Abstract
The serotonin transporter (SERT) initiates the reuptake of extracellular serotonin in the synapse to terminate neurotransmission. The cryogenic electron microscopy structures of SERT bound to ibogaine and the physiological substrate serotonin resolved in different states have provided a glimpse of the functional conformations at atomistic resolution. However, the conformational dynamics and structural transitions to intermediate states are not fully understood. Furthermore, the molecular basis of how serotonin is recognized and transported remains unclear. In this study, we performed unbiased microsecond-long simulations of the human SERT to investigate the structural dynamics to various intermediate states and elucidated the complete substrate import pathway. Using Markov state models, we characterized a sequential order of conformational-driven ion-coupled substrate binding and transport events and calculated the free energy barriers of conformation transitions associated with the import mechanism. We find that the transition from the occluded to inward-facing state is the rate-limiting step for substrate import and that the substrate decreases the free energy barriers to achieve the inward-facing state. Our study provides insights on the molecular basis of dynamics-driven ion-substrate recognition and transport of SERT that can serve as a model for other closely related neurotransmitter transporters.
Significance
Serotonin reuptake by the serotonin transporter (SERT) is critical for regulating serotonergic signaling in neurons. Neurotransmitter reuptake transporters, including SERT, are major drug targets for treating psychiatric disorders. SERT adopts three main conformational states, named outward facing, occluded, and inward facing, to transport substrates across the cell membrane. In this study, all-atom molecular dynamics simulations and statistical modeling were employed to characterize the transitions between SERT functional states and elucidate the mechanism of how serotonin is transported into the cell at atomistic resolution. Our results show that the binding of the substrate modulates the conformational landscape to lower the free energy barriers and promote transitions to the inward-facing state for substrate release.
Introduction
The serotonin transporter (SERT) terminates synaptic transmission by catalyzing the reuptake of extracellular serotonin from the synapse. Reuptake is critical for normal serotonergic signaling in the brain with implications for mood, cognition, behavior, and appetite (1). Consequently, improper SERT function is associated with numerous neurological disorders, including depression, autism, and bipolar disorder (2). Additionally, SERT is expressed in platelet membranes and regulates blood coagulation throughout the circulatory system (3). Given its medical importance, SERT is a major molecular target for therapeutic drugs and drugs of abuse (4,5). Similar to other members of the solute carrier 6 (SLC6) neurotransmitter transporter family, SERT-mediated serotonin (5-hydroxytryptamine, 5-HT) translocation from the synapse and surrounding area is coupled to favorable ion co-transport of one Na+ with a Cl− ion dependence, and export of one K+ to complete an overall electroneutral cycle (6, 7, 8, 9, 10). Other conduction states and stoichiometries with unclear physiological significance may occur under different conditions (11, 12, 13, 14, 15).
SERT, and the closely related dopamine transporter (DAT) and norepinephrine transporter (NET), belongs to a class of monoamine transporters in the neurotransmitter:sodium symporter (NSS) family. These members share a distinct 12-transmembrane (TM) helix architecture known as the leucine transporter (LeuT) fold, with TM1-5 and TM6-10 forming inverted pentahelical repeats around a pseudo 2-fold axis of symmetry (16,17). Cysteine labeling studies on SERT revealed that the 5-HT binding site is accessible from both extracellular and intracellular sides of the membrane, providing the first glimpse of evidence of an alternating access model (18). Quick and Javitch developed a proteomic approach to characterize the sodium-dependent substrate transport mechanism in the tyrosine transporter Tyt1 (19). These biochemical studies elucidated that the NSS family of transporters function based on the principle of an alternating access mechanism (20). Crystal structures of the bacterial NSS homolog LeuT obtained in three functional states, outward-facing (OF), occluded (OC), and inward-facing (IF) states, have validated the NSS transport process is by an alternating access mechanism, in which the substrate and ions first access their central binding sites via an open extracellular vestibule, and then are released within the cell through the sequential closure of an extracellular gate and opening of an intracellular exit pathway (17,20, 21, 22, 23). Historically, LeuT has served as a structural template to study monoamine transporters (24, 25, 26, 27, 28) and based on in-depth studies of bacterial transporters, including electron paramagnetic resonance (EPR) spectroscopy (29, 30, 31), molecular modeling (32,33), and single-molecule fluorescence resonance energy transfer (smFRET) experiments (28,34), substrate permeation through the NSS family transporters is facilitated by reorientation of helices around the central axis, in particular the movement of TM1a away from the helical bundle to open an intracellular vestibule for substrate release (21,35,36). Despite low sequence similarity with human NSS transporters, these efforts paved the way for early rational drug design for treating various psychiatric disorders (37, 38, 39).
Structural investigations into human NSS transporters have further benefited from the more recent resolution of eukaryotic monoamine transporters Drosophila DAT (dDAT), human SERT (hSERT) (40, 41, 42, 43), and human glycine transporter T1 (hGlyT1) (44). The screening and docking studies using these crystal structures have provided a structural basis of antidepressant recognition and inhibition (45, 46, 47, 48, 49, 50). Cryogenic electron microscopy (cryo-EM) structures of hSERT complexed with the psychedelic non-competitive inhibitor ibogaine reveal the occluded and IF states with similar structural arrangements as seen in LeuT (17,23,51,52). Most recently, structures of hSERT bound with serotonin have illuminated the role of the allosteric site in serotonin binding (53). However, given the structural discrepancies between SERT and other NSS structural models, the molecular basis of transitions between the intermediate states remains unknown. Closure of the extracellular vestibule is coordinated by helix motions of TM1b and TM6a where Arg104 and Glu493 are proposed to serve as extracellular gating residues to stabilize the OC and IF states. Further inspection the cryo-EM structure shows that the helix orientation of TM1b in the SERT OC conformation is closely aligned to that of OF LeuT (51). Moreover, among the current SERT OF structures, the distance between the guanidinium group of Arg104 and carboxyl of Glu493 varies from 4.4 Å to 7.4 Å, while, in the OC and IF states, this distance is 7.2 Å and 4.6 Å, respectively. As a result, the role of these gating residues and their interactions during conformational transitions is unclear. Furthermore, the N-terminal residues preceding TM1a and its interactions with TM6 and TM8 regulates the helix motion of TM1a during substrate release and acts as an intracellular gate (36,54,55). Hydrogen-deuterium exchange (HDX) experiments have provided an alternative approach to understanding the conformational dynamics within the NSS family and have shown that ion-substrate binding facilitates changes in dynamics in TM1a, TM6, and TM7 (24,56, 57, 58). Intricate loop dynamics, specifically motions of extracellular loops (EL) 3 and 4, fluctuate significantly during substrate transport (24,56, 57, 58). The combined structural and biochemical studies have provided invaluable insights into the functional mechanism of the NSS family. However, the realistic motions of structural transitions at atomistic resolution are not fully known to understand the conformational-driven substrate transport cycle.
In this study, we performed unbiased all-atom molecular dynamics (MD) simulations to obtain a comprehensive view of the import mechanism for the physiological substrate serotonin in hSERT. Our study shows the key binding and transport events, including substrate interactions at an extracellular allosteric site, neurotransmitter binding within subsite B, coordination of two metal ions, and a single symported sodium ion being displaced into the cytosol by the movement of serotonin into the exit pathway. Using a Markov state model (MSM)-based adaptive sampling approach to explore the conformational landscape, our simulations show a preferred sequential order of the ion-substrate binding, conformational transitions, and transport processes consistent with the current understanding of NSS substrate transport mechanism. The conformational free energy landscapes reveal that the structural isomerization from OC to IF is a rate-limiting step for import and is further facilitated by the import of 5-HT to the intracellular space. Our results provide an in-depth perspective into the molecular recognition and transport of 5-HT in SERT and may aid in the development of conformational selective inhibitors for the treatment of psychiatric disorders.
Methods
MD simulation setup
The OF-SERT crystal structure (PDB: 5I73) was used as the starting model for MD simulation (45). Thermostable mutations in the crystal structure Ala110, Ala291, and Ser439 were reverted to wild-type Tyr110, Ile291, and Thr439, respectively. Glu508 was protonated according to pKa predicted by PROPKA (59). The protein was embedded in a phosphatidylcholine (POPC) bilayer with CHARMM-GUI (60) and solvated with TIP3P water molecules (61). 150 mM NaCl was added to neutralize the system and mimic physiological conditions. Terminal chains were capped with acetyl and methyl amide groups. Overall, the final Na+-SERT system contained 66,801 atoms which consisted of 1 SERT monomer, 121 POPC molecules, 123 Na+ ions, 135 Cl− ions, and 13,582 TIP3P water molecules in a periodic box volume of 77 × 77 × 113 Å3. The MD system was built using the tleap module of AMBER18 (62) using Amber ff14SB (63) force field combined with GAFF force field (64). The MD system was minimized for 20,000 steps using the conjugate gradient method, heated from 0 to 300 K at NVT, and equilibrated for 40 ns under NPT conditions. An Na+ bound in the Na1 and Na2 site OF-SERT structure, obtained from Na+-SERT simulations, was used as the starting model to capture the mechanism of 5-HT import. One 5-HT molecule was randomly added to the simulation box and equilibrated under the same conditions described previously. Simulations were implemented using the Amber18 package (62,65, 66, 67) at constant NPT conditions (300K, 1 atm) and periodic boundary conditions. Temperature was maintained using Langevin dynamics with 1.0 ps−1 damping coefficient. Pressure was maintained using a Monte Carlo barostat with an update interval of 100 steps. Electrostatic interactions were treated with the partial mesh Ewald method (68), and hydrogen bonds were constrained using the SHAKE algorithm (69). Nonbonded distance cutoff was set at 10 Å. An integration timestep of 4 fs with hydrogen mass repartitioning (70) was used for all simulations. Snapshots were saved every 100 ps during production simulations. Simulations were performed on NVIDIA Tesla K20X GPUs on the Blue Waters supercomputer or NVIDIA GeForce GTX 980 GPUs on our local computing cluster.
