Abstract
Xylanases are the enzymes that catalyze the breakdown of the main hemicellulose present in plant cell walls. They have attracted attention due to their biotechnological potential for the preparation of industrially interesting products from lignocellulose. While many xylanases have been characterized from bacteria and filamentous fungi, information on yeast xylanases is scarce and no yeast xylanase belonging to glycoside hydrolase (GH) family 30 has been described so far. Here, we cloned, expressed and characterized GH30 xylanase SlXyn30A from the yeast Sugiyamaella lignohabitans. The enzyme is active on glucuronoxylan (8.4 U/mg) and rhodymenan (linear β-1,4-1,3-xylan) (3.1 U/mg) while its activity on arabinoxylan is very low (0.03 U/mg). From glucuronoxylan SlXyn30A releases a series of acidic xylooligosaccharides of general formula MeGlcA2Xyln. These products, which are typical for GH30-specific glucuronoxylanases, are subsequently shortened at the non-reducing end, from which xylobiose moieties are liberated. Xylobiohydrolase activity was also observed during the hydrolysis of various xylooligosaccharides. SlXyn30A thus expands the group of glucuronoxylanases/xylobiohydrolases which has been hitherto represented only by several fungal GH30-7 members.
Keywords: xylanase, glucuronoxylanase, xylobiohydrolase, xylan, glycoside hydrolase family 30, GH30-7 subfamily, Sugiyamaella lignohabitans, yeast
1. Introduction
Endo-β-1,4-xylanases (EXs, EC 3.2.1.8) are main xylan depolymerizing enzymes cleaving the polysaccharide backbone to xylooligosaccharides (XOs) of various lengths. Most EXs are classified in glycoside hydrolase (GH) families 10 and 11 (www.cazy.org, accessed on 1 December 2021) [1]; however, they are also found in GH families 5, 8, 30, 43, 98 and 141. During the last few years, great attention has been paid to the EXs from GH30 family [2,3,4,5,6,7,8,9,10,11,12,13]. Prokaryotic EXs are grouped into subfamily GH30-8, while eukaryotic xylanases are members of GH30-7 subfamily. Catalytic properties of the GH30-8 subfamily enzymes are quite uniform and most of them are specific glucuronoxylanases (EC 3.2.1.136) requiring glucuronic or 4-O-methyl-glucuronic acid (MeGlcA) substitution of the main chain for their action [13,14,15]. On the other hand, catalytic properties of the GH30-7 subfamily representatives are diverse and include specific glucuronoxylanases, xylobiohydrolases, non-specific endoxylanases, endoxylanases/xylobiohydrolases and xylanases releasing xylose from the reducing end of the substrate [11,12]. All characterized GH30-7 xylanases come from filamentous fungi and so far no yeast GH30 EX has been described.
Compared to bacterial and fungal xylanases, yeast xylanases have not been studied so extensively, presumably due to much lower levels of xylanase production by the yeasts. The best characterized yeast xylanases are from yeast-like fungus Aureobasidium pullulans [16,17] and from several Cryptococcus species [18,19,20], but EXs were purified and characterized also from Pichia (Scheffersomyces), Pseudozyma and Blastobotrys genera [21,22,23]. All of them belong to either the GH10 or GH11 family. Bioinformatic mining in 332 yeast genomes from the phylum Ascomycota has revealed only a few putative yeast xylanases: one from the GH11 family (Blastobotrys mokoenii), five from the GH10 family (two from Spencermartinsiella europaea, two from Sugiyamaella lignohabitans and one from B. peoriensis) and three from the GH30-7 subfamily (Sp. europaea, Su lignohabitans, B. mokoenii) [23].
The genus Sugiyamaella, which belongs to the family Trichomonascaceae, was established by Kurtzman and Robnett [24]. The members of Sugiyamaella clade are mostly found in a wood environment. Sugiyamaella (Candida) lignohabitans strains were isolated from tenebrionid beetles inhabiting a rotten log or from decayed wood [25]. A total of 16 yeast isolates belonging to the genus Sugiyamaella recovered from rotting wood and sugarcane bagasse samples in different Brazilian regions were studied in relation to d-xylose fermentation, xylitol production, and xylanase activities [26]. All of them exhibited xylanase activity and almost all were able to produce ethanol and xylitol. Among them, S. lignohabitans exhibited the highest xylanase activity and the best xylitol yield and productivity after 24 h [26]. This yeast has been also described as a host for the production of organic acids from lignocellulosic hydrolysates [27]. Yeast strains able to degrade polysaccharides and simultaneously convert their constituents to desired products are interesting for possible applications in lignocellulosic biorefining.
To expand the knowledge on yeast xylanases, we have cloned, expressed and characterized GH30 xylanase SlXyn30A from S. lignohabitans. It was found to be a glucuronoxylanase with auxiliary xylobiohydrolase activity.
2. Results
2.1. Sequence Analysis of SlXyn30A
SlXyn30A (GenBank: ANB12318) is composed of 470 amino acids, including signal peptide which is 16 amino acids long. The mature protein has a theoretical molecular mass of 49,539 Da. BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 2 November 2021) showed the highest similarity (68.4%, identity 53.7%) to a putative GH30 xylanase from Lineolata rhizophorae (GenBank: KAF2460202.1). Of the characterized GH30 enzymes, SlXyn30A showed the highest similarity (65.9%) and identity (47.7%) to xylanase C from Talaromyces purpureogenus, but other hits showed a similar level of homology and identity: glucuronoxylanase/xylobiohydrolase TtXyn30A from Thermothelomyces thermophila (65.5% similarity, 47.5% identity), glucuronoxylanase/xylobiohydrolase TcXyn30B from Talaromyces cellulolyticus (63.4% similarity, 42.1% identity), xylanase XylD from Bispora (61.9% similarity, 42.7% identity), and glucuronoxylanase TrXynVI from Trichoderma reesei (59.7% similarity, 44.3% identity).
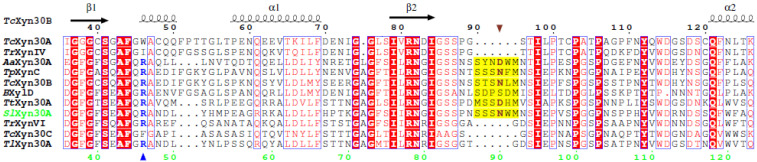
Based on amino acid sequence alignment (Figure S1, Supplementary Materials), two catalytic residues were identified: Glu199 as an acid/base and Glu292 as a nucleophile. The alignment also revealed that SlXyn30A displays structural features of the GH30-7 enzymes including a longer β2-α2 loop, a lack of α6-helix, and a presence of β-strands β8A and β8B [12]. Moreover, SlXyn30A contains several structural elements typical for GH30-7 glucuronoxylanases/xylobiohydrolases (Figure 1). The first is a presence of Arg46 which was suggested to be responsible for a MeGlcA recognition and glucuronoxylanase activity of TcXyn30B [4]. Although the 3D structure of another GH30-7 glucuronoxylanase, TtXyn30A, did not confirm such a role of the arginine, mutational studies of TtXyn30A indicated its importance [9]. The second structural feature considered to be responsible for xylobiohydrolase activity is the length and amino acid sequence of the β2-α2 loop. This loop is of the same length in SlXyn30A, TcXyn30B and TtXyn30A, and is longer than in other GH30-7 enzymes (e.g., TrXynVI; Figure 1). Asn93 in TcXyn3B and Asp78 in TtXyn30A in this loop were shown to interact with Xylp residue accommodated in the −2a subsite [4,9]. The corresponding residue in SlXyn30A is Asn90 which may play a similar role.
