Abstract
Background
The COVID-19 pandemic affected healthcare systems worldwide. Predictive models developed by Artificial Intelligence (AI) and based on timely, centralized and standardized real world patient data could improve management of COVID-19 to achieve better clinical outcomes. The objectives of this manuscript are to describe the structure and technologies used to construct a COVID-19 Data Mart architecture and to present how a large hospital has tackled the challenge of supporting daily management of COVID-19 pandemic emergency, by creating a strong retrospective knowledge base, a real time environment and integrated information dashboard for daily practice and early identification of critical condition at patient level. This framework is also used as an informative, continuously enriched data lake, which is a base for several on-going predictive studies.
Methods
The information technology framework for clinical practice and research was described. It was developed using SAS Institute software analytics tool and SAS® Vyia® environment and Open-Source environment R ® and Python ® for fast prototyping and modeling. The included variables and the source extraction procedures were presented.
Results
The Data Mart covers a retrospective cohort of 5528 patients with SARS-CoV-2 infection. People who died were older, had more comorbidities, reported more frequently dyspnea at onset, had higher d-dimer, C-reactive protein and urea nitrogen. The dashboard was developed to support the management of COVID-19 patients at three levels: hospital, single ward and individual care level.
Interpretation
The COVID-19 Data Mart based on integration of a large collection of clinical data and an AI-based integrated framework has been developed, based on a set of automated procedures for data mining and retrieval, transformation and integration, and has been embedded in the clinical practice to help managing daily care. Benefits from the availability of a Data Mart include the opportunity to build predictive models with a machine learning approach to identify undescribed clinical phenotypes and to foster hospital networks. A real-time updated dashboard built from the Data Mart may represent a valid tool for a better knowledge of epidemiological and clinical features of COVID-19, especially when multiple waves are observed, as well as for epidemic and pandemic events of the same nature (e. g. with critical clinical conditions leading to severe pulmonary inflammation). Therefore, we believe the approach presented in this paper may find several applications in comparable situations even at region or state levels. Finally, models predicting the course of future waves or new pandemics could largely benefit from network of DataMarts.
1. Introduction
An unexpected rapid spread of SARS-CoV-2, the agent of the coronavirus disease 2019 (COVID-19), had been observed in China since January 2020, which resulted in a pandemic [1]. On January 5th 2022, more than 200 million people were found positive to SARS-CoV-2 globally and more than 5 million died [2].
Since the beginning, a relevant percentage of people with COVID-19 had clinical deterioration requiring hospitalization or intensive care admission and national health care systems were profoundly challenged in a very short time [3]. Hospitals have a critical role in the response to the pandemic [4]; however, many countries early saturated their intensive care bed capacities. During the different phases of the pandemic, Italy has been among the most impacted countries at global level.
Facing the challenge of the emergency, availability of a timely, centralized, standardized, and reliable patient dataset is of high priority. The development of diagnostic and prognostic predictive models through the application of advanced statistical modeling and machine learning techniques (Artificial Intelligence – AI) have the potential to improve patient outcomes [5], [6]. To name three main purposes of an AI framework from real-world data (RWD), COVID-19 archives may, at the same time, be used as a data classifier, as an approach to stratify patients into risk categories and as resource to train predictive tools. A real-time acquisition, centralization, and constant update of a COVID-19 Data Mart with information collected in healthcare systems of patients affected by COVID-19, and the availability of user-oriented data visualization tools, is a valuable source of information to support clinical practice and research on the pandemic.
The primary objective of this manuscript is to describe the structure and technologies used to construct the COVID-19 Data Mart architecture. The secondary objective is to present a pragmatic response to urgent needs due to the COVID-19 pandemic, particularly, creating a strong retrospective knowledge base, a real-time environment and integrated information dashboard for daily practice and early identification of critical condition at patient level.
