Abstract
Depression and obesity are major public health concerns, and there is mounting evidence that they share etiopathophysiological mechanisms. The neurobiological pathways involved in both mood and energy balance regulation are complex, multifactorial and still incompletely understood. As a coactivator of the pleiotropic transcription factor cAMP response element-binding protein (CREB), CREB-regulated transcription coactivator 1 (CRTC1) has recently emerged as a novel regulator of neuronal plasticity and brain functions, while CRTC1 dysfunction has been associated with neurodegenerative and psychiatric diseases. This review focuses on recent evidence emphasizing the critical role of CRTC1 in the neurobiology of depression and comorbid obesity. We discuss the role of CRTC1 downregulation in mediating chronic stress-induced depressive-like behaviors, and antidepressant response in the light of the previously characterized Crtc1 knockout mouse model of depression. The putative role of CRTC1 in the alteration of brain energy homeostasis observed in depression is also discussed. Finally, we highlight rodent and human studies supporting the critical involvement of CRTC1 in depression-associated obesity.
Keywords: major depressive disorder, obesity, circadian rhythms, neuroplasticity, CREB, CRTC1, BDNF
Introduction
According to the World Health Organization, more than 300 million people suffer from depression worldwide (World Health Organization, 2017). This mood disorder is now the leading cause of disability and a major contributor to the overall global burden of disease. A high suicide risk is associated with severe depression and more than 800,000 people commit suicide each year. Current antidepressant therapies are mostly targeting monoamines neurotransmission with a limited efficacy. Several weeks of treatment are needed before mood improvement occurs, and approximately 30% of patients do not respond to at least two consecutive antidepressant treatments (Caraci et al., 2018). Treatment-resistant depression may benefit from the recent development of rapid-acting antidepressants targeting glutamate neurotransmission, such as the N-methyl-D-aspartate (NMDA) receptor blocker ketamine (Duman et al., 2016, 2019b; Zanos and Gould, 2018; Krystal et al., 2019; Shinohara et al., 2021). However, the dissociative and potentially addictive side effects of ketamine have been stimulating the development of new antidepressant drugs based on ketamine, but without its drawbacks (Ragguett et al., 2019). Unfortunately, many clinical trials failed to show a significant advantage for antidepressant medication over inert placebo (Kraus et al., 2019; Wilkinson and Sanacora, 2019). These failures might also reflect insufficient clinical subtyping of major depressive disorder (MDD), which would certainly benefit from a better understanding of the neurobiology of depression that remains elusive despite decades of intensive research (Akil et al., 2018).
Overweight and obesity are major public health problems that continue to increase both in developing and developed countries. Epidemiological evidence strongly supports the existence of a bidirectional relationship between depression and abdominal (visceral) obesity, which is the main risk factor for metabolic syndrome (Milaneschi et al., 2019). Clinically, obesity and metabolic syndrome are associated with the atypical depression subtype of MDD characterized by neurovegetative symptoms consisting of lethargy, fatigue, excessive sleepiness, increased food intake, weight gain and depressive symptoms that are lowest in the morning and worsen as the day progresses (Gold, 2015). Among the shared biological pathways that may mechanistically explain the atypical depression–obesity link, increased chronic inflammation is emerging as the central pathophysiological process involved (Milaneschi et al., 2019).
The monoamine hypothesis of depression is based on a deficit of monoamines (serotonin and noradrenaline) that would be corrected by antidepressants. However, current antidepressants require weeks of treatment to produce a clinical response, which suggests that their effects do not only rely on enhanced monoamines transmission, but rather to long-term changes in monoamines signaling and neuronal circuitry. Therefore, research efforts have been focusing on the long-term molecular changes that underlie depression and antidepressant treatments, and led to the neurotrophic and network hypotheses of depression, which proposed that impaired mechanisms of neuroplasticity are a core pathophysiological feature of MDD. Neuroplasticity is a fundamental mechanism of neuronal adaptation involving several forms of plasticity, such as adult hippocampal neurogenesis, neuronal survival and maturation, synaptogenesis, and structural plasticity (e.g., number of spines and complexity of dendritic arborization). Decreased neuroplasticity in the hippocampus and prefrontal cortex, and the resulting problems in information processing within relevant neural networks might thus underlie mood disorders (Castren, 2005; Pittenger and Duman, 2008; Marsden, 2013; Boku et al., 2018). Indeed, chronic stress strongly contributes to the development of MDD by affecting neuroplasticity and connectivity at several levels (Pittenger and Duman, 2008; Albert, 2019; Duman et al., 2019a). The transcription factor cAMP response element-binding protein (CREB) and one of its target genes, brain-derived neurotrophic factor (BDNF) are critically involved in the concept of altered neuroplasticity in MDD (Carlezon et al., 2005; Blendy, 2006; Castren and Hen, 2013; Levy et al., 2018; Umemori et al., 2018; Xiao et al., 2018; Castren and Monteggia, 2021). Moreover, there is evidence in rodents and humans that the CREB-BDNF pathway is also implicated in obesity (Rios, 2013; Lin et al., 2014; Marosi and Mattson, 2014; Xu and Xie, 2016; Amare et al., 2017). BDNF and several other neuroplasticity genes are regulated by CREB and a coactivator called CREB-Regulated Transcription Coactivator 1 (CRTC1) (Zhou et al., 2006; Kovacs et al., 2007). Recent studies suggest that CRTC1 dysregulation may be involved in the etiopathogenesis of many brain disorders, including MDD (Saura and Cardinaux, 2017). This review will focus on the increasing evidence of the key role of the transcription coactivator CRTC1 in the central control of mood and eating behavior, and its possible involvement in depression and comorbid obesity.
CREB-Regulated Transcription Coactivator 1 in Brain Functions and Disorders
The CRTC family includes three members in mammals, with CRTC2 and CRTC3 expressed in most tissues, and CRTC1 mainly found in the brain (Kovacs et al., 2007; Li et al., 2009; Watts et al., 2011). Several comprehensive reviews already provided a detailed description of the signaling pathways regulating CRTC1 and its roles in brain physiology and pathology (Altarejos and Montminy, 2011; Escoubas et al., 2017; Saura and Cardinaux, 2017; Uchida and Shumyatsky, 2018; Parra-Damas and Saura, 2019). In this section, we summarize the mechanisms of CRTC1 regulation and its main functions in the brain, with a particular emphasis on CRTC1’s role in neuroplasticity.
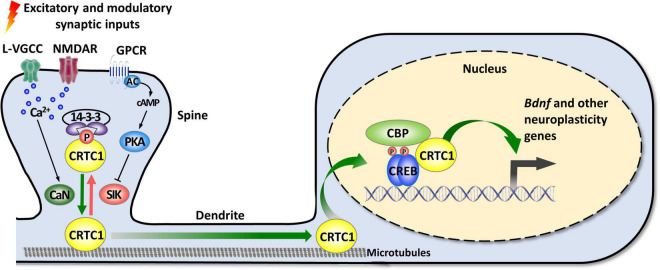
As mentioned in the introduction, CRTC1 is a coactivator of CREB, a transcription factor playing pleiotropic roles in the nervous system (Carlezon et al., 2005; Barco and Marie, 2011). The canonical mode of CREB activation involves its phosphorylation at Ser133 by multiple signaling pathways, and the ensuing recruitment of CBP/p300 coactivators that activate transcription by acetylating nucleosomal histones and by interacting with factors of the general transcription machinery (Mayr and Montminy, 2001; Lonze and Ginty, 2002; Carlezon et al., 2005; Josselyn and Nguyen, 2005; Benito and Barco, 2010). However, this model was challenged by several studies suggesting that CREB can be activated without Ser133 phosphorylation (Brindle et al., 1995; Impey et al., 1996; Briand et al., 2015). The discovery of the CRTC family of coactivators brought elements of explanation about how CREB-mediated transcription could be enhanced independently of Ser133 phosphorylation, and suggested the existence of a non-canonical, alternative way of CREB target genes activation (Conkright et al., 2003; Iourgenko et al., 2003; Altarejos and Montminy, 2011; Parra-Damas et al., 2017b; Feldmann et al., 2019). De facto, the N-terminal domain of CRTCs interacts with the dimerization and DNA-binding bZIP domain of CREB in a phosphorylation-independent manner. Once recruited to gene promoters, CRTCs strongly activate CREB-mediated transcription through their C-terminal transactivation domain. CREB is nonetheless not constitutively activated by CRTCs, as their phosphorylation state regulates their cytoplasmic versus nuclear localization (Bittinger et al., 2004; Screaton et al., 2004; Sonntag et al., 2017). In resting conditions, they are phosphorylated and sequestered in the cytoplasm by scaffolding 14-3-3 proteins and their nuclear translocation requires the concomitant activation of calcium and cAMP signaling pathways (Figure 1). Upon elevated intracellular calcium levels, CRTCs are dephosphorylated by the Ca2+-dependent protein phosphatase 2B (PP2B)/calcineurin leading to their dissociation from 14–3–3 proteins and subsequent nuclear translocation. Coincidently, increased cAMP levels stimulate calcium-mediated dephosphorylation by inhibiting salt-inducible kinases (SIKs) that phosphorylate CRTCs (Altarejos and Montminy, 2011).
FIGURE 1.
Activity-dependent synapse-to-nucleus translocation of CRTC1 mediates the activation of neuroplasticity gene transcription. CRTC1 is sequestered in dendritic spines under basal conditions via a phosphorylation-dependent association with 14-3-3 proteins. Simultaneous activation of calcium and cAMP pathways (by L-VGCC or NMDAR, and GPCR-activated AC) triggers release from 14-3-3 proteins by, respectively, activating the phosphatase calcineurin (CaN) and inhibiting kinases of the AMPK family (SIK). Dephosphorylated CRTC1 migrates into the nucleus and is recruited to the promoter via an interaction with the bZIP domain of CREB, thus promoting expression of Bdnf and other neuroplasticity genes. AC, adenylate cyclase; AMPK, AMP-activated protein kinase; CaN, calcineurin; CBP, CREB-binding protein; CREB, cAMP response-element binding protein; CRTC1, CREB-regulated transcription coactivator 1; GPCR, G protein-coupled receptor; L-VGCC, L-type voltage-gated calcium channels; NMDAR, N-methyl-D-aspartate receptor; PKA, protein kinase A; SIK, salt-inducible kinase.