Adaptive sampling
Obtaining sufficient sampling is a recurring challenge in simulating complex biological processes. To overcome this issue, we adopted an MSM-based adaptive sampling methodology to efficiently explore the conformational landscape (71,72). In each round of adaptive sampling, multiple short MD simulations are conducted in parallel. The simulation data are clustered using the K-means algorithm (73) based on a designated metric, and starting structures are chosen from the least populated states to seed the subsequent rounds of simulation. The sampling bias introduced from least-count selection is eliminated during the construction of the MSM by estimating the reverse transition probability matrix for transition between all conformational states. In the limit of long timescales, the sampling errors are expected to be small. For Na+-SERT simulations, the adaptive sampling and clustering were guided based on the distances of the extracellular and intracellular gating residues. For 5-HT-SERT simulations, the extracellular and intracellular gating distances, in addition to the z-components of 5-HT, the bound chloride ion, and symported Na2 ion, were used as adaptive sampling metrics. The number of trajectories for each adaptive sampling round is detailed in Tables S3 and S4 for Na+-SERT and 5-HT-SERT simulations, respectively. A total of 356 μs of Na+ and 519 μs of 5-HT import simulation data were obtained. The progression of the changes in the gating distances and 5-HT translocation is shown in Figs. S1–S3. Multiple substrate binding and transport events were captured as a direct advantage of the MSM-based adaptive sampling approach (Fig. S4). The entire MD dataset was used for MSM construction and analysis.
Markov state modeling
MSMs were constructed using the pyEMMA Python package (74). MSMs were constructed for both Na+ and 5-HT transport datasets. Sixteen residue-residue pair distances surrounding the permeation pathway were used for clustering (Fig. S5). Additionally, the z-components of the two binding Na+ ions were incorporated with the 16 residue-residue pair distances as featurization metrics for Na+-SERT simulations. The z-components of 5-HT, Cl−, and the symported Na+ ion were added along with the 16 distances to characterize the 5-HT import process. Time-structure independent component analysis (tICA) was performed on the feature matrix to reduce the dimension space by obtaining the slowest-relaxing degrees of freedom as a linear combination of the features (75). The optimum number of clusters and time-independent components (tICs) were based on the convergence of the implied timescales and the set that yielded the greatest sum of the eigenvalues of the transition matrix, also known as the VAMP1 score. Six-hundred clusters and three tICs were used to construct the MSM for Na+-SERT simulations. Five-hundred clusters and four tICs were used for 5-HT-SERT MSM. (Fig. S6 A). Projection of the simulation data on the tICA space map shows the first two tICs, which correspond to the two slowest processes, correlate with motions involved in opening and closure of the transporter and 5-HT diffusion through the transporter (Figs. S7–S9 and Table S5). The lag time of 12 ns was determined for MSM construction from implied timescale plots (Fig. S6 B). The Chapman-Kolmogorov test, which validates the Markovian behavior of the MSM (76), was performed on five macrostates implemented in pyEMMA (Figs. S10 and S11). MSM states were further clustered into macrostates and visualized with Visual Molecular Dynamics (VMD) (77) and PyMOL (Schrödinger).
Trajectory analysis
The CPPTRAJ and pytraj (78) modules in AmberTools and MDTraj Python library (79) were used for post-processing the trajectory data. The predicted fraction deuteration was calculated according to Adhikary et al. (24) on 20,000 OF and IF structures randomly extracted from Na+-SERT and 5-HT-SERT simulations (see supporting material for more details). The 5-HT-residue interactions were obtained from Python scripts implemented from the GetContacts package (https://getcontacts.github.io/). To calculate the root mean square fluctuation (RMSF) between transitions, 20,000 structures of OC and IF states were randomly extracted from the conformational landscape and measured with respect to the cryo-EM structure of the prior conformational state (i.e., OF-OC: 20,000 OC structures with respect to OF cryo-EM structure). Channel pore radius was calculated using the HOLE program (80). Cross-correlation analysis of Na+-SERT and 5-HT-SERT was performed using the Bio3D R package (81).
Generation of conformational free energy landscape
MSM-weighted simulation data were plotted on the coordinates of the gating distances, specifically the distances between the closest heavy atom between Arg104 and Glu493 of the extracellular gate, and the closest heavy atom between groups of residues Val86-Ser91, Tyr350 (TM1a, TM6b) and Val274-Trp282, Glu444 (TM5, TM8) for the intracellular gate (Fig. S12). Standard error of the free energy landscapes was calculated using the bootstrap method by randomly selecting 80% of the trajectories for 500 independent samples. The resulting error values were projected on the previously described metrics of the gating distances (Fig. S13). In-house scripts and the matplotlib Python library were used to generate plots.
Results and discussion
Substrate binding decreases the free energy barrier for SERT conformational transitions to the IF state
To understand the effects of substrate-induced SERT dynamics, the entire import process of 5-HT was studied using MD simulations. An MSM-based adaptive sampling approach was used to explore the entire accessible conformational space of SERT (71). Simulations were initiated from the OF crystal structure of hSERT (PDB: 5I73) and a total of 356 μs of 5-HT-free SERT (referred to as Na+-SERT) were obtained. Na+-bound SERT in an OF conformation obtained from Na+-SERT simulations, with the Na1 and Na2 sites occupied, was used to seed simulations of the 5-HT import process (referred to as 5-HT-SERT). One 5-HT molecule was placed in the simulation box and a total of 519 μs of data were collected. All simulation data were used to construct an MSM, which parses the simulation data into kinetically relevant states and calculates the transition probabilities between the states (see section “methods” for additional details). MSM-weighted simulation data were projected onto a coordinate system defined by distances between extracellular and intracellular gating residues (Figs. 1 and S12).
Figure 1.
Conformational free energy landscapes of SERT obtained from MD simulations. Relative free energies from MSM-weighted simulation data plotted against the distances between extracellular and intracellular gates for (A) Na+-SERT and (B) 5-HT-SERT. The OF SERT crystal structure (PDB: 5I73) was used as the starting structure for MD simulations and transitioned to OC and IF states. An HG state, in which both gates are open, was also observed. The SERT structure is represented as cartoon with TM1, 5, 6, 8, and 10 colored in teal, green, magenta, yellow, and orange respectively. (C) Cross section through SERT conformational states viewed from the membrane plane, shown as surface representations. The channel pore volume across the transporter is depicted as dark blue spheres and extracellular gates Arg104 and Glu493 are shown as teal and orange sticks, respectively. To see this figure in color, go online.
The conformational landscape plots reveal that, despite the absence of 5-HT binding, Na+-SERT may undergo transitions from the OF state to the IF state (Fig. 1 A). Extracellular gating residues Arg104 (TM1b) and Glu493 (TM10) can separate to 10 Å, enlarging the extracellular permeation pathway. The equivalent charged residues in the bacterial transporter LeuT (Arg30 and Asp404) have been previously implicated in the gating mechanism (17,82). The OF states are stable, with a relative free energy of ∼1.5 kcal/mol. The distance between gating residues Arg104-Glu493 decreases to 3 Å and is associated with electrostatic interactions (Figs. 1 and S14), forming OC conformations that are more stable than the OF state. Previous studies have shown that binding of Na+ stabilizes the extracellular open conformation in SERT and related transporters, agreeing with the free energy landscapes calculated from MD simulation (28,29,83, 84, 85). Distance distribution plots reveal that the extracellular surface remains open while the intracellular vestibule is closed in OF and OC states, and vice versa in the IF state (Fig. S15). Closure of the extracellular pathway as SERT isomerizes from the OF to OC state weakens contacts on the intracellular side of the transporter, creating an energetically accessible pathway toward a partial IF state, in which the intracellular pathway measured by the opening of TM1a and TM5 extends to ∼6 Å. The free energy barrier for transition from the OC-IF state in Na+-SERT is estimated as ∼3 kcal/mol, which is higher compared with the OF-OC transition (∼1.5 kcal/mol). Biophysical investigations of LeuT also reveal that IF states are less populated and that transitions to IF states are facilitated by the presence of the substrate (28,29,31,34). Formation of the IF state is associated with the partial unwinding of the cytoplasmic base of TM5, the breakage of electrostatic interactions between Arg79 (N-Term) and Asp452 (TM8) and Glu80 (N-Term) and Lys275 (TM5) at the intracellular gate, and increased dynamics of the flanking loops (Fig. S14).
The substrate-present conformational landscape plot exhibits deviations in the relative free energies of conformational states and reduced free energy barriers between states (Fig. 1 B). Binding of 5-HT in the entrance pathway stabilizes the OF states to a greater extent compared with Na+-SERT simulations (Figs. 1 and S15 C) (28). The gating residues form alternative interactions with Gln332 (TM6) and Lys490 (TM10), thereby widening the extracellular vestibule (Fig. S14). Similar to observations seen in human DAT simulations (40), the diffusion of 5-HT to the orthosteric (S1) site via the allosteric (S2) site promotes the inward closure of extracellular gating helices TM1b, TM6a, and TM10 to facilitate formation of the OC state (Fig. S16). The OF-OC transition has a free energy barrier of ∼1.5 kcal/mol, similar to Na+-SERT. The “downward” movement of 5-HT facilitates opening of the intracellular gate and isomerization to the IF state. The free energy barrier for the OC-IF structural transition is estimated as ∼2 kcal/mol. The presence of 5-HT in the intracellular pathway stabilizes SERT in a greater IF state, with a relative free energy of 2 kcal/mol compared with 3 kcal/mol in Na+-SERT. Compared with Na+-SERT, the intracellular vestibule in the IF state now extends to 6–8 Å, thus allowing for the substrate to be released, and is associated with the breakage of hydrogen bonding networks between intracellular gating helices (Figs. S14 and S17–S20). Terry et al. demonstrated that the selective substrate-driven conformational transition to the IF state in LeuT is highly favorable compared with non-selective substrates and ion binding alone (28). Furthermore, cysteine accessibility measurements demonstrate that the binding of the substrate promotes the opening of the intracellular pathway more than that of just Na+ alone (86). Using transition path theory, we estimated the mean first passage time (MFPT) for different transitions observed in the simulations. We found that the rate of transition between OC-IF states is rate limiting compared with transitions between OF-OC states both in the presence and absence of 5-HT (Fig. S21).