2. Methods
A competence center named Generator, based on Data Integration, Analytics and AI was built at the Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy. The main purpose of the Generator program is to centralize and integrate previously decentralized, and heterogeneous (structured and unstructured) healthcare data, stored daily in the hospital's Data Warehouse (DWH) or archives of individual departments, using high-quality ontology-based systems and effective information technology (IT) procedures, while respecting data ownership and patient privacy. Generator has the goal to build standardized and structured archives (called Data Marts) for a specific area or disease to conduct research projects, quality assessments and to develop a rapid-learning framework through the daily feeding of data sources and the periodic re-train and validation of predictive models on new data [7], [8].
2.1. A COVID-19 real time analysis framework
An integrated framework for clinical practice and research for COVID-19 disease was implemented at the Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy, a 1450-bed hospital where, since March 1st, two Centers were dedicated to the care of patients with COVID-19: the Gemelli Hospital (FPG) and the Columbus Hospital (CIC) – in the highest peak moments of the spread of the disease, additional temporary care units were made available. The COVID-19 IT framework is available to all care units to serve three main goals:
-
•
A real-time Data Mart covering patients hospitalized at Policlinico Gemelli, refreshed on daily basis with new inpatients, discharged and deceased patients, update of clinical data for the ones still in charge. Thus, providing a patient-centered longitudinal view on the disease and treatment progression.
-
•
A library of Data Visualization dashboards, profiled for the different wards treating Covid-19 patients, showing the evolution of the cared cohort for the ward and for each patient on daily basis, and allowing to drill down the most critical cases with their key indicators of the disease status (including hospitalization history).
-
•
A set of AI-based modeling tools that currently support ongoing research studies from several clinical teams.
The framework was designed through the following steps a) the cross-disciplinary group of clinicians from all domains (Gemelli against Covid Group) identified the list of variables of interest sharing knowledge and evidences from the daily practice in treating COVID-19; the focus has been to capture the widest set of data to be able to follow all relevant clinical aspects in the context of a rapidly evolving scenario of the disease; b) IT specialists and data scientists have developed the interoperability procedures to extract from hospital clinical workflows the data variables as agreed with the clinician team (in structured and unstructured format) for all hospitalized and incoming patients; c) standard procedures have been developed for data treatment and the daily updated of the integrated Data Mart; these include natural language processing (NLP) algorithms to map medical reports into categorical variables, validation procedures for data quality and consistency, semantic control steps; d) Iterative design and development of profiled visualization dashboard for data analytics and daily exploitation from the clinical teams; e) lastly, the platform provides a series of tools for the development of predictive models directly used by clinicians in clinical practice through user-friendly interfaces.
The framework was developed using SAS Institute software analytics tool and SAS® Vyia® environment and Open-Source environment R ® and Python ® for fast prototyping and modeling.
2.2. Identification of variables of interest and data archives
As a result of this process of clinical variable selection and semantic control, the following variable groups have been identified to feed the Data Mart and support clinical activities: demographic, reverse-transcriptase polymerase chain reaction (RT-PCR) nasopharyngeal test for SARS-CoV-2, SARS-CoV-2 serology, respiratory isolated pathogens other than SARS-CoV-2, laboratory parameters at admission and during the hospitalization, comorbidities and treatments before the hospital admission, symptoms, arterial blood gasses parameters, respiratory support, therapies for COVID-19, anticoagulant therapies, other drugs, radiology findings, intensive care measures, complications during the hospitalization, length of hospitalization and outcomes (needs of oxygen therapy, mechanical ventilation and death)
The detailed description of the data available for each category is available in Table 1 in the online Appendix.
2.3. Data and source extraction procedures
The selected variables have been extracted from the corresponding data sources through the implementation of a standard extract, transform and load (ETL) procedure. This procedure has made it possible to integrate data from different applications, including data cleaning and standardization to the target structure – a relational database (the COVID-19 Data Mart) with a general structure able to support the daily practice and research activities. Where necessary, the procedures include a transformation step to transform unstructured information into useful structured data. Therefore, this ETL procedures consisted of several components as briefly described below and summarized in Fig. 1 .
-
1.
Daily update of the cohort of patients currently hospitalized, including only those patients who have carried out at least one positive RT-PCR nasopharyngeal test for SARS-CoV-2 and who have passed through one of the COVID dedicated wards in the hospital; patients discharged (full recovered or transferred to a pre-discharge structure) or deceased become part of the retrospective cohorts for statistical analysis and research studies.
