ABSTRACT
Replicative aging is an underexplored field of research in medical mycology. Cryptococcus neoformans (Cn) and Candida glabrata (Cg) are dreaded fungal pathogens that cause fatal invasive infections. The fungal cell wall is essential for yeast viability and pathogenesis. In this study, we provide data characterizing age-associated modifications to the cell wall of Cn and Cg. Here, we report that old yeast cells upregulate genes of cell wall biosynthesis, leading to cell wall reorganization and increased levels of all major components, including glucan, chitin, and its derivatives, as well as mannan. This results in a significant thickening of the cell wall in aged cells. Old-generation yeast cells exhibited drastic ultrastructural changes, including the presence of abundant vesicle-like particles in the cytoplasm, and enlarged vacuoles with altered pH homeostasis. Our findings suggest that the cell wall modifications could be enabled by augmented intracellular trafficking. This work furthers our understanding of the cell phenotype that emerges during aging. It highlights differences in these two fungal pathogens and elucidates mechanisms that explain the enhanced resistance of old cells to antifungals and phagocytic attacks.
KEYWORDS: Candida glabrata, Cryptococcus neoformans, cell wall, replicative lifespan, vesicle trafficking
INTRODUCTION
Fungal pathogens are an emerging threat to global health due to the rise of immunosuppressed patients (1). Their prevalence may even be affected by the evolving climate change (2, 3). Among the human fungal pathogens, Candida and Cryptococcus account for most of the invasive fungal infections (4). Cryptococcus neoformans (Cn), a member of the phylum Basidiomycota, is the leading agent of fungal meningitis and affects mostly patients living with advanced HIV disease (5). Candida glabrata (Cg), a member of the phylum Ascomycota, is the second most common fungal pathogen that causes bloodstream infections in North America and is associated with rapidly acquired antifungal resistance (6, 7). Currently, there are no vaccines for fungal diseases, and antifungal therapeutic options are still limited (8, 9).
The fungal cell wall is an ideal target for antifungal chemotherapies since it is essential for fungal viability and virulence but absent in human cells (10). The fungal cell wall is composed of a complex matrix of glucose, proteins, lipids, and pigments (11). The cell wall of Cn is mainly composed of polymers of glucose (α- and β-glucans), N-acetylglucosamine (GlcNAc) (chitin and chitooligomers), glucosamine (chitosan), and glycoproteins (mannoproteins) (12). Similarly, the cell wall of Cg is mainly composed of α- and β-glucans, mannoproteins, and chitin (10).
Glucans and chitin are the most important structural components of fungal cell walls (13). Chitin can be degraded to chitooligomers, chitin-derived structures composed of 3 to 20 residues of GlcNAc (14). Chitooligomers are found around nascent buds and participate in connecting the cryptococcal cell wall to the capsule (14, 15). Also, chitin can be enzymatically deacetylated to chitosan (16). Most fungal pathogens expose chitin, but Cn is one of the few species that can evade the host immune system by replacing the chitin in its cell walls with chitosan (17). Mannose residues by N- or O-linkages can be found in association with proteins and are highly immunogenic (11).
The fungal cell wall is a dynamic structure that regulates cellular morphology (18) and remodels its components in response to environmental stresses, morphogenesis, cellular growth, and cell division (19). Aging is the consequence of cell division and a conserved natural process among eukaryotic cells. Replicative aging is the result of asymmetric cell divisions and is an important contributor to pathogenesis (20). During replicative aging, the fungal cell wall becomes thicker, which is associated with increased resistance to antifungals and phagocytic uptake (21, 22). However, the changes in cell wall composition and architecture during aging have not been elucidated. In this study, we investigate cell wall remodeling during aging in Cn and Cg. We report the increased expression of essential genes related to fungal cell wall biosynthesis. These transcriptome changes concur with our observation of enhanced levels of significant cell wall components (chitin, chitooligomers, and glucans) and enhanced cell wall thickness. Cell wall remodeling during aging is accompanied by formation of intracellular vesicles and multivesicular bodies, which suggests intensification of vesicular trafficking. In addition, our data indicate modifications in the vacuole morphology and pH, underscoring the importance of this organelle in the dynamics of progressive aging. Our data provide insights into the complex aging-related changes in fungal pathogens.
RESULTS
Regulation of cell wall biosynthesis genes is altered in older-generation cells.
Old yeast cells previously biotin-labeled and conjugated to streptavidin were separated when they reached the desired generation using a magnetic field, while nonlabeled young cells were washed off from the system. After isolation, we first performed a focused transcriptome analysis on known essential cell wall biosynthesis genes. Of the eight chitin synthases in Cn and four in Cg, CHS3 generates the majority of chitin (16, 23). CHS3 was upregulated in old Cn (3.3-fold change, P ≤ 0.0001) (Fig. 1A) but not in old Cg (Fig. 1B). Likewise, we observed upregulations of CHI22, an endochitinase involved in chitooligomer formation (24) in older Cn (2.38-fold change, P ≤ 0.0001) (Fig. 1A) and CTS1, an endochitinase (2.66-fold change, P ≤ 0.0001), in older Cg (Fig. 1B).
FIG 1.
Increased expression levels of genes related to cell wall biosynthesis in old C. neoformans (Cn) and C. glabrata (Cg). (A and B) Gene expression was analyzed by qRT-PCR in young (blue bars) and old (red bars) cells of RC2 Cn (A) and BG2 Cg (B). The data were normalized according to the gene expression of young cells, and the gene actin 1 (ACT1) was used as an internal control. The dotted lines represent 2-fold up or down of the respective genes. All experiments were done in technical triplicates and repeated at least two times independently. Error bars correspond to the standard deviations of at least two independent experiments (n = 2). A t test was performed to determine the P value (**, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
In Cn, chitin can be converted to chitosan by four putative chitin deacetylases (CDAs) (17, 25). We found that CDA1 transcription was downregulated (0.45-fold change, P ≤ 0.0001), whereas both CDA3 and CDA4 were upregulated (2.27-fold change and 2.08-fold change, respectively, P ≤ 0.001) in old Cn (Fig. 1A). In Candida species, chitosan is present only in the chlamydospore cell wall and is generated by two genes (CDA1 and CDA2) (26), of which CDA2 was upregulated (>2-fold change, P ≤ 0.0001) in old Cg (Fig. 1B). This is different from the Cn, where CDA2 was not upregulated in the old cells (Fig. 1A).
Transcription of AGS1, which encodes an α-1,3-glucan synthase, was upregulated in old Cn (9-fold change, P ≤ 0.0001) (Fig. 1A). In contrast, transcription of α-1,4-glucan synthase (GLC3) of Cg was not significantly altered Fig. 1B). Genes relevant for β-glucan synthesis, FKS1 and SKN1, did not significantly change in old Cn (Fig. 1A) but were downregulated in old Cg, genes FKS2 (0.27-fold change, P ≤ 0.05) and SKN1 (0.46-fold change, P ≤ 0.0001). FKS3 was increased (2.48-fold change, P ≤ 0.0001), whereas KRE2 and KRE5 remained not markedly altered in older Cg (Fig. 1B).
Yeast cell wall architecture is reshaped during aging.
Next, we assessed the cell wall main components by fluorescence microscopy (Fig. 2). In old Cn, chitin staining (by calcofluor white [CFW]) was enhanced surrounding the cell surface compared to in younger cells (Fig. 2A). In old Cg, chitin staining was enhanced but remained localized to bud scars in the cell wall (Fig. 2B). A similar pattern was noted with the binding of wheat germ lectin (WGA), which binds to chitooligomers. Again, it was abundant throughout the cell wall of most old Cn cells. In young Cn, chitooligomers are located in the area of the emerging developing buds (Fig. 2C). The latter pattern was similar to chitooligomer staining in young Cg. WGA staining of old Cg is also bound only to the numerous persistent bud scars (Fig. 2D), similar to the calcofluor binding to chitin. The deacetylated form of chitin, chitosan, binds specifically to the anionic dye eosin Y (25). Chitosan is uniformly enhanced and distributed through old Cn cell walls (Fig. 2E). In contrast, chitosan was not detected in Cg cell walls (data not shown).
FIG 2.
