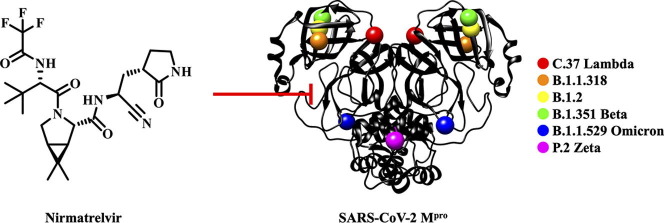
Graphical abstract
Keywords: SARS-CoV-2, Main protease, Variants, Nirmatrelvir, Inhibitors
Abstract
The COVID-19 pandemic continues to be a public health threat. Multiple mutations in the spike protein of emerging variants of SARS-CoV-2 appear to impact on the effectiveness of available vaccines. Specific antiviral agents are keenly anticipated but their efficacy may also be compromised in emerging variants. One of the most attractive coronaviral drug targets is the main protease (Mpro). A promising Mpro inhibitor of clinical relevance is the peptidomimetic nirmatrelvir (PF-07321332). We expressed Mpro of six SARS-CoV-2 lineages (C.37 Lambda, B.1.1.318, B.1.2, B.1.351 Beta, B.1.1.529 Omicron, P.2 Zeta), each of which carries a strongly prevalent missense mutation (G15S, T21I, L89F, K90R, P132H, L205V). Enzyme kinetics reveal that these Mpro variants are catalytically competent to a similar degree as the wildtype. We show that nirmatrelvir has similar potency against the variants as the wildtype. Our in vitro data suggest that the efficacy of the specific Mpro inhibitor nirmatrelvir is not compromised in current COVID-19 variants.
Since its emergence in late 2019,1 COVID-19 has significantly impacted on societies worldwide.2 Over 5.7 million deaths have been attributed to COVID-19, with the number of confirmed SARS-CoV-2 infections surpassing 400 million.3 The outbreak of SARS-CoV-2 prompted multiple successful vaccine development campaigns.4 Currently approved vaccines, such as viral vector or mRNA vaccines, successfully limited the pandemic’s impact on global health.5, 6 Most COVID-19 vaccines function by stimulating an immune response against the SARS-CoV-2 spike protein (S)7, 8, 9 but, as the spike gene has gathered considerable genetic variability,10, 11 it is a concern if the effectiveness of existing vaccines is affected by variants of SARS-CoV-2.5, 6, 10, 12 At the time of writing, the World Health Organization (WHO) lists five variants of concern (VOC; Alpha, Beta, Gamma, Delta, Omicron) and two variants of interest (VOI; Lambda, Mu).13 A possible reformulation of the vaccines adjusted to currently circulating lineages of SARS-CoV-2 is being investigated.14, 15, 16 The deployment of vaccines clearly remains the best public health measure to control the spread of SARS-CoV-2 and the severe health effects of COVID-19.17, 18
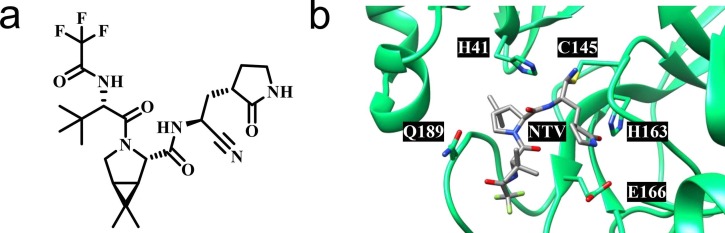
Complementary to preventive vaccines, antiviral drugs are urgently needed to combat COVID-19.19 Since the discovery of SARS-CoV-1 in 2003,20 several coronaviral drug targets have been identified,21 including the RNA-dependent RNA polymerase (RdRp, nsp12),22 the helicase (nsp13),23 the papain-like protease (PLpro, part of nsp3)24 and the main protease (Mpro, 3CLpro, nsp5).25 Despite this, treatment options for COVID-19 are limited. Recombinant neutralizing monoclonal antibodies (mAbs) are employed in the clinical management of COVID-19, but resistance of the SARS-CoV-2 Omicron variant is a major concern.26 The orally active drugs molnupiravir (MK-4482, EIDD-2801, Lagevrio™) and nirmatrelvir (PF-07321332, Paxlovid™ as combination drug with ritonavir as booster) were first approved for emergency use in the United Kingdom and the United States in late 2021. Molnupiravir targets RdRp by acting as a nucleoside analogue prodrug, but was originally developed against different RNA viruses.27 Nirmatrelvir is an orally available peptidomimetic targeting Mpro, employing a nitrile warhead to covalently bind the catalytic cysteine residue in the active site of the protease (Fig. 1 ).28
Fig. 1.
