14ARF positively regulates the tumor suppressor p53, thus preventing an excess load of damaged DNA in the cell [9].
CDKN2A mutations increase the risk of malignant melanoma itself, yet certain pathogenic CDKN2A variants are associated with other cancer types, such as pancreatic cancer, lung cancer, breast cancer, sarcoma, and, in rare cases, mesothelioma and esophageal squamous cell cancer [13,18-21].
The most common CDKN2A variant-associated melanoma-dominant tumor syndrome is called familial atypical multiple mole-melanoma (FAMMM) and is rarely associated with CDK4 and MITF (microphthalmia-associated transcription factor) alterations [1,13]. The typical phenotypic manifestations of FAMMM are a high nevi count over 50 and multiple precancerous dysplastic nevi [13]. In this tumor syndrome, affected patients tend to regularly develop melanoma on unaffected and healthy skin tissue, but not from nevi. As with hereditary cancers in general, the onset of disease is also early in FAMMM [1].
Patients with Melanoma-Astrocytoma syndrome have a higher incidence of nervous system tumors (NSTs), such as astrocytoma, which can occur before or after the development of melanoma [1]. Here, the main gene involved is CDKN2A/ARF, which encodes p14ARF, although there have been rare associations with more comprehensive 9p21 chromosome alterations affecting the genetic cluster of CDKN2A, CDKN2B, and CDKN2BAS until the gene MLLT3 [1,22].
For nearly two decades, CDKN2A and CDK4 were the only two genes tested in a clinical context when hereditary melanoma was suspected [23].
Pathways affected by CDKN2A and other genes with high-penetrance variants associated with melanoma susceptibility are shown in Figure 1, whereas Figure 2 depicts a gene network analysis based on genes with both high-and medium-penetrance variants.
Figure 1.
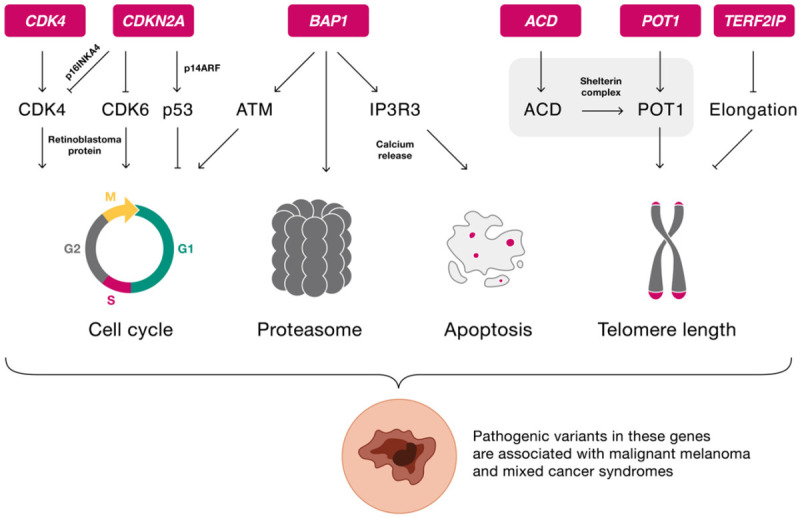
Physiological pathways of genes with possible pathogenic high-penetrance variants associated with hereditary melanoma. ACD: adrenocortical dysplasia; ATM: ataxia telangiectasia-mutated signaling pathway; BAP1: BRCA1-associated protein-1; CDK4: cyclin-dependent kinase 4; CDK6: cyclin-dependent kinase 6; CDKN2A: cyclin-dependent kinase inhibitor 2 a; IP3R3: receptor for inositol 1,4,5-trisphosphate; p14ARF: p14 alternate reading frame; p16INKA4: p16 inhibitor of cyclin-dependent kinase 4; p53: tumor protein 53; POT1: protection of telomeres 1; TERF2IP: telomeric repeat binding factor 2 interacting protein.
Figure 2.
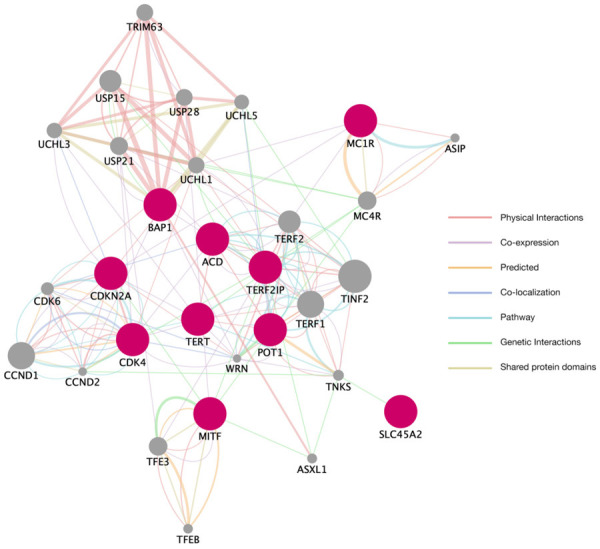
Established melanoma susceptibility genes with high and medium-penetrance variants and their most closely related genes. Utilizing the GeneMania framework (Version 3.5.2) in Cytoscape (Version 3.8.2), we generated a network analysis based on the established hereditary melanoma susceptibility genes, usually bearing high-and medium-penetrance pathogenic variants. These are shown in red. The most closely related genes are shown in gray. Genes were visually grouped according to molecular function. ACD: adrenocortical dysplasia; BAP1: BRCA1-associated protein-1; CDK4: cyclin-dependent kinase 4; CDKN2A: cyclin-dependent kinase inhibitor 2 a; MC1R: melanocortin 1 receptor gene; MITF: microphthalmia-associated transcription factor; POT1: protection of telomeres 1; SLC45A2: solute carrier family 45 member 2; TERF2IP: telomeric repeat binding factor 2 interacting protein; TERT: telomerase reverse transcriptase.
CDK4 (Cyclin-dependent kinase 4)
CDK4 is a gene with rare pathogenic variants found on chromosome 12q4 [8,9,23]. Its identically named translational product is part of the same signaling pathway of the cell cycle as the gene products of CDKN2A [9]. Pathogenic variants of CDK4 associated with malignant melanoma are uncommon, and only 18 families have been identified worldwide, as well as a single case of a male patient in Italy, leading to a subsequent need for further studies with more significant populations to solidify the effect of pathogenic variants of CDK4 on the development of hereditary melanoma [9,24].
CDK4 and CDK6 are physiologically needed to transition to the S-phase of the cell cycle from the G1-phase by phosphorylating retinoblastoma protein (RB) [9]. To date, only one specific locus with specific pathogenic variants in CDK4 has been found: Arg24, in exon 2, codon 24 [24]. There, three pathogenic variants were identified. The arginine can be interchanged for either cysteine (Arg24Cys), histidine (Arg24His), or leucine (Arg24Leu with only one described case) [9]. This eliminates the binding domain of p16INKA4, leading to reduced p16INK4A inhibition of CDK4 kinase activity, subsequently deregulating CDK4, resulting in uncontrolled continuation of the cell cycle [18,24].
As a result, patients carrying pathogenic CDK4 variants were susceptible to an earlier onset of malignant melanoma with a median age of 39 years at the time of diagnosis, and a lifetime risk of 74% among the studied individuals [9].
The implications of pathogenic CDK4 variants are, due to the same pathway, similar to the ones of CDKN2A. Pathogenic CDK4 gene variants are associated with a higher risk of cutaneous melanoma, pancreatic cancer, multiple primary melanomas, and atypical nevi [18,23]. Pathogenic CDK4 variations are also associated with the development of FAMMM syndrome [13].
BAP1 (BRCA1-associated protein-1)
BAP1 is a high-penetrance gene found on chromosome 3p21 [8,9]. Studies have found that pathogenic variations are responsible for only up to 1% of cutaneous melanoma (CM), but up to 4% of uveal melanoma cases (UM, also known as ocular melanoma) [25].
Its encoded protein is a deubiquitinating enzyme found in the nucleus, mitochondria, and cytosol of the cells [26]. It is mainly a transcription-regulating tumor suppressor—as part of the ubiquitin-proteasome complex. It also participates in regulating the cell cycle, apoptosis, and gluconeogenesis [27].
As a result, pathogenic variants can lead to decreased DNA repair mechanisms via the ataxia telangiectasia-mutated (ATM) signaling pathway, proliferation through uncontrolled cell cycles, and eventually, tumorigenesis [25,27]. BAP1 also plays a role in regulating the receptor for inositol 1,4,5-trisphosphate (IP3R3 receptor), which is part of the apoptosis mechanism through intracellular calcium release [25].
Patients harboring pathogenic BAP1 variants can show specific lesions with distinct morphology, immunohistochemistry, and early onset in young adults [9,25]. These so-called BAPomas, or melanocytic BAP1-mutated atypical intradermal tumors (MBAIT), atypical Spitzoid nevi, or nowadays, BAP1-inactivated melanocytic tumors (BIMT) are similar to Spitzoid tumors, yet do not fulfill all given criteria and usually can be seen as skin-colored to pink nodules and papules, varying in size by up to one cm in diameter [9,25].
BAP1 variants are associated with a tumor syndrome named BAP1-tumor predisposition syndrome (BAP1-TPDS), which is dominated by mesothelioma and uveal melanoma [23,27]. Other types of cancer are associated with pathogenic BAP1 variants. These show varying frequencies depending on the cancer subtype, thus concluding that in this case, cancer development might depend on a broader array of genetic or epigenetic factors. These further associated cancers include cutaneous melanoma, basal cell carcinoma, mesothelioma, clear cell renal cell carcinoma, cholangiocarcinoma, and possibly additional unknown tumor types [23,27].
POT1 (Protection of telomeres 1)
POT1 is a high-penetrance gene on chromosome 7 with rare pathogenic variants [8,28]. The gene itself codes for the POT1 protein, one of six proteins forming the shelterin complex. The complex regulates and protects the single-stranded DNA telomere regions at the end of each chromosome from degradation and chromosomic fusion [29].
Most pathogenic variants have a negative effect on the binding sites—called oligonucleotide/oligosaccharide-binding fold domains (OB fold domains)—needed to adhere the shelterin complex to the single-stranded DNA [9].