The cell wall architecture of C. neoformans (Cn) and C. glabrata (Cg) is remodeled during aging. Total chitin (blue) was stained with calcofluor white (CFW) (A and B), exposed chitooligomers (orange) with tetramethylrhodamine-labeled wheat germ agglutinin (WGA-TRITC) (C and D), chitosan (green) with eosin Y (EY) (E), β-glucan (blue) with Fc-dectin 1/DyLight 405 goat anti-human IgG + IgM (F), and mannan (red) with concanavalin-Texas Red (CoA-TR) (G and H). Stained cells were image by fluorescence microscopy, using the following channels: DAPI (CFW), DsRed (WGA- TRITC and CoA-TR), and GFP (EY). DIC, differential interference contrast. Imaging of at least 100 cells of Cn (RC2) and Cg (BG2) was performed at 100× magnification in a Zeiss Axio Observer microscope and a Zeiss Axiovert 200M microscope (Thornwood, NY), respectively. Images were processed using ImageJ/Fiji software. Scale bars correspond to 5 μm for Cn and 2 μm for Cg.
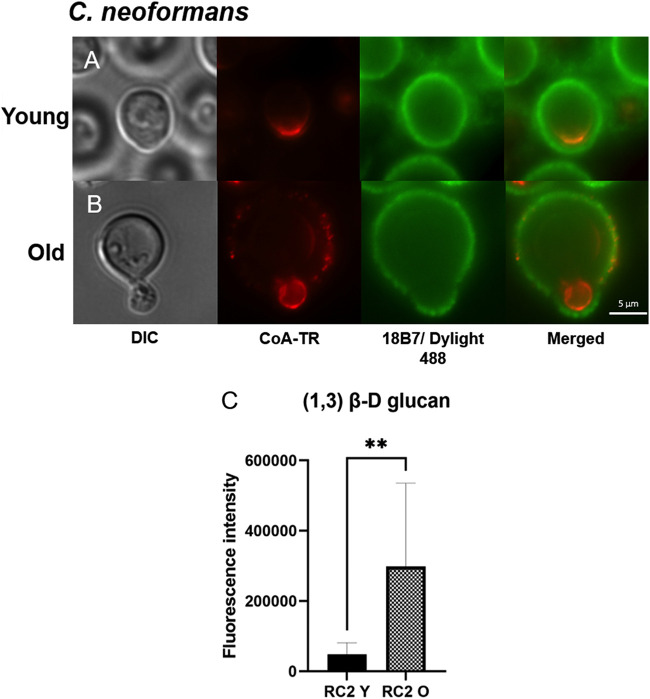
The Cn polysaccharide capsule prevents binding of the monoclonal antibodies (MAbs) MOPC-104E (α-glucan) and Fc-dectin (β-glucan); hence, like others, we were not able to assess α- and β-glucan levels (27, 28). Both young and old Cg exhibited comparable β-glucan staining of the cell surface (Fig. 2F). Finally, we found that mannoproteins were enriched in the region next to the developing buds in both young and old Cn. However, old Cn presented slightly greater intensity than young cells (Fig. 2G). Furthermore, mannoproteins of old cells also displayed a dotted staining pattern and colocalized in the capsule (Fig. 3B), which was not observed in young cells (Fig. 3A). In Cg, continuous staining of mannoproteins on the outer surface of the cell wall of young and old Cg cells was comparable (Fig. 2H).
FIG 3.
Colocalization of mannan in the polysaccharide capsule and enhanced content of β-glucan in the cell wall of old C. neoformans (Cn). (A and B) Images represent microscopy analysis of RC2 Cn young (A) and old (B) cells. DIC (differential interference contrast), followed by mannan (red) was stained with concanavalin A conjugated to Texas Red (CoA-TR), and GXM (green) in the capsule was stained with primary monoclonal Ab 18B7 and secondary Ab anti-IgG conjugated to DyLight 488 (18B7/DyLight 488). (C) The 1,3-β-d-glucan content of young cells (RC2 Y) and old Cn cells (RC2 O) was verified using biochemical assay with aniline blue. Experiments were done in biological triplicates, and statistical analysis was done by unpaired t test (**, P = 0.0062).
Old yeast cells presented increased levels of cell wall components.
Flow cytometry was used to quantify the relative levels of cell wall carbohydrates in both young and old Cn and Cg (Fig. S1). Mean fluorescence intensity (MFI) levels confirmed increased chitin levels in old Cn (P < 0.0001; Fig. 4A) and Cg (P = 0.014; Fig. 4B) compared to young cells. In agreement with microscopic findings, both older Cn and Cg exhibited significantly more chitooligomer quantity (P < 0.0001 and P = 0.0008, respectively; Fig. 4C and D). Although chitooligomers can be quantified by labeling with WGA, CFW penetrates the cell surface more efficiently, and it also binds to other chitin derivates (28). As expected, chitosan levels were also significantly higher in old Cn compared to young cells (P = 0.0058; Fig. 4E).
FIG 4.

Old C. neoformans (Cn) and C. glabrata (Cg) presented increased cell wall content levels. (A to H) By flow cytometry, analysis of levels in young (RC2 Y and BG2 Y) and old cells of Cn (RC2 O) and Cg (BG2 O) of total chitin (A and B), exposed chitooligomers (C and D), chitosan (E), β- 1,3-glucan (F), and mannoproteins (G and H). Unstained cells were sorted as controls to determined positive labeling. For each group, a total of 10,000 events were gated, and levels of chitin (blue; calcofluor white [CFW]), chitooligomers (orange; WGA-TRITC), chitosan (green; eosin Y [EY]), β-glucan (light blue; Fc-dectin1/DyLight 405), and mannan (red; concanavalin A, Texas Red conjugated) were represented by the mean fluorescence intensity (MFI). The following lasers were used: violet, 405 nm (for CFW and DyLight 405); YelGreen, 561 nm (for WGA-TRITC and CoA-TR); blue, 488 nm (for EY). All experiments were done in biological triplicates, and statistical analyses were performed by t test (****, P ≤ 0.0001; *** P ≤ 0.001; ** P ≤ 0.01; * P ≤ 0.05).
Flow cytometry analysis of young and old C. neoformans and C. glabrata cells. At least 10,000 events were gated in the FSC/SSC plots and are represented as histograms with mean fluorescence on the x axis and cell counts on the y axis. Unstained young (red) and old (green) cells were sorted as controls to determine positive labeling of young (blue) and old (orange) cells. CFW (calcofluor white, chitin), tetramethylrhodamine-labeled wheat germ agglutinin, chitooligomers (WGA-TRITC), eosin Y, chitosan (EY), Concanavalin A conjugated to Texas Red, mannan (CoA-Texas red), Fc-dectin1/DyLight 405 (β-glucan). Data were analyzed with FlowJo v10 software, and the image is representative of one experiment producing similar results that were obtained in three independent experiments. Download FIG S1, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Regarding the mannoprotein content, old Cn and Cg presented higher levels compared to younger cells (P = 0.0019 and P = 0.0015, respectively; Fig. 4G and H). β-d-glucan content in the Cn cell wall could only be quantified using a biochemical assay. These data indicated old Cn contained increased levels in comparison to young cells (P = 0.0062; Fig. 3C). Also, a higher level of β-glucan content was found in the cell wall of old Cg compared to that in young cells (P = 0.0025; Fig. 4F), which could not be documented by microscopy.
Old yeast cells developed thicker and robust cell walls, giant vacuoles, and increased multivesicular body-like structures.
As expected, transmission electron microscopy (TEM) analysis showed that older Cn had a significantly thicker cell wall compared to that of younger cells (235.9 versus 123.7 nm; P < 0.0001). Specifically, we found in young Cn that the inner cell wall layer measured on average 66.76 nm, and the outer layer measured 91.29 nm. In old cells, the average thickness of the inner layer was 103.4 nm and grew to 138.3 nm (P < 0.0001). These results suggest that both cell wall layers enlarged equally during replicative aging. Old Cg also showed notable growth in the total thickness of the cell wall compared to young cells (132.5 versus 93.11 nm; P < 0.0001). Here, the inner cell wall layer was thicker than the outer. Specifically, in old Cg the inner layer measured 103.5 nm versus 78.21 nm in young cell (P = 0.0007). Conversely, the outer cell layer of young Cg was thicker than that of old Cg (34.75 nm versus 25.39 nm; P = 0.0017) (Fig. 5B).
FIG 5.
Old yeast cells present thicker cell walls and an accumulation of vesicle-like structures in the cytosol. (A and B) Quantification of the thickness of the inner (squares), outer (triangles), and total (inner + outer) (circles) layers of young (blue) and old (red) C. neoformans (Cn) (A) and C. glabrata (Cg) (B). Data represent the analysis of 30 individual cells for each group by ImageJ/Fiji software. An unpaired t test with Welch’s correction was used to compare the pairs (young and old) of each group, and error bars represent the standard deviation (****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; ####, P ≤ 0.0001; ##, P ≤ 0.01). The asterisk symbol (*) represents differences between the groups young and old, whereas the pound symbol (#) represents differences within the groups young or old. (A to J) Representative electron micrographs showing the ultrastructural changes of young and old cell walls of Cn (RC2) (C to F) and Cg (BG2) (G to J). White arrows point to vesicle-like particles in the cytoplasm of old yeast cells.
