Abstract
Background
Salmonella enterica in cattle has long been problematic and suspected to be transmitted by wildlife in Tokachi, Hokkaido, a major cattle farming area in Japan. Understanding the role of wildlife in S. enterica transmission would be helpful for developing control strategies of bovine salmonellosis.
Objectives
We aimed to elucidate the possibility of S. enterica transmission between sympatric wildlife, including raccoons and crows and cattle, in Tokachi from 2008 to 2018 by analysing S. enterica detection records, and the genetic relatedness of serotypes shared between wildlife and cattle.
Methods
S. enterica detection records were based on the results of a field survey and existing cattle records at relevant organisations, including clinical reports, a monitoring survey and quarantine for introduced calves at growing farms and public calving farms. S. enterica was identified by polymerase chain reaction assay and serotyped by agglutination assay. The detection records were organised chronologically to investigate whether common serotypes in wildlife and cattle were detected in the same year. The isolates corresponding to detection records were assessed for their genetic patterns by pulsed‐field gel electrophoresis.
Results
The prevalence of S. enterica in raccoons and crows was 10.7% (17/159) and 5.7% (55/967), respectively. The following serotypes were detected from both wildlife and cattle: Braenderup, Dublin, Infantis, Mbandaka, Montevideo, 4,[5],12:i:‐ and Typhimurium. Genetically similar isolates for S. Braenderup, S. Dublin, S. Montevideo and S. 4,[5],12:i:‐ were detected from both species in the same year.
Conclusions
Our long‐term retrospective observations supported that S. enterica was shared between wildlife and cattle. Wildlife invasions should be controlled at farms to prevent inter‐species transmission of S. enterica from livestock farms.
Keywords: cattle, pulsed‐field gel electrophoresis, Salmonella enterica, wildlife
Salmonella seemed to be shared between wildlife and cattle in cattle farming area, which was based on the occurrence records and genetic analysis.

1. INTRODUCTION
Salmonella enterica is one of the most common and burdensome foodborne pathogens worldwide (EFSA, 2019a; Lee & Yoon, 2021). It is carried by various vertebrates, including livestock (Markey et al., 2013). The global yearly estimate of Salmonella gastroenteritis is 95.1 million cases, with 50,771 deaths in 2017 (Stanaway et al., 2019), which may come from livestock products including poultry, eggs, pork, beef and dairy products (Pires et al., 2014; Scallan et al., 2011). Therefore, understanding S. enterica ecology around livestock will help in controlling human salmonellosis.
Various surveys have shown the prevalence of S. enterica in wildlife (Bondo et al., 2016; Janecko et al., 2015; Skov et al., 2008), and genetic analyses have implied the association of S. enterica between wildlife and livestock (Carlson et al., 2015; Horton et al., 2013; Mentaberre et al., 2013). Accordingly, it has been postulated that wildlife carriers might transmit S. enterica from contaminated farms to other farms, which makes S. enterica prevention more difficult on farms in a large area. However, livestock farms strongly and unwittingly attract wildlife by providing them with feed and resting spaces (Tsukada et al., 2010); therefore, the protection measures demand great efforts, for which they may not be motivated. Further investigation of the transmission of S. enterica between wildlife and cattle will aid the decision‐making in applying protection measures.
Tokachi district, a major cattle farming area in Hokkaido, Japan, has 1400 dairy farms and 800 beef farms rearing 450,000 cattle (Ministry of Agriculture, Forestry and Fisheries, 2018); cattle salmonellosis has occurred here every year (Hokkaido Prefectural Government, 2020). Livestock products are notable food sources for human foodborne salmonellosis (Pires et al., 2014); thus, S. enterica prevention in cattle farms in Tokachi is critical for both farmers’ health in this district, and for food safety nationwide. A previous study (Fujii et al., 2012) revealed that wildlife was infected with the same serotypes of S. enterica as cattle in Tokachi, which suggested that the wildlife was potentially a source of bovine salmonellosis. Due to this background, farmers are concerned about S. enterica in both wildlife and cattle. S. enterica surveys such as monitoring surveys using wildlife carcasses and environmental samples at cattle farms have been performed in some municipalities in Tokachi. However, the survey results have not been sufficiently well‐organised to investigate inter‐species transmission of S. enterica. Comprehensive knowledge of S. enterica from wildlife and cattle in Tokachi, including genetic characteristics of isolates, will be helpful to evaluate the possibility of transmission of S. enterica between wildlife and cattle.
The objective of this study was to elucidate whether S. enterica has been shared between wildlife and cattle. Then, we aimed to organise retrospectively S. enterica detection information derived from survey results and cattle clinical records by serotype between 2008 and 2018 in Tokachi, and assess the trends of detection of each serotype in each species, and similarity of the genetic characteristics in the corresponding S. enterica serotypes from wildlife and cattle. In this study, we tested the following working hypotheses: (1) wildlife and cattle inhabiting the same area were infected with the same S. enterica serotypes in the same time period and (2) S. enterica serotypes from wildlife and cattle had similar genetic patterns [pulsed‐field gel electrophoresis (PFGE) patterns]. In Tokachi, the previous study showed that jungle crows (Corvus macrophalynchos), carrion crows (Corvus corone) and raccoons (Procyon lotor) were infected with S. enterica (Fujii et al., 2012). These species are known to often invade livestock farms in search of feed and resting sites (Takeda et al., 2015; Yamaguchi et al., 2020), which probably results in S. enterica transmission between wildlife and cattle via faeces or direct contact. Accordingly, we surveyed these wildlife species as high‐risk species for S. enterica transmission between cattle farms in Tokachi.
2. MATERIALS AND METHODS
2.1. Study area
We investigated S. enterica in wildlife, consisting of raccoons and crows, and cattle in three municipalities in Tokachi retrospectively between 2008 and 2018, using the past monitoring survey results for wildlife and cattle environments, diagnosis records for individual cattle and testing records for introduced calves. Three adjacent municipalities, towns A, B and C, were selected as study areas to survey wildlife and cattle sympatrically inhabiting Tokachi. The number of cattle farms in these municipalities ranged from 50 to 200, and tended to decrease as the study period progressed. The average herd size ranged from 150 to 200 in dairy farms and from 700 to 2300 in beef farms in 2015, and has recently tended to increase.
2.2. Collection of wildlife samples
To identify S. enterica serotypes in wildlife, first of all, we collected the individuals exterminated in towns A, B and C between 2008 and 2018: the details for each species are shown in Table 1. Crows were collected only in town A. The individuals were caught by traps and sacrificed by CO2 gas or gun shots under pest control programs. Because samples from wildlife were dependent on the program, samples could not be collected evenly across years, species and towns. Among the tested individuals, approximately 60% of raccoons and 90% of crows were captured at cattle farms. The other capture sites included riverside, cropfields and yards of human residences for raccoons and landfill for crows. Rectal/cloacal swabs were collected from all individuals. Considering the possibility of mechanical transmission of S. enterica by wildlife, footpad swabs were collected from 60% to 90% of individuals in each species. We did not test the footpad swabs provided with other specimens in a single bag, because of the possibility of cross‐contamination. After collection, the swab samples were immediately put into pre‐enrichment or enrichment broth for culture tests.
TABLE 1.
