Abstract
One of the most fascinating questions in chemistry is why nature chose CGAT as the alphabet of life. Very likely, such selection was the result of multiple factors and a long period of refinement. Here, we explore how the intermolecular interactions influenced such process, by characterizing the formation of dimers between adenine, theobromine and 4‐aminopyrimidine. Using a combination of mass‐resolved excitation spectroscopy and DFT calculations, we determined the structure of adenine‐theobromine and 4‐aminopyrimidine‐theobromine dimers. The binding energy of these dimers is very close to the canonical adenine‐thymine nucleobases. Likewise, the dimers are able to adopt Watson‐Crick conformations. These findings seem to indicate that there were many options available to build the first versions of the informational polymers, which also had to compete with other molecules, such as 4‐aminopyrimidine, which does not have a valid attaching point for a saccharide. For some reason, nature did not select the most strongly‐bonded partners or if it did, such proto‐bases were later replaced by the nowadays canonical CGAT.
Keywords: aggregation, computational chemistry, laser spectroscopy, noncovalent interactions, prebiotic chemistry
Exploiting the synergy between laser spectroscopy and calculations, the interaction potential energy surfaces of the dimers between adenine and 4‐aminopyrimidine with theobromine have been explored. Both systems form stable aggregates with two strong hydrogen bonds, with binding energy values resembling the Watson‐Crick analogue interactions. An energetic comparison suggests how intermolecular interactions influenced the evolutionary sequence.

Introduction
One of the most exciting questions in science is the origin of DNA and the reasons for its chemical composition. Why nature selected cytosine, guanine, adenine and thymine (CGAT) as the “alphabet of life” is still an open question. There is a general agreement in that they are the result of evolutionary pressure at several different levels. [1]
The initial molecules from which CGAT have evolved may have been substantially different from the present nucleobases. Among other reasons, the physico‐chemical conditions in the primal broth were probably very different from those in present days. The oldest rocks found are 3.6 billion years old and already contain rests of cellular organisms.[ 2 , 3 , 4 ] This means that life appeared in the first 400 million years of existence of our planet. [5] At those times, no free oxygen existed and very likely, the atmosphere was not strongly reducing. [6] Depending on the iron redox chemistry and on the speed at which iron sunk to the core, the atmosphere could have been from very rich to very poor in methane. [7] With time, the atmosphere might have evolved to contain mainly CO, CO2, H2O H2 and N2, but no ammonia.[ 1 , 8 ] Under such conditions, the rich chemistry of the condensation of HCN in presence of H2CO may have contributed to the appearance of the first prebiotic molecules.[ 2 , 9 ] Among the results of such condensation not only CGAT, but also many other purines, pyrimidines as well as other heterocyclic compounds are formed.[ 2 , 10 , 11 , 12 ] Perhaps, chemical stability could contribute to produce an enrichment of the medium in some of those compounds. However, studies on half‐life of purines and pyrimidines demonstrate that this was not the determinant (or at least the only determinant) factor for the selection of CGAT, as there are compounds, such as 2,6‐diaminopurine or 2,4‐diaminopyrimidine that are more stable or at least equally stable than the canonical nucleobases.[ 13 , 14 , 15 ]
An important selection factor could be light irradiation. Most authors agree that the primal atmosphere was substantially more permeable to VUV [16] and therefore, it may have helped to both drive synthetic reactions and select the most stable products. The pioneering works by de Vries, [17] Crespo‐Hernández, [18] and Hobza, among others,[ 19 , 20 , 21 , 22 ] have demonstrated that the canonical nucleobases present very short electronic excited state lifetimes. This fact has been taken as a protection against photodamage. However, other prebiotic compounds also present very short lifetimes, even shorter than the nucleobases. For example, hypoxanthine has an excited state lifetime of 0.13 ps, [23] compared to adenine (0.18 ps) [24] or guanine (0.16 ps). [25]
One of the problems to understand why nature selected the canonical nucleobases is that they were not selected in the first instants of life formation. Or that they continued evolving in parallel to the evolution of the first biopolymers. In a compelling review, Rios and Tor identify several stages at which such selection may have taken place. [1] According to these authors, the rich variety of compounds produced by the prebiotic chemistry was later refined during the assembly of the first informational polymers, the emergence and early evolution of life and the formation of the last universal common ancestor (LUCA). Actually, some authors speculate with the idea of an initial set of nucleobases containing more than just four units. In favor of such hypothesis is the existence of modified nucleobases in ancestral organisms, such as bacteriophages,[ 26 , 27 ] or the possibility of creating expanded alphabets using synthetic chemistry.[ 28 , 29 ]
Regarding the second stage on the evolutionary path depicted above, one of the conditions that a molecule must fulfil to become a valid nucleobase is to present the appropriate collection of intermolecular interactions. The nucleobases must selectively couple with their complementary base, but at the same time, they must present additional functional groups for the interaction with other molecules (proteins) that are part of the complex transductional machinery of the cell. Following these thoughts, we are engaged in the study of the interaction landscape of the nucleobases and other structurally related biomolecules, but that were not chosen to form part of DNA/RNA, even when some of them are found in living beings. Deepening the knowledge on the differences and similarities in the aggregation preferences of these compounds, is the first step in the process of deciphering the evolution of the nucleobases.