SARS-CoV-2 Mpro inhibitor nirmatrelvir (PF-07321332).28 (a) Chemical structure of nirmatrelvir. (b) X-ray co-crystal structure of SARS-CoV-2 Mpro in complex with nirmatrelvir (NTV) indicating the catalytic dyad and key interacting residues as proposed by Owen et al. (PDB: 7RFW).28
SARS-CoV-2 Mpro is a homodimeric cysteine protease, which processes the majority of the viral polyproteins pp1a and pp1ab encoded by the ORF1a/b gene.25, 29 Inhibition of Mpro thus ultimately hinders the assembly of the replication and transcription complexes (RTCs).25, 30 The protease has a distinct recognition motif, with – in the Schechter-Berger notation31 – preference for leucine in P 2 and especially strong preference for glutamine in P 1.25, 32 Human host proteases have different substrate specificities and it is therefore anticipated that selective inhibitors have limited off-target effects.25
Previous research on SARS-CoV-1 Mpro (which is 96% identical in amino acid sequence to SARS-CoV-2 Mpro)25 demonstrated that missense point mutations can influence protease activity. Mutants have been identified with slightly enhanced (S284, T285, I286)33, 34 and slightly or severely reduced catalytic activity (G11, N28, S139, F140, E166, N214, R298).33, 35, 36, 37, 38, 39 Specifically the R298A mutation has become a tool to study the protease in its monomeric form, since it inactivates the protease by disrupting the Mpro dimer.36 The present study assesses the Mpro mutants of emerging SARS-CoV-2 lineages. We analyzed the most widespread amino acid substitutions in SARS-CoV-2 Mpro, characterized them by enzyme kinetics and assessed their susceptibility to inhibition by nirmatrelvir.
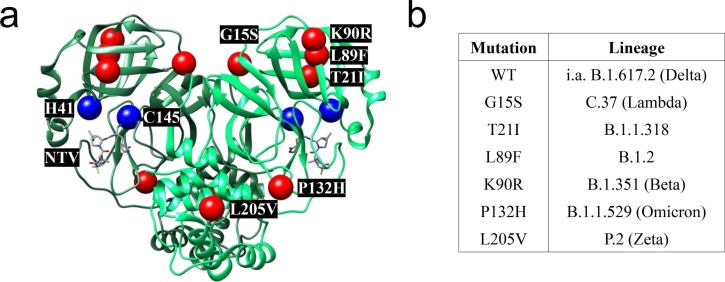
Utilizing the Outbreak.info database by Scripps Research,40 which partially operates with data provided by the GISAID Initiative,41 we performed an analysis of the genomes of SARS-CoV-2 lineages, including the VOC and VOI. The WIV04 sequence (EPI_ISL_402124)42 acted as wildtype (WT) reference genome. Lineage comparison43 of VOCs and VOIs revealed three missense mutations in the Mpro section of the ORF1a/b gene with > 20% frequency of occurrence. The mutations are G15S, which is > 85% prevalent44 in the Lambda VOI (or C.37, using PANGO nomenclature)45, K90R, which is > 95% prevalent46 in the Beta VOC (B.1.351) and P132H, which is > 95% prevalent47 in the Omicron VOC (B.1.1.529). The Delta VOC (B.1.617.2), which was the dominant lineage for most of the second half of 2021,48, 49 did not display any particularly prevalent (>20%)43 missense mutations within the Mpro part of ORF1a/b, implying that its Mpro is identical to that of the WT. Furthermore, we chose to investigate three additional abundant Mpro mutations to cover a larger variety of lineages: T21I, which is > 90% prevalent50 in B.1.1.318, a WHO variant under monitoring (VUM),13 L89F, which is > 95% prevalent51 in the B.1.2 lineage, and L205V, which is > 95% prevalent52 in the former VOI Zeta (P.2) (Fig. 2 b). Hence, we selected the six mutations G15S, T21I, L89F, K90R, P132H and L205V for further investigations.
Fig. 2.