Germline POT1 variants lead to an autosomal dominantly inherited syndrome consisting of several tumor types, predominantly angiosarcoma and superficially spreading melanoma, but also malignant glioma, anaplastic astrocytoma, thyroid cancer, and colorectal cancer [8,29,30]. As driver mutations, somatic POT1 mutations are often found in chronic lymphocytic leukemia (CLL) [29].
TERF2IP (Telomeric repeat binding factor 2 interacting protein) and ACD (Adrenocortical dysplasia)
ACD and TERF2IP, otherwise known as adrenocortical dysplasia protein homolog, are both classified as genes with rare pathogenic high-penetrance variants. Together with POT1, they form a gene group with protein products needed to assemble the shelterin complex, thus regulating the telomeric end of the linear chromosomes [8].
ACD contains a POT1 protein binding domain and is therefore involved in preventing an early degradation of the single-stranded DNA by adhering the shelterin complex to the DNA [9].
TERF2IP has a contrary effect on DNA regulation by preventing excessive elongation of the telomeric region. For pathogenic TERF2IP variants, early melanoma onset as young as 15 years of age has been described [9]. Generally, an early onset and multiple primary melanomas, are typical for pathogenic ACD and TERF2IP variants [8].
Associated cancers, other than superficial spreading melanoma and lentigo maligna melanoma, are breast cancer, ovarian cancer, cervical cancer, uterine cancer, thyroid cancer, colon cancer, lung cancer, renal cancer, urinary cancer, prostate cancer, esophageal cancer, lymphoma, leukemia, and possibly others [8,30].
Genes with medium-penetrance variants
Historically, there has been a general perception that there is a clear distinction between hereditary tumors with singular high-penetrance pathogenic variants and spontaneous somatic mutations. Several studies, including Lu et al. (2014), have shown that there is an underlying polygenic inheritance even for spontaneous tumors, including non-hereditary melanoma [31]. Genes with medium-penetrance variants might be a part of this polygenic inheritance, not by causing cancer development directly, but in a combination reaching a threshold that could lead to tumorigenesis. Furthermore, medium-and low-penetrance gene variants are much more common than their high-penetrance counterparts [9].
TERT (Telomerase reverse transcriptase)
TERT is a medium-penetrance gene on chromosome 5p15 [8,9,27]. There have only been a few findings of hereditary pathogenic variants, mainly in the promoter region of TERT. In contrast, up to 70% of somatic mutations in this gene lead to tumorigenesis [9,30].
It is thought that genes protecting and regulating telomeric regions contribute to approximately 1% of hereditary melanoma cases [27]. TERT is a part of these genes. The combined protein product of TERT and TERC (telomerase RNA component) is the telomerase reverse transcriptase as part of the telomerase complex. This complex is needed to manage the length of the telomeric regions at each end of the chromosome to prevent early replicative senescence [9,30].
Pathogenic variants in TERT and TERC lead to disproportional elongation of the telomeric region, thus increasing melanoma and various cancer risks. They are also distinctly associated with superficial spreading and nodular melanoma [32].
Other associated cancers include breast, bronchial, ovarian, bladder, and renal cell cancer [30]. In addition to TERT and TERC, the two genes OBFC1 and RTEL1 are also discussed in the spectrum of telomere regulation and increased cancer risk [32].
MC1R (Melanocortin 1 receptor gene)
MC1R is a medium-penetrance gene found on chromosome 16. Usually, the inheritance shows an autosomal recessive pattern; however, a dominant-negative effect on wild type alleles has also been described. It has been shown that inheriting any single pathogenic variant in MC1R can lead to a 28% higher risk of developing melanoma [9].
A common feature shared by MC1R, MITF (microphthalmia-associated transcription factor) and SLC45A2 (solute carrier family 45 member 2) is that they are all part of a wider gene subgroup affecting tanning ability and skin pigmentation [9].
As its name indicates, MC1R encodes the melanocortin-1 receptor, which is a G protein-coupled receptor, triggering a signaling cascade when activated through the binding of α-melanocyte stimulating hormone (α-MSH) or UV radiation [9,30]. This cascade runs via adenylate cyclase, increasing intracellular cAMP levels, microphthalmia-associated transcription factor (MITF), and tyrosinase. At the cellular level, this leads to the growth of melanocytes as well as melanization by shifting the production to darker UVB-ray-protecting eumelanin instead of brighter non-UVB-protecting pheomelanin. Other effects include dendrite formation, the development of the specific dendritic shape of melanocytes, which is needed to distribute melanins to neighboring keratinocytes, and roles in DNA repair [9].
Variants can lead to a reduced receptor density on the cells, thus also leading to a specific phenotype with the traits of red hair, fair skin, freckles, reduced tanning ability, and consequently, an increased risk of melanoma due to UV sensitivity [30]. There are two types of variants. R alleles are highly affiliated with the red hair color (RHC) phenotype, with which r alleles are less strongly associated [9].
A reduced tanning ability would be the simplest explanation for increased tumor rates. However, Caucasian patients with darker skin pigmentation have a higher melanoma risk when inheriting a pathogenic MC1R variant than their red hair colored counterparts. This leads to the conclusion that skin pigmentation processes are not the only drivers in this scenario, but also other regulatory mechanisms in the cell that are mediated by MC1R [9].
In addition to the upper body, which is usually a primary site of melanoma, it has been observed that the upper extremities are an affected site if pathogenic MC1R variants are present [30]. In addition, pathogenic variants of MC1R have been found to predispose to congenital melanocytic nevi, subsequently leading to an increased risk of melanoma as a complication [33].
MITF (Microphthalmia-associated transcription factor)
MITF is a gene found on chromosome 3 with a single medium-penetrance pathogenic variant, p.E318K [8,9]. Approximately 1% of Europeans have inherited this variant, which is believed to increase melanoma susceptibility by 3 to 5-fold or 8 to 31-fold if the family anamnesis is positive for pancreatic or renal cell cancer, respectively [30].
MITF is a part of the Myc proto-oncogene group and a key gene in the regulation and growth of melanocytes. The same-named protein product is believed to be a transcription factor for 37 genes in this cell type [9,30]. MITF and other transcription factors are regulated by small-ubiquitin-like modifier proteins (SUMO) [9].
Research has shown that a specific pathogenic variant, MITF p.E318K, which leads to the substitution of glutamic acid with lysine at position 318, reduces the ability to bind to SUMO proteins, thus increasing cell cycle activities such as differentiation, proliferation, and survival of the melanocytic cells, making it a gain-of-function mutation [30]. The risk of melanoma is increased because the variant has a regulatory effect on 17 of the 37 genes regulated by MITF [9].
A specific phenotypic observation for the MITF p.E318K variant is non-blue eye color, darker hair, yet fair skin [9,30]. Familial atypical multiple mole-melanoma (FAMMM) and an elevated number of nevi are also associated with this variant [13].
Next to malignant melanoma, which may be nodular, amelanotic, and more thickened than usual, the pathogenic MITF variant is also associated with renal cell cancer, as MITF also activates hypoxia-inducible factor 1A (HIF1A) [30].
SLC45A2 (Solute carrier family 45, member 2)
SLC45A2 is a gene found on chromosome 5 with medium-penetrance variants linked to cutaneous melanoma (CM) [9].
With seven exons, the gene codes for a 530 amino acid protein membrane-associated transporter of the melanosome—cellular organelles for the production of melanins [3,34]. This transporter is thought to regulate, process, and transport proteins needed in the melanosome, for example, tyrosinase [9].
In addition to findings in regards to hereditary melanoma, SLC45A2 variants also frequently undergo research in terms of general skin pigmentation and oculocutaneous albinism [35].
In various cases, the cutaneous melanoma-associated variant rs16891982 (p.L374F) has been associated with a protective role against the disease. Furthermore, the variant is associated with a phenotype of olive up to darker skin color, yet it also maintains its protective role for persons with fair skin coloration. The variant is found mostly in Southern Europe, with a decreasing gradient towards Northern Europe [9]. This finding is in discussion as other SLC45A2 variants have been proposed as melanoma risk factors [34,36,37].
Affected genes in melanoma-subordinate syndromes
Pathogenic variants in melanoma-associated genes can predispose not only to melanoma, but also to a variety of specific cancers, which often derive from a specific tumor cluster and thus can be defined as a mixed cancer syndrome (MCS) or melanoma tumor syndrome [1]. These syndromes can manifest themselves as melanoma-dominant, with pathogenic variants in CDKN2A, CDK4, BAP1, MITF, and POT1, or as melanoma-subordinate syndromes. Table 1 provides a summary of the latter [18].
Table 1.
Melanoma-subordinate syndromes
| Syndrome name | Main pathology | Affected genes | Reference |
|---|---|---|---|
| Werner syndrome | Accelerated aging | WRN | [57,58] |
| BRCA1- and BRCA2-associated hereditary breast and ovarian cancer syndrome (HBOC) | Breast and ovarian cancer | BRCA1, BRCA2 | [59] |
| PTEN hamartoma tumor syndrome (PHTS) with its subform Cowden syndrome | Hamartoma, macrocephaly, gastrointestinal polyposis, lipoma, intellectual disabilities, disorders of the autism spectrum and increased cancer risk | PTEN | [18,60] |
| Lynch syndrome Alternatively, hereditary non-polyposis colorectal cancer syndrome (HNPCC) | Colorectal, endometrial, and ovarian cancer | MLH1, MSH2, MSH6, PMS2, EPCAM | [57,61] |
| Li-Fraumeni syndrome (LFS) Alternatively, sarcoma, breast, leukemia, and adrenal gland syndrome (SBLA) | Adrenocortical carcinoma, breast cancer, central nervous system tumors, osteosarcoma, soft-tissue sarcoma | TP53 | [57,62] |
| Xeroderma pigmentosum | Non-melanoma skin cancers | XPA, XPB, XPC, XPD, XPF, XPG, POLH | [18,63] |
Candidate genes
Only a few genes with high-penetrance variants associated with malignant melanoma have been identified, leading to the presumption that most cases of hereditary melanoma carry various low-risk gene alterations and other genetic modifiers [9].
Aside from the high-and medium-penetrance variants in the established genes described above, several genome-wide association studies (GWAS), for example mentioned in the study of Landi et al. (2020), have found a high number of loci associated with melanoma and its phenotypic associations such as pigmentation, tanning ability, and nevus count [9,38]. Affected genes can also participate in DNA repair or regulation of the telomeric regions [38,39]. Table 2 provides an overview of candidate genes that can harbor single or multiple low-penetrance germline variants associated with melanoma and its risk factors. These candidate genes were also visually displayed as a gene network analysis in Figure 3.
