Abstract
Human induced pluripotent stem cell-derived brain organoids have shown great potential for studies of human brain development and neurological disorders. However, quantifying the evolution of the electrical properties of brain organoids during development is currently limited by the measurement techniques, which cannot provide long-term stable three-dimensional (3D) bioelectrical interfaces with developing brain organoids. Here, we report a cyborg brain organoid platform, in which “tissue-like” stretchable mesh nanoelectronics are designed to match the mechanical properties of brain organoids and to be folded by the organogenetic process of progenitor or stem cells, distributing stretchable electrode arrays across the 3D organoids. The tissue-wide integrated stretchable electrode arrays show no interruption to brain organoid development, adapt to the volume and morphological changes during brain organoid organogenesis, and provide long-term stable electrical contacts with neurons within brain organoids during development. The seamless and non-invasive coupling of electrodes to neurons enables long-term stable, continuous recording and captures the emergence of single-cell action potentials from early-stage brain organoid development.
Keywords: Bioelectronics, brain organoids, neural interface, electrophysiology, nanoelectronics, stretchable electronics
Graphical Abstract

Building cyborg brain organoids through the integration of stretchable mesh nanoelectronics with human induced pluripotent stem cell-derived progenitors and neurons through organogenesis. The stretchable mesh nanoelectronics can adapt the morphogenesis of organoids and record organoid-wide electrophysiology at single-cell, millisecond resolution, chronically and non-invasively. Emergence and evolution of single-unit action potentials over the time course of brain organoid development were measured.
1. Introduction
The ability to record tissue-wide, millisecond-timescale single-cell electrophysiology over the time course of human brain development is important to understand the emergence of orchestrated neuronal activities[1] and elucidate the origin of neurodevelopmental diseases[2, 3]. This ability has not yet been achieved due to the inaccessibility of the human brain at early developmental stages. Recent breakthroughs in the development of human induced pluripotent stem cells (hiPSCs) have introduced techniques to grow brain organoids from in vitro cultured stem cells that can proliferate, differentiate, and self-assemble[4–6] into three-dimensional (3D) tissues, resembling the cellular architecture, diversity, and electrophysiology of the human brain at early stages[7, 8]. Brain organoids thus provide a reliable and easily accessible platform to study human brain development and neurodevelopmental diseases[9–12], bridging the gap between animal research and human clinical study.
However, long-term stable recording of single-cell electrophysiology in developing brain organoids is still a challenge. The recording technology not only needs to form minimally-invasive and long-term stable electrical interfaces with individual neurons 3D distributed across brain organoids but also needs to accommodate the rapid volume change occurring during the organoid organogenesis and cortical expansion. Optical imaging coupled with fluorescence dyes[13] or calcium indicators[14] has been used to visualize the neuron activities in 3D. They, however, are limited by temporal resolution, penetration depth, and long-term signal stability. Electrical measurement techniques such as 2D multielectrode arrays (MEA)[15, 16] and patch-clamp[17, 18] have been applied to measure the functional development of brain organoids, but they can only capture the activities from the bottom surface of brain organoids[1, 19, 20] or assay one cell at a time with cell membrane disruption. The recent development of 3D bioelectronics enables 3D interfaces with brain organoids[21–27]. However, they either only contact organoids at the surface by flexible electronics[21–23], where non-correlated and 3D distributed single-unit action potentials cannot be recorded, or penetrate organoids invasively by rigid probes[25], which cannot further accommodate volume and morphological changes of brain organoids during development. It has also been shown that organoids can grow around a suspended array of electrodes[26, 27], but the electrodes cannot deform to adapt to the morphological changes of the organoid. To date, it is still a challenge to non-invasively probe neuron activity at single-cell, single-spike spatiotemporal resolution across the 3D volume of brain organoids and over the time course of development. This constraint prevents further understanding of the functional development in brain organoids and standardizing culture conditions and protocols for brain organoid generation based on their electrical functions.
Recently, we developed a cyborg organoid platform by integrating “tissue-like” stretchable mesh nanoelectronics with 2D stem cell sheets. Leveraging the 2D-to-3D reconfiguration during organoid development, 2D stem cell sheets fold and embed stretchable mesh nanoelectronics with electrodes throughout the entire 3D organoid. The embedded electronics can then enable continuous electrical recording[24]. Here, we design stretchable mesh nanoelectronics, mimicking the mechanical and structural properties of brain organoids to build cyborg human brain organoids. Using the 3D embedded stretchable electrodes, we achieved reliable long-term electrical recording of the same hiPSC-derived neural tissue at single-cell, millisecond spatiotemporal resolution for 6 months, revealing the evolution of the tissue-wide single-cell electrophysiology over hiPSC-derived neuron development. Applying this technology to brain organoids at early developmental stages, we traced the gradually emerging single-cell action potentials and network activities.
2. Results
2.1. Design of stretchable mesh nanoelectronics for brain organoid integration
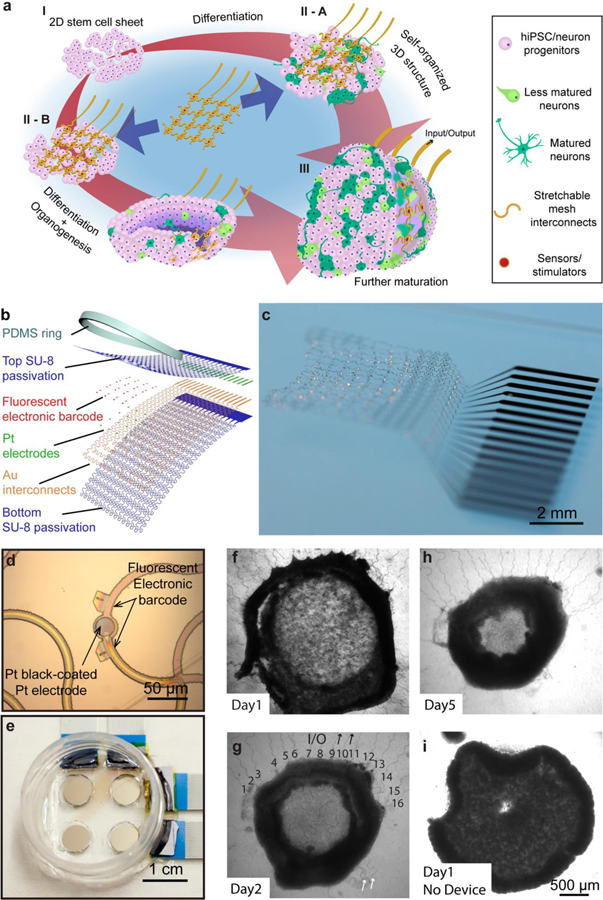
To obtain long-term stable 3D bioelectrical interfaces with developing brain organoids, we integrated the stretchable mesh nanoelectronics with hiPSC-derived neurons or hiPSCs, which can self-organize into 3D neural tissues and human brain organoids (Figure 1a). Briefly, for 3D hiPSC-derived neural tissue culture, the hiPSCs (Stage I) were first induced for neuronal differentiation. After confirming the spontaneous action potentials from the hiPSC-derived neurons by 2D microelectrode arrays, cells were dissociated, integrated with the stretchable mesh nanoelectronics, and induced to self-organize into 3D structures (Stage II-A). For human brain organoid culture, hiPSCs were cultured on the stretchable mesh nanoelectronics/Matrigel hybrid structure to form a 3D structure and then induced for neuronal differentiation (Stage II-B). The 2D-to-3D reconfiguration during organoid development folds the 2D stem cell plate/nanoelectronics hybrids into 3D structures with stretchable mesh electrodes seamlessly distributed across 3D brain organoids. Ultimately, the 3D embedded electrodes (Stage III) were connected with an amplification and data acquisition system to continuously monitor the electrical signals from neural progenitors and neurons.