Comparison of Mpro mutants. (a) X-ray co-crystal structure of SARS-CoV-2 Mpro in complex with nirmatrelvir (NTV) (PDB: 7RFW).28 The sites of mutations (red) and the catalytic dyad (blue) in the two protomers (green) are indicated. (b) List of prevalent Mpro mutations and their corresponding SARS-CoV-2 lineage.
X-ray crystal structures of WT SARS-CoV-2 Mpro (e.g. PDB: 6Y2E, 6LU7)53, 54 indicate that the residues G15, T21, K90 and P132 are solvent-exposed, whereas the hydrophobic residues L89 and L205 are buried within the protease. Except for the mutations T21I and P132H, the mutations introduce no major changes in the chemical character of the side-chains, as indicated by low – or, in the case of T21I and P132H, moderate – values of Miyata’s distances.55 The mutations G15S, T21I, L89F and K90R are located in domain I, whereas the mutations P132H and L205V are in domains II and III, respectively (Fig. 2a).25 To the best of our knowledge, these residues participate neither in the active site nor the allosteric binding sites of SARS-CoV-2 Mpro discussed by Günther et al.56
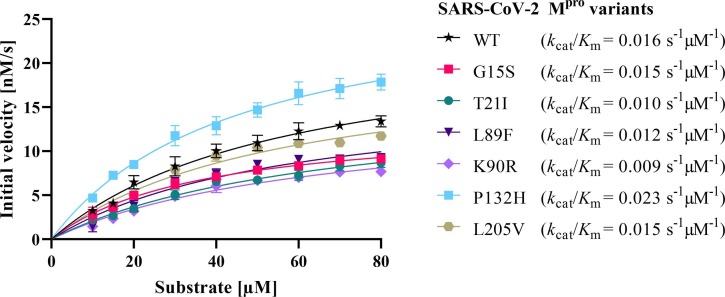
WT SARS-CoV-2 Mpro and the mutants G15S, T21I, L89F, K90R, P132H and L205V were expressed in E. coli and purified. An established Förster resonance electron transfer (FRET) in vitro assay of Mpro activity54, 57 was employed to determine initial velocities of the proteolytic activity at various substrate concentrations. The data confirmed that all mutants are enzymatically active, which was expected25, 58 as a dysfunctional Mpro would prevent replication of SARS-CoV-2. The seven Mpro variants exhibited turnover numbers (k cat) between 0.54 and 1.03 s− 1, and Michaelis constants (K m) ranging from 37 to 67 µM (Table S1 ). The catalytic efficiencies (k cat/K m) calculated for the mutants (0.009 to 0.023 s− 1 µM− 1) are similar to that of WT Mpro (0.016 s−1µM−1), confirming that all Mpro variants are equally competent with regard to their proteolytic activities (Fig. 3 , Table S1).
Fig. 3.
Michaelis-Menten kinetics of SARS-CoV-2 Mpro variants specifying their catalytic efficiency (kcat/Km).
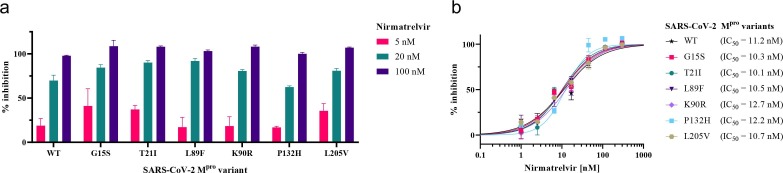
Following the kinetic analysis of the SARS-CoV-2 Mpro variants, the clinical candidate nirmatrelvir (also known as PF-07321332; Fig. 1) was used to assess the potential impact of Mpro mutations on the drug’s efficacy (Fig. 4 ). The inhibition constant (K i) of nirmatrelvir against SARS-CoV-2 WT Mpro has been reported to be 3.1 nM.28 Our FRET assay confirmed that nirmatrelvir inhibits the activity of Mpro variants at nanomolar inhibitor concentrations. Furthermore, the extent of inhibition was similar across the different protease variants. An initial screening at three selected concentrations showed that the compound displayed below 50% inhibition at 5 nM, over 50% inhibition at 20 nM and fully inhibited the enzymatic activity of all mutants and the WT at 100 nM (Fig. 4 a). Subsequently, we determined IC50 values of nirmatrelvir against the mutants and WT, ranging from 10 nM to 13 nM with overlapping confidence intervals (Fig. 4b, Table S2).