Next, we tested if replicative aging affected cell wall strength. Cell wall integrity is vital for maintaining cell wall functionality and structure (29). We assessed the sensitivity of old cells to cell wall stressors, including Congo red (inhibits assembly of cell wall polymers such as chitin), caffeine (interferes with cell wall-related signal transduction pathways), and sodium dodecyl sulfate (disrupts plasma membrane) (28, 30). These data showed aging had no impact on resistance of the cell wall in old Cn and Cg (Fig. S2).
No impact on cell wall integrity was observed in old C. neoformans (Cn) and C. glabrata (Cg) cells. Young and old cells of Cn and Cg were serially diluted and spotted onto YPD with the addition of several cell wall stressors (caffeine, FB 28 [fluorescent brightener 28], Congo red, and sodium dodecyl sulfate [SDS]) at 30°C and 37°C for 48 h. This experiment was repeated twice with similar results. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The ultramicroscopic images showed that old cells of both yeasts exhibited expanded vacuoles with amorphous lumen content, occupying a significant fraction of the total cell (Fig. 5E and I). Interestingly, the cytoplasm and cell wall (Fig. S3) of old cells contained a large number of electron-dense particles (Fig. 5E and I), suggestive of vesicles derived from the conventional secretion pathway. The vacuoles in old Cn (Fig. 6D) and Cg (Fig. 6H) contained vesicles resembling multivesicular bodies (MVBs). MVB-like structures accumulated in 67.31% of old Cn, whereas they were found in only 24.53% of young cells. In a similar fashion in old Cg, MVB-like structures were present in 33.96% of old cells and only in 4% in young cells (Table 1).
FIG 6.
Alterations of vacuolar morphologies in young and old C. neoformans (Cn) and C. glabrata (Cg). Representative transmission electron microscopy (TEM) images of young and old cells of Cn in the upper panel (A to D) and Cg in the lower panel (E to H). Boxed areas illustrating vacuoles (ordinary or oversized, with or without intravacuolar vesicles) (left panels), were magnified in the right panels. Scale bars represent 500 nm.
TABLE 1.
Distribution of multivesicular body (MVB)-like structures in young and old cells of C. neoformans (Cn) and C. glabrata (Cn)a
| Strain | Vacuole | Young (%) (n) | Old (%) (n) |
|---|---|---|---|
| Cn (RC2) | Ordinary | 64.15 (34) | 32.69 (17) |
| Oversized, MVB-like | 24.53 (13) | 67.31 (35) | |
| Other | 11.32 (6) | 0 | |
| Cg (BG2) | Ordinary | 80 (40) | 49.05 (26) |
| Oversized, MVB-like | 4 (1) | 33.96 (18) | |
| Other | 16 (8) | 16.99 (9) |
Transmission electron microscopy (TEM) images of at least 50 cells from each group were analyzed according to the size of the vacuole and the presence of MVB-like structures.
Ultramicroscopic image of old C. neoformans cells with numerous vesicle-like particles in the cytosol, inside the vacuole, and crossing the cell wall indicated by white arrows. Scale bar represents 500 nm. Download FIG S3, TIF file, 0.5 MB (492.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Old Cn accumulated intracellular GXM and upregulated secretion gene SEC14.
Cn extracellular vesicles (EVs) contain glucuronoxylomannan (GXM) (31). Analysis of ultrathin sections of both young and old Cn labeled with MAb 18B7 for GXM (32) revealed a significant increase in GXM detection in the intracellular space in old compared to young Cn (Fig. 7A to E). Immunogold-labeled antibodies detected GXM in the vacuole (Fig. 7D) and also bound to vesicle-like structures in the cell wall (Fig. 7E) of old yeast. We also tested transcription of two genes essential for secretion, SEC14 and SAV1 (33–35). SEC14 transcription was upregulated (3.09-fold change), whereas SAV1 transcription was not significantly altered in old Cn (Table 2).
FIG 7.
Intracellular GXM was enriched in old C. neoformans (Cn). (A and B) Quantitative analysis of immunogold labeling to detect intracellular GXM with monoclonal antibody to GXM (MAb 18B7) in young (A) and old (B) Cn (RC2). In panel C, error bars represent standard deviations. Statistical significance was evaluated using an unpaired t test (****, P ≤ 0.0001). For each group (young and old), 50 cells were analyzed. A significantly increased amount of intracellular GXM was detected in old Cn compared to young cells. (D and E) Illustrative images of an old Cn cell showing the association of GXM inside vacuole (D) and with vesicular structures in the cell wall (E). Right panels in D (pink) and E (blue) represent magnifications of the boxed areas in left panel B.
TABLE 2.
Expression of genes relevant for conventional secretion mechanisms, regulation of vacuole pH, and calcium homeostasis in young and old cells of C. neoformans
| Gene ID | Gene name | Young (fold-change) | Old (fold-change) |
|---|---|---|---|
| CNAG_02817 | SAV1 | 1.034 | 0.517 |
| CNAG_03153 | SEC14 | 1.065 | 3.099a |
| CNM_02200 | ALL2 | 1.041 | 1.800 |
| CNAG_01704 | CMR1 | 1.121 | 4.846a |
Statistical difference was considered when the fold-change was greater than 2-fold.
The number, size, and pH of vacuoles are affected in old yeast cells.
In fungi, the vacuole participates in secretory pathways, Ca2+ storage, and pH regulation (36, 37). Because reduced vacuolar acidity has been reported in aged Saccharomyces cerevisiae (38), we further investigated modifications in vacuole morphology and pH in the old cells. First, we stained the vacuolar membranes with FM 4-64 dye (Fig. 8A and D). The average vacuole area was significantly larger in old compared to young cells, corresponding to an increase of 3.46-fold for Cn (0.7481 μm2 versus 0.2162 μm2, P < 0.0001) and 4-fold in Cg (0.04 nm2 versus 0.01 nm2, P < 0.0001). The surface area of old cells was 1.82-fold increased in Cn (41.38 μm2 versus 22.68 μm2, P < 0.0001) and 1.63-fold in Cg (12.93 nm2 versus 7.89 nm2, P < 0.0001) (Fig. S4). Calculations of the ratio of vacuole and cell size confirmed disproportional vacuolar growth in old Cn (P = 0.0155; Fig. 8B) and Cg (P < 0.0001; Fig. 8E). The average number of vacuoles in young cells was higher in Cn (5.9 versus 7.8 for old and young cells, respectively, P = 0.0231; Fig. 8C) and Cg (3.9 versus 6.4 for old and young, respectively, P < 0.001; Fig. 8F). We hypothesize that the enlargement of vacuoles in aging cells could be the result of the fusion of multiple vacuoles.
FIG 8.
Vacuole morphology and pH are affected in yeast old cells. (A and D) Images depicting vacuolar morphology (white arrows) stained with FM4-64 in young (Y) and old (O) cells of C. neoformans (Cn) (A) and C. glabrata (Cg) (D). DIC, differential interference contrast. (B and E) The ratio of vacuolar/cell area was significantly increased in old (red) Cn (B) and Cg (E) compared to younger cells (blue). An unpaired t test with Welch’s correction was used to generate the P-values (*, P = 0.0245; ****, P < 0.0001). (C and F) The average number of vacuoles per yeast cell was significantly lower in old (red) Cn (C) and Cg (F) compared to the respective young cells (blue). An unpaired t test with Welch’s correction was used (**, P = 0.0098; ****, P < 0.0001). For all analyses, 25 cells of each group were measured using ImageJ/Fiji software. Measurement of quinacrine levels with a spectrophotometer showed a significant decrease in fluorescence in aged Cn compared to young cells (G). This was confirmed with microscopy. In contrast, significantly increased fluorescence was measured in old cells of Cg compared to the young cells (H). This was also confirmed with microscopy. The assay was done in biological triplicate and an unpaired t test with Welch’s correction was used to generate the P values (*, P = 0.0155; ***, P = 0.0007).
