Abstract
Background
Cutaneous human papillomaviruses (cuHPV) and polyomaviruses (HPyV) have been implicated in skin cancers; however, interpretation of findings across studies is complicated by limited understanding of the natural history of these infections across normal tissue types.
Methods
In total, 675 eyebrow hair (EBH) and skin swab (SSW) samples were collected from 71 skin cancer screening patients every 6 months over 2 years and measured for presence of β-HPV, γ-HPV, and HPyV. Incidence, persistence, and clearance of cuHPV/HPyV were estimated, and risk factors associated with infection were examined.
Results
Prevalence, incidence, and persistence of β-HPV, γ-HPV, and HPyV were consistently higher in SSW than in EBH, with types 5, 24, 49, 76 and Merkel cell polyomavirus (MCPyV) having incidence rates greater than 20 per 1000 person-months. Prevalent γ-HPV EBH infections persisted more often in women (P = .024), incident β-HPV EBH infections persisted less often among individuals with history of blistering sunburn (P = .019), and prevalent MCPyV SSW infections persisted more often in those with a history of skin cancer (P = .033).
Conclusions
Incidence and persistence of cuHPV/HPyV were observed in SSW and EBH; however, none of the risk factors examined were commonly associated with cuHPV/HPyV infections across normal tissue types.
Keywords: basal cell carcinoma, cutaneous HPV, incidence, natural history, persistence, polyomavirus, risk factors, squamous cell carcinoma
Cutaneous human papillomavirus and polyomavirus infection incidence and persistence were consistently higher in skin swabs than in eyebrow hairs. Our findings suggest that the normal tissue type of the infection may play a role in the viral natural history.
Human papillomaviruses (HPV) are ubiquitous DNA viruses with tropism for cutaneous and mucosal epithelium [1, 2]. Classification of the more than 200 existing HPV types [3, 4] is based on the L1 gene sequence with mucosal types belonging to the genera alpha and cutaneous types included in alpha, beta, gamma, mu, and nu genera [5]. Persistent mucosal HPV infection is causally associated with 91% of cervical cancers and between 63% and 91% of oropharyngeal, penile, and anal cancers [6, 7]. The role for cutaneous HPV (cuHPV) infection in skin cancer etiology is less clear, with some studies suggesting that ultraviolet radiation (UVR) exposure cooperates with β-HPV types to indirectly promote skin carcinogenesis [8–10].
Similar to HPV, human polyomaviruses (HPyV) are small viruses with double-stranded DNA [11]. In contrast to the large number of HPV types, only 14 HPyV types have been identified to date [12], including 6 HPyV types detected on the skin: Merkel cell polyomavirus (MCPyV), HPyV6, HPyV7, HPyV9, Lyon IARC PyV, and trichodysplasia spinulosa polyomavirus (TSV) [13]. All HPyVs encode for the large and small T-antigen which are multifunctional proteins expressed during the early stage of viral replication [14, 15]. MCPyV infection is thought to be causally associated with Merkel cell carcinoma (MCC), with some studies reporting 80% of MCC tumors to be MCPyV positive with evidence of viral integration [16]. While MCPyV and other HPyVs have also been detected in keratinocyte carcinoma tumors, particularly cutaneous squamous cell carcinomas (cuSCC) [17–19], their precise role in cuSCC carcinogenesis is less clear.
Previous epidemiologic studies of cuHPV and HPyV infections in relation to skin cancer have used a variety of biomarkers to measure infection in normal tissues, including the presence of viral DNA in eyebrow hairs (EBH) and skin swabs (SSW). Some studies incorporating viral DNA in EBH have observed positive associations with cuSCC [10, 20–22] while others reported null associations [18, 23]. Our own recent study was the first to examine viral DNA in SSW in relation to skin cancer risk and observed positive associations between cuSCC and HPV [24] and null associations for HPyV [18]. Comparison of findings across these studies is complicated by differences in study design, types of viral markers used, and the number of virus types analyzed. Interpretation of differences across studies is further complicated by limited information available on the natural history of HPV/HPyV across different types of normal tissues.
Previous studies have examined the persistence of cuHPV types using various markers such as viral DNA in SSW [25–28], EBH [25, 29], and skin biopsies [30]. However, these studies were limited by short follow-up periods [25, 27], sampling frequency [26, 30], and sample size [28]. To our knowledge, only 2 studies have examined HPyV persistence [31, 32], and very limited literature exists on the incidence and clearance of cuHPV and HPyV infections [25, 31]. The HPV Infection in Men (HIM) substudy [25, 31] examined the natural history of both cuHPV and HPyV infection in normal skin of healthy men but, to our knowledge, no studies have examined the natural history of these viruses in a population at higher risk for skin cancer.
To examine the natural history of cuHPV and HPyV infections among a population of skin cancer screening patients at elevated risk for skin cancer, we sought to describe the incidence, persistence, and clearance of cuHPV/HPyV infection in EBH and SSW samples obtained every 6 months over a 2-year period from a subcohort of individuals enrolled in the Viruses in Skin Cancer (VIRUSCAN) Study. Furthermore, we aimed to assess factors associated with measures of incident and persistent cuHPV/HPyV infection, including repeated measures of UVR exposure. The risk of cuSCC associated with incident and persistent cuHPV/HPyV infection was also explored.
METHODS
Study Design and Population
The VIRUSCAN Study methods have been previously reported in detail [33]. Briefly, patients undergoing routine skin cancer screening exams were recruited between 2014 and 2017 from the University of South Florida Dermatology Clinic and were eligible to enroll in the study if they were 60 years or older and had not had a history of both cuSCC and basal cell carcinoma (BCC) at the time of study enrollment, thus ensuring all participants were naive to at least 1 type of keratinocyte carcinoma. Participants were followed for up to 4 years through September 2018, with follow-up visits occurring every 6–12 months. Study visits were coupled with patients’ clinical appointments for routine total body skin cancer screening examinations (TBSE).
VIRUSCAN participants were followed for different periods of time based on their time of cohort enrollment [33]. A total of 1008 participants returned to the clinic for a TBSE at least once over the 4-year follow-up period with a mean follow-up time of 788 days. Of these patients, 485 returned every year within the first 2 years of enrollment. To facilitate comparison with previous studies of the natural history of mucosal HPV infections, most of which estimated incidence and persistence using 6-month repeated samples [34], we restricted the current analysis to the subcohort of 71 participants who contributed an EBH and/or a SSW sample every 6 months for 5 consecutive study visits over a 2-year period. No significant differences were observed between the subcohort and the full VIRUSCAN cohort with respect to age, sex, race, or other baseline risk factors (Table 1). Among these 71 participants, 66 contributed an EBH sample and 69 contributed a SSW sample at 5 consecutive visits. Participants who did not have assay-specific β-globin amplified in their baseline EBH or SSW sample and those with 2 consecutive β-globin–negative samples after baseline were excluded from the corresponding analysis. Therefore, the final sample size for each of the following assay-specific analyses was 66 for β-HPV in EBH, 66 for γ-HPV in EBH, 66 for HPyV in EBH, 64 for β-HPV in SSW, 62 for γ-HPV in SSW, and 63 for HPyV in SSW. The study methods were approved by the University of South Florida Institutional Review Board, and all participants signed an informed consent prior to enrollment.
Table 1.