Figure 1.
Amino acid sequence alignment of SlXyn30A with GH30-7 xylanases Talaromyces cellulolyticus TcXyn30B (GAM36763), TcXyn30C (GAM40414.1), TcXyn30A (GAM43270), Thermothelomyces thermophila TtXyn30A (AEO55025), Talaromyces purpureogenus (Penicillium purpurogenum) TpXynC (AKH40280), Bispora sp. BXylD (ADG62369.1), Trichoderma reesei TrXynVI (EGR45006.1), T. reesei TrXynIV (AAP64786.1), Acremonium alcalophilum AaXyn30A [7], and Talaromyces leycettanus TlXyn30A [10]. The secondary structure elements and numbering of TcXyn30B are shown on top, numbering of SlXyn30A is at the bottom. Arginine suggested to be responsible for the recognition of MeGlcA substitution is shown in blue and is marked by a blue up triangle. Longer β2-α2 loop in xylobiohydrolases is highlighted in yellow and the residue interacting with Xylp moiety occupying the −2a subsite is brown and marked by a brown down triangle.
2.2. Recombinant Strain Selection
Four clones with integrated pPICZαA vector carrying the Slxyn30A gene were selected from the ZeocinTM (250 mg/L) plate. The clones were individually cultivated in shake flasks, and after 120 h of induction, supernatant was screened by SDS-PAGE electrophoresis for the presence of 50 kDa protein which should correspond to a mature enzyme. The expected protein was confirmed in three transformants (Figure S2). The molecular mass of the enzyme was, however, a little bit higher (58 kDa), presumably due to glycosylation. According to NetNGlyc server, the amino acid sequence of SlXyn30A contains 4 potential N-glycosylation sites (N86, N114, N252, N306). The first one is conserved in TcXyn30B where is actually glycosylated [2]. Three transformants were then tested for xylanase activity and the transformant with the highest activity (transformant 4) was used for the determination of the catalytic properties of SlXyn30A.
2.3. Thermal and pH Optima and Stability
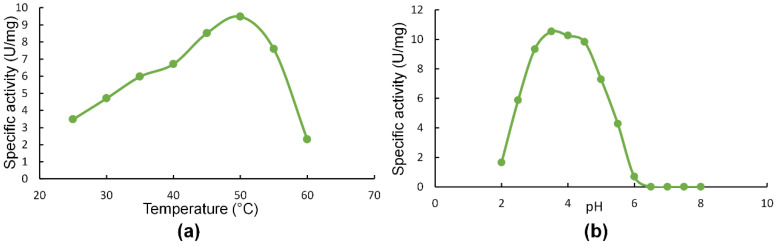
SlXyn30A showed a temperature optimum of 50 °C, showing 71% and 24% of maximal activity at 40 °C and 60 °C, respectively (Figure 2a). pH optimum was around 3.5 and the enzyme was active in acidic range of pH, keeping only 6.6% of the maximal activity at pH 6 (Figure 2b). The enzyme was stable at temperatures up to 50 °C, while at 60 °C it completely lost its activity within 30 min.
Figure 2.
Effect of temperature (a) and pH (b) on activity of SlXyn30A.
2.4. Hydrolysis of Polysaccharides
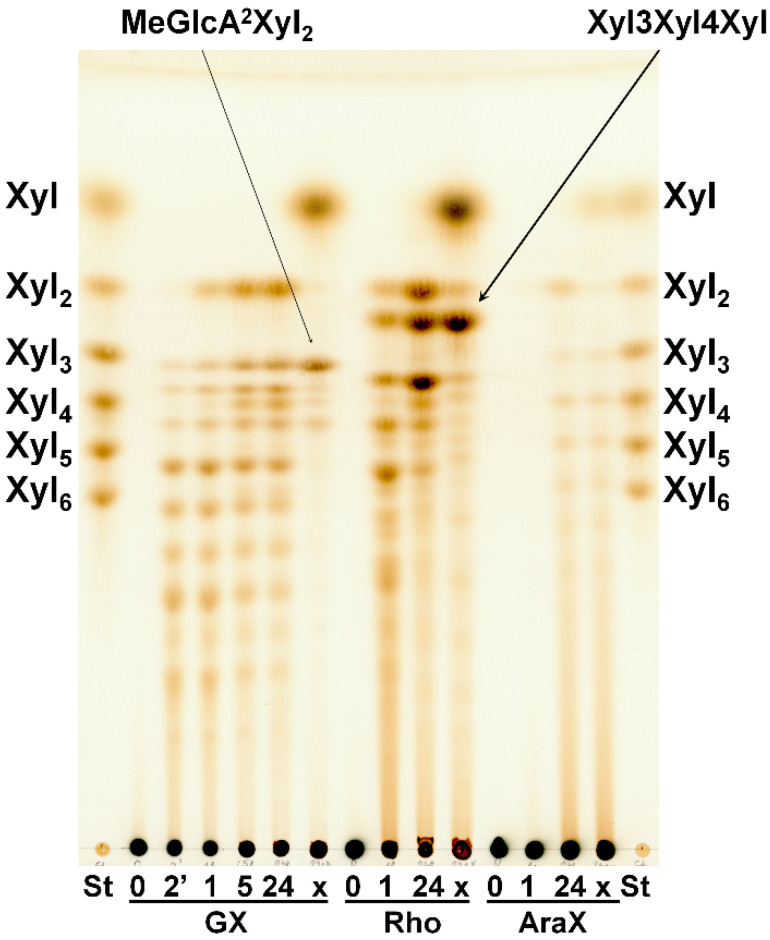
SlXyn30A showed the highest specific activity on glucuronoxylan (GX) (8.4 U/mg), while activity on linear β-1,3-1,4-xylan (rhodymenan, Rho) was about 2.5 times lower (3.1 U/mg). Specific activity on arabinoxylan was extremely low (0.03 U/mg). TLC analysis of hydrolysis products showed that GX was initially cleaved to a series of acidic xylooligosaccharides (XOs) (Figure 3) which were partly shortened after a prolonged incubation.
Figure 3.
TLC analysis of hydrolysis products released from GX, Rho and AraX by SlXyn30A after 2 min, 1 h, 5 h, 24 h and after treatment with β-xylosidase (x). St—standards of linear XOs.