Number of tested samples from wildlife for Salmonella enterica between 2008 and 2018, in Tokachi, Hokkaido, Japan
| Species | Town | Year | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | ||
| Raccoon | A, B, C | – | 39 (39) | 11 (11) | 15 (15) | 14 (7) | 6 (2) | – | – | – | 70 (14) | 4 (4) |
| Jungle Crow | A | – | 41 (41) | 37 (37) | 73 (73) | 54 (11) | 23 (0) | – | 9 (9) | 176 (176) | 151 (151) | 41 (34) |
| Carrion Crow | A | 14 (12) | 74 (74) | 19 (19) | 37 (37) | 50 (20) | 25 (0) | – | 1 (1) | 100 (100) | 32 (32) | 10 (8) |
Note: The number outside of parenthesis refers to the number of individuals from which rectal/cloacal swabs were taken. The number in parenthesis refers to the number of individuals from which footpad swabs were also taken.
‘S. enterica‐positive’ in each individual was defined as isolating S. enterica from either or both sample types and counted as one detection by serotype. The prevalence of S. enterica in wildlife was calculated by species and sample type. We tested the difference in prevalence by species and sample type using Fisher's exact test in R version 4.0.2 (R Core Team, 2020).
In organising the prevalence and serotypes of S. enterica in each species, the data in wildlife between 2008 and 2010 referred to the previous report (Fujii et al., 2012). Ten isolates, including S. Typhimurium, S. Braenderup and S. Infantis, reported by Fujii et al. (2012) were assessed genetically by PFGE analysis for the first time in this study.
2.3. Collection of cattle samples
To determine S. enterica serotypes in cattle, we collected S. enterica detection records between 2008 and 2018 in the following four collection schemes. A brief explanation of the records collected in each scheme follows: (1) clinical signs: S. enterica detection using faeces or blood samples from cattle presenting clinical signs suspected to be salmonellosis in dairy and beef farms in towns A, B and C; (2) introduction test: S. enterica detections in quarantine in faeces samples from calves introduced into beef growing farms or public calving farms in towns A, B and C; (3) annual monitoring: results of monitoring for S. enterica dissemination using environmental samples taken from manure storage and alleys for lactating cattle at all dairy farms in town A once or twice a year and (4) monthly monitoring: results of monthly monitoring for S. enterica dissemination using environmental samples by the same method as with annual monitoring at nine dairy farms sending calves to public calving farms in town A between 2017 and 2018. In the clinical signs and introduction tests, the number of detections of S. enterica serotypes shared between wildlife and cattle was available in each town, but the total number of tests was not available. The details of each scheme are described in the Appendix.
S. enterica detection records were collected from the Union for Livestock Infectious Diseases Prevention (ULIDP), comprised local livestock farming‐related organisations, and from Tokachi Livestock Hygiene Service Center (LHSC), a public organisation for diagnosis of livestock diseases. Collection of culture test samples and S. enterica testing for each sampling scheme were performed by multiple organisations including ULIDP, LHSC and the Animal Research Center (see Table S1).
2.4. Salmonella enterica culture test and serotyping of isolates
S. enterica culture tests were performed according to the previous study (Fujii et al., 2012). The footpad swabs from wildlife and environmental samples were incubated at 37°C for 18–24 h in pre‐enrichment broth. The cloacal/rectal swabs from wildlife were inoculated in enrichment broth, skipping the pre‐enrichment stage. For pre‐enrichment, 10 ml of buffered peptone water (BPW) (Oxoid) and 9 ml of Enterobacteriaceae Enrichment Mannitol (EEM) (Eiken) broth per sample were used in 2008 and in 2009 to 2018, respectively. In enrichment, 1 ml of the pre‐enrichment culture or rectal/cloacal swab sample was added to 10 ml of enrichment broth and incubated at 42°C for 18–24 h. For enrichment, tetrathionate broth (Eiken) and Rappaport‐Vassiliadis broth (Eiken or Atect) were used in 2008 and in 2009 to 2018, respectively. After enrichment, the culture was plated on both desoxycholate hydrogen sulfide lactose (Eiken) agar containing 20 μg novobiocin/ml (n‐DHL; Sigma‐Aldrich Japan) and ES Salmonella Agar II (Eiken) and incubated at 37°C for 24–48 h. For S. enterica detections from cattle in the sampling schemes for clinical signs and introduction tests, only the detection records with their serotypes were provided by LHSC.
A colony suspected to be S. enterica, black‐coloured on n‐DHL agar or pink‐coloured on ES Salmonella II, was picked up from each agar and cultivated on the same kind of agar to obtain a pure culture. The isolates were identified serologically using polyvalent O‐ and O1‐grouping antisera (Denka Seiken) and confirmed by PCR assay by detecting the invA gene of S. enterica. The template DNA for PCR was extracted from a purely cultivated colony using Instagene matrix (BioRad Inc.). The primers are as follows, referring to Chiu and Ou (1996): INVA‐1 (5′‐ACAGTGCTCGTTTACGACCTGAAT‐3′) and INVA‐2 (5′‐AGACGACTGGTACTGATCGATAAT‐3′). The total 25 μl volume of PCR reaction mixture constituted 12.5 μl of KAPA2G™ Fast HotStart ReadyMix (Kapa Biosystems) and 1.25 μl of each 10μM primer and 2.5 μl template. The thermal protocol was modified according to manufacturer recommendations and involved 3 min of initial denaturation at 95°C, followed by 35 cycles of denaturation (95°C, 15 s), primer annealing (60°C, 15 s), extension (72°C, 5 s) and a final extension (72°C, 1 min) using i Cycler Thermal cycler (BioRad Inc.). DNA extracted from a known S. enterica Typhimurium colony by the same method as the samples was used as a positive control. Amplified PCR products were electrophoresed on 1.5% agarose gel to verify the presence of amplicons of the expected size (224 bp). S. enterica isolates were serotyped using commercial O and H antisera (Denka Seiken) based on the Kauffmann‐White scheme (Grimont & Weill, 2007). Isolates determined to be monophasic variants of S. Typhimurium, that is S. 4,[5],12:i:‐, were considered separate from S. Typhimurium.
2.5. Organising Salmonella enterica detection records
All S. enterica detection records were arranged chronologically by species and serotype to investigate whether the same serotype was detected from both wildlife and cattle in the same year. The month‐level data were also investigated for the relevant detection events caused by genetically similar isolates between wildlife and cattle. S. enterica detection was recorded individually in wildlife and at farm level in cattle. Multiple detections of the same serotype in cattle in the same farm in a year were considered to be a sequential outbreak and counted as one detection. Because cattle sample collection methods and the subject serotype in town A were different from those in towns B and C, the detection records from cattle in town A were also arranged by sampling schemes independently. This study retrospectively analysed the existing sampling records, which were not initially recorded with the aim of comparing the prevalence between species in the long term; therefore, the numbers of collected records were irregular and uneven among species. Then, we focused on the trends of detection of each serotype among detected S. enterica.
2.6. Pulsed‐field gel electrophoresis (PFGE) analysis
The serotypes detected from both wildlife and cattle were assessed by PFGE analysis to determine the genetic diversity and relatedness among isolates and their corresponding detection records. One isolate recovered at each S. enterica detection event was subjected to PFGE analysis. Multiple detections of the same serotype from the same species caught at the same place in a year were considered to be derived from a single epidemic strain with the same genetic characteristics; isolates from the first individual identified to be positive for S. enterica in each serotype and species in a year were subjected to PFGE analysis. Without a significant difference in S. enterica prevalence between the crow species, they were regarded as the same species ‘crow’ in selecting the isolate for PFGE analysis.