Comparison of Baseline Characteristics Among VIRUSCAN Study Participants Who Did and Did Not Have 5 Consecutive Visits
| Baseline Characteristics | Participants Without 5 Consecutive Visits, Full Cohort (n = 937)a | Participants With 5 Consecutive Visits, Subcohort (n = 71)a | P Valueb |
|---|---|---|---|
| n (%) | |||
| Age, y | |||
| Mean (SD) | 69.0 (6.2) | 69.9 (6.7) | .232 |
| 60–69 | 538 (57.4) | 38 (53.5) | |
| 70–79 | 334 (35.6) | 28 (39.4) | |
| 80–89 | 65 (6.9) | 5 (7.0) | .602 |
| Sex | |||
| Female | 488 (52.1) | 30 (42.3) | .114 |
| Male | 449 (47.9) | 41 (57.7) | |
| Race | |||
| White | 904 (96.6) | 71 (100.0) | .126 |
| Multiple/other | 32 (3.4) | 0 (0.0) | |
| Ethnicity | |||
| Non-Hispanic | 893 (95.3) | 68 (95.8) | .999 |
| Hispanic | 44 (4.7) | 3 (4.2) | |
| Smoking status | |||
| Never | 469 (50.9) | 38 (53.5) | .719 |
| Ever | 452 (49.1) | 33 (46.5) | |
| Sunburn | |||
| No | 230 (24.9) | 21 (29.6) | .391 |
| Yes | 692 (75.1) | 50 (70.4) | |
| Sun job | |||
| No | 644 (69.8) | 46 (64.8) | .42 |
| Yes | 278 (30.2) | 25 (35.2) | |
| Skin color | |||
| Level 1 | 228 (26.8) | 17 (25.0) | .402 |
| Level 2 | 412 (48.4) | 29 (42.6) | |
| Level 3 | 157 (18.4) | 18 (26.5) | |
| Level 4–10 | 54 (6.3) | 4 (5.9) | |
| Autoimmune disease | |||
| No | 850 (92.1) | 65 (91.5) | .988 |
| Yes | 73 (7.9) | 6 (8.5) | |
| Skin cancer | |||
| No | 491 (53.2) | 33 (46.5) | .306 |
| Yes | 432 (46.8) | 38 (53.5) | |
| UVR indicator as continuous, mean (SD) | 13.0 (5.2) | 13.8 (5.0) | .224 |
Abbreviations: SD, standard deviation; UVR, ultraviolet radiation; VIRUSCAN, Viruses in Skin Cancer.
Data are shown as n (%) except where indicated.
P values were calculated using Barnard test for binary baseline characteristics, Cochran-Armitage test for ordinal baseline characteristics, and Wilcoxon rank-sum test for continuous age and the UVR indicator.
Data and Sample Collection
Biospecimen Collection
As described previously, participants contributed EBH and SSW samples at baseline and at each follow-up visit [33]. Plucked EBH with attached follicles were snap frozen in liquid nitrogen and stored at −70°C until further processing. Normal SSW were collected from the top of the forearm for each participant. SSW were placed in a separate vial and stored at 4°C until further processing.
Viral DNA Measurement
All EBH and SSW samples were processed and analyzed by the International Agency for Research on Cancer as previously described [33, 35]. Multiplex polymerase chain reaction/Luminex assays [36, 37] were used for the measurement of viral DNA, corresponding to 46 β-HPV types (species 1 [types 5, 8, 12, 14, 19, 20, 21, 24, 25, 36, 47, 93, 98, 99, 105, 118, 124, 143, 152], species 2 [types 9, 15, 17, 22, 23, 37, 38, 80, 100, 104, 107, 110, 111, 113, 120, 122, 145, 151, 159, 174], species 3 [types 49, 75, 76, 115], species 4 [type 92], and species 5 [types 96, 150]); 52 γ-HPV types (4, 48, 50, 60, 65, 88, 95, 101, 103, 108, 109, 112, 116, 119, 121, 123, 126, 127, 128, 129, 130, 131, 132, 133, 134, 148, 149, 156, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 175, 178, 179, 180, 184, 197, 199, 200, 201, 202, and SD2); and 5 HPyVs (HPyV6, HPyV7, HPyV9, MCPyV, and TSV).
Two primers for the amplification of β-globin were included as a positive control for template DNA quality and assay-specific results were considered valid if the particular assay amplified β-globin. A Luminex reader was used to quantify the reporter florescence on the bead expressed as the median florescence intensity of at least 100 beads per set.
Questionnaire and UVR Exposure Measurement
Participants completed a baseline electronic questionnaire with information on demographics, medical history, and skin cancer risk factors [33]. To facilitate the measurement of recent UVR exposure at baseline and at each study visit, a Konica Minolta CM-600D spectrophotometer was used to measure skin pigmentation as described previously [33, 38]. Spectrophotometer readings were obtained at the inner arm and the top of the forearm. The difference in skin pigmentation readings obtained at the sun-exposed forearm and sun-unexposed inner arm was used as an indicator of recent UVR exposure, also as previously described [33, 38]
Incident cuSCC Case Ascertainment
Study coordinators routinely reviewed clinic pathology reports to identify incident cases of cuSCC and to document the associated clinical and pathologic characteristics. Participants were considered cuSCC cases if they developed at least 1 incident cuSCC, basosquamous or SCC/BCC merged tumor during follow-up, while participants who did not develop an incident cuSCC during follow-up were considered noncases. Participants with multiple incident cuSCC were censored at the date of the first cuSCC biopsy, and noncases were censored on the date of the last TBSE.
Statistical Analysis
HPV/HPyV infection status at each study visit was defined based on the presence of viral DNA in EBH and SSW samples collected at 5 visits (enrollment, 6 months, 12 months, 18 months, and 24 months), with 0 indicating a virus-negative sample, 1 indicating a virus-positive sample, and N indicating a β-globin-negative sample. Patterns of infection across the 5 visits were described using a string of 5 binary digits or N (eg, 101N0). To maximize the sample size, the last observation carried forward method was applied to impute the viral infection status for the follow-up samples where β-globin was not amplified (N). If β-globin was not amplified on 2 or more consecutive visits, the patient was excluded from the assay-specific analysis (1 case in each of the SSW β-HPV, γ-HPV, and HPyV assays). Figure 1 illustrates the 32 possible combinations of the 5-digit string and how the coding scheme was used to define viral infection incidence and persistence using HPV 38 in SSW as an example. The same coding schema was applied to each of the 46 β-HPV, 52 γ-HPV, and 5 HPyV types separately in EBH and SSW.
Figure 1.
Patterns of 6-month repeated cutaneous HPV infection. The figure depicts the patterns of repeated cutaneous HPV infection using HPV 38 viral DNA in skin swabs as an example. Boxes 2, 4, and 5 correspond to how prevalent infections, persistence of prevalent infections, and clearance of prevalent infections were coded, respectively. Boxes 8, 9, and 10 together correspond to incident infections, box 15 corresponds to persistence of incident infections, and box 16 corresponds to clearance of incident infections. Abbreviation: HPV, human papillomavirus.
The 5-digit strings were used to classify infections as prevalent or incident, having persisted or not and having cleared or not, as describe in Figure 1. The prevalence, incidence, and persistence of HPV/HPyV was first described at the type level, and subsequently summarized at the branch, species, and genus level separately for EBH and SSW. To examine the internal validity of the coding schema, the relationship between type-specific prevalence, incidence, and clearance was examined by plotting prevalence as a function of the product of incidence and duration [39] (estimated by 1 minus clearance) and robust linear regression with bisquare weighting was used to estimate the relationship across types.
The viral exposures (prevalent infection, prevalent persistent infection, incident infection, and incident persistent infection) were described for categories of each skin cancer risk factor and compared using Barnard test, Cochran-Armitage test, or Wilcoxon rank-sum test as appropriate. P values were adjusted for multiple testing using the false discovery rate method. Additionally, to estimate the impact of changes in UVR exposure on the persistence and clearance of viral infection, participants with consecutive UVR exposure measured at baseline and at all 4 follow-up visits were stratified by patterns of UVR exposure. The 2 major patterns identified were defined as consistent (UVR measurements obtained during follow-up remained relatively consistent with the baseline UVR measurement) and inconsistent (UVR measurements obtained during follow-up were different than the baseline UVR measurement). At the patient level, the consistent pattern was further categorized into consistently low (all UVR measures were ≤ 13.33 [median of the baseline UVR measure]) and consistently high (all UVR measures were > 13.33). Unadjusted logistic regression was used to estimate the associations between consistently low versus consistently high UVR exposure and persistence or clearance of viral infection measured by EBH and SSW.