(A to D) Cell surface and vacuolar areas are significantly increased in old C. glabrata (A and B) and C. neoformans (C and D). Fungal cells were stained with FM4-64, and analyses of 25 cells for each group (young and old) and strain were measured using ImageJ/Fiji software. Statistical analyses were performed by t test with Welch’s correction (****, P ≤ 0.0001). Download FIG S4, TIF file, 0.6 MB (639.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, the vacuole pH of young and old cells was assessed with quinacrine staining, a fluorescent probe that increases under acidic conditions (39). In Cn, higher pH was observed in the vacuoles of old compared to young cells (2.98 × 106 MFI versus 3.85 106 MFI, P = 0.0155) (Fig. 8G). Consistent with this finding ALL2 expression, which is linked to the homeostasis of intracellular pH in Cn (40), trended up, and CMR1, which promotes calcium homeostasis (41), was significantly upregulated (4.84-fold increase, P < 0.005) (Table 2). In contrast, old Cg cells exhibited lower pH in vacuoles compared to young cells (3.6 × 106 MFI versus 2.6 × 106 MFI, P = 0.0007) (Fig. 8H).
A simplified illustration model summarizes the phenotypic characteristics observed in this study during replicative aging in Cn and Cg in Fig. 9.
FIG 9.
(A and B) Illustrative summary of phenotypic modifications observed during replicative aging in the pathogenic yeast C. neoformans (A) and C. glabrata (B). During aging, cell wall increases in thickness produce enlarged vacuoles and more multivesicular bodies (MVBs). Blue shapes indicate total chitin, dark green shapes indicate chitosan, orange shapes indicate chitooligomers, black shapes indicate melanin, red pentagons indicate mannan, and blue squares and green circles indicate β-glucan. The illustration was created using BioRender.
DISCUSSION
The cell wall remains an attractive target for developing fungal vaccines (9) and antifungal drugs (10) because it is essential for fungal virulence (11). This study demonstrates that critical genes for cell wall synthesis are upregulated during replicative aging of Cn and Cg. The development of a thicker cell wall in old cells is probably facilitated by increased intracellular trafficking, associated with the formation of oversized vacuoles that contain MVB-like structures. Additionally, our results indicate that the morphology and pH of vacuoles are affected in old yeast cells. The observed reorganization of the cell wall partially explains the enhanced resistance to phagocytosis and antifungals, which has been observed in Cn, Cg, Candida albicans and Candida auris (21, 22, 42, 43).
Higher chitin levels result from budding and are implicated in increasing antifungal resistance, albeit not in all fungal species (44–46). For instance, chitin content is associated with caspofungin resistance in Cn (45), but not in Cg (44). Specifically, increasing chitin levels were noted throughout the life span in both yeasts, similar to Saccharomyces cereviseae (47). Staining patterns for chitooligomers mimicked the chitin staining pattern with more diffuse staining throughout the cell wall of old Cn and are strictly bud scar-associated in Cg. In Cn, binding of chitooligomer-specific antibodies increases fungicidal efficacy of amphotericin B (48). As chitooligomers are fungi-specific, this novel therapeutic approach could be explored to treat the enhanced antifungal tolerance of old Cn and possibly Cg.
In Cryptococcus species, chitosan is a critical virulence factor, and its levels can exceed those of chitin (16, 49, 50). It is produced from chitin by deacetylating enzymes (25), and CDA3 transcription was upregulated in old Cn. Cda3 activity was more important during infection by Cryptococcus gattii (49); however, Cda1 (51) and Cda2 were the main regulators for Cn pathogenesis under host conditions (52). Chitosan inhibits protective Th1-type adaptive immune responses (53), maintains cell integrity, and allows melanization, thereby enhancing antifungal resistance (54–56). Our data suggest that higher chitosan content in the cell wall promotes enhanced melanization of old Cn cells during replicative aging (21). A common architecture for chitosan-containing elements in the fungal cell wall from ascomycetes to basidiomycetes has been proposed based on nuclear magnetic resonance (NMR) studies (57). Although Cg produces a melanin-like pigment (58), it is unknown if it accumulates during replicative aging. Chitosan was not detected in any Cg cells, similar to other Candida species, where expression is limited to the chlamydospore (26).
Higher β-1,3-d-glucan levels were confirmed in old Cn by biochemical assays and in old Cg by fluorescent staining. Two of the three FKS genes involved in the synthesis were found to be upregulated in old Cg (42). However, FKS1 levels in old Cn were not significantly increased. Transcript and protein levels do not always correlate, and unstable mRNA can be translated more efficiently (59). In Cn lacking posttranscriptional gene regulator PUF4, the FKS1 mRNA and Fks1p levels did not correspond (45). Unfortunately, we were unable to quantify α-glucan with the available methods, but the α-glucan synthase AGS was upregulated in old Cn, indicating that this glucan is likely also increased. Nevertheless, it is still unclear whether the quantification of the cell wall content may be influenced by the enlargement of old cells. Further analysis, such as high-performance liquid chromatography, would be required to resolve this issue (60, 61). However, the low yield of old cells isolated from yeast culture is still currently a limitation for performing this technique.
Fungal pathogens can evade host immune recognition by masking β-glucans at the cell wall surface (62, 63). The exposure of β-glucans is reduced by Cn’s capsule (60) and by mannan in the outer cell wall layer in Candida species (44, 64). Mannan is present in Cn polysaccharide capsule (65, 66); thus, we propose that the observed accumulation of mannan could be due to augmented vesicle export. Alternatively, an altered permeability in the polysaccharide capsule in old cells (67) may efficiently expose the mannan epitopes. Altered mannoprotein content may contribute to the poor recognition of old yeast by host phagocytes (21, 22), which is suggested by data from C. gattii (68) and C. albicans (69).
Comparisons of TEM images of Cn and Cg identified differential growth of their inner and outer cell wall layer. The outer cell wall layer was thicker in old Cn, although the inner cell wall layer dominated in Cg. Chitinous structures and chitosan are evenly distributed across the cell wall of Cn but must be flexible enough to permit even expansion of the cell wall. We hypothesize that the heterogeneous composition of the inner cell wall in Cg results from chitin breaks in the bud scars, similar to S. cerevisiae (47), and causes the wavy cell wall. It is conceivable that the mannan layer in old Cg is more compactly arranged than in young cells. Reduced fibril length on the cell surface has been correlated with increased exposure on β-1,3-glucan (44). Alternatively, the outer layer of aged Cg can be more fragile and could have partially detached during ultrastructural processing. The increase in the inner cell wall layer thickness avoids a hyperosmotic shock and favors fungal cell survival (70). Most importantly, analogous to S. cerevisiae (47), old and young cells exhibit comparable tolerance to common cell wall stressors. Furthermore, comparable doubling times of 10th-generation (10GEN) old Cn as well as 14GEN old Cg cells (22, 71) indicate that old cells are equally fit in the host environment.
Another important discovery is the development of oversized vacuoles with multiple vesicle-like structures in the cytoplasm of old yeast cells. Enlarged vacuoles have been described in old as well as nutrient-deprived S. cerevisiae cells (37, 38, 72). Vacuoles play a role in membrane trafficking (73) and protein sorting (37). Secretion mechanisms can also be influenced by autophagic processes (74). The autophagosome can merge first with MVBs and then with the cell membrane to deliver heterogeneous cargo outside the cell (75). In Cn, MVBs participate in laccase trafficking (76), a cell wall-associated virulence factor (77); in addition, GXM and melanin are exported through EVs (31, 78). Cytoplasmic MVBs can originate fungal EVs (79). Cell wall architecture proteins were found in EVs from Cn (80), Cg (81), S. cerevisiae (82), Histoplasma capsulatum (83), and Aspergillus fumigatus (84). The cloning of a “mother cell enriching mutant” in Cn and Cg, similar to the one developed in S. cerevisiae (85), would permit prolonged growth of old cells, with isolation and analysis of their EV content.
The enlarged vacuole in old Cn and Cg cells is most likely the result of the fusion of smaller vacuoles. Vacuolar fusion in S. cerevisiae is necessary and sufficient for life span extension (86). Vacuolar acidification is also a critical determinant of replicative life span and declines continuously with age, thereby limiting the replicative life span of a cell (87, 88). In C. albicans, vacuolar pH is a regulator of fission and fusion of vacuoles (89). We demonstrated loss of vacuolar acidity in old compared to young Cn similar to S. cerevisiae (38) but different from old Cg, where the vacuole pH was more acidic. Different factors, including glucose starvation, can also affect vacuolar pH (90). It is possible that Cg is experiencing more carbon starvation and has not advanced in its life span as much as Cn. Differences in pH regulation during aging and carbon starvation have been described in different Cn strains (91). Cn has a unique protein (All2p) involved in pH homeostasis (92), which accumulates during aging in low-glucose conditions (40). More extensive analysis will be necessary to better understand the mechanisms involved in age-associated regulation of intracellular pH in these yeast cells, including the use of other pH probes that consistently colocalize within the vacuoles (93). It is noteworthy that these studies were conducted on a single strain for each pathogen, and therefore, further work including several strains of different genotypes is required.