Table 2.
Melanoma susceptibility candidate genes
| Affected mechanism or pathology | Candidate genes | Reference |
|---|---|---|
| Pigmentation and tanning ability | ADAM15, AGR3, ASIP, CDKAL1, CYP1B1, DSTYK, FOXD3, GBA, GPR98, HDGFL1, IRF4, KIAA0930, MAFF, MCF2L, MED13L, MFSD12, MSC, MX2, OCA2, HERC2, PLA2G6, PPARGC1B, RP11-383H13.1, SOX6, TYR, TYRP1, ZBTB7B | [8,9,38,39,42] |
| Nevus count | AGR3, AKAP12, ASAP1, ATM, CASP8, CBWD1, CDH1, CYP1B1, DTNB, DTNBP1, FNM1, FTO, GDI2, HDAC4, HDGFL1, IRF4, IRX6, KIAA0930, KIAA1239, KLF4, MAFF, MFSD12, MKLN1, MSC, MTAP, OBFC1, PARP1, PLA2G6, PPARGC1B, RAPGEF1, RP11-383H13.1, SOX6, SYNE2, TFAP2B, TMEM38B (intergenic), TP53, ZFP36L1 | [9,38,39] |
| Dysplastic nevi | CDK6, XRCC1 | [64] |
| Epidermal development | CASP8 | [42] |
| DNA repair | ATM, ERCC2, ERCC4, PARP1 | [9,39,42,65] |
| Telomere length and maintenance | CLMPT1L, MPHOSPH6, OBFC1, RETL1, TERT, TINF2 | [38,42,66] |
| Cell-cycle progression | ATM, CCND1, CDK10 | [42] |
| Uveal melanoma | ATM, BRCA1, BRCA2, CHEK2, CLPTM1L, CTNNA1, MDB4, MLH1, MSH3, MSH6, PALB2, PMS1, SMARCE1, TDP1, TP53 | [26,67-71] |
| Single primary melanomas (SPM) | CYP1B1 | [72] |
| Multiple primary melanomas (MPM) | MGMT (protective), PIK3CA, SPI1 (protective) | [72-74] |
| Melanoma (not further specified) | ACD, ACTRT3, ADTRP, AHNAK, AKAP12, APOBEC3A/3B, ARHGEF40, ARNT/SETDB1, ATM, BACH2, BRCA2, BRD9, BRIP1, CASP8, CBWD1, CCND1, CDC91L1, CDH23, CDKAL1, CDKN2B, CTSK, DLG2, DNAJB4, DOT1L, EBF3, EZH2, FABP2, FAM160B2, FAT3, FTO, FZD4, GATA2, GNA11, GOLM1, HAL, HERC2, HLA-DQB2, IKZF2, IL1RN, IRF/EXOC2, LASS2ANXA9, LMO3, LMO7, MAP3K1, MARK3, MCL1, MET, MGMT, MSH2, MTAP, MTH7B, MX2, MXI1, NEK11, NGLY1, NIPAL3, OCA2, PAH, PALB2, PARP1, PIGU, PLA2G6, POLE, PPFIBP2, PRKDC, PROSER2, PRSS23, PTPN14, RAD50, RAD51B, RAPGEF5, RASA3, RB1, RP11-256L6.3, RREB1, SDHA, SLC24A5, SLCO4C1, STK11, TMEM135, TMEM136, TMEM163, TYR, TYRP1, WRN, XRCC3 | [8,9,36,39,42,54,65,72,75-97] |
| Copy number variation (CNV)-associated susceptibility: ACBD3, ACDK3, AK055856, ANGPT1, BC032899, BC039356, CABC1, CDC42BPA, CDKN2A, CLP-36, CXCR4, E2F1, GBE1, GCNT2, IDH1, ITPKB, HIST1H1B, KIAA1296, LIN9, MIXL1, PARP1, PDE5A, PDLIM1, PSEN2, SORBS1, SPOPL, STUM, ZNF517 | [98-100] |
Figure 3.
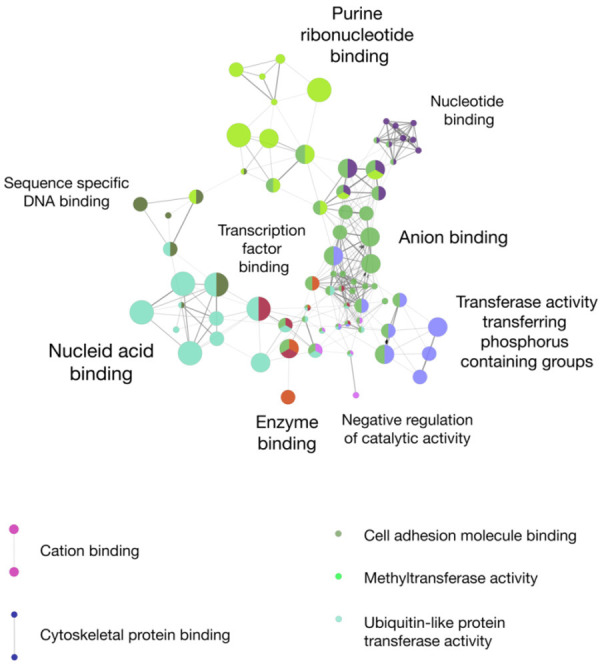
Melanoma susceptibility candidate genes grouped by molecular function. We used the ClueGO framework (Version 2.5.8) in Cytoscape (Version 3.8.2) to visualize a gene network analysis of melanoma susceptibility candidate genes [101,102]. Colors represent affiliations to gene groups with similar molecular functions. The node size and leading GO term were based on the gene quantity per term. GO term fusion was applied.
Polygenic risk scores (PRS)
Several genome-wide association studies (GWAS) have identified thousands of possible pathogenic loci with low susceptibility to melanoma. This can lead to possible embedding of polygenic risk scores with their weighted sum of low-risk variants into clinical application to improve risk prediction [40,41]. Several polygenic risk scores have been developed for melanoma, which show increased sensitivity compared to regular phenotypic risk scores comprising of skin, eye and hair color, freckle and nevi number, environmental factors, and monogenetic predisposition. So far, these proposed polygenic risk scores are made up of 11-204 SNVs, with most of the scores containing between 11 and 45 examined SNV locations [42].
Noteworthy PRS findings due to their high odds ratios (OR) for high-risk groups were published by Potjer et al. (2021) and Bakshi et al. (2021), with maximum OR values of 5.70 and 3.66, respectively [43,44]. The PRS was significantly higher in patients with multiple primary melanomas (MPM) than in those with single primary melanomas (SPM). However, PRS values were slightly, although not significantly lower in families with a higher than average cancer burden, probably due to the possible presence of a more dominant, still unidentified, family-specific single pathogenic variant [44]. Graff et al. (2021) also found a link between melanoma and cancer of the oral cavity and pharynx, while analyzing several cancer PRS and their possible pleiotropy [45].
PRS for skin cancers are not currently in clinical use, although some show equally good sensitivity rates as polygenic risk scores for prostate, lung, or breast cancer, which are commonly included in regular gene panel testing [40,42]. It has to be mentioned that until now, GWAS for melanoma have only been conducted in Western populations and that participating patient numbers were limited [42]. In this context, Zhang et al. (2020) predicted that considerably increasing sample numbers could lead to a 40% reduction in missing hereditability of GWAS and at the same time, improve PRS quality [41]. Another effort to advance research in the field of low-penetrance variant risk prediction is found in the “Cancer PRSweb” project, a database systematically collecting publicly available PRS data and currently evaluating it against the biobanks of Michigan Genomics Initiative and UK Biobank [46].
Gene interactions
The epistasis of gene variants can affect melanoma susceptibility. It has been found that pathogenic MC1R variants have a direct positive effect on the penetrance of pathogenic CDK2NA variants. There have been other results on epistatic interactions between SLC45A2 and VDR (vitamin D receptor), MC1R and TYR (tyrosinase), as well as TERF1 (telomeric repeat binding factor 1) and AFAP1L2 (actin filament associated protein 1 like 2), with the last-mentioned pair further confirming a connection between telomere length and melanoma risk [9,39]. Sangalli et al. (2017) discovered a sex-specific genetic interaction. Men who carry the RNASEL rs486907 A allele, as well as the C allele of miR-146a rs2910164, have a higher risk of developing malignant melanoma [47]. Wu et al. (2018) suggested two clusters with five and 17 genes, respectively, leading to an increased melanoma risk due to their epistatic interactions although their findings need further statistical validation [48]. Furthermore, while doing a study on a polygenic risk score (PRS) for a Dutch population, Potjer et al. (2020) discovered, that in order to develop melanoma, carriers of the MITF p.E318K variant require additional genetic risk factors and a high PRS may represent these. They hypothesized that this might also be true for other medium-penetrance variants [44].
Constitutional epigenetics
As a newer field in genetics, epigenetic alterations have been widely researched with regard to somatic melanoma mutations. In the last few years, there has also been an effort to prove a statistical association between constitutional epigenetic changes associated with hereditary melanoma, either with or without present pathogenic DNA variants [49]. The focus of these studies was mostly on hypo- and hypermethylation. Other mechanisms of epigenetic change may include chromatin remodeling, histone modifications and regulation by non-coding RNAs [12,50-52].
The most recent genome-wide association study (GWAS) by Salgado et al. (2020) did not find any support for heritable epimutations as a cause of familial melanoma. However, they studied a small group, which spanned only five families from the Netherlands where melanoma occurred throughout the ancestral tree [52]. In addition, no specific promoter methylation events were found by Boru et al. (2019) regarding uveal melanoma in melanoma-prone families or by Hyland et al. (2013) for the promoter region of LINE-1 (long interspersed nuclear element-1) in cutaneous melanoma [49,50].