The coded strings were subsequently censored by the incident cuSCC date for participants who developed cuSCC during follow-up period, and by the date of the last TBSE for patients who did not develop cuSCC during follow-up period. The censored strings were categorized into the viral exposures using similar methods described above. Age- and sex-adjusted Cox regression was used to calculate the proportional hazards ratio (HR) and 95% confidence intervals (CIs) for associations between cuSCC and each of the viral exposures. Due to sparse data, a shrinkage adjustment was applied to all Cox models to control extreme estimates [40].
RESULTS
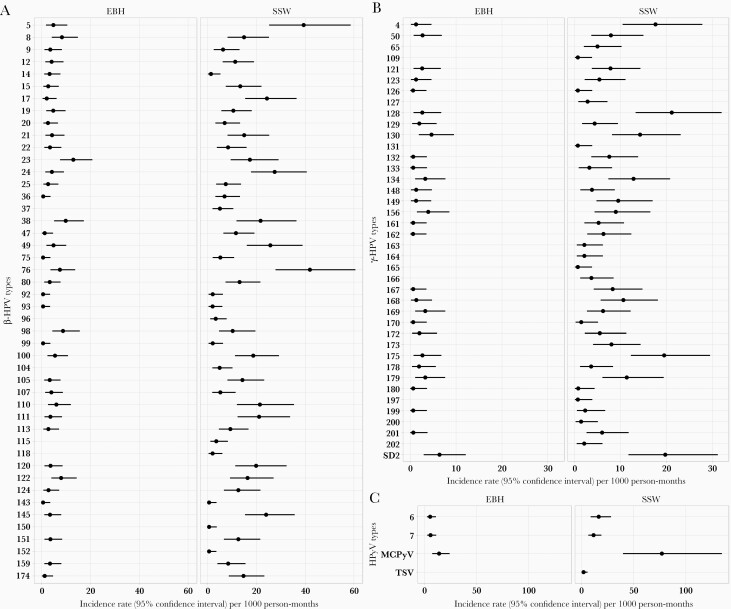
The VIRUSCAN subcohort was composed of participants with 5 consecutive visits, of whom 57.7% were male, 100.0% were white, and 4.2% were non-Hispanic (Table 1). The type-specific prevalence, incidence, persistence, and clearance for each virus type examined are presented in Supplementary Tables 1 and 2. Overall cuHPV type-specific incidence rates were higher in the SSW than in the EBH (Figure 2A and 2B). CuHPV types 23, 38, and 98 in EBH were the top 3 types with the highest incidence rates, ranging from 8.8 to 13.0 cases per 1000 person-months, while the top 3 most commonly occurring HPV infections in the SSW were 5, 76, and 24, ranging in incidence from 27.6 to 41.9 cases per 1000 person-months (Figure 2A). For HPyV infections, MCPyV was detected most often in both EBH (14.2 cases per 1000 person-months) and SSW (77 cases per 1000 person-months) (Figure 2C). Interestingly, when plotting the incidence × duration by the prevalence of cuHPV infection at the type level, a positive association was observed between incidence × duration and prevalence for β-HPV in EBH and SSW, as well as for γ-HPV in SSW (Figure 3A–3D), consistent with prevalence being a function of incidence and duration [39].
Figure 2.
Incidence rates of cutaneous HPV and HPyV. The type-specific incidence rate of (A) β-HPV types (B) γ-HPV types and (C) HPyV types as measured in EBH and SSW. Overall, type-specific incidence rates were higher as measured in SSW than that in EBH. Types with zero incidence in both EBH and SSW were excluded from the figure (γ-HPV: 48, 60, 88, 95, 101, 103, 108, 112, 116, 119, 171, and 184, and HPyV 6). Abbreviations: EBH, eyebrow hair; HPV, human papillomavirus; HPyV, human polyomavirus; SSW, skin swab.
Figure 3.
Correlation between prevalence, incidence, and duration of cutaneous HPV infection. Type-specific scatterplot showing the relationship between prevalence and incidence × duration for (A) β-HPV types measured in EBH, (B) β-HPV types measured in SSW, (C) γ-HPV types measured in EBH, and (D) γ-HPV types measured in SSW. Duration was estimated as 1 minus clearance and robust linear regression was used to summarize the relationship across types. Abbreviations: EBH, eyebrow hair; HPV, human papillomavirus; SSW, skin swab.
Type-specific infections were summarized by genus at the patient level to examine associations with baseline risk factors (Figure 4). The percent of patients with at least 1 prevalent infection, as well as the percentage of patients with at least 1 prevalent infection that persisted, were greater in the SSW than in the EBH for β-HPV, γ-HPV, and HPyV (Figure 4). For example, at the patient level, β-HPV prevalence and the persistence of prevalent β-HPV infection were 51.5% and 79.4%, respectively, in EBH and 96.9% and 95.2%, respectively, in SSW. Regarding clearance, patients with at least 1 prevalent β-HPV infection were more likely to clear the infection in the SSW (75.8%) than in the EBH (50.0%), while the opposite pattern was observed for γ-HPV and HPyV infections (Figure 4). The percent of patients with at least 1 incident infection or an incident infection that persisted was also greater in SSW than EBH for β-HPV, γ-HPV, and HPyV, particularly for β-HPV (84.4% in SSW vs 65.2% in EBH; Figure 4). The percent of patients that cleared at least 1 incident β-HPV and γ-HPV infection tended to be higher in SSW compared to EBH, while patients were more likely to clear an incident HPyV infection in the EBH than in the SSW (Figure 4).
Figure 4.
Prevalence, incidence, persistence, and clearance of cutaneous HPV and HPyV at the patient level. The viral infection status across 5 visits over a 2-year period summarized at patient level: prevalent, the percent of patients who had infection at baseline among all patients; prevalent that persisted, the percent of patients who had at least 1 prevalent infection that persisted among those who had prevalent infections; percent that cleared, the percent of patients who had at least 1 prevalent infection that cleared among those who had prevalent infection; incident, the percent of patients who had at least 1 incident infection among all patients; incident that persisted, the percent of patients who had at least 1 of incident infection that persisted among those who had incident infection; incident that cleared, the percent of patients who had at least 1 incident infection that cleared among patients who had incident infection. Abbreviations: EBH, eyebrow hair; HPyV, human polyomavirus; HPV, human papillomavirus; SSW, skin swab.
No associations were observed between baseline characteristics and prevalence or persistence of cuHPV in SSW; however, persistence of prevalent γ-HPV infection in EBH was significantly more likely to occur in women (100% women vs 0% men; P value = .024; Supplementary Table 3). Persistence of incident β-HPV in EBH was inversely associated with history of blistering sunburn (53.3% no vs 46.7% yes; P value = .019; Table 2), while no associations were observed between incidence and persistence of HPyV infections (Table 3). Persistence of prevalent MCPyV infections in SSW was more likely to occur in individuals with a history of skin cancer (31.7% no vs 68.3% yes; P value = .033; Supplementary Table 4).
Table 2.