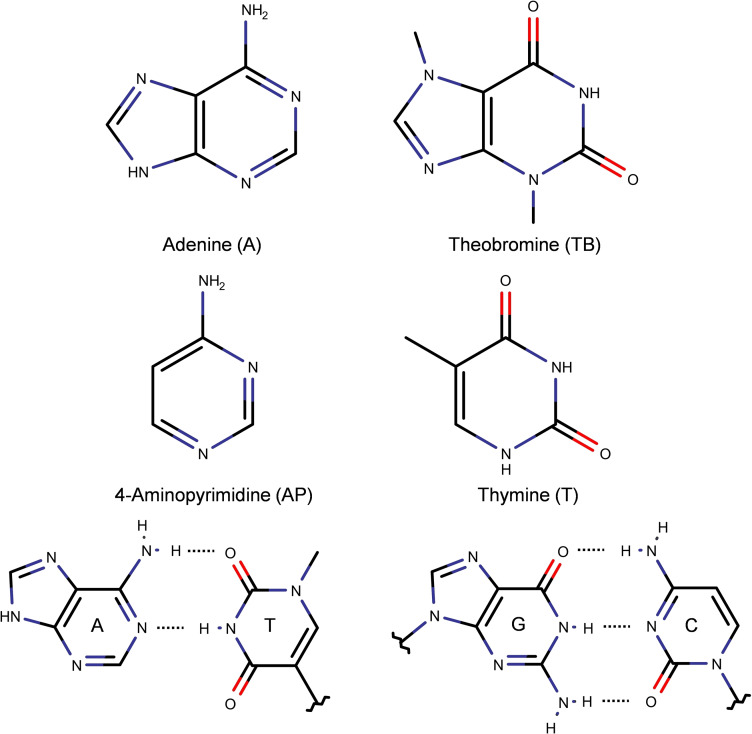
In order to elaborate on this topic, we present here a joined experimental and computational analysis of the interactions between theobromine (TB), adenine (A) and 4‐aminopyridine (AP, see Scheme 1).
Scheme 1.
Structure of theobromine, adenine, 4‐aminopyrimidine, thymine, and Watson‐Crick (WC) interactions for the canonical nucleobases.
Xanthines are found in the human body as part of the guanine metabolism. [30] Some of their methylated derivatives, like TB, present biological activity due to the similarity of their structure to that of other metabolites. Certainly, its framework is a mixture between the CONHCO motif of thymine and the purinic scaffold of A. Previous spectroscopic studies on TB demonstrated that its interaction with water is similar to those of the canonical nucleobases. Water attaches to the CONH side forming two hydrogen bonds (HBs) of similar strength as in A‐water. Besides, its tautomeric equilibrium is altered by the interaction with the water molecule in the same way as in the nucleobases. [31]
In this study, we used a supersonic expansion to form theobromine‐adenine (TB+A) dimers and explored their spectroscopy using mass‐resolved excitation techniques. Comparison between the physical observables obtained with spectroscopic techniques with the predictions from computational chemistry led us to propose an assignment for the recorded spectra. The determination of the structure of these aggregates allowed us to compare with the aggregation preferences of adenine‐thymine (A+T). The study was complemented with the characterization of the theobromine‐4‐aminopyrimidine dimer (TB+AP). AP has the same NH2CN motif as A, and therefore one would expect the aggregates formed by this dimer to resemble those of A+T, but with the purinic‐pyrimidinic structure reversed. Therefore, comparison of the structure and binding energy of TB+AP with A+T allows us to understand the role of the same A+T interactions in other nucleobase candidates. Finally, we examined the results obtained on these aggregates on the light of those in the literature regarding the canonical WC nucleobase pairs.