PFGE analysis was performed to determine the genetic patterns of S. enterica isolates referring to the following protocol (Ribot et al., 2006). Bacterial DNA prepared in agar plug was digested with 30 units of XbaI (Nippon gene). The plugs were run off in 22 h using a CHEF Mapper XA system (Bio‐Rad Laboratories) in 0.5 × Tris‐borate‐EDTA buffer (Nippon gene) supplemented with 20 μM thiourea at 14°C and 6 V/cm, while the pulse switch time was increased from 2.2 to 63.8 s. Lambda PFG Ladder N0341 (New England Biolabs Japan Inc.) was used as a molecular reference marker. The fragment patterns were automatically analysed, comparing them with the pattern of the reference marker using GelJ software (Heras et al., 2015). Dendrograms for each serotype were constructed using the unweighted pair group matching algorithm (UPGMA) using Dice coefficients with 2% band position tolerance. Based on the criteria for interpreting PFGE patterns (Tenover et al., 1995), the same Clade ID was given to isolates with PFGE band differences ≤ 3, designating them as closely related strains.
3. RESULTS
3.1. Salmonella enterica detections in wildlife
Prevalence of S. enterica at the individual level between 2008 and 2018 was 10.7% (17/159, 95% CI: 6.4–16.5%) in raccoons, 5.8% (35/605, 95% CI: 4.1–8.0%) in jungle crows and 5.5% (20/362, 95% CI: 3.4–8.4%) in carrion crows (Table 2). In raccoons and jungle crows, S. enterica was detected more frequently from rectal/cloacal swabs than from footpad swabs (p = 0.02, p < 0.01, respectively). In carrion crows, the frequency of S. enterica in cloacal swabs did not significantly differ from that in footpad swabs (p = 0.2). While the prevalence did not significantly differ between the crow species (p = 1), S. enterica was more prevalent in raccoons than in crows (p = 0.02). Isolated serotypes were Agona, Braenderup, Dublin, Infantis, Manhattan, Mbandaka, Montevideo, Nigeria, 4,[5],12:i:‐, 4,[5],12:HUT (untypable H antigen), Stanley, Thompson and Typhimurium (Table 2). The most frequently isolated serotypes were Braenderup and Thompson in raccoons, and Infantis and Mbandaka in both crow species (Table 2).
TABLE 2.
Prevalence and serotypes of Salmonella isolated from swab samples from wildlife in Tokachi, Hokkaido, Japan, between 2008 and 2018
| Species | Rectum/cloaca | Footpad | Individual b | ||
|---|---|---|---|---|---|
| Raccoon |
16/159 (10.1%) |
Braenderup (5) Infantis (1) Stanley (2) Thompson (4) 4,[5],12:i:‐ (2) Typhimurium (2) |
2/92 (2.2%) |
Braenderup (1) Stanley (1) |
17/159 (10.7%) |
| Jungle crow |
32/605 (5.3%) |
Agona (2) Dublin (2) Infantis (12) Manhattan (1) Mbandaka (10) 4,[5],12:i:‐ (1) Typhimurium (4) |
7/532 (1.3%) |
Infantis (4) Montevideo (2) Typhimurium (1) |
35/605 (5.8%) |
| Carrion crow |
16/362 (4.4%) |
Agona (2) Infantis (5) Mbandaka (6) Montevideo (1) Nigeria (1) 4,[5],12:HUT b (1) |
7/303 (2.3%) |
Braenderup (1) Infantis (2) Mbandaka (1) Montevideo (3) |
20/362 (5.5%) |
Salmonella‐positive individual was defined as isolating Salmonella from either or both sample types.
HUT: untypable H antigen.
3.2. Salmonella enterica detections in cattle
In town A, 29 detections of S. enterica of nine serotypes, Anatum, Braenderup, Dublin, Muenster, 4,[5],12:d:‐, 4,[5],12:i:‐, Reading, Typhimurium and Wangata, were recorded in dairy farms and one detection of S. Dublin was recorded in a beef farm (Table 3). Twenty‐one detections of the same serotype as in the wildlife isolates were recorded in dairy cattle in Town A throughout the study period: Braenderup, Dublin, 4,[5],12:i:‐ and Typhimurium. Table S2 shows the detected serotypes in town A by year and each scheme. In towns B and C, 60 detections of the same serotype as the wildlife isolates were recorded throughout the study period: Braenderup, Dublin, Infantis, Mbandaka, Montevideo, 4,[5],12:i:‐ and Typhimurium (Table 3).
TABLE 3.
Serotypes of Salmonella isolated from cattle and wildlife animals in Tokachi, Hokkaido, Japan, between 2008 and 2018
| Year | Cattle | Wildlife | ||||
|---|---|---|---|---|---|---|
| Dairy cattle | Beef cattle | Dairy and Beef cattle a | Raccoon | Jungle crow | Carrion crow | |
| Town A | Town A | Town B and C | Town A, B, C | Town A | Town A | |
| 2008 |
Anatum(2) 4,[5],12:d:‐(1) Typhimurium(2) |
None |
Braenderup(1) Infantis(1) Typhimurium(1) |
ND c | ND | Braenderup(1) |
| 2009 | Typhimurium(1) | None |
Braenderup(2) Typhimurium(1) |
Braenderup(1) Thompson(2) |
Infantis(4) Typhimurium(1) |
Infantis(4) |
| 2010 | None b | None |
Infantis(1) Typhimurium(1) |
Braenderup(1) Thompson(1) Typhimurium(2) |
Infantis(3) |
Infantis(1) |
| 2011 | 4,[5],12:d:‐(1) | None |
Braenderup(1) Infantis(1) Montevideo(1) |
Braenderup(2) Thompson(1) |
Montevideo(2) |
Infantis(1) Montevideo(3) |
| 2012 | Braenderup(1) | None |
Mbandaka(1) 4,[5],12:i:‐(1) |
Braenderup(2) |
Infantis(1) Manhattan(1) Typhimurium(1) |
4,[5],12:HUT(1) |
| 2013 | 4,[5],12:i:‐(1) | None | 4,[5],12:i:‐(4) |
None |
Infantis(2) |
None |
| 2014 |
Braenderup(1) 4,[5],12:i:‐(1) |
None | 4,[5],12:i:‐(2) | ND | ND | ND |
| 2015 | Dublin(1) 4,[5],12:i:‐(1) | Dublin(1) | 4,[5],12:i:‐(10) | ND |
Infantis(2) Dublin(1) |
None |
| 2016 |
Dublin(3) Muenster(1) 4,[5],12:i:‐(1) , Wangata(1) |
None |
Dublin(1) 4,[5],12:i:‐(12) |
ND |
Agona(1) Dublin(1) Mbandaka(3) 4,[5],12:i:‐(1) Typhimurium(3) |
Agona(2) Mbandaka(6) |
| 2017 |
Dublin(1) Muenster(1) 4,[5],12:i:‐(3) |
None |
Dublin(1) Mbandaka(1) 4,[5],12:i:‐(7) |
Infantis(1) 4,[5],12:i:‐(2) Stanley(2) |
Mbandaka(6) |
None |
| 2018 |
Dublin(2) 4,[5],12:i:‐(1) Reading(1) Typhimurium(1) |
None |
Dublin(1) Mbandaka(1) 4,[5],12:i:‐(7) |
None |
Agona(1) Mbandaka(1) |
Nigeria(1) |
Note: The number in parenthesis in cattle refers to the number of farms in which each serotype was detected. The number in parenthesis in wildlife refers to the number of individuals in which each serotype was detected. Serotypes in bold characters were common to both wildlife and cattle. Underlined serotypes were common to both wildlife and cattle in each year.