Associations Between Baseline Characteristics and Incident and Persistent Cutaneous HPV DNA in EBH and SSW
| EBHa | SSWa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incident Any β-HPV | Incident Any γ-HPV | Incident Any β-HPV | Incident Any γ-HPV | |||||||||||||
| Baseline Characteristics | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent |
| n (%) | 18 (27.3) | 48 (72.7) | 16 (34.8) | 30 (65.2) | 25 (37.9) | 41 (62.1) | 18 (52.9) | 16 (47.1) | 0 (0) | 64 (100) | 10 (15.6) | 54 (84.4) | 1 (1.6) | 61 (98.4) | 20 (32.8) | 41 (67.2) |
| Age, y | ||||||||||||||||
| 60–69 | 10 (55.6) | 25 (52.1) | 9 (56.2) | 15 (50) | 12 (48) | 23 (56.1) | 10 (55.6) | 9 (56.2) | 0 (0) | 35 (54.7) | 4 (40) | 31 (57.4) | 1 (100) | 33 (54.1) | 9 (45) | 24 (58.5) |
| 70–79 | 8 (44.4) | 18 (37.5) | 6 (37.5) | 12 (40) | 12 (48) | 14 (34.1) | 6 (33.3) | 6 (37.5) | 0 (0) | 25 (39.1) | 5 (50) | 20 (37) | 0 (0) | 24 (39.3) | 10 (50) | 14 (34.1) |
| 80–89 | 0 (0) | 5 (10.4) | 1 (6.2) | 3 (10) | 1 (4) | 4 (9.8) | 2 (11.1) | 1 (6.2) | 0 (0) | 4 (6.2) | 1 (10) | 3 (5.6) | 0 (0) | 4 (6.6) | 1 (5) | 3 (7.3) |
| P valueb | .587 | .755 | 1.000 | .885 | .681 | .771 | .915 | |||||||||
| Sex | ||||||||||||||||
| Female | 11 (61.1) | 16 (33.3) | 6 (37.5) | 9 (30) | 10 (40) | 17 (41.5) | 8 (44.4) | 6 (37.5) | 0 (0) | 27 (42.2) | 3 (30) | 24 (44.4) | 0 (0) | 26 (42.6) | 9 (45) | 17 (41.5) |
| Male | 7 (38.9) | 32 (66.7) | 10 (62.5) | 21 (70) | 15 (60) | 24 (58.5) | 10 (55.6) | 10 (62.5) | 0 (0) | 37 (57.8) | 7 (70) | 30 (55.6) | 1 (100) | 35 (57.4) | 11 (55) | 24 (58.5) |
| P value | .315 | .807 | 1.000 | .885 | .871 | .771 | .924 | |||||||||
| Race | ||||||||||||||||
| White | 18 (100) | 48 (100) | 16 (100) | 30 (100) | 25 (100) | 41 (100) | 18 (100) | 16 (100) | 0 (0) | 64 (100) | 10 (100) | 54 (100) | 1 (100) | 61 (100) | 20 (100) | 41 (100) |
| Multiple/other | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | ||||||||||||||||
| Ethnicity | ||||||||||||||||
| Non-Hispanic | 16 (88.9) | 47 (97.9) | 16 (100) | 29 (96.7) | 24 (96) | 39 (95.1) | 17 (94.4) | 15 (93.8) | 0 (0) | 61 (95.3) | 10 (100) | 51 (94.4) | 1 (100) | 58 (95.1) | 18 (90) | 40 (97.6) |
| Hispanic | 2 (11.1) | 1 (2.1) | 0 (0) | 1 (3.3) | 1 (4) | 2 (4.9) | 1 (5.6) | 1 (6.2) | 0 (0) | 3 (4.7) | 0 (0) | 3 (5.6) | 0 (0) | 3 (4.9) | 2 (10) | 1 (2.4) |
| P value | .355 | .974 | 1.000 | 1.000 | .871 | 1.000 | .915 | |||||||||
| Smoking status | ||||||||||||||||
| Never | 13 (72.2) | 22 (45.8) | 8 (50) | 12 (40) | 13 (52) | 22 (53.7) | 9 (50) | 9 (56.2) | 0 (0) | 36 (56.2) | 6 (60) | 30 (55.6) | 0 (0) | 34 (55.7) | 11 (55) | 23 (56.1) |
| Ever | 5 (27.8) | 26 (54.2) | 8 (50) | 18 (60) | 12 (48) | 19 (46.3) | 9 (50) | 7 (43.8) | 0 (0) | 28 (43.8) | 4 (40) | 24 (44.4) | 1 (100) | 27 (44.3) | 9 (45) | 18 (43.9) |
| P value | .315 | .755 | 1.000 | .885 | 1.000 | .771 | .963 | |||||||||
| Sunburn | ||||||||||||||||
| No | 3 (16.7) | 18 (37.5) | 1 (6.2) | 16 (53.3) | 4 (16) | 17 (41.5) | 5 (27.8) | 9 (56.2) | 0 (0) | 19 (29.7) | 3 (30) | 16 (29.6) | 0 (0) | 19 (31.1) | 5 (25) | 14 (34.1) |
| Yes | 15 (83.3) | 30 (62.5) | 15 (93.8) | 14 (46.7) | 21 (84) | 24 (58.5) | 13 (72.2) | 7 (43.8) | 0 (0) | 45 (70.3) | 7 (70) | 38 (70.4) | 1 (100) | 42 (68.9) | 15 (75) | 27 (65.9) |
| P value | .315 | .019 | .375 | .402 | 1.000 | .771 | .915 | |||||||||
| Sun job | ||||||||||||||||
| No | 14 (77.8) | 28 (58.3) | 11 (68.8) | 15 (50) | 18 (72) | 24 (58.5) | 10 (55.6) | 7 (43.8) | 0 (0) | 42 (65.6) | 9 (90) | 33 (61.1) | 1 (100) | 40 (65.6) | 14 (70) | 26 (63.4) |
| Yes | 4 (22.2) | 20 (41.7) | 5 (31.2) | 15 (50) | 7 (28) | 17 (41.5) | 8 (44.4) | 9 (56.2) | 0 (0) | 22 (34.4) | 1 (10) | 21 (38.9) | 0 (0) | 21 (34.4) | 6 (30) | 15 (36.6) |
| P value | .354 | .689 | .881 | .885 | .437 | .771 | .915 | |||||||||
| Skin color | ||||||||||||||||
| Level 1 | 4 (23.5) | 11 (23.9) | 5 (33.3) | 5 (17.2) | 6 (26.1) | 9 (22.5) | 4 (22.2) | 3 (20) | 0 (0) | 16 (26.2) | 2 (22.2) | 14 (26.9) | 0 (0) | 15 (25.9) | 5 (25) | 10 (26.3) |
| Level 2 | 7 (41.2) | 20 (43.5) | 5 (33.3) | 14 (48.3) | 8 (34.8) | 19 (47.5) | 9 (50) | 8 (53.3) | 0 (0) | 26 (42.6) | 3 (33.3) | 23 (44.2) | 0 (0) | 27 (46.6) | 9 (45) | 18 (47.4) |
| Level 3 | 5 (29.4) | 12 (26.1) | 5 (33.3) | 7 (24.1) | 6 (26.1) | 11 (27.5) | 5 (27.8) | 3 (20) | 16 (26.2) | 4 (44.4) | 12 (23.1) | 1 (100) | 13 (22.4) | 5 (25) | 8 (21.1) | |
| Level 4–10 | 1 (5.9) | 3 (6.5) | 0 (0) | 3 (10.3) | 3 (13) | 1 (2.5) | 0 (0) | 1 (6.7) | 3 (4.9) | 0 (0) | 3 (5.8) | 0 (0) | 3 (5.2) | 1 (5) | 2 (5.3) | |
| P value | .942 | .689 | 1.000 | .885 | .871 | .771 | .924 | |||||||||
| Autoimmune disease | ||||||||||||||||