Figure 1. Stretchable mesh nanoelectronics for brain organoid integration.
a, Schematics illustrate the stepwise integration of stretchable mesh nanoelectronics into 3D hiPSC-derived neural tissues through cell self-organization and brain organoids through organogenesis. (I) Human induced pluripotent stem cells (hiPSCs) were seeded with Matrigel. (II) Lamination of stretchable mesh nanoelectronics onto the two-dimensional (2D) cell sheet after neuronal differentiation into neural progenitors (II-A) or lamination of stretchable mesh nanoelectronics onto the hiPSCs before neuronal differentiation (II-B). (III) The 2D-to-3D self-organization folds the 2D cell sheet/nanoelectronics hybrid into a 3D structure. 3D embedded sensors are connected to external recording electronics to keep monitoring the electrophysiology of hiPSC-derived neurons and neural progenitors. b, Exploded view of the stretchable mesh nanoelectronics design consisting of (from top to bottom) an 800-nm-thick top SU-8 encapsulation layer, a 50-nm-thick platinum (Pt) electrode layer electroplated with Pt black, a 40-nm-thick gold (Au) interconnects layer, and an 800-nm-thick bottom SU-8 encapsulation layer. The serpentine layout of interconnects is designed to enable stretchability. A polydimethylsiloxane (PDMS) ring is bonded around the device as a chamber to define the size and initial cell number in the seeded hiPSC sheet. c, Optical photograph of stretchable mesh nanoelectronics released from the substrate and floating in the saline solution. d, Optical bright-field (BF) microscopic image of stretchable mesh nanoelectronics before released from the fabrication substrate shows a single Pt electrode coated with Pt black. e, Optical photograph of a 2×2 devices well, with a single culture chamber for 4 cyborg brain organoids cultured simultaneously. f-h, Optical phase images of hiPSC-derived neurons integrated with stretchable mesh nanoelectronics from Day 1 to Day 5 show that the 2D cell sheet with embedded stretchable mesh nanoelectronics self-folded into a 3D cyborg brain organoid. Black numbers and arrows indicate the input/output (I/O) stretchable connectors for the 16-channel electrode array (g). White arrows highlight the stretchable anchors used to keep the stretchable mesh nanoelectronics unfolded on the substrate, which were released after seeding with cells (g). i, Optical phase images of organoid without nanoelectronics integrated at Day 1 of culture as control showing minimal interruption from the integration with stretchable mesh nanoelectronics to the organogenesis of brain organoids.
Considering the significant mechanical and structural differences between cardiac and brain organoids, we have focused on the design of the unit block of stretchable mesh nanoelectronics that is structurally and mechanically compatible with neural tissues and the brain organoid organogenetic process. In addition, neuronal action potentials are much faster (2–3 ms) and smaller (10–100 μV) than cardiac signals (ca. 100 ms - 1 s and 100–1,000 μV), which makes high signal-to-noise ratio (SNR) recordings of brain organoids more challenging[24]. We further improved the interfacial impedance stability of electrodes and the dielectric performance of the interconnects in stretchable mesh nanoelectronics to enable the chronically stable neural recording in dynamic developing tissue. Finally, brain organoid culture protocol requires the adjustment of mesh nanoelectronics dimensions to control the initial number of stem cells. The intrinsic heterogeneity between brain organoids requires the careful control of culture conditions across samples for a longitudinal study.
Given all of these parameters, first, our biomimetic design exploits a serpentine layout with an overall filling ratio of less than 7% and an in-plane stretchability of up to 30% (Figure 1b–d, Supporting Information Figure S1–S3)[28]. This design allows the structure of nanoelectronics to be compressed and folded through the buckling of ribbons in the mesh network[29]. Also, these multiscale deformations can accommodate the compression, folding, and expansion during brain organoid organogenesis[30]. Second, brain organoids are softer than cardiac organoids24, given that brain tissues (elastic modulus of a few kPa)[31, 32] are softer than cardiac tissues (elastic modulus of a few tens of kPa)[33]. Therefore, we downscaled the width and serpentine pitch of the mesh nanoelectronics (ribbon width/thickness = 7.5/1.6 μm) by 25% and 50%, respectively. As a result, the effective bending stiffness is 6.7×10−16 N·m2 (or flexural rigidity of 0.090 N·m), which is >50 times lower than that of the previous design[24] and comparable to the mechanical properties of brain tissues[32]. Third, to control the initial cell numbers for brain organoids culture, a polydimethylsiloxane (PDMS) ring (thickness of 100–200 μm, diameter of 6.5 mm) was cast around the mesh nanoelectronics to define the initial region of the stem cell sheet, which ultimately controls the cell number and the size of the 3D organoid. Fourth, brain organoids take months to years to develop and mature; therefore, to enable a stable single-unit action potential electrical recording, we designed the electrode with a diameter of 25 μm, comparable to or even smaller than the electrodes used for the in vitro and in vivo single-unit action potential recording[22, 32]. We also used platinum (Pt) black to modify Pt electrodes instead of PEDOT:PSS due to its long-term stability and low impedance compared to other electrode modification methods[34] (Figure 1d, Supporting Information Figure S4). The electrochemical impedance of the Pt black-coated sensors (diameter of 25 μm) has an initial average impedance modulus of (1.40 ± 0.50) × 105 Ω (mean ± S.D., n = 16) at 1 kHz frequency, which only slightly increased to (3.00 ± 0.33) × 105 Ω after 180 days of incubation in 1x phosphate-buffered saline (PBS) at 37 °C. Importantly, to reduce the noise level, we used thicker dielectric layers (800 nm SU-8 2000.5) to encapsulate the serpentine interconnects, which ensure the noise is less than 15–20 μV between 100–6,000Hz for signals around 20–100 μV. Notably, only devices showing a uniform electrochemical impedance modulus below 4.00 × 105 Ω at 1 kHz frequency and no crosstalk between channels (see Methods) were used for brain organoid culture and recording. Last, to simultaneously culture multiple brain organoids under the same conditions, each reactor contains four independent 16-channel devices. Electrodes from each device can be individually connected to the voltage amplifier through flip chip-bonded anisotropic conductive film/flat flexible cables (Figure 1e). Notably, we used photolithography to define a pair of center-symmetric and unique binary fluorescence electronic barcodes (E-barcode) for each sensor by doping Rhodamine 6G (R6G) in SU-8 precursors[35]. The fluorescence E-barcode (Figure 1d) will be used to determine the 3D position of each sensor within the brain organoids by post hoc tissue clearing, staining, and confocal microscopic imaging after electrophysiological recording.
Stretchable mesh nanoelectronics were then released from the substrate with stretchable anchors maintaining their 2D structures. A layer of Matrigel was cured underneath the device to form a mesh nanoelectronics/Matrigel hybrid. Figure 1f–h shows that hiPSC-derived neurons were dissociated and cultured on the nanoelectronics/Matrigel hybrid to form a continuous cell/nanoelectronics/Matrigel sheet. From day 1 to 5 of assembly. Bright-field (BF) phase images (Figure 1f–h) showed that hiPSC-derived neurons self-organize into a neuroepithelium-like structure. After releasing the anchors that hold meshes on the substrate (Figure 1f–h), the cell sheet embedded with stretchable mesh nanoelectronics gradually folded into a 3D structure. The 16 metal lines embedded in the polymer mesh were still connected to the substrate after the 3D re-organization. Furthermore, the comparison to an organoid without device integrated at Day 1 of culture as control shows that the stretchable mesh nanoelectronics does not affect the size and shape of the 3D culture (Figure 1i). These results proved that the mechanical and structural properties of stretchable mesh nanoelectronics allow for effective embedding and integration into brain organoids through the organogenetic process.