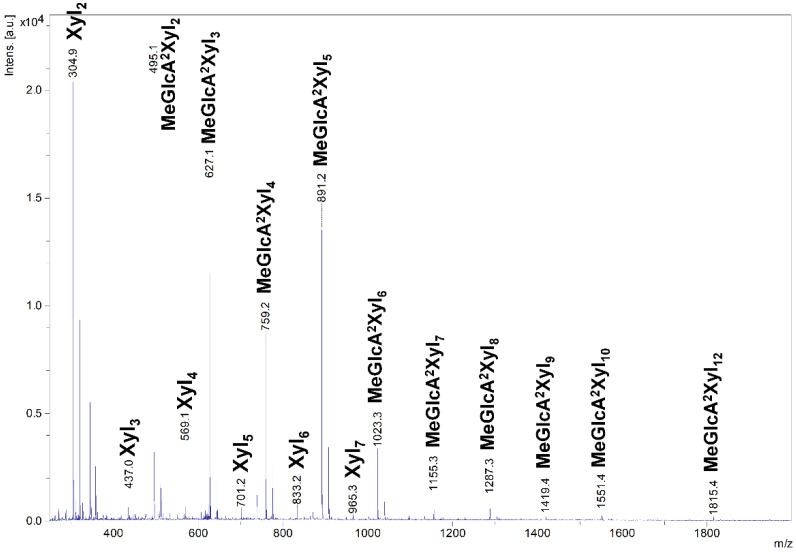
The acidic XOs shortening was accompanied by xylobiose (Xyl2) formation, which was detectable after 1 h, indicating xylobiohydrolase activity of the enzyme. Later, after 5 h, xylotetraose (Xyl4) also appeared as a result of transglycosylation reaction. After an application of β-xylosidase to 24 h hydrolysate, the acidic XOs were hydrolyzed to MeGlcA2Xyl2 which indicates that all acidic XOs had MeGlcA substitution on the second xylopyranosyl (Xylp) residue from the reducing end. This mode of GX hydrolysis is typical for the GH30 glucuronoxylanases [13,14,15]. Glucuronoxylanase activity accompanied by xylobiohydrolase activity has been already described for other GH30-7 enzymes TcXyn30B and TtXyn30A [2,3]. Our results indicate that SlXyn30A might also be a glucuronoxylanase/xylobiohydrolase. However, the acidic XOs in the 24 h hydrolysate were not shortened exclusively to MeGlcA2Xyl2 and MeGlcA2Xyl3, as was observed in the case of TtXyn30A [3], but longer acidic products remained in the hydrolysate even after 5-day incubation or after an addition of fresh enzyme. In this regard, SlXyn30A resembles much more TcXyn30B than TtXyn30A. MALDI-ToF analysis of the 5-day hydrolysate confirmed the presence of a broad spectrum of acidic XOs from MeGlcAXyl2 to MeGlcAXyl12 (MeGlcAXyl2—MeGlcAXyl5 prevailing) (Figure 4). Xyl2 was predominant neutral XO but traces of longer XOs up to DP10 were also observed. Kinetic parameters determined for GX were Km 16.8 mg/mL, kcat 20.2 s−1 and kcat/Km 1.2 mL/mg·s. Rho was hydrolyzed to a mixture of β-1,4-linked XOs and β-1,3-1,4-XOs, among which the most predominant were Xyl2, Xyl4, Xylβ1-3Xylβ1-4Xyl and Xylβ1-4Xylβ1-3Xylβ1-4Xyl (Figure 3). β-Xylosidase added to the 24 h-hydrolysate of Rho cleaved all β-1,4-linked XOs to Xyl and all the β-1,3-1,4-linked XOs to Xylβ1-3Xylβ1-4Xyl. The final extent of hydrolysis seems to be higher for Rho than GX. Hydrolysis of AraX was very weak (Figure 3). In this case, Xyl2 was a predominant product accompanied by a few Ara-substituted XOs, most probably having the Ara substitution on the non-reducing end (they were not attacked by β-xylosidase).
Figure 4.
MALDI-ToF MS analysis of 5-day hydrolysate of GX by SlXyn30A.
2.5. Hydrolysis of Oligosaccharides
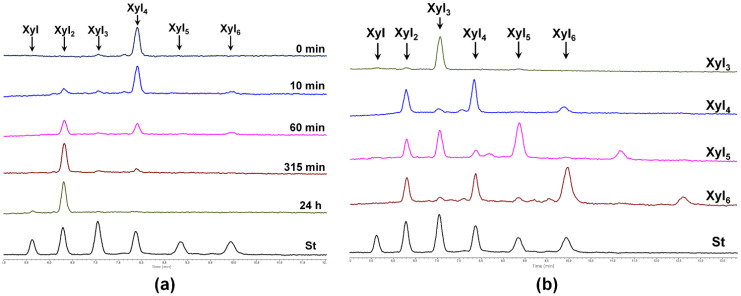
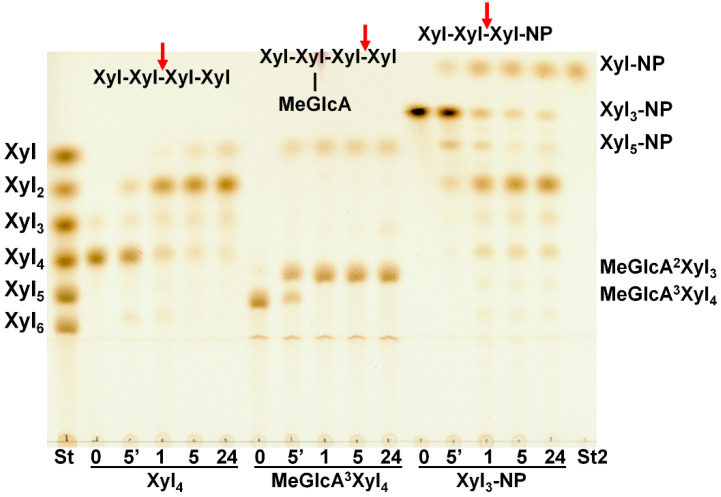
Xylobiohydrolase activity of SlXyn30A was also observed on various XOs. The main hydrolysis product released from Xyl4 was Xyl2, but Xyl6 was also formed, presumably through transglycosylation reaction (Figure 5a). Xyl5 was mainly cleaved to Xyl2 and Xyl3, and Xyl6 to Xyl2 and Xyl4. In both cases, the transglycosylation products with DP higher by two than the substrate were observed during the early stages of reaction (Figure 5b). Xyl3 was the worst substrate, being cleaved to Xyl2 and Xyl only after a prolonged incubation. Transglycosylation products were also formed during the action of SlXyn30A on 4-nitrophenyl glycosides of β-1,4-xylobiose (Xyl2-NP) and β-1,4-xylotriose (Xyl3-NP). Xyl2 release was accompanied by a liberation of 4-nitrophenol from Xyl2-NP and 4-nitrophenyl xyloside from Xyl3-NP. This mode of action has unambiguously confirmed that xylobiose moiety is released from the non-reducing end of the substrates. Compared to linear β-1,4-XOs and the corresponding NP-glycosides, MeGlcA-substituted XOs of the same DP were cleaved faster (Figure 6). About 60% of MeGlcA3Xyl4 was hydrolyzed after 5 min of the reaction, when only about 10% of Xyl4 or Xyl3-NP were converted. Specific activities were 0.037 U/mg for Xyl3, 0.222 U/mg for Xyl4, 1.9 U/mg for MeGlcA3Xyl4 and 56.7 U/mg for MeGlcA3Xyl3 showing the preference of the enzyme for MeGlcA-substituted substrates. It should be noted that the transglycosylation reaction was not observed during the processing of the acidic XOs MeGlcA3Xyl4 and MeGlcA3Xyl3.
Figure 5.
HPLC analysis of hydrolysis of XOs by SlXyn30A. (a) Time course of Xyl4 hydrolysis; (b) Analysis of hydrolysis products generated from Xyl3-Xyl6 after 30 min of reaction.
Figure 6.
TLC analysis of hydrolysis products released from Xyl4, MeGlcA3Xyl4 and Xyl3-NP by SlXyn30A after 5 min, 1 h, 5 h and 24 h. St—standards of linear XOs, St2—standard of 4-nitrophenyl β-d-xylopyranoside.