Only detections of the same serotype with wildlife isolates are described.
ND means Salmonella test was not done.
None means no Salmonella was detected.
3.3. Common serotypes in wildlife and cattle
Seven serotypes were detected from both wildlife and cattle in towns A, B and C: Braenderup, Dublin, Infantis, Mbandaka, Montevideo, 4,[5],12:i:‐ and Typhimurium (Table 3). Comparing the number of detections of each serotype by species, S. Dublin (wildlife: 2, cattle: 11) and S. 4,[5],12:i:‐ (wildlife: 3, cattle: 51) were frequently found in cattle. In contrast, S. Infantis (wildlife: 19, cattle: 3) and S. Mbandaka (wildlife: 16, cattle: 3) were more frequently isolated from wildlife. S. Typhimurium (wildlife: 7, cattle: 7) and S. Braenderup (wildlife: 7, cattle: 6) were isolated from all species at a similar frequency. S. Montevideo was detected in a cattle farm and from five crows caught in the same trap on the same day in 2011. Out of the five crows, one crow was positive for S. Montevideo in both cloacal and footpad samples, while the others were positive only in footpad samples; therefore, these crows were probably contaminated with S. Montevideo in the trap mechanically. The seven serotypes were detected from wildlife and cattle in the same year at least once during the study period.
3.4. Similarity of PFGE patterns
Similarity of PFGE patterns between wildlife and cattle isolates varied by serotype (Figures 1, 2, 3, 4, 5, 6, 7). For S. Braenderup, S. Dublin, S. Montevideo, S. 4,[5],12:i:‐ and S. Typhimurium, a part of the PFGE clade included both wildlife and cattle isolates. For S. Braenderup, S. Dublin, S. Montevideo and S. 4,[5],12:i:‐, some of the wildlife and cattle isolates grouped into the same PFGE clade were recovered in the same year. For S. Braenderup, raccoon isolates were recovered 4 months before and 4 months after the cattle isolates in 2009, and 1 month after the cattle isolates in 2012. Month data were missing for cattle isolates in 2011. For S. Dublin, the crow isolate was recovered 1 month after the recovery of the cattle isolate in 2015, and the crow and cattle isolates were recovered in the same month, and cattle isolates were also recovered 2 months before and 1 month after in 2016. For S. Montevideo, the cattle isolate data at the level of month were not available. For S. 4,[5],12:i:‐, cattle isolates were intermittently recovered throughout the year in 2016 and 2017, in which wildlife isolates were recovered. For S. Infantis and S. Mbandaka, PFGE clades for wildlife and cattle isolates were different from each other. Four isolates were not able to be assessed for PFGE analysis: one isolate each of S. Braenderup, S. Typhimurium, S. 4,[5],12:i:‐ and S. Dublin.
FIGURE 1.

Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Braenderup isolates. Isolates from raccoons are marked with circles. The isolate from the crow is marked with a square
FIGURE 2.
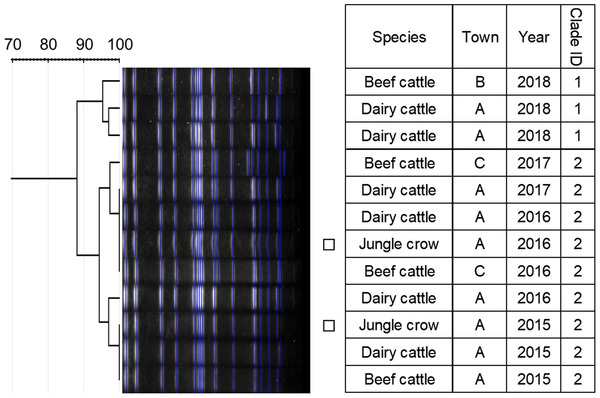
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Dublin isolates. Isolates from crows are marked with squares
FIGURE 3.
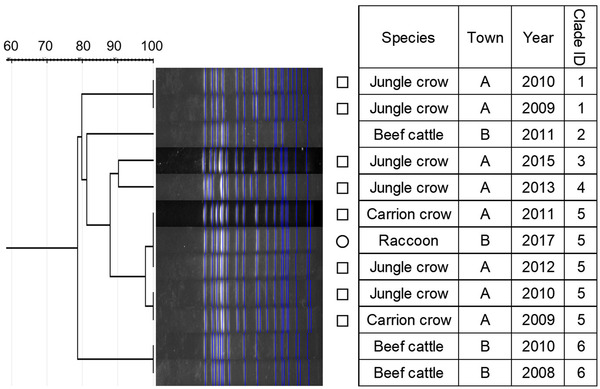
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Infantis isolates. The isolate from the raccoon is marked with a circle. Isolates from crows are marked with squares
FIGURE 4.
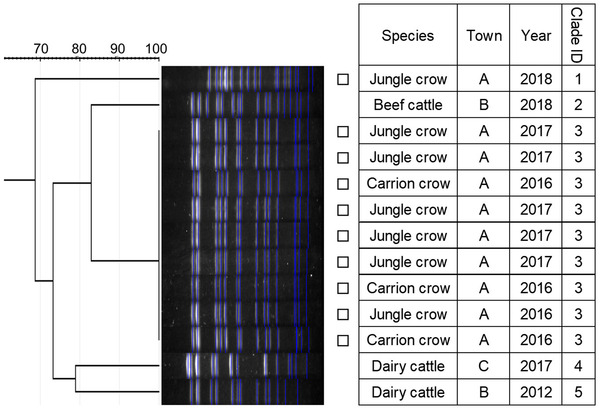
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Mbandaka isolates. Isolates from crows are marked with squares
FIGURE 5.
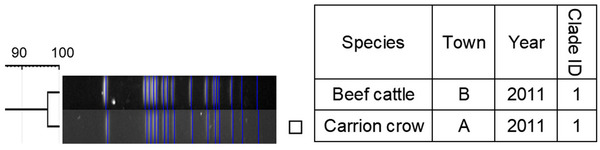
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Montevideo isolates. The isolate from the crow is marked with a square
FIGURE 6.
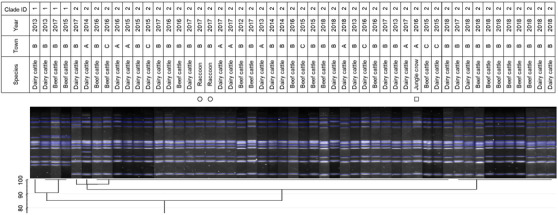
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella 4,[5],12:i:‐ isolates. Isolates from raccoons are marked with circles. The isolate from the crow is marked with a square
FIGURE 7.