In Na+-SERT simulations, although the extracellular gating residues Arg104 and Glu493 maintain an electrostatic interaction, this does not propagate to the full closure of the extracellular helices toward the scaffold. Only in the presence of the substrate do the simulated helix rearrangements involved in opening and closing of the transporter agree most with the cryo-EM structures and other NSS structures (Figs. S22 and S23). We also observed partial OF-IF like conformations, which we have termed an hourglass-like (HG) state in which both gates are open but constricted at the junction of the two rocking bundles; i.e., the Na2 site (Fig. S24 D). Terry et al. identified intermediate conformations in LeuT that are open at both extracellular and cytoplasmic halves and may enable the transport of Na+ and substrate molecules (28). The HG states may serve as water conducting states that exist as transient, short-lived intermediates during the transport cycle, similarly observed in various membrane transporters (87). The conformational landscape plots reveal that these states are less stable (>3 kcal/mol) than canonical OF, OC, and IF states. Therefore, any release of Na+ ions and substrate molecules through HG states is a more energetically demanding process than the traditional substrate transport via the IF state. In Na+-SERT simulations, we observed uncoupled ion leaks; in contrast, Na+ binds tightly to its respective site and no ion leaks were observed in 5-HT-SERT (Fig. S25). This state has been observed in other membrane transporters (41,88, 89, 90), but transitions from HG to other intermediate states are restricted as the free energy barriers are high and the physiological relevance of this state in SERT remains unclear.
The calculated RMSF plots illustrate that, in both Na+ and 5-HT-SERT simulations, EL2 and EL4 are highly flexible during the OF to OC transitions. Alternatively, the loop regions are more stable during OC to IF in 5-HT-SERT simulations compared with Na+ simulations (Fig. 2). HDX mass spectrometry studies hint that EL2 and EL4 regions are destabilized and show increased deuterium exchange upon ion and substrate binding (24,56,58,90). Furthermore, EL2 exhibits higher deuterium uptake kinetics in the OF state compared with the IF state in LeuT (24) and is consistent with our observations from OC to IF transitions in 5-HT-SERT simulations. In 5-HT-SERT simulations, EL4 shows less pronounced fluctuations during OC to IF transitions. The experimental results from HDX show EL4 regions are more stabilized during K+ uptake, which is hypothesized to stabilize the IF state based on LeuT studies (58,91). The increased deuterium uptake of TM1a in the presence of K+ agrees with the large fluctuations we observed for transitions to the IF state. The comparison of the calculated deuterium exchange fraction of Na+ and 5-HT-SERT simulation data is further consistent with previous HDX studies of LeuT (Fig. S26) (24). The comparison of the calculated deuterium exchange fraction of Na+- and 5-HT-SERT simulation data further agrees with previous HDX studies of LeuT. Simulations of SERT predicted an overall higher deuteration fraction for segments in the N terminus, EL3, and TM6, and decreased fraction for TM7. For segments in TM1a and EL4, both the LeuT and SERT computed deuteration fraction are qualitatively agree with experimental values, but ultimately underestimated the deuteration fraction.
Figure 2.
MD analysis of global fluctuations during conformational transitions. RMSF of Na+ (A and B) and 5-HT transport (C and D) for OF to OC transition and OC to IF transition mapped to the SERT structure. Tube thickness corresponds to the RMSF values of each residue. To calculate the RMSF between transitions, 20,000 structures of OC and IF states were randomly extracted from the conformational landscape and measured with respect to the cryo-EM structure of the prior conformational state. The simulations show fluctuations involving EL4, EL6, and TM5 to be coupled with substrate import. More specifically, EL4 and EL6 experience greater dynamics in the presence of the 5-HT when SERT transitions from OF to OC. Compared with Na+-SERT simulations, the EL dynamics are less pronounced. In OC to IF transitions of 5-HT-SERT, EL2 is stabilized while the fluctuations at cytoplasmic base of TM5 increase to form the intracellular exit pathway. Alternatively, in Na+-SERT, EL2 is destabilized and TM5 dynamics are reduced. To see this figure in color, go online.
Buried glutamates in the hydrophobic pocket stabilize the fold for substrate transport
We note that the presence of Glu136 and Glu508 buried within hydrophobic TM regions is highly unusual; in this case, Glu136 forms hydrogen bonds to the exposed backbone N–H groups to support the unwinding of TM6 near the central binding site. These two glutamate residues are highly conserved among the SLC6 family, with the exception of Glu508, where it is substituted by threonine in the proline transporter and methionine in the glycine transporter 1C (Fig. S27). Mutations of Glu136 in SERT to either alanine or glutamine has been shown to abolish 5-HT uptake activity, while Glu136Asp retained activity but at a reduced rate (92). Similar observations were reported when the equivalent glutamate residue in DAT (93,94) and GABA transporter GAT-1 (95) were mutated to charge-neutral residues. However, in NET (96), the substitution to aspartate resulted in a complete loss of transport activity, whereas glutamine retained similar transport properties compared with the wild type. pKa calculations in conjunction with computational modeling (97) have suggested Glu508 and its equivalent residues to be protonated or that a proton is shared between the two glutamate residues. Nevertheless, these functional studies emphasize the role of these conserved residues in maintaining proper transporter function.
In our simulations, we find Glu136 to interact with the backbone amine of Gly340 on TM6 and the carboxyl side chain of Glu508 on TM10 (Fig. 3). Distance distribution plots show that Glu136 and Gly340 maintain a hydrogen bond across the three conformational states, whereas Glu136 and Glu508 side-chain distances are more likely to increase as SERT transitions to the IF state (Fig. S28 A and B). As a result, the number of water molecules within this region increases, thus rearranging the hydrogen bonding network to enable transitions of TM6 for substrate release (Figs. 3 A, B, and S28 C). To observe how Glu508 protonation affects the structural fold of this region, we simulated SERT with Glu508 in its deprotonated form. As anticipated, the negative charges from opposing glutamate residues repel one another and expand this buried pocket (Fig. S28 B). Interestingly, in some simulations, the Na+ ion from the Na1 site migrates to this cavity where it is then coordinated by the carboxylates of Glu136 and Glu508 and the sulfur of Met135 through water molecules (Fig. 3 C). A third metal ion site (the Na3 site, to be consistent with prior nomenclature) has not previously been described in SERT, but computational modeling, biochemical analysis, and electrophysiology recordings indicate that equivalent residues of the neuronal glycine transporter 2 (GlyT2) have been suggested to form this third Na+ site (98,99). However, in SERT simulations when Glu508 was protonated, the binding of a Na+ ion to the Na3 site was not observed within the simulated timescales.
Figure 3.
(A and B) MD snapshots of the hydrogen bonding network involving buried glutamate residues and water molecules when SERT adopts the OF state (A) and the IF state (B). TM helices 2, 6, and 10 colored as pale blue, magenta, and orange respectively. (C) A third Na+ ion binding site buried beneath the orthosteric pocket as a result of Glu508 deprotonation. MD snapshot (TM2 colored pale blue; TM6, magenta; TM10, orange) superimposed with the SERT crystal structure (PDB: 5I73, gray), with electron density shown at 0.5 σ. To see this figure in color, go online.
The substrate translocation process of SERT
Using transition path theory, the highest flux pathway for conformational change and 5-HT import can be determined from the MSM and used to predict an ordered sequence of binding events and structural changes. SERT undergoes complete transitions to the IF state in the simulations, with permeation toward the intracellular side upon binding of substrates in the order following Na+, 5-HT, and Cl− ions (Fig. 4). We describe each step of the import process in detail.
Figure 4.
The major flux pathway and MFPTs for SERT conformational transitions and 5-HT import determined from transition path theory. The transport process begins with the binding of two Na+ ions to the Na1 and Na2 sites in the OF state (2, 3). 5-HT diffuses to the orthosteric site (4). Next, a Cl− ion, accompanied by an additional Na+ (not shown), enters the transporter and binds (5). The accompanying Na+ ion dissociates back to the extracellular space (see Fig. S33 for additional details). The binding of the substrate and ions facilitates the closure of the extracellular gate to form the OC state (6). Isomerization to the IF state is associated with the release of Na+ from the Na2 site and 5-HT diffuses out (6–9). Arrow thickness represents relative flux between transitions. To see this figure in color, go online.
The import process begins with the binding of Na+ to the Na1 site, followed by a second Na+ binding the Na2 site (Figs. S29 and S30). These two sites are well described in the SERT crystal structure and the literature (23,45,47,100). Na+ bound at the Na1 site couples activity between the ion and substrate binding sites, whereas computational studies of related transporters have indicated that Na+ coordinated at the Na2 site dissociates during the transporter cycle to become the symported metal ion (40,101,102). The importance of the Na2 site is underscored by its conservation in distantly related secondary transporters (103). Na+ ions entering the transporter interact with Asp328 and Asn112 at the extracellular surface, then rapidly diffuse into the allosteric site (Figs. S29 A and S30 A). Here, the Na+ ions interact with Glu493 and Glu494, and a rotameric shift in Glu493 enables the ions to descend past the extracellular gate to their central binding sites (Figs. S29 B, C, S30 B, and C). Na1 is stabilized by Ala96, Asn101, Ser336, and Asn368 (Fig. S29 D), while Na2 is coordinated by backbone carbonyls of Gly94, Val97, Leu434, and side chains of Asp437 and Ser438 (Fig. S30 D). Previous simulations of SERT have shown Asp437 as a key residue for maintaining Na2-ion interactions (102), and mutations of Asp437 and Ser438 have confirmed their role in coordination and dissociation of Na2 (102,104). Similar to other transporters in the NSS family (29,83,84), the binding of Na+ ions to their respective sites stabilizes SERT in the OF state while neutralizing the polar cavity to allow protonated 5-HT diffusion.