Results and Discussion
Exploration of the conformational space
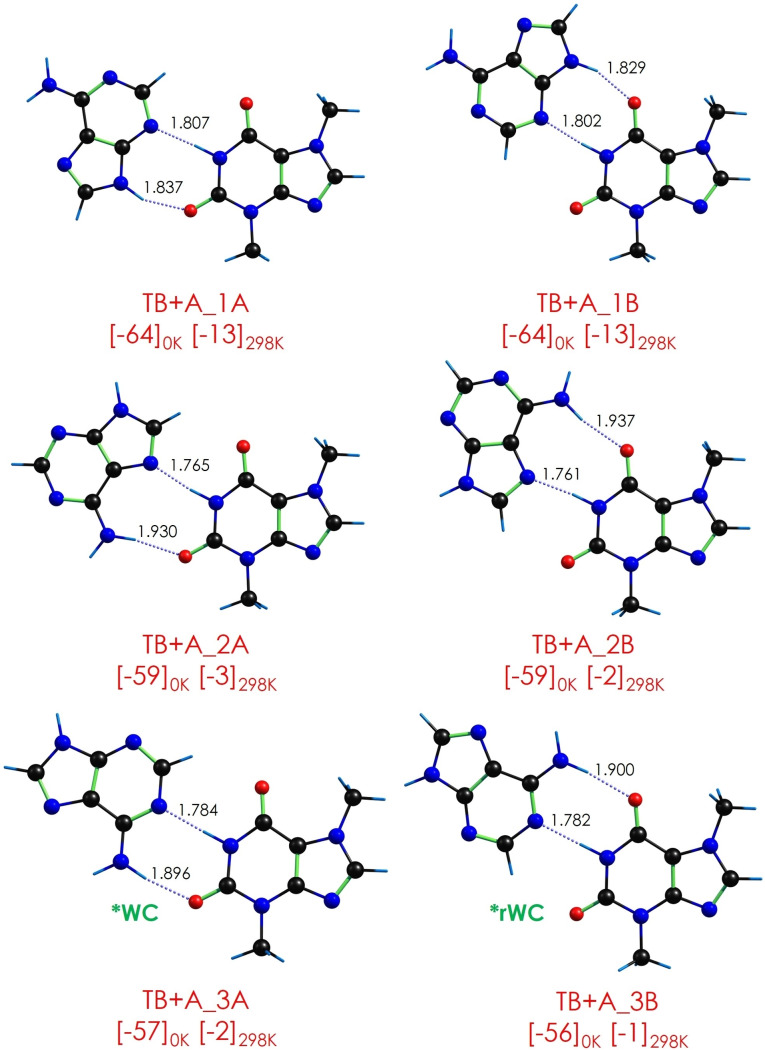
The presence of several proton donor and acceptor groups in the interacting molecules results in a rich collection of conformational isomers. Consequently, a careful exploration of the potential energy surface is required to locate all the relevant minima. Figure 1 summarizes the six most stable isomers found for theobromine‐adenine (TB+A), together with their binding energy at 0 and 298 K. Figures S1–S4 of the Supporting Information collect all the structures found for both dimers, together with their relative stability computed at M06‐2X/6‐311+G(d,p) and B3LYP‐ED=GD3BJ/def2TZVP levels.
Figure 1.
The six most stable conformational isomers of theobromine‐adenine, together with their binding energy at 0 and 298 K. The binding energy value (kJ/mol) is the average of the two values computed at M06‐2X/6‐311+G(d,p) and B3LYP‐ED=GD3BJ/def2TZVP. The original values obtained with each theory level may be found in the Supporting Information. Hydrogen bond distances are those computed at the B3LYP‐ED=GD3BJ/def2TZVP level. Distances in Å.
An exhaustive exploration of the interaction potential energy surface (iPES) revealed three different families of interactions: (i) structures containing two strong HBs, (ii) single HB structures and (iii) isomers mainly attached by π⋅⋅⋅π stacking interactions. It is worth noting that due to the low flexibility of the systems, relative stability and binding energy are directly related.
The most favorable interaction in TB+A is the formation of a double C=O⋅⋅⋅HN/N−H⋅⋅⋅N HB. Such interaction can be established with either of the two carbonyl groups in TB, resulting in pairs of (almost) degenerate structures (labelled as A and B in Figure 1). Due to the similarity of those pairs, they are indistinguishable in our experimental system, as they present a very similar spectroscopic signature. Therefore, we will refer to each pair as a single isomer, although both structures can be present in the expansion.
In tight competition with the above‐mentioned interaction, the formation of C=O⋅⋅⋅HNH/N−H⋅⋅⋅N HBs is also possible, with a small reduction in the binding energy. Depending on the relative orientation of the two molecules, the resulting structure resembles the WC complex of A+T. Following the WC nomenclature, isomer TB+A_3A would correspond to a WC conformation, while TB+A_3B would correspond to a reverse WC conformation (rWC, see Figure 1).
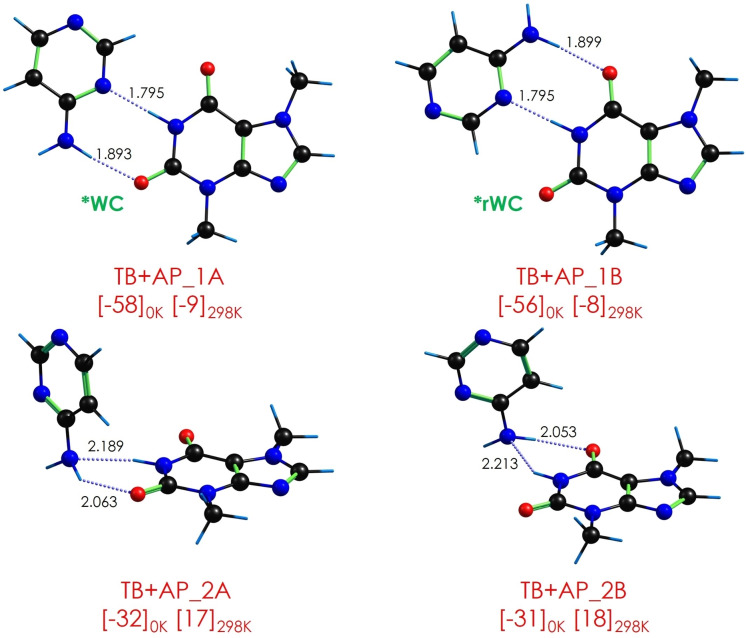
Transition from a puric to a pyrimidinic base resulted in a substantially simpler conformational landscape. Thus, two quasi‐degenerate structures dominate the iPES of theobromine‐4‐aminopyridine (TB+AP, Figure 2). Interestingly, both isomers but specially TB+AP_1A reproduce the WC configuration of A+T.