-
2.
Daily extraction, validation, and Data Mart update for structured clinical variables (e. g. laboratory data) for each hospitalized patient (defined as ETL 1 + ETL 3 in Fig. 1); baseline data for patients just hospitalized were included in this step. In the case of a structured source, an identification code has been associated with each field. The codes used were referred to national and international standard such as: the International Classification of Disease (ICD) version 9 ICD9, Diagnosis Related Group Classification. Where none of these standards were available, specific coding for hospital legacy applications were used.
-
3.
Daily extraction, transformation, validation, and Data Mart update for unstructured clinical variables (e. g. text extracted from medical reports and converted into structured clinical data) for each patient (defined as ETL 2 + ETL 3 in Fig. 1). To obtain structural information from unstructured texts (such as clinical diary, radiology reports etc.) NLP algorithms have been applied, based on text mining procedures such as: sentences/words tokenization; rule-based approach supported by annotations defined by the clinical SMEs, and using semantic / syntactic corrections where necessary.
Fig. 1.
Generator infrastructure to create and automatically update the COVID-19 Data Marts from Operational Data Warehouses and Production Databases.
To ensure the development of a framework that respects privacy by design, specific procedures for pseudonymization have been included. The resulting COVID-19 Data Mart is a relational database, where the primary key is a patient identifier in pseudonymized form, that provides a longitudinal, comprehensive view of the disease status for each hospitalized patient allowing to drill down on symptoms, vital signs, oxygenation status, comorbidities. This is built on the base of chained ETL procedures on an incremental basis and includes robust and reliable error management through the creation of a Log file.
Once identified the sensitive data associated with each patient (such as hospital code and hospitalization code), the cryptographic hash function MD5 has been used for each identification code. The conversion table is saved into a dedicated and safe area for separation of duty and privacy reasons.
A simplified view of the relational database generated is shown as an example in Fig. 2 . Statistical differences between died and survived patients was evaluated by Pearson's chi-square for categorical variables and Mann-Whitney test for the numerical ones.
Fig. 2.
simplified view of patient-centered COVID-19 Data Mart in place at Policlinico Gemelli.
2.4. Ethical aspects
The Fondazione Policlinico Universitario A. Gemelli IRCCS Institutional Review Board approved the study protocol (IRB 3447).
3. Results
3.1. A COVID-19 data mart cohort description
As of January 5th 2022, the Data Mart covers a retrospective cohort of 5528 patients with SARS-CoV-2 infection. We excluded from the analysis 210 who were currently being hospitalized and under care.
Table 1 shows a summary view of the main characteristics of the retrospective cohort, on a subset of patients’ and clinical data selected from the larger set available in the Data Mart shown in Appendix. The data confirm several findings from previous research, e. g. people who died were older, had more comorbidities, were more frequently dyspnoic at onset, had higher d-dimer, C-reactive protein and urea nitrogen.
Table 1.
Characteristic of the study population as of January 5st 2022.