2.2. Long-term electrical recording
We first integrated the stretchable mesh nanoelectronics with hiPSC-derived neurons to demonstrate the stable long-term electrical recording of neuronal activities. We differentiated hiPSCs into neurons on a 2D substrate until they acquired spontaneous electrical activities. Their 2D development and differentiation were assessed by the BF phase and fluorescence imaging (Supporting Information Figure S5a–d). After 4 months of differentiation, the 2D hiPSC-derived neurons were dissociated and seeded on a 2D mesh electrode array (of the same structural design as the stretchable mesh nanoelectronics) for electrophysiological recording. Spontaneous extracellular single-unit action potentials and bursts could be reliably detected and susceptible to glutamate receptor antagonists. Specifically, the application of 20 μM (2R)-amino-5-phosphonopentanoate (D-AP5) and 20 μM cyanquixaline (CNQX) could significantly reduce the number of spikes and bursts (Supporting Information Figure S6). The 3D brain organoids (Supporting Information Figure S5e) were also placed on a 2D mesh electrode array to compare their electrical activities with previous studies[1, 19, 20]. Spontaneous extracellular single-unit action potentials could be reliably detected from the bottom layer of brain organoids (Supporting Information Figure S7). Both experiments confirmed the spontaneous electrical activities from the hiPSC-derived neurons and brain organoids generated through our protocol as well as the ability to record single-unit action potential by the mesh nanoelectronics.
Next, the hiPSC-derived neurons were dissociated and seeded on a stretchable mesh nanoelectronics/Matrigel hybrid. The releasing of the anchors allowed the cell sheet to gradually form a 3D structure with electrodes fully embedded (Figure 2a, Supporting Information Figure S8). After one-month post-assembly, we can detect spontaneous local field potentials (LFPs) and single-unit action potentials from the tissue-wide embedded mesh electrodes. To confirm that signals were from neurons, the cyborg 3D hiPSC-derived neural tissues were first exposed to 30 mM potassium chloride (KCl) solution to induce neuronal membrane depolarizations. A statistically significant increase of the electrical activity (1,128 ± 1,894%, mean ± S.D., signal root mean square (RMS) amplitude, n = 12 channels, p<0.01, two-tail, paired t-test) could be recorded from 3D hiPSC-derived neural tissues (Figure 2b, Supporting Information Figure S9). Then, 3D hiPSC-derived neural tissues were exposed to glutamate receptor antagonists (20 μM CNQX and 20 μM D-AP5 solution). A statistically significant decrease (50.4 ± 18.6%, mean ± S.D., decrease in signal RMS amplitude, n = 12 channels, p<0.01, two-tail, paired t-test) of the electrical activity could be recorded (Figure 2c, Supporting Information Figure S9).
Figure 2. Long-term tracking neural activities by stretchable mesh nanoelectronics.
a, Schematic of the stepwise assembly of mesh nanoelectronics with hiPSC-derived neurons. b, Injection of KCl produces a significant increase in spiking rate of signals detected by multiple electrodes. c, Injection of CNQX and D-AP5 produces a significant decrease in spiking rate of signals detected by multiple electrodes (n = 8, bar plots show mean ± S.D.). * P<0.05, two-tailed, paired t-test. d, Raw voltage traces of a 16-channel device showing neural activities at month 5 of differentiation (i.e., month 1 of assembly). e, Zoom-in panels from the red dashed box (d). f, Single-spike waveforms (mean ± S.D.) extracted from the voltage traces filtered by 100 Hz-3000 band-pass filter, for each cluster detected by spike sorting. Vertical scale bars, 20μV. g, Full width at half minimum (FWHm) of depolarization of each neuron detected on hiPSC-derived neural tissues as a function of culture time (mean ± S.D., * P< 0.05 using one-way ANOVA with the month 5 group as control). h, Normalized phase plot of single-unit action potentials (inset) detected on the same channel at months 6 and 7 post-differentiation, showing an increase in the depolarization’s speed. i, Single-unit action potentials (top) and corresponding raster plots (bottom) detected from the same channel at 5, 6, and 10 months of culture.
Both burst firing and individual action potentials could be reliably detected from the tissue-embedded electrodes (Supporting Information Figure S10), which were consistent with previous results from 2D MEA[1], demonstrating the ability of tissue-embedded stretchable mesh nanoelectronics to capture the activities from the coordinated neural network and individual neurons. Notably, representative voltage traces showed temporal delays between signals from different channels, suggesting the ability of tissue-wide distributed electrodes to record the signals from different neurons in 3D hiPSC-derived neural tissues instead of recording the same neuron signals by multiple electrodes (Figure 2d, e). We carried out statistical analyses on the single-unit action potentials to study the evolution of neuronal activities over the time course of 3D hiPSC-derived neural tissue development. We filtered the signals by 100–3,000 Hz bandpass filter and applied spike sorting to extract single spikes for analysis. Multiple neurons can be detected from each channel (Figure 2f). We noticed that the total duration of single-unit action potentials from neural tissues was slower (2- to 3-ms duration) compared with the duration of action potentials detected by mesh nanoelectronics from the adult animal’s brain (1- to 2-ms duration)[36, 37], which may suggest the immature nature of neurons in the hiPSC-derived neural tissues. The proximity and high density of neurons surrounding the tissue-embedded sensors allowed us to observe that the field potentials emerge from collective neuronal activities. As a result, we can observe a strong correlation between field potential events and spiking bursts (Supporting Information Figure S11), which suggests that the 3D neural tissue-embedded electrodes could detect cross-frequency coupling and oscillatory waves reported by Trujillo et al.[1], in a 3D fashion.
To confirm the capability of mesh nanoelectronics to chronically record the functional development of 3D hiPSC-derived neural tissues at single-cell resolution, we conducted a 6-month electrical recording from the same hiPSC-derived neural tissues (n = 4) and performed statistical analysis of single-neuron signals. The long-term recording showed that the average full width at half minimum (FWHm) of the depolarization of action potential decreased from 0.57±0.23 ms in month 5 of differentiation (n = 27 single units, mean ± S.D.) to 0.37±0.16 ms in month 10 of differentiation (n = 11 single units, mean ± S.D.) (Figure 2g), while the one-way analysis of the variance (ANOVA) suggested that this change is not statistically significant. Similarly, we found that the spiking rates were not significantly changing over time (Supporting Information Figure S12). Using the phase-space[38] analysis to characterize the evolution of single-unit action potential waveform, the result suggests that the membrane depolarization rate has been increased throughout the development of 3D hiPSC-derived neural tissues (Figure 2h). In addition, the waveforms clustering from representative channels shows that the detected number of single-unit action potentials can increase during development (Figure 2i). The electrical measurement from the 3D hiPSC-derived neural tissue system proved the capability of tissue-embedded stretchable mesh nanoelectronics to continuously record 3D neural signals from developing neural tissues with minimal change in the noise level during the dynamic reconfiguration of the mesh nanoelectronics over the tissue development (Supporting Information Figure S13). The overall chronic stability of single neuron recordings further suggested a minimal interruption of tissue-wide neuronal activities by embedded stretchable mesh nanoelectronics during the 6-month 3D culture of hiPSC-derived neural tissues.