Fig. 4.
Inhibition of SARS-CoV-2 Mpro variants by nirmatrelvir. (a) Inhibition at 5 nM, 20 nM, and 100 nM. (b) Dose-response curves.
In summary, we identified the currently most prevalent Mpro variants (G15S, T21I, L89F, K90R, P132H, L205V) in different lineages of SARS-CoV-2 (C.37 Lambda, B.1.1.318, B.1.2, B.1.351 Beta, B.1.1.529 Omicron, P.2 Zeta) and found that, in a biochemical assay, they are catalytically competent to a similar degree as the wildtype. In addition, we confirmed that nirmatrelvir maintains effective inhibition of all these Mpro variants in vitro. This suggests that the inhibitory effect of nirmatrelvir and potentially other specific SARS-CoV-2 Mpro inhibitors would at present not be compromised for these virus variants. It must be noted, however, that widespread use of Mpro inhibitors may challenge SARS-CoV-2 to develop Mpro mutations that overcome these inhibitors, as previously experienced for, e.g., HIV protease inhibitors.59 Despite these challenges, protease inhibitors have revolutionized antiviral treatment for viral infectious diseases, including HIV and HCV.60 It can thus be expected that Mpro inhibitors will have a similar impact on the future development of the COVID-19 pandemic.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
C.N. thanks the Australian Research Council (ARC) for a Discovery Early Career Research Award (DE190100015) and Discovery Project funding (DP200100348). G.O. thanks the ARC for a Laureate Fellowship (FL170100019) and acknowledges support by the ARC Centre of Excellence for Innovations in Peptide & Protein Science (CE200100012). This study was supported by a RAMR (MAWA) grant awarded to S.U. and C.N.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmcl.2022.128629.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu N.A., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiscott J., Alexandridi M., Muscolini M., et al. The global impact of the coronavirus pandemic. Cytokine Growth Factor Rev. 2020;53:1–9. doi: 10.1016/j.cytogfr.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8(2):153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbarao K. The success of SARS-CoV-2 vaccines and challenges ahead. Cell Host Microbe. 2021;29(7):1111–1123. doi: 10.1016/j.chom.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tregoning J.S., Flight K.E., Higham S.L., Wang Z., Pierce B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol. 2021;21(10):626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19(10):667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 8.Teijaro J.R., Farber D.L. COVID-19 vaccines: modes of immune activation and future challenges. Nat Rev Immunol. 2021;21(4):195–197. doi: 10.1038/s41577-021-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont L., Snell L.B., Graham C., et al. Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern. Nat Microbiol. 2021;6(11):1433–1442. doi: 10.1038/s41564-021-00974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao K., Tzou P.L., Nouhin J., et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey W.T., Carabelli A.M., Jackson B., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatsi E.-B., Filippatos F., Michos A. SARS-CoV-2 variants and effectiveness of vaccines: a review of current evidence. Epidemiol Infect. 2021;1–24 doi: 10.1017/s0950268821002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. Tracking SARS-CoV-2 variants; 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ Accessed 10 February 2022.