| Characteristics | All patients (N = 5528) | Alive (n = 4675) | Died (n = 853) | p-value | |
|---|---|---|---|---|---|
| Demographics | Age, median(SD) | 65.0 (19.0) | 62.0 (18.8) | 80.0 (11.8) | |
| Male | 3066 (55.5%) | 2566 (54.9%) | 500 (58.6%) | 0.02 | |
| BMI, median (IQR) | 25.9 (23.4; 28.7) | 25.9 (23.5; 28.7) | 25.7 (23.4; 28.4) | 0.1 | |
| Coexisting Conditions | Any | 3158 (57%) | 2507 (54%) | 651 (76%) | <0.01 |
| Current or Former Smoker | 108 (2.0%) | 101 (2.2%) | 7 (0.8%) | 0.01 | |
| Arteriopathy | 61 (1.1%) | 39 (0.8%) | 22 (2.6%) | <0.01 | |
| Chronic Liver Disease | 55 (1.0%) | 47 (1.0%) | 8 (0.9%) | 1 | |
| Cirrhosis | 46 (0.8%) | 33 (0.7%) | 13 (1.5%) | 0.02 | |
| Dyslipidemia | 373 (6.7%) | 310 (6.6%) | 63 (7.4%) | 0.4 | |
| HIV | 94 (1.7%) | 85 (1.8%) | 9 (1.1%) | 0.1 | |
| Myocardial Infarction | 665 (12.0%) | 481 (10.3%) | 184 (21.6%) | <0.01 | |
| Kidney Failure | 323 (5.8%) | 207 (4.4%) | 116 (13.6%) | <0.01 | |
| Hypertension | 2013 (36.4%) | 1628 (34.8%) | 385 (45.1%) | <0.01 | |
| Autoimmune Disease | 245 (4.4%) | 210 (4.5%) | 35 (4.1%) | 0.6 | |
| Hematologic Neoplasm | 87 (1.6%) | 60 (1.3%) | 27 (3.2%) | <0.01 | |
| Neurologic Impairment | 486 (8.8%) | 309 (6.6%) | 177 (20.8%) | <0.01 | |
| Pancreatitis | 37 (0.7%) | 28 (0.6%) | 9 (1.1%) | 0.2 | |
| Cardiovascular Pathology | 865 (15.6%) | 609 (13.0%) | 256 (30.0%) | <0.01 | |
| Lung Pathology | 531 (9.6%) | 372 (8.0%) | 159 (18.6%) | <0.01 | |
| Heart Failure | 245 (4.4%) | 148 (3.2%) | 97 (11.4%) | <0.01 | |
| Hepatic Ulcer | 106 (1.9%) | 69 (1.5%) | 37 (4.3%) | ||
| Symptoms At Admission | Any | 4102 (74.2%) | 3454 (73.9%) | 648 (76.0%) | 0.2 |
| Cough | 1523 (27.6%) | 1381 (29.5%) | 142 (16.6%) | <0.01 | |
| Dyspnea | 2535 (45.9%) | 2051 (43.9%) | 484 56.7%) | <0.01 | |
| Fever | 3393 (61.4%) | 2910 (62.2%) | 483 (56.6%) | <0.01 | |
| Nausea or Vomiting | 247 (4.5%) | 221 (4.7%) | 26 (3.0%) | 0.03 | |
| Diarrhea | 372 (6.7%) | 338 (7.2%) | 34 (4.0%) | <0.01 | |
| Time From Symptom Onset to Admission, median (IQR) | 8 (3; 366) | 9 (4; 366) | 5 (2; 11) | <0.01 | |
| Vital Signs on the Day of Admission, median (IQR) | Temperature, °C | 36.4 (36.0; 37.5) | 36.4 (36 0.0; 37.6) | 36.3 (36.0; 37.5) | 0.01 |
| Systolic Blood Pressure, mm Hg | 129.0 (115.0; 140.0) | 130.0 (116.0; 140.0) | 130.0 (116.0; 140.0) | <0.01 | |
| Laboratory Findings on the Day of Admission, median (IQR) | White Blood Cell Count, /µL | 7.4 (5.4; 10.4) | 7.3 (5.4; 10.1) | 8.4 (5.8; 12.0) | <0.01 |
| Lymphocyte Count, /µL | 1.1 (0.7; 1.5) | 1.1 (0.8; 1.5) | 0.9 (0.6; 1.3) | <0.01 | |
| Hemoglobin Level, g/dL | 13.4 (11.8; 14.7) | 13.5 (12.0; 14.8) | 12.6 (10.8; 14.0) | <0.01 | |
| Platelets, µL | 208.0 (161.0; 272.0) | 211.0 (164.0; 274.0) | 191.0 (142.0; 259.5) | <0.01 | |
| Creatinine Level, mg/dL | 0.9 (0.7; 1.1) | 0.8 (0.7; 1.1) | 1.1 (0.8; 1.8) | <0.01 | |
| D-dimer level, ng/mL | 846.0 (460.0; 1874.0) | 764.0 (427.0; 1620.0) | 1606.5 (794.5; 3686.0) | <0.01 | |
| C-reactive protein level, mg/L | 60.1 (23.9; 121.4) | 52.9 (21.1; 110.7) | 99.3 (50.2; 160.6) | <0.01 | |
| Urea Nitrogen, mg/dL | 18.0 (13.0; 27.0) | 17.0 (13.0; 23.0) | 30.0 (21.0; 47.0) | <0.01 | |
| Albumin, g/L | 32.0 (29.0; 35.0) | 33.0 (29.0; 36.0) | 29.0 (25.0; 32.0) | <0.01 | |
| Vitamin D, ng/mL | 17.1 (12.1; 27.7) | 17.3 (12.4; 27.7) | 15.2 (8.6; 27.3) | 0.2 | |
| P/F | 232.1 (152.8; 333.3) | 247.5 (162.9; 340.0) | 163.3 (109.5; 266.7) | <0.01 | |
Leveraging the wealth of information available from the data mart updated on daily basis, we are currently analyzing in detail a variety of correlation index which are pre-requisite to build accurate predictors for critical outcome.