2.3. Tracing of electrophysiology during early brain development
We integrated stretchable mesh nanoelectronics with early-stage brain organoids, aiming to demonstrate the capability to capture the emergence of single-neuron action potentials. The stretchable mesh nanoelectronics was integrated with hiPSCs before neural differentiation (Figure 1a) to build the cyborg brain organoids. BF images showed that the embedded mesh nanoelectronics do not affect the organoid morphologies (Supporting Information Figure S14). To investigate the effects of the embedded mesh nanoelectronics on cell differentiation, the cyborg brain organoids at different differentiation stages were fixed, sectioned, and immunostained for stage-specific protein marker expressions and compared to control brain organoids without mesh nanoelectronics embedding (Supporting Information Figure S15a). From day 40 to 90 of differentiation in brain organoids, the density of neurons, cortical progenitors, and neural progenitors were consistent across different samples, as shown by the consistent expression levels of Hexaribonucleotide Binding Protein-3 (NeuN) and beta Tubulin 3 (Tuj1), Paired-box 6 (Pax6), and Nestin, respectively. Cortical neuron density statistically significantly increased during development, as shown by the increase of cells expressing T-box brain protein 1 (TBR1), which suggests the continuous differentiation and development of neurons over time. Importantly, statistical results showed no significant difference in different types of cells between cyborg and control organoids, confirming the minimal interruptions from the implanted mesh nanoelectronics to the neural differentiation in brain organoids (Supporting Information Figure S15b).
To further investigate the effects of the embedded mesh nanoelectronics on cell differentiation, cell-type composition, and gene expression during the brain organoid development, we performed single-cell RNA sequencing (scRNA-seq) on 10,846 single cells from both cyborg (6,420 cells) and control brain organoids (4,426 cells) at 7-month post differentiation. After filtering the high-quality single cells, 9220 cells were retained for principal component analysis (PCA) and unsupervised clustering analysis by Seurat V4.0.5 R packages[39] (see Methods). Uniform Manifold Approximation and Projection (UAMP) was then used to visualize the gene expression for each cluster (Figure 3a, 3b). We compared the differentially expressed genes for each cluster with previous human brain organoids and identified 9 groups of cells (Figure 3b–d)[8,40–44 ], including the neuron group expressing PAX6 and RELN, the astroglia group expressing AQP4 and GFAP, and the mesenchymal group expressing COL1A1 and COL3A1. We also identified the cycling progenitors for the neuron, astroglia, and mesenchymal group that express TOP2A and MKI67 in addition to their cell type markers. In addition, we also observed other populations such as the choroid plexus expressing TTR and HTR2C, the early neuron group expressing STMN2, and the apoptotic cell populations expressing ATF3 and DDIT3. The results demonstrate that the cyborg and control brain organoids show highly consistent gene expression patterns across all different cell types from the UMAP plot (Figure 3a–b and Supporting Information Figure S16) and contain the same cell-type composition (Figure 3d).
Figure 3. Single-cell RNA sequencing of cyborg and control human brain organoids.
a, UMAP visualization of single-cell RNA expression in cyborg and control organoids. Cells are colored by cell identities. (n = 9,920 cells. 5,240 and 3,980 cells from cyborg and control brain organoids, respectively). b, UMAP visualization of single-cell RNA expression in cyborg and control brain organoids. Cells are colored by the cell type assignment. c, Violin plots of marker genes expressions across cell types in brain organoids. Colors correspond to cell types, and the colored area indicates the density distribution of each gene. d, Heatmap showing the row z-scored expression of the markers for each cell type from cyborg and control brain organoids (left). Cell-type compositions in cyborg and control organoids (right). Colors correspond to cell types. e, Violin plot of neuron marker gene expression in cyborg and control brain organoids. Colors correspond to their identities. (two-tailed, unpaired t-test). f, UMAP visualization of single-cell RNA expression from neuron populations in cyborg (up) and control brain organoid (down). Cells are colored by the pseudotime value obtained by Monocle3. g, Neuron marker gene expressions along pseudotime from (f) in cyborg (up) and control brain organoid (down). Colors correspond to cell identities. Ellipse draws a 95% confidence level for a multivariate t-distribution. h, Distribution plot (up) and boxplot (down) of pseudotime from neurons in cyborg and control brain organoids (two-tailed, unpaired t-test). Colors correspond to cell identities.
We then compared the marker gene expression in neuron groups between cyborg and control organoids. First, the violin plots show highly consistent expression profiles between cyborg and control brain organoids with no significant difference (Figure 3e). Second, we computationally inferred the pseudotime trajectory by Monocle 3[45] (see Methods) for neuron lineage analysis, which includes neurons and cycling neurons, to further analyze the temporal change in gene expression pattern. The cycling neuron progenitors were specified as the starting state when inferring the pseudotime[8]. The inferred pseudotime in neuron lineage shows a highly consistent pattern between cyborg and control brain organoids in the UMAP plot (Figure 3f). The distributions of pseudotime show no significant difference (Figure 3h). Third, we examined the expression level of neuronal marker gene RELN and PAX6 along inferred pseudotime (Figure 3g). Both RELN and PAX6 show an increased expression level along pseudotime with a consistent expression between cyborg and control brain organoids, demonstrating the minimal interruptions from the tissue-wide embedded stretchable mesh nanoelectronics to the neural differentiation in brain organoids.
In addition to the neuron lineage, we also compared the marker gene expression in the astroglia lineage between cyborg and control organoids. First, the violin plots show highly consistent expression profiles between cyborg and control brain organoids with no significant difference (Supporting Information Figure S17a). Second, the computationally inferred pseudotime in astroglia lineage (including astroglia and cycling astroglia) shows highly consistent patterns between cyborg and control brain organoids in the UMAP plot (Supporting Information Figure S17b). The distributions of pseudotime show no significant difference (Supporting Information Figure S17d). Third, we examined the expression level of astroglia marker genes AQP4 and GFAP along the inferred pseudotime (Supporting Information Figure S17c). Both AQP4 and GFAP show increased expression levels along pseudotime with consistent expressions between cyborg and control brain organoids. Taken together, the scRNA-seq data confirm the minimal interruptions from the tissue-wide embedded stretchable mesh nanoelectronics to cell development in the different cell lineage.
We then tracked neural activities from cyborg brain organoids (n=7 organoids) after one-month differentiation and organogenesis, when the 3D organoids were formed (Figure 4a). A gradually increased activity can be detected from brain organoids over the first 3 months post-differentiation (Figure 4b). To analyze the single-neuron signals from early developmental stages, we filtered voltage traces in the range of 100–3,000 Hz to retain slow spikes detected at months 1 and 2 post-differentiation (Supporting Information Figure S18). The results showed that both signal amplitude and the number of single-unit action potentials detected per channel increased over time (Figure 4b). Moreover, spectral analysis from representative channels showed an increase in power between 0 and 1.5 kHz during the first three months of differentiation (Figure 4c, d). Statistical analysis of all the cyborg brain organoids showed a significant increase in the power at 300 Hz from month 1 to 3 post-differentiation (n=16 channels, p<0.01, one-way ANOVA) (Figure 4e), suggesting gradually increased neuronal activities and functional development of brain organoids, which agrees with the protein marker staining results (Supporting Information Figure S15) and gene expression analysis (Figure 3).
Figure 4. Electrical recording of human brain organoids during early development.
a, Schematics of the stepwise assembly of mesh nanoelectronics with hiPSCs for cyborg brain organoids. b, Raw voltage traces at 1, 2, and 3 months after cortical differentiation c, Spectrograms at 1, 2, and 3 months of differentiation for channel 3 showing a strong increase in power between 0 to 1 kHz after 3 months of differentiation. * Denotes voltage artifacts. d, Corresponding power spectrums to (c). e, Signal power at 300 Hz for electrodes with detected neural activity. f, Normalized phase plot of single-unit action potentials and its corresponding waveforms (inset) detected from the same channel at 2 and 3 months of differentiation, showing an increase in the rate of depolarization. g-h, FWHm of depolarization (g), and spike count per neurons per 2-min recording (h) at 1, 2, and 3 months of differentiation. i, Spiking rate per neuron detected as a function of the inverse of the FWHm of depolarization, showing that the single-cell action potential spikes from neurons evolve towards shorter spike width and higher spiking rate over the time course of brain organoid development. In panels (e, g), value=mean ± S.D., ** P<0.01, one-way ANOVA with “Month 1” group as control. j, Injection of bicuculline (BCC) produces a significant increase in the spiking rate of signals detected by multiple electrodes. k, Injection of CNQX and D-AP5 produces a significant decrease in spiking rate of signals detected by multiple electrodes (n = 4 electrodes including data from p = 2 different cyborg brain organoids, bar plots show mean ± S.D.). * P<0.05, two-tailed, paired t-test.