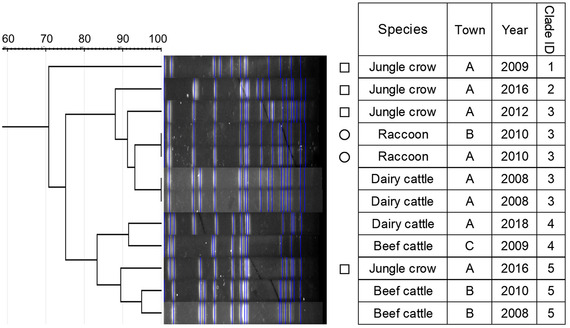
Dendrogram of pulsed‐field gel electrophoresis (PFGE) gel band patterns for Salmonella Typhimurium isolates. Isolates from raccoons are marked with circles. Isolates from crows are marked with squares
4. DISCUSSION
Our retrospective study organised the S. enterica detection records in Tokachi stored in relevant organisations in order to elucidate the distribution and genetic relatedness of S. enterica serotypes in wildlife and cattle over the long term (10 years). We observed detections of the same serotype of S. enterica from wildlife and cattle in the same year. For S. Braenderup, S. Dublin, S. Montevideo and S. 4,[5],12:i:‐, the isolates from both species in the same year were genetically similar to each other (Figures 1, 2, 5 and 6), which demonstrated that S. enterica was shared between wildlife and cattle. These serotypes included critical ones for livestock hygiene and public health: S. Dublin, highly pathogenic in cattle (Harvey et al., 2018), and S. 4,[5],12:i:‐, which has recently been widespread worldwide in livestock and humans (Arai et al., 2018; EFSA, 2019b). All isolates of S. Dublin and S. 4,[5],12:i:‐ from wildlife and one S. Braenderup isolate from a raccoon were recovered in the same month or, 1 month before or after the cattle isolates of the same serotype. Such detection patterns indicate that S. enterica probably transmitted between species directly or indirectly in the detection period. Prevalence in jungle crows and carrion crows was not significantly different and the typical serotypes were S. Mbandaka and S. Infantis in both types of crows (Table 2). Such similarity in prevalence and typical serotypes in the crows may be caused by horizontal transmission between crows due to shared feeding grounds and mixed‐species flocks.
Although it was difficult to compare the frequency using test data based on uneven sampling methods, S. Dublin and S. 4,[5],12:i:‐ were infrequently detected from wildlife, unlike from cattle (Table 3). Additionally, these serotypes were detected from wildlife only in years with outbreaks of each serotype in cattle (Table 3). In contrast, S. Infantis and S. Mbandaka isolates that were not genetically similar between each species were detected frequently from wildlife, unlike from cattle (Figures 3 and 4 and Table 3). Considering these observations of S. enterica detection, the shared isolates for S. Dublin and S. 4,[5],12:i:‐ likely originated from cattle or other unsurveyed species; wildlife did not appear to play a role in the source of infection for transmission. Inherent characteristics of S. Dublin and S. 4,[5],12:i:‐ also corroborate this suggestion. S. Dublin is a cattle specific serotype (Harvey et al., 2018); detection of this serotype from wildlife is limited to a few studies in wild mammals including mice and foxes (Glawischnig et al., 2017; Jones & Twigg, 1976; Tablante & Lane, 1989), and the present study shows the first detection from wild birds. In the previous reports, S. Dublin was detected from wildlife in the vicinity of S. Dublin epidemics in cattle as in this study, which implied that S. Dublin seemed to be transmitted from cattle to wildlife (Glawischnig et al., 2017; Jones & Twigg, 1976). A monophasic variant of S. Typhimurium, S. 4,[5],12:i:‐, has been increasingly reported as the causative agent of salmonellosis in livestock and humans around the world, including Japan (EFSA, 2018; Kijima et al., 2019; Soyer et al., 2009). Previous studies suggested that a certain epidemic clone was dominant in recent S. 4,[5],12:i:‐ isolates in Europe and Japan (Arai et al., 2018; Petrovska et al., 2016). In this study, detections of S. 4,[5],12:i:‐ isolates with the same genetic patterns have rapidly increased since 2012, which probably implies that the S. 4,[5],12:i:‐ strains originated in the same epidemic clone spreading globally in humans and domestic animals. Therefore, the S. 4,[5],12:i:‐ strains in Tokachi might not be mainly sourced from wildlife, but rather from cattle or other livestock animals and humans, which were not surveyed here. The genetically similar S. Typhimurium strains were shared between wildlife and cattle, but not detected in the same year. S. Typhimurium is a common serotype in both wildlife and cattle (Kijima et al., 2019 Simpson et al., 2018; Tizard, 2004). Considering the year of detection of isolates in each species for clade IDs 3 and 5, to which both wildlife and cattle isolates belong (Figure 7), cattle strains were possibly transmitted to wildlife and then maintained in the population.
Many pathogens including S. enterica have been shown to be potentially transmitted between wildlife and livestock at their interface (Gortázar et al., 2016; Miller et al., 2013). In the case of S. enterica, pathogens seem to be transmitted between wildlife and livestock via feeding (Carlson et al., 2011a; Davies & Wales, 2013) and the number of wild birds invading cattle farms could be a risk factor for S. enterica contamination of the farms (Carlson et al., 2011b). On the other hand, S. enterica transmission from livestock to wildlife has also been implicated (Mentaberre et al., 2013; Skov et al., 2008); our obtained results for S. Dublin and S. 4,[5],12:i:‐ seemed to be similar to the latter. Raccoons and crows use various types of habitat including livestock farms, cities and natural environments (Ikeda et al., 2004; Morishita et al., 2003; Takeda et al., 2015; Zeveloff, 2002), which may allow them to spread pathogens into various types of environments, resulting in a hub of transmission between livestock and other environments. The serotypes detected from wildlife, comprised Infantis, Stanley, Typhimurium, Thompson and 4,[5],12:i:‐, were also identified as common serotypes of foodborne S. enterica in Japan (National Institute of Infectious Diseases Japan, 2018). Accordingly, the importance of preventing S. enterica spreading by wildlife in Tokachi is heightened not only for livestock hygiene but also for food safety of farm products. Recently, antimicrobial resistant (AMR) S. enterica has become a growing concern in human and animal health worldwide (Hernando‐Amado et al., 2019); wildlife is also infected with AMR organisms (Torres et al., 2020; Vittecoq et al., 2016). Livestock farms have been implicated in playing an important role in producing AMR organisms because of their frequent antimicrobial use (Silbergeld et al., 2008). Because wildlife around cattle farms might contribute to spreading AMR. S. enterica sourced from cattle, the degree of AMR of the isolates from wildlife would be critical for considering the risk of S. enterica infection in wildlife in the study site in future research.
This study had the following limitations. First, because our study aimed to investigate the S. enterica distribution retrospectively using existing detection records, which were based on unique sampling schemes in each municipality, detection records were uneven and irregular in each species. Furthermore, only one isolate per serotype per year in each species was chosen for PFGE analysis, which might lead to underestimation of the genetic diversity of S. enterica. Because previous studies suggested that S. enterica distribution in wildlife, cattle and the natural environment was seasonal (Bondo et al., 2016; Nielsen et al., 2004; Thomas et al., 2013), S. enterica potentially failed to be detected because some sampling in this study was done infrequently (e.g. annual monitoring in the third scheme). Additionally, because S. enterica can survive in farm environments for months (Alegbeleye et al., 2018), we should consider the possibility that the cattle farm had been contaminated for several months prior to the detection by the monitoring survey when assessing the relationship of the timing of S. enterica detection in wildlife and cattle on a monthly basis. Accordingly, investigation based on an appropriate strategy with regular and even sampling and genetic analysis is essential for evaluating the possibility of S. enterica transmission quantitatively and precisely in the future. Second, PFGE analysis might be inappropriate for distinguishing the isolates genetically because the patterns of some isolates for S. Braenderup and S. Infantis were stable over 7 years (Figures 1 and 3), which made it difficult to envision an epidemiological association between the isolates after such a long time. For better understanding of genetic relationships between S. enterica isolates, genetic analyses with high discriminatory power like whole genome analysis (Tang et al., 2019) would be required. Third, it was possible that S. enterica was transmitted from a common source to both wildlife and cattle; however, the present study focused on wildlife and cattle and environment contaminated with cattle dung, which made it difficult to assess it. There are some important sources of S. enterica, including cattle feed (Van Metre et al., 2008), which were not surveyed in this study. S. enterica surveillance in other possible sources would also be helpful to reveal S. enterica transmission dynamics.