| No | 16 (88.9) | 44 (91.7) | 14 (87.5) | 29 (96.7) | 21 (84) | 39 (95.1) | 17 (94.4) | 16 (100) | 0 (0) | 59 (92.2) | 9 (90) | 50 (92.6) | 1 (100) | 55 (90.2) | 17 (85) | 38 (92.7) |
| Yes | 2 (11.1) | 4 (8.3) | 2 (12.5) | 1 (3.3) | 4 (16) | 2 (4.9) | 1 (5.6) | 0 (0) | 0 (0) | 5 (7.8) | 1 (10) | 4 (7.4) | 0 (0) | 6 (9.8) | 3 (15) | 3 (7.3) |
| P value | .942 | .689 | .541 | .885 | 1.000 | 1.000 | .915 | |||||||||
| Skin cancer | ||||||||||||||||
| No | 8 (44.4) | 23 (47.9) | 6 (37.5) | 17 (56.7) | 8 (32) | 23 (56.1) | 13 (72.2) | 9 (56.2) | 0 (0) | 29 (45.3) | 2 (20) | 27 (50) | 1 (100) | 27 (44.3) | 8 (40) | 19 (46.3) |
| Yes | 10 (55.6) | 25 (52.1) | 10 (62.5) | 13 (43.3) | 17 (68) | 18 (43.9) | 5 (27.8) | 7 (43.8) | 0 (0) | 35 (54.7) | 8 (80) | 27 (50) | 0 (0) | 34 (55.7) | 12 (60) | 22 (53.7) |
| P value | .942 | .689 | .375 | .885 | .437 | .771 | .915 | |||||||||
| UVR indicator dichotomizedc | ||||||||||||||||
| ≤ 13.33 | 12 (66.7) | 20 (43.5) | 8 (50) | 11 (39.3) | 13 (52) | 19 (48.7) | 6 (35.3) | 11 (73.3) | 0 (0) | 31 (50) | 6 (66.7) | 25 (47.2) | 0 | 30 (50) | 13 (65) | 17 (42.5) |
| 6 (33.3) | 26 (56.5) | 8 (50) | 17 (60.7) | 12 (48) | 20 (51.3) | 11 (64.7) | 4 (26.7) | 0 (0) | 31 (50) | 3 (33.3) | 28 (52.8) | 0 | 30 (50) | 7 (35) | 23 (57.5) | |
| P value | .315 | .755 | 1.000 | .321 | .681 | .915 | ||||||||||
| UVR indicator as continuous | ||||||||||||||||
| Mean (SD) | 12.7 (4.6) | 14.2 (5.2) | 13.6 (5.4) | 14.7 (5.2) | 13.3 (4.9) | 14.1 (5.1) | 14.6 (4.8) | 12 (4.7) | 0 (0) | 14.2 (5) | 12.5 (7.4) | 14.4 (4.5) | 14.3 (4.9) | 13.5 (5) | 14.7 (4.8) | |
| P value | .453 | .755 | 1.000 | .321 | .437 | .915 | ||||||||||
Abbreviations: EBH, eyebrow hairs; HPV, human papillomavirus; SSW, skin swabs; UVR, ultraviolet radiation.
Data are shown as n (%) except where indicated.
P values were calculated using Barnard test for binary baseline characteristics, Cochran-Armitage test for ordinal baseline characteristics, and Wilcoxon rank-sum test for continuous forearm spectrophotometer readings. P values were adjusted for multiple testing using false discovery rate method.
The baseline UVR indicator was dichotomized by the median value of 13.33, which was calculated using baseline spectrophotometer readings of all patients who were included in the repeated viral DNA analysis.
Table 3.
Associations Between Baseline Characteristics and Incident and Persistent HPyV DNA in EBH and SSW
| EBHa | SSWa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incident Any HPyV | Incident MCPyV | Incident Any HPyV | Incident MCPyV | |||||||||||||
| Baseline Characteristics | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent | No | Yes | Not Persistent | Persistent |
| n (%) | 42 (63.6) | 24 (36.4) | 17 (85.0) | 3 (15.0) | 31 (70.5) | 13 (29.5) | 9 (81.8) | 2 (18.2) | 31 (49.2) | 32 (50.8) | 12 (41.4) | 17 (58.6) | 2 (14.3) | 12 (85.7) | 4 (33.3) | 8 (66.7) |
| Age, y | ||||||||||||||||
| 60–69 | 24 (57.1) | 11 (45.8) | 7 (41.2) | 2 (66.7) | 16 (51.6) | 7 (53.8) | 6 (66.7) | 1 (50) | 18 (58.1) | 16 (50) | 6 (50) | 8 (47.1) | 1 (50) | 7 (58.3) | 3 (75) | 4 (50) |
| 70–79 | 16 (38.1) | 10 (41.7) | 8 (47.1) | 1 (33.3) | 13 (41.9) | 5 (38.5) | 2 (22.2) | 1 (50) | 12 (38.7) | 13 (40.6) | 5 (41.7) | 7 (41.2) | 1 (50) | 4 (33.3) | 1 (25) | 3 (37.5) |
| 80–89 | 2 (4.8) | 3 (12.5) | 2 (11.8) | 0 (0) | 2 (6.5) | 1 (7.7) | 1 (11.1) | 0 (0) | 1 (3.2) | 3 (9.4) | 1 (8.3) | 2 (11.8) | 0 (0) | 1 (8.3) | 0 (0) | 1 (12.5) |
| P valueb | .686 | .653 | .997 | 1.000 | 1.000 | .845 | 1.000 | .686 | ||||||||
| Sex | ||||||||||||||||
| Female | 16 (38.1) | 11 (45.8) | 8 (47.1) | 0 (0) | 13 (41.9) | 6 (46.2) | 5 (55.6) | 0 (0) | 12 (38.7) | 15 (46.9) | 8 (66.7) | 7 (41.2) | 0 (0) | 7 (58.3) | 2 (50) | 5 (62.5) |
| Male | 26 (61.9) | 13 (54.2) | 9 (52.9) | 3 (100) | 18 (58.1) | 7 (53.8) | 4 (44.4) | 2 (100) | 19 (61.3) | 17 (53.1) | 4 (33.3) | 10 (58.8) | 2 (100) | 5 (41.7) | 2 (50) | 3 (37.5) |
| P value | .873 | .527 | .997 | .729 | 1.000 | .840 | .772 | .898 | ||||||||
| Race | ||||||||||||||||
| White | 42 (100) | 24 (100) | 17 (100) | 3 (100) | 31 (100) | 13 (100) | 9 (100) | 2 (100) | 31 (100) | 32 (100) | 12 (100) | 17 (100) | 2 (100) | 12 (100) | 4 (100) | 8 (100) |
| Multiple/other | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | ||||||||||||||||
| Ethnicity | ||||||||||||||||
| Non-Hispanic | 39 (92.9) | 24 (100) | 17 (100) | 3 (100) | 30 (96.8) | 13 (100) | 9 (100) | 2 (100) | 30 (96.8) | 30 (93.8) | 12 (100) | 16 (94.1) | 2 (100) | 12 (100) | 4 (100) | 8 (100) |
| Hispanic | 3 (7.1) | 0 (0) | 0 (0) | 0 (0) | 1 (3.2) | 0 (0) | 0 (0) | 0 (0) | 1 (3.2) | 2 (6.2) | 0 (0) | 1 (5.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | .686 | .997 | 1.000 | .845 | ||||||||||||