- 14.Prévost J., Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29(3):322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Zyoud W., Haddad H. Dynamics prediction of emerging notable spike protein mutations in SARS-CoV-2 implies a need for updated vaccines. Biochimie. 2021;191:91–103. doi: 10.1016/j.biochi.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cevik M., Grubaugh N.D., Iwasaki A., Openshaw P. COVID-19 vaccines: keeping pace with SARS-CoV-2 variants. Cell. 2021;184(20):5077–5081. doi: 10.1016/j.cell.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grubaugh N.D., Hodcroft E.B., Fauver J.R., Phelan A.L., Cevik M. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184(5):1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viana J., van Dorp C.H., Nunes A., et al. Controlling the pandemic during the SARS-CoV-2 vaccination rollout. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-23938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murgolo N., Therien A.G., Howell B., et al. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLOS Pathog. 2021;17(2):e1009225. doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drosten C., Günther S., Preiser W., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 21.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt A.N., Gallazzi F., Quinn T.P., Lorson C.L., Sönnerborg A., Singh K. Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opin Ther Pat. 2021;31(4):339–350. doi: 10.1080/13543776.2021.1884224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudvand S., Shokri S. Interactions between SARS coronavirus 2 papain-like protease and immune system: a potential drug target for the treatment of COVID-19. Scand J Immunol. 2021;94(4) doi: 10.1111/sji.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30(17):127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann M., Krüger N., Schulz S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185(3):447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Painter G.R., Natchus M.G., Cohen O., Holman W., Painter W.P. Developing a direct acting, orally available antiviral agent in a pandemic: the evolution of molnupiravir as a potential treatment for COVID-19. Curr Opin Virol. 2021;50:17–22. doi: 10.1016/j.coviro.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen D.R., Allerton C.M.N., Anderson A.S., et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 29.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e910. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J Biol Chem. 2020;295(37):12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schechter I., Berger A. On the size of the active site in proteases I Papain. Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 32.Rut W., Groborz K., Zhang L., et al. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat Chem Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 33.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273(5):1035–1045. doi: 10.1111/ejb.2006.273.issue-510.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim L., Shi J., Mu Y., Song J., Fraternali F. Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS ONE. 2014;9(7):e101941. doi: 10.1371/journal.pone.0101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S., Hu T., Zhang J., et al. Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations. J Biol Chem. 2008;283(1):554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J Virol. 2008;82(9):4620–4629. doi: 10.1128/jvi.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrila J., Gabelli S.B., Bacha U., Amzel L.M., Freire E. Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CLpro. Biochemistry. 2010;49(20):4308–4317. doi: 10.1021/bi1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu T., Zhang Y.u., Li L., et al. Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology. 2009;388(2):324–334. doi: 10.1016/j.virol.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng S.-C., Chang G.-G., Chou C.-Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys J. 2010;98(7):1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullen JL, Tsueng G, Abdel Latif A, et al. Outbreak.info - a standardized, open-source database of COVID-19 resources and epidemiology data 2020. https://outbreak.info/. Accessed 7 November 2021.
- 41.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhukova A., Blassel L., Lemoine F., Morel M., Voznica J., Gascuel O. Origin, evolution and global spread of SARS-CoV-2. C R Biol. 2021;344(1):57–75. doi: 10.5802/crbiol.29. [DOI] [PubMed] [Google Scholar]
- 43.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - lineage comparison; 2021. https://outbreak.info/compare-lineages. Accessed 8 November 2021.
- 44.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:G3278S mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AG3278S. Accessed 8 November 2021.
- 45.Rambaut A., Holmes E.C., O’Toole Á., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:K3353R mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AK3353R. Accessed 8 November 2021.
- 47.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:P3395H mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AP3395H. Accessed 18 December 2021.
- 48.He X., He C., Hong W., Zhang K., Wei X. The challenges of COVID-19 Delta variant: prevention and vaccine development. MedComm. 2021;2(4):846–854. doi: 10.1002/mco2.v2.410.1002/mco2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - variant report by location over time; 2021. https://outbreak.info/location-reports?loc. Accessed 21 December 2021.
- 50.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:T3284I mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AT3284I. Accessed 8 November 2021.
- 51.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:L3352F mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AL3352F. Accessed 8 November 2021.
- 52.Abdel Latif A, Mullen JL, Alkuzweny M, et al. Outbreak.info - ORF1a:L3468V mutation report; 2021. https://outbreak.info/situation-reports?pango&muts=ORF1a%3AL3468V. Accessed 8 November 2021.
- 53.Jin Z., Du X., Xu Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyata T., Miyazawa S., Yasunaga T. Two types of amino acid substitutions in protein evolution. J Mol Evol. 1979;12(3):219–236. doi: 10.1007/bf01732340. [DOI] [PubMed] [Google Scholar]
- 56.Günther S., Reinke P.Y.A., Fernández-García Y., et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372(6542):642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ullrich S., Sasi V.M., Mahawaththa M.C., et al. Challenges of short substrate analogues as SARS-CoV-2 main protease inhibitors. Bioorg Med Chem Lett. 2021;50:128333. doi: 10.1016/j.bmcl.2021.128333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeldovich K.B., Chen P., Shakhnovich E.I. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104(41):16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wensing A.M.J., van Maarseveen N.M., Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antivir Res. 2010;85(1):59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 60.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D.A. Proteases and protease inhibitors in infectious diseases. Med Res Rev. 2018;38(4):1295–1331. doi: 10.1002/med.2018.38.issue-410.1002/med.21475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.