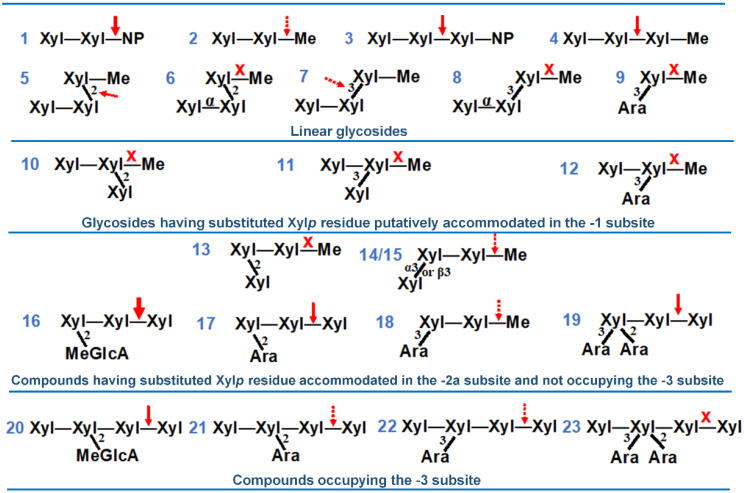
The activity of SlXyn30A was further tested on oligosaccharides containing β-1,3- or β-1,2-linkages or arabinosyl substitution (Figure 7, compounds 1–23). The enzyme was able to slowly release methanol from methyl β-1,4-xylobioside (2) indicating that Xylp unit accommodated in the +1 subsite is not indispensable for enzyme activity. Methyl β-1,4-xylotrioside (4) was hydrolyzed much faster than (2), and exclusively to Xyl2 and Xyl-Me in accordance with xylobiohydrolase activity of the enzyme. SlXyn30A was able to cleave very slowly also β-1,2- and β-1,3-linkages in X4X2XMe (5) and X4X3XMe (7), the former being cleaved faster but significantly slower than (4). However, if Xylp residues at the non-reducing end are connected by α-1,4-linkage, the substances (6,8) are not hydrolyzed. The compounds, in which the substitution would occur on Xylp unit accommodated in the −1 subsite, were not hydrolyzed (9–12). On the other hand, some substitutions of Xylp in the −2a subsite were tolerated. The tolerance was influenced by three factors: (1) position of the decoration (2 and/or 3); (2) the nature of the substituent; and (3) whether the compound was further elongated at the non-reducing end, i.e., it carried additional Xylp residue that was accommodated in the subsite −3. If the Xylp unit accommodated in the −2a subsite is decorated at position 2 by the MeGlcA, the substrate (MeGlcA3Xyl3, 16) is readily hydrolyzed. When the MeGlcA substitution is replaced by α-l-arabinofuranose, the substrate (A2X4X4X, 17) is slowly attacked, but a change for β-d-xylopyranose (X2X4XMe, 13) caused a resistance to the enzyme attack. The tolerance at position 3 is greater since A3X4XMe (18) and X3X4XMe (14) as well as Xα3X4XMe (15) were slowly processed. SlXyn30A was also able to cleave the substrate (19) with doubly 2,3-O-arabinosylated Xylp residue accommodated in the −2a subsite. However, if the compounds with the substitution on Xylp residue in the −2a subsite are by one Xylp longer at the non-reducing end (i.e., they occupy also the −3 subsite), the hydrolysis is slowed down (20–22) or even abolished (23).
Figure 7.
Various XOs tested as the substrates for SlXyn30A. The site of cleavage is denoted by an arrow, X marks compounds which were not attacked.
3. Discussion
Many xylanases have been isolated and characterized from bacteria and filamentous fungi while a number of characterized yeast xylanases is limited and no yeast GH30 xylanase has been described so far. Catalytic properties of eukaryotic GH30 xylanases belonging to GH30-7 subfamily, where SlXyn30A is also grouped, are diverse. It was, therefore, interesting to determine the specificity of the yeast xylanase, which may be related to a biotechnological potential of the yeast due to its reported ability to convert xylose to xylitol, ethanol or organic acids [26,27]. The GH30 xylanase SlXyn30A was cloned, expressed, and characterized. SlXyn30A showed the highest amino acid similarity to glucuronoxylanases/xylobiohydrolases TtXyn30A and TcXyn30B. SlXyn30A, similarly to these fungal enzymes, contains Arg46 which was shown to play a role in MeGlcA recognition [4,9]. Another aspect of similarity between the three enzymes is the exact length of β2-α2 loop, which may affect the occupation of the −3 subsite. Moreover, Asn90 in this loop corresponds to Asn93 in TcXyn3B and Asp78 in TtXyn30A which are supposed to play a role in xylobiohydrolase activity of the GH30-7 glucuronoxylanases [4,9]. All these structural features of SlXyn30A are in accordance with its biochemical properties. Glucuronoxylan was the best substrate among the heteroxylans studied and during its hydrolysis, the MeGlcA residue was accommodated in the −2b subsite of the enzyme, yielding acidic XOs of general formula MeGlcA2Xyln. Such a hydrolysis of GX is typical for GH30 glucuronoxylanases [13,14,15]. Later, the acidic XOs were partially shortened by SlXyn30A through a liberation of Xyl2 from the non-reducing end, similarly to GH30 xylobiohydrolases [7,8] and glucuronoxylanses/xylobiohydrolases [2,3]. The shortening of the acidic XOs was not complete and the aldouronic acids of a medium size persisted in the SlXyn30A hydrolysate. From this point of view, SlXyn30A most resembles TcXyn30B, which shows an essentially identical end-stage hydrolysis profile [2], and slightly differs from TtXyn30A, which liberated the acidic XOs and Xyl2 simultaneously, and Xyl2, MeGlcA2Xyl2 and MeGlcA2Xyl3 were the only final products of GX hydrolysis [3]. In addition, kinetic parameters of SlXyn30A on GX are also similar to those of TcXyn30B [2].
The xylobiohydrolase activity of SlXyn30A was even more pronounced during the hydrolysis of rhodymenan. It was reflected in an accumulation of not only Xyl2, but also isomeric xylotriose Xylβ1-3Xylβ1-4Xyl and isomeric xylotetraose Xylβ1-4Xylβ1-3Xylβ1-4Xyl. The release of Xylβ1-3Xylβ1-4Xyl is in agreement with the ability of SlXyn30A to cleave X3X4XMe (Figure 7, 14) to X3X4X and methanol. Hydrolysis of X4X3XMe is very slow (not finished after 145 h of hydrolysis) indicating a very limited ability of the enzyme to hydrolyze β-1,3-xylosidic linkage. This is in accordance with the presence of Xylβ1-4Xylβ1-3Xylβ1-4Xyl in 24 h hydrolysate of Rho. In contrast, xylobiohydrolases AaXyn30A and HcXyn30A are able to cleave β-1,3-linkages much more efficiently and therefore Xylβ1-4Xylβ1-3Xylβ1-4Xyl was not accumulated but cleaved to two molecules of Xyl2 in their hydrolysates [7,8].
Linear β-1,4-XOs were processed by SlXyn30A in the same way as was described for TcXyn30B and TtXyn30A [2,3]. Xyl2 was the main product and XOs longer by two xylose units were formed via transglycosylation. For SlXyn30A, Xyl3 was a much worse substrate than Xyl4, suggesting that an occupation of the +2 subsite has a significant positive effect on the enzyme activity.