Figure 2.
The four most stable conformational isomers of theobromine‐4‐aminopyrimidine, together with their binding energy at 0 and 298 K. The binding energy value (kJ/mol) is the average of the two values computed at M06‐2X/6‐311+G(d,p) and B3LYP‐ED=GD3BJ/def2TZVP. The original values obtained with each theory level may be found in the Supporting Information. Hydrogen bond distances are given for B3LYP‐ED=GD3BJ/def2TZVP level. Distances in Å.
Spectroscopic characterization of the aggregates
One of the main difficulties in the study of these systems is the short lifetimes of their electronic excited states,[ 32 , 33 , 34 ] which result in low signal intensity. Furthermore, the transfer of these molecules into gas phase requires of sophisticated set‐ups, such as laser desorption systems, that also increase the signal's noise. The mass‐resolved excitation spectra of TB and A may be found in Figure S5. While the spectrum of TB presents a large number of vibronic transitions due to the activation of several out of plane vibrations, [35] the spectrum of A in the 36000–36700 cm−1 region contains only three bands built over a background. The short lifetime of A, 180 fs, [24] makes the two‐photon ionization process very inefficient, seriously reducing the overall sensibility of the experiment. Likewise, AP presents a short excited electronic state lifetime (400 fs) [36] hampering the recording of its electronic excitation spectrum. The study of the dimer was possible using TB as chromophore to record the IR/UV spectrum of the TB+AP dimer.
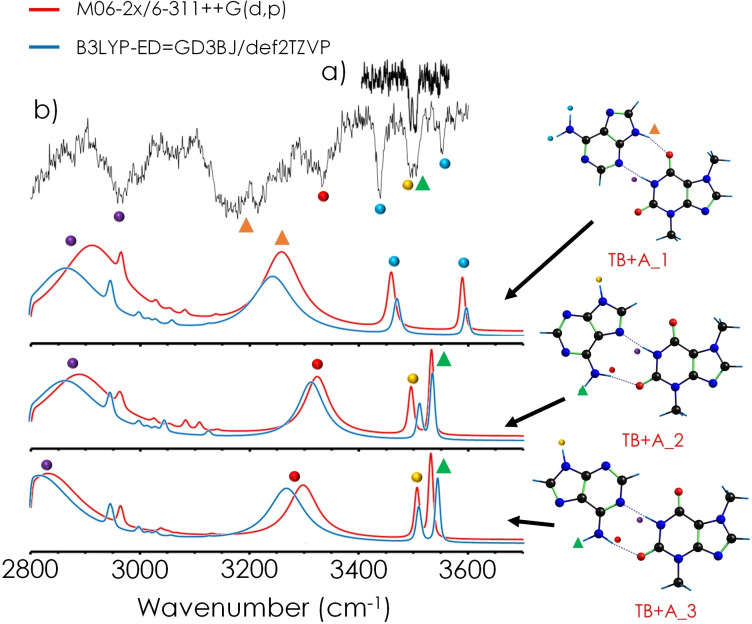
Figures 3 and 4 present the IR/UV traces of TB+A and TB+AP respectively, together with the predictions for the most stable isomers, computed at the two levels of theory. The wavelengths used to record the IR spectra can be found in Figure S5. The unresolved nature of the excitation spectra precluded performing hole‐burning experiments to identify the contribution from different isomers. The spectra in Figure 3 clearly show the presence of at least two isomers. It is interesting to note that some of the bands are broad absorptions, probably due to the formation of strong HBs or even to the possibility of a concerted double proton transfer between the two molecules.[ 37 , 38 , 39 , 40 ]
Figure 3.
Comparison between the experimental spectrum of TB+A and the predictions for some selected computed structures. Experimental traces were recorded at a) 35,097 cm−1 and b) 35,769 cm−1 excitation wavelengths. The predictions for the two most stable structures fit very well with the experimental observation. The rest of the predicted spectra may be found in the Supporting Information.
Figure 4.
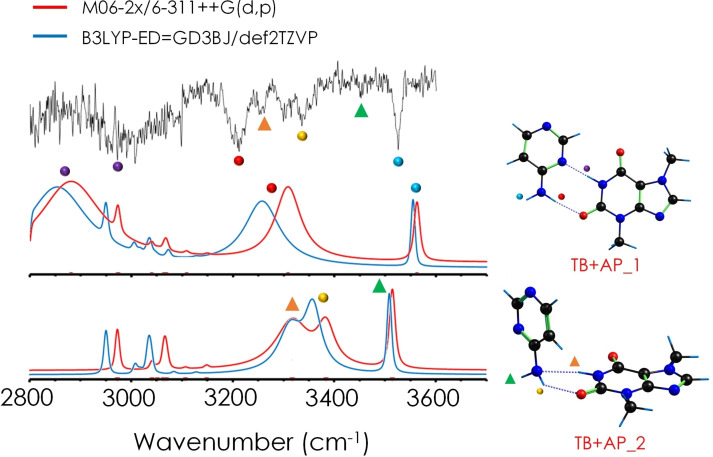
Comparison between the experimental spectrum (recorded at 34,794 cm−1) of TB+AP and the predictions for two computed structures, whose spectra are compatible with the experimental trace. The rest of the predicted spectra may be found in the Supporting Information.