In contrast, several other studies have shown significant changes in methylation patterns in peripheral blood cells, as surrogate cells, and subsequently proposed a possible link between epigenetic alterations and the risk of melanoma [11,12]. Pergoli et al. (2014) studied peripheral blood mononuclear cells (PBMCs) as non-tumor substitution cells. They suggested a possible link between germline (constitutional) epimutations and the risk of cutaneous melanoma, although they could not exclude the roles of environmental factors and somatic epimutations. They observed that controls carrying dysplastic nevi were more likely to show decreased methylation levels of TNF-α and hTERT. Cutaneous melanoma itself was associated with increased methylation of the TNF-α promoter and the transposable element ALU. Increased methylation of CDKN2A/p16 and MLH1 in PBMCs was found to be associated with cutaneous melanoma [12]. Cappetta et al. (2015) discovered a statistically viable link between DNA hypomethylation in leukocytes, melanoma, and breast cancer [53]. Hyland et al. (2014) found statistical evidence that constitutional hypomethylation and increased expression of TNFRSF10C in blood DNA might be associated with cutaneous melanoma. Their study included CDKN2A variant positive and negative cases [11]. In contrast to Pergoli et al., there was no evidence that constitutional epigenetic changes in CDKN2A might play a role in cutaneous melanoma susceptibility, however, interestingly TNFRSF10C hypomethylation only occurred in CDKN2A pathogenic variant-free probands [11]. The first epigenome-wide association study (EWAS) related to melanoma risk factors was performed by Roos et al. (2017), who showed that several differentially methylated regions are associated with a higher nevus count and melanoma risk. The top-ranked regions involved strong enhancers in melanocyte biology such as RAF1 and CTC1. In cis, differentially methylated regions in known GWAS SNVs, for example, in PLA2G6 and NID1 were associated with increased nevus count, and in MC1R, MX2, and TERT/CLPTM1L with melanoma risk [54]. The epigenetic findings from the studies mentioned above are shown in Table 3 and Figure 4.
Table 3.
Constitutional epigenetic changes of specific genes and melanoma susceptibility
| Study | Gene methylation associated with melanoma risk factors | Gene methylation associated with cutaneous melanoma | Reference |
|---|---|---|---|
| Pergoli et al. (2014) | DYSPLASTIC NEVI | Positive: CDKN2A/p16, MLH1, TNF-α and with lower nevi count (<20), ICAM-1 and ALU can also be associated with cutaneous melanoma | [12] |
| Positive: ALU | Negative (protective): CDK4, yet positively associated with higher nevi count (>20) | ||
| Negative (protective): hTERT, TNF-α | |||
| Hyland et al. (2014) | - | Hypomethylation of TNFRSF10C (occurred only in probands without CDKN2A pathogenic variants) | [11] |
| Cappetta et al. (2015) | - | Widespread non-gene specific DNA hypomethylation in leukocytes (also valid for breast cancer) | [53] |
| Roos et al. (2017) | HIGH NEVUS COUNT | DNA methylation changes in ACTRT3, ANXA9, ARNT, ASIP, ATM, CASP8, CDC91L1, DKN2A, CLPTM1L, CTSK, DOCK3, EYS, FTO, LASS2, MC1R, MCL1, MX2, NR, PARP1, PLA2G6, SETDB1, SLC45A2, TERT, TET2, TYR | [54] |
| Hypermethylation: ARRDC1, FAM107B, KCNN4 | |||
| Hypomethylation: CTC1, GABRB3 | |||
| Unspecified DNA methylation changes: NID1, PLA2G6, RAF1, STUM, ZSWIM2 |
Figure 4.
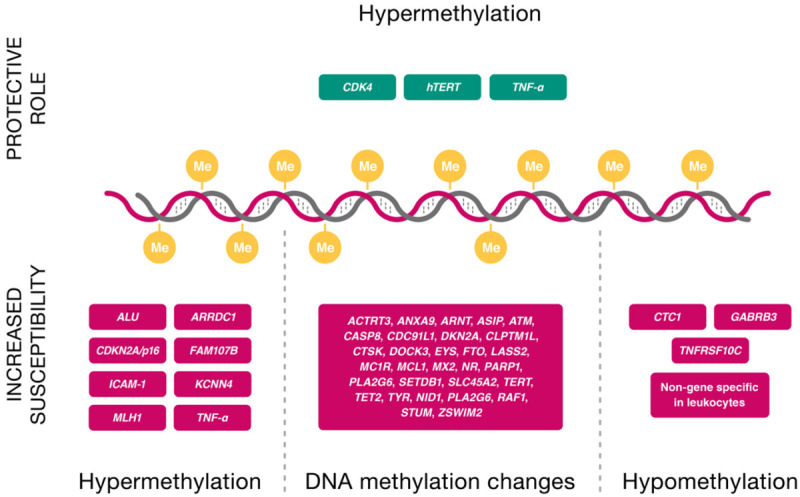
Constitutional epigenetic changes of specific genes associated with melanoma and its risk factors. Me: methyl group.
Overall epigenetic research in melanoma has advanced more towards markers for detection and management of malignant melanoma. However, for treatment prognosis, Cortellini et al. (2018) showed that patients with a variety of hereditary cancers, including melanoma, had a significantly better response to treatment, an increased time to treatment failure, and a better overall survival rate [55].
Discussion
In analyzing scientific work, mostly from the last five years, this review aimed to collect and summarize all available up-to-date knowledge on genetic and epigenetic pathomechanisms of melanoma susceptibility.
Findings and recommendations indicate that the scientific achievements of the previous years can be used in two ways; either to reduce mortality rates through better genetic and thus clinical screening for this cancer type, or, second, to increase chances of better treatment outcome through personalized medicine, relying on genetic and epigenetic markers.
We expected to find genome-wide association studies resulting in the discovery of several high-or medium-penetrance germline variants. However, research has shown that malignant melanoma is a cancer that is most likely to develop not only due to high-penetrance variants but also due to polygenic inheritance patterns, leaving no clear division between the hereditary and sporadic development of tumors [9,31]. As a result, there were noticeable early changes in genetic testing. Previously, only a few single genes were tested clinically. Today, polygenic risk scores (PRS) for malignant melanoma are in development, showing equally good sensitivity rates to those for prostate, lung, or breast cancer, which are commonly included in regular gene panel testing. These polygenic risk scores contain, in addition to high-penetrance variants, a wide array of pathogenic low-and medium-penetrance germline variants that are being tested [31,40,42]. To make polygenic risk scores for melanoma viable for clinical use, larger biobanking, studies on wider and higher variety of populations, and validation of risk prediction reliability are necessary [42].
Genetic testing for melanoma risk could be especially beneficial to teenagers and young adults, as intensified screening can detect lesions early [7,56]. In addition, a negative test result after genetic testing can provide partial relief to especially anxious family members [1].
Unsurprisingly, several pathogenic variants increase susceptibility to melanoma in fair-skinned patients. Despite this, specific germline variants, which are associated with increased melanoma risk, have been found in patients with a darker skin type [9]. Generally, risk group patients harboring pathogenic germline alterations should undergo more frequent dermatological check-ups, as better treatment outcomes often follow early lesion detection. It is a common motivation to support intensified genetic testing, as this could be a method to increase the productive years of affected individuals and possibly lower the financial burden on healthcare systems.
Limitations to findings in studies on melanoma susceptibility are the few studies, small population sizes, and the lack of ethnic diversity. Studies are mostly undertaken in Western countries of the Northern Hemisphere, as well as in Australia.
In conclusion, further studies with larger populations and a wider variety of ethnicities and diverse geographical areas are recommended. It would also be advisable to endeavor this in the form of partnerships between several institutions to create bigger datasets, thus hopefully finding additional genetic patterns with increased accuracy. As previously mentioned, this could lead to improved prediction of melanoma susceptibility, which is most likely applicable through advanced polygenic risk scores. The use of advanced risk scores, consisting of PRS, genetic, epigenetic, and phenotypic risk factors, could be favorable, especially for countries that lack infrastructure for satisfactory melanoma screening programs [40].
Disclosure of conflict of interest
None.