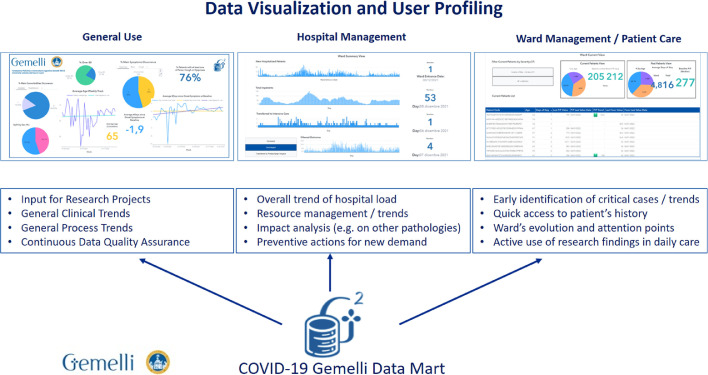
3.2. Data visualization utilities and process view
With the COVID-19 Data Mart online, the team has developed an extensive library of visualization dashboards, using SAS® Vyia® functionalities, to enable information at bedside for the clinical teams engaged in the daily care of infected patients. These dashboards cover cumulative views for hospital management level, ward management and patient care level. A conceptual view on how this is exploited is shown in Fig. 3 .
Fig. 3.
Conceptual view of Dashboard profiling for General Use, Hospital Management, Ward and Patient Care.
3.3. Dashboard views: hospital management level
Figs. 4A and 4B show two examples of what is available online for the hospital care management in terms of day-to-day view of the inpatients, discharged, intensive care unit and other wards’ loads, as well as evolution of outcomes at overall hospital level.
Fig. 4.
A. Weekly retrospective overview of cohort, split by wards, compared with non-COVID-19 inpatients evolution. Fig. 4B Daily view of new hospitalized patients, total patients in charge, filtered view of outcomes.
Some of the dashboard views show median age of hospitalized patients and comparison among COVID-19 waves, the evolution of oxygenation parameters such as PO2/FiO2 ratio (P/F) for the arterial blood gasses at admission and during the hospitalization course, the average length of hospitalization or the rate and timing of admission to Intensive Care. This can help understanding several factors influencing the progression of the disease and comparative view of the different waves, which is relevant given that COVID-19 has shown mutable features in different stages.
Figs. 5A to 5C show another set of cumulative dashboards, showing the overall cohort (retrospective, discharged patients and currently patients) with focus on evolution of clinical parameters. Dashboard 5A is focused on providing one integrated snapshot of the evolution of main clinical features along the timeline of the disease spread (wave 1, wave 2). It includes average age of inpatients overtime, impact of comorbidities, symptoms etc. This dashboard also focuses how the number of days for symptoms onset evolves on average. Dashboard 5B-5C show how this integrated framework can be used to investigate to which extent different clinical parameters at baseline can translate into early predictors and provide real-time comparison for the most recently hospitalized patients (examples shown cover d-dimer and IL-6 but the same are available for all critical laboratory data). These dashboards are useful to explore in real-time the possible correlations among included variables and clinical outcomes (Fig. 6).