Despite the rich conformational landscape of the TB+A dimer, a first look at the experimental IR spectra clearly discards two of the observed interaction families: no structures with single HB can explain both free and bound NH bands, and the absence of the free NH bands of TB discards all the stacking possibilities. Further information of the predicted spectra of all conformers can be found in the Supporting Information (Figures S6 and S7).
Comparison between the IR/UV spectrum of TB+A and the predictions for the most stable computed structures shows that the spectrum of the global minimum fits very well with the experimental trace, but the presence of an additional isomer is required to explain those transitions marked with yellow, green and red dots in Figure 3. Actually, the prediction for the second most stable isomer faithfully reproduces the position of the second set of bands. The prediction for the WC conformer is also consistent with the spectrum of the second experimental conformer, but it is higher in energy and therefore, the most likely explanation is that the two formed isomers correspond to the two most stable structures.
The IR/UV spectrum of TB+AP contains less and broader bands. The spectral signatures of the formation of strong HBs are also clearly visible in the red shift of the transitions. These features are well predicted in the computed spectrum of the two most stable isomers. The computed spectrum of the WC isomer (TB+AP_1) reproduces very well the position of the stretching frequency of the free NH and part of the observed broad transitions, but, apparently, there are some extra bands (yellow and orange dots) that require of a second isomer for their assignment. In this second isomer, AP would adopt a position perpendicular to the plane of TB. Such structure is ∼26 kJ/mol less stable than the global minimum, suggesting the presence of high barriers for isomerization.
Comparison with other systems
The two main conclusions that one can extract from the assigned structures are that both TB+A and TB+AP systems present very similar interactions to those found in the canonical nucleobases in an environment that is free from external perturbations. In the case of TB+AP, the global minimum is of the WC type, against what was observed in gas phase for CGAT. [41] However, one must keep in mind that AP is not a valid molecule to act as nucleobase.
Table 1 collects the binding energy values of the species detected and identified in this work and compares them with the computed values for similar systems studied by other authors. To carry out such comparison, the structures reported in the literature were optimized at B3LYP‐ED=GD3BJ/def2TZVP level. The computed structures of the dimers reported in Table 1 are illustrated in Figure S10.
Table 1.
Binding energies for several dimers mimicking the WC interactions computed at the B3LYP‐ED=GD3BJ/def2TZVP level.
|
Complex |
Binding energy 0 K[a] |
Binding energy 298 K[a] |
|---|---|---|
|
TB+A |
61/60 |
8/6 |
|
TB+AP |
61/59 |
10/9 |
|
T+A |
62/60 |
8/7 |
|
AP+1‐methylthymine[b] |
62/60 |
12/10 |
|
AP+6‐methyl‐4‐pyrimidinone (M4PMN)[b] |
68 |
18 |
|
9‐methylamine+2‐pyridinone (9 mA+2PY)[c] |
68 |
17 |
|
AP+2‐pyrimidinone (AP+2PY) |
68 |
18 |
|
TB+2,4‐aminopyridine (2,4AP)[c] |
73/73 |
18/19 |
|
G+C |
117 |
60 |
[a] Binding energy values given in kJ/mol. For the case of two different values, they correspond to the WC and rWC structures, respectively. [b] Ref. [9]. [c] Ref. [37]. [d] Ref. [39].
The binding energy values were computed at two different temperatures: 0 and 298 K. The former is more relevant to compare with the IR spectra recorded in the beam, while the latter may highlight the existence of structures particularly favored by entropy, and very likely, is closer to the temperature at which the first informational polymers were formed. The exact conditions at which life was first formed are still subject to debate, but they should allow the molecular building blocks to have a reasonable lifetime and to form molecular aggregates, as storage and interpretation of the information on the biopolymers requires of intermolecular interactions to be stable.
The clear correlation between the values computed at both temperatures shows that no particular isomer was significantly more stable at room temperature.
Close examination of the binding energies of the systems in Table 1 shows that the interaction energy values of TB+A and TB+AP are of comparable strength to that of T+A, both at 0 and 298 K. However, the interaction energy values for G+C are nearly twice stronger than those for T+A dimers and analogue interactions, due to presence of an extra HB, in good agreement with previous theoretical studies about DNA base pairs. [42] In between are the values for several similar systems that could have existed in the prebiotic environment. Binding energies for WC‐like interactions in AP+M4PMN, 9 mA+2PY and AP+2PY are slightly larger than those predicted for TB+A and TB+AP.
From a mechanistic point of view,[ 43 , 44 ] the contributions of the electrostatic, exchange‐repulsion, induction and dispersion are comparable for all the dimers studied, with the exception of AP+M4PMN, which presents a reduced contribution of all the terms (Table S3). As it is commonly observed in systems attached by strong hydrogen bonds, the most important contribution is the electrostatic term, with the induction and dispersion terms presenting similar contributions.