References
- 1.Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: update on syndromes and management: genetics of familial atypical multiple mole melanoma syndrome. J Am Acad Dermatol. 2016;74:395–407. doi: 10.1016/j.jaad.2015.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- 3.Shimizu H. (Malignant) melanoma. In: Shimizu H, editor. Shimizu’s dermatology. 2nd edition. Chichester: John Wiley & Sons, Ltd.; 2017. pp. 521–527. [Google Scholar]
- 4.Guy GP, Ekwueme DU. Years of potential life lost and indirect costs of melanoma and non-melanoma skin cancer: a systematic review of the literature. Pharmacoeconomics. 2011;29:863–874. doi: 10.2165/11589300-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Brustugun OT, Moller B, Helland A. Years of life lost as a measure of cancer burden on a national level. Br J Cancer. 2014;111:1014–1020. doi: 10.1038/bjc.2014.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarrett SG, D’Orazio JA. Hormonal regulation of the repair of UV photoproducts in melanocytes by the melanocortin signaling axis. Photochem Photobiol. 2017;93:245–258. doi: 10.1111/php.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neill CH, Scoggins CR. Melanoma. J Surg Oncol. 2019;120:873–881. doi: 10.1002/jso.25604. [DOI] [PubMed] [Google Scholar]
- 8.Pastorino L, Andreotti V, Dalmasso B, Vanni I, Ciccarese G, Mandala M, Spadola G, Pizzichetta MA, Ponti G, Tibiletti MG, Sala E, Genuardi M, Chiurazzi P, Maccanti G, Manoukian S, Sestini S, Danesi R, Zampiga V, La Starza R, Stanganelli I, Ballestrero A, Mastracci L, Grillo F, Sciallero S, Cecchi F, Tanda ET, Spagnolo F, Queirolo P, Italian Melanoma I, Goldstein AM, Bruno W, Ghiorzo P. Insights into genetic susceptibility to melanoma by gene panel testing: potential pathogenic variants in ACD, ATM, BAP1, and POT1. Cancers (Basel) 2020;12:1007. doi: 10.3390/cancers12041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Read J, Wadt KA, Hayward NK. Melanoma genetics. J Med Genet. 2016;53:1–14. doi: 10.1136/jmedgenet-2015-103150. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JH, Taylor JC, Bright C, Harland M, Dunning AM, Akslen LA, Andresen PA, Avril MF, Azizi E, Bianchi Scarra G, Brossard M, Brown KM, Debniak T, Elder DE, Friedman E, Ghiorzo P, Gillanders EM, Gruis NA, Hansson J, Helsing P, Hocevar M, Hoiom V, Ingvar C, Landi MT, Lang J, Lathrop GM, Lubinski J, Mackie RM, Molven A, Novakovic S, Olsson H, Puig S, Puig-Butille JA, van der Stoep N, van Doorn R, van Workum W, Goldstein AM, Kanetsky PA, Pharoah PD, Demenais F, Hayward NK, Newton Bishop JA, Bishop DT, Iles MM GenoMEL Consortium. Fine mapping of genetic susceptibility loci for melanoma reveals a mixture of single variant and multiple variant regions. Int J Cancer. 2015;136:1351–1360. doi: 10.1002/ijc.29099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyland PL, Burke LS, Pfeiffer RM, Rotunno M, Sun D, Patil P, Wu X, Tucker MA, Goldstein AM, Yang XR. Constitutional promoter methylation and risk of familial melanoma. Epigenetics. 2014;9:685–692. doi: 10.4161/epi.28151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pergoli L, Favero C, Pfeiffer RM, Tarantini L, Calista D, Cavalleri T, Angelici L, Consonni D, Bertazzi PA, Pesatori AC, Landi MT, Bollati V. Blood DNA methylation, nevi number, and the risk of melanoma. Melanoma Res. 2014;24:480–487. doi: 10.1097/CMR.0000000000000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middlebrooks CD, Stacey ML, Li Q, Snyder C, Shaw TG, Richardson-Nelson T, Rendell M, Ferguson C, Silberstein P, Casey MJ, Bailey-Wilson JE, Lynch HT. Analysis of the CDKN2A gene in FAMMM syndrome families reveals early age of onset for additional syndromic cancers. Cancer Res. 2019;79:2992–3000. doi: 10.1158/0008-5472.CAN-18-1580. [DOI] [PubMed] [Google Scholar]
- 14.Debniak T, van de Wetering T, Scott R, Nagay L, Cybulski C, Gorski B, Jakubowska A, Gronwald J, Masojc B, Huzarski T, Byrski T, Nej-Wolosiak K, Kladny J, Maleszka R, Lubinski J. Low prevalence of CDKN2A/ARF mutations among early-onset cancers of breast, pancreas and malignant melanoma in Poland. Eur J Cancer Prev. 2008;17:389–391. doi: 10.1097/CEJ.0b013e3282f75eb1. [DOI] [PubMed] [Google Scholar]
- 15.Erlandson A, Appelqvist F, Enerback C. Epigenetic mutations in CDKN2A in western Swedish families with hereditary malignant melanoma. Mol Med Rep. 2008;1:89–91. [PubMed] [Google Scholar]
- 16.Gensini F, Sestini R, Piazzini M, Vignoli M, Chiarugi A, Brandani P, Ghiorzo P, Salvini C, Borgognoni L, Palli D, Bianchi-Scarra G, Carli P, Genuardi M. The p. G23S CDKN2A founder mutation in high-risk melanoma families from Central Italy. Melanoma Res. 2007;17:387–392. doi: 10.1097/CMR.0b013e3282f1d328. [DOI] [PubMed] [Google Scholar]
- 17.Law MH, Aoude LG, Duffy DL, Long GV, Johansson PA, Pritchard AL, Khosrotehrani K, Mann GJ, Montgomery GW, Iles MM, Cust AE, Palmer JM, Melanoma GC, Shannon KF, Spillane AJ, Stretch JR, Thompson JF, Saw RPM, Scolyer RA, Martin NG, Hayward NK, MacGregor S. Multiplex melanoma families are enriched for polygenic risk. Hum Mol Genet. 2020;29:2976–2985. doi: 10.1093/hmg/ddaa156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leachman SA, Lucero OM, Sampson JE, Cassidy P, Bruno W, Queirolo P, Ghiorzo P. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017;36:77–90. doi: 10.1007/s10555-017-9661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betti M, Aspesi A, Biasi A, Casalone E, Ferrante D, Ogliara P, Gironi LC, Giorgione R, Farinelli P, Grosso F, Libener R, Rosato S, Turchetti D, Maffe A, Casadio C, Ascoli V, Dianzani C, Colombo E, Piccolini E, Pavesi M, Miccoli S, Mirabelli D, Bracco C, Righi L, Boldorini R, Papotti M, Matullo G, Magnani C, Pasini B, Dianzani I. CDKN2A and BAP1 germline mutations predispose to melanoma and mesothelioma. Cancer Lett. 2016;378:120–130. doi: 10.1016/j.canlet.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Jouenne F, Chauvot de Beauchene I, Bollaert E, Avril MF, Caron O, Ingster O, Lecesne A, Benusiglio P, Terrier P, Caumette V, Pissaloux D, de la Fouchardiere A, Cabaret O, N’Diaye B, Velghe A, Bougeard G, Mann GJ, Koscielny S, Barrett JH, Harland M, Newton-Bishop J, Gruis N, Van Doorn R, Gauthier-Villars M, Pierron G, Stoppa-Lyonnet D, Coupier I, Guimbaud R, Delnatte C, Scoazec JY, Eggermont AM, Feunteun J, Tchertanov L, Demoulin JB, Frebourg T, Bressac-de Paillerets B. Germline CDKN2A/P16INK4A mutations contribute to genetic determinism of sarcoma. J Med Genet. 2017;54:607–612. doi: 10.1136/jmedgenet-2016-104402. [DOI] [PubMed] [Google Scholar]
- 21.van der Wilk BJ, Noordman BJ, Atmodimedjo PN, Dinjens WNM, Laheij RJF, Wagner A, Wijnhoven BPL, van Lanschot JJB. Development of esophageal squamous cell cancer in patients with FAMMM syndrome: two clinical reports. Eur J Med Genet. 2020;63:103840. doi: 10.1016/j.ejmg.2020.103840. [DOI] [PubMed] [Google Scholar]
- 22.Chan AK, Han SJ, Choy W, Beleford D, Aghi MK, Berger MS, Shieh JT, Bollen AW, Perry A, Phillips JJ, Butowski N, Solomon DA. Familial melanoma-astrocytoma syndrome: synchronous diffuse astrocytoma and pleomorphic xanthoastrocytoma in a patient with germline CDKN2A/B deletion and a significant family history. Clin Neuropathol. 2017;36:213–221. doi: 10.5414/NP301022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalmasso B, Ghiorzo P. Evolution of approaches to identify melanoma missing heritability. Expert Rev Mol Diagn. 2020;20:523–531. doi: 10.1080/14737159.2020.1738221. [DOI] [PubMed] [Google Scholar]
- 24.Bottillo I, La Starza R, Radio FC, Molica C, Pedace L, Pierini T, De Bernardo C, Stingeni L, Bargiacchi S, Paiardini A, Janson G, Mecucci C, Grammatico P. A novel germline mutation in CDK4 codon 24 associated to familial melanoma. Clin Genet. 2018;93:934–935. doi: 10.1111/cge.13129. [DOI] [PubMed] [Google Scholar]
- 25.Zhang AJ, Rush PS, Tsao H, Duncan LM. BRCA1-associated protein (BAP1)-inactivated melanocytic tumors. J Cutan Pathol. 2019;46:965–972. doi: 10.1111/cup.13530. [DOI] [PubMed] [Google Scholar]
- 26.Toomey CB, Protopsaltis NJ, Phou S, Bakhoum MF, Thorson JA, Ediriwickrema LS, Korn BS, Kikkawa DO, Goldbaum MH, Lin JH. Prevalence of mismatch repair gene mutations in uveal melanoma. Retina. 2020;40:2216–2220. doi: 10.1097/IAE.0000000000002732. [DOI] [PubMed] [Google Scholar]