| Smoking status | ||||||||||||||||
| Never | 18 (42.9) | 17 (70.8) | 11 (64.7) | 3 (100) | 14 (45.2) | 11 (84.6) | 8 (88.9) | 2 (100) | 16 (51.6) | 19 (59.4) | 7 (58.3) | 12 (70.6) | 1 (50) | 8 (66.7) | 3 (75) | 5 (62.5) |
| Ever | 24 (57.1) | 7 (29.2) | 6 (35.3) | 0 (0) | 17 (54.8) | 2 (15.4) | 1 (11.1) | 0 (0) | 15 (48.4) | 13 (40.6) | 5 (41.7) | 5 (29.4) | 1 (50) | 4 (33.3) | 1 (25) | 3 (37.5) |
| P value | .347 | .650 | .175 | 1.000 | 1.000 | .845 | 1.000 | .898 | ||||||||
| Sunburn | ||||||||||||||||
| No | 12 (28.6) | 9 (37.5) | 7 (41.2) | 1 (33.3) | 11 (35.5) | 4 (30.8) | 3 (33.3) | 1 (50) | 8 (25.8) | 11 (34.4) | 6 (50) | 5 (29.4) | 0 (0) | 3 (25) | 2 (50) | 1 (12.5) |
| Yes | 30 (71.4) | 15 (62.5) | 10 (58.8) | 2 (66.7) | 20 (64.5) | 9 (69.2) | 6 (66.7) | 1 (50) | 23 (74.2) | 21 (65.6) | 6 (50) | 12 (70.6) | 2 (100) | 9 (75) | 2 (50) | 7 (87.5) |
| P value | .873 | .913 | .997 | 1.000 | 1.000 | .845 | .954 | .686 | ||||||||
| Sun job | ||||||||||||||||
| No | 28 (66.7) | 14 (58.3) | 11 (64.7) | 0 (0) | 18 (58.1) | 10 (76.9) | 8 (88.9) | 0 (0) | 22 (71) | 19 (59.4) | 9 (75) | 7 (41.2) | 2 (100) | 6 (50) | 2 (50) | 4 (50) |
| Yes | 14 (33.3) | 10 (41.7) | 6 (35.3) | 3 (100) | 13 (41.9) | 3 (23.1) | 1 (11.1) | 2 (100) | 9 (29) | 13 (40.6) | 3 (25) | 10 (58.8) | 0 (0) | 6 (50) | 2 (50) | 4 (50) |
| P value | .873 | .381 | .997 | .183 | 1.000 | .840 | .772 | 1.000 | ||||||||
| Skin color | ||||||||||||||||
| Level 1 | 9 (23.1) | 6 (25) | 3 (17.6) | 1 (33.3) | 7 (25) | 3 (23.1) | 2 (22.2) | 1 (50) | 8 (27.6) | 8 (25.8) | 2 (18.2) | 5 (29.4) | 0 (0) | 4 (36.4) | 0 (0) | 4 (50) |
| Level 2 | 14 (35.9) | 13 (54.2) | 10 (58.8) | 1 (33.3) | 10 (35.7) | 7 (53.8) | 5 (55.6) | 0 (0) | 13 (44.8) | 14 (45.2) | 7 (63.6) | 7 (41.2) | 1 (100) | 4 (36.4) | 2 (66.7) | 2 (25) |
| Level 3 | 13 (33.3) | 4 (16.7) | 3 (17.6) | 1 (33.3) | 10 (35.7) | 3 (23.1) | 2 (22.2) | 1 (50) | 6 (20.7) | 8 (25.8) | 2 (18.2) | 4 (23.5) | 0 (0) | 3 (27.3) | 1 (33.3) | 2 (25) |
| Level 4–10 | 3 (7.7) | 1 (4.2) | 1 (5.9) | 0 (0) | 1 (3.6) | 0 (0) | 0 (0) | 0 (0) | 2 (6.9) | 1 (3.2) | 0 (0) | 1 (5.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| P value | .686 | .913 | .997 | 1.000 | 1.000 | .845 | 1.000 | .686 | ||||||||
| Autoimmune disease | ||||||||||||||||
| No | 38 (90.5) | 22 (91.7) | 17 (100) | 3 (100) | 27 (87.1) | 12 (92.3) | 9 (100) | 2 (100) | 28 (90.3) | 29 (90.6) | 12 (100) | 14 (82.4) | 2 (100) | 9 (75) | 4 (100) | 5 (62.5) |
| Yes | 4 (9.5) | 2 (8.3) | 0 (0) | 0 (0) | 4 (12.9) | 1 (7.7) | 0 (0) | 0 (0) | 3 (9.7) | 3 (9.4) | 0 (0) | 3 (17.6) | 0 (0) | 3 (25) | 0 (0) | 3 (37.5) |
| P value | 1.000 | .997 | 1.000 | .840 | .954 | .686 | ||||||||||
| Skin cancer | ||||||||||||||||
| No | 19 (45.2) | 12 (50) | 10 (58.8) | 2 (66.7) | 16 (51.6) | 7 (53.8) | 5 (55.6) | 1 (50) | 13 (41.9) | 15 (46.9) | 7 (58.3) | 7 (41.2) | 1 (50) | 7 (58.3) | 4 (100) | 3 (37.5) |
| Yes | 23 (54.8) | 12 (50) | 7 (41.2) | 1 (33.3) | 15 (48.4) | 6 (46.2) | 4 (44.4) | 1 (50) | 18 (58.1) | 17 (53.1) | 5 (41.7) | 10 (58.8) | 1 (50) | 5 (41.7) | 0 (0) | 5 (62.5) |
| P value | .895 | .913 | .997 | 1.000 | 1.000 | .845 | 1.000 | .557 | ||||||||
| UVR indicator dichotomizedc | ||||||||||||||||
| ≤ 13.33 | 20 (50) | 12 (50) | 8 (47.1) | 0 (0) | 16 (51.6) | 7 (53.8) | 5 (55.6) | 0 (0) | 16 (51.6) | 15 (50) | 7 (58.3) | 8 (47.1) | 2 (100) | 5 (41.7) | 2 (50) | 3 (37.5) |
| 20 (50) | 12 (50) | 9 (52.9) | 3 (100) | 15 (48.4) | 6 (46.2) | 4 (44.4) | 2 (100) | 15 (48.4) | 15 (50) | 5 (41.7) | 9 (52.9) | 0 (0) | 7 (58.3) | 2 (50) | 5 (62.5) | |
| P value | 1.000 | .527 | .997 | .729 | 1.000 | .845 | .772 | .898 | ||||||||
| UVR indicator as continuous | ||||||||||||||||
| Mean (SD) | 14 (4.7) | 13.5 (5.6) | 14.5 (5.7) | 16.2 (0.6) | 13.5 (4.9) | 14 (5.3) | 14.1 (6.1) | 16.5 (0.6) | 14.6 (4.9) | 13.5 (5.2) | 12.8 (4.4) | 13.3 (5.1) | 10.7 (2.6) | 14.4 (5.6) | 15.7 (5.6) | 13.7 (5.8) |
| P value | .873 | .718 | .997 | 1.000 | 1.000 | .845 | .879 | .898 | ||||||||
Abbreviations: EBH, eyebrow hairs; HPyV, human polyomavirus; SSW, skin swabs; UVR, ultraviolet radiation.
Data are shown as n (%) except where indicated.
P values were calculated using Barnard test for binary baseline characteristics, Cochran-Armitage test for ordinal baseline characteristics, and Wilcoxon rank-sum test for continuous forearm spectrophotometer readings. P values were adjusted for multiple testing using false discovery rate method.
The baseline UVR indicator was dichotomized by the median value of 13.33, which was calculated using baseline spectrophotometer readings of all patients who were included in the repeated viral DNA analysis.