It is also important to note that the values in Table 1 reflect the natural aggregation preferences of the molecules under isolated conditions, while life most likely appeared in solution or even in other complex media. However, these values are a good indication of the interaction strength as it is expected that solution or the presence of salts would affect all the aggregates in the same way, due to the similarity of their chemical structure.
The data in Table 1 offers a complex panorama. Indeed, A‐T WC‐analogs interaction energies are very close, which points out a possible competition for binding to adenine. If the interaction energy were the only selection pressure, only those systems able to establish three HBs, such as G+C would have survived this selection stage. Clearly, in an environment with a rich chemistry and a large variety of species, the ability to form strong HBs would have offered a clear advantage. Perhaps, these results indicate that the first biomolecules were formed by single strands.[ 45 , 46 ] The limiting factor to form a successful single strand would be the probability of formation of a given species and its stability in the medium. However, in modern organisms, the binding energy of the base pairs is a compromise between the affinity for the partner (C→G/A→T) and the interaction with the amino acids of the proteins that take part in the translation of the gene information into protein production. The affinity for the partner must be high enough, but not as high as for the proteins to be unable to unzip the two DNA strands. Therefore, the initial molecules chosen to build the information polymers were, very likely, refined at subsequent stages until they resulted in the today's well‐known CGAT sequence.
Conclusion
An experimental and computational study of the formation of dimers between theobromine, adenine and 4‐amino pyrimidine has been reported. Interpretation of the IR/UV spectra on the light of the computational predictions shows that TB+AP tends to form WC‐like structures. AP cannot act as nucleobase, at least in the present form of DNA, because it does not have a valid point to attach the ribose, but still, it can compete for the interaction with the canonical nucleobases. The two systems studied here are able to form stable aggregates with two strong intermolecular hydrogen bonds, with binding energy values similar to those of A+T. Comparison with the values for similar systems, including those formed by the canonical nucleobases shows a wide range of values, with G+C forming the strongest dimers. According to the studies of formation of nucleobases in prebiotic environments, the same reactions that originate CGAT also yield TB, AP and other similar compounds. This means, on one hand, that election of T and A as nucleobases was not based (or at least not exclusively based) on the strength of the intermolecular interactions, and on the other, that the canonical nucleobases had to compete with other interacting partners for the formation of the proto‐information polymers.
Experimental Section
Experimental details: The experimental setup employed in this work was described in detail in previous works.[ 47 , 48 ] It consists of a supersonic expansion chamber coupled to an in‐house laser ablation system. This configuration allows us to vaporize thermolabile molecules. The supersonic expansion chamber is also coupled to a time‐o‐flight (TOF) mass spectrometer, to carry out mass‐selected detection. The supersonic expansion was produced using a pulsed valve (General Valve Series 9, 10 Hz) with a backing pressure of 10 bar (Ar, >99,9 %, Praxair). Theobromine (Sigma Aldrich, 98 %) and adenine (Sigma Aldrich, 98 %) or 4‐aminopyrimidine (Sigma Aldrich, 98 %) was mixed with carbon nanotubes (MWCNT, Cheap Tubes) to enhance laser desorption (Quantel, Brilliant‐b, 1064 nm). The ablation laser was synchronized with the opening of the valve, so that the desorbed molecules were picked up by the supersonic expansion. Collisions with the buffer gas cool the molecules and create the conditions required to form stable, isolated molecular aggregates. The molecular beam was then probed using a combination of tunable UV (Quantel Q‐Scan, Coumarin 540 A, 500 μJ/pulse) and IR (LaserVision OPO/A, ∼7 mJ/pulse) lasers to record REMPI and IDIRS spectra.
Computational details: Detailed description of the computational protocol may be found in Refs. [31,49]. The procedure started with a conformational search carried out using three different force fields: MMFFs, [50] AMBER,[ 51 , 52 ] and OPLS3e. [53] The generated structures were grouped by a clustering process, and the selected candidates were optimized at M06‐2X [54] /6‐311++G(d,p) level to find the global minimum. Then, a careful analysis of all the optimized structures was performed, in order to build the iPES diagram of the complexes, labelling and classifying structures depending on their interaction type. The interaction energy of the dimers was computed including the ZPE and BSSE corrections. Theoretical simulations of the IR spectra were generated by applying an experimentally determined scaling factor [49] to the normal modes obtained from the Gaussian16 program package. For the M06‐2X/6‐311++G(d,p) level, scaling factors of 0.963 and 0.951 were used for CH and NH stretches, respectively. In order to simulate the experimental trace, the predicted frequencies were represented with a Lorentzian function, with a variable width depending on the type and strength of the interaction. Additionally, the effect of the laser on the shape of the transition was also taken into account by the convolution of a Gaussian function (FMHW=6 cm−1) with the simulated spectra.
Finally, all the structures optimized with the M06‐2X method were also re‐optimized at the B3LYP‐ED=GD3BJ [55] /def2TZVP level. For this level of theory, a single scaling factor of 0.963 was used to account for anharmonicity for both CH and NH stretches.