5-HT is recognized by Tyr107, Ile108, Gln111, and Asp328 at the extracellular vestibule to initiate the binding in the OF state. Ile108 forms hydrophobic contacts with the indole ring of 5-HT, while other residues form polar interactions with the substrate that favors binding. 5-HT then diffuses inside the translocation pore and binds to the allosteric site (Fig. 5 B). The substrate is stabilized by aromatic ring packing against Phe335 and Arg104, and a hydrogen bonding network with Asp328, Gln332, and Glu494 (Fig. 5 B). The antidepressant molecule vilazodone binds to the allosteric site where it forms similar aromatic interactions to the endogenous substrate (105) (Fig. S31 A). Furthermore, previous mutations of residues in the allosteric site have been shown to alter inhibitor potency (106). 5-HT undergoes a 90° rotation by rapidly exchanging its polar interactions and shifts toward the orthosteric site (Fig. 5C). The switching of amine group interactions to Glu494 triggers the movement of 5-HT from the allosteric site to the orthosteric site. 5-HT rotates such that the conformation becomes perpendicular to the membrane, which is further favored by polar and hydrophobic interactions by Asp328, Gln332, Leu502, and Ala331. The extracellular gating residues form a salt bridge interaction and enlarge the binding cavity such that substrate can escape to the primary binding site. This substrate binding site in the NSS family contains three well-studied subsites and has served as the basis for designing various tricyclic antidepressant molecules (107). In the orthosteric site, the indole moiety forms aromatic interactions with Tyr95 and Phe341, while the protonated amine moiety of 5-HT interacts with Asp98 of subsite A, disrupting the hydrogen bond between Asp98 and Tyr176 (Fig. S32). Tyr176 is highly conserved among the SLC6 family, whereas Asp98 is uniquely conserved in only monoamine transporters; the majority of SLC6 transporters contain glycine in this position. This network of polar interactions involving Asp98, Tyr176, and the amine moiety of the substrate is critical for substrate binding and transport function (45). The mutation of Tyr176 to phenylalanine severs the hydrogen bond with Asp98 and reduces 5-HT transport (86). The phenylalanine substitution of the cognate tyrosine residue in hDAT (Tyr156) and LeuT (Tyr108) also results in decreased transport activity of their respective substrates (86,108). Further site-directed mutagenesis and computational docking studies have emphasized the significance of Asp98 in 5-HT recognition and transport (109,110), while biochemical studies show that the disruption of aromatic interactions with the substrate leads to a loss of function or decreased potency of antidepressants (106,111, 112, 113).
Figure 5.
MD snapshots of the simulated mechanism for SERT-catalyzed 5-HT import. (A) Overlaid MD snapshots of 5-HT translocation, from when 5-HT enters the extracellular vestibule (blue) to its cytosolic exit (red). The positions of ions in the OC state are shown as spheres. TM helices 1, 5, 6, 8, and 10 are colored in teal, green, magenta, yellow, and orange, respectively. (B) 5-HT enters the transporter by binding in the allosteric site. (C) From the allosteric site, 5-HT rotates down the transport pathway to the orthosteric site. (D) Initial Cl− recognition is assisted by Asp98, Arg104, Tyr175, Phe335, and indole of 5-HT. To see this figure in color, go online.
5-HT mediates the binding of an electrogenic Cl− ion
The permeation of the Cl− into the extracellular vestibule is accompanied by an additional extracellular Na+ ion. As the ions approach the extracellular gating residues, the Na+ ion disrupts the salt bridge by interacting with Glu493 and Glu494 to allow Cl− to form polar interactions with Arg104 and Tyr176. The shift of the guanidinium group of Arg104 then facilitates the diffusion of the Cl− ion into the central cavity (Fig. S33 B). The accompanying Na+ ion then diffuses out to the extracellular space as the orthosteric cavity is neutralized by the substrate and Na+ ions in the Na1 and Na2 site. Interestingly, we observed the binding of the Cl− ion to occur solely in 5-HT-SERT simulations. Despite Cl− ions being present in the solvent of Na+-SERT simulations, we only observe the binding of Na+ ions to the transporter within the simulated timescales (Fig. S34). To further investigate this phenomenon, we calculated the electrostatic potential using the adaptive Poisson-Boltzmann solver (APBS) (114). Projection of the electrostatic potential of the SERT three-dimensional structure shows that the extracellular vestibule of SERT is highly electronegative and remains electronegative after the binding of the two Na+ ions, as observed in Na+-SERT simulations (Fig. S35 A and B). When 5-HT is bound in the orthosteric site, the extracellular vestibule is then neutralized to allow for favorable diffusion of Cl−. The additional interaction of Cl− with the indole-NH of 5-HT further stabilizes the ion in the exposed extracellular recognition site (Fig. 5 D). The indole ring of 5-HT occupies subsite C of the primary binding site, which in turn favors the transition of the Cl− ion to its binding site (Fig. S31 B). The simulated Na1, Na2, and Cl− sites concur with their respective sites observed in crystal structures of SERT, LeuT, and DAT (23,45,47,100) (Fig. S36).
The binding of ions and the substrate to their respective sites leads to the closure of the extracellular gates to obtain the OC state where the permeation pathway is closed at both ends. The decrease of the pore results in shifting the aromatic ring of 5-HT from subsite C to subsite B within the orthosteric binding pocket. The amine moiety of the neurotransmitter remains bound to Asp98, Tyr95, and the C-terminal pole of TM1a within subsite A (Fig. 6 A and B). The simulated configuration of 5-HT in subsite B agrees with the crystal structure of dopamine-bound dDAT (100) (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 F). The indole-NH ring of 5-HT interacts with Thr473 and other hydrophobic contacts by Ala169, Ala441, Gly442, and Leu443 residues stabilizing the substrate in subsite B. Additionally, 5-HT may enter into subsite B in which the indole-NH is oriented toward Phe341 adopting a binding pose also reported in previous modeling studies (110,115). Previous experimental studies indicate that mutations of these residues can decrease 5-HT transport (106,111), and the binding pose is further supported by several studies showing that interactions in subsite B are critical for inhibitor potency (107,111,112).
Figure 6.
MD snapshots of 5-HT in the orthosteric binding site and intracellular release. TM helices 1, 5, 6, 8, and 10 are colored in teal, green, magenta, yellow, and orange, respectively. Substrate color relates to progression of 5-HT-import as shown in Fig. 5A. (A and B) While 5-HT is in subsites C and B, the amine moiety of 5-HT at the orthosteric site interacts with Asp98, disrupting the hydrogen bonding interaction between Tyr176-Asp98. (C) The rotameric flip of the phenol ring of Tyr95 initiates the opening of the intracellular vestibule and allows for permeation of 5-HT toward the intracellular exit pathway. (D) 5-HT translocation through the exit pathway between TM1a and TM5. To see this figure in color, go online.
Figure 7.
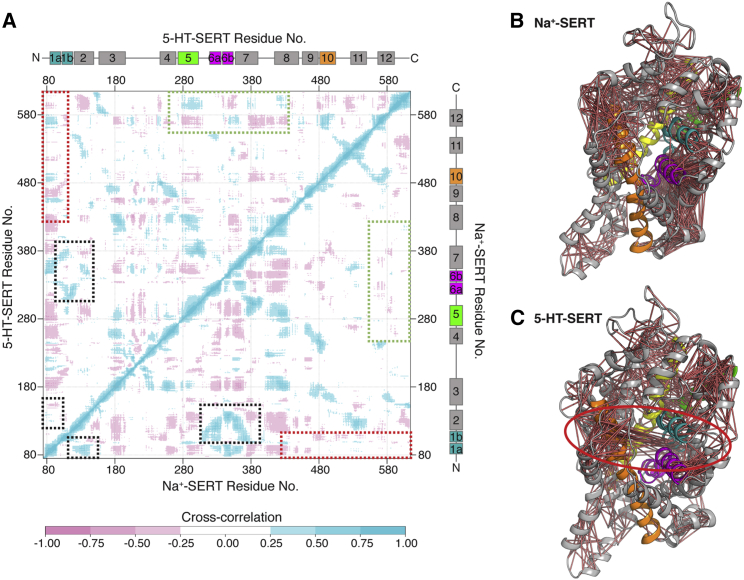
Cross-correlation analysis of Na+-SERT and 5-HT-SERT simulations. (A) The cross-correlation matrix depicts coupled motion between structural elements of the transporter. The upper left triangle of the matrix contains correlation values in 5-HT-SERT simulations, while the lower right triangle is from Na+-SERT simulations. The presence of the substrate increases the cooperative dynamics of the gating helices, specifically the motions of TM1a and TM10 (circled in red), while these motions are less pertinent in Na+-SERT. Additionally, greater correlated motions of TM12 with TM5 and TM7 are present with the addition of the substrate (circled in green), while motions of TM2 show higher correlations without substrate (circled in magenta). (B and C) Positive cross-correlation relationships between residues plotted as pale red lines on the SERT tertiary structure. The correlation of the gating helices in the presence of the substrate is circled in bold red in (C). To see this figure in color, go online.
Mechanism of 5-HT translocation down the exit pathway
The conformational free energy landscape suggests that structural isomerization to the IF state is limited by a large free energy barrier that is decreased in the presence of the substrate. The prolonged binding of 5-HT in subsite B weakens Na+ interactions in the Na2 site and results in its dissociation. Na+ loses its interaction with the backbone carbonyl of Leu434 and enters the intracellular vestibule, thereby initiating structural transitions from the OC to IF state. Koldsø et al. have shown that solvation of the intracellular vestibule allows Asp437 to rotate Na2 toward the intracellular pathway and facilitates transitions to the IF conformation (102). Similarly, we also observed that the rotameric shift in Asp437 initiates the dissociation of Na+ from the Na2 site (Fig. S37 A and B). The Na+ ion then interacts with Ser91 and is further stabilized by Phe347 and Phe440 via cation-π interactions (Fig. S37 C). Finally, the Na+ ion engages with Asp87 and diffuses into the intracellular space (Fig. S23 D). Koldsø et al. also predicted that Glu444 interacts with Na2 in the exit pathway, (102) yet, in our simulations, Glu444 forms an ionic interaction with Arg462 and does not interact with Na2. Simulations show that the release of the Na+ ion from the Na2 site increases the probability of opening the intracellular exit pathway (Table S1). Furthermore, as the intracellular pathway is formed, the interactions of TM1a and TM8 that coordinate the Na2 site are decoupled, further facilitating the outward motion of TM1a from TM5 and TM8. At this juncture, the intracellular gating residues (Arg79 and Asp452) still hold their hydrogen bond interactions, and Na+ can access the intracellular pathway without the breakage of ionic contacts. However, in Na+-SERT simulations, the Na2 ion has a tendency to rebind to the Na2 site after release (Fig. S38). Such intracellular binding events were not observed in 5-HT-SERT simulations. The coupling of 5-HT import to the cytoplasmic release of Na+ from the Na2 site explains the 1:1 sodium to neurotransmitter stoichiometry of the transport cycle.