5. CONCLUSION
Our study demonstrated that S. Braenderup, S. Dublin, S. Montevideo and S. 4,[5],12:i:‐ with similar genetic patterns were shared between wildlife and cattle in Tokachi. In particular, for S. Dublin and S. 4,[5],12:i:‐, the trends of detection and the epidemiological characteristics of each species suggested that wildlife was not the initial host of the shared isolates, but rather that the source was probably other species including cattle. Therefore, limiting invasions of these wildlife into case farms, possibly the source of infection for wildlife, seems to be particularly critical for preventing dissemination of S. enterica at a regional level. Control strategies should be developed based on the role of wildlife in S. enterica ecology in livestock areas, which would contribute not only to livestock hygiene but also to public health, in accordance with a One Health concept.
FUNDING INFORMATION
This work was partially financed by KAKENHI, Grant Number 20880033 from Japan Society for the Promotion of Science.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
ETHICS STATEMENT
This study was approved by the Animal Ethical Committee and the Animal Care and Use Committee of Obihiro University, and Hokkaido Animal Research Center.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.685.
AUTHOR CONTRIBUTIONS
Emi Yamaguchi: Conceptualisation; data curation; formal analysis; investigation; methodology; project administration; visualisation; writing‐original draft. Kei Fujii: Conceptualisation; data curation; funding acquisition; investigation; methodology. Mitsunori Kayano: Conceptualisation; writing‐review & editing. Yoshie Sakurai: Data curation; investigation; methodology. Atsuko Nakatani: Data curation; formal analysis; investigation; methodology. Motoki Sasaki: Investigation. Julia A. Hertl: Writing‐review & editing. Yrjo Grohn: Conceptualisation; supervision; writing‐review & editing.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
Cattle isolates were kindly provided by Tokachi Livestock Hygiene Service Center. Wildlife samples were collected with the cooperation of municipalities and farmers. Some raccoons were captured under trapping permission granted by Tokachi General Subprefecture Bureau of the Hokkaido government (authorisation no. Tokansei‐2534 in 2012, Tokansei‐1255 in 2013, Tokansei‐349 in 2014). Antimicrobial paper disks with ceftiofur and marbofloxacin were kindly provided by Zoetis Japan Inc. and Meiji Seika Pharma Co., Ltd. We thank all co‐operators of this study.
DETAILS OF FOUR SCHEMES FOR SALMONELLA ENTERICA DETECTION FROM CATTLE
Clinical signs: Cattle presenting clinical signs such as diarrhoea and fever suspected to be salmonellosis by farmers and veterinarians were diagnosed by culture tests using cattle faeces or blood samples. Salmonella enterica (Salmonella) detection in clinical signs was recorded by serotype at farm level in dairy and beef farms in towns A, B and C. Records of Salmonella detection in clinical signs in town A were collected regardless of serotype from ULIDP in town A. In contrast, the records in towns B and C were based on the collection data of Salmonella isolates at LHSC, limited to the same serotypes detected from wildlife. We were only able to collect detection records, and the total number of Salmonella tests for cattle with clinical signs was unknown.
Introduction test: Cattle introduced into beef growing farms or public calving farms in towns A, B and C were tested for Salmonella individually with faeces samples when they were introduced into each farm. Samples were sent to private laboratories for Salmonella detection, and LHSC identified the serotype of Salmonella isolates. In each municipality, one public calving farm and fewer than 40 beef growing farms operated. Public calving farms had contracted for taking in and looking after milking calves from dairy farms in the same town until the first parturition or earlier. Salmonella detection in the introduction test was recorded by serotype at supplier farm level. Though some cattle were introduced from sites not included in the study, we also counted Salmonella detection from such cattle because we regarded them as Salmonella introductions into the study site by positive introduced cattle. In cases where the same serotype was detected from multiple supplier farms in the same beef growing farms or public calving farms in a year, the first isolates from introduced cattle by each municipality were selected for genetic analyses. Records of introduction tests were collected as well as clinical signs; that is, the total number of introduction tests was unknown and Salmonella detection records in towns B and C were limited to the same serotypes detected from wildlife.
Annual monitoring: Salmonella dissemination was monitored using environmental samples at all dairy farms in town A, approximately 40 farms, once in summer (July to August) between 2008 and 2013, and once in each summer and autumn (October to November) between 2014 and 2018. We cannot show the specific number of monitored farms, which is equal to the number of dairy farms in town A and would lead to identification of the municipality. Salmonella detection in annual monitoring was recorded by serotype at farm level. The environmental samples were obtained from – two to three locations per farm at manure storage and alleys for lactating cattle, including dung from many cattle. The samples were taken using sterilised swabs and kept in zipped plastic bags at 4°C before culture tests were performed, detailed in Section 2. Annual monitoring had been conducted by the ULIDP and we undertook the culture tests for samples. At positive farms, all individuals were tested to detect the carriers regardless of serotype. Isolates from individuals were used for genetic analyses. Even if the subsequent individual tests could not detect positive cattle, isolates from environmental samples were assessed.
Monthly monitoring: At all nine dairy farms sending calves to public calving farms in town A, Salmonella dissemination was monitored every month between 2017 and 2018 by the same method as with annual monitoring. The tested farms were a part of the tested farms in annual monitoring; therefore, only the results in annual monitoring were adopted in months for annual monitoring to avoid duplication of Salmonella detection.