The ability of SlXyn30A to recognize MeGlcA substitution was confirmed by a comparison of specific activities on linear XOs and the corresponding XOs decorated by MeGlcA. Specific activity on MeGlcA3Xyl3 was about 1500 times higher than on Xyl3. On the other hand, MeGlcA3Xyl4 was only about 8 times better substrate than Xyl4. MeGlcA3Xyl3 was hydrolyzed about 30 times faster than MeGlcA3Xyl4. These data clearly indicate the preference of the enzyme for the MeGlcA-decorated substrates and for the substrates not occupying the −3 subsite of the enzyme. The latter preference was confirmed also during the hydrolysis of various methyl glycosides and arabinoxylooligosaccharides when elongation of the XO chain to the −3 subsite of the enzyme (Figure 7, 16 vs. 20, 17 vs. 21, 18 vs. 22, 19 vs. 23) caused a slowdown or an abolishment of the reaction. The evaluation of these compounds allowed us to draw the following conclusions on the requirement of SlXyn30A on the structure of the substrates. First, SlXyn30A does not tolerate any substitution on Xylp residue accommodated in the −1 subsite. Xylose residue accommodated in the −2a subsite may be substituted at position 2 by MeGlcA, which improves the activity, and by α-l-arabinofuranose but not by β-d-xylopyranose. Position 3 may be both arabinosylated and xylosylated, but the activity on such substrates is lower compared to unsubstituted XOs. The elongation of the substrate main chain that results in an occupation of the −3 subsite may dramatically decrease the hydrolysis rate.
4. Materials and Methods
4.1. Substrates, Standards and Enzymes
Beechwood 4-O-methylglucuronoxylan (GX), aldotetraouronic acid MeGlcA3Xyl3 and aldopentaouronic acid MeGlcA3Xyl4 were prepared as described earlier [28,29]. Rhodymenan, an algal linear β-1,3-β-1,4-xylan from Palmaria palmata, was a kind gift of Prof. M. Claeyssens (University of Ghent, Ghent, Belgium). Wheat arabinoxylan (Ara:Xyl 38:62, medium viscosity), 4-nitrophenyl glycosides of xylose, xylobiose and xylotriose, linear β-1,4-xylooligosaccharides (Xyl2-Xyl6) and arabinoxylooligosaccharides (Figure 7) A2X4X4X α-l-Araf-1,2-β-d-Xylp-1,4-β-d-Xylp-1,4-d-Xyl, (17), A3[A2]X4X4X (α-l-Araf-1,3-[α-l-Araf-1,2]-β-d-Xylp-1,4-β-d-Xylp-1,4-d-Xyl, (19), X4[A3]X4X4X (β-d-Xylp-1,4-[α-l-Araf-1,3]-β-d-Xylp-1,4-β-d-Xylp-1,4-d-Xyl, (18), X4[A2][A3]X4X4X (β-d-Xylp-1,4-[α-l-Araf-1,2][α-l-Araf-1,3]-β-d-Xylp-1,4-β-d-Xylp-1,4-d-Xyl, (23) and a mixture of X4[A3]X4X4X and X4[A2]X4X4X (β-d-Xylp-1,4-[α-l-Araf-1,2]-β-d-Xylp-1,4-β-d-Xylp-1,4-d-Xyl, (22,21) were purchased from Megazyme International (Bray, Ireland). Methyl glycosides of (arabino)xylooligosaccharides (Figure 7)—A3XMe (α-l-Araf-1,3-β-d-Xylp-O-Me, (9), X4[A3]XMe (β-d-Xylp-1,4-[α-l-Araf-1,3]-β-d-Xylp-O-Me, (12), X4XMe (β-d-Xylp-1,4-β-d-Xylp-O-Me, (2), X4X4XMe (β-d-Xylp-1,4-β-d-Xylp-1,4-β-d-Xylp-O-Me, (4), X4X3XMe (β-d-Xylp-1,4-β-d-Xylp-1,3-β-d-Xylp-O-Me, (7), X4X2XMe (β-d-Xylp-1,4-β-d-Xylp-1,2-β-d-Xylp-O-Me, (5), X3X4XMe (β-d-Xylp-1,3-β-d-Xylp-1,4-β-d-Xylp-O-Me, (14), Xα3X4XMe (α-d-Xylp-1,3-β-d-Xylp-1,4-β-d-Xylp-O-Me, (15), X2X4XMe (β-d-Xylp-1,2-β-d-Xylp-1,4-β-d-Xylp-O-Me, (13), X4[X3]XMe (β-d-Xylp-1,4-[β-dxylp-1,3]-β-d-Xylp-O-Me, (11), X4[X2]XMe (β-d-Xylp-1,4-[β-d-Xylp-1,2]-β-d-Xylp-O-Me, (10), Xα4X3XMe (α-d-Xylp-1,4-β-d-Xylp-1,3-β-d-Xylp-O-Me, (8), Xα4X2XMe (α-d-Xylp-1,4-β-d-Xylp-1,2-β-d-Xylp-O-Me, (6)—were synthesized previously [30,31,32,33,34,35] and were generously supplied by Dr. Ján Hirsch (Institute of Chemistry, Slovak Academy of Sciences, Bratislava, Slovakia). β-Xylosidase was a recombinant Aspergillus niger enzyme from GH3 family expressed in Saccharomyces cerevisiae [36].
4.2. Amino Acid Sequence Comparison
Amino acid sequence of SlXyn30A (GenBank: ANB12318) was aligned with amino acid sequences of Talaromyces cellulolyticus TcXyn30B (GAM36763), TcXyn30C (GAM40414.1), TcXyn30A (GAM43270), Thermothelomyces thermophila TtXyn30A (AEO55025), Talaromyces purpureogenus (Penicillium purpurogenum) TpXynC (AKH40280), Bispora sp. BXylD (ADG62369.1), Trichoderma reesei TrXynVI (EGR45006.1), T. reesei TrXynIV (AAP64786.1), Acremonium alcalophilum AaXyn30A [7], and Talaromyces leycettanus TlXyn30A [10] using Clustal Omega server [37] and visualized by ESPript server [38].
4.3. Recombinant Strain Preparation
In this work, P. pastoris KM71H (MutS strain) was used. The strain preparation and cultivation conditions for production process are described in Rosenbergová et al. [39]. Briefly, the microorganism was cultivated on YPD (P. pastoris) plates with 2% (w/v) agar and 50 mg/L ZeocinTM (InvivoGen, San Diego, CA, USA). For flask cultivations of P. pastoris, BMGY [Buffered Glycerol-complex Medium; 1% (w/v) yeast extract, 2% (w/v) peptone, 1.34% (w/v) YNB, 4 × 10−5% (w/v) biotin, 1% (v/v) glycerol, and 100 mM potassium phosphate (pH 6)] and BMMH [Buffered Minimal Methanol Medium; 1.34% (w/v) YNB, 4 × 10−5% (w/v) biotin, 0.5% (v/v) methanol, and 100 mM potassium phosphate (pH 6)] media were used.