Energy decomposition into electrostatic, exchange‐repulsion, induction and dispersion was done using symmetry‐adapted perturbation theory (SAPT). Calculations were used with the second‐order intramonomer correlation correction and third‐order intermonomer dispersion corrections, SAPT2+(3), [56] with the aug‐cc‐pVDZ basis set, as implemented in PSI4.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We thank MICIU (PGC2018‐098561‐B‐C21) and the Basque Government (IT1162‐19) for financial support. A.C. thank the Basque Government for a pre‐doctoral fellowship. Computational and laser resources of the SGIKER (UPV/EHU, MICIU‐FEDER) were used in this work.
A. Camiruaga, I. Usabiaga, C. Calabrese, I. Lamas, F. J. Basterretxea, J. A. Fernández, Chem. Eur. J. 2022, 28, e202103636.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Rios A. C., Tor Y., Isr. J. Chem. 2013, 53, 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joyce G. F., Nature 1989, 338, 217–224. [DOI] [PubMed] [Google Scholar]
- 3. Walter M. R., in Earth's Earliest Biosphere (Eds:Schopf J. W.,), Princeton University Press, 1983, p. 197. [Google Scholar]
- 4. Buick R., Dunlop J. S. R., Groves D. I., Alcheringa. 1981, 5, 161–181.< [Google Scholar]
- 5. Schidlowski M., Appel P. W. U., Eichmann R., Junge C. E., Geochim. Cosmochim. Acta 1979, 43, 189–199. [Google Scholar]
- 6. Hayes J. M., in Earth's Earliest Biosphere (Eds:Schopf J. W.,), Princeton University Press, 1983, p. 291. [Google Scholar]
- 7. Chang S., Des Marais D. J., Mack R., Miller S. L., Strathearn G., in Earth's Earliest Biosphere (Eds: Schopf J. W.), Princeton University Press, 1983, p. 53. [Google Scholar]
- 8. Cafferty B. J., Hud N. V., Isr. J. Chem. 2015, 55, 891–905. [Google Scholar]
- 9. Nosenko Y., Kunitski M., Stark T., Göbel M., Tarakeshwar P., Brutschy B., Phys. Chem. Chem. Phys. 2013, 15, 11520–11530. [DOI] [PubMed] [Google Scholar]
- 10. Kolb V. M., Dworkin J. P., Miller S. L., J. Mol. Evol. 1994, 38, 549–557. [DOI] [PubMed] [Google Scholar]
- 11. Saladino R., Botta G., Pino S., Costanzo G., Di Mauro E., Chem. Soc. Rev. 2012, 41, 5526–5565. [DOI] [PubMed] [Google Scholar]
- 12. Siegel J. S., Tor Y., Org. Biomol. Chem. 2005, 3, 1591–1592. [DOI] [PubMed] [Google Scholar]
- 13. Robertson M. P., Levy M., Miller S. L., J. Mol. Evol. 1996, 43, 543–550. [DOI] [PubMed] [Google Scholar]
- 14. Levy M., Miller S. L., Proc. Nat. Acad. Sci. 1998, 95, 7933 LP-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. House C. H., Miller S. L., Biochemistry 1996, 35, 315–320. [DOI] [PubMed] [Google Scholar]
- 16. Sagan C., J. Theor. Biol. 1973, 39, 195–200. [DOI] [PubMed] [Google Scholar]
- 17. Boldissar S., de Vries M. S., Phys. Chem. Chem. Phys. 2018, 20, 9701–9716. [DOI] [PubMed] [Google Scholar]
- 18. Middleton C. T., de La Harpe K., Su C., Law Y. K., Crespo-Hernández C. E., Kohler B., Annu. Rev. Phys. Chem. 2009, 60, 217–239. [DOI] [PubMed] [Google Scholar]
- 19. Kang H., Lee K. T., Jung B., Ko Y. J., Kim S. K., J. Am. Chem. Soc. 2002, 124, 12958–12959. [DOI] [PubMed] [Google Scholar]
- 20. Brister M. M., Gustavsson T., Crespo-Hernández C. E., Mol. 2020, 25, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barbatti M., Aquino A. J. A., Szymczak J. J., Nachtigallová D., Hobza P., Lischka H., Proc. Nat. Acad. Sci. 2010, 107, 21453 LP-21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shukla M. K., Leszczynski J., in Radiation Induced Molecular Phenomena in Nucleic Acids (Eds: Shukla M. K., Leszczynski J.), Springer Netherlands, 2008, p. 369. [Google Scholar]
- 23. Chen J., Kohler B., Phys. Chem. Chem. Phys. 2012, 14, 10677–10682. [DOI] [PubMed] [Google Scholar]
- 24. Cohen B., Hare P. M., Kohler B., J. Am. Chem. Soc. 2003, 125, 13594–13601. [DOI] [PubMed] [Google Scholar]
- 25. Villabona-Monsalve J. P., Noria R., Matsika S., Peón J., J. Am. Chem. Soc. 2012, 134, 7820–7829. [DOI] [PubMed] [Google Scholar]
- 26. Gommers-Ampt J. H., Borst P., FASEB J. 1995, 9, 1034–1042. [DOI] [PubMed] [Google Scholar]
- 27. Rae P. M. M., Steele R. E., BioSystems 1978, 10, 37–53.566131 [Google Scholar]