The spectrophotometer-based UVR exposure readings obtained from participants at each study visit during follow-up are shown in Supplementary Figure 1A. When examining the associations between repeated measurements of viral infection and the patterns of UVR exposure, we observed that participants with consistently high UVR exposure were more likely to have persistent HPyV infection in EBH and SSW compared with participants with consistently low UVR exposure, although certain risk estimates were extreme due to sparse data. For example, prevalent and persistent HPyV infection in EBH was positively associated with consistently high UVR exposure (HR = 3.75; 95% CI, .35–53.50) as was prevalent and persistent MCPyV infection in EBH (HR = 8.00; 95% CI, .63–229.77) (Supplementary Figure 1B).
Among the 71 participants with repeated viral infection measurements, 20 participants developed at least 1 incident keratinocyte carcinoma during follow-up (15 cuSCC and 15 BCC). No evidence of increased cuSCC risk with incident or persistent cuHPV/HPyV infection was observed among participants (Supplementary Table 5 and 6). Of note, only 4 of the 20 participants who developed keratinocyte carcinoma developed a virus-positive tumor, including 1 cuSCC positive for HPV 111, another cuSCC positive for HPyV 6, and 2 BCCs positive for MCPyV. However, none of the viruses detected in the tumors were consistently present in SSW/EBH collected prior to the tumor development, except for HPyV 6 which was consistently observed in both SSW and EBH prior to cuSCC diagnosis.
DISCUSSION
Among skin cancer screening patients recruited to the VIRUSCAN Study whose EBH and SSW samples were obtained every 6 months over a 2-year period, the incidence and persistence of cuHPV was higher in SSW than EBH, with β-HPV types having a higher incidence and persistence than γ-HPV types, consistent with previous studies [25, 26]. Similarly, the incidence and persistence of HPyV tended to be higher in SSW than in EBH, also consistent with previous findings from the HIM study [31]. When comparing the 25 β-HPV, 12 γ-HPV, and 5 HPyV types common across the VIRUSCAN and HIM studies no systematic difference was observed in the type-specific incidence rates, with type-specific rates being higher in 1 study versus the other except for γ-HPV in SSW where the incidence rate was higher among HIM participants [25]. Differences in the type-specific incidence rates across studies could be due to underlying differences in the study population as the HIM study included younger male participants while VIRUSCAN included an older population at higher risk for skin cancer.
We did not observe any associations between baseline characteristics and incident β-HPV or γ-HPV infection in SSW, consistent with the HIM study [25]. However, blistering sunburn was inversely associated with incident and persistent β-HPV in EBH, differing from the null associations observed among HIM participants [25]. Of note, the HIM study included only men, and in the present VIRUSCAN study, prevalent γ-HPV infections were more likely to persist in women.
The VIRUSCAN study incorporated a spectrophotometer-based measure of UVR exposure as an indicator of recent UVR exposure, previously shown to correlate with sun exposure in the past week [33]. This enabled us to easily obtain quantifiable and repeated measures of recent UVR exposure throughout the 2-year follow-up period. No associations were found between incident and persistent β-HPV infection in EBH and SSW and consistently high recent UVR exposure compared to consistently low recent UVR exposure. However, participants with consistently high recent UVR exposure tended to have persistent HPyV infection in both EBH and SSW, although our sample size was too small to show statistical significance. These findings are inconsistent with our previous analysis among VIRUSCAN Study participants demonstrating that recent UVR exposure was positively associated with prevalent viral infection in both SSW and EBH at baseline [38]. The lack of association with recent UVR exposure and β-HPV infection in this study could be due to differences in the number of viral types examined or possibly due to sample size. Alternatively, measurements of lifetime UVR exposure may be needed if there is a lag-time between the UVR exposure and an individual’s infection status.
To our knowledge, we are the first to examine the association between incident and persistent cuHPV/HPyV infection measured by viral DNA and future development of cuSCC. However, we found no evidence of an association between incident or persistent β-HPV in EBH or SSW and subsequent cuSCC in our study. While the study sample size was limited, the 2-year follow-up period and 6-month sampling allowed us to accurately assess natural history of infection. Our exploratory analysis of the association between infection and cuSCC development was underpowered; however, as participant follow-up continues, additional cuSCC end points maybe observed. The overall generalizability of our findings may be restricted to an older population at a higher risk for skin cancer; however, skin cancer screening patients represent the ideal population that would be targeted for novel skin cancer prevention or personalized screening regimens. Despite some limitations, our study is the first to report on natural history of cuHPV and HPyV in both men and women, incorporating more virus types than previous studies, and also the first study to include repeated spectrophotometer-based measures of recent UVR exposure at each time point at which infections were measured.
In conclusion, incidence and persistence was higher in SSW than in EBH for HPV and HPyV types. Persistence of β-HPV infection in SSW was inversely associated with history of blistering sunburn while persistence of MCPyV in SSW was positively associated with a history of skin cancer. Taken together, our data suggest that the normal tissue type of the infection (EBH versus SSW) may play a role in the natural history of these viruses. Larger studies are needed to further understand the HPV/HPyV natural history and assess the interplay of UVR exposure, incidence/persistence of these cutaneous viruses, and subsequent skin cancer development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The funders were not involved in the study design, data collection/analysis, or manuscript preparation. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Financial support. This work was supported by the National Cancer Institute at the National Institutes of Health (grant number R01 CA177586 to D. E. R.); and in part by the Tissue Core and the Participant Research, Interventions, and Measurement Core at the H. Lee Moffitt Cancer Center and Research Institute, a comprehensive cancer center designated by the National Cancer Institute and funded in part by a Moffitt Cancer Center support grant (grant number P30 CA076292).
Contributor Information
Rossybelle P Amorrortu, Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, Florida, USA.
Yayi Zhao, Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, Florida, USA.
Neil A Fenske, Department of Dermatology and Cutaneous Surgery, University of South Florida College of Medicine, Tampa, Florida, USA.
Basil S Cherpelis, Department of Dermatology and Cutaneous Surgery, University of South Florida College of Medicine, Tampa, Florida, USA.
Jane L Messina, Department of Anatomic Pathology, Moffitt Cancer Center, Tampa, Florida, USA; Department of Cutaneous Oncology, Moffitt Cancer Center, Tampa, Florida, USA.
Anna R Giuliano, Center for Immunization and Infection Research in Cancer, Moffitt Cancer Center, Tampa, Florida, USA.
Vernon K Sondak, Department of Cutaneous Oncology, Moffitt Cancer Center, Tampa, Florida, USA.
Michael J Schell, Biostatistics and Bioinformatics Shared Resource, Moffitt Cancer Center, Tampa, Florida, USA.
Sandrine Mckay-Chopin, International Agency for Research on Cancer, World Health Organization, Lyon, France.
Tarik Gheit, International Agency for Research on Cancer, World Health Organization, Lyon, France.
Tim Waterboer, Infections and Cancer Epidemiology, German Cancer Research Center, Heidelberg, Germany.
Massimo Tommasino, International Agency for Research on Cancer, World Health Organization, Lyon, France.
Dana E Rollison, Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, Florida, USA.