Gene coding for SlXyn30A (GenBank: ANB12318.1) was codon-optimized for P. pastoris and purchased from Generay Biotech Co., Ltd. (Shanghai, China). Plasmid pPICZαA with ligated xyn30A was linearized with SacI (Fast Digest, ThermoFisher Scientific, Waltham, MA, USA). Approximately 5 µg of linearized plasmid was electroporated to P. pastoris KM71H competent cells (prepared according to Lin-Cereghino et al. [40]). Transformed cells were plated on YPD with 100, 150, 200 and 250 mg/L of ZeocinTM and cultivated at 30 °C for 48 h.
Four transformants from YPD plates with 250 mg/L of ZeocinTM were selected and screened for a recombinant SlXyn30A production. Additionally, 500 mL shake-flasks with 100 mL of BMGY medium were inoculated with a single P. pastoris colony and cultivated at 30 °C and 200 rpm for 22 h. The induction of enzyme was carried out as reported previously [41]. The cells were harvested by centrifugation (7197× g, 10 °C, 5 min), resuspended in 6 mL of sterile distilled water and transferred to 100 mL of BMMH medium with 0.5% (v/v) methanol. The cells were then cultivated at 30 °C and 200 rpm for 120 h, and methanol (100 µL) was added 2 times per day. After termination of cultivation, biomass was centrifuged (7197× g, 10 °C, 5 min) and the supernatants were concentrated and desalted on Microcon centrifugal filter devices (10 kDa cut-off, Millipore) and used for enzyme characterization. Protein concentration was determined by Bradford method using BSA as a standard [42].
4.4. Determination of pH and Temperature Optimum and Temperature Stability
pH optimum was determined at 50 °C using 10 mg·mL−1 solution of GX in 40 mM Britton–Robinson buffer (pH 2.0–8.0), 15 min incubation, and 64.4 nM SlXyn30A. Temperature optimum was determined in the same manner in 50 mM sodium acetate buffer, pH 3.5, and temperatures ranging from 23 °C to 60 °C. Temperature stability was tested in 50 mM sodium acetate buffer, pH 3.5, at 40–60 °C for up to 5 h. During the incubation, aliquots were taken at different time points and the residual activity was immediately determined as described above (10 mg·mL−1 GX, pH 3.5, 50 °C, 15 min).
4.5. Hydrolysis of Polysaccharides and Oligosaccharides
For specific activity determination, solutions (10 mg·mL−1) of GX, Rho and AraX in 50 mM sodium acetate buffer, pH 3.5, were mixed with 64.4 nM (GX and Rho) or 644 nM (AraX) SlXyn30A and incubated at 50 °C. At time intervals (5–30 min) 100 µL aliquots were taken for a determination of released reducing sugars by Somogyi–Nelson procedure [43]. One unit of activity is defined as the amount of the enzyme liberating in 1 min 1 μmol of reducing sugars expressed as an equivalent of xylose. All reactions were performed at least in triplicate. Kinetic parameters for GX hydrolysis were determined at 50 °C in 50 mM sodium acetate, pH 3.5, using 2.5–30 mg·mL−1 substrate concentration and 64.4 nM SlXyn30A. Kinetic constants were calculated by a non-linear regression using Origin 6.0 program (OriginLab Corp., Northampton, MA, USA).
For TLC (thin-layer chromatography) analysis, the same polysaccharide solutions (GX, Rho, AraX) were incubated with 1.6 µM SlXyn30A at 50 °C. Aliquots of 5 µL were spotted on silica-gel-coated aluminum sheets (Merck, Darmstadt, Germany) after 2 min, 1 h, 5 h, and 24 h of hydrolysis of GX, and 1 h and 24 h of hydrolysis of Rho and AraX. The reaction was terminated after 24 h by heating the mixtures at 100 °C for 5 min. Subsequent treatment with β-xylosidase (1 U·mL−1) was performed overnight at 50 °C. TLC plates were developed twice in the solvent system ethyl acetate/acetic acid/2-propanol/formic acid/water 25:10:5:1:15 (v/v) and the sugars were visualized with orcinol reagent (0.5% orcinol in 5% sulphuric acid in ethanol). For TLC analysis of Xyl4, MeGlcA3Xyl4, Xyl3-NP and methyl glycosides hydrolysis, 2.5 mM substrates were incubated with 1.6 μM SlXyn30A at 50 °C. Aliquots of 2 µL were spotted on TLC plate after 5 min, 1 h, 5 h and 24 h of hydrolysis. The plate was developed in a solvent system of n-butanol/ethanol/water 10:8:5 (v/v), and the sugars were visualized with the orcinol reagent.
For HPLC analysis, 5 mM Xyl3–Xyl6 solutions in 50 mM sodium acetate buffer, pH 3.5, were incubated with 1.3 µM SlXyn30A at 50 °C. At time intervals, 10 μL aliquots were taken and heated at 95 °C for 5 min. The samples were mixed with acetonitrile (1:4) and analyzed on a chromatographic apparatus Dionex UltiMate 3000 UHPLC system (ThermoFisher Scientific, Germering, Germany) equipped with a solvent degasser, quaternary pump, autosampler and thermostatic column compartment coupled to a Corona Veo RS detector (Thermo Fisher Scientific, Germering, Germany). Data processing was carried out with Chromeleon 7.2 SR3 software (Thermo Fisher Scientific, Waltham, MA, USA). Nitrogen gas was supplied by Peak nitrogen generator and Peak air compressor (Peak Scientific Instruments Ltd., Inchinnan, Renfrewshire, Scotland). The CAD device settings was as follows: data collection was set to 50.0 Hz at a filter constant of 3.6 s, power function for response and signal correction was set to 1.00 and evaporator temperature was set to 60 °C. Chromatographic separation was conducted on ARION HILIC Plus column (100 Å, 3.0 μm, 150 mm × 4.6 mm) maintained at 30 °C. Mobile phase A consisted of 0.5% acetic acid adjusted to pH 6.97 with NH4OH (25%, NH3 water solution), and mobile phase B was 100% acetonitrile. The elution was isocratic at a flow rate of 0.5 mL·min−1 with a mixture of mobile phases A and B in a ratio of 30:70. Specific activities were determined on 1 mM Xyl3, Xyl4, MeGlcA3Xyl4 and MeGlcA3Xyl3 in 50 mM sodium acetate buffer, pH 3.5, at 50 °C using 1.3 µM SlXyn30A, and calculated on the basis of the amount of liberated Xyl2 (linear XOs, in the case of Xyl4 divided by two) or Xyl (MeGlcA3Xyl4 and MeGlcA3Xyl3).
4.6. MALDI-ToF MS
The hydrolysates were decationized by Dowex 50 (H+ form) and 1 µL was mixed with 1 µL of the matrix (1% solution of 2,5-dihydroxybenzoic acid in 30% acetonitrile) directly on MS target plate. After air-drying, the samples were analyzed by UltrafleXtreme MALDI ToF/ToF mass spectrometer (Bruker Daltonics, Bremen, Germany) operating in reflectron positive mode.
5. Conclusions
The first GH30 xylanase originating from a yeast has been cloned, expressed and characterized. The enzyme SlXyn30A from S. lignohabitans is a glucuronoxylanase with auxiliary xylobiohydrolase activity. In addition to hardwood glucuronoxylan, it efficiently depolymerizes linear β-1,3-β-1,4-xylan but not cereal arabinoxylan. Its amino acid sequence has the highest similarity to the fungal bifunctional GH30-7 enzymes TcXyn30B and TtXyn30A which also display glucuronoxylanase and xylobiohydrolase activities. Catalytic properties of SlXyn30A also resemble those of TcXyn30B and TtXyn30A including the recognition of MeGlcA side chain in the −2b subsite, no substitution of xylose occupying the subsite −1, and certain flexibility of decoration of xylopyranosyl unit bound in the −2a subsite. Further characterization of new xylanases from different yeast species will help us to reveal how the yeasts cope with xylan degradation in nature and to better evaluate their biotechnological potential. The crystal structure of SlXyn30A with appropriate ligands would improve our knowledge of how GH30-7 glucuronoxylanases/xylobiohydrolases switch between endo- and exo-activities which is not yet fully understood.