In neurons, synaptic activity-induced CRTC1 translocation from synapses to the nucleus is critical for the transcription-dependent phase of neuronal plasticity (Zhou et al., 2006; Kovacs et al., 2007; Li et al., 2009; Finsterwald et al., 2010; Ch’ng et al., 2012, 2015; Nonaka et al., 2014; Parra-Damas et al., 2014, 2017a). Activity-dependent transport of CRTC1 from dendritic spines to the nucleus requires local elevation of calcium triggered by activation of glutamate receptors and L-type voltage-gated calcium channels, leading to calcineurin activation and dephosphorylation of three conserved serine residues (S64, S151, and S245) in the amino-terminal third of CRTC1, which contains a nuclear localization signal (Ch’ng et al., 2015). Dephosphorylated CRTC1 is released from 14-3-3 proteins and actively translocated to the nucleus via a dynein motor protein-mediated retrograde transport along microtubules. Nuclear CRTC1 upregulates the expression of genes that have been involved in synaptic plasticity and memory formation. These neuroplasticity genes include, among others, the neurotrophic factor Bdnf, the immediate-early genes c-fos, Zif268/Egr1, and Arc, the orphan nuclear receptors Nr4a1 and Nr4a2, the brain-specific growth factor Fgf1, and autophagy genes involved in synaptic turnover and late-phase long-term synaptic depression (Zhou et al., 2006; Espana et al., 2010; Breuillaud et al., 2012; Ch’ng et al., 2012, 2015; Nonaka et al., 2014; Parra-Damas et al., 2014, 2017a; Fukuchi et al., 2015; Uchida et al., 2017; Pan et al., 2021). In agreement with CRTC1’s role in the induction of these neuroplasticity genes, accumulating evidence supports its involvement in neuronal plasticity processes, such as activity- and BDNF-induced dendritic growth of developing cortical neurons (Li et al., 2009; Finsterwald et al., 2010), maintenance of hippocampal late-phase LTP (Zhou et al., 2006; Kovacs et al., 2007; Uchida et al., 2017), and long-term memory formation (Sekeres et al., 2012; Nonaka et al., 2014; Parra-Damas et al., 2014, 2017a; Uchida et al., 2017; Zhang et al., 2019; Shu et al., 2020; Yan et al., 2021). Of particular importance, increasing evidence suggests that CRTC1 dysregulation may be implicated in the etiopathogenesis of neurodegenerative diseases, such as Huntington’s, Parkinson’s, and Alzheimer’s disease, as well as psychiatric disorders (Boulting et al., 2021), including mood disorders [reviewed in Saura and Cardinaux (2017)].
Emerging Role of CREB-Regulated Transcription Coactivator 1 in the Pathophysiology of Major Depressive Disorder
In line with CRTC1’s role in neuroplasticity processes, as well as the involvement of CREB and BDNF in rodent models of depression, increasing evidence suggests that CRTC1 is implicated in the pathogenesis of MDD (summarized in Table 1). Originally, we developed a CRTC1 knockout mouse model (Breuillaud et al., 2009), and performed its in-depth phenotypic characterization (Breuillaud et al., 2012). This led us to provide the first preclinical evidence of the involvement of CRTC1 in mood regulation and antidepressant response. Indeed, Crtc1–/– mice exhibit depressive-like neurobehavioral endophenotypes, including increased behavioral despair in the forced swim test (FST) and in the repeated open-space forced swim test (OSFST), social withdrawal, decreased sexual motivation, psychomotor retardation, and increased emotional response to stressful events (Breuillaud et al., 2012). High-performance liquid chromatography (HPLC) monitoring of the levels of monoamines and metabolites in the prefrontal cortex, hippocampus, hypothalamus, and nucleus accumbens revealed significantly lower levels of the dopamine metabolites 3,4 dihydroxy-phenylacetic acid (DOPAC) and homovanillic acid (HVA), as well as of the serotonin metabolite 5-hydroxyindole acetic acid (5-HIAA) in the prefrontal cortex of Crtc1–/– male mice as compared to wild-type littermates. Decreased levels of these metabolites are thought to reflect a reduced dopamine and serotonin release in the prefrontal cortex that was previously associated with aggressive behaviors, impulsivity, and depressive-like behaviors. Moreover, Crtc1–/– male mice have a decreased expression of several neuroplasticity genes in the prefrontal cortex and hippocampus, including Bdnf and its receptor TrkB, as well as the nuclear receptors Nr4a1-3, thus suggesting impaired neuroplasticity processes in these brain structures. Interestingly, we showed that Crtc1–/– mice are resistant to the chronic antidepressant effects of the selective serotonin reuptake inhibitor fluoxetine, as well as the tricyclic antidepressant desipramine, in a behavioral despair paradigm (OSFST), which suggests that CRTC1 is required for conventional antidepressant therapy (Breuillaud et al., 2012; Meylan et al., 2016b). Supporting the blunted behavioral response to chronic desipramine, this antidepressant does not increase Bdnf expression in the prefrontal cortex of Crtc1–/– mice in contrast to its upregulation in wild-type mice. Moreover, we showed that chronic systemic administration of the histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA, also known as vorinostat) partially rescues the depressive-like behavior of Crtc1–/– mice and restores Bdnf expression in the prefrontal cortex (Meylan et al., 2016b). The rationale behind this epigenetic intervention was that CRTC1 helps phosphorylated CREB to recruit the histone acetyltransferase CBP and that the absence of CRTC1 decreases CBP recruitment and histone acetylation of neuroplasticity gene promoters involved in mood regulation. Reduction of CBP recruitment and histone acetylation in Crtc1–/– mice can thus be partially overcome by HDAC inhibition, which restores the expression of a subset of genes by acting downstream of CRTC1. These findings are in line with the current understanding of the involvement of epigenetic processes in the pathogenesis of MDD and antidepressant action, as well as with the recent evidence of the therapeutic potential of HDAC inhibitors (Vialou et al., 2013; Misztak et al., 2018; Uchida et al., 2018).
TABLE 1.
Summary of studies implicating CRTC1 in the pathophysiology of major depressive disorder (MDD).
| References | System | Findings |
| Breuillaud et al., 2012 | Crtc1–/– mice | Depressive-like behaviors in Crtc1–/– males: increased behavioral despair (FST, OSFST), decreased sexual motivation, anhedonia, social withdrawal. Blunted antidepressant response to the selective serotonin reuptake inhibitor fluoxetine. Decreased dopamine and serotonin turnover in the prefrontal cortex (PFC). Decreased expression of neuroplasticity genes, including Bdnf, its receptor TrkB, the nuclear receptors Nr4a1-3, and several other CREB-regulated genes in the hippocampus (HIP) and PFC. |
| Meylan et al., 2016a | Crtc1–/– mice |
Crtc1–/– male mice not responding to the tricyclic antidepressant desipramine in a behavioral despair paradigm (OSFST). Crtc1–/– male mice responding to the antidepressant effects of chronic SAHA administration (HDAC inhibitor). Epigenetic rescue of Bdnf expression in the PFC. |
| Meylan et al., 2016b | Crtc1–/– mice | Upregulation of the agmatine-degrading enzyme agmatinase in the HIP and PFC of Crtc1–/– male and female mice. Acute agmatine and ketamine treatments comparably improved the behavioral despair (FST) of Crtc1–/– male and female mice. NMDA receptor antagonist properties of agmatine possibly underlying its fast-acting antidepressant effect. |
| Jiang B. et al., 2019 | CSDS and CUMS mouse models of depression | Increased expression of hippocampal SIK2 by chronic stress (CSDS and CUMS), leading to reduced CRTC1 nuclear translocation and Bdnf expression, as well as decreased levels of hippocampal CRTC1. AAV-mediated overexpression of SIK2 in the HIP of non-stressed mice induced depressive-like behaviors in the FST, tail suspension test, sucrose preference test, and social interaction test. Hippocampal SIK2 overexpression reduced nuclear and total CRTC1 levels, downregulated BDNF and its signaling cascade, and decreased adult hippocampal neurogenesis. AAV-shRNA-mediated knockdown of hippocampal SIK2 or genetic knockout of Sik2 protected mice in CSDS and CUMS models of depression with antidepressant-like effects depending on the CRTC1-CREB-BDNF pathway. Fluoxetine, venlafaxine and mirtazapine, belonging to three different antidepressant classes, all reversed the effects of CUMS and CSDS on hippocampal SIK2 and CRTC1, and their antidepressant actions were fully abolished by hippocampal CRTC1 silencing. |
| Ni et al., 2019 | LPS-induced mouse model of depression | AAV-shRNA-mediated downregulation of CRTC1 induced depressive-like behaviors in naive mice and a decreased expression of BDNF and VGF in the ventral HIP. AAV-mediated CRTC1 overexpression in the ventral HIP prevented LPS-induced depressive-like behaviors, and restored BDNF and VGF levels. Upregulation of CRTC1 in the ventral HIP decreased LPS-induced pro-inflammatory cytokines (IL-6, IL1β, TNFα). |
| Liu et al., 2020 | CSDS and CUMS mouse models of depression | ARN-3236, a selective inhibitor of SIK2, induced significant antidepressant-like effects in both the CSDS and CUMS models of depression by acting on the hippocampal CRTC1-CREB-BDNF pathway and adult hippocampal neurogenesis. |
| Wang et al., 2020 | 3 chronic stress mouse models of depression | Imipramine, a tricyclic antidepressant, reversed the down-regulating effects of chronic restraint stress (CRS), CUMS and CSDS on CRTC1 expression in the medial PFC but not in the HIP. AAV-shRNA-mediated downregulation of CRTC1 in the medial PFC fully abolished the antidepressant-like actions of imipramine in the FST, tail suspension test, and sucrose preference test. |
| Cherix et al., 2020 | Crtc1–/– mice |
In vivo neuroimaging techniques, such as high field magnetic resonance imaging (MRI), spectroscopy (MRS) and positron emission tomography (PET) revealed an altered glucose metabolism and low energetic status in the HIP of Crtc1–/– male mice resulting in excessive GABAergic neurotransmitter cycling and depressive-like behaviors. Restoration of hippocampal energy balance with ebselen, an energy-boosting mood-stabilizer, rescued the behavioral despair (OSFST) of Crtc1–/– male mice. |
| Si et al., 2021 | Prenatally stressed male offspring rats | Depressive-like behaviors (FST and sucrose preference test) of adult male offspring rats from mothers exposed to CRS during pregnancy were associated with decreased levels of total CRTC1, nuclear CRTC1, calcineurin, BDNF and c-fos in the HIP and PFC. Chronic fluoxetine treatment (from postnatal days 30–51) reversed both the depressive-like behaviors and the downregulation of the CRTC1-BDNF pathway of prenatally stressed rats. |