Fig. 5.
A. Cumulative clinical dashboard displaying incidence of comorbidities, average age for inpatients over time, symptoms and evolution of average days of symptoms onset. Fig. 5B Drill down for the historical cohort of COVID-19 patients with evolution over time of IL-6 at baseline and comparison with most recent hospitalized patients. Fig. 5C. Same dashboard as 5B for D-Dmer.
Fig. 6.
A. Summary ward dashboard: newly hospitalized, total in charge, transfer to ICU, split by outcome. Fig. 6B Drill-down at war: historical compare with current average PaO2/FiO2 ratio and length of stay; list of patients with alert flag for most critical cases. Fig. 6C. Patient drill-down dashboard example, providing history of PaO2/FiO2 ratio and fever, last outcome of TC, swab sequence and days from symptom onset.
3.4. Dashboard views: ward management level
A real-time situation for each ward (number of patients, severity scoring, new admissions, number of patients who died or who have been transferred to Intensive Care Unit the day before) are also available to support the daily management. Dashboards 6A-6C show the views that are made available on daily basis at ward level to support their overall management and prioritization:
The view in 6A aims at giving in one-page a summary of the situation of the ward with respect to the COVID-19 trend i.e., newly hospitalized patients, total patients in charge, number of patients transferred to intensive care unit (ICU). The dashboard allows to also filter the discharged patients by outcomes (recovery; transfer to a pre-discharge facility; death). Views 6B-6C are designed to support clinicians at bedside in their daily activity: 6B gives an overall summary of hospitalized patients. Besides providing compare for key average parameters (such as P/F) to have a one-shot summary of how critical the ward situation is, more relevant is the list of hospitalized patients with a high-level scoring of their severity condition, in terms of age, days from first symptom onset, length of stay, current P/F and the P/F trend.
3.5. Dashboard views: patient management level
In dashboard 6C a detail for a specific patient is provided. This includes the history and the trend of both P/F and fever along the hospitalization period; result of RT-PCR nasopharyngeal test; features of most recent chest X-ray or chest computed tomography (CT).
4. Discussion
The burden of the COVID-19 pandemic on the daily life of people and the impact on healthcare systems all over the world are still impressive. The high number of people with COVID-19 admitted to emergency rooms, general wards and ICUs critically stressed hospitals [9]. Preparedness for the pandemic has been largely suboptimal [10]. In particular, the onset of several multiple waves of pandemic, with continuously mutable condition, is an additional challenge that requires flexible and comprehensive tools for data analysis and understanding. In fact, clinical and epidemiological data may be significantly different among patients from the different waves and therefore healthcare needs may vary even in very short time. Among strategies to respond to a pandemic such as that caused by SARS-CoV-2, we experienced the need to evolve from manual data sharing towards building a health data infrastructure (the so-called “health data superhighway”) [11] that facilitates automatic, interoperable data exchange and use. The potential insights provided from a very comprehensive Data Mart integrating clinical data and RWD for COVID-19 patients along with the whole natural history of the disease combined with AI methods is significant.
We presented basic assumptions and the description of a Data Mart architecture developed at the Fondazione Policlinico Universitario A. Gemelli IRCCS based on the Generator infrastructure just after the onset of the SARS-CoV-2 pandemic. An extensive amount of data was quickly available for data monitoring, data analysis and clustering. Variables collected and integrated with the Data Mart are those commonly collected in daily practice [12], [13], [14], [15], [16], [17], [18] and already available in electronic medical records (EMRs); the Data Mart can be augmented with multi-dimensional or external data. Details on technical aspects and a list of variables included in the COVID-19 Data Mart were shared to ensure reproducibility.