Acknowledgments
The authors are grateful to Marc Claeyssens (University of Ghent, Ghent, Belgium) for supplementation of rhodymenan. The authors also thank Pavol Kováč (NIH National Institute of Diabetes & Digestive & Kidney Diseases, National Institutes of Health, Bethesda, MD, USA) and Ján Hirsch (Institute of Chemistry, Slovak Academy of Sciences) for a generous gift of methyl glycosides of various oligosaccharides.
Abbreviations
| EX | endo-β-1,4-xylanase |
| XOs | xylooligosaccharides |
| GH | glycoside hydrolase |
| GX | 4-O-methylglucuronoxylan |
| MeGlcA | 4-O-methylglucuronic acid |
| AraX | arabinoxylan |
| Rho | rhodymenan (linear β-1,3-1,4-xylan) |
| Xylp | xylopyranosyl |
| Xyln | linear xylooligosaccharide composed of n d-xylopyranosyl residues linked by β-1,4-linkages |
| MeGlcAzXyln | Xyln wherein zth xylopyranosyl residue counted from reducing end is α-glycosylated at position 2 by MeGlcA |
| NP | 4-nitrophenyl |
| YPD | yeast extract peptone dextrose |
| YNB | yeast nitrogen base |
Supplementary Materials
The following supporting information can be downloaded online, Figure S1: Multiple sequence alignment of GH30 catalytic domain of SlXyn30A with selected GH30-7 xylanases, Figure S2: SDS-PAGE electrophoresis of recombinant P. pastoris fermentation broths after induction with methanol.
Author Contributions
Conceptualization, K.Š. and M.R.; methodology, K.Š., A.C., Z.H. and M.R.; investigation, K.Š., A.C., V.P., Z.H. and M.R.; resources, M.R., A.C. and V.P.; writing—original draft preparation, K.Š.; writing—review and editing, M.R., A.C., Z.H. and V.P.; visualization, K.Š. and V.P.; project administration, V.P.; funding acquisition, K.Š., M.R. and V.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Scientific Grant Agency under the contract No. 2/0171/22, and by the Slovak Research and Development Agency under the Contract no. APVV-20-0591. This work was funded by the Agency for supporting research and development, according to Agreement Nr. APVV-18-0254. This article is based upon work from COST Action “Nonconventional yeasts for the production of bioproducts” (YEAST4BIO) CA18229, supported by COST (European Cooperation in Science and Technology, www.cost.eu, accessed on 13 December 2021).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Sample Availability
Samples of the compounds are available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamichi Y., Fouquet T., Ito S., Watanabe M., Matsushika A., Inoue H. Structural and functional characterization of a GH30-7 xylanase B from the filamentous fungus Talaromyces cellulolyticus. J. Biol. Chem. 2019;294:4065–4078. doi: 10.1074/jbc.RA118.007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katsimpouras C., Dedes G., Thomaidis N.S., Topakas E. A novel fungal GH30 xylanase with xylobiohydrolase auxiliary activity. Biotechnol. Biofuels. 2019;12:120. doi: 10.1186/s13068-019-1455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamichi Y., Watanabe M., Matsushika A., Inoue H. Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus. FEBS Open Bio. 2020;10:1180–1189. doi: 10.1002/2211-5463.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamichi Y., Fouquet T., Ito S., Matsushika A., Inoue H. Mode of action of GH30-7 reducing-end xylose-releasing exoxylanase A (Xyn30A) from the filamentous fungus Talaromyces cellulolyticus. Appl. Environ. Microbiol. 2019;85:e00552-19. doi: 10.1128/AEM.00552-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamichi Y., Fujii T., Fouquet T., Matsushika A., Inoue H. GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus. Appl. Environ. Microbiol. 2019;85:e0144219. doi: 10.1128/AEM.01442-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Šuchová K., Puchart V., Spodsberg N., Mørkeberg Krogh K.B.R., Biely P. A novel GH30 xylobiohydrolase from Acremonium alcalophilum releasing xylobiose from the non-reducing end. Enzyme Microb. Technol. 2020;134:109484. doi: 10.1016/j.enzmictec.2019.109484. [DOI] [PubMed] [Google Scholar]
- 8.Šuchová K., Puchart V., Biely P. A novel bacterial GH30 xylobiohydrolase from Hungateiclostridium clariflavum. Appl. Microbiol. Biotechnol. 2021;105:185–195. doi: 10.1007/s00253-020-11023-x. [DOI] [PubMed] [Google Scholar]
- 9.Nikolaivits E., Pentari C., Kosinas C., Feiler C.G., Spiliopoulou M., Weiss M.S., Dimarogona M., Topakas E. Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies. Carbohydr. Polym. 2021;273:118553. doi: 10.1016/j.carbpol.2021.118553. [DOI] [PubMed] [Google Scholar]
- 10.Šuchová K., Spodsberg N., Mørkeberg Krogh K.B.R., Biely P., Puchart V. Non-Specific GH30_7 Endo-β-1,4-xylanase from Talaromyces leycettanus. Molecules. 2021;26:4614. doi: 10.3390/molecules26154614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Šuchová K., Puchart V., Spodsberg N., Mørkeberg Krogh K.B.R., Biely P. Catalytic Diversity of GH30 Xylanases. Molecules. 2021;26:4528. doi: 10.3390/molecules26154528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puchart V., Šuchová K., Biely P. Xylanases of glycoside hydrolase family 30—An overview. Biotechnol. Adv. 2021;47:107704. doi: 10.1016/j.biotechadv.2021.107704. [DOI] [PubMed] [Google Scholar]
- 13.Biely P., Puchart V., Stringer M.A., Mørkeberg Krogh K.B.R. Trichoderma reesei XYN VI–a novel appendage dependent eukaryotic glucuronoxylan hydrolase. FEBS J. 2014;281:3894–3903. doi: 10.1111/febs.12925. [DOI] [PubMed] [Google Scholar]
- 14.Vršanská M., Kolenová K., Puchart V., Biely P. Mode of action of glycoside hydrolase family 5 glucuronoxylan xylanohydrolase from Erwinia chrysanthemi. FEBS J. 2007;274:1666–1677. doi: 10.1111/j.1742-4658.2007.05710.x. [DOI] [PubMed] [Google Scholar]
- 15.St. John F.J., Rice J.D., Preston J.F. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J. Bacteriol. 2006;188:8617–8626. doi: 10.1128/JB.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leathers T.D. Color variants of Aureobasidium pullulans overproduce xylanase with extremely high specific activity. Appl. Environ. Microbiol. 1986;52:1026–1030. doi: 10.1128/aem.52.5.1026-1030.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X.L., Zhang Z.Q., Dean J.F.D., Eriksson K.E.L., Ljungdahl L.G. Purification and characterization of a new xylanase (APX-II) from the fungus Aureobasidium pullulans Y-2311-1. Appl. Environ. Microbiol. 1993;59:3212–3218. doi: 10.1128/aem.59.10.3212-3218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biely P., Vršanská M., Krátky Z. Xylan-degrading enzymes of the yeast Cryptococcus albidus: Identification and cellular localization. Eur. J. Biochem. 1980;108:313–321. doi: 10.1111/j.1432-1033.1980.tb04725.x. [DOI] [PubMed] [Google Scholar]