In summary, these data provide insights regarding the dynamics of cell wall rearrangement during aging of Cn and Cg. The data highlight the divergence between these pathogens in bud scar formation throughout the life span and uncover differences such as cell wall composition, especially concerning mannan and chitosan. This work opens new avenues of investigation, such as the impact of cell wall reorganization in old yeast cells on the host innate response and sensitivity to cell wall-targeting antifungals. Further work is also needed to elucidate the implications of EV secretion during aging, including cargo and “cross talk” between young and old pathogenic yeast cells and explore the association among aging, intracellular pH, and mitochondrial function.
MATERIALS AND METHODS
Strains and media.
The fungal strains Cn RC2 (ATCC 24067 variant) and Cg BG2 were kept on yeast extract-peptone-dextrose (YPD) agar (Difco BD) plates. Synthetic medium (SM) (43) was used for cultivation of yeast cells.
Isolation of young and old Cn and Cg cells.
Yeast cells (108) from an overnight culture were washed with phosphate-buffered saline (PBS) and labeled with 8 mg/mL sulfosuccinimidyl-6-(biotin-amido) hexanoate (Sulfo-NHS-LC-LC-Biotin, 21338; Thermo Fisher Scientific) for 30 min at room temperature (RT). Subsequently, cells were washed with PBS and grown for 5 to 7 doubling times. Then, 100 μL of magnetic streptavidin microbeads (130-048-101; Miltenyi Biotec) were added to every 108 cells/mL, following incubation for 15 min at 4°C. Biotin-labeled yeasts were isolated using autoMACS magnetic columns (130-021-101; Miltenyi Biotec) and the autoMACS Pro separator (Miltenyi Biotec). The biotin-streptavidin-labeled older cells were passed through a magnet where they get stuck, while the younger unlabeled population flowed through. The labeled cells were then recovered after the magnetic field was removed. The positive cell fraction was grown again in SM to the desired generation (10GEN Cn, 14GEN Cg) and isolated as outlined above. Young cells washed off from the magnetic columns were kept as controls. The purity of fractions was verified by microscopy. In this work, “old” Cn are 10 generations old, and “old” Cg are 14 generations old.
RNA isolation and quantitative reverse transcriptase PCR (RT-qPCR).
RNA isolation and RT-qPCR were done as described (22). Briefly, the RNAeasy kit (Qiagen) was used with on-column DNase digestion following the manufacturer’s instructions. The concentration of total RNA was measured using a NanoDrop instrument (Eppendorf). The quantity of RNA was adjusted to 250 ng and converted to cDNA using the Verso cDNA synthesis kit (Thermo Scientific). cDNA was diluted and used in a mix that contained Power SYBR green mastermix (Applied Biosystems), together with oligo(dT) primers (Table S1). The experiment was performed using the high-performance real-time PCR (Roche) and LightCycler 480 systems (Roche). Gene expression was computed using ΔΔCT. ACT1 was used to normalize the gene expression, and the gene expression in young cells was used as the reference. Values above 2-fold were significant. Threshold cycle (CT) values of ACT1 in old and young cells were similar to one another, signifying that ACT1 gene expression does not vary between the young and old cells. Each experiment was repeated at least twice on two separate days.
Sequence of primers (5′-3′) used in this study. Download Table S1, PDF file, 0.1 MB (78.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cell wall staining.
Young and old cells (106) were fixed in 4% paraformaldehyde (PFA) for 15 min at RT, washed with PBS, and stained with calcofluor white (CFW; cell wall staining of chitin; 5 μg/mL for 10 min at 37°C), concanavalin A, Texas Red conjugate (ConA-Texas Red; cell wall staining of mannoproteins; 50 μg/mL for 45 min at 37°C), eosin Y (EY, cell wall staining of chitosan; 300 μg/mL for 10 min at 37°C) (94), or tetramethylrhodamine-labeled wheat germ agglutinin (TRITC-WGA; cell wall staining of chitooligomers; 5 μg/mL for 30 min at 37°C) (14).
To stain β-glucan and GXM, fungal cells were blocked with 1% bovine serum albumin (BSA) for 1 h at 37°C and incubated, respectively, with the primary antibody Fc-dectin 1 (AdipoGen; AG-40B-0138) (5 μg/mL; 40 min at 4°C) (28) or MAb 18B7 (10 μg/mL; 1 h at 37°C) (32). After washing with PBS, cells were incubated with secondary antibody DyLight 405 goat anti-human IgG + IgM (5 μg/mL; 30 min at 4°C) or with an anti-murine IgG DyLight 488 (10 μg/mL; 30 min at 37°C) (Jackson ImmunoResearch).
Prior to analysis using a BD LRSFortessa flow cytometer, cells were washed and resuspended in PBS. The following lasers were used: blue, 488 nm (for EY); YelGreen, 561 nm (for ConA-Texas Red and WGA-TRITC); violet, 405 nm (for CFW and 405 DyLight). Data were analyzed using BD FACSDiva or FlowJo v10.1 software. A total of 10,000 events were gated in the forward scatter/side scatter (FSC/SSC) plots and represented as histograms with mean fluorescence intensity (MFI) on the x axis and cell counts on the y axis. Unstained cells were used as negative controls. For fluorescence microscopy, the following channels were used: fluorescein isothiocyanate (FITC) (EY and DyLight 488), DAPI (4′,6-diamidino-2-phenylindole) (CFW and DyLight 405), and DsRed (Texas Red and TRITC). Imaging was performed at 100× magnification in a Zeiss Axio Observer or a Zeiss Axiovert 200M (Thornwood, NY). The same exposure time was used to image young and old cells. The images were processed using ImageJ software.
(1,3) β-d-glucan quantification.
Cn cells (4 × 106) were washed with Tris-EDTA buffer (pH 8.0) and suspended in 500 μL of Tris-EDTA and 56 μL of NaOH (10 M) to achieve a final concentration of 1 M. To solubilize (1,3) β-d-glucan, cells were incubated at 80°C for 30 min. Then, 2.1 mL of aniline blue solution (0.03% aniline blue, 0.18 M HCl, 0.49 M glycine/NaOH, pH 9.5) was added to the cells to interact with linear (1,3) β-d-glucan. Following homogenization, cells were incubated for 30 min at 50°C and for 30 min at RT. The fluorescence was measured using a plate reader SpectraMax i3X (Molecular Probes), with excitation at 400 nm and emission at 460 nm (95).
Ultramicroscopy.
Cells were fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, at 4°C overnight. Then, cells were washed 3 times with 0.1 M sodium cacodylate buffer and dispersed in ultra-low-temperature agarose at 4°C for 30 min. Subsequently, the agarose samples were cut into small cubes, and the pieces were postfixed in aqueous 1% potassium permanganate (KMNO4) for 1 h at RT. Blocks were rinsed with distilled water (dH2O) and treated with 0.5% sodium meta-periodate for 15 min at RT to allow infiltration. Next, blocks were rinsed with dH2O and dehydrated through a graded ethanol series. Samples were treated with 100% propylene oxide, embedded with Spurr resin, and polymerized in an oven at 60°C for 24 h. Ultrathin sections (80 nm) were cut using a Leica EM UC7 ultramicrotome, placed on 300-mesh copper grids, and counterstained with uranyl acetate and lead citrate. Images were acquired at magnifications of 18,500× and 49,000× using an FEI TeCnai 12 BioTwinG2 TEM. Digital images were acquired with an AMT XR 60 CCD digital camera system. At least 30 young and old cells were analyzed, and the thickness of cell wall layers was measured with ImageJ software.
Intracellular glucuronoxylomannan (GXM) of Cn.
Cn cells were fixed for 45 min at RT in fixate solution (synthetic medium + 4% PFA + 0.1% glutaraldehyde + 0.1 M cacodylate buffer). Samples were washed twice with 0.1 M cacodylate buffer, suspended in 0.1 M cacodylate buffer, and left overnight at 4°C. The cells were washed with PBS, placed in gelatin, cut into cubes of 1 mm, and postfixed in 0.1 M sodium cacodylate containing 2.0% paraformaldehyde. Samples were then dehydrated through a graded series of ethanol, with a progressive lowering of the temperature to −50°C in a Leica EMAFS, embedded in Lowicryl HM-20 mono-step resin (Electron Microscopy Sciences), and polymerized using UV light. Ultrathin sections were cut on a Leica EMUC7, and immunolabeled with MAb 18B7 (50 μg/mL) and gold-labeled anti-mouse IgG. Control systems included ultrathin sections incubated without the MAb 18B7, followed by the gold-labeled anti-mouse IgG. Samples were stained with uranyl acetate and viewed on a JEOL 1400 Plus transmission electron microscope at 80 kv. Gold particles in electron micrographs were quantified as described (96). For quantitative analysis of GXM labeling, 50 cells of the young group and 47 cells from the old group were analyzed. For normalization, the average number of gold particles counted in control systems (no MAb 18B7 labeling) was subtracted from the total number of gold particles of each MAb 18B7-labeled section, as described reference 97.