Yamaguchi, E. , Fujii, K. , Kayano, M. , Sakurai, Y. , Nakatani, A. , Sasaki, M. , Hertl, J. A. , & Grohn, Y. T. (2022). Is Salmonella enterica shared between wildlife and cattle in cattle farming areas? An 11‐year retrospective study in Tokachi district, Hokkaido, Japan. Veterinary Medicine and Science, 8, 758–770. 10.1002/vms3.685
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- Alegbeleye, O. O. , Singleton, I. , & Sant'Ana, A. S. (2018). Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiology, 73, 177–208. 10.1016/j.fm.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai, N. , Sekizuka, T. , Tamamura, Y. , Tanaka, K. , Barco, L. , Izumiya, H. , Kusumoto, M. , Hinenoya, A. , Yamasaki, S. , Iwata, T. , Watanabe, A. , Kuroda, M. , Uchida, I. , & Akiba, M. (2018). Phylogenetic characterization of Salmonella enterica Serovar Typhimurium and its monophasic variant isolated from food animals in Japan revealed replacement of major epidemic clones in the last 4 decades. Journal of Clinical Microbiology, 56(5). 10.1128/JCM.01758-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondo, K. J. , Pearl, D. L. , Janecko, N. , Boerlin, P. , Reid‐Smith, R. J. , Parmley, J. , & Jardine, C. M. (2016). Epidemiology of Salmonella on the paws and in the faeces of free‐ranging raccoons (Procyon Lotor) in Southern Ontario, Canada. Zoonoses and Public Health, 63(4), 303–310. 10.1111/zph.12232 [DOI] [PubMed] [Google Scholar]
- Carlson, J. C. , Engeman, R. M. , Hyatt, D. R. , Gilliland, R. L. , Deliberto, T. J. , Clark, L. , Bodenchuk, M. J. , & Linz, G. M. (2011a). Efficacy of European starling control to reduce Salmonella enterica contamination in a concentrated animal feeding operation in the Texas panhandle. BMC Veterinary Research, 7(1), 9. 10.1186/1746-6148-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J. C. , Franklin, A. B. , Hyatt, D. R. , Pettit, S. E. , & Linz, G. M. (2011b). The role of starlings in the spread of Salmonella within concentrated animal feeding operations. Journal of Applied Ecology, 48(2), 479–486. 10.1111/j.1365-2664.2010.01935.x [DOI] [Google Scholar]
- Carlson, J. C. , Hyatt, D. R. , Bentler, K. , Mangan, A. M. , Russell, M. , Piaggio, A. J. , & Linz, G. M. (2015). Molecular characterization of Salmonella enterica isolates associated with starling‐livestock interactions. Veterinary Microbiology, 179(1–2), 109–118. 10.1016/j.vetmic.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Chiu, C. H. , & Ou, J. T. (1996). Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture‐multiplex PCR combination assay. Journal of Clinical Microbiology, 34(10), 2619–2622. 10.1128/JCM.34.10.2619-2622.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. H. , & Wales, A. D. (2013). Salmonella contamination of cereal ingredients for animal feeds. Veterinary Microbiology, 166(3–4), 543–549. 10.1016/j.vetmic.2013.07.003 [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA) . (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2017. EFSA Journal, 16(12). 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA) . (2019a). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA Journal, 17(2). 10.2903/j.efsa.2019.5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA) . (2019b). The European Union One Health 2018 zoonoses report. EFSA Journal, 17(12). 10.2903/j.efsa.2019.5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, K. , Onoe, S. , Sashika, M. , Kobayashi, K. , Imai, K. , Yamaguchi, H. , & Senna, K. (2012). Survey of Salmonella enterica in wildlife in and around cattle farms in Hokkaido, Japan. Journal of the Japan Veterinary Medical Association, 65(2), 118–121. 10.12935/jvma.65.118 [DOI] [Google Scholar]
- Glawischnig, W. , Lazar, J. , Wallner, A. , & Kornschober, C. (2017). Cattle‐derived Salmonella enterica serovar dublin infections in red foxes (Vulpes vulpes) in Tyrol, Austria. Journal of Wildlife Diseases, 53(2), 361–363. 10.7589/2016-04-087 [DOI] [PubMed] [Google Scholar]
- Gortázar, C. , Ruiz‐Fons, J. F. , & Höfle, U. (2016). Infections shared with wildlife: an updated perspective. European Journal of Wildlife Research, 62(5), 511–525. 10.1007/s10344-016-1033-x [DOI] [Google Scholar]
- Grimont, & Weill, F. ‐ X. (2007). Antigenic formulae of the Salmonella serovars. In World Health Organization Collaborating Centre for Reference and Research on Salmonella (9th ed.). Paris, France: Pasteur Institute. [Google Scholar]
- Harvey, R. R , Friedman, C. R. , Crim, S. M. , Judd, M. , Barrett, K. A. , Tolar, B. , Folster, J. P. , Griffin, P. M. , & Brown, A. C. (2018). Epidemiology of Salmonella enterica Serotype Dublin Infections among Humans, United States, 1968 –2013. Emerging Infectious Diseases, 23(9), 1493–1501. 10.3201/eid2309.170136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras, J. , Domínguez, C. , Mata, E. , Pascual, V. , Lozano, C. , Torres, C. , & Zarazaga, M. (2015). GelJ – A tool for analyzing DNA fingerprint gel images. BMC Bioinformatics, 16(1), 270. 10.1186/s12859-015-0703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando‐Amado, S. , Coque, T. M. , Baquero, F. , & Martínez, J. L. (2019). Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nature Microbiology, 4(9), 1432–1442. 10.1038/s41564-019-0503-9 [DOI] [PubMed] [Google Scholar]
- Hokkaido Prefectural Government . (2020) Status of outbreaks of livestock infectious diseases in Hokkaido. Retrieved from http://www.pref.hokkaido.lg.jp/ns/tss/kachikueisei/densenseisippei.htm
- Horton, R. A. , Wu, G. , Speed, K. , Kidd, S. , Davies, R. , Coldham, N. G. , & Duff, J. P. (2013). Wild birds carry similar Salmonella enterica serovar Typhimurium strains to those found in domestic animals and livestock. Research in Veterinary Science, 95(1), 45–48. 10.1016/j.rvsc.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Ikeda, T. , Asano, M. , Matoba, Y. , & Abe, G. (2004). Present status of invasive alien raccoon and its impact in Japan. Global Environmental Research, 8, 125–131. [Google Scholar]
- Janecko, N. , Čížek, A. , Halová, D. , Karpíšková, R. , Myšková, P. , & Literák, I. (2015). Prevalence, characterization and antibiotic resistance of Salmonella isolates in large corvid species of Europe and North America between 2010 and 2013. Zoonoses and Public Health, 62(4), 292–300. 10.1111/zph.12149 [DOI] [PubMed] [Google Scholar]
- Jones, P. W. , & Twigg, G. I. (1976). Salmonellosis in wild mammals. Journal of Hygiene, 77(1), 51–54. 10.1017/S0022172400055509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima, M. , Shirakawa, T. , Uchiyama, M. , Kawanishi, M. , Ozawa, M. , & Koike, R. (2019). Trends in the serovar and antimicrobial resistance in clinical isolates of Salmonella enterica from cattle and pigs between 2002 and 2016 in Japan. Journal of Applied Microbiology, 127(6), 1869–1875. 10.1111/jam.14431 [DOI] [PubMed] [Google Scholar]
- Lee, H. , & Yoon, Y. (2021). Etiological agents implicated in foodborne illness world wide. Food Science of Animal Resources, 41(1), 1–7. 10.5851/KOSFA.2020.E75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey, B. , Leonard, F. , Archambault, M. , Cullinane, A. , & Maguire, D. (2013). Enterobacteriaceae (2nd ed., pp. 239–274). Markey B., Leonard F., Archambault M., Cullinane A. & Maguire D., Clinical veterinary microbiology. London: Mosby Elsevie; r. [Google Scholar]
- Mentaberre, G. , Porrero, M. C. , Navarro‐Gonzalez, N. , Serrano, E. , Domínguez, L. , & Lavín, S. (2013). Cattle drive Salmonella infection in the wildlife‐livestock interface. Zoonoses and Public Health, 60(7), 510–518. 10.1111/zph.12028 [DOI] [PubMed] [Google Scholar]