Using a cDNA microarray approach, we identified differentially expressed genes in the cortex of Crtc1–/– mice, among which we found an upregulation of agmatinase, the enzyme degrading the arginine-decarboxylation product agmatine (Meylan et al., 2016a). Increasing evidence suggests that agmatine is an important neuromodulator, possibly even serving as a neurotransmitter, as it is stored in synaptic vesicles and released upon depolarization, selectively recaptured and finally degraded by agmatinase to form putrescine, a key precursor for polyamine synthesis (Li et al., 1994; Reis and Regunathan, 2000; Uzbay, 2012; Piletz et al., 2013). Agmatine is primarily found in neurons of brain regions that subserve cognition, processing of emotions, visceral and neuroendocrine control, and pain perception, as the distribution of agmatine-containing neurons is concentrated in the cerebral cortex, hippocampus, amygdala, septum, bed nucleus of the stria terminalis, periventricular thalamic and hypothalamic nuclei, as well as several areas of the midbrain and brainstem (Reis and Regunathan, 2000). Agmatine has been colocalized with other neurotransmitters, notably glutamate in synaptic terminals in hippocampal CA1 region (Seo et al., 2011). It also binds with high affinity several postsynaptic membrane receptors, such as α2-adrenergic receptors, imidazoline I1 and I2 receptors, nicotinic receptors and serotoninergic 5HT-2A and 5HT-3 receptors, and blocks glutamate NMDA receptor channels in a similar way as the rapid-acting antidepressant ketamine (Yang and Reis, 1999; Reis and Regunathan, 2000; Halaris and Piletz, 2007; Uzbay, 2012; Neis et al., 2016a,b, 2018; Barua et al., 2019; Camargo and Rodrigues, 2019; Valverde et al., 2021). Moreover, agmatine is a key modulator of nitric oxide and polyamine overproduction during the “Polyamine Stress Response” that is important for proper adaptive responses of quiescent cells to stressful conditions (Piletz et al., 2013). Accumulating evidence implicates the agmatinergic system in the etiopathogenesis of mood disorders (Laube and Bernstein, 2017; Weiss et al., 2018; Watts et al., 2019) and other central nervous system disorders (Neis et al., 2017). Post-mortem studies showed a strong upregulation of agmatinase levels in hippocampal GABAergic interneurons of brain tissues from depressed subjects (Bernstein et al., 2012), and decreased agmatine levels in the cortex of individuals who died by suicide (Chen et al., 2018). Acute and chronic stress reduce agmatine levels in rodent forebrain and exogenous agmatine administration has rapid antidepressant effect in animal models of depression (Zomkowski et al., 2002; Freitas et al., 2016). Consistent with their common property to block glutamate NMDA receptors, we found that acute agmatine and ketamine treatments comparably improve the depressive-like phenotype of male and female Crtc1–/– mice in the forced swim test (Meylan et al., 2016a). Current understanding of the mechanisms underlying ketamine’s rapid antidepressant action involves direct and indirect downstream consequences of NMDA glutamate receptor antagonism (Duman et al., 2019b; Krystal et al., 2019). The indirect hypothesis of the antidepressant actions of ketamine implies that ketamine initially blocks NMDA receptors on GABA inhibitory interneurons leading to “indirect” disinhibition of glutamate transmission and enhanced stimulation of AMPA glutamate receptors on excitatory neurons. AMPA receptor activation triggers a signaling cascade that raises BDNF levels. TrkB receptors activated by the local release of BDNF stimulate the activation of the molecular target of rapamycin complex 1 (mTORC1), which, in turn, catalyzes local protein synthesis necessary for increasing dendritic spine formation and restoring synaptic connectivity. The second hypothesis of ketamine’s action is based on the direct inhibition of NMDA receptors located on excitatory neurons, and the involvement of the eukaryotic elongation factor 2 (eEF2) kinase pathway. Ketamine blockade of NMDA receptors at rest leads to suppression of eEF2 kinase activity and subsequent decreased eEF2 phosphorylation that activates molecular target of rapamycin (mTOR)-regulated local protein synthesis. The resulting increased synthesis of BDNF and other postsynaptic proteins stimulates the shuttling of AMPA glutamate receptors to the synapse, which enhances synaptic efficacy (Autry et al., 2011). In our study, we found that agmatine rapidly increases BDNF levels only in the prefrontal cortex of wild-type females, and decreases eEF2 phosphorylation in the prefrontal cortex of male and female wild-type mice, indicating that agmatine might function as a fast-acting antidepressant with NMDA receptor antagonist properties. In contrast to wild-type mice, the acute antidepressant effects of agmatine do not seem to rely on eEF2 dephosphorylation-dependent increase of BDNF synthesis in Crtc1–/– mice. These findings support a causal role of the deregulated agmatinergic system in the Crtc1–/– mouse model of depression, as it has been suggested for human MDD. On the other hand, the mechanistic relationship between agmatinase upregulation and Crtc1 deficiency is not clear yet and deserves more investigation.
Recently, several studies highlighted the importance of CRTC1 in stress- and inflammation-induced depressive-like behaviors, thus extending our pioneering findings on the role of CRTC1 in MDD (Davis, 2019; Jiang B. et al., 2019; Ni et al., 2019; Liu et al., 2020; Wang et al., 2020; Si et al., 2021). In a quite extensive study, the SIK2-CRTC1-CREB-BDNF pathway was proved highly instrumental in mediating chronic stress-induced depressive-like behaviors and antidepressant response in C57BL/6J mice (Jiang B. et al., 2019). Both chronic unpredictable mild stress (CUMS) and chronic social defeat stress (CSDS) markedly increase hippocampal Sik2 expression, which reduces CRTC1 nuclear translocation and Bdnf expression, as well as total protein levels of hippocampal CRTC1. Adeno-associated virus (AAV)-mediated overexpression of SIK2 in the hippocampus of non-stressed mice induces depressive-like behaviors in the FST, tail suspension test, sucrose preference test, and social interaction test, thus mimicking the effects of chronic stress. Hippocampal SIK2 overexpression lowers nuclear and total CRTC1 levels, downregulates BDNF and its signaling cascade, and decreases adult hippocampal neurogenesis. Conversely, AAV-short hairpin RNA (shRNA)-mediated knockdown of hippocampal SIK2 or genetic knockout of Sik2 protect mice against both chronic stress models of depression with antidepressant-like effects that require the downstream CRTC1-CREB-BDNF pathway. Finally, fluoxetine, venlafaxine and mirtazapine, belonging to three different antidepressant classes, all reverse the effects of CUMS and CSDS on hippocampal SIK2 and CRTC1, and their antidepressant actions are fully abolished by hippocampal CRTC1 silencing. Two follow-up studies from the same laboratory then showed that the antidepressant effects of imipramine depend on CRTC1 in the medial prefrontal cortex (Wang et al., 2020), and that ARN-3236, a selective inhibitor of SIK2, induced significant antidepressant-like effects in both the CSDS and CUMS models of depression by acting on the hippocampal CRTC1-CREB-BDNF pathway (Liu et al., 2020). Moreover, it was recently shown that prenatally stressed offspring male rats display depressive-like behaviors associated with decreased levels of CRTC1 and BDNF in the hippocampus and prefrontal cortex, which are restored by a chronic fluoxetine treatment (Si et al., 2021). Taken together, these findings strengthen the pivotal role of hippocampal and prefrontal CRTC1 in the pathogenesis of MDD and in antidepressant response.
Another study convincingly highlighted CRTC1’s key role in lipopolysaccharide (LPS)-induced depressive-like behaviors (Ni et al., 2019). Systemic administration of LPS causes chronic neuroinflammation and depressive-like behaviors in mice (Dantzer et al., 2008). This animal model is relevant for the human neuroinflammation hypothesis of MDD that is associated with the atypical depression subtype (Woelfer et al., 2019). Starting from the observation that a concentration of LPS inducing depressive-like behaviors in ICR mice also decreases hippocampal CRTC1 protein levels, Ni et al. (2019) used AAV-mediated silencing or upregulation of CRTC1 in the ventral hippocampus to determine its role in LPS-induced depressive-like behaviors. AAV-shRNA-mediated downregulation of CRTC1 induces depressive-like behaviors in naive mice and a decreased expression of BDNF and VGF in the ventral hippocampus. Conversely, ventral hippocampal AAV-mediated CRTC1 overexpression prevents depressive-like behaviors induced by a single i.p. injection of LPS, and restores BDNF and VGF levels. Interestingly, upregulation of CRTC1 in the ventral hippocampus interferes with neuroinflammatory processes, as shown by the dampened accumulation of LPS-induced pro-inflammatory cytokines, such as interleukin-6, interleukin 1-β and tumor necrosis factor α. These compelling findings further support the essential role of CRTC1 in MDD pathogenesis, and suggest that CRTC1 coactivates genes controlling neuroinflammation through still unknown mechanisms (Davis, 2019; Ni et al., 2019).
Lately, ketamine has attracted much attention for its rapid antidepressant effects in patients with treatment-resistant depression (Kraus et al., 2019). Ketamine and a few other compounds acting on glutamate neurotransmission are the only antidepressant drugs in development that are able to relieve MDD symptoms within hours after a single dose, the effects of which lasting for up to a week. Unfortunately, ketamine is also a well-known psychedelic drug with an addictive potential, and almost nothing is known about the risks of its long-term use. Current research is thus focusing on the development of ketamine-like molecules having the same fast-acting antidepressant benefits without the undesirable side effects (Zanos et al., 2018; Ragguett et al., 2019; Wilkinson and Sanacora, 2019). This preclinical development of novel pharmacotherapies requires a better understanding of the molecular and cellular mechanisms underlying the antidepressant actions of ketamine. A recent study showed that ketamine’s acute antidepressant effects precede its action on spine formation in the prefrontal cortex, indicating that spinogenesis is not required for the rapid (less than 12 h) behavioral response (Moda-Sava et al., 2019). In contrast, prefrontal cortical restoration of lost spines by a single ketamine injection is necessary for its sustained (2–7 days) antidepressant effects. BDNF and its receptor TrkB, as well as VGF, have been implicated in ketamine’s antidepressant effects (Autry et al., 2011; Bjorkholm and Monteggia, 2016; Ma et al., 2017; Song et al., 2017; Zanos and Gould, 2018; Jiang C. et al., 2019). However, their respective function in the acute and/or sustained effects of ketamine is not completely understood. The involvement of CRTC1 in the regulation of BDNF and VGF expression (Zhou et al., 2006; Parra-Damas et al., 2014; Fukuchi et al., 2015; Ni et al., 2019), as well as the decreased brain BDNF levels in Crtc1–/– mice (Breuillaud et al., 2012; Meylan et al., 2016a), suggest that ketamine’s antidepressant effects might rely on CRTC1, as well. We previously showed that the immobility of Crtc1–/– males and females is significantly decreased 30 min after a single injection of ketamine (3 mg/kg) in the FST paradigm [Supplementary Figure 2 in Meylan et al. (2016a)], thus indicating that CRTC1 is not required for the acute antidepressant effects of ketamine. Future investigations are nevertheless needed to determine whether CRTC1 is involved in the neuroplasticity-related sustained long-term antidepressant effects of ketamine. In summary, our findings, as well as several subsequent studies, highlight the pivotal role of the CRTC1-CREB-BDNF pathway in preclinical animal models of MDD, and provide evidence of its involvement in antidepressant response.