The rotameric shift of Tyr95 results in permeation of 5-HT to the exit pathway (Fig. 6 C). The flipping of the indole ring of 5-HT displaces the ionic interactions with Asp98, resulting in the aromatic ring of 5-HT being positioned between Tyr95 and Val343. The downward movement of 5-HT disrupts contacts between TM1a and TM5, specifically the hydrogen bonding interactions between Asp87 and Trp282 (Fig. S20). The distance between Asp87 and Trp282 increases up to 16 Å, and thus drives the dissociation of 5-HT to the cytoplasmic half of the transporter. Additionally, the intracellular salt bridge network between Glu80-Lys279 and Arg79-Asp452 weakens as the cytoplasmic base of TM5 unwinds, further resulting in the opening of the intracellular pathway. 5-HT shifts to the intracellular vestibule and occupies the Na2 site (Fig. 6 C). The amine group forms strong polar contacts with residues in the Na2 site, and the indole ring is lodged between Tyr95, Phe347, and Phe440. 5-HT further diffuses down; however, the amine group of 5-HT still forms interactions with Tyr95, and the indole-NH forms additional interactions with Ser91. Finally, 5-HT leaves the transporter through a widened intracellular pathway surrounded by TM1a and TM5. Our results show that the rotation of Tyr95 propagates the opening of TM1a and mediates substrate transport to the cytoplasmic half. The conditional probability of an opened intracellular exit pathway given a sole event of either Na2 unbinding, Tyr95 rotating, TM5 unfolding, or 5-HT exiting the orthosteric pocket shows lower probabilities compared with when all listed events occur together. The probability of 50% for opening the intracellular pathway given these events supports our conclusions of a higher free energy barrier in order to transition to the IF state (Table S2). Computing the conditional probability of 5-HT exiting the orthosteric site shows lower probabilities when only Na2 is unbound or the intracellular exit pathway is opened, approximately 27% and 39%, respectively (Table S2). Compared with other conditions, 5-HT is more likely to exit the orthosteric pocket when Tyr95 is rotated, signifying the mechanistic importance of the Tyr95 in enabling 5-HT release. The probability of the intracellular release of 5-HT is further increased to 81% when the events of Na2 unbinding, Tyr95 flipping, TM5 unfolding, and the intracellular exit pathway opening are simultaneously occurring. The equivalent residue in hDAT, Phe76, has been shown to undergo a similar rotameric transition to allow for substrate release (40). It is well known that these regions play a crucial role in regulating SERT transport activity (84). We observed that the cytoplasmic base of TM5 unwinds and shifts outward by ∼8–10 Å to facilitate the cytoplasmic opening of the exit pathway (Fig. 6 D), as similarly seen in the IF SERT cryo-EM structure (51) and other NSS members (58,116).
Conclusions
In this study, we present an atomistic view of the substrate import process in SERT as well as characterizing the thermodynamics of key states involved in substrate transport. By implementing an MSM-based adaptive sampling protocol to sample the conformational landscape, we investigated global transitions from OF to IF for both 5-HT-free and 5-HT-importing SERT. MSM-weighted conformational free energy landscapes show the OF and OC states are relatively stable, and transitions to and from OF and OC states are relatively low energy. Transitions from OC to IF are substantially higher, with energy barriers of ∼3 kcal/mol in Na+-SERT; however, the presence of 5-HT not only lowers the free energy barrier of OF to OC transitions but further stabilizes SERT in the IF state compared with SERT in the absence of 5-HT. The dependence of the IF state on the presence of substrate has experimentally been observed for the bacterial LeuT transporter, where addition of extracellular substrate promotes dynamics at the intracellular gate (82).
The simulated structures show similar helix orientations at the extracellular and intracellular gates with respect to experimental SERT structures (23,45,47). The comparison of Na+ and 5-HT-SERT data reveal that the structural transitions from OF to OC involve minimal helix movements in TM1b, TM6a, and TM10, while OC to IF transitions show higher fluctuations of intracellular helix tips of TM1a, TM5, and TM7 to facilitate opening of the intracellular vestibule (Fig. S22). Biochemical experiments in conjunction with computational modeling have shown that cholesterol binding to a conserved site comprising TM1a, TM5, and TM7 inhibits these helix movements and may act as a regulatory mechanism for SERT function (47,117,118). Deviations were observed in the opening of the intracellular vestibule of the IF state between the cryo-EM and predicted MD structures, specifically the outward motion of TM1a. As such, the observed exit pathway is lined between TM1a and TM5, as opposed to an alternative pathway between TM1a and TM6b as proposed based on resolved IF structures (23,51,116). This might reflect the loss of lateral pressure following detergent extraction; the membrane is anticipated to constrain the extent to which TM1a can move away from the helical bundle. Nevertheless, our results show that partial opening of the intracellular pathway is sufficient for substrate transport. Experimental studies of LeuT also indicate that a partially open IF conformation is suitable for substrate transport (28).
Conducting simulations of both serotonin-free and serotonin-present conditions allows us to examine the finer details of the substrate-induced conformational transitions and the cooperative nature between the transporter and the substrate. Motions of TM2 were highly correlated with TM1b in Na+-SERT, whereas the binding of 5-HT decreases the cooperativity as TM1b rotates and closes the extracellular cavity to open the exit pathway (Fig. 7 A). The conformation dynamics examined through HDX studies reveal that TM2 undergoes less fluctuation when bound to Na+, in contrast to when the transporter is substrate bound (56). Similar trends were observed between TM6a and TM2; more specifically, the presence of 5-HT destabilizes this region and favors the conformational change required for substrate transport (Fig. 7 A). Gating residues Arg104-Glu493 remain open until the ions and 5-HT bind to the primary binding site, and later close the extracellular cavity as the exit pathway opens to allow the release of substrate molecules (Fig. S16). At the binding site, Tyr176 has the tendency to form a polar interaction with Asp98 in the OF and OC states. Interactions by the amine moiety of the substrate sever the Tyr176-Asp98 interaction, which is further disrupted when the opening of the intracellular cavity induces a rotameric shift in Tyr176 away from Asp98 (Fig. S32). Most significantly, the closure of the extracellular allosteric cavity is coupled to opening of the intracellular helix TM1a when serotonin is bound (Fig. 7 C). Experimental studies show that the substrate binding increases dynamics between EL4 and TM12 (24,56, 57, 58). In simulations, EL4 inserts deep inside the extracellular cavity, which is further facilitated by the movement of TM12 (Fig. S39). Furthermore, HDX studies on DAT support the coupled motions between TM7 and TM12 observed in simulations when the substrate is bound, compared with just Na+ binding (Fig. 7 A).
Our simulations show an ordered sequence of binding and transport events that agree with the 1:1 substrate:Na+ stoichiometry as previously characterized for the NSS family. First, Na+ ions favorably bind to the Na1 and Na2 sites, followed by 5-HT and Cl− ion to their respective binding sites. The binding of ions and the substrate triggers the closure of the extracellular permeation pathway and subsequent opening of the intracellular gating helices. Finally, the release of Na+ from the Na2 site decouples the interactions between TM1a, TM5, and TM8, increasing the solvation of the intracellular permeation pathway and allowing TM1a to open for substrate release. As unbiased MD simulations sample events with low free energy barriers, we have the highest likelihood of observing the most probable binding sequence of ions and substrate. However, we do not disregard that different sequences of ion and substrate binding events with higher associated free energy barriers are possible and may alter the kinetics of the SERT transport cycle. For example, as both Na+ and Cl− ions were present in the solution in Na+-SERT simulations, we observed the rapid diffusion and binding of Na+ ions to their binding sites instead of Cl− ion binding (Fig. S34). Simulations conducted by Szöllősi and Stockner showed how, if Cl− is initially bound to SERT, a stronger negative electrostatic potential is generated to allow more favorable Na+ binding compared with a completely apo transporter (119). From 5-HT-SERT simulations, Cl− entering the transporter is enabled by Na+ ions breaking the ionic interactions between Arg104 and Glu493 and further facilitated by the substrate in the orthosteric cavity. While the binding of 5-HT neutralizes the electronegative binding cavity, Cl− binding may be possible without the substrate but with higher free energy barriers. A recent study has shown that binding of Na+ and Cl− alone can trigger the closure of the extracellular pathway in the glycine transporter GlyT1b (120). Projection of the electrostatic potential of hGlyT1b shows the extracellular vestibule to be closer to neutral than hSERT, which may favor Cl− binding without the substrate (Fig. S40). Recent discussions regarding the role of the Cl− ion in SERT suggest that Cl− is not released into the intracellular space but remains bound to the transporter (121,122). Although SERT is capable of transporting 5-HT without Cl− present, the uptake rates are heavily diminished (123), possibly alluding to higher free energy barriers. Thus, Cl− could be considered a requirement to facilitate necessary substrate-ion coupling for substrate translocation. Nevertheless, the role of the Cl− ion and its implications on the stoichiometry, electroneutrality, ion-substrate coupling, and conformational dynamics highlight discrepancies in the current understanding of substrate transport in the NSS family.