- 27.Potrony M, Badenas C, Aguilera P, Puig-Butille JA, Carrera C, Malvehy J, Puig S. Update in genetic susceptibility in melanoma. Ann Transl Med. 2015;3:210. doi: 10.3978/j.issn.2305-5839.2015.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, Nagore E, Avril MF, Caporaso NE, McMaster ML, Cullen M, Wang Z, Zhang X NCI DCEG Cancer Sequencing Working Group; NCI DCEG Cancer Genomics Research Laboratory; French Familial Melanoma Study Group. Bruno W, Pastorino L, Queirolo P, Banuls-Roca J, Garcia-Casado Z, Vaysse A, Mohamdi H, Riazalhosseini Y, Foglio M, Jouenne F, Hua X, Hyland PL, Yin J, Vallabhaneni H, Chai W, Minghetti P, Pellegrini C, Ravichandran S, Eggermont A, Lathrop M, Peris K, Scarra GB, Landi G, Savage SA, Sampson JN, He J, Yeager M, Goldin LR, Demenais F, Chanock SJ, Tucker MA, Goldstein AM, Liu Y, Landi MT. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46:482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen E, Xiu J, Lopez GY, Bentley R, Jalali A, Heimberger AB, Bainbridge MN, Bondy ML, Walsh KM. POT1 mutation spectrum in tumour types commonly diagnosed among POT1-associated hereditary cancer syndrome families. J Med Genet. 2020;57:664–670. doi: 10.1136/jmedgenet-2019-106657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toussi A, Mans N, Welborn J, Kiuru M. Germline mutations predisposing to melanoma. J Cutan Pathol. 2020;47:606–616. doi: 10.1111/cup.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Ek WE, Whiteman D, Vaughan TL, Spurdle AB, Easton DF, Pharoah PD, Thompson DJ, Dunning AM, Hayward NK, Chenevix-Trench G Q-MEGA and AMFS Investigators; ANECS-SEARCH; UKOPS-SEARCH; BEACON Consortium. Macgregor S. Most common ‘sporadic’ cancers have a significant germline genetic component. Hum Mol Genet. 2014;23:6112–6118. doi: 10.1093/hmg/ddu312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iles MM, Bishop DT, Taylor JC, Hayward NK, Brossard M, Cust AE, Dunning AM, Lee JE, Moses EK, Akslen LA AMFS Investigators; Andresen PA, Avril MF, Azizi E, Scarra GB, Brown KM, Debniak T, Elder DE, Friedman E, Ghiorzo P, Gillanders EM, Goldstein AM, Gruis NA, Hansson J, Harland M, Helsing P, Hocevar M, Hoiom V IBD investigators; Ingvar C, Kanetsky PA, Landi MT, Lang J, Lathrop GM, Lubinski J, Mackie RM, Martin NG, Molven A, Montgomery GW, Novakovic S, Olsson H, Puig S, Puig-Butille JA QMEGA and QTWIN Investigators; Radford-Smith GL, Randerson-Moor J SDH Study Group; van der Stoep N, van Doorn R, Whiteman DC, MacGregor S, Pooley KA, Ward SV, Mann GJ, Amos CI, Pharoah PD, Demenais F, Law MH, Newton Bishop JA, Barrett JH GenoMEL Consortium. The effect on melanoma risk of genes previously associated with telomere length. J Natl Cancer Inst. 2014;106:dju267. doi: 10.1093/jnci/dju267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinsler VA, O’Hare P, Bulstrode N, Calonje JE, Chong WK, Hargrave D, Jacques T, Lomas D, Sebire NJ, Slater O. Melanoma in congenital melanocytic naevi. Br J Dermatol. 2017;176:1131–1143. doi: 10.1111/bjd.15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibarrola-Villava M, Hu HH, Guedj M, Fernandez LP, Descamps V, Basset-Seguin N, Bagot M, Benssussan A, Saiag P, Fargnoli MC, Peris K, Aviles JA, Lluch A, Ribas G, Soufir N. MC1R, SLC45A2 and TYR genetic variants involved in melanoma susceptibility in southern European populations: results from a meta-analysis. Eur J Cancer. 2012;48:2183–2191. doi: 10.1016/j.ejca.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Nathan V, Johansson PA, Palmer JM, Howlie M, Hamilton HR, Wadt K, Jonsson G, Brooks KM, Pritchard AL, Hayward NK. Germline variants in oculocutaneous albinism genes and predisposition to familial cutaneous melanoma. Pigment Cell Melanoma Res. 2019;32:854–863. doi: 10.1111/pcmr.12804. [DOI] [PubMed] [Google Scholar]
- 36.Reis LB, Bakos RM, Vianna FSL, Macedo GS, Jacovas VC, Ribeiro-Dos-Santos AM, Santos S, Bakos L, Ashton-Prolla P. Skin pigmentation polymorphisms associated with increased risk of melanoma in a case-control sample from southern Brazil. BMC Cancer. 2020;20:1069. doi: 10.1186/s12885-020-07485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ransohoff KJ, Wu W, Cho HG, Chahal HC, Lin Y, Dai HJ, Amos CI, Lee JE, Tang JY, Hinds DA, Han J, Wei Q, Sarin KY. Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget. 2017;8:17586–17592. doi: 10.18632/oncotarget.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landi MT, Bishop DT, MacGregor S, Machiela MJ, Stratigos AJ, Ghiorzo P, Brossard M, Calista D, Choi J, Fargnoli MC, Zhang T, Rodolfo M, Trower AJ, Menin C, Martinez J, Hadjisavvas A, Song L, Stefanaki I, Scolyer R, Yang R, Goldstein AM, Potrony M, Kypreou KP, Pastorino L, Queirolo P, Pellegrini C, Cattaneo L, Zawistowski M, Gimenez-Xavier P, Rodriguez A, Elefanti L, Manoukian S, Rivoltini L, Smith BH, Loizidou MA, Del Regno L, Massi D, Mandala M, Khosrotehrani K, Akslen LA, Amos CI, Andresen PA, Avril MF, Azizi E, Soyer HP, Bataille V, Dalmasso B, Bowdler LM, Burdon KP, Chen WV, Codd V, Craig JE, Debniak T, Falchi M, Fang S, Friedman E, Simi S, Galan P, Garcia-Casado Z, Gillanders EM, Gordon S, Green A, Gruis NA, Hansson J, Harland M, Harris J, Helsing P, Henders A, Hocevar M, Hoiom V, Hunter D, Ingvar C, Kumar R, Lang J, Lathrop GM, Lee JE, Li X, Lubinski J, Mackie RM, Malt M, Malvehy J, McAloney K, Mohamdi H, Molven A, Moses EK, Neale RE, Novakovic S, Nyholt DR, Olsson H, Orr N, Fritsche LG, Puig-Butille JA, Qureshi AA, Radford-Smith GL, Randerson-Moor J, Requena C, Rowe C, Samani NJ, Sanna M, Schadendorf D, Schulze HJ, Simms LA, Smithers M, Song F, Swerdlow AJ, van der Stoep N, Kukutsch NA, Visconti A, Wallace L, Ward SV, Wheeler L, Sturm RA, Hutchinson A, Jones K, Malasky M, Vogt A, Zhou W, Pooley KA, Elder DE, Han J, Hicks B, Hayward NK, Kanetsky PA, Brummett C, Montgomery GW, Olsen CM, Hayward C, Dunning AM, Martin NG, Evangelou E, Mann GJ, Long G, Pharoah PDP, Easton DF, Barrett JH, Cust AE, Abecasis G, Duffy DL, Whiteman DC, Gogas H, De Nicolo A, Tucker MA, Newton-Bishop JA GenoMEL Consortium; Q-MEGA and QTWIN Investigators; ATHENS Melanoma Study Group; 23andMe; SDH Study Group; IBD Investigators; Essen-Heidelberg Investigators; AMFS Investigators; MelaNostrum Consortium. Peris K, Chanock SJ, Demenais F, Brown KM, Puig S, Nagore E, Shi J, Iles MM, Law MH. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law MH, Bishop DT, Lee JE, Brossard M, Martin NG, Moses EK, Song F, Barrett JH, Kumar R, Easton DF, Pharoah PDP, Swerdlow AJ, Kypreou KP, Taylor JC, Harland M, Randerson-Moor J, Akslen LA, Andresen PA, Avril MF, Azizi E, Scarra GB, Brown KM, Debniak T, Duffy DL, Elder DE, Fang S, Friedman E, Galan P, Ghiorzo P, Gillanders EM, Goldstein AM, Gruis NA, Hansson J, Helsing P, Hocevar M, Hoiom V, Ingvar C, Kanetsky PA, Chen WV GenoMEL Consortium; Essen-Heidelberg Investigators; SDH Study Group; Q-MEGA and QTWIN Investigators; AMFS Investigators; ATHENS Melanoma Study Group. Landi MT, Lang J, Lathrop GM, Lubinski J, Mackie RM, Mann GJ, Molven A, Montgomery GW, Novakovic S, Olsson H, Puig S, Puig-Butille JA, Qureshi AA, Radford-Smith GL, van der Stoep N, van Doorn R, Whiteman DC, Craig JE, Schadendorf D, Simms LA, Burdon KP, Nyholt DR, Pooley KA, Orr N, Stratigos AJ, Cust AE, Ward SV, Hayward NK, Han J, Schulze HJ, Dunning AM, Bishop JAN, Demenais F, Amos CI, MacGregor S, Iles MM. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soura E, Stratigos A. Implementing polygenic risk scores in skin cancer: a step towards personalized risk prediction. Br J Dermatol. 2019;181:1117–1118. doi: 10.1111/bjd.18324. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YD, Hurson AN, Zhang H, Choudhury PP, Easton DF, Milne RL, Simard J, Hall P, Michailidou K, Dennis J, Schmidt MK, Chang-Claude J, Gharahkhani P, Whiteman D, Campbell PT, Hoffmeister M, Jenkins M, Peters U, Hsu L, Gruber SB, Casey G, Schmit SL, O’Mara TA, Spurdle AB, Thompson DJ, Tomlinson I, De Vivo I, Landi MT, Law MH, Iles MM, Demenais F, Kumar R, MacGregor S, Bishop DT, Ward SV, Bondy ML, Houlston R, Wiencke JK, Melin B, Barnholtz-Sloan J, Kinnersley B, Wrensch MR, Amos CI, Hung RJ, Brennan P, McKay J, Caporaso NE, Berndt SI, Birmann BM, Camp NJ, Kraft P, Rothman N, Slager SL, Berchuck A, Pharoah PDP, Sellers TA, Gayther SA, Pearce CL, Goode EL, Schildkraut JM, Moysich KB, Amundadottir LT, Jacobs EJ, Klein AP, Petersen GM, Risch HA, Stolzenberg-Solomon RZ, Wolpin BM, Li D, Eeles RA, Haiman CA, Kote-Jarai Z, Schumacher FR, Al Olama AA, Purdue MP, Scelo G, Dalgaard MD, Greene MH, Grotmol T, Kanetsky PA, McGlynn KA, Nathanson KL, Turnbull C, Wiklund F Breast Cancer Association Consortium (BCAC); Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON); Colon Cancer Family Registry (CCFR); Transdisciplinary Studies of Genetic Variation in Colorectal Cancer (CORECT); Endometrial Cancer Association Consortium (ECAC); Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Melanoma Genetics Consortium (GenoMEL); Glioma International Case-Control Study (GICC); International Lung Cancer Consortium (ILCCO); Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium; International Consortium of Investigators Working on Non-Hodgkin’s Lymphoma Epidemiologic Studies (InterLymph); Ovarian Cancer Association Consortium (OCAC); Oral Cancer GWAS; Pancreatic Cancer Case-Control Consortium (PanC4); Pancreatic Cancer Cohort Consortium (PanScan); Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL); Renal Cancer GWAS; Testicular Cancer Consortium (TECAC) Chanock SJ, Chatterjee N, Garcia-Closas M. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat Commun. 2020;11:3353. doi: 10.1038/s41467-020-16483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts MR, Asgari MM, Toland AE. Genome-wide association studies and polygenic risk scores for skin cancer: clinically useful yet? Br J Dermatol. 2019;181:1146–1155. doi: 10.1111/bjd.17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakshi A, Yan M, Riaz M, Polekhina G, Orchard SG, Tiller J, Wolfe R, Joshi A, Cao Y, McInerney-Leo AM, Yanes T, Janda M, Soyer HP, Cust AE, Law MH, Gibbs P, McLean C, Chan AT, McNeil JJ, Mar VJ, Lacaze P. Genomic risk score for melanoma in a prospective study of older individuals. J Natl Cancer Inst. 2021;113:1379–1385. doi: 10.1093/jnci/djab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potjer TP, van der Grinten TWJ, Lakeman IMM, Bollen SH, Rodriguez-Girondo M, Iles MM, Barrett JH, Kiemeney LA, Gruis NA, van Asperen CJ, van der Stoep N. Association between a 46-SNP polygenic risk score and melanoma risk in Dutch patients with familial melanoma. J Med Genet. 