Deregulation of Brain Energy Homeostasis in Major Depressive Disorder: Possible Role of CREB-Regulated Transcription Coactivator 1
Several lines of evidence suggest that altered energy metabolism underlies mood disorders, both at the brain and peripheral level (Ostergaard et al., 2018). The rationale being that normal neuronal function is challenged in MDD due to an imbalance in energy homeostasis, through either an impaired glycolytic (Videbech, 2000) or mitochondrial ATP production (Klinedinst and Regenold, 2015). This imbalance is exacerbated by excessive energy demand, typically occurring during stress exposure (Picard et al., 2018). Several magnetic resonance spectroscopy (MRS) and positron-emission tomography (PET) studies have identified key brain regions with abnormal metabolic activity. However, it appears that systemic metabolic mechanisms play a role as well, associating several endocrine dysregulations and inflammatory processes with impaired neuroenergetics. An example of this association is the relation of metabolic syndrome to atypical depression, as highlighted by their high comorbidity (Marazziti et al., 2014). Metabolic syndrome is a cluster of health conditions including abdominal obesity, high blood pressure and insulin resistance, which reflect impaired energy utilization and storage. In fact, MDD has been associated with impaired glucose homeostasis, and in particular insulin resistance [for a systematic review and meta-analysis see Kan et al. (2013)], which can occur in the brain itself (Watson et al., 2018; Lyra et al., 2019). Brain insulin resistance has thus become a therapeutic target for treating MDD and its associated symptoms (Hamer et al., 2019). For instance, insulin sensitizers have proven antidepressant efficacy in animal models (Zhao et al., 2016) and patients (Sepanjnia et al., 2012; Kemp et al., 2014) leading to improved glucose metabolism (Lin et al., 2015). Among the numerous possible causes, stress appears to be a key modulator, if not initiator, of such metabolic disturbances. Notably, insulin resistance is a recognized consequence of allostatic overload triggered by stress (Rasgon and McEwen, 2016) and has been associated with depressive phenotype (Li et al., 2013; van der Kooij et al., 2018; Yang et al., 2018).
Control of energy homeostasis is complex and involves several energy-sensing molecules. Intracellular detectors like AMPK (AMP-activated kinase), SIRT1 (Sirtuin 1) or HIF (Hypoxia-inducible factor), provide a rapid response to fluctuations in ATP, NADH and oxygen concentrations, and have been implicated in MDD (Ostergaard et al., 2018). Intercellular signaling, in a paracrine, endocrine or neurotransmission fashion (e.g., through monoamines, glucocorticoids or insulin), is another key mechanism for regulating metabolic activity based on energy demand. As such, the cAMP-CREB pathway is central to metabolic homeostasis by stimulating tissue-specific gene expression, through its extracellular metabotropic signaling sensitivity. CRTCs, as co-activators of CREB, play a central modulatory role in this cascade (Altarejos and Montminy, 2011). Hepatic CRTC2 promotes gluconeogenic response through opposite action of insulin or glucagon on SIK2. During short-term fasting, glucagon-induced rise in hepatic cAMP triggers the dephosphorylation of CRTC2, whereas upon feeding, the insulin-dependent activation of Akt leads to CRTC2 phosphorylation and cytoplasmic retention (Altarejos and Montminy, 2011). CRTC2 also holds a regulatory role on insulin secretion in the β-cells of the pancreatic islets (Screaton et al., 2004). In contrast, CRTC3 is mainly found in adipose tissue, where it enhances insulin resistance and obesity (Song et al., 2010). Finally, in skeletal muscles, the CRTC-CREB pathway seems to play a role in mitochondrial biogenesis through the expression of peroxisome proliferator activated receptor-γ (PPARγ) co-activator 1α (PGC1α) (Wu et al., 2006). CRTC1 is primarily expressed in the central nervous system and particularly abundant in limbic areas of mammalian brain. Neuronal CRTC-1 is controlling mitochondrial metabolism in Caenorhabditis elegans (Burkewitz et al., 2015). Likewise, the SIK-CRTC1 pathway was identified as an important cascade for controlling neuronal energy homeostasis in Drosophila (Choi et al., 2011; Shen et al., 2016). Studies in rodents highlight the importance of the neuronal CRTC1-CREB pathway in translating metabotropic neurotransmission signaling into synaptic plasticity (Saura and Cardinaux, 2017). While it seems clear that synapse formation and plasticity require stimulation of energy metabolism, the mechanistic pathway involving CRTC1 remains to be discovered in the mammalian brain. The role of CRTC1 in controlling energy homeostasis appears to be complex and multifactorial, involving both peripheral and central control. In fact, despite being primarily expressed in the brain, CRTC1 plays a role in peripheral tissues as well (Kim et al., 2015; Kim, 2016; Schumacher et al., 2016; Gao et al., 2018; Morhenn et al., 2019). Deletion of Crtc1 gene in mice induces insulin resistance and obesity, together with a depressive-like phenotype (Altarejos et al., 2008; Breuillaud et al., 2009, 2012; Rossetti et al., 2017). Using in vivo MRS at high field, we have observed that this phenotype translates into measurable neurochemical alterations in the hippocampus of Crtc1–/– mice (Cherix et al., 2020). Noteworthy, we identified lower levels of phosphocreatine relative to creatine, an indication that ATP production or consumption is impaired. These results suggest that CRTC1 is required for normal energy homeostasis in the brain and could mechanistically underlie similar observations arising from MRS studies in human depression (Moore et al., 1997; Harper et al., 2017). Finally, the strong resemblance between the Crtc1–/– mouse phenotype and metabolic syndrome might provide new avenues for exploring how energy deregulations relate to human MDD (Rasgon and McEwen, 2016; Watson et al., 2018).
CREB-Regulated Transcription Coactivator 1 in Major Depressive Disorder-Associated Obesity
Epidemiological evidence clearly suggests a comorbid association between depression and obesity (Amare et al., 2017; Milaneschi et al., 2019). Indeed, recent cross-sectional and longitudinal meta-analyses have demonstrated a positive and bidirectional relationship between these two pathological conditions. Moreover, these meta-analyses indicate that the MDD-obesity comorbidity occurs in both adulthood and adolescence, that it is not related to sociodemographic and lifestyle factors, and cannot be completely explained by the body weight increase induced by antidepressant medications. Compared to the general depression-obesity association, even stronger comorbidity was observed in atypical depressed patients, a subgroup of MDD patients showing sustained food intake during depressive episodes, increased visceral fat deposition and adverse metabolic profile (Xu et al., 2011). In keeping with this observation, a recent investigation suggests that the association between atypical depressive symptoms and obesity-related traits may arise from shared pathophysiological mechanisms (Milaneschi et al., 2017).
The wide distribution of CRTC1 in the rodent brain has been related to several functions spanning from synaptic plasticity, learning and memory, emotional processing and central energy-balance regulation (Saura and Cardinaux, 2017). In accordance with this broad function, the genetic suppression of CRTC1 in the mouse was found to induce depressive-like symptoms and obesity (Altarejos et al., 2008; Breuillaud et al., 2009, 2012; Rossetti et al., 2017). Although the human CRTC1 polymorphisms studied so far were not clearly associated with depression, they were associated with obesity markers [body mass index (BMI), fat mass] in psychiatric cohorts and in individuals with MDD (Choong et al., 2013; Quteineh et al., 2016). With regard to obesity, further human genetic investigations showed that the CRTC1 locus links fat mass to cardiometabolic diseases (Lu et al., 2016) and that genetic and epigenetic control of CRTC1 transcription affects fat distribution and eating behavior (Booij, 2019; Delacretaz et al., 2019; Rohde et al., 2019; Wang et al., 2021). Altogether, these observations strengthen the hypothesis that CRTC1 may represent a pivotal transcription coactivator regulating both MDD and obesity etiological pathways (see Tables 1, 2).
TABLE 2.
Summary of studies implicating CRTC1 in obesity.
| References | System | Findings |
| Altarejos et al., 2008 | Crtc1–/– mice |
Crtc1–/– mice are hyperphagic, obese and infertile. Hypothalamic CRTC1 phosphorylated and inactive in leptin-deficient ob/ob mice, and leptin administration increased dephosphorylated nuclear CRTC1. CRTC1 regulates the Cartpt and Kiss1 genes, which encode hypothalamic neuropeptides that mediate leptin’s effects on satiety and fertility. |
| Breuillaud et al., 2009 | Crtc1–/– mice | Crtc1–/– mice are obese, but not infertile. No alteration of hypothalamic Kiss1 gene expression and plasma luteinizing hormone levels. |
| Kim et al., 2015 | Streptozotocin-induced (STZ) diabetic Crtc1+/− mice | Leptin improved diabetic glucose metabolism through Crtc1-dependent and independent mechanisms. Leptin reduced diabetic hyperglycemia, hepatic gluconeogenic gene expression and selectively increased glucose disposal to brown adipose tissue and heart, in STZ-diabetic WT mice but not Crtc1+/– mice. Leptin promoted CRTC1 nuclear translocation in pro-opiomelanocortin (Pomc) and non-Pomc neurons within the hypothalamic arcuate nucleus, and leptin-induced Pomc gene expression was blunted in STZ-diabetic Crtc1+/– mice. |
| Rossetti et al., 2017 | Crtc1–/– mice | Gender difference in the homeostatic regulation of energy balance. Crtc1–/– males are hyperphagic and rapidly develop obesity on normal chow diet. Crtc1–/– females exhibit mild late-onset obesity without hyperphagia. Alterations in the expression of several orexigenic and anorexigenic hypothalamic genes in mutant males. Crtc1–/– males’ hyperphagic behavior is restricted to the diurnal (resting) phase of the light cycle during which they have a higher locomotor activity. |
| Matsumura et al., 2021 | Sf1-cre – Crtc1loxP/loxP mice | Mice with a ventromedial hypothalamus (VMH)-specific knockdown of Crtc1 are sensitive to high-fat diet-induced obesity, exhibiting hyperphagia and increased body weight gain. Unlike Crtc1–/– mice, VMH-specific Crtc1 deletion did not affect body weight gain or food intake in normal chow feeding. |
| Hu et al., 2021 | Crtc1–/– mice | New Crtc1–/– mouse model generated by the CRISPR/Cas9 system exhibiting an obese phenotype, but apparently independent of alterations in food intake or energy expenditure. Crucial role of CRTC1 in regulating lipid metabolism in adipose tissue during development. |
| Choong et al., 2013 | Human, psychiatric patients and general population | First study showing an association of CRTC1 polymorphisms with body mass index (BMI) and fat mass in humans. |
| Quteineh et al., 2016 | Human, general population samples |
CRTC1 polymorphisms seem to play a role with obesity markers in individuals diagnosed with lifetime MDD rather than non-depressive individuals. No direct association of CRTC1 polymorphisms with MDD in the three samples tested. |
| Lu et al., 2016 | Human, meta-analysis in a large sample | CRTC1 locus found in the 12 loci reaching genome-wide significance in the genome-wide association meta-analysis of body fat percentage in more than 100,000 individuals. |
| Rohde et al., 2019 | Human, general population samples | DNA methylation levels of a CpG within the CRTC1 rs7256986 polymorphism and in a neighboring CpG were allele/genotype-dependent, suggesting a methylation quantitative trait locus (meQTL) in whole blood and adipose tissue. The presence of the SNP and/or DNA methylation correlated with CRTC1 gene expression, which in turn, related to BMI and fat distribution. |
| Delacretaz et al., 2019 | Human, psychiatric patients with psychotropic treatment | Significant methylation changes observed in three CRTC1 CpG sites in the blood of patients with early and important weight gain. One of these 3 CpG sites was significantly associated with early weight gain in patients carrying the G allele of rs4808844A > G, a SNP associated with this methylation site. |
| Wang et al., 2021 | Human, 4 obese patients and 4 controls | Genome-wide DNA methylation analysis and pyrosequencing confirmation revealed that the methylation levels of 2 CpG sites in CRTC1 were significantly changed in patients with obesity compared with normal controls. |
Several human studies evaluated the association of CRTC1 single nucleotide polymorphisms (SNPs) with obesity markers in the general population. To our knowledge, this association was first studied by Choong et al. (2013), who showed no significant association of the CRTC1 SNP rs6510997 (a proxy of the rs3746266 G allele) with BMI, weight or waist circumference in the general population. Similarly, CRTC1 locus did not reach genome-wide significance for BMI in the study of Locke et al. (2015), despite the very large sample size. Conversely, CRTC1 SNP rs757318 was found, for the first time, to be associated with Body Fat percentage (BF%) and BMI in a genome-wide association study (GWAS) aiming at establishing a link between adiposity and cardiometabolic disease risk (Lu et al., 2016). This study reported that CRTC1 SNP rs757318 had a more pronounced effect on BF% than on BMI, suggesting that BMI is an heterogeneous and less precise marker for adiposity, as it depends on both lean and fat mass. Interestingly, CRTC1 locus showed a significant sex-specific interaction with a two to threefold larger effect in women than in men. However, the BF%-increasing CRTC1 rs757318 allele was not associated with any of the cardiometabolic traits analyzed in this study. An interaction between CRTC1 SNP and sex was also observed in a human study that examined whether the CRTC1 polymorphism was associated with obesity markers in subjects with lifetime depression. Here, the CRTC1 SNP rs6510997 was found to be negatively associated with BMI in women with a lifetime diagnosis of depression (Quteineh et al., 2016). Taken together these observations suggest that: (1) the association of CRTC1 locus with obesity markers is stronger when reliable markers of adiposity (such as BF%) are used, (2) it may exist a sex-specific effect, even though, the use of different CRTC1 SNPs and different sample populations (general population versus psychiatric cohorts) complicate the interpretation of these GWAS studies. Moreover, due to the lack of knowledge about the biological effect of the tested SNPs on CRTC1’s function in humans (enhancement or decrease of CRTC1 activity in the brain), it is difficult to know whether the sex difference in energy balance regulation observed in Crtc1–/– mice is a specific feature of rodents.