References
- 1. de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology 2013; 445:2–10. [DOI] [PubMed] [Google Scholar]
- 2. Van Doorslaer K. Evolution of the papillomaviridae. Virology 2013; 445:11–20. [DOI] [PubMed] [Google Scholar]
- 3. Kocjan BJ, Bzhalava D, Forslund O, Dillner J, Poljak M.. Molecular methods for identification and characterization of novel papillomaviruses. Clin Microbiol Infect 2015; 21:808–16. [DOI] [PubMed] [Google Scholar]
- 4. Mühr LSA, Eklund C, Dillner J.. Towards quality and order in human papillomavirus research. Virology 2018; 519:74–6. [DOI] [PubMed] [Google Scholar]
- 5. Egawa N, Doorbar J.. The low-risk papillomaviruses. Virus Res 2017; 231:119–27. [DOI] [PubMed] [Google Scholar]
- 6. Shiels MS, Kreimer AR, Coghill AE, Darragh TM, Devesa SS.. Anal cancer incidence in the United States, 1977–2011: distinct patterns by histology and behavior. Cancer Epidemiol Biomarkers Prev 2015; 24:1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arbyn M, Bosch X, Cuzick J,. et al. IARC monographs on the evaluation of carcinogenic risks to humans: human papillomaviruses. Vol. 90. Lyon, France: World Health Organization, 2007. [Google Scholar]
- 8. Karagas MR, Nelson HH, Sehr P,. et al. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst 2006; 98:389–95. [DOI] [PubMed] [Google Scholar]
- 9. Viarisio D, Decker KM, Aengeneyndt B, Flechtenmacher C, Gissmann L, Tommasino M.. Human papillomavirus type 38 E6 and E7 act as tumour promoters during chemically induced skin carcinogenesis. J Gen Virol 2013; 94:749–52. [DOI] [PubMed] [Google Scholar]
- 10. Bouwes Bavinck JN, Neale RE, Abeni D,. et al. Multicenter study of the association between Betapapillomavirus infection and cutaneous squamous cell carcinoma. Cancer Res 2010; 70:9777–86. [DOI] [PubMed] [Google Scholar]
- 11. DeCaprio JA, Garcea RL.. A cornucopia of human polyomaviruses. Nat Rev Microbiol 2013; 11:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gheit T, Dutta S, Oliver J,. et al. Isolation and characterization of a novel putative human polyomavirus. Virology 2017; 506:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prado JCM, Monezi TA, Amorim AT, Lino V, Paladino A, Boccardo E.. Human polyomaviruses and cancer: an overview. Clinics (Sao Paulo) 2018; 73:e558s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moens U, Ludvigsen M, Van Ghelue M.. Human polyomaviruses in skin diseases. Patholog Res Int 2011; 2011:123491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheu JC, Tran J, Rady PL, Dao H Jr, Tyring SK, Nguyen HP.. Polyomaviruses of the skin: integrating molecular and clinical advances in an emerging class of viruses. Br J Dermatol 2019; 180:1302–11. [DOI] [PubMed] [Google Scholar]
- 16. Becker JC, Kauczok CS, Ugurel S, Eib S, Bröcker EB, Houben R.. Merkel cell carcinoma: molecular pathogenesis, clinical features and therapy. J Dtsch Dermatol Ges 2008; 6:709–19. [DOI] [PubMed] [Google Scholar]
- 17. Rollison DE, Giuliano AR, Messina JL,. et al. Case-control study of Merkel cell polyomavirus infection and cutaneous squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2012; 21:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amorrortu RP, Zhao Y, Messina JL,. et al. Association between human polyomaviruses and keratinocyte carcinomas: a prospective cohort study. Cancer Epidemiol Biomarkers Prev 2021; 30:1761–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dworkin AM, Tseng SY, Allain DC, Iwenofu OH, Peters SB, Toland AE.. Merkel cell polyomavirus in cutaneous squamous cell carcinoma of immunocompetent individuals. J Invest Dermatol 2009; 129:2868–74. [DOI] [PubMed] [Google Scholar]
- 20. Struijk L, Bouwes Bavinck JN, Wanningen P,. et al. Presence of human papillomavirus DNA in plucked eyebrow hairs is associated with a history of cutaneous squamous cell carcinoma. J Invest Dermatol 2003; 121:1531–5. [DOI] [PubMed] [Google Scholar]
- 21. Iannacone MR, Gheit T, Pfister H,. et al. Case-control study of genus-beta human papillomaviruses in plucked eyebrow hairs and cutaneous squamous cell carcinoma. Int J Cancer 2014; 17:28552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hampras SS, Michel A, Schmitt M,. et al. Merkel cell polyomavirus (MCV) T-antigen seroreactivity, MCV DNA in eyebrow hairs, and squamous cell carcinoma. Infect Agent Cancer 2015; 10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neale RE, Weissenborn S, Abeni D,. et al. Human papillomavirus load in eyebrow hair follicles and risk of cutaneous squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2013; 22:719–27. [DOI] [PubMed] [Google Scholar]
- 24. Rollison DE, Amorrortu RP, Zhao Y, et al. Cutaneous human papillomaviruses and the risk of keratinocyte carcinomas. Cancer Res 2021; 81:4628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hampras SS, Giuliano AR, Lin HY,. et al. Natural history of cutaneous human papillomavirus (HPV) infection in men: the HIM study. PLoS One 2014; 9:e104843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazard K, Karlsson A, Andersson K, Ekberg H, Dillner J, Forslund O.. Cutaneous human papillomaviruses persist on healthy skin. J Invest Dermatol 2007; 127:116–9. [DOI] [PubMed] [Google Scholar]
- 27. Bolatti EM, Chouhy D, Hošnjak L,. et al. Natural history of human papillomavirus infection of sun-exposed healthy skin of immunocompetent individuals over three climatic seasons and identification of HPV209, a novel Betapapillomavirus. J Gen Virol 2017; 98:1334–48. [DOI] [PubMed] [Google Scholar]
- 28. Weissenborn SJ, De Koning MN, Wieland U, Quint WG, Pfister HJ.. Intrafamilial transmission and family-specific spectra of cutaneous betapapillomaviruses. J Virol 2009; 83:811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Koning MN, Struijk L, Bavinck JN,. et al. Betapapillomaviruses frequently persist in the skin of healthy individuals. J Gen Virol 2007; 88:1489–95. [DOI] [PubMed] [Google Scholar]
- 30. Berkhout RJ, Bouwes Bavinck JN, ter Schegget J.. Persistence of human papillomavirus DNA in benign and (pre)malignant skin lesions from renal transplant recipients. J Clin Microbiol 2000; 38:2087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hampras SS, Giuliano AR, Lin HY,. et al. Natural history of polyomaviruses in men: the HPV infection in men (HIM) study. J Infect Dis 2015; 211:1437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hashida Y, Higuchi T, Tanaka M,. et al. Prevalence and viral loads of cutaneous human polyomaviruses in the skin of patients with chronic inflammatory skin diseases. J Infect Dis 2019; 219:1564–73. [DOI] [PubMed] [Google Scholar]
- 33. Amorrortu RP, Fenske NA, Cherpelis BS,. et al. Viruses in skin cancer (VIRUSCAN): study design and baseline characteristics of a prospective clinic-based cohort study. Cancer Epidemiol Biomarkers Prev 2020; 29:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rositch AF, Koshiol J, Hudgens MG,. et al. Patterns of persistent genital human papillomavirus infection among women worldwide: a literature review and meta-analysis. Int J Cancer 2013; 133:1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rollison DE, Schell MJ, Fenske NA,. et al. Cutaneous viral infections across 2 anatomic sites among a cohort of patients undergoing skin cancer screening. J Infect Dis 2019; 219:711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gheit T, Billoud G, de Koning MN,. et al. Development of a sensitive and specific multiplex PCR method combined with DNA microarray primer extension to detect Betapapillomavirus types. J Clin Microbiol 2007; 45:2537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gheit T, Landi S, Gemignani F,. et al. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J Clin Microbiol 2006; 44:2025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Y, Amorrortu RP, Fenske NA,. et al. Cutaneous viral infections associated with ultraviolet radiation exposure. Int J Cancer 2020; 148:448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szklo M, Nieto FJ.. Epidemiology: beyond the basics. Sudbury, MA: Jones & Bartlett Publishers, 2014. [Google Scholar]
- 40. Greenland S, Mansournia MA, Altman DG.. Sparse data bias: a problem hiding in plain sight. BMJ 2016; 352:i1981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.