This study has explored the conformational dynamics and substrate import of a monomeric unit of SERT; however, fluorescent microscopy has shown SERT to form functional oligomers in the membrane (124,125). The LeuT crystal structure has been resolved as a dimeric unit, and while recent modeling studies have explored possible dimeric SERT units (126), the exact interface of SERT oligomerization and the effects of coupled dynamics remain unclear. The set of simulations conducted in this study were initiated from the SERT crystal structure without the full-length N and C termini. Indeed, the cytoplasmic terminal domains are important elements of the NSS structure. Notably, protein-protein interactions of the SNARE protein syntaxin 1A and the N terminus are involved in regulating surface expression, transport activity, membrane trafficking, and phosphorylation of various NSS transporters (14,127, 128, 129, 130). In hDAT, phosphatidylinositol 4,5-biphosphate (PIP2) directly interacts with the N terminus to induce inward open conformations (54,131). Not only are the terminal residues highly diverse in sequence and length across the entire human NSS family but the lack of structural data of the termini presents inherent challenges to accurately initialize and validate a full-length SERT structural model. Nevertheless, Fenollar-Ferrer et al. combined de novo modeling with biophysical and spectroscopic techniques to provide initial insights into how the SERT N and C termini may structurally influence the conformational cycle and regulatory interaction partners (132). While truncation of the N-terminal residues retains serotonin transport function (133), the transport kinetics may be altered and therefore the rates calculated by the MSM may not represent exact in vivo conditions. Further investigation is required into understanding how in vivo regulation further affects the kinetics and conformational landscape of SERT.
Data availability
Representative structures from the MSM are available on Zenodo at https://zenodo.org/record/4707727. MD simulation dataset generated from this study are available from the corresponding author upon reasonable request.
Author contributions
D.S. designed the research. M.C.C. and B.S. performed simulations. All authors analyzed data. M.C.C., B.S., and H.J.Y. wrote the manuscript with input from E.P. and D.S.
Acknowledgments
This research is part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) in the State of Illinois, and, as of December, 2019, the National Geospatial-Intelligence Agency. Blue Waters is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications.
This work was funded by NSF MCB 18-45606 to D.S. E.P. was supported by R21 MH113155 from NIMH.
Editor: Lucie Delemotte.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.01.024.
Supporting material
References
- 1.Berger M., Gray J.A., Roth B.L. The expanded biology of serotonin. Annu. Rev. Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy D.L., Lerner A., et al. Lesch K.-P. Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol. Interv. 2004;4:109. doi: 10.1124/mi.4.2.8. [DOI] [PubMed] [Google Scholar]
- 3.Ni W., Watts S.W. 5-Hydroxytryptamine in the cardiovascular system: focus on the serotonin transporter (SERT) Clin. Exp. Pharmacol. Physiol. 2006;33:575–583. doi: 10.1111/j.1440-1681.2006.04410.x. [DOI] [PubMed] [Google Scholar]
- 4.Gether U., Andersen P.H., et al. Schousboe A. Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol. Sci. 2006;27:375–383. doi: 10.1016/j.tips.2006.05.003. http://www.sciencedirect.com/science/article/pii/S0165614706001271 [DOI] [PubMed] [Google Scholar]
- 5.Rudnick G., Wall S.C. The molecular mechanism of ”ecstasy” [3,4-methylenedioxy-methamphetamine (MDMA)]: serotonin transporters are targets for MDMA-induced serotonin release. Proc. Natl. Acad. Sci. U S A. 1992;89:1817–1821. doi: 10.1073/pnas.89.5.1817. https://www.pnas.org/content/89/5/1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson P.J., Rudnick G. The role of chloride ion in platelet serotonin transport. J. Biol. Chem. 1982;257:6151–6155. http://www.jbc.org/content/257/11/6151 [PubMed] [Google Scholar]
- 7.Rudnick G., Nelson P.J. Platelet 5-hydroxytryptamine transport, an electroneutral mechanism coupled to potassium. Biochemistry. 1978;17:4739–4742. doi: 10.1021/bi00615a021. [DOI] [PubMed] [Google Scholar]
- 8.Rudnick G. Active transport of 5-hydroxytryptamine by plasma membrane vesicles isolated from human blood platelets. J. Biol. Chem. 1977;252:2170–2174. [PubMed] [Google Scholar]
- 9.Hasenhuetl P.S., Freissmuth M., Sandtner W. Electrogenic binding of intracellular cations defines a kinetic decision point in the transport cycle of the human serotonin transporter. J. Biol. Chem. 2016;291:25864–25876. doi: 10.1074/jbc.M116.753319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson P.J., Rudnick G. Coupling between platelet 5-hydroxytryptamine and potassium transport. J. Biol. Chem. 1979;254:10084–10089. [PubMed] [Google Scholar]
- 11.Lin F., Lester H.A., Mager S. Single-channel currents produced by the serotonin transporter and analysis of a mutation affecting ion permeation. Biophys. J. 1996;71:3126–3135. doi: 10.1016/S0006-3495(96)79506-1. http://www.sciencedirect.com/science/article/pii/S0006349596795061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mager S., Min C., et al. Lester H.A. Conducting states of a mammalian serotonin transporter. Neuron. 1994;12:845–859. doi: 10.1016/0896-6273(94)90337-9. http://www.sciencedirect.com/science/article/pii/0896627394903379 [DOI] [PubMed] [Google Scholar]
- 13.Ruchala I., Cabra V., et al. Eltit J.M. Electrical coupling between the human serotonin transporter and voltage-gated Ca2+ channels. Cell Calcium. 2014;56:25–33. doi: 10.1016/j.ceca.2014.04.003. http://www.sciencedirect.com/science/article/pii/S0143416014000712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quick M.W. Regulating the conducting states of a mammalian serotonin transporter. Neuron. 2003;40:537–549. doi: 10.1016/s0896-6273(03)00605-6. http://www.sciencedirect.com/science/article/pii/S0896627303006056 [DOI] [PubMed] [Google Scholar]
- 15.Keyes S.R., Rudnick G. Coupling of transmembrane proton gradients to platelet serotonin transport. J. Biol. Chem. 1982;257:1172–1176. [PubMed] [Google Scholar]
- 16.Forrest L.R. Structural symmetry in membrane proteins. Annu. Rev. Biophys. 2015;44:311–337. doi: 10.1146/annurev-biophys-051013-023008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita A., Singh S.K., et al. Gouaux E. Crystal structure of a bacterial homologue of Na +/Cl- -dependent neurotransmitter transporters. Nature. 2005;437:215. doi: 10.1038/nature03978. https://www.nature.com/articles/nature03978 [DOI] [PubMed] [Google Scholar]
- 18.Chen J.-G., Rudnick G. Permeation and gating residues in serotonin transporter. Proc. Natl. Acad. Sci. U S A. 2000;97:1044–1049. doi: 10.1073/pnas.97.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quick M., Javitch J.A. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. U S A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180:134–136. doi: 10.1038/180134a0. http://www.nature.com/articles/180134a0 [DOI] [PubMed] [Google Scholar]
- 21.Forrest L.R., Zhang Y.-W., et al. Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. U S A. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. https://www.pnas.org/content/105/30/10338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969. doi: 10.1038/211969a0. https://www.nature.com/articles/211969a0 [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy H., Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. https://www.nature.com/articles/nature10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adhikary S., Deredge D.J., et al. Singh S.K. Conformational dynamics of a neurotransmitter:sodium symporter in a lipid bilayer. Proc. Natl. Acad. Sci. U S A. 2017;114:E1786–E1795. doi: 10.1073/pnas.1613293114. https://www.pnas.org/content/114/10/E1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gur M., Zomot E., et al. Bahar I. Energy landscape of LeuT from molecular simulations. J. Chem. Phys. 2015;143:243134. doi: 10.1063/1.4936133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L., Quick M., et al. Javitch J.A. The mechanism of a neurotransmitter:sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. http://www.sciencedirect.com/science/article/pii/S1097276508003596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S.K., Pal A. In: Methods in Enzymol. Shukla A.K., editor. Academic Press; 2015. Chapter nine - biophysical approaches to the study of LeuT, a prokaryotic homolog of neurotransmitter sodium symporters; pp. 167–198.http://www.sciencedirect.com/science/article/pii/S0076687915000038 volume 557 of Membrane Proteins–Engineering, Purification and Crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terry D.S., Kolster R.A., et al. Blanchard S.C. A partially-open inward-facing intermediate conformation of LeuT is associated with Na + release and substrate transport. Nat. Commun. 2018;9:230. doi: 10.1038/s41467-017-02202-y. https://www.nature.com/articles/s41467-017-02202-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claxton D.P., Quick M., et al. Mchaourab H.S. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 2010;17:822–829. doi: 10.1038/nsmb.1854. https://www.nature.com/articles/nsmb.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazmier K., Sharma S., et al. Mchaourab H.S. Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1. Proc. Natl. Acad. Sci. U S A. 2014;111:14752–14757. doi: 10.1073/pnas.1410431111. https://www.pnas.org/content/111/41/14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazmier K., Sharma S., et al. Mchaourab H.S. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat. Struct. Mol. Biol. 2014;21:472–479. doi: 10.1038/nsmb.2816. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4050370/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Focke P.J., Wang X., Larsson H.P. Neurotransmitter transporters: structure meets function. Structure. 2013;21:694–705. doi: 10.1016/j.str.2013.03.002. http://www.sciencedirect.com/science/article/pii/S0969212613000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaikh S.A., Tajkhorshid E. Modeling and dynamics of the inward-facing state of a Na+/Cl- dependent neurotransmitter transporter homologue. PLoS Comput. Biol. 2010;6:e1000905. doi: 10.1371/journal.pcbi.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y., Terry D., et al. Javitch J.A. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465:188–193. doi: 10.1038/nature09057. https://www.nature.com/articles/nature09057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng M., Bahar I. Coupled global and local changes direct substrate translocation by neurotransmitter-sodium symporter ortholog LeuT. Biophys. J. 2013;105:630–639. doi: 10.1016/j.bpj.2013.06.032. http://www.sciencedirect.com/science/article/pii/S0006349513007406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng M.H., Bahar I. Complete mapping of substrate translocation highlights the role of LeuT N-terminal segment in regulating transport cycle. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003879. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4191883/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen J., Stuhr-Hansen N., et al. Strømgaard K. Molecular determinants for selective recognition of antidepressants in the human serotonin and norepinephrine transporters. Proc. Natl. Acad. Sci. U S A. 2011;108:12137–12142. doi: 10.1073/pnas.1103060108. https://www.pnas.org/content/108/29/12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortensen O.V., Kortagere S. Designing modulators of monoamine transporters using virtual screening techniques. Front. Pharmacol. 2015;6 doi: 10.3389/fphar.2015.00223/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlessinger A., Geier E., et al. Sali A. Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc. Natl. Acad. Sci. U S A. 2011;108:15810–15815. doi: 10.1073/pnas.1106030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng M., Bahar I. Molecular mechanism of dopamine transport by human dopamine transporter. Structure. 2015;23:2171–2181. doi: 10.1016/j.str.2015.09.001. http://www.sciencedirect.com/science/article/pii/S0969212615003718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng M.H., Kaya C., Bahar I. Quantitative assessment of the energetics of dopamine translocation by human dopamine transporter. J. Phys. Chem. B. 2018;122:5336–5346. doi: 10.1021/acs.jpcb.7b10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razavi A.M., Khelashvili G., Weinstein H. A Markov state-based quantitative kinetic model of sodium release from the dopamine transporter. Sci. Rep. 2017;7:40076. doi: 10.1038/srep40076. https://www.nature.com/articles/srep40076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng M.H., Bahar I. Monoamine transporters: structure, intrinsic dynamics and allosteric regulation. Nat. Struct. Mol. Biol. 2019;26:545–556. doi: 10.1038/s41594-019-0253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shahsavar A., Stohler P., et al. Nissen P. Structural insights into the inhibition of glycine reuptake. Nature. 2021;591:677–681. doi: 10.1038/s41586-021-03274-z. [DOI] [PubMed] [Google Scholar]
- 45.Coleman J.A., Green E.M., Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532:334–339. doi: 10.1038/nature17629. https://www.nature.com/articles/nature17629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabrielsen M., Ravna A.W., et al. Sylte I. Substrate binding and translocation of the serotonin transporter studied by docking and molecular dynamics simulations. J. Mol. Model. 2012;18:1073–1085. doi: 10.1007/s00894-011-1133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penmatsa A., Wang K.H., Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. https://www.nature.com/articles/nature12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue W., Yang F., et al. Zhu F. What contributes to serotonin–norepinephrine reuptake inhibitors’ dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018;9:1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 49.Wang H., Goehring A., et al. Gouaux E. Structural basis for action by diverse antidepressants on biogenic amine transporters. Nature. 2013;503:141–145. doi: 10.1038/nature12648. https://www.nature.com/articles/nature12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coleman J.A., Gouaux E. Structural basis for recognition of diverse antidepressants by the human serotonin transporter. Nat. Struct. Mol. Biol. 2018;25:170. doi: 10.1038/s41594-018-0026-8. https://www.nature.com/articles/s41594-018-0026-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coleman J.A., Yang D., et al. Gouaux E. Serotonin transporter–ibogaine complexes illuminate mechanisms of inhibition and transport. Nature. 2019;569:141–145. doi: 10.1038/s41586-019-1135-1. http://www.nature.com/articles/s41586-019-1135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malinauskaite L., Quick M., et al. Nissen P. A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat. Struct. Mol. Biol. 2014;21:1006–1012. doi: 10.1038/nsmb.2894. https://www.nature.com/articles/nsmb.2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D., Gouaux E. Illumination of serotonin transporter mechanism and role of the allosteric site. Sci. Adv. 2021;7:eabl3857. doi: 10.1126/sciadv.abl3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khelashvili G., Stanley N., et al. Weinstein H. Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem. Neurosci. 2015;6:1825–1837. doi: 10.1021/acschemneuro.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kniazeff J., Shi L., et al. Gether U. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J. Biol. Chem. 2008;283:17691–17701. doi: 10.1074/jbc.M800475200. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2427322/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Möller I.R., Slivacka M., et al. Rand K.D. Conformational dynamics of the human serotonin transporter during substrate and drug binding. Nat. Commun. 2019;10:1687. doi: 10.1038/s41467-019-09675-z. https://www.nature.com/articles/s41467-019-09675-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen A.K., Möller I.R., et al. Loland C.J. Substrate-induced conformational dynamics of the dopamine transporter. Nat. Commun. 2019;10:2714. doi: 10.1038/s41467-019-10449-w. https://www.nature.com/articles/s41467-019-10449-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merkle P.S., Gotfryd K., et al. Rand K.D. Substrate-modulated unwinding of transmembrane helices in the NSS transporter LeuT. Sci. Adv. 2018;4:eaar6179. doi: 10.1126/sciadv.aar6179. https://advances.sciencemag.org/content/4/5/eaar6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsson M.H., Søndergaard C.R., et al. Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 60.Jo S., Kim T., et al. Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 61.Jorgensen W.L., Chandrasekhar J., et al. Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 62.Case D.A., Ben-Shalom I., et al. Kollman P.A. University of California; 2018. AMBER 2018. [Google Scholar]
- 63.Maier J.A., Martinez C., et al. Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Wolf R.M., et al. Case D.A. Development and testing of a general amber force field. J. Chem. Theory Comput. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 65.Gotz A.W., Williamson M.J., et al. Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salomon-Ferrer R., Gotz A.W., et al. Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 67.Le Grand S., Götz A.W., Walker R.C. SPFP: speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 68.York D.M., Darden T.A., Pedersen L.G. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the Ewald and truncated list methods. J. Chem. Phys. 1993;99:8345–8348. doi: 10.1063/1.465608. [DOI] [Google Scholar]
- 69.Kräutler V., Gunsteren W.F.V., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. https://onlinelibrary.wiley.com/doi/abs/10.1002/1096-987X%2820010415%2922%3A5%3C501%3A%3AAID-JCC1021%3E3.0.CO%3B2-V [Google Scholar]
- 70.Hopkins C.W., Le Grand S., et al. Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 71.Bowman G.R., Ensign D.L., Pande V.S. Enhanced modeling via network theory: adaptive sampling of Markov state models. J. Chem. Theory Comput. 2010;6:787–794. doi: 10.1021/ct900620b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selvam B., Mittal S., Shukla D. Free energy landscape of the complete transport cycle in a key bacterial transporter. ACS Cent. Sci. 2018;4:1146–1154. doi: 10.1021/acscentsci.8b00330. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6161048/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sculley D. Proceedings of the 19th International Conference on World Wide Web - WWW ’10. ACM Press; 2010. Web-scale k-means clustering; p. 1177.http://portal.acm.org/citation.cfm?doid=1772690.1772862 [Google Scholar]
- 74.Scherer M.K., Trendelkamp-Schroer B., et al. Noé F. PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 75.Naritomi Y., Fuchigami S. Slow dynamics in protein fluctuations revealed by time-structure based independent component analysis: the case of domain motions. J. Chem. Phys. 2011;134:02B617. doi: 10.1063/1.3554380. [DOI] [PubMed] [Google Scholar]
- 76.Prinz J.-H., Wu H., et al. Noé F. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 2011;134:174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 77.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. http://www.sciencedirect.com/science/article/pii/0263785596000185 [DOI] [PubMed] [Google Scholar]
- 78.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 79.McGibbon R., Beauchamp K., et al. Pande V. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. http://www.sciencedirect.com/science/article/pii/S0006349515008267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smart O.S., Neduvelil J.G., et al. Sansom M.S.P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. http://www.sciencedirect.com/science/article/pii/S026378559700009X [DOI] [PubMed] [Google Scholar]
- 81.Grant B.J., Rodrigues A.P., et al. Caves L.S. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 82.Zhao Y., Terry D.S., et al. Javitch J.A. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. https://www.nature.com/articles/nature09971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quick M., Yano H., et al. Javitch J.A. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter: sodium symporter from Fusobacterium nucleatum. J. Biol. Chem. 2006;281:26444–26454. doi: 10.1074/jbc.M602438200. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y.-W., Rudnick G. The cytoplasmic substrate permeation pathway of serotonin transporter. J. Biol. Chem. 2006;281:36213–36220. doi: 10.1074/jbc.M605468200. [DOI] [PubMed] [Google Scholar]
- 85.Bjerregaard H., Severinsen K., et al. Sinning S. A dualistic conformational response to substrate binding in the human serotonin transporter reveals a high affinity state for serotonin. J. Biol. Chem. 2015;290:7747–7755. doi: 10.1074/jbc.M114.573477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y.-W., Tavoulari S., et al. Rudnick G. Structural elements required for coupling ion and substrate transport in the neurotransmitter transporter homolog LeuT. Proc. Natl. Acad. Sci. U S A. 2018;115:E8854–E8862. doi: 10.1073/pnas.1716870115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J., Shaikh S.A., et al. Tajkhorshid E. Transient formation of water-conducting states in membrane transporters. Proc. Natl. Acad. Sci. U S A. 2013;110:7696–7701. doi: 10.1073/pnas.1218986110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun P., Li J., et al. Deng D. Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel. J. Biol. Chem. 2018 doi: 10.1074/jbc.RA118.003876. http://www.jbc.org/content/early/2018/10/17/jbc.RA118.003876 jbc.RA118.003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao Y., Jin X., et al. Zhou M. Crystal structure of a potassium ion transporter, TrkH. Nature. 2011;471:336–340. doi: 10.1038/nature09731. https://www.nature.com/articles/nature09731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Machtens J.-P., Kortzak D., et al. Fahlke C. Mechanisms of anion conduction by coupled glutamate transporters. Cell. 2015;160:542–553. doi: 10.1016/j.cell.2014.12.035. http://www.sciencedirect.com/science/article/pii/S0092867414016419 [DOI] [PubMed] [Google Scholar]