Vacuole morphology and pH analysis.
Vacuoles from 106 fungal cells were stained at 37°C with FM 4-64 (Thermo Fisher) (10 μg/mL) for 10 min following manufacturer’s protocol. Another set of 106 fungal cells were labeled with quinacrine in separate tubes at a final concentration of 200 μM and incubated for 20 min at 37°C. Cells were washed three times with PBS and imaged at 100× using a deconvolution microscope (Zeiss Axiovert 200M), using the filters Texas Red (for FM4-64) and green fluorescent protein (GFP; for quinacrine). Then, 25 cells from each group and strain were analyzed using ImageJ software.
Also, 200 μL of yeast suspensions (106) labeled with quinacrine were plated in triplicates in a black 96-well plate, and the fluorescence was measured with a Spectramax i3X spectrophotometer (Molecular Probes), using excitation at 436 nm and emission at 525 nm. The values of unstained cells were subtracted from the values obtained from the stained cells. All experiments were repeated three times independently.
Cell wall stressor assay.
Serial dilutions of yeast suspensions (final concentrations 105 to 102 cells) were spotted on YPD agar containing each of the following stressors: fluorescent brightener 28 (FB 28), 1.5 mg/mL; Congo red, 0.5%; caffeine, 1 mg/mL or 0.5 mg/mL; sodium dodecyl sulfate (SDS), 0.01% or 0.0025%. Next, the plates were incubated at 30°C and 37°C for 48 h. Control conditions included cells added on YPD agar without the stressors. For Congo red, the stressor was added to the YPD medium prior to autoclaving, whereas for caffeine, FB 28, and SDS, stock solutions were prepared, filter-sterilized, and added to YPD after autoclaving (28).
Statistical analysis.
Statistical differences between young and old cells were performed using a t test with Welch’s correction. All statistical tests were performed with Prism 9.0 software (GraphPad Software, Inc.).
ACKNOWLEDGMENTS
TEM sample preparation and imaging were performed at the C-MIC: Central Microscopy Imaging Center at Stony Brook University, Stony Brook, New York, and alternatively, at the Analytical Imaging Facility, Albert Einstein College of Medicine, Bronx, New York. We also are thankful to the Flow Cytometry Research Core Facility at Stony Brook Hospital and Del Poeta for allowing us to use the fluorescent microscope. MAb 18b7 was a gift from Arturo Casadevall.
This work was financially supported by the National Institutes of Health (NIH 1R01-AI127704-01A1 to B.C.F.). B.C.F. is also supported by VA Merrit I01BX003741-01A2. The contents of this article do not represent the views of the Veterans Administration (VA) or the United States government.
Footnotes
This article is a direct contribution from Bettina C. Fries, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Stuart Levitz, University of Massachusetts Medical School, and Peter Williamson, National Institutes of Health.
Contributor Information
Bettina C. Fries, Email: Bettina.Fries@stonybrookmedicine.edu.
J. Andrew Alspaugh, Duke University Medical Center
REFERENCES
- 1.Vallabhaneni S, Mody RK, Walker T, Chiller T. 2016. The global burden of fungal diseases. Infect Dis Clin North Am 30:1–11. doi: 10.1016/j.idc.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Nnadi NE, Carter DA. 2021. Climate change and the emergence of fungal pathogens. PLoS Pathog 17:e1009503. doi: 10.1371/journal.ppat.1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A. 2020. Climate change brings the specter of new infectious diseases. J Clin Invest 130:553–555. doi: 10.1172/JCI135003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel, Switzerland) 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Healey KR, Perlin DS. 2018. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J Fungi (Basel) 4:105. doi: 10.3390/jof4030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. 2018. Invasive candidiasis. Nat Rev Dis Primers 4:18026. doi: 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- 8.Perfect JR. 2017. The antifungal pipeline: a reality check. Nat Rev Drug Discov 16:603–616. doi: 10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira LVN, Wang R, Specht CA, Levitz SM. 2021. Vaccines for human fungal diseases: close but still a long way to go. NPJ Vaccines 6:33. doi: 10.1038/s41541-021-00294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibe C, Munro CA. 2021. Fungal cell wall: an underexploited target for antifungal therapies. PLoS Pathog 17:e1009470. doi: 10.1371/journal.ppat.1009470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N. 2020. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front Microbiol 10. doi: 10.3389/fmicb.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gow NAR, Latge JP, Munro CA. 2017. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr 5:5.3.01. doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Herrera J, Ortiz-Castellanos L. 2019. Cell wall glucans of fungi. a review. Cell Surf 5:100022. doi: 10.1016/j.tcsw.2019.100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues ML, Alvarez M, Fonseca FL, Casadevall A. 2008. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell 7:602–609. doi: 10.1128/EC.00307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonseca FL, Guimarães AJ, Kmetzsch L, Dutra FF, Silva FD, Taborda CP, Araujo GDS, Frases S, Staats CC, Bozza MT, Schrank A, Vainstein MH, Nimrichter L, Casadevall A, Rodrigues ML. 2013. Binding of the wheat germ lectin to Cryptococcus neoformans chitooligomers affects multiple mechanisms required for fungal pathogenesis. Fungal Genet Biol 60:64–73. doi: 10.1016/j.fgb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hembach L, Bonin M, Gorzelanny C, Moerschbacher BM. 2020. Unique subsite specificity and potential natural function of a chitosan deacetylase from the human pathogen Cryptococcus neoformans. Proc Natl Acad Sci USA 117:3551–3559. doi: 10.1073/pnas.1915798117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Free SJ. 2013. Fungal cell wall organization and biosynthesis. Adv Genet 81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 19.Sherrington SL, Sorsby E, Mahtey N, Kumwenda P, Lenardon MD, Brown I, Ballou ER, MacCallum DM, Hall RA. 2017. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog 13:e1006403. doi: 10.1371/journal.ppat.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya S, Bouklas T, Fries BC. 2020. Replicative aging in pathogenic fungi. J Fungi (Basel) 7:6. doi: 10.3390/jof7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orner EP, Bhattacharya S, Kalenja K, Hayden D, Del Poeta M, Fries BC. 2019. Cell wall-associated virulence factors contribute to increased resilience of old Cryptococcus neoformans cells. Front Microbiol 10:2513. doi: 10.3389/fmicb.2019.02513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouklas T, Alonso-Crisóstomo L, Székely T, Jr, Diago-Navarro E, Orner EP, Smith K, Munshi MA, Del Poeta M, Balázsi G, Fries BC. 2017. Generational distribution of a Candida glabrata population: resilient old cells prevail, while younger cells dominate in the vulnerable host. PLoS Pathog 13:e1006355. doi: 10.1371/journal.ppat.1006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlet R, Pruvost Y, Tumba G, Istel F, Poulain D, Kuchler K, Sendid B, Jawhara S. 2018. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci Rep 8:3316. doi: 10.1038/s41598-018-21422-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker LG, Specht CA, Lodge JK. 2009. Chitinases are essential for sexual development but not vegetative growth in Cryptococcus neoformans. Eukaryot Cell 8:1692–1705. doi: 10.1128/EC.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bemena LD, Min K, Konopka JB, Neiman AM. 2021. A conserved machinery underlies the synthesis of a chitosan layer in the Candida chlamydospore cell wall. mSphere 6:e00080-21. doi: 10.1128/mSphere.00080-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese AJ, Doering TL. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol 50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- 28.Esher SK, Ost KS, Kohlbrenner MA, Pianalto KM, Telzrow CL, Campuzano A, Nichols CB, Munro C, Wormley FL, Jr, Alspaugh JA. 2018. Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition. PLoS Pathog 14:e1007126. doi: 10.1371/journal.ppat.1007126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Oliveira HC, Rossi SA, García-Barbazán I, Zaragoza Ó, Trevijano-Contador N. 2021. Cell wall integrity pathway involved in morphogenesis, virulence and antifungal susceptibility in Cryptococcus neoformans. J Fungi (Basel) 7:831. doi: 10.3390/jof7100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram AFJ, Klis FM. 2006. Identification of fungal cell wall mutants using susceptibility assays based on calcofluor white and Congo red. Nat Protoc 1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, Kozel TR, Lendvai N, Mukherjee J, Pirofski LA, Rivera J, Rosas AL, Scharff MD, Valadon P, Westin K, Zhong Z. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother 42:1437–1446. doi: 10.1128/AAC.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoneda A, Doering TL. 2009. An unusual organelle in Cryptococcus neoformans links luminal pH and capsule biosynthesis. Fungal Genet Biol 46:682–687. doi: 10.1016/j.fgb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chayakulkeeree M, Johnston SA, Oei JB, Lev S, Williamson PR, Wilson CF, Zuo X, Leal AL, Vainstein MH, Meyer W, Sorrell TC, May RC, Djordjevic JT. 2011. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol Microbiol 80:1088–1101. doi: 10.1111/j.1365-2958.2011.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneda A, Doering TL. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell 17:5131–5140. doi: 10.1091/mbc.e06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Herman PK, Emr SD. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veses V, Richards A, Gow NAR. 2008. Vacuoles and fungal biology. Curr Opin Microbiol 11:503–510. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Henderson KA, Hughes AL, Gottschling DE. 2014. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife 3:e03504. doi: 10.7554/eLife.03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierzyńska-Mach A, Janowski PA, Dobrucki JW. 2014. Evaluation of acridine orange, LysoTracker Red, and quinacrine as fluorescent probes for long-term tracking of acidic vesicles. Cytometry A 85:729–737. doi: 10.1002/cyto.a.22495. [DOI] [PubMed] [Google Scholar]
- 40.Orner EP, Zhang P, Jo MC, Bhattacharya S, Qin L, Fries BC. 2019. High-throughput yeast aging analysis for cryptococcus (HYAAC) microfluidic device streamlines aging studies in Cryptococcus neoformans. Commun Biol 2:256. doi: 10.1038/s42003-019-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao C, Wang Y, Husain S, Soteropoulos P, Xue C. 2019. A mechanosensitive channel governs lipid flippase-mediated echinocandin resistance in Cryptococcus neoformans. mBio 10:e01952-19. doi: 10.1128/mBio.01952-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya S, Fries BC. 2018. Enhanced efflux pump activity in old Candida glabrata cells. Antimicrob Agents Chemother 62:e02227-17. doi: 10.1128/AAC.02227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. 2009. Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryot Cell 8:858–866. doi: 10.1128/EC.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker LA, Munro CA. 2020. Caspofungin induced cell wall changes of Candida species influences macrophage interactions. Front Cell Infect Microbiol 10:164. doi: 10.3389/fcimb.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalem MC, Subbiah H, Leipheimer J, Glazier VE, Panepinto JC. 2021. Puf4 mediates post-transcriptional regulation of cell wall biosynthesis and caspofungin resistance in Cryptococcus neoformans. mBio 12:e03225-20. doi: 10.1128/mBio.03225-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Rubio R, Hernandez RY, Clear A, Healey KR, Shor E, Perlin DS. 2021. Critical assessment of cell wall integrity factors contributing to in vivo echinocandin tolerance and resistance in Candida glabrata. Front Microbiol 12:702779–702779. doi: 10.3389/fmicb.2021.702779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powell CD, Quain DE, Smart KA. 2003. Chitin scar breaks in aged Saccharomyces cerevisiae. Microbiology (Reading) 149:3129–3137. doi: 10.1099/mic.0.25940-0. [DOI] [PubMed] [Google Scholar]
- 48.Figueiredo ABC, Fonseca FL, Kuczera D, Conte FP, Arissawa M, Rodrigues ML. 2021. Monoclonal antibodies against cell wall chitooligomers as accessory tools for the control of cryptococcosis. Antimicrob Agents Chemother 65:e0118121. doi: 10.1128/AAC.01181-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam WC, Upadhya R, Specht CA, Ragsdale AE, Hole CR, Levitz SM, Lodge JK. 2019. Chitosan biosynthesis and virulence in the human fungal pathogen Cryptococcus gattii. mSphere 4:e0118121. doi: 10.1128/mSphere.00644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker LG, Specht CA, Lodge JK. 2011. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot Cell 10:1264–1268. doi: 10.1128/EC.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Upadhya R, Baker LG, Lam WC, Specht CA, Donlin Maureen J, Lodge JK. 2018. Cryptococcus neoformans Cda1 and its chitin deacetylase activity are required for fungal pathogenesis. mBio 9:e02087-18. doi: 10.1128/mBio.02087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Upadhya R, Lam WC, Hole CR, Parchment D, Lee CK, Specht CA, Levitz SM, Lodge JK. 2021. Cryptococcus neoformans Cda1 and Cda2 coordinate deacetylation of chitin during infection to control fungal virulence. Cell Surf 7:100066. doi: 10.1016/j.tcsw.2021.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Upadhya R, Lam WC, Maybruck B, Specht CA, Levitz SM, Lodge JK. 2016. Induction of protective immunity to cryptococcal infection in mice by a heat-killed, chitosan-deficient strain of Cryptococcus neoformans. mBio 7:e00547-16. doi: 10.1128/mBio.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nosanchuk JD, Stark RE, Casadevall A. 2015. Fungal melanin: what do we know about structure? Front Microbiol 6:1463. doi: 10.3389/fmicb.2015.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Duin D, Casadevall A, Nosanchuk JD. 2002. Melanization of Cryptococcus neoformans and Histoplasma capsulatum reduces their susceptibilities to amphotericin B and caspofungin. Antimicrob Agents Chemother 46:3394–3400. doi: 10.1128/AAC.46.11.3394-3400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chrissian C, Camacho E, Fu MS, Prados-Rosales R, Chatterjee S, Cordero RJB, Lodge JK, Casadevall A, Stark RE. 2020. Melanin deposition in two Cryptococcus species depends on cell-wall composition and flexibility. J Biol Chem 295:1815–1828. doi: 10.1074/jbc.RA119.011949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chrissian C, Lin CP, Camacho E, Casadevall A, Neiman AM, Stark RE. 2020. Unconventional constituents and shared molecular architecture of the melanized cell wall of C. neoformans and spore wall of S. cerevisiae. J Fungi (Basel) 6:329. doi: 10.3390/jof6040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almeida-Paes R, Figueiredo-Carvalho MH, da Silva LB, Gerfen G, S Araújo G.Rd, Frases S, Zancopé-Oliveira RM, Nosanchuk JD. 2021. Candida glabrata produces a melanin-like pigment that protects against stress conditions encountered during parasitism. Future Microbiol 16:509–520. doi: 10.2217/fmb-2020-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy B, Jacobson A. 2013. The intimate relationships of mRNA decay and translation. Trends Genet 29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukaremera L, Lee KK, Wagener J, Wiesner DL, Gow NAR, Nielsen K. 2018. Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. The Cell Surface 1:15–24. doi: 10.1016/j.tcsw.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, Lee CG, Elias JA, Nielsen JN, Boulware DR, Bohjanen PR, Jenkins MK, Levitz SM, Nielsen K. 2015. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog 11:e1004701. doi: 10.1371/journal.ppat.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim J, Coates CJ, Seoane PI, Garelnabi M, Taylor-Smith LM, Monteith P, Macleod CL, Escaron CJ, Brown GD, Hall RA, May RC. 2018. Characterizing the mechanisms of nonopsonic uptake of cryptococci by macrophages. J Immunol 200:3539–3546. doi: 10.4049/jimmunol.1700790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J 24:1277–1286. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasim S, Allison DP, Retterer ST, Hopke A, Wheeler RT, Doktycz MJ, Reynolds TB. 2016. β-(1,3)-Glucan unmasking in some Candida albicans mutants correlates with increases in cell wall surface roughness and decreases in cell wall elasticity. Infect Immun 85:e00601-16. doi: 10.1128/IAI.00601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang ZA, Li LX, Doering TL. 2018. Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 28:719–730. doi: 10.1093/glycob/cwy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jesus MD, Nicola AM, Chow SK, Lee IR, Nong S, Specht CA, Levitz SM, Casadevall A. 2010. Glucuronoxylomannan, galactoxylomannan, and mannoprotein occupy spatially separate and discrete regions in the capsule of Cryptococcus neoformans. Virulence 1:500–508. doi: 10.4161/viru.1.6.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cordero RJB, Pontes B, Guimarães AJ, Martinez LR, Rivera J, Fries BC, Nimrichter L, Rodrigues ML, Viana NB, Casadevall A. 2011. Chronological aging is associated with biophysical and chemical changes in the capsule of Cryptococcus neoformans. Infect Immun 79:4990–5000. doi: 10.1128/IAI.05789-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reuwsaat JCV, Motta H, Garcia AWA, Vasconcelos CB, Marques BM, Oliveira NK, Rodrigues J, Ferrareze PAG, Frases S, Lopes W, Barcellos VA, Squizani ED, Horta JA, Schrank A, Rodrigues ML, Staats CC, Vainstein MH, Kmetzsch L. 2018. A predicted mannoprotein participates in Cryptococcus gattii capsular structure. mSphere 3:e00023-18. doi: 10.1128/mSphere.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bie X, Zhang S, Luo X, Qi RQ. 2019. Candida albicans cell wall mannoprotein synergizes with lipopolysaccharide to affect RAW264.7 proliferation, phagocytosis and apoptosis. Microb Pathog 131:98–105. doi: 10.1016/j.micpath.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 70.Ene IV, Walker LA, Schiavone M, Lee KK, Martin-Yken H, Dague E, Gow NAR, Munro CA, Brown AJP. 2015. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986-15. doi: 10.1128/mBio.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouklas T, Pechuan X, Goldman DL, Edelman B, Bergman A, Fries BC. 2013. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. mBio 4:e00455-13. doi: 10.1128/mBio.00455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Egilmez NK, Chen JB, Jazwinski SM. 1990. Preparation and partial characterization of old yeast cells. J Gerontol 45:B9–B17. doi: 10.1093/geronj/45.1.B9. [DOI] [PubMed] [Google Scholar]