Besides these human studies, the greatest contribution to the understanding of the regulatory role of CRTC1 in energy balance comes from animal experiments. A first study described the effects of the suppression of CRTC1 in the central control of food intake and energy expenditure (Altarejos et al., 2008). Crtc1–/– mice exhibited hyperphagia and body weight gain from early adult age with consequent development of obesity. The obese state of these mutant mice was also accompanied by reduced locomotor activity, lower energy expenditure and impaired sensitivity to leptin and insulin. However, it should be emphasized that there are gender differences regarding the vulnerability to develop obesity in this mouse model. Indeed, we showed that Crtc1–/– females are not hyperphagic and are less prone to gain weight as compared to mutant males, whose hyperphagia appears to be related to altered circadian locomotor activity (Rossetti et al., 2017). Moreover, CRTC1 seems necessary to protect against hepatic steatosis, which is strongly associated with obesity and metabolic syndrome, and to modulate leptin’s glucoregulatory actions in insulin-dependent diabetes (Kim et al., 2015; Kim, 2016). Whether CRTC1 is solely involved in the central control of energy balance or whether it also plays a role in peripheral organs is still unclear. A new Crtc1–/– mouse model generated by the CRISPR/Cas9 system also exhibits an obese phenotype, but it appears to be independent of alterations in food intake or energy expenditure (Hu et al., 2021). In this complete knockout mouse model, CRTC1 seems to be rather implicated in the regulation of lipid metabolism in adipose tissue during development. The creation of Crtc1 conditional knockout (cKO) mouse models should greatly help to decipher the specific role of CRTC1 in various brain regions and cell types. To date, there has been only one article reporting the use of a Crtc1 cKO mouse model (Matsumura et al., 2021). This study showed that mice with a ventromedial hypothalamus-specific knockdown of Crtc1 are sensitive to high-fat diet-induced obesity, exhibiting hyperphagia and increased body weight gain, but have a normal feeding behavior with a control chow diet.
Energy balance regulation depends on the interaction of many brain pathways that finely adapt food intake and energy expenditure to maintain energy homeostasis. After feeding, the release of leptin from adipocytes promotes satiety by acting on arcuate neurons in the hypothalamus (Myers et al., 2009; Pan and Myers, 2018). The activity of leptin depends on the stimulation of hypothalamic neurons releasing anorexigenic peptides and on the concomitant inhibition of neurons that secrete orexigenic peptides (Schwartz et al., 2000; Williams and Elmquist, 2012; Yeo and Heisler, 2012). The current working hypothesis suggests that CRTC1 contributes to leptin anorexigenic activity by modulating the expression of genes that participate in energy homeostasis. Accordingly, experimental evidence shows that leptin is able to activate the CRTC1/CREB pathway through different manners. This adipokine promotes CREB phosphorylation through the activation of the JAK2/STAT3 intracellular cascade (Catalano et al., 2009). In addition, leptin enhances CRTC1 nuclear translocation (Altarejos et al., 2008), possibly through the inhibition of AMP-activated kinase (Minokoshi et al., 2004) and increased activity of anorexigenic POMC neurons (Cowley et al., 2001). On the other hand, disrupted leptin signaling in leptin–deficient ob/ob mice is associated with increased amounts of phosphorylated, inactive CRTC1 in the cytoplasm of hypothalamic neurons (Altarejos et al., 2008).
Among the CRTC1/CREB-regulated genes, Cart (cocaine-amphetamine related transcript) is of particular importance for the suppression of food consumption, as it mediates leptin activity in the hypothalamus. In line with these molecular interactions, obese leptin-deficient ob/ob mice have a lower expression of Cart in the hypothalamus, and peripheral leptin injection normalizes hypothalamic CART peptide levels in these obese mice (Duan et al., 2007). Moreover, the capacity of CRTC1 to mediate the stimulating effect of leptin on Cart expression is supported by the increased plasmatic levels of leptin, reduced leptin receptor (LepRb) mRNA expression and lower Cart transcription in the arcuate nucleus of obese Crtc1–/– mice. Conversely, younger hyperphagic but not obese Crtc1–/– mice that are still leptin-sensitive, show comparable number of phospho-STAT3 positive cells in the arcuate nucleus after leptin infusion and similar leptin receptor (LepRb) and Cart mRNA levels as compared to lean wild-type mice (Altarejos et al., 2008; Rossetti et al., 2017). These data validate the functional interplay between leptin and CRTC1/CREB pathway, but point out the fact that the impaired Cart expression is probably just a consequence of leptin resistance and obesity, and that the lack of CRTC1 has likely broader effects on energy balance regulation by affecting other relevant genes.
In this regard, BDNF is an interesting candidate. Although first implicated in the etiology of MDD, this member of the neurotrophin family of growth factors was later recognized as a key component of mechanisms regulating energy intake and expenditure (Rios, 2013; Marosi and Mattson, 2014; Xu and Xie, 2016). According to a physiological role of BDNF in energy control, the expression of Bdnf and its receptor TrkB is sensitive to the nutritional state. Indeed, food deprivation reduces BDNF mRNA expression in the rat ventromedial hypothalamus, whereas glucose injection rapidly induces Bdnf and TrkB transcription in the same region (Xu et al., 2003; Unger et al., 2007). Complementary studies showed that the central injections of BDNF in rodents (i.c.v. or directly in the ventromedial or paraventricular hypothalamus) lead to reduced food intake, increased energy expenditure and body weight loss (Toriya et al., 2010). On the contrary, different genetic manipulations in mice, resulting in lower expression of either Bdnf or its TrkB receptor, cause hyperphagia and induce signs of metabolic syndrome including leptin and insulin resistance, dyslipidemia and hyperglycemia (Kernie et al., 2000; Rios et al., 2001). Finally, BDNF is also playing a role in the hedonic regulation of food intake involving the mesolimbic dopamine system. Consumption of palatable, high-fat food (HFF) influences Bdnf and TrkB expression in the dopaminergic neurons of the ventral tegmental area (VTA), whereas BDNF depletion in the VTA affects the reward function of the mesolimbic dopamine system and leads to excessive intake of palatable HFF, but not of standard chow, and to increased body weight under HFF conditions (Cordeira et al., 2010). This study suggests that an alteration of BDNF signaling may interfere with the activity of the mesolimbic dopamine pathway, leading to reward deficiency and compensatory overeating of palatable food.
In parallel to this preclinical research, human genetic studies also support the involvement of BDNF in energy balance regulation. The functional loss of one copy of the BDNF gene reduces serum BDNF concentration, increase ad libitum food intake and provokes severe early-onset obesity (Gray et al., 2006). Moreover, a missense mutation in the NTRK2 gene (human TrkB receptor gene) that prevents the regular activity of the receptor was identified in patients exhibiting overweight and severe obesity (Yeo et al., 2004). Finally, a functional polymorphism of the BDNF gene (BDNF Val66Met), which impedes the correct secretion and signaling of BDNF, was correlated with obesity predisposition in children and adolescents (Beckers et al., 2008; Skledar et al., 2012).
Consistent with the importance of the CRTC1/CREB pathway for Bdnf transcription, Crtc1–/– mice have lower mRNA levels of both Bdnf and TrkB in different brain regions as compared to wild-type mice. Precisely, the analysis of the multiple Bdnf splice variants in the hypothalamus of these mice revealed a significant reduction in the Bdnf mRNA for the exon I and IV (Breuillaud et al., 2012). A recent study using mice with selective knockdown of BDNF production from either promoter I, II, IV, or VI showed that disruption of BDNF from promoter I or II, but not IV or VI, induces hyperphagic obesity (McAllan et al., 2018). The energy balance dysregulation observed in Crtc1–/– mice might thus be related to a decrease of Bdnf exon I mRNA in the hypothalamus. Collectively, these findings suggest that BDNF could represent one of the most important CRTC1/CREB downstream genes that contribute to the energy homeostasis. However, a direct causality between Crtc1 deficiency and reduced hypothalamic Bdnf transcripts in the development of obesity is still elusive, and therefore, further molecular and behavioral researches are required.
Although less studied than Bdnf, another potential interesting CRTC1/CREB target gene is the neuron-derived orphan nuclear receptor 1 (Nor-1, also known as Nr4a3). This gene controls food intake in the arcuate nucleus by integrating leptin and glucocorticoid signaling (Kim et al., 2013). Interestingly, its expression in the arcuate nucleus is downregulated both in young and old Crtc1–/– mice (Rossetti et al., 2017).