Next, we analyzed the spike waveform of single-unit action potentials. Phase-space analysis of individual spikes detected from the same electrode showed an increase in depolarization rate from months 2 and 3 (Figure 4f). Statistical analysis of FWHm and spiking rate of each single-unit spike (n = 7 organoids) showed the narrowing of spikes and increase in spiking rate over time (Figure 4g, h). FWHm decreased significantly from 1.05±0.63 ms in month 1 (n = 9 single units, mean ± S.D.) to 0.38±0.22 ms in month 3 (n = 15 single units, mean ± S.D.). Plotting spiking rate per neuron as a function of FWHm−1 further revealed the positive correlation between narrowing of single-unit action potentials and increasing of spike rate over the time course of development (Figure 4i). While the increase in mean firing rate and signal power has been previously reported[1, 20, 22], the narrowing of spikes’ depolarization in hiPSC-derived brain organoids is reported for the first time. This result also agrees with previous findings for hiPSC-induced 2D neuron cultures[46].
To confirm that spiking activity arises from synaptic transmissions and diverse types of neurons throughout development, we applied GABAergic (bicuculline) and glutamatergic (CNQX and D-AP5) receptor antagonists to block inhibitory and excitatory synaptic transmission. Results showed that application of 10 μM bicuculline (BCC) can introduce a statistically significant increase of 2,870 ± 2,575% (mean ± S.D.) in spontaneous spiking rate (n=4 organoids, p<0.05, two-tailed, paired t-test), while application of 20 μM CNQX and 20 μM D-AP5 can introduce a statistically significant decrease of 91 ± 16% (mean ± S.D.) in spontaneous spiking rate (n = 4 organoids, p<0.05, two-tailed, paired t-test) (Figure 4j, 4k, Supporting Information Figure S19). These results suggest that both inhibitory and excitatory connections have been built in the brain organoids after 3-month neuronal differentiation.
We applied the intact organoid clearing[47, 48] and imaging to confirm the seamless integration of the stretchable mesh electrodes (Supporting Information Figure S20) with cyborg brain organoids. As the cyborg brain organoids were cultured on Matrigel substrate instead of suspended in the culture medium, the brain organoids showed relative flat morphology. After removal of the phospholipids of cell membranes during tissue clearing, the organoids become flattered due to the change of mechanical properties. Each electrode’s position in 3D and its position relative to the surrounding neurons, however, could still be identified by identifying the fluorescent E-barcodes (Figure 5a). In cyborg organoids 3-month post-differentiation, we recorded LFP, non-correlated individual action potentials, and spontaneous bursting activity (Figure 4b, Supporting Information Figure S21). We combined the spike sorting analysis with the fluorescent imaging, generating the 3D map of single-unit action potential recorded from sensors distributed across the organoid (Figure 5b, c). We applied a 4–8Hz bandpass filter to extract theta-wave, which showed a clear temporal delay among channels distributed across the 3D organoids (Figure 5d), illustrating the global field potential propagation across the organoid. Finally, statistical analysis showed that the FWHm of spikes’ depolarization of cyborg brain organoids at 1-month post-differentiation (1.05±0.63 ms, n = 6 organoids, mean ± S.D.) is significantly longer than other samples, while the FWHm of brain organoids at 3 months post-differentiation (0.38±0.22 ms, n = 6 organoids, mean ± S.D.) is not statistically different from the FWHm of 3D hiPSC-derived neural tissues at 5 months post-differentiation (0.57±0.23 ms, n = 5 3D hiPSC-derived neural tissues, mean ± S.D. two-tailed, unpaired, t-test) (Figure 5e). These results suggest that the cyborg brain organoid platform can be used to quantify the electrophysiological evolution of neurons during the development phase and detect variations across samples prepared by different protocols.
Figure 5. Electrophysiology of human brain organoids during the early developmental stage.
a, 3D views of reconstructed fluorescence images of tissue cleared, immunostained cyborg brain organoids at month 3 of differentiation. Red, green, and blue colors correspond to Device, Tuj 1, and 4′,6-diamidino-2-phenylindole (DAPI), respectively. White arrows highlight the position of the representative sensors. Channel number was read out through the fluorescence electronic barcode identification. (b, c) 3D positions of the 16 sensors in the cyborg brain organoid from (a) in two different planes. Average waveforms (± S.D.) detected at Month 3 of integration are indicated for each sensor. Vertical scale bars, 25 μV. d, Theta oscillations (4–8 Hz band) measured in the cyborg brain organoid shown in panel (a), at Month 3 of integration. The vertical lines show a clear dephasing between the different electrodes. e, Comparison of the full width at half minimum (FWHm) of neuron’s depolarization at months 1 and 3 of early-stage (cyborg brain organoids, n=6) and month 5 (cyborg hiPSC-derived neural tissues, n=5) of long-term electrophysiological recordings (mean ± S.D., ** P<0.01 and **** P<0.0001, one-way ANOVA with the group “cyborg brain organoids, month 1” as control).
3. Conclusion
We have demonstrated a cyborg brain organoid platform that can stably record 3D brain organoid-wide, millisecond-timescale, and single-neuron electrophysiology over the time course of brain organoid development. Long-term stable electrophysiological measurements of neural activity throughout the development of brain organoids reveal the increased number of neurons that can be detected from electrodes and millisecond-timescale evolution of the single-unit action potential waveform over 3D development. In addition, electrophysiological measurements during early brain organoid development reveal that single-unit action potentials can be detected from brain organoids after 3-month neuronal differentiation. The increase of action potential amplitude and firing rate and narrowing of action potential duration suggest changes in neural network connectivity and cellular ion channel expression level. We envision that further integration of cyborg brain organoids with organoid-wide connectomics and in situ sequencing will further test this hypothesis[35, 49].
Cyborg brain organoid technology can potentially become a useful tool for quantifying the functional development of brain organoids and standardizing the culture conditions across different types of protocols by tracing organoid-wide tissue and single-cell electrical activity during the entire organoid development. It will also be useful for studies of developmental neuroscience and drug screening. As the stretchable mesh nanoelectronics are fabricated by the standard lithographic process, this method is scalable by integrating high-density sensors and stimulators through the integration of multiplexing integrated circuits[50–52]. This high-density stretchable electrode array could address any potential concerns related to the single-unit action potential identification through spike-sorting algorithm by recording the same neurons simultaneously with multiple electrodes [53, 54]. Further integration of multifunctional sensors and stimulators will offer the potential of combined multimodal interrogation (e.g., mechanical and chemical) and intervention (e.g., optogenetics) capabilities to the cyborg brain organoids platform for human brain developmental studies.