- Miller, R. S. , Farnsworth, M. L. , & Malmberg, J. L. (2013). Diseases at the livestock‐wildlife interface: Status, challenges, and opportunities in the United States. Preventive Veterinary Medicine, 110(2), 119–132. 10.1016/j.prevetmed.2012.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Agriculture, Forestry and Fisheries . (2018). World census of agriculture and forestry in Japan. Retrieved from https://www.maff.go.jp/j/tokei/census/afc/
- Morishita, E. , Itao, K. , Sasaki, K. , & Higuchi, H. (2003). Movements of crows in urban areas, based on PHS tracking. Global Environmental Research, 7(2), 181–191. [Google Scholar]
- National Institute of Infectious Diseases Japan . (2018). Salmonella serovars from human sources, 2014–2018. Retrieved from Infectious Agents Surveillance Report website: https://www.niid.go.jp/niid/ja/iasr.html
- Nielsen, L. R. , Schukken, Y. H. , Gröhn, Y. T. , & Ersbøll, A. K. (2004). Salmonella Dublin infection in dairy cattle: Risk factors for becoming a carrier. Preventive Veterinary Medicine, 65(1–2), 47–62. 10.1016/j.prevetmed.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Petrovska, L. , Mather, A. E. , Abuoun, M. , Branchu, P. , Harris, S. R. , Connor, T. , Hopkins, K. L. , Underwood, A. , Lettini, A. A. , Page, A. , Bagnall, M. , Wain, J. , Parkhill, J. , Dougan, G. , Davies, R. , & Kingsley, R. A. (2016). Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerging Infectious Diseases, 22(4), 617–624. 10.3201/eid2204.150531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, S. M. , Vieira, A. R. , Hald, T. , & Cole, D. (2014). Source attribution of human salmonellosis: An overview of methods and estimates. Foodborne Pathogens and Disease, 11(9), 667–676. 10.1089/fpd.2014.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing. Version 4.0. 2 (Taking Off Again). Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ribot, E. M. , Fair, M. A. , Gautom, R. , Cameron, D. N. , Hunter, S. B. , Swaminathan, B. , & Barrett, T. J. (2006). Standardization of pulsed‐field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease, 3(1), 59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- Scallan, E. , Hoekstra, R. M. , Angulo, F. J. , Tauxe, R. V. , Widdowson, M.‐A. , Roy, S. L. , Jones, J. L. , & Griffin, P. M. (2011). Foodborne illness acquired in the United States – Major pathogens. Emerging Infectious Diseases, 17(1), 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbergeld, E. K. , Graham, J. , & Price, L. B. (2008). Industrial food animal production, antimicrobial resistance, and human health. Annual Review of Public Health, 29(1), 151–169. 10.1146/annurev.publhealth.29.020907.090904 [DOI] [PubMed] [Google Scholar]
- Simpson, K. M. J. , Hill‐Cawthorne, G. A. , Ward, M. P. , & Mor, S. M. (2018). Diversity of Salmonella serotypes from humans, food, domestic animals and wildlife in New South Wales, Australia. BMC Infectious Diseases, 18(1), 1–11. 10.1186/s12879-018-3563-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov, M. N. , Madsen, J. J. , Rahbek, C. , Lodal, J. , Jespersen, J. B. , Jørgensen, J. C. , Dietz, H. H. , Chriél, M. , & Baggesen, D. L. (2008). Transmission of Salmonella between wildlife and meat‐production animals in Denmark. Journal of Applied Microbiology, 105(5), 1558–1568. 10.1111/j.1365-2672.2008.03914.x [DOI] [PubMed] [Google Scholar]
- Soyer, Y. , Moreno Switt, A. , Davis, M. A. , Maurer, J. , Mcdonough, P. L. , Schoonmaker‐Bopp, D. J. , Dumas, N. B. , Root, T. , Warnick, L. D. , GröHn, Y. T. , & Wiedmann, M. (2009). Salmonella enterica Serotype 4,5,12:i:‐, an emerging Salmonella serotype that represents multiple distinct clones. Journal of Clinical Microbiology, 47(11), 3546–3556. 10.1128/JCM.00546-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanaway, J. D. , Parisi, A. , Sarkar, K. , Blacker, B. F. , Reiner, R. C. , Hay, S. I. , Nixon, M. R. , Dolecek, C. , James, S. L. , Mokdad, A. H. , Abebe, G. , Ahmadian, E. , Alahdab, F. , Alemnew, B. T. T. , Alipour, V. , Allah Bakeshei, F. , Animut, M. D. , Ansari, F. , Arabloo, J. , … Crump, J. A. (2019). The global burden of non‐typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. The Lancet Infectious Diseases, 19(12), 1312–1324. 10.1016/S1473-3099(19)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tablante, N. L. , & Lane, V. M. (1989). Wild mice as potential reservoirs of Salmonella Dublin in a closed dairy herd. The Canadian Veterinary Journal, 30(7), 590–592. [PMC free article] [PubMed] [Google Scholar]
- Takeda, T. , Aoyama, M. , & Sugita, S. (2015). Seasonal variation in movement distances by jungle crows (Corvus macrorhychos) and incoming frequencies to use of livestock barns. Nihon Chikusan Gakkaiho, 86(2), 191–199. 10.2508/chikusan.86.191 [DOI] [Google Scholar]
- Tang, S. , Orsi, R. H. , Luo, H. , Ge, C. , Zhang, G. , Baker, R. C. , Stevenson, A. , & Wiedmann, M. (2019). Assessment and comparison of molecular subtyping and characterization methods for Salmonella . Frontiers in Microbiology, 10(JULY). 10.3389/fmicb.2019.01591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover, F. C. , Arbeit, R. D. , Goering, R. V. , Mickelsen, P. A. , Murray, B. E. , Persing, D. H. , & Swaminathan, B. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed‐field gel electrophoresis: Criteria for bacterial strain typing. Journal of Clinical Microbiology, 33(9), 2233–2239. 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. L. , Slawson, R. M. , & Taylor, W. D. (2013). Salmonella serotype diversity and seasonality in urban and rural streams. Journal of Applied Microbiology, 114(3), 907–922. 10.1111/jam.12079 [DOI] [PubMed] [Google Scholar]
- Tizard, I. (2004). Salmonellosis in wild birds. Seminars in Avian and Exotic Pet Medicine, 13(2), 50–66. 10.1053/j.saep.2004.01.008 [DOI] [Google Scholar]
- Torres, R. T. , Fernandes, J. , Carvalho, J. , Cunha, M. V. , Caetano, T. , Mendo, S. , Serrano, E. , & Fonseca, C. (2020). Wild boar as a reservoir of antimicrobial resistance. Science of the Total Environment, 717(November), 135001. 10.1016/j.scitotenv.2019.135001 [DOI] [PubMed] [Google Scholar]
- Tsukada, H. , Takeuchi, M. , Fukasawa, M. , & Shimizu, N. (2010). Depredation of concentrated feed by wild mammals at a stock farm in Japan. Mammal Study, 35(4), 281–287. 10.3106/041.035.0408 [DOI] [Google Scholar]
- Van Metre, D. C. , Tennant, B. C. , & Whitlock, R. H. (2008). Infectious diseases of the gastrointestinal tract. In Divers T. J. & Peek S. F., Rebhun's diseases of dairy cattle (2nd ed., pp. 200–294). St. Louis, MO: Saunders Elsevier. [Google Scholar]
- Vittecoq, M. , Godreuil, S. , Prugnolle, F. , Durand, P. , Brazier, L. , Renaud, N. , Arnal, A. , Aberkane, S. , Jean‐Pierre, H. , Gauthier‐Clerc, M. , Thomas, F. , & Renaud, F. (2016). REVIEW: Antimicrobial resistance in wildlife. Journal of Applied Ecology, 53(2), 519–529. 10.1111/1365-2664.12596 [DOI] [Google Scholar]
- Yamaguchi, E. , Kadohira, M. , Fujii, K. , Kobayashi, K. , & Takada, M. (2020). Utilization of municipality records for the early‐stage management of introduced raccoons in Japan. Management of Biological Invasions, 11(2), 306–324. 10.3391/mbi.2020.11.2.09 [DOI] [Google Scholar]
- Zeveloff, S. I. (2002). Raccoons: A natural history. Washington, DC., USA: Smithsonian Books. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Research data are not shared.