Over the last decade, a compelling number of studies showed that chronodisruption (or circadian rhythm alteration) in humans severely increases the risk for developing both psychiatric and metabolic diseases (Barandas et al., 2015; Albrecht, 2017; Logan and McClung, 2019; Xie et al., 2019). Accordingly, the deletion of genes belonging to the mouse circadian clock machinery, notably Bmal1 and Clock, leads to hyperphagia, metabolic alterations and obesity (Rudic et al., 2004; Turek et al., 2005; Albrecht, 2012). Many environmental and genetic factors contribute to the maintenance of the circadian rhythm and allow the optimization of multiple biological functions along the daily night and day cycle (Dietrich and Horvath, 2013; Challet, 2019). It is long time known that photic inputs activate the CREB pathway in neurons of the suprachiasmatic nucleus (SCN) of the hypothalamus. This effect is likely due to variations in Ca2+ and cAMP levels in the SCN neurons induced by increased glutamate release from the fibers of the retino-hypothalamic tract that connects neural cells in the retina to the SCN cells (Ginty et al., 1993; Obrietan et al., 1999). This hypothesis is in accordance with the fact that both cAMP and Ca2+ levels oscillate within the SCN (Prosser and Gillette, 1991; Ikeda et al., 2003). In line with the role of CREB in the light-evoked circadian clock entrainment in SCN neurons, CREB-phosphorylation oscillates along the day in the mouse SCN and is higher during the first part of the light cycle (Travnickova-Bendova et al., 2002). More recently, it was shown that the dephosphorylation of CRTC1 and its subsequent nuclear accumulation peaks during the light part of the daily cycle (Table 3). This effect was specific for CRTC1, because CRTC2 did not show any variation (Sakamoto et al., 2013). Besides, the interaction of the CRTC1/CREB pathway with the circadian system is bidirectional, because complementary evidence exists about the control of some core clock genes by this transcriptional pathway. The suppression of CREB functionality in a tetracycline-inducible CREB repressor mouse strain leads to a significant reduction of both the expression of the circadian clock proteins PERIOD1 and PERIOD2 and the clock output hormones AVP and VIP in the SCN (Lee et al., 2010). Moreover, CRTC1 is involved in the negative feedback of the clock-resetting process that follows a light phase shift (jet lag) (Jagannath et al., 2013). In the SCN, light-activated CRTC1 increases SIK1 that phosphorylates CRTC1 and inactivates it. With this feedback mechanism, CRTC1 limits the transcription of Period1 that promotes clock resetting. Thereby, CRTC1 may slow down clock resetting and prevent an abrupt desynchrony between the master clock located in the SCN and the other peripheral clocks. This buffering system could protect the SCN clock (master clock) from inappropriate phase shifting caused not only by exposure to sudden changes in light intensity during the night, but also by abnormal Zeitgeber stimuli (such as food intake, physical activity, and temperature changes) that are all known to affect its resetting. Because the SCN master clock controls the other clocks located in other brain regions and in peripheral organs, an abrupt shift of the SCN clock would severely compromise normal physiological functions of the organism [see discussion in Jagannath et al. (2013)]. More recently, Jagannath et al. (2021) also highlighted the role of the CREB/CRTC1 pathway in adenosine-mediated integration of both light and sleep signaling to regulate circadian timing in mice. Another study in Drosophila showed that CRTC mutation affects the circadian oscillation of clock genes expression producing a phase delay especially in PERIOD and TIMLESS proteins. This work highlights that the interaction between CRTC homologs and the circadian clock machinery has likely an ancestral origin and selectively evolved among different species (Kim et al., 2016).
TABLE 3.
Summary of studies implicating CRTC1 in circadian rhythms regulation.
| References | System | Findings |
| Sakamoto et al., 2013 | C57BL/6 mice | Rhythmic expression of CRTC1 in the suprachiasmatic nucleus (SCN). CRTC1 expression was detected in the middle of the subjective day, with limited expression during early night, and late night expression levels intermediate between mid-day and early night levels. During early and late subjective night, a brief light pulse induced strong nuclear accumulation of CRTC1 in the SCN. Evidence of CRTC1-mediated Per1 gene regulation. |
| Jagannath et al., 2013 | C57BL/6 mice | In the SCN, light-activated CRTC1 increases SIK1 levels, which in turn phosphorylate and inactivate CRTC1. This feedback mechanism limits the transcription of Per1 that promotes clock resetting. CRTC1 slows down clock-resetting and prevents an abrupt desynchrony between the master clock located in the SCN and the other peripheral clocks. |
| Kim et al., 2016 | Drosophila | Light-independent role of Drosophila CRTC in sustaining circadian behaviors. Crtc null mutation dampens light-independent oscillations of TIMELESS (and not PERIOD) in the clock neurons. |
| Rossetti et al., 2017 | Crtc1–/– mice | Crtc1–/– males have a hyperphagic behavior that is restricted to the diurnal (resting) phase of the light cycle during which they have a higher locomotor activity possibly due to circadian rhythms alteration. |
| Jagannath et al., 2021 | C57BL/6 mice | Adenosine, encoding sleep history, acts upon the circadian clockwork via adenosine A1/A2A receptor signaling through the activation of the Ca2+-ERK-AP-1 and CREB/CRTC1-CRE pathways to regulate the clock genes Per1 and Per2. These signaling pathways converge upon and inhibit the same pathways activated by light. Circadian entrainment by light is thus systematically modulated on a daily basis by sleep history. |
Although the effective participation of CRTC1 in the entrainment of circadian clock in vivo and the consequent effect on the circadian energy homeostasis are not completely understood, the behavioral observation of Crtc1–/– mice suggested that the obesity of mutant males is due to overeating during the resting phase of the light cycle and to circadian alteration of spontaneous locomotor activity (Rossetti et al., 2017). Remarkably, the depressive-like phenotype of Crtc1–/– mice and their concomitant altered circadian rhythms and feeding behavior strongly suggest that CRTC1 is an important player in the central regulation of mood, circadian rhythms, and energy balance – all of which are dysregulated in MDD (see Figure 2).
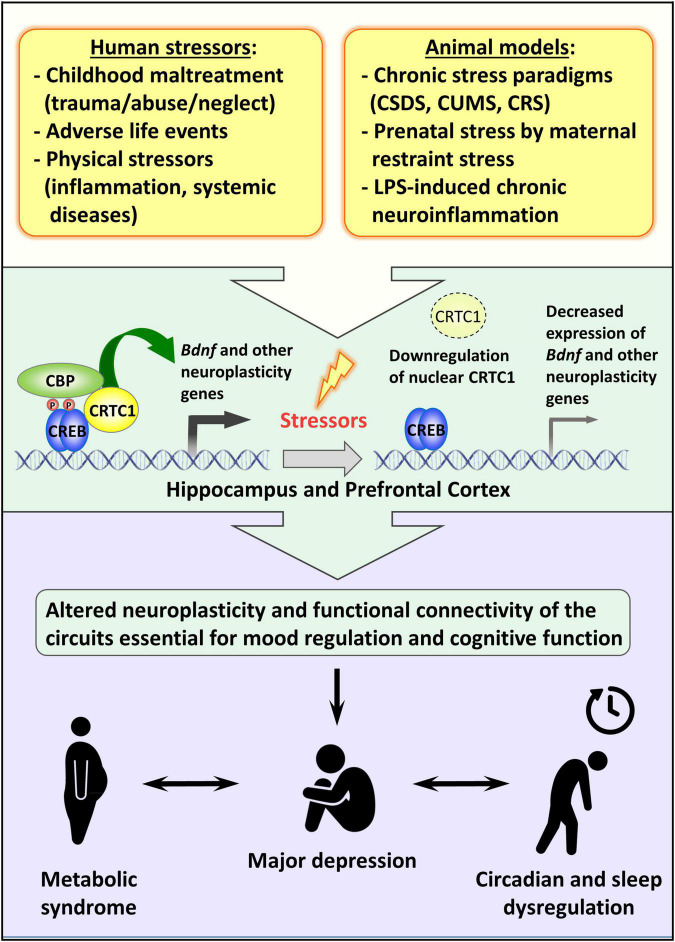
FIGURE 2.
Possible role of CRTC1 in the pathogenesis of depression and associated disorders. Preclinical animal models of depression suggest that life stressors decrease CRTC1 levels in the prefrontal cortex and hippocampus, which triggers an altered neuroplasticity and functional connectivity related to the pathogenesis of depression and associated metabolic syndrome and chronodisruption.
Conclusion and Perspectives
This review highlighted the increasing evidence of the pivotal role of the transcription coactivator CRTC1 in MDD and comorbid obesity. Mice lacking CRTC1 exhibit depressive-like endophenotypes (Breuillaud et al., 2012; Meylan et al., 2016a,b) and an altered regulation of feeding behavior resulting in hyperphagic obesity only in males (Rossetti et al., 2017). Interestingly, this gender-specific alteration of energy balance is associated with a dysregulated circadian locomotor activity. More research is needed to understand why Crtc1–/– female mice are less affected and whether the altered circadian activity of the males correlates, for instance, with a reduced average duration of sleep and modified sleep patterns. Possibly related to these metabolic and circadian alterations, MRS studies with Crtc1–/– male mice suggest that CRTC1 plays a critical role in regulating brain neuroenergetics. A possible limitation to the face validity of the Crtc1 knockout mouse model of depression should, however, be considered, as the constitutive lack of CRTC1 may have neurodevelopmental effects that may influence the adulthood phenotypes. Future studies with Crtc1 conditional knockout mouse models that may be combined with viral approaches should address this issue and assess a putative role of CRTC1 during neurodevelopment. Human CRTC1 polymorphisms were associated with obesity markers (BMI, fat mass and distribution, eating behavior) in psychiatric cohorts and in the general population (Choong et al., 2013; Lu et al., 2016; Quteineh et al., 2016; Rohde et al., 2019). However, these polymorphisms were not directly associated with depression and further investigations are required to find a possible link between CRTC1 and MDD in humans.
Recent reports highlighted the instrumental role of ventral hippocampal CRTC1 in mediating chronic stress- and LPS-induced depressive-like behaviors, and antidepressant response in mice (Jiang B. et al., 2019; Ni et al., 2019). Jiang B. et al. (2019) compellingly shed light on the specific increased hippocampal expression of SIK2 triggered by chronic stress in two different paradigms (CUMS and CSDS). The higher levels of SIK2 increase CRTC1 phosphorylation and its retention in the cytoplasm, thus leading to a less active CRTC1-CREB-BDNF pathway and depressive-like symptoms. In the LPS-induced sickness behavior model of depression, Ni et al. (2019), uncovered the important regulatory role of hippocampal CRTC1 in controlling neuroinflammation and the CRTC1-CREB-BDNF-VGF pathway. Future research should characterize the molecular mechanisms of SIK2 induction in various chronic stress paradigms. The effects of CRTC1, BDNF, and VGF downregulation on hippocampal neuroplasticity should also be better defined and correlated with depressive-like behaviors. We and others showed that CRTC1 is required for conventional antidepressants response, but it is still unclear whether CRTC1 is involved in the sustained long-term effects of the novel rapid-acting antidepressants targeting glutamate neurotransmission. Finally, it remains to be determined whether the same CRTC1-regulated genes are jointly involved in MDD-related neuroplasticity processes, brain energy metabolism, circadian rhythms, and central control of energy balance, or whether different subsets of CRTC1 target genes are independently implicated. The transcription factor CREB is considered as a primary hub for activity-driven neuronal gene expression (Benito et al., 2011). Likewise, as a coactivator of CREB and possibly other transcription factors, CRTC1 could be a primary hub for a network of genes involved in the central regulation of mood, circadian rhythms, and energy balance. Future studies aiming at deciphering the underlying mechanisms through which downregulated CRTC1 functions commonly impinge on mood, circadian rhythms and energy balance regulation should lead to better insight into the etiopathogenesis of MDD and comorbid obesity, and may provide new therapeutic targets.