- 28. Kim H.-J., Leal N. A., Benner S. A., Bioorg. Med. Chem. 2009, 17, 3728–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dien V. T., Holcomb M., Romesberg F. E., Biochemistry 2019, 58, 2581–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marro P. J., Baumgart S., Delivoria-Papadopoulos M., Zirin S., Corcoran L., McGaurn S. P., Davis L. E., Clancy R. R., Pediatr. Res. 1997, 41, 513–520. [DOI] [PubMed] [Google Scholar]
- 31. Usabiaga I., Camiruaga A., Calabrese C., Veloso A., D'mello V. C., Wategaonkar S., Fernández J. A., Phys. Chem. Chem. Phys. 2020, 22, 15759–15768. [DOI] [PubMed] [Google Scholar]
- 32. Schultz T., Samoylova E., Radloff W., Hertel I. V., Sobolewski A. L., Domcke W., Science 2004, 306, 1765 LP - 1768. [DOI] [PubMed] [Google Scholar]
- 33. Sobolewski A. L., Domcke W., Hättig C., Proc. Natl. Acad. Sci. USA 2005, 102, 17903 LP-17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gengeliczki Z., Callahan M. P., Svadlenak N., Pongor C. I., Sztáray B., Meerts L., Nachtigallová D., Hobza P., Barbatti M., Lischka H., de Vries M. S., Phys. Chem. Chem. Phys. 2010, 12, 5375–5388. [DOI] [PubMed] [Google Scholar]
- 35. Camiruaga A., Usabiaga I., D'mello V. C., García G. A., Wategaonkar S., Fernández J. A., Phys. Chem. Chem. Phys. 2019, 21, 26430–26437. [DOI] [PubMed] [Google Scholar]
- 36. Zechmann G., Barbatti M., Int. J. Quantum Chem. 2008, 108, 1266–1276. [Google Scholar]
- 37. Frey J. A., Müller A., Frey H.-M., Leutwyler S., J. Chem. Phys. 2004, 121, 8237–8245. [DOI] [PubMed] [Google Scholar]
- 38. Callahan M. P., Gengeliczki Z., Svadlenak N., Valdes H., Hobza P., de Vries M. S., Phys. Chem. Chem. Phys. 2008, 10, 2819–2826. [DOI] [PubMed] [Google Scholar]
- 39. Gengeliczki Z., Callahan M. P., Kabeláč M., Rijs A. M., de Vries M. S., J. Phys. Chem. A 2011, 115, 11423–11427. [DOI] [PubMed] [Google Scholar]
- 40. Frey J. A., Ottiger P., Leutwyler S., J. Phys. Chem. B 2014, 118, 682–691. [DOI] [PubMed] [Google Scholar]
- 41. Plützer C., Hünig I., Kleinermanns K., Nir E., de Vries M. S., ChemPhysChem 2003, 4, 838–842. [DOI] [PubMed] [Google Scholar]
- 42. Jurečka P., Hobza P., J. Am. Chem. Soc. 2003, 125, 15608–15613. [DOI] [PubMed] [Google Scholar]
- 43. Emamian S., Lu T., Kruse H., Emamian H., J. Comput. Chem. 2019, 40, 2868–2881. [DOI] [PubMed] [Google Scholar]
- 44. Su G., Zhang H., Zhang X., ChemRxiv. 2021, 10.26434/chemrxiv.14101487.v1. [DOI] [Google Scholar]
- 45. von Kiedrowski G., Angew. Chem. Int. Ed. 1986, 25, 932–935; [Google Scholar]; Angew. Chem. 1986, 98, 934–935. [Google Scholar]
- 46. Joyce G. F., Szostak J. W., Cold Spring Harbor Perspect. Biol. 2018, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Usabiaga I., González J., Arnáiz P. F., León I., Cocinero E. J., Fernández J. A., Phys. Chem. Chem. Phys. 2016, 18, 12457–12465. [DOI] [PubMed] [Google Scholar]
- 48. Camiruaga A., Usabiaga I., Insausti A., León I., Fernández J. A., Phys. Chem. Chem. Phys. 2017, 19, 12013–12021. [DOI] [PubMed] [Google Scholar]
- 49. Usabiaga I., Camiruaga A., Calabrese C., Maris A., Fernández J. A., Chem. Eur. J. 2019, 25, 14230–14236. [DOI] [PubMed] [Google Scholar]
- 50. Halgren T. A., J. Comput. Chem. 1996, 17, 490–519. [Google Scholar]
- 51. Case D. A., Cheatham T. E., Darden T., Gohlke H., Luo R., Merz K. M., Onufriev A., Simmerling C., Wang B., Woods R. J., J. Comput. Chem. 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kirschner K. N., Yongye A. B., Tschampel S. M., Gonzalez-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J., J. Comput. Chem. 2008, 29, 622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., Friesner R. A., J. Chem. Theory Comput. 2016, 12, 281–296. [DOI] [PubMed] [Google Scholar]
- 54. Zhao Y., Truhlar D. G., Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- 55. Grimme S., Antony J., Ehrlich S., Krieg H., J. Chem. Phys. 2010, 132, 154104. [DOI] [PubMed] [Google Scholar]
- 56. Hohenstein E. G., Sherrill C. D., J. Chem. Phys. 2010, 133, 14101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.