2021;58:760–766. doi: 10.1136/jmedgenet-2020-107251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graff RE, Cavazos TB, Thai KK, Kachuri L, Rashkin SR, Hoffman JD, Alexeeff SE, Blatchins M, Meyers TJ, Leong L, Tai CG, Emami NC, Corley DA, Kushi LH, Ziv E, Van Den Eeden SK, Jorgenson E, Hoffmann TJ, Habel LA, Witte JS, Sakoda LC. Cross-cancer evaluation of polygenic risk scores for 16 cancer types in two large cohorts. Nat Commun. 2021;12:970. doi: 10.1038/s41467-021-21288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsche LG, Patil S, Beesley LJ, VandeHaar P, Salvatore M, Ma Y, Peng RB, Taliun D, Zhou X, Mukherjee B. Cancer PRSweb: an online repository with polygenic risk scores for major cancer traits and their evaluation in two independent biobanks. Am J Hum Genet. 2020;107:815–836. doi: 10.1016/j.ajhg.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sangalli A, Orlandi E, Poli A, Maurichi A, Santinami M, Nicolis M, Ferronato S, Malerba G, Rodolfo M, Gomez Lira M. Sex-specific effect of RNASEL rs486907 and miR-146a rs2910164 polymorphisms’ interaction as a susceptibility factor for melanoma skin cancer. Melanoma Res. 2017;27:309–314. doi: 10.1097/CMR.0000000000000360. [DOI] [PubMed] [Google Scholar]
- 48.Wu M, Huang J, Ma S. Identifying gene-gene interactions using penalized tensor regression. Stat Med. 2018;37:598–610. doi: 10.1002/sim.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyland PL, Burke LS, Pfeiffer RM, Mirabello L, Tucker MA, Goldstein AM, Yang XR. LINE-1 methylation in peripheral blood and the risk of melanoma in melanoma-prone families with and without CDKN2A mutations. Melanoma Res. 2013;23:55–60. doi: 10.1097/CMR.0b013e32835adc51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boru G, Grosel TW, Pilarski R, Stautberg M, Massengill JB, Jeter J, Singh A, Marino MJ, McElroy JP, Davidorf FH, Cebulla CM, Abdel-Rahman MH. Germline large deletion of BAP1 and decreased expression in non-tumor choroid in uveal melanoma patients with high risk for inherited cancer. Genes Chromosomes Cancer. 2019;58:650–656. doi: 10.1002/gcc.22752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mervis JS, McGee JS. DNA methylation and inflammatory skin diseases. Arch Dermatol Res. 2020;312:461–466. doi: 10.1007/s00403-019-02005-9. [DOI] [PubMed] [Google Scholar]
- 52.Salgado C, Gruis N, Consortium B, Heijmans BT, Oosting J, van Doorn R. Genome-wide analysis of constitutional DNA methylation in familial melanoma. Clin Epigenetics. 2020;12:43. doi: 10.1186/s13148-020-00831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cappetta M, Berdasco M, Hochmann J, Bonilla C, Sans M, Hidalgo PC, Artagaveytia N, Kittles R, Martinez M, Esteller M, Bertoni B. Effect of genetic ancestry on leukocyte global DNA methylation in cancer patients. BMC Cancer. 2015;15:434. doi: 10.1186/s12885-015-1461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roos L, Sandling JK, Bell CG, Glass D, Mangino M, Spector TD, Deloukas P, Bataille V, Bell JT. Higher nevus count exhibits a distinct DNA methylation signature in healthy human skin: implications for melanoma. J Invest Dermatol. 2017;137:910–920. doi: 10.1016/j.jid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortellini A, Bersanelli M, Buti S, Gambale E, Atzori F, Zoratto F, Parisi A, Brocco D, Pireddu A, Cannita K, Iacono D, Migliorino MR, Gamucci T, De Tursi M, Sidoni T, Tiseo M, Michiara M, Papa A, Angius G, Tomao S, Fargnoli MC, Natoli C, Ficorella C. Family history of cancer as surrogate predictor for immunotherapy with anti-PD1/PD-L1 agents: preliminary report of the FAMI-L1 study. Immunotherapy. 2018;10:643–655. doi: 10.2217/imt-2017-0167. [DOI] [PubMed] [Google Scholar]
- 56.Pilarski R, Carlo M, Cebulla C, Abdel-Rahman M. BAP1 tumor predisposition syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1993-2021. [PubMed] [Google Scholar]
- 57.Hamid RN, Akkurt ZM. Hereditary tumor syndromes with skin involvement. Dermatol Clin. 2019;37:607–613. doi: 10.1016/j.det.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 58.Oshima J, Sidorova JM, Monnat RJ Jr. Werner syndrome: clinical features, pathogenesis and potential therapeutic interventions. Ageing Res Rev. 2017;33:105–114. doi: 10.1016/j.arr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1993-2021. [Google Scholar]
- 60.Schultz KAP, Rednam SP, Kamihara J, Doros L, Achatz MI, Wasserman JD, Diller LR, Brugieres L, Druker H, Schneider KA, McGee RB, Foulkes WD. PTEN, DICER1, FH, and their associated tumor susceptibility syndromes: clinical features, genetics, and surveillance recommendations in childhood. Clin Cancer Res. 2017;23:e76–e82. doi: 10.1158/1078-0432.CCR-17-0629. [DOI] [PubMed] [Google Scholar]
- 61.Weber MF, Marshall HM, Rankin N, Duffy S, Fong KM, Dunlop K, Humphries L, Smit AK, Cust AE, Taylor N, Mitchell G, Kang YJ, Tucker K, Jenkins M, Macrae F, Lockart I, Danta M, Armstrong BK, Howe M. Cancer screening in Australia: future directions in melanoma, Lynch syndrome, and liver, lung and prostate cancers. Public Health Res Pract. 2019;29:e2921910. doi: 10.17061/phrp2921910. [DOI] [PubMed] [Google Scholar]
- 62.Schneider K, Zelley K, Nichols KE, Garber J. Li-Fraumeni Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1993-2021. [Google Scholar]
- 63.Schierbeck J, Vestergaard T, Bygum A. Skin cancer associated genodermatoses: a literature review. Acta Derm Venereol. 2019;99:360–369. doi: 10.2340/00015555-3123. [DOI] [PubMed] [Google Scholar]
- 64.Liang X, Pfeiffer RM, Li WQ, Brossard M, Burke LS, Wheeler W, Calista D, Fargnoli MC, Ghiorzo P, Peris K, Bianchi-Scarra G, Chaudru V, Zelenika D, Maeder D, Burdette L, Yeager M, Chanock S, Landi MT, Demenais F, Tucker MA, Goldstein AM, Yang XR. Association of genetic variants in CDK6 and XRCC1 with the risk of dysplastic nevi in melanoma-prone families. J Invest Dermatol. 2014;134:481–487. doi: 10.1038/jid.2013.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilmott JS, Johansson PA, Newell F, Waddell N, Ferguson P, Quek C, Patch AM, Nones K, Shang P, Pritchard AL, Kazakoff S, Holmes O, Leonard C, Wood S, Xu Q, Saw RPM, Spillane AJ, Stretch JR, Shannon KF, Kefford RF, Menzies AM, Long GV, Thompson JF, Pearson JV, Mann GJ, Hayward NK, Scolyer RA. Whole genome sequencing of melanomas in adolescent and young adults reveals distinct mutation landscapes and the potential role of germline variants in disease susceptibility. Int J Cancer. 2019;144:1049–1060. doi: 10.1002/ijc.31791. [DOI] [PubMed] [Google Scholar]
- 66.He H, Li W, Comiskey DF, Liyanarachchi S, Nieminen TT, Wang Y, DeLap KE, Brock P, de la Chapelle A. A truncating germline mutation of TINF2 in individuals with thyroid cancer or melanoma results in longer telomeres. Thyroid. 2020;30:204–213. doi: 10.1089/thy.2019.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdel-Rahman MH, Sample KM, Pilarski R, Walsh T, Grosel T, Kinnamon D, Boru G, Massengill JB, Schoenfield L, Kelly B, Gordon D, Johansson P, DeBenedictis MJ, Singh A, Casadei S, Davidorf FH, White P, Stacey AW, Scarth J, Fewings E, Tischkowitz M, King MC, Hayward NK, Cebulla CM. Whole exome sequencing identifies candidate genes associated with hereditary predisposition to uveal melanoma. Ophthalmology. 2020;127:668–678. doi: 10.1016/j.ophtha.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derrien AC, Rodrigues M, Eeckhoutte A, Dayot S, Houy A, Mobuchon L, Gardrat S, Lequin D, Ballet S, Pierron G, Alsafadi S, Mariani O, El-Marjou A, Matet A, Colas C, Cassoux N, Stern MH. Germline MBD4 mutations and predisposition to uveal melanoma. J Natl Cancer Inst. 2021;113:80–87. doi: 10.1093/jnci/djaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hajkova N, Hojny J, Nemejcova K, Dundr P, Ulrych J, Jirsova K, Glezgova J, Ticha I. Germline mutation in the TP53 gene in uveal melanoma. Sci Rep. 2018;8:7618. doi: 10.1038/s41598-018-26040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson PA, Nathan V, Bourke LM, Palmer JM, Zhang T, Symmons J, Howlie M, Patch AM, Read J, Holland EA, Schmid H, Warrier S, Glasson W, Hoiom V, Wadt K, Jonsson G, Olsson H, Ingvar C, Mann G, Brown KM, Hayward NK, Pritchard AL. Evaluation of the contribution of germline variants in BRCA1 and BRCA2 to uveal and cutaneous melanoma. Melanoma Res. 2019;29:483–490. doi: 10.1097/CMR.0000000000000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomsen H, Chattopadhyay S, Hoffmann P, Nothen MM, Kalirai H, Coupland SE, Jonas JB, Hemminki K, Forsti A. Genome-wide study on uveal melanoma patients finds association to DNA repair gene TDP1. Melanoma Res. 2020;30:166–172. doi: 10.1097/CMR.0000000000000641. [DOI] [PubMed] [Google Scholar]