The dashboard provides several functions. At hospital level, it covers cumulative views, provide a visual trend of evolution of critical parameters in the different waves, gives a snapshot of the evolution of main clinical features overtime, allows the knowledge in real-time of total number of inpatients in charge, discharged patients, those admitted and transferred in ICU and other wards’ loads, as well as evolution of outcomes at hospital level. Moreover, the dashboard allows to investigate early predictors of unfavorable outcomes and provide real-time comparison of clinical characteristics and laboratory parameters for the most recently hospitalized patients.
At the ward level, the dashboard gives information listing hospitalized patients with a high-level scoring of their severity condition, older, with longer hospitalization stay, the current P/F and its trend.
Finally, at the patient level, the dashboard provides a capture for a specific patient including days from symptom onset, evolution of P/F and fever along the hospitalization period, results of polymerase chain reaction–positive nasopharyngeal tests and most recent chest X-ray or CT.
Many benefits are stemming from the availability of a Data Mart focused on COVID-19. First of all, this wide knowledge base can be exploited to build diagnostic and prognostic predictive models. Such advanced predictive models may be a great support to the most impacted wards in early alerting of critical evolution and most severe outcomes. The identification of the individual risk for each patient can also ease a more personalized approach. From a clinical point of view, such a tool may enhance the early implementation of supporting and therapeutic measures, to differentiate levels of needed healthcare resources (low, medium or high intensity) or may ease the identification of predictors of chronic lung damage in the follow-up. Moreover, different combinations of clinical variables and features may cluster into previously undescribed phenotypes and define different risks for poor outcomes. From a public health point of view, the prediction model may support optimal resource use. For instance, predictive models identify clinical criteria and laboratory values to safely allocate a person to common wards or to be discharged at home or to be de-isolated when probability of a COVID-19 diagnosis is poor [19]. This may result in saving hospital resources (beds, nurse and physician staff, personal protective equipment, disinfectant materials). In addition, we are currently working to connect this approach with tools for early prediction of critical evolution within the practice of base medicine and territory health management. In this regard, a hospital-based Data Mart may also be integrated with patient self-reported or personal data (through addressed App or IoT) to improve the diagnosis or to identify specific pattern of onset, progression or recovery for COVID-19.
Of course, machine learning techniques to personalize treatment plans are not peculiar to COVID-19: machine learning has previously been used to improve diagnostic algorithms [20], [21], predicting outcomes for patients with different diseases [22] or conditions [23]. Building a Data Mart for patients with COVID-19 may be reproduced for other clinical and epidemiological scenarios.
Our framework has possible limitations. First, currently only routinely available clinical data of electronic health records are used. A generalization of the rules used to apply text mining techniques, based on the recognition of reports drawn up in national language would be necessary. An external and international ontology validation is mandatory and finally this workflow has only been implemented and used by a single hospital.
In conclusion, a large Data Mart, including numerous structured and unstructured variables, gives the opportunity to realize a real-world, readily available, interactive dashboard and to build sophisticated and advanced predictive models [24]. Networks and pan-cohorts promoting collaboration across health centers, disciplines, and institutions represent crucial instruments to respond to pandemics or global health events. Therefore, complex integration of a large volume of clinical, radiological and laboratory data in an advanced architecture could be useful to quickly and reliably test new predictive models or therapeutic agents active against SARS-CoV-2 or innovative regimens.
5. Conclusion
The experience that can be produced by the application and exploitation of a COVID DataMart can be paradigmatic for a wider application such as that of an entire region or state. Lastly, models predicting the course of future waves or new pandemics could largely benefit from dataMart networks like the one presented here.
Author contributions
R.M., C.M., S.P. and V.V conceived of the presented idea and drafted the manuscript.
C.M., S.P., A.D., A.M., P.D.A.S. extracted and analysed the data.
All other authors contributed equally, discussed the results and concurred to the final manuscript.
Declarations of Competing Interest
None.
Acknowledgment
We are grateful to all the healthcare workers of SITRA (Servizio Infermieristico, Tecnico e Riabilitativo Aziendale del Policlinico Gemelli). We wish to thank Franziska Lohmeyer for her English language assistance. We also want to acknowledge the professional services support of SAS Analytics ® team who was instrumental for the Data Mart build, automated procedures, quality assurance and semantic consistency process.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpb.2022.106655.