4. Methods
4.1. Device fabrication
Fabrication of the ultra-thin, stretchable mesh nanoelectronics made of SU-8 negative photoresist was based on methods described previously[24]. The key steps (Supporting Information Figure S1) included: (1) Cleaning a glass wafer (500- μ m thickness) with acetone, isopropyl alcohol, and water. (2) Depositing 100-nm-thick nickel (Ni) using a thermal evaporator (Sharon) as a sacrificial layer. (3) Spin-coating SU-8 precursor (SU-8 2000.5, MicroChem, 800-nm or 400-nm thickness for, respectively, a spinning at 1000 or 3000 rpm), followed by pre-baked at (65 °C, 95 °C) for 2 min each, exposed to 365 nm ultra-violet (UV) for 200 mJ/cm2, post-baked at (65 °C, 95 °C) for 2 min each, developed using SU-8 developer (MicroChem) for 60 s, and baked at 180 °C for 40 min to define mesh SU-8 patterns (800-nm or 400-nm thickness for, respectively, spinning at 1,000 or 3,000 rpm) for bottom encapsulation. To define the fluorescent electronic barcode, 0.005 wt% of Rhodamine 6G powder (Sigma-Aldrich) was added into the SU-8 precursor. (4) Spin-coating LOR3A photoresist (MicroChem) at 4,000 rpm, followed by pre-baking at 180 °C for 5 min; spin-coating S1805 photoresist (MicroChem) at 4000 rpm, followed by pre-backing at 115 °C for 1 min; exposed to 405 nm UV for 40 mJ/cm2, and developed using CD-26 developer (Micropost) for 70 s to define interconnects patterns. (5) Depositing 5/40/5-nm-thick chromium/gold/chromium (Cr/Au/Cr) by electron-beam evaporator (Denton), followed by a standard lift-off procedure in remover PG (MicroChem) overnight to define the Au interconnects. (6) Repeating Step (4) to define sensors tip patterns in LOR3A/S1805 bilayer photoresists; (7) Depositing 5/50-nm-thick chromium/platinum (Cr/Pt) by electron-beam evaporator (Denton), followed by a standard lift-off procedure in remover PG (MicroChem) overnight to define the Pt electrodes. (8) Repeating Step (3) for top SU-8 encapsulation and SU-8 barcodes (containing R6G at a concentration of 50 μg/mL). (9) Electroplating Pt black on the Pt electrode using the recipe described below. (10) Soldering a 16-channel flexible flat cable (Molex) onto the input/output pads using a flip-chip bonder (Finetech Fineplacer). (11) A 500-μm-thick PDMS ring was attached around each device after 2-min oxygen plasma (Anatech 106 oxygen plasma barrel asher) treatment at 50W. (12) Gluing a chamber onto the substrate wafer to completely enclose a 2×2 mesh device array using a bio-compatible adhesive (Kwik-Sil, WPI). (13) Treating the surface of the device with light oxygen plasma (Anatech 106 oxygen plasma barrel asher), followed by adding 3 mL of Ni etchant (type TFB, Transene) into the chamber for 2 to 4 hours to completely release the mesh electronics from the substrate wafer. The device was then ready for subsequent sterilization steps before cell culture.
4.2. Materials preparation
We used an SP-150 potentiostat from Bio-logic© alongside its commercial software EC-lab in voltage or current control for electrodeposition. Electrodes from devices were connected to the working electrode. The counter electrode was a platinum wire, also serving as a voltage reference, which was immersed in the precursor solution. For Pt black electrodeposition, the precursor (0.8 wt% Chloroplatinic acid solution) solution was applied by the current at 1 mA/cm2 for 5–10 minutes.
4.3. Device impedance characterization
A three-electrodes setup was used to measure the electrochemical impedance spectrum of the electrodes from each device. Platinum wire (300 μm in diameter, 1.5 cm in length immersed) and a standard silver/silver chloride electrode were used as counter electrode and reference electrode, respectively. The device was immersed in a 1xPBS solution during measurement. The SP-150 potentiostat (Bio-logic) along with its commercial software EC-lab was used to perform the measurements. For each measurement, at least three frequency sweeps were measured from 1 MHz down to 1Hz to obtain statistical results. A sinusoidal voltage of 100 mV peak-to-peak was applied. For each data point, the response to 10 consecutive sinusoids (spaced out by 10% of the period duration) was accumulated and averaged. Optical images of the measurement setup can be found in our previous work[24]. Crosstalk between channels was evaluated at 1 kHz using a Blackrock CerePlex Direct voltage amplifier. All channels of all devices implanted were tested for electrochemical impedance and crosstalk at 1 kHz before implantation.
4.4. Electrophysiological recordings
A Blackrock CerePlex Direct voltage amplifier along with a 32 channels Blackrock μ digital headstage connected to the device was used to record electrical activity (5 min long recordings) from organoids. The headstage-to-device connector (16 channels) was homemade. The organoid culture medium was grounded to the earth and a reference electrode was also inserted in the media, far from the device (distance above 1 cm). Platinum wires were used as ground and reference electrodes. A sampling rate of 30000 samples per second was used. Band-pass filters (Butterworth, 4th order) were applied depending on the analysis performed. MATLAB codes provided by Blackrock were used to convert raw data files into an accessible format. Data were then transferred to Graphpad Prism for post-processing.
4.5. Data Analysis
We developed our own MATLAB code to analyze neural signals (code is available at https://github.com/CyBrainOrg). The threshold for spikes detection was set a −5*standard deviation of the filtered (300 – 6000 Hz or 100 – 6000 Hz bandpass) time series and principal component analysis was used for dimension reduction. MATLAB’s “kmeans” function was used to cluster the extracted waveforms and exclude noise artifacts. Only clusters with more than 25 waveforms were kept for analysis. Phase-space analysis. Discrete derivatives of average waveforms were calculated. To exclude amplitude variations from the analysis, the average waveform and derivative of the average waveform were normalized by their minimum value.
4.6. Organoid culture
HiPSC line hiPSCs-(IMR90)-1 were obtained from WiCell Research Institute (Madison, WI, USA) and cultured in a 6-well plate coated with Matrigel (Corning) in Essential 8 medium (Life Technologies). Authentication and test for the free of mycoplasma were performed by WiCell Research Institute. Neuron differentiation was based on methods described previously[7] with minor modification. Briefly, hiPSCs at 2D or 3D hiPSC cyborg organoids were induced for cortical neuron differentiation for 11 days in the induction medium containing DMEM/F12 (50%) and Neurobasal (50%) medium. From Day0 to Day11, 1% N-2 supplement (Life Technologies), 2% B27 (Life Technologies), SB431542 (10 μM, Selleckchem), and LDN193189 (100 nM, Selleckchem) were applied. The cells were maintained in Neurobasal medium including BDNF (20 ng mL−1, PeproTech), GDNF (10 ng mL−1, PeproTech), L-ascorbic acid (200 μM, Sigma), dibutyryl-cAMP (0.5 mM, Santa Cruz Biotechnology), and 2.5 μM DAPT (Selleckchem) after Day11 of differentiation. For 2D neurons dissociating and replating, the hiPSC-derived neurons were treated with 5 μM Rock inhibitor at 37 °C, 5% CO2 for at least 1 hour. Then the 2D neurons were treated with 0.05% Trypsin for 5–10 minutes at 37 °C and dissociated into single cells. The non-released microelectrode array was coated with Poly-D-lysine hydrobromide (Sigma) and Matrigel solution (Corning) overnight, respectively, before plating the cells. No medium change on the first three-day after cell seeding, then the medium changes half every other day. 5 μM Rock inhibitor was added in the medium for the first three-day culture.
4.7. Cyborg organoids integration
Briefly, the released mesh nanoelectronics was rinsed with DI water and then immersed in 70% ethanol at room temperature for 15 minutes to sterilize. The device was sequentially coated with Poly-D-lysine hydrobromide (Sigma) and Matrigel solution (Corning). Finally, about 20 μL liquid Matrigel (10 mg/mL) was added to each well in the cell culture chamber from the device-free side on ice. The cell culture chamber was incubated at 37 °C for at least 30 minutes to cure the Matrigel layer. hiPSCs or hiPSC-derived neurons (1×106 cells) were suspended in a mixture of E8 or neural maturation medium, and then transferred onto the cured Matrigel in each well of the cell culture chamber and cultured at 37 °C, 5% CO2. For the cyborg brain tissues culture, there was no medium change on the first three days after cell seeding. 5 μM Rock inhibitor was added in the medium for the first three-day culture. Then the medium changed half every other day.