- 91.Billesbølle C.B., Mortensen J.S., et al. Loland C.J. Transition metal ion FRET uncovers K+ regulation of a neurotransmitter/sodium symporter. Nat. Commun. 2016;7:12755. doi: 10.1038/ncomms12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Korkhov V.M., Holy M., et al. Sitte H.H. The conserved glutamate (Glu136) in transmembrane domain 2 of the serotonin transporter is required for the conformational switch in the transport cycle. J. Biol. Chem. 2006;281:13439–13448. doi: 10.1074/jbc.M511382200. http://www.jbc.org/content/281/19/13439 [DOI] [PubMed] [Google Scholar]
- 93.Sen N., Shi L., et al. Javitch J.A. A pincer-like configuration of TM2 in the human dopamine transporter is responsible for indirect effects on cocaine binding. Neuropharmacology. 2005;49:780–790. doi: 10.1016/j.neuropharm.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 94.Chen N., Vaughan R.A., Reith M.E. The role of conserved tryptophan and acidic residues in the human dopamine transporter as characterized by site-directed mutagenesis. J. Neurochem. 2001;77:1116–1127. doi: 10.1046/j.1471-4159.2001.00312.x. [DOI] [PubMed] [Google Scholar]
- 95.Keshet G.I., Bendahan A., et al. Kanner B.I. Glutamate-101 is critical for the function of the sodium and chloride-coupled GABA transporter GAT-1. FEBS Lett. 1995;371:39–42. doi: 10.1016/0014-5793(95)00859-8. [DOI] [PubMed] [Google Scholar]
- 96.Sucic S., Paczkowski F.A., et al. Bryan-Lluka L.J. Functional significance of a highly conserved glutamate residue of the human noradrenaline transporter. J. Neurochem. 2002;81:344–354. doi: 10.1046/j.1471-4159.2002.00826.x. [DOI] [PubMed] [Google Scholar]
- 97.Forrest L.R., Tavoulari S., et al. Honig B. Identification of a chloride ion binding site in Na+/Cl–dependent transporters. Proc. Natl. Acad. Sci. U S A. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Subramanian N., Scopelitti A.J., et al. Vandenberg R.J. Identification of a 3rd Na+ binding site of the Glycine transporter, GlyT2. PLoS One. 2016;11:e0157583. doi: 10.1371/journal.pone.0157583. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0157583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benito-Muñoz C., Perona A., et al. López-Corcuera B. Modification of a putative third sodium site in the Glycine transporter GlyT2 influences the chloride dependence of substrate transport. Front. Mol. Neurosci. 2018;11:347. doi: 10.3389/fnmol.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang K.H., Penmatsa A., Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015;521:322–327. doi: 10.1038/nature14431. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4469479/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stolzenberg S., Li Z., et al. Shi L. The role of transmembrane segment 5 (TM5) in Na2 release and the conformational transition of neurotransmitter:sodium symporters toward the inward-open state. J. Biol. Chem. 2017;292:7372–7384. doi: 10.1074/jbc.M116.757153. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5418039/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koldsø H., Noer P., et al. Schiøtt B. Unbiased simulations reveal the inward-facing conformation of the human serotonin transporter and Na+ ion release. PLoS Comput. Biol. 2011;7:e1002246. doi: 10.1371/journal.pcbi.1002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khafizov K., Perez C., et al. Forrest L.R. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. Proc. Natl. Acad. Sci. U S A. 2012;109:E3035–E3044. doi: 10.1073/pnas.1209039109. https://www.pnas.org/content/109/44/E3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Felts B., Pramod A.B., et al. Henry L.K. The two Na+ sites in the human serotonin transporter play distinct roles in the ion coupling and electrogenicity of transport. J. Biol. Chem. 2014;289:1825–1840. doi: 10.1074/jbc.M113.504654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plenge P., Yang D., et al. Loland C.J. The antidepressant drug vilazodone is an allosteric inhibitor of the serotonin transporter. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-25363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andersen J., Stuhr-Hansen N., et al. Kristensen A.S. Molecular basis for selective serotonin reuptake inhibition by the antidepressant agent fluoxetine (Prozac) Mol. Pharmacol. 2014;85:703–714. doi: 10.1124/mol.113.091249. http://molpharm.aspetjournals.org/content/85/5/703 [DOI] [PubMed] [Google Scholar]
- 107.Sørensen L., Andersen J., et al. Kristensen A.S. Interaction of antidepressants with the serotonin and norepinephrine transporters: mutational studies of the S1 substrate binding pocket. J. Biol. Chem. 2012;287:43694–43707. doi: 10.1074/jbc.M112.342212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beuming T., Kniazeff J., et al. Loland C.J. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat. Neurosci. 2008;11:780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barker E.L., Moore K.R., et al. Blakely R.D. Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J. Neurosci. 1999;19:4705–4717. doi: 10.1523/JNEUROSCI.19-12-04705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Celik L., Sinning S., et al. Schiøtt B. Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation. J. Am. Chem. Soc. 2008;130:3853–3865. doi: 10.1021/ja076403h. [DOI] [PubMed] [Google Scholar]
- 111.Andersen J., Olsen L., et al. Kristensen A.S. Mutational mapping and modeling of the binding site for (S)-citalopram in the human serotonin transporter. J. Biol. Chem. 2010;285:2051–2063. doi: 10.1074/jbc.M109.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koldsø H., Severinsen K., et al. Sinning S. The two enantiomers of citalopram bind to the human serotonin transporter in reversed orientations. J. Am. Chem. Soc. 2010;132:1311–1322. doi: 10.1021/ja906923j. [DOI] [PubMed] [Google Scholar]
- 113.Barker E.L., Perlman M.A., et al. Blakely R.D. High affinity recognition of serotonin transporter antagonists defined by species-scanning mutagenesis an aromatic residue in transmembrane domain I dictates species-selective recognition of citalopram and mazindol. J. Biol. Chem. 1998;273:19459–19468. doi: 10.1074/jbc.273.31.19459. [DOI] [PubMed] [Google Scholar]
- 114.Jurrus E., Engel D., et al. Baker N.A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2017;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hellsberg E., Ecker G.F., et al. Forrest L.R. A structural model of the human serotonin transporter in an outward-occluded state. PLoS One. 2019;14:e0217377. doi: 10.1371/journal.pone.0217377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Malinauskaite L., Quick M., et al. Nissen P. A mechanism for intracellular release of Na+ by neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 2014;21:1006–1012. doi: 10.1038/nsmb.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeppelin T., Ladefoged L.K., et al. Schiøtt B. A direct interaction of cholesterol with the dopamine transporter prevents its out-to-inward transition. PLoS Comput. Biol. 2018;14:e1005907. doi: 10.1371/journal.pcbi.1005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laursen L., Severinsen K., et al. Sinning S. Cholesterol binding to a conserved site modulates the conformation, pharmacology, and transport kinetics of the human serotonin transporter. J. Biol. Chem. 2018;293:3510–3523. doi: 10.1074/jbc.M117.809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Szöllősi D., Stockner T. Investigating the mechanism of sodium binding to SERT using direct simulations. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.673782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y.-W., Uchendu S., et al. Rudnick G. Chloride-dependent conformational changes in the GlyT1 glycine transporter. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2017431118. e2017431118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rudnick G., Sandtner W. Serotonin transport in the 21st century. J. Gen. Physiol. 2019;151:1248–1264. doi: 10.1085/jgp.201812066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rudnick G. Forty four years with Baruch Kanner and the chloride ion. Neurochem. Res. 2021 doi: 10.1007/s11064-021-03330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Henry L.K., Iwamoto H., et al. Blakely R.D. A conserved asparagine residue in transmembrane segment 1 (TM1) of serotonin transporter dictates chloride-coupled neurotransmitter transport. J. Biol. Chem. 2011;286:30823–30836. doi: 10.1074/jbc.M111.250308. http://www.jbc.org/content/286/35/30823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Anderluh A., Klotzsch E., et al. Schütz G.J. Single molecule analysis reveals coexistence of stable serotonin transporter monomers and oligomers in the live cell plasma membrane. J. Biol. Chem. 2014;289:4387–4394. doi: 10.1074/jbc.M113.531632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anderluh A., Hofmaier T., et al. Schütz G.J. Direct PIP2 binding mediates stable oligomer formation of the serotonin transporter. Nat. Commun. 2017;8:14089. doi: 10.1038/ncomms14089. https://www.nature.com/articles/ncomms14089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Periole X., Zeppelin T., Schiøtt B. Dimer interface of the human serotonin transporter and effect of the membrane composition. Sci. Rep. 2018;8:5080. doi: 10.1038/s41598-018-22912-7. https://www.nature.com/articles/s41598-018-22912-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cervinski M., Foster J., Vaughan R. Syntaxin 1A regulates dopamine transporter activity, phosphorylation and surface expression. Neuroscience. 2010;170:408–416. doi: 10.1016/j.neuroscience.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deken S.L., Beckman M.L., et al. Quick M.W. Transport rates of GABA transporters: regulation by the N-terminal domain and syntaxin 1A. Nat. Neurosci. 2000;3:998–1003. doi: 10.1038/79939. [DOI] [PubMed] [Google Scholar]
- 129.Niambi Horton M.W.Q. Syntaxin 1A up-regulates GABA transporter expression by subcellular redistribution. Mol. Membr. Biol. 2001;18:39–44. doi: 10.1080/09687680010029383. [DOI] [PubMed] [Google Scholar]
- 130.Beckman M.L., Bernstein E.M., Quick M.W. Protein kinase C regulates the interaction between a GABA transporter and syntaxin 1A. J. Neurosci. 1998;18:6103–6112. doi: 10.1523/jneurosci.18-16-06103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hamilton P.J., Belovich A.N., et al. Galli A. PIP2 regulates psychostimulant behaviors through its interaction with a membrane protein. Nat. Chem. Biol. 2014;10:582–589. doi: 10.1038/nchembio.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fenollar-Ferrer C., Stockner T., et al. Forrest L.R. Structure and regulatory interactions of the cytoplasmic terminal domains of serotonin transporter. Biochemistry. 2014;53:5444–5460. doi: 10.1021/bi500637f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kern C., Erdem F.A., et al. Sucic S. The N terminus specifies the switch between transport modes of the human serotonin transporter. J. Biol. Chem. 2017;292:3603–3613. doi: 10.1074/jbc.m116.771360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Representative structures from the MSM are available on Zenodo at https://zenodo.org/record/4707727. MD simulation dataset generated from this study are available from the corresponding author upon reasonable request.