Appendix. Supplementary materials
References
- 1.WHO Director-General's opening remarks at the media briefing on COVID19, March 11th 2020.
- 2.http://opendatadpc.maps.arcgis.com/apps/opsdashboard/index.html#/b0c68bce2cce478eaac82fe38d4138b1.
- 3.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395(10231):1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adalja A.A., Toner E., Inglesby T.V. Priorities for the US Health Community Responding to COVID-19. JAMA. 2020;323(14):1343–1344. doi: 10.1001/jama.2020.3413. [DOI] [PubMed] [Google Scholar]
- 5.Raita Y., Goto T., Faridi M.K., Brown D.F.M., Camargo C.A., Jr., Hasegawa K. Emergency department triage prediction of clinical outcomes using machine learning models. Crit. Care. 2019;23(1):64. doi: 10.1186/s13054-019-2351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdick H., Pino E., Gabel-Comeau D., et al. Effect of a sepsis prediction algorithm on patient mortality, length of stay and readmission: a prospective multicentre clinical outcomes evaluation of real-world patient data from US hospitals. BMJ Health Care Inform. 2020;27(1) doi: 10.1136/bmjhci-2019-100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambin P., Roelofs E., Reymen B., et al. Rapid Learning health care in oncology–an approach towards decision support systems enabling customised radiotherapy. Radiother. Oncol. 2013;109:159–164. doi: 10.1016/j.radonc.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Meldolesi E., van Soest J., Dinapoli N., et al. An umbrella protocol for standardized data collection (SDC) in rectal cancer: a prospective uniform naming and procedure convention to support personalized medicine. Radiother. Oncol. 2014;112:59–62. doi: 10.1016/j.radonc.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z., McGoogan J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, doi: 10.1001/jama.2020.2648. [DOI] [PubMed]
- 10.Smith N., Fraser M. Straining the System: novel Coronavirus (COVID-19) and Preparedness for Concomitant Disasters. Am. J. Public Health. 2020;110(5):648–649. doi: 10.2105/AJPH.2020.305618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Council of State and Territorial Epidemiologists. Driving Public Health in the Fast Lane: The Urgent Need for a 21st Century Data Superhighway. Available at http://resources.cste.org/datasuperhighway/mobile/index.html. Accessed May 21, 2020.
- 12.Liu F., Li L., Xu M., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R., Liang W., Jiang M., et al. Risk Factors of Fatal Outcome in Hospitalized Subjects With Coronavirus Disease 2019 From a Nationwide Analysis in China. Chest. 2020;S0012-3692(20) doi: 10.1016/j.chest.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassford C.R., Krucien N., Ryan M., et al. U.K. Intensivists’ Preferences for Patient Admission to ICU: evidence From a Choice Experiment. Crit. Care Med. 2019;47(11):1522–1530. doi: 10.1097/CCM.0000000000003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X., Yu Y., Xu J., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasselli G., Zangrillo A., Zanella A., et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seymour C.W., Kennedy J.N., Wang S., et al. Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis. JAMA. 2019;321(20):2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murri R., Lenkowicz J., Masciocchi C., et al. A machine-learning parsimonious multivariable predictive model of mortality risk in patients with Covid-19. Sci. Rep. 2021;11:21136. doi: 10.1038/s41598-021-99905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P., Berzin T.M., Glissen Brown J.R., et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68(10):1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehteshami Bejnordi B., Veta M., Johannes van Diest P., et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poplin R., Varadarajan A.V., Blumer K., et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018;2:158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 23.Guedalia J., Lipschuetz M., Novoselsky Persky M., et al. Real-time data analysis using a machine learning model significantly improves prediction of successful vaginal deliveries. Am. J. Obstet. Gynecol. 2020;S0002-9378(20) doi: 10.1016/j.ajog.2020.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Damiani A., Masciocchi C., Lenkowicz J. et al. Building an Artificial Intelligence Laboratory Based on Real World Data: the Experience of Gemelli Generator. Front Comput Sci in press. doi: 10.3389/fcomp.2021.768266, 2022. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.