4.8. Drug test
The brain organoids were recorded in the neural maturation medium. For drug test[1], 30 mM KCl, 20 μM CNQX, 20 μM D-AP5, 10 μM bicuculline, and 1 μM TTX were applied. In the measurement, baseline recording was performed before and after the addition of chemicals. The sample was rinsed three times with DPBS every time after the drug test before adding a fresh medium. 5 minutes long recordings were performed to obtain average values of RMS amplitude and spiking rates.
4.9. Imaging of organoids
For whole organoids imaging, procedures were adapted from tissue clearing techniques CLARITY[47] and passive clarity technique (PACT)[48]; the organoids were fixed with 4% PFA at 4 °C overnight and incubated with hydrogel solution (0.25% (w/v) VA-044 and 4% (w/v) acrylamide in PBS) at 4 °C for 24 hours. The samples were placed in an X-CLARITY hydrogel polymerization device for 3–4 hours at 37 °C with −90 kPa vacuum, followed by a wash in PBS overnight before electrophoretic lipid extraction for 24 hours in the X-CLARITY electrophoretic tissue clearing (ETC) chamber. Then immunostaining was performed by staining the primary antibodies, Tuj1 (BioLegend Cat# 801201) or TBR1(Abcam Cat# ab31940), for 3–5 days and the secondary antibodies for 2–4 days, respectively. The samples were submerged in optical clearing solution overnight and embedded in 2% agarose gel before imaging using Leica TCS SP8 confocal microscopy. For the characterization of cyborg brain organoids, the organoids at day 40 and month 3 were fixed with 4% PFA at 4 °C overnight and immersed in 30% sucrose for at least 12 hours. Then the samples were embedded in optimal cutting temperature (OCT) compound and cryostat section of 30-μm-thick slices. Brain organoids without device integration were used as control. For staining, the first antibodies, NeuN (Abcam Cat# Ab177487), Tuj1, Pax6 (BioLegend Cat# 901301), Nestin (BioLegend Cat# 809801), and TBR1, were incubated at 4 °C overnight and the secondary antibodies were stained at room temperature (RT) for 3–4 hours. For 2D, the cells were fixed with 4% paraformaldehyde (PFA) at RT for 15 minutes. Cells were incubated with primary antibodies at 4°C overnight and the secondary antibodies were stained at RT for 1–2 hours. Finally, 4’,6-diamidino phenylindole 8 (DAPI) were stained for 10 minutes. All samples were imaged by Leica TCS SP8 confocal microscopy. Imaging was analyzed by Leica Application Suite X (LAS X) and Fiji. Fluorescence intensity was calculated by Fiji. Data analysis and statistical tests were performed by Graphpad Prism.
4.10. scRNA-seq of brain organoids
Cyborg brain organoid and control brain organoid were dissociated into single cells as described previously[8]. The single cells were resuspended in 1X DPBS (without Ca2+ and Mg2+) containing 0.04% BSA (Sigma) at a concentration of 1,000 cells/μl. The library preparation and sequencing were performed at the Bauer Sequencing Core facility, Harvard University.
4.11. scRNA-seq data analysis
The Cell Ranger 6.1.1 pipeline (10x Genomics) was used to perform read alignments to the reference human genome GRCh38. Default parameters were used to align reads and to count UMI to generate gene-by-cell count matrices. We then used Seurat R package V4.0.5 to perform downstream analysis[39]. Cells were then filtered to retain only higher-quality cells (mitochondrial reads < 15%, genes detected > 2000). For both cyborg and control brain organoid samples, the UMI counts were normalized with total expression, followed by multiplying a scaling factor of the median number of UMI counts and log-transformed. Seurat’s default method was used for finding most 2,000 variable genes. We then integrated cyborg and control organoid data using SelectIntegrationFeatures, FindIntegrationAnchors, and IntegrateData from Seurat. The integrated data were then scaled with the ScaleData function followed by PCA. 15 PCs were chosen to construct k-nearest-neighbors graph for Louvain clustering with a resolution of 0.5. UMAP visualization was performed with RunUMAP in Seurat. The differentially expressed genes were identified with FindAllMarkers in Seurat using Wilconxon Rank Sum test. To perform cell type annotations, genes with a minimum expression in 25% of the cells and a minimum 0.25 log-fold change were taken into consideration. The marker genes were then compared with previous publications for identifying cell types for each cluster[8,40–44,55,56].
Pseudotime analysis was performed for each different lineage (including neuron and astroglia) using the Monocle package 3[45] with default parameters. The CellDataSet object was converted using as.cell_data_set from SeuratWrappers. When ordering the cells along this trajectory, cells previously assigned to the cycling progenitors (cycling neuron, cycling astroglia, and cycling mesenchymal) were specified as the starting state[8].
Supplementary Material
Acknowledgements
We acknowledge the discussion and assistance from all Liu Group members. We acknowledge the support from the NIH/NIMH 1RF1MH123948; NSF through the Harvard University Materials Research Science and Engineering Center Grant #DMR-2011754; Harvard University Center for Nanoscale Systems supported by the NSF; Aramont Fund for Emerging Science Research; and the William F. Milton Fund; We thank The Bauer Core Facility at Harvard University for help with scRNA-seq library preparation and sequencing.
Footnotes
Conflict of interests
The authors declare no financial or commercial conflicts of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.