Author Contributions
CR, AC, LG, and J-RC drafted and edited the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank present and past lab members for their scientific contribution to the content of this review. We are also grateful to P. J. Magistretti, O. Halfon, and K. J. von Plessen for their support. We apologize to authors whose original work could not be cited because of space limitations.
Funding
This work was supported by a grant from the Swiss National Science Foundation (31003A-170126).
References
- Akil H., Gordon J., Hen R., Javitch J., Mayberg H., Mcewen B., et al. (2018). Treatment resistant depression: a multi-scale, systems biology approach. Neurosci. Biobehav. Rev. 84 272–288. 10.1016/j.neubiorev.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. R. (2019). Adult neuroplasticity: a new “cure” for major depression? J. Psychiatry Neurosci. 44 147–150. 10.1503/jpn.190072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U. (2012). Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 74 246–260. 10.1016/j.neuron.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Albrecht U. (2017). Molecular mechanisms in mood regulation involving the circadian clock. Front. Neurol. 8:30. 10.3389/fneur.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos J. Y., Goebel N., Conkright M. D., Inoue H., Xie J., Arias C. M., et al. (2008). The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat. Med. 14 1112–1117. 10.1038/nm.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos J. Y., Montminy M. (2011). CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12 141–151. 10.1038/nrm3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amare A. T., Schubert K. O., Klingler-Hoffmann M., Cohen-Woods S., Baune B. T. (2017). The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl. Psychiatry 7:e1007. 10.1038/tp.2016.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry A. E., Adachi M., Nosyreva E., Na E. S., Los M. F., Cheng P. F., et al. (2011). NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 475 91–95. 10.1038/nature10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandas R., Landgraf D., Mccarthy M. J., Welsh D. K. (2015). Circadian clocks as modulators of metabolic comorbidity in psychiatric disorders. Curr. Psychiatry Rep. 17:98. 10.1007/s11920-015-0637-2 [DOI] [PubMed] [Google Scholar]
- Barco A., Marie H. (2011). Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol. Neurobiol. 44 330–349. 10.1007/s12035-011-8209-x [DOI] [PubMed] [Google Scholar]
- Barua S., Kim J. Y., Kim J. Y., Kim J. H., Lee J. E. (2019). Therapeutic effect of agmatine on neurological disease: focus on ion channels and receptors. Neurochem. Res. 44 735–750. 10.1007/s11064-018-02712-1 [DOI] [PubMed] [Google Scholar]
- Beckers S., Peeters A., Zegers D., Mertens I., Van Gaal L., Van Hul W. (2008). Association of the BDNF Val66Met variation with obesity in women. Mol. Genet. Metab. 95 110–112. 10.1016/j.ymgme.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Benito E., Barco A. (2010). CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 33 230–240. 10.1016/j.tins.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Benito E., Valor L. M., Jimenez-Minchan M., Huber W., Barco A. (2011). cAMP response element-binding protein is a primary hub of activity-driven neuronal gene expression. J. Neurosci. 31 18237–18250. 10.1523/JNEUROSCI.4554-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. G., Stich C., Jager K., Dobrowolny H., Wick M., Steiner J., et al. (2012). Agmatinase, an inactivator of the putative endogenous antidepressant agmatine, is strongly upregulated in hippocampal interneurons of subjects with mood disorders. Neuropharmacology 62 237–246. 10.1016/j.neuropharm.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Bittinger M. A., Mcwhinnie E., Meltzer J., Iourgenko V., Latario B., Liu X., et al. (2004). Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 14 2156–2161. 10.1016/j.cub.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Bjorkholm C., Monteggia L. M. (2016). BDNF - a key transducer of antidepressant effects. Neuropharmacology 102 72–79. 10.1016/j.neuropharm.2015.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy J. A. (2006). The role of CREB in depression and antidepressant treatment. Biol. Psychiatry 59 1144–1150. 10.1016/j.biopsych.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Boku S., Nakagawa S., Toda H., Hishimoto A. (2018). Neural basis of major depressive disorder: beyond monoamine hypothesis. Psychiatry Clin. Neurosci. 72 3–12. 10.1111/pcn.12604 [DOI] [PubMed] [Google Scholar]
- Booij L. (2019). Genetic and epigenetic regulation of CRTC1 in human eating behaviour and fat distribution: Methodological and clinical insights and considerations. EBioMedicine 45 15–16. 10.1016/j.ebiom.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting G. L., Durresi E., Ataman B., Sherman M. A., Mei K., Harmin D. A., et al. (2021). Activity-dependent regulome of human GABAergic neurons reveals new patterns of gene regulation and neurological disease heritability. Nat. Neurosci. 24 437–448. 10.1038/s41593-020-00786-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuillaud L., Halfon O., Magistretti P. J., Pralong F. P., Cardinaux J. R. (2009). Mouse fertility is not dependent on the CREB coactivator Crtc1. Nat. Med. 15 989–990; authorrely991. 10.1038/nm0909-989 [DOI] [PubMed] [Google Scholar]
- Breuillaud L., Rossetti C., Meylan E. M., Merinat C., Halfon O., Magistretti P. J., et al. (2012). Deletion of CREB-regulated transcription coactivator 1 induces pathological aggression, depression-related behaviors, and neuroplasticity genes dysregulation in mice. Biol. Psychiatry 72 528–536. 10.1016/j.biopsych.2012.04.011 [DOI] [PubMed] [Google Scholar]
- Briand L. A., Lee B. G., Lelay J., Kaestner K. H., Blendy J. A. (2015). Serine 133 phosphorylation is not required for hippocampal CREB-mediated transcription and behavior. Learn. Memory 22 109–115. 10.1101/lm.037044.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle P., Nakajima T., Montminy M. (1995). Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc. Natl. Acad. Sci. U.S.A. 92 10521–10525. 10.1073/pnas.92.23.10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K., Morantte I., Weir H. J., Yeo R., Zhang Y., Huynh F. K., et al. (2015). Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 160 842–855. 10.1016/j.cell.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo A., Rodrigues A. L. S. (2019). Novel targets for fast antidepressant responses: possible role of endogenous neuromodulators. Chronic Stress (Thousand Oaks) 3:2470547019858083. 10.1177/2470547019858083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraci F., Calabrese F., Molteni R., Bartova L., Dold M., Leggio G. M., et al. (2018). International union of basic and clinical pharmacology CIV: the neurobiology of treatment-resistant depression: from antidepressant classifications to novel pharmacological targets. Pharmacol. Rev. 70 475–504. 10.1124/pr.117.014977 [DOI] [PubMed] [Google Scholar]
- Carlezon W. A., Jr., Duman R. S., Nestler E. J. (2005). The many faces of CREB. Trends Neurosci. 28 436–445. 10.1016/j.tins.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Castren E. (2005). Is mood chemistry? Nat. Rev. Neurosci. 6 241–246. 10.1038/nrn1629 [DOI] [PubMed] [Google Scholar]
- Castren E., Hen R. (2013). Neuronal plasticity and antidepressant actions. Trends Neurosci. 36 259–267. 10.1016/j.tins.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren E., Monteggia L. M. (2021). Brain-derived neurotrophic factor signaling in depression and antidepressant action. Biol. Psychiatry 90 128–136. 10.1016/j.biopsych.2021.05.008 [DOI] [PubMed] [Google Scholar]
- Catalano S., Giordano C., Rizza P., Gu G., Barone I., Bonofiglio D., et al. (2009). Evidence that leptin through STAT and CREB signaling enhances cyclin D1 expression and promotes human endometrial cancer proliferation. J. Cell Physiol. 218 490–500. 10.1002/jcp.21622 [DOI] [PubMed] [Google Scholar]
- Ch’ng T. H., Desalvo M., Lin P., Vashisht A., Wohlschlegel J. A., Martin K. C. (2015). Cell biological mechanisms of activity-dependent synapse to nucleus translocation of CRTC1 in neurons. Front. Mol. Neurosci. 8:48. 10.3389/fnmol.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng T. H., Uzgil B., Lin P., Avliyakulov N. K., O’dell T. J., Martin K. C. (2012). Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell 150 207–221. 10.1016/j.cell.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E. (2019). The circadian regulation of food intake. Nat. Rev. Endocrinol. 15 393–405. 10.1038/s41574-019-0210-x [DOI] [PubMed] [Google Scholar]
- Chen G. G., Almeida D., Fiori L., Turecki G. (2018). Evidence of reduced agmatine concentrations in the cerebral cortex of suicides. Int. J. Neuropsychopharmacol. 21 895–900. 10.1093/ijnp/pyy058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherix A., Poitry-Yamate C., Lanz B., Zanoletti O., Grosse J., Sandi C., et al. (2020). Deletion of Crtc1 leads to hippocampal neuroenergetic impairments associated with depressive-like behavior. bioRxiv [Preprint]. 10.1101/2020.11.05.370221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Kim W., Chung J. (2011). Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J. Biol. Chem. 286 2658–2664. 10.1074/jbc.C110.119222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong E., Quteineh L., Cardinaux J. R., Gholam-Rezaee M., Vandenberghe F., Dobrinas M., et al. (2013). Influence of CRTC1 polymorphisms on body mass index and fat mass in psychiatric patients and the general adult population. JAMA Psychiatry 70 1011–1019. 10.1001/jamapsychiatry.2013.187 [DOI] [PubMed] [Google Scholar]
- Conkright M. D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J. B., et al. (2003). TORCs: transducers of regulated CREB activity. Mol. Cell 12 413–423. 10.1016/j.molcel.2003.08.013 [DOI] [PubMed] [Google Scholar]
- Cordeira J. W., Frank L., Sena-Esteves M., Pothos E. N., Rios M. (2010). Brain-derived neurotrophic factor regulates hedonic feeding by acting on the mesolimbic dopamine system. J. Neurosci. 30 2533–2541. 10.1523/JNEUROSCI.5768-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M. A., Smart J. L., Rubinstein M., Cerdan M. G., Diano S., Horvath T. L., et al. (2001). Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411 480–484. 10.1038/35078085 [DOI] [PubMed] [Google Scholar]
- Dantzer R., O’connor J. C., Freund G. G., Johnson R. W., Kelley K. W. (2008). From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9 46–56. 10.1038/nrn2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L. (2019). Understanding depression: the hippocampus might hold the answer in a CREB-regulated transcription coactivator: an editorial for ‘AAV-mediated over-expression of CRTC1 in the hippocampal dentate gyrus ameliorates lipopolysaccharide-induced depression-like behavior in mice’ on page 111. J. Neurochem. 149 9–11. 10.1111/jnc.14659 [DOI] [PubMed] [Google Scholar]
- Delacretaz A., Glatard A., Dubath C., Gholam-Rezaee M., Sanchez-Mut J. V., Graff J., et al. (2019). Psychotropic drug-induced genetic-epigenetic modulation of CRTC1 gene is associated with early weight gain in a prospective study of psychiatric patients. Clin. Epigenetics 11:198. 10.1186/s13148-019-0792-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M. O., Horvath T. L. (2013). Hypothalamic control of energy balance: insights into the role of synaptic plasticity. Trends Neurosci. 36 65–73. 10.1016/j.tins.2012.12.005 [DOI] [PubMed] [Google Scholar]