- 72.De Summa S, Lasorella A, Strippoli S, Giudice G, Guida G, Elia R, Nacchiero E, Azzariti A, Silvestris N, Guida M, Guida S, Tommasi S, Pinto R. The genetic germline background of single and multiple primary melanomas. Front Mol Biosci. 2020;7:555630. doi: 10.3389/fmolb.2020.555630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferguson R, Archambault A, Simpson D, Morales L, Chat V, Kazlow E, Lax R, Yoon G, Moran U, Shapiro R, Pavlick A, Polsky D, Osman I, Kirchhoff T. Immunomodulatory germline variation associated with the development of multiple primary melanoma (MPM) Sci Rep. 2019;9:10173. doi: 10.1038/s41598-019-46665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Summa S, Guida M, Tommasi S, Strippoli S, Pellegrini C, Fargnoli MC, Pilato B, Natalicchio I, Guida G, Pinto R. Genetic profiling of a rare condition: co-occurrence of albinism and multiple primary melanoma in a Caucasian family. Oncotarget. 2017;8:29751–29759. doi: 10.18632/oncotarget.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aoude LG, Heitzer E, Johansson P, Gartside M, Wadt K, Pritchard AL, Palmer JM, Symmons J, Gerdes AM, Montgomery GW, Martin NG, Tomlinson I, Kearsey S, Hayward NK. POLE mutations in families predisposed to cutaneous melanoma. Fam Cancer. 2015;14:621–628. doi: 10.1007/s10689-015-9826-8. [DOI] [PubMed] [Google Scholar]
- 76.Arbesman J, Ravichandran S, Funchain P, Thompson CL. Melanoma cases demonstrate increased carrier frequency of phenylketonuria/hyperphenylalanemia mutations. Pigment Cell Melanoma Res. 2018;31:529–533. doi: 10.1111/pcmr.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Artomov M, Stratigos AJ, Kim I, Kumar R, Lauss M, Reddy BY, Miao B, Daniela Robles-Espinoza C, Sankar A, Njauw CN, Shannon K, Gragoudas ES, Marie Lane A, Iyer V, Newton-Bishop JA, Timothy Bishop D, Holland EA, Mann GJ, Singh T, Daly MJ, Tsao H. Rare variant, gene-based association study of hereditary melanoma using whole-exome sequencing. J Natl Cancer Inst. 2017;109:djx083. doi: 10.1093/jnci/djx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bartsch DK, Langer P, Habbe N, Matthai E, Chaloupka B, Sina M, Hahn SA, Slater EP. Clinical and genetic analysis of 18 pancreatic carcinoma/melanoma-prone families. Clin Genet. 2010;77:333–341. doi: 10.1111/j.1399-0004.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 79.Belpinati F, Malerba G, Dal Toe M, Ceccuzzi L, Rodolfo M, Poli A, Turco A, Vergani E, Sangalli A, Gomez-Lira M. Enhancer of zeste 2 polycomb repressive complex 2 subunit polymorphisms in melanoma skin cancer risk. Exp Dermatol. 2020;29:980–986. doi: 10.1111/exd.14163. [DOI] [PubMed] [Google Scholar]
- 80.Bertrand JU, Steingrimsson E, Jouenne F, Bressac-de Paillerets B, Larue L. Melanoma risk and melanocyte biology. Acta Derm Venereol. 2020;100:adv00139. doi: 10.2340/00015555-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campos C, Fragoso S, Luis R, Pinto F, Brito C, Esteves S, Pataco M, Santos S, Machado P, Vicente JB, Rosa JC, Cavaco BM, Moura C, Pojo M. High-throughput sequencing identifies 3 novel susceptibility genes for hereditary melanoma. Genes (Basel) 2020;11:403. doi: 10.3390/genes11040403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Casula M, Paliogiannis P, Ayala F, De Giorgi V, Stanganelli I, Mandala M, Colombino M, Manca A, Sini MC, Caraco C, Ascierto PA, Satta RR Melanoma Unit of Sassari (MUS); Lissia A, Cossu A, Palmieri G Italian Melanoma Intergroup (IMI) Germline and somatic mutations in patients with multiple primary melanomas: a next generation sequencing study. BMC Cancer. 2019;19:772. doi: 10.1186/s12885-019-5984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christodoulou E, van Doorn R, Visser M, Teunisse A, Versluis M, van der Velden P, Hayward NK, Jochemsen A, Gruis N. NEK11 as a candidate high-penetrance melanoma susceptibility gene. J Med Genet. 2020;57:203–210. doi: 10.1136/jmedgenet-2019-106134. [DOI] [PubMed] [Google Scholar]
- 84.Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, Gast A, Thirumaran RK, Planelles D, Hemminki K, Schadendorf D, Kumar R. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutat Res. 2010;702:8–16. doi: 10.1016/j.mrgentox.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Fuiten AM, Fankhauser RG, Smit DJ, Stark MS, Enright TF, Wood MA, DePatie NA, Pivik K, Sturm RA, Berry EG, Kulkarni RP. Genetic analysis of multiple primary melanomas arising within the boundaries of congenital nevi depigmentosa. Pigment Cell Melanoma Res. 2021 doi: 10.1111/pcmr.12979. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 86.Goldstein AM, Xiao Y, Sampson J, Zhu B, Rotunno M, Bennett H, Wen Y, Jones K, Vogt A, Burdette L, Luo W, Zhu B, Yeager M, Hicks B, Han J, De Vivo I, Koutros S, Andreotti G, Beane-Freeman L, Purdue M, Freedman ND, Chanock SJ, Tucker MA, Yang XR. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum Mol Genet. 2017;26:4886–4895. doi: 10.1093/hmg/ddx368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hall MJ, Bernhisel R, Hughes E, Larson K, Rosenthal ET, Singh NA, Lancaster JM, Kurian AW. Germline pathogenic variants in the ataxia telangiectasia mutated (ATM) gene are associated with high and moderate risks for multiple cancers. Cancer Prev Res (Phila) 2021;14:433–440. doi: 10.1158/1940-6207.CAPR-20-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, Scott AD, Krassowski M, Cherniack AD, Houlahan KE, Jayasinghe R, Wang LB, Zhou DC, Liu D, Cao S, Kim YW, Koire A, McMichael JF, Hucthagowder V, Kim TB, Hahn A, Wang C, McLellan MD, Al-Mulla F, Johnson KJ Cancer Genome Atlas Research Network. Lichtarge O, Boutros PC, Raphael B, Lazar AJ, Zhang W, Wendl MC, Govindan R, Jain S, Wheeler D, Kulkarni S, Dipersio JF, Reimand J, Meric-Bernstam F, Chen K, Shmulevich I, Plon SE, Chen F, Ding L. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–370. e314. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen J, Alexander T, Jiang H, Hill N, Abdullaev Z, Pack SD, Hsu AP, Holland SM, Hickstein DD, Engels EA, Brownell I. Melanoma in patients with GATA2 deficiency. Pigment Cell Melanoma Res. 2018;31:337–340. doi: 10.1111/pcmr.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ponti G, Losi L, Pellacani G, Wannesson L, Cesinaro AM, Venesio T, Petti C, Seidenari S. Malignant melanoma in patients with hereditary nonpolyposis colorectal cancer. Br J Dermatol. 2008;159:162–168. doi: 10.1111/j.1365-2133.2008.08575.x. [DOI] [PubMed] [Google Scholar]
- 91.Potrony M, Puig-Butille JA, Farnham JM, Gimenez-Xavier P, Badenas C, Tell-Marti G, Aguilera P, Carrera C, Malvehy J, Teerlink CC, Puig S. Genome-wide linkage analysis in Spanish melanoma-prone families identifies a new familial melanoma susceptibility locus at 11q. Eur J Hum Genet. 2018;26:1188–1193. doi: 10.1038/s41431-018-0149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salgado C, Kwesi-Maliepaard EM, Jochemsen AG, Visser M, Harland M, van Leeuwen F, van Doorn R, Gruis N. A novel germline variant in the DOT1L gene co-segregating in a Dutch family with a history of melanoma. Melanoma Res. 2019;29:582–589. doi: 10.1097/CMR.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 93.Stolarova L, Jelinkova S, Storchova R, Machackova E, Zemankova P, Vocka M, Kodet O, Kral J, Cerna M, Volkova Z, Janatova M, Soukupova J, Stranecky V, Dundr P, Foretova L, Macurek L, Kleiblova P, Kleibl Z. Identification of germline mutations in melanoma patients with early onset, double primary tumors, or family cancer history by NGS analysis of 217 genes. Biomedicines. 2020;8:404. doi: 10.3390/biomedicines8100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teerlink CC, Huff C, Stevens J, Yu Y, Holmen SL, Silvis MR, Trombetti K, Zhao H, Grossman D, Farnham JM, Wen J, Facelli JC, Thomas A, Babst M, Florell SR, Meyer L, Zone JJ, Leachman S, Cannon-Albright LA. A nonsynonymous variant in the GOLM1 gene in cutaneous malignant melanoma. J Natl Cancer Inst. 2018;110:1380–1385. doi: 10.1093/jnci/djy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tuominen R. Stockholm: Karolinska Institutet-Department of Oncology-Pathology; 2015. Genetic and epigenetic alterations in melanoma (doctoral dissertation) p. 9. [Google Scholar]
- 96.Vengoechea J, Tallo C. A germline deletion of 9p21.3 presenting as familial melanoma, astrocytoma and breast cancer: clinical and genetic counselling challenges. J Med Genet. 2017;54:682–684. doi: 10.1136/jmedgenet-2017-104690. [DOI] [PubMed] [Google Scholar]
- 97.Yepes S, Tucker MA, Koka H, Xiao Y, Jones K, Vogt A, Burdette L, Luo W, Zhu B, Hutchinson A, Yeager M, Hicks B, Freedman ND, Chanock SJ, Goldstein AM, Yang XR. Using whole-exome sequencing and protein interaction networks to prioritize candidate genes for germline cutaneous melanoma susceptibility. Sci Rep. 2020;10:17198. doi: 10.1038/s41598-020-74293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fidalgo F, Rodrigues TC, Silva AG, Facure L, de Sa BC, Duprat JP, Achatz MI, Rosenberg C, Carraro DM, Krepischi AC. Role of rare germline copy number variation in melanoma-prone patients. Future Oncol. 2016;12:1345–1357. doi: 10.2217/fon.16.22. [DOI] [PubMed] [Google Scholar]
- 99.Rocca MS, Benna C, Mocellin S, Rossi CR, Msaki A, Di Nisio A, Opocher G, Foresta C. E2F1 germline copy number variations and melanoma susceptibility. J Transl Med. 2019;17:181. doi: 10.1186/s12967-019-1933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi J, Zhou W, Zhu B, Hyland PL, Bennett H, Xiao Y, Zhang X, Burke LS, Song L, Hsu CH, Yan C, Chen Q, Meerzaman D, Dagnall CL, Burdette L, Hicks B, Freedman ND, Chanock SJ, Yeager M, Tucker MA, Goldstein AM, Yang XR. Rare germline copy number variations and disease susceptibility in familial melanoma. J Invest Dermatol. 2016;136:2436–2443. doi: 10.1016/j.jid.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]