References
- [1].Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, Domissy A, Vandenberghe M, Devor A, Yeo GW, Voytek B, Muotri AR, Cell Stem Cell 2019, 25, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McPartland JC, Curr. Opin. Neurol. 2016, 29, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jeste SS, Frohlich J, Loo SK, Curr. Opin. Neurol. 2015, 28, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kelava I, Lancaster MA, Dev. Biol. 2016, 420, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Koo B, Choi B, Park H, Yoon KJ, Mol. Cells 2019, 42, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rossi G, Manfrin A, Lutolf MP, Nat. Rev. Genet. 2018, 19, 671. [DOI] [PubMed] [Google Scholar]
- [7].Yoon S-J, Elahi LS, Pașca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, Huguenard JR, Geschwind DH, Pașca SP, Nat. Methods 2019, 16, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P, Nature 2019, 570, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lancaster MA, Knoblich JA, Science 2014, 345, 1247125. [DOI] [PubMed] [Google Scholar]
- [10].Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA, Nature 2013, 501, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Di Lullo E, Kriegstein AR, Nat. Rev. Neurosci. 2017, 18, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tambalo M, Lodato S, Brain Res. 2020, 1746, 147028. [DOI] [PubMed] [Google Scholar]
- [13].Puppo F, Sadegh S, Trujillo CA, Thunemann M, Campbell EP, Vandenberghe M, Shan X, Akkouh I, Miller E, Bloodgood BL, Front. Cell. Neurosci. 2021, 15, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nguyen JP, Shipley FB, Linder AN, Plummer GS, Liu M, Setru SU, Shaevitz JW, Leifer AM, Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thomas C Jr, Springer P, Loeb G, Berwald-Netter Y, Okun L, Exp. Cell Res. 1972, 74, 61. [DOI] [PubMed] [Google Scholar]
- [16].James CD, Spence AJH, Dowell-Mesfin NM, Hussain RJ, Smith KL, Craighead HG, Isaacson MS, Shain W, Turner IEEE JN. Trans. Biomed. Eng. 2004, 51, 1640. [DOI] [PubMed] [Google Scholar]
- [17].Hamill OP, Marty A, Neher E, Sakmann B, Sigworth F, Pflügers Archiv 1981, 391, 85. [DOI] [PubMed] [Google Scholar]
- [18].Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, Sandberg R, Tolias AS, Nat. Biotechnol. 2016, 34, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shim C, Jo Y, Cha HK, Kim MK, Kim H, Kook G, Kim K, Son GH, Lee HJ, “Highly Stretchable Microelectrode Array for Free-form 3D Neuronal Tissue”, presented at 2020 IEEE 33rd International Conference on Micro Electro Mechanical Systems (MEMS), 18–22 Jan. 2020, 2020. [Google Scholar]
- [20].Fair SR, Julian D, Hartlaub AM, Pusuluri ST, Malik G, Summerfied TL, Zhao G, Hester AB, Ackerman W. E. t., Hollingsworth EW, Ali M, McElroy CA, Buhimschi IA, Imitola J, Maitre NL, Bedrosian TA, Hester ME, Stem Cell Rep. 2020, 15, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kalmykov A, Reddy JW, Bedoyan E, Wang Y, Garg R, Rastogi SK, Cohen-Karni D, Chamanzar M, Cohen-Karni T, J. Neural Eng 2020, 18, 055005. [DOI] [PubMed] [Google Scholar]
- [22].Park Y, Franz CK, Ryu H, Luan H, Cotton KY, Kim JU, Chung TS, Zhao S, Vazquez-Guardado A, Yang DS, Li K, Avila R, Phillips JK, Quezada MJ, Jang H, Kwak SS, Won SM, Kwon K, Jeong H, Bandodkar AJ, Han M, Zhao H, Osher GR, Wang H, Lee K, Zhang Y, Huang Y, Finan JD, Rogers JA, Sci. Adv. 2021, 7, eabf9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ryu H, Park Y, Luan H, Dalgin G, Jeffris K, Yoon HJ, Chung TS, Kim JU, Kwak SS, Lee G, Adv. Mater. 2021, 33, 2100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Q, Nan K, Le Floch P, Lin Z, Sheng H, Blum TS, Liu J, Nano Lett. 2019, 19, 5781. [DOI] [PubMed] [Google Scholar]
- [25].Sharf T, van der Molen T, Guzman E, Glasauer SM, Luna G, Cheng Z, Audouard M, Ranasinghe KG, Kudo K, Nagarajan S, Available at SSRN 3797268 2021.
- [26].McDonald M, Sebinger D, Brauns L, Gonzalez-Cano L, Menuchin-Lasowski Y, Mierzejewski M, Psathaki O-E, Stumpf A, Wickham J, Rauen T, Schöler H, Jones PD, (Preprint) bioRxiv:2020.09. 02.279125,v2, Submitted Jan 2021. [DOI] [PubMed]
- [27].Soscia DA, Lam D, Tooker AC, Enright HA, Triplett M, Karande P, Peters SKG, Sales AP, Wheeler EK, Fischer NO, Lab Chip 2020, 20, 901. [DOI] [PubMed] [Google Scholar]
- [28].Fan JA, Yeo W-H, Su Y, Hattori Y, Lee W, Jung S-Y, Zhang Y, Liu Z, Cheng H, Falgout L, Bajema M, Coleman T, Gregoire D, Larsen RJ, Huang Y, Rogers JA, Nat. Commun. 2014, 5, 3266. [DOI] [PubMed] [Google Scholar]
- [29].Xu S, Yan Z, Jang K-I, Huang W, Fu H, Kim J, Wei Z, Flavin M, McCracken J, Wang R, Science 2015, 347, 154. [DOI] [PubMed] [Google Scholar]
- [30].Tallinen T, Chung JY, Rousseau F, Girard N, Lefèvre J, Mahadevan L, Nat. Phys. 2016, 12, 588. [Google Scholar]
- [31].Gefen A, Margulies SS, J. Biomech 2004, 37, 1339. [DOI] [PubMed] [Google Scholar]
- [32].Hong G, Lieber CM, Nat. Rev. Neurosci. 2019, 20, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jacot JG, Martin JC, Hunt DL, Biomech J. 2010, 43, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mandal HS, Knaack GL, Charkhkar H, McHail DG, Kastee JS, Dumas TC, Peixoto N, Rubinson JF, Pancrazio JJ, Acta Biomater. 2014, 10, 2446. [DOI] [PubMed] [Google Scholar]
- [35].Li Q, Lin Z, Liu R, Tang X, Huang J, He Y, Zhou H, Sheng H, Shi H, Wang X, Liu J, (Preprint) bioRxiv 2021, 2021.04.22.440941.
- [36].Liu J, Fu T-M, Cheng Z, Hong G, Zhou T, Jin L, Duvvuri M, Jiang Z, Kruskal P, Xie C, Nature Nanotechnol. 2015, 10, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang X, Zhou T, Zwang TJ, Hong G, Zhao Y, Viveros RD, Fu T-M, Gao T, Lieber CM, Nature Mater. 2019, 18, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sasaki T, Matsuki N, Ikegaya Y, Science 2011, 331, 599. [DOI] [PubMed] [Google Scholar]
- [39].Hao Y, Hao S, Andersen-Nissen E, Mauck III WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, and Hoffman P, Cell 2021, 184, 3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Yang SM, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, and Boyden ES, Nature 2017, 545, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, and Knoblich JA, Proc. Natl. Acad. Sci. U.S A. 2015, 112, 15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].López-Tobón A, Villa CE, Cheroni C, Trattaro S, Caporale N, Conforti P, Iennaco R, Lachgar M, Rigoli MT, de la Cruz BM, and Riso PL, Stem Cell Reports 2019, 13, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, and Lancaster MA, Science 2020, 369, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Miura Y, Li MY, Birey F, Ikeda K, Revah O, Thete MV, Park JY, Puno A, Lee SH, Porteus MH, and Pașca SP, Nat. Biotechnol. 2020, 38, 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, and Trapnell C, Nature 2019, 566, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pre D, Nestor MW, Sproul AA, Jacob S, Koppensteiner P, Chinchalongporn V, Zimmer M, Yamamoto A, Noggle SA, Arancio O, PLoS One 2014, 9, e103418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Nature 2013, 497, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V, Cell 2014, 158, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K, Science 2018, 361, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fang H, Yu KJ, Gloschat C, Yang Z, Song E, Chiang C-H, Zhao J, Won SM, Xu S, Trumpis M, Zhong Y, Han SW, Xue Y, Xu D, Choi SW, Cauwenberghs G, Kay M, Huang Y, Viventi J, Efimov IR, Rogers JA, Nat. Biomed. Eng. 2017, 1, 0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jiang Y, Li X, Liu B, Yi J, Fang Y, Shi F, Gao X, Sudzilovsky E, Parameswaran R, Koehler K, Nair V, Yue J, Guo K, Fang Y, Tsai H-M, Freyermuth G, Wong RCS, Kao C-M, Chen C-T, Nicholls AW, Wu X, Shepherd GMG, Tian B, Nat. Biomed. Eng. 2018, 2, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jiang Y, Tian B, Nat. Rev. Mater. 2018, 3, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydin Ç, Barbic M, Nature 2017, 551, 7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tolias AS, Ecker AS, Siapas AG, Hoenselaar A, Keliris GA, Logothetis NK, J. Neurophysiol 2007, 98, 6. [DOI] [PubMed] [Google Scholar]
- [55].Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, Su K, Li S, Lu L, Jacob F, and Nguyen PT, Cell Stem Cell 2020, 26, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, and Fiddes IT, Cell 2019, 176, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.