- Duan J., Choi Y. H., Hartzell D., Della-Fera M. A., Hamrick M., Baile C. A. (2007). Effects of subcutaneous leptin injections on hypothalamic gene profiles in lean and ob/ob mice. Obesity (Silver Spring) 15 2624–2633. 10.1038/oby.2007.314 [DOI] [PubMed] [Google Scholar]
- Duman R. S., Aghajanian G. K., Sanacora G., Krystal J. H. (2016). Synaptic plasticity and depression: new insights from stress and rapid-acting antidepressants. Nat. Med. 22 238–249. 10.1038/nm.4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R. S., Sanacora G., Krystal J. H. (2019a). Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron 102 75–90. 10.1016/j.neuron.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R. S., Shinohara R., Fogaca M. V., Hare B. (2019b). Neurobiology of rapid-acting antidepressants: convergent effects on GluA1-synaptic function. Mol. Psychiatry 24 1816–1832. 10.1038/s41380-019-0400-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas C. C., Silva-Garcia C. G., Mair W. B. (2017). Deregulation of CRTCs in aging and age-related disease risk. Trends Genet. 33 303–321. 10.1016/j.tig.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espana J., Valero J., Minano-Molina A. J., Masgrau R., Martin E., Guardia-Laguarta C., et al. (2010). beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 30 9402–9410. 10.1523/JNEUROSCI.2154-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann K. G., Chowdhury A., Becker J. L., Mcalpin N., Ahmed T., Haider S., et al. (2019). Non-canonical activation of CREB mediates neuroprotection in a Caenorhabditis elegans model of excitotoxic necrosis. J. Neurochem. 148 531–549. 10.1111/jnc.14629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterwald C., Fiumelli H., Cardinaux J. R., Martin J. L. (2010). Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. J. Biol. Chem. 285 28587–28595. 10.1074/jbc.M110.125740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas A. E., Neis V. B., Rodrigues A. L. S. (2016). Agmatine, a potential novel therapeutic strategy for depression. Eur. Neuropsychopharmacol. 26 1885–1899. 10.1016/j.euroneuro.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Fukuchi M., Tabuchi A., Kuwana Y., Watanabe S., Inoue M., Takasaki I., et al. (2015). Neuromodulatory effect of Galphas- or Galphaq-coupled G-protein-coupled receptor on NMDA receptor selectively activates the NMDA receptor/Ca2+/calcineurin/cAMP response element-binding protein-regulated transcriptional coactivator 1 pathway to effectively induce brain-derived neurotrophic factor expression in neurons. J. Neurosci. 35 5606–5624. 10.1523/JNEUROSCI.3650-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W. W., Tang H. V., Cheng Y., Chan C. P., Chan C. P., Jin D. Y. (2018). Suppression of gluconeogenic gene transcription by SIK1-induced ubiquitination and degradation of CRTC1. Biochim. Biophys. Acta 1861 211–223. 10.1016/j.bbagrm.2018.01.021 [DOI] [PubMed] [Google Scholar]
- Ginty D. D., Kornhauser J. M., Thompson M. A., Bading H., Mayo K. E., Takahashi J. S., et al. (1993). Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260 238–241. 10.1126/science.8097062 [DOI] [PubMed] [Google Scholar]
- Gold P. W. (2015). The organization of the stress system and its dysregulation in depressive illness. Mol. Psychiatry 20 32–47. 10.1038/mp.2014.163 [DOI] [PubMed] [Google Scholar]
- Gray J., Yeo G. S., Cox J. J., Morton J., Adlam A. L., Keogh J. M., et al. (2006). Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes 55 3366–3371. 10.2337/db06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaris A., Piletz J. (2007). Agmatine : metabolic pathway and spectrum of activity in brain. CNS Drugs 21 885–900. 10.2165/00023210-200721110-00002 [DOI] [PubMed] [Google Scholar]
- Hamer J. A., Testani D., Mansur R. B., Lee Y., Subramaniapillai M., Mcintyre R. S. (2019). Brain insulin resistance: a treatment target for cognitive impairment and anhedonia in depression. Exp. Neurol. 315 1–8. 10.1016/j.expneurol.2019.01.016 [DOI] [PubMed] [Google Scholar]
- Harper D. G., Jensen J. E., Ravichandran C., Perlis R. H., Fava M., Renshaw P. F., et al. (2017). Tissue type-specific bioenergetic abnormalities in adults with major depression. Neuropsychopharmacology 42 876–885. 10.1038/npp.2016.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lv J., Fang Y., Luo Q., He Y., Li L., et al. (2021). Crtc1 deficiency causes obesity potentially via regulating PPARgamma pathway in white adipose. Front. Cell Dev. Biol. 9:602529. 10.3389/fcell.2021.602529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Sugiyama T., Wallace C. S., Gompf H. S., Yoshioka T., Miyawaki A., et al. (2003). Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38 253–263. 10.1016/s0896-6273(03)00164-8 [DOI] [PubMed] [Google Scholar]
- Impey S., Mark M., Villacres E. C., Poser S., Chavkin C., Storm D. R. (1996). Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron 16 973–982. 10.1016/s0896-6273(00)80120-8 [DOI] [PubMed] [Google Scholar]
- Iourgenko V., Zhang W., Mickanin C., Daly I., Jiang C., Hexham J. M., et al. (2003). Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 100 12147–12152. 10.1073/pnas.1932773100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath A., Butler R., Godinho S. I., Couch Y., Brown L. A., Vasudevan S. R., et al. (2013). The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell 154 1100–1111. 10.1016/j.cell.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath A., Varga N., Dallmann R., Rando G., Gosselin P., Ebrahimjee F., et al. (2021). Adenosine integrates light and sleep signalling for the regulation of circadian timing in mice. Nat. Commun. 12:2113. 10.1038/s41467-021-22179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B., Wang H., Wang J. L., Wang Y. J., Zhu Q., Wang C. N., et al. (2019). Hippocampal salt-inducible kinase 2 plays a role in depression via the CREB-regulated transcription coactivator 1-cAMP response element binding-brain-derived neurotrophic factor pathway. Biol. Psychiatry 85 650–666. 10.1016/j.biopsych.2018.10.004 [DOI] [PubMed] [Google Scholar]
- Jiang C., Lin W. J., Salton S. R. (2019). Role of a VGF/BDNF/TrkB autoregulatory feedback loop in rapid-acting antidepressant efficacy. J. Mol. Neurosci. 68 504–509. 10.1007/s12031-018-1124-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn S. A., Nguyen P. V. (2005). CREB, synapses and memory disorders: past progress and future challenges. Curr. Drug Targets CNS Neurol. Disord. 4 481–497. 10.2174/156800705774322058 [DOI] [PubMed] [Google Scholar]
- Kan C., Silva N., Golden S. H., Rajala U., Timonen M., Stahl D., et al. (2013). A systematic review and meta-analysis of the association between depression and insulin resistance. Diabetes Care 36 480–489. 10.2337/dc12-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. E., Schinagle M., Gao K., Conroy C., Ganocy S. J., Ismail-Beigi F., et al. (2014). PPAR-gamma agonism as a modulator of mood: proof-of-concept for pioglitazone in bipolar depression. CNS Drugs 28 571–581. 10.1007/s40263-014-0158-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernie S. G., Liebl D. J., Parada L. F. (2000). BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 19 1290–1300. 10.1093/emboj/19.6.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. H., Szabo A., King E. M., Ayala J., Ayala J. E., Altarejos J. Y. (2015). Leptin recruits Creb-regulated transcriptional coactivator 1 to improve hyperglycemia in insulin-deficient diabetes. Mol. Metab. 4 227–236. 10.1016/j.molmet.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. (2016). The transcription cofactor CRTC1 protects from aberrant hepatic lipid accumulation. Sci. Rep. 6:37280. 10.1038/srep37280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Lee H., Hur J.-H., Choe J., Lim C. (2016). CRTC potentiates light-independent timeless transcription to sustain circadian rhythms in Drosophila. Sci. Rep. 6:32113. 10.1038/srep32113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Lee B., Kim D. H., Kim J., Lee S., Lee S. K., et al. (2013). Control of energy balance by hypothalamic gene circuitry involving two nuclear receptors, neuron-derived orphan receptor 1 and glucocorticoid receptor. Mol. Cell Biol. 33 3826–3834. 10.1128/MCB.00385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinedinst N. J., Regenold W. T. (2015). A mitochondrial bioenergetic basis of depression. J. Bioenerg. Biomembr. 47 155–171. 10.1007/s10863-014-9584-6 [DOI] [PubMed] [Google Scholar]
- Kovacs K. A., Steullet P., Steinmann M., Do K. Q., Magistretti P. J., Halfon O., et al. (2007). TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 104 4700–4705. 10.1073/pnas.0607524104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus C., Kadriu B., Lanzenberger R., Zarate C. A., Jr., Kasper S. (2019). Prognosis and improved outcomes in major depression: a review. Transl. Psychiatry 9:127. 10.1038/s41398-019-0460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal J. H., Abdallah C. G., Sanacora G., Charney D. S., Duman R. S. (2019). Ketamine: a paradigm shift for depression research and treatment. Neuron 101 774–778. 10.1016/j.neuron.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube G., Bernstein H. G. (2017). Agmatine: multifunctional arginine metabolite and magic bullet in clinical neuroscience? Biochem. J. 474 2619–2640. 10.1042/BCJ20170007 [DOI] [PubMed] [Google Scholar]
- Lee B., Li A., Hansen K. F., Cao R., Yoon J. H., Obrietan K. (2010). CREB influences timing and entrainment of the SCN circadian clock. J. Biol. Rhythms 25 410–420. 10.1177/0748730410381229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M. J. F., Boulle F., Steinbusch H. W., Van Den Hove D. L. A., Kenis G., Lanfumey L. (2018). Neurotrophic factors and neuroplasticity pathways in the pathophysiology and treatment of depression. Psychopharmacology (Berl) 235 2195–2220. 10.1007/s00213-018-4950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Regunathan S., Barrow C. J., Eshraghi J., Cooper R., Reis D. J. (1994). Agmatine: an endogenous clonidine-displacing substance in the brain. Science 263 966–969. 10.1126/science.7906055 [DOI] [PubMed] [Google Scholar]
- Li L., Li X., Zhou W., Messina J. L. (2013). Acute psychological stress results in the rapid development of insulin resistance. J. Endocrinol. 217 175–184. 10.1530/JOE-12-0559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang C., Takemori H., Zhou Y., Xiong Z. Q. (2009). TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J. Neurosci. 29 2334–2343. 10.1523/JNEUROSCI.2296-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. W., Wroolie T. E., Robakis T., Rasgon N. L. (2015). Adjuvant pioglitazone for unremitted depression: clinical correlates of treatment response. Psychiatry Res. 230 846–852. 10.1016/j.psychres.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Hales C. M., Garber K., Jin P. (2014). Fat mass and obesity-associated (FTO) protein interacts with CaMKII and modulates the activity of CREB signaling pathway. Hum. Mol. Genet. 23 3299–3306. 10.1093/hmg/ddu043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tang W., Ji C., Gu J., Chen Y., Huang J., et al. (2020). The Selective SIK2 