- 19.Petrescu I., Lamotte-Brasseur J., Chessa J.-P., Ntarima P., Claeyssens M., Devreese B., Marino G., Gerday C. Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles. 2000;4:137–144. doi: 10.1007/s007920070028. [DOI] [PubMed] [Google Scholar]
- 20.Parachin N.S., Siqueira S., de Faria F.P., Torres F.A.G., de Moraesa L.M.P. Xylanases from Cryptococcus flavus isolate I-11: Enzymatic profile, isolation and heterologous expression of CfXYN1 in Saccharomyces cerevisiae. J. Mol. Catal. B Enzym. 2009;59:52–57. doi: 10.1016/j.molcatb.2008.12.018. [DOI] [Google Scholar]
- 21.Ding C., Li M.X., Hu Y. High-activity production of xylanase by Pichia stipitis: Purification, characterization, kinetic evaluation and xylooligosaccharides production. Int. J. Biol. Macromol. 2018;117:72–77. doi: 10.1016/j.ijbiomac.2018.05.128. [DOI] [PubMed] [Google Scholar]
- 22.Adsul M.G., Bastawde K.B., Gokhale D.V. Biochemical characterization of two xylanases from yeast Pseudozyma hubeiensis producing only xylooligosaccharides. Bioresour. Technol. 2009;100:6488–6495. doi: 10.1016/j.biortech.2009.07.064. [DOI] [PubMed] [Google Scholar]
- 23.Ravn J.L., Engqvist M.K.M., Larsbrink J., Geijer C. CAZyme prediction in ascomycetous yeast genomes guides discovery of novel xylanolytic species with diverse capacities for hemicellulose hydrolysis. Biotechnol. Biofuels. 2021;14:150. doi: 10.1186/s13068-021-01995-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtzman C.P., Robnett C.J. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res. 2007;7:141–151. doi: 10.1111/j.1567-1364.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- 25.Houseknecht J.L., Hart E.L., Suh S.O., Zhou J.J. Yeasts in the Sugiyamaella clade associated with wood-ingesting beetles and the proposal of Candida bullrunensis sp. nov. Int. J. Syst. Evol. Microbiol. 2011;61:1751–1756. doi: 10.1099/ijs.0.026427-0. [DOI] [PubMed] [Google Scholar]
- 26.Sena L.M., Morais C.G., Lopes M.R., Santos R.O., Uetanabaro A.P., Morais P.B., Vital M.J., de Morais M.A., Jr., Lachance M.A., Rosa C.A. d-Xylose fermentation, xylitol production and xylanase activities by seven new species of Sugiyamaella. Antonie Leeuwenhoek. 2017;110:53–67. doi: 10.1007/s10482-016-0775-5. [DOI] [PubMed] [Google Scholar]
- 27.Bellasio M., Mattanovich D., Sauer M., Marx H. Organic acids from lignocellulose: Candida lignohabitans as a new microbial cell factory. J. Ind. Microbiol. Biotechnol. 2015;42:681–691. doi: 10.1007/s10295-015-1590-0. [DOI] [PubMed] [Google Scholar]
- 28.Ebringerová A., Kramár A., Rendoš F., Domanský R. Stepwise extraction of hemicellulose from wood of white beech (Carpinus betulus L.) Holzforschung. 1967;21:74–77. doi: 10.1515/hfsg.1967.21.3.74. [DOI] [Google Scholar]
- 29.Biely P., Vršanská M., Tenkanen M., Kluepfel D. Endo-β-1,4-xylanases: Differences in catalytic properties. J. Biotechnol. 1997;57:151–166. doi: 10.1016/S0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch J., Kováč P. Alternative synthesis of methylated sugars. 21. Synthesis of 2 isomeric methyl β-d-xylotriosides containing a (1-2)-β-linkage. Carbohydr. Res. 1979;77:241–244. doi: 10.1016/S0008-6215(00)83812-4. [DOI] [Google Scholar]
- 31.Hirsch J., Petráková E. Sequential synthesis and C-13 NMR-spectra of methyl 3-O and 2-O-(β-d-xylobiosyl)-β-d-xylopyranosides. Chem. Pap. 1984;38:409–417. [Google Scholar]
- 32.Hirsch J., Petráková E., Schraml J. Stereoselective synthesis and 13C-N.m.r. spectra of two isomeric methyl β-glycosides of trisaccharides related to arabinoxylan. Carbohydr. Res. 1984;131:219–226. doi: 10.1016/0008-6215(84)85243-X. [DOI] [Google Scholar]
- 33.Kováč P. Alternative synthesis of methylated sugars. 17. Synthesis of methyl 3,4-di-O-(β-d-xylopyranosyl)-β-d-xylopyranoside, a methyl β-xylotrioside related to branched xylans. Collect. Czechoslov. Chem. Commun. 1979;44:928–932. doi: 10.1135/cccc19790928. [DOI] [Google Scholar]
- 34.Kováč P., Hirsch J. Alternative synthesis of methylated sugars. 23. Stepwise synthesis of methyl 4-O-[3-O-(β-d-xylopyranosyl)-β-Dxylopyranosyl]-β-d-xylopyranoside. Carbohydr. Res. 1980;79:303–307. doi: 10.1016/S0008-6215(00)83844-6. [DOI] [Google Scholar]
- 35.Kováč P., Hirsch J. Alternative synthesis of methylated sugars. 24. Sequential synthesis and 13C-N.M.R spectra of methyl β-glycosides of (1-4)-β-d-xylo-oligosaccharides. Carbohydr. Res. 1982;100:177–193. doi: 10.1016/S0008-6215(00)81034-4. [DOI] [Google Scholar]
- 36.Biely P., Hirsch J., la Grange D.C., van Zyl W.H., Prior B.A. A chromogenic substrate for a β-xylosidase-coupled assay of α-glucuronidase. Anal. Biochem. 2000;286:289–294. doi: 10.1006/abio.2000.4810. [DOI] [PubMed] [Google Scholar]
- 37.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., et al. The EMBLEBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenbergová Z., Kántorová K., Šimkovič M., Breier A., Rebroš M. Optimisation of recombinant myrosinase production in Pichia pastoris. Int. J. Mol. Sci. 2021;22:3677. doi: 10.3390/ijms22073677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin-Cereghino J., Wong W.W., Xiong S., Giang W., Luong L.T., Vu J., Johnson S.D., Lin-Cereghino G.P. Condensed Protocol for Competent Cell Preparation and Transformation of the Methylotrophic Yeast Pichia pastoris. Biotechniques. 2005;38:44–48. doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markošová K., Weignerová L., Rosenberg M., Křen V., Rebroš M. Upscale of recombinant α-l-rhamnosidase production by Pichia pastoris MutS strain. Front. Microbiol. 2015;6:1140. doi: 10.3389/fmicb.2015.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 43.Paleg L.G. Citric acid interference in the estimation of reducing sugars with alkaline copper reagents. Anal. Chem. 1959;31:1092–1094. doi: 10.1021/ac60155a072. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.