- 73.Richards A, Gow NAR, Veses V. 2012. Identification of vacuole defects in fungi. J Microbiol Methods 91:155–163. doi: 10.1016/j.mimet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Ding H, Caza M, Dong Y, Arif AA, Horianopoulos LC, Hu G, Johnson P, Kronstad JW. 2018. ATG genes influence the virulence of Cryptococcus neoformans through contributions beyond core autophagy functions. Infect Immun 86:e00069-18. doi: 10.1128/IAI.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fader CM, Colombo MI. 2009. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ 16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 76.Park Y-D, Chen SH, Camacho E, Casadevall A, Williamson PR. 2020. Role of the ESCRT pathway in laccase trafficking and virulence of Cryptococcus neoformans. Infect Immun 88:e00954-19. doi: 10.1128/IAI.00954-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu X, Gibbons J, Garcia-Rivera J, Casadevall A, Williamson PR. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect Immun 69:5589–5596. doi: 10.1128/IAI.69.9.5589-5596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. 2009. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology (Reading, Engl) 155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Casadevall A, Nosanchuk JD, Williamson P, Rodrigues ML. 2009. Vesicular transport across the fungal cell wall. Trends Microbiol 17:158–162. doi: 10.1016/j.tim.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rizzo J, Wong SSW, Gazi AD, Moyrand F, Chaze T, Commere P-H, Novault S, Matondo M, Péhau-Arnaudet G, Reis FCG, Vos M, Alves LR, May RC, Nimrichter L, Rodrigues ML, Aimanianda V, Janbon G. 2021. Cryptococcus extracellular vesicles properties and their use as vaccine platforms. J Extracell Vesicles 10:e12129. doi: 10.1002/jev2.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karkowska-Kuleta J, Kulig K, Karnas E, Zuba-Surma E, Woznicka O, Pyza E, Kuleta P, Osyczka A, Rapala-Kozik M, Kozik A. 2020. Characteristics of extracellular vesicles released by the pathogenic yeast-like fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells 9:1722. doi: 10.3390/cells9071722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao K, Bleackley M, Chisanga D, Gangoda L, Fonseka P, Liem M, Kalra H, Al Saffar H, Keerthikumar S, Ang C-S, Adda CG, Jiang L, Yap K, Poon IK, Lock P, Bulone V, Anderson M, Mathivanan S. 2019. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun Biol 2:305. doi: 10.1038/s42003-019-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. 2008. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol 10:1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzo J, Chaze T, Miranda K, Roberson RW, Gorgette O, Nimrichter L, Matondo M, Latgé JP, Beauvais A, Rodrigues ML. 2020. Characterization of extracellular vesicles produced by Aspergillus fumigatus protoplasts. mSphere 5:e00476-20. doi: 10.1128/mSphere.00476-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lindstrom DL, Gottschling DE. 2009. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics 183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wuttke D, de Magalhães JP. 2012. Osh6 links yeast vacuolar functions to lifespan extension and TOR. Cell Cycle 11:2419–2419. doi: 10.4161/cc.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes AL, Gottschling DE. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aufschnaiter A, Büttner S. 2019. The vacuolar shapes of ageing: from function to morphology. Biochim Biophys Acta Mol Cell Res 1866:957–970. doi: 10.1016/j.bbamcr.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 89.Rane HS, Hayek SR, Frye JE, Abeyta EL, Bernardo SM, Parra KJ, Lee SA. 2019. Candida albicans Pma1p contributes to growth, pH homeostasis, and hyphal formation. Front Microbiol 10:1012. doi: 10.3389/fmicb.2019.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martínez-Muñoz GA, Kane P. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem 283:20309–20319. doi: 10.1074/jbc.M710470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bouklas T, Masone L, Fries BC. 2018. Differences in sirtuin regulation in response to calorie restriction in Cryptococcus neoformans. J Fungi (Basel) 4:26. doi: 10.3390/jof4010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jain N, Bouklas T, Gupta A, Varshney AK, Orner EP, Fries BC. 2016. ALL2, a homologue of ALL1, has a distinct role in regulating pH homeostasis in the pathogen Cryptococcus neoformans. Infect Immun 84:439–451. doi: 10.1128/IAI.01046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harrison TS, Chen J, Simons E, Levitz SM. 2002. Determination of the pH of the Cryptococcus neoformans vacuole. Med Mycology 40:329–332. doi: 10.1080/mmy.40.3.329.332. [DOI] [PubMed] [Google Scholar]
- 94.O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4:e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fuchs BB, Tegos GP, Hamblin MR, Mylonakis E. 2007. Susceptibility of Cryptococcus neoformans to photodynamic inactivation is associated with cell wall integrity. Antimicrob Agents Chemother 51:2929–2936. doi: 10.1128/AAC.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.García-Rivera J, Chang YC, Kwon-Chung KJ, Casadevall A. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot Cell 3:385–392. doi: 10.1128/EC.3.2.385-392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rizzo J, Colombo AC, Zamith-Miranda D, Silva VKA, Allegood JC, Casadevall A, Del Poeta M, Nosanchuk JD, Kronstad JW, Rodrigues ML. 2018. The putative flippase Apt1 is required for intracellular membrane architecture and biosynthesis of polysaccharide and lipids in Cryptococcus neoformans. Biochim Biophys Acta Mol Cell Res 1865:532–541. doi: 10.1016/j.bbamcr.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry analysis of young and old C. neoformans and C. glabrata cells. At least 10,000 events were gated in the FSC/SSC plots and are represented as histograms with mean fluorescence on the x axis and cell counts on the y axis. Unstained young (red) and old (green) cells were sorted as controls to determine positive labeling of young (blue) and old (orange) cells. CFW (calcofluor white, chitin), tetramethylrhodamine-labeled wheat germ agglutinin, chitooligomers (WGA-TRITC), eosin Y, chitosan (EY), Concanavalin A conjugated to Texas Red, mannan (CoA-Texas red), Fc-dectin1/DyLight 405 (β-glucan). Data were analyzed with FlowJo v10 software, and the image is representative of one experiment producing similar results that were obtained in three independent experiments. Download FIG S1, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
No impact on cell wall integrity was observed in old C. neoformans (Cn) and C. glabrata (Cg) cells. Young and old cells of Cn and Cg were serially diluted and spotted onto YPD with the addition of several cell wall stressors (caffeine, FB 28 [fluorescent brightener 28], Congo red, and sodium dodecyl sulfate [SDS]) at 30°C and 37°C for 48 h. This experiment was repeated twice with similar results. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Ultramicroscopic image of old C. neoformans cells with numerous vesicle-like particles in the cytosol, inside the vacuole, and crossing the cell wall indicated by white arrows. Scale bar represents 500 nm. Download FIG S3, TIF file, 0.5 MB (492.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A to D) Cell surface and vacuolar areas are significantly increased in old C. glabrata (A and B) and C. neoformans (C and D). Fungal cells were stained with FM4-64, and analyses of 25 cells for each group (young and old) and strain were measured using ImageJ/Fiji software. Statistical analyses were performed by t test with Welch’s correction (****, P ≤ 0.0001). Download FIG S4, TIF file, 0.6 MB (639.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence of primers (5′-3′) used in this study. Download Table S1, PDF file, 0.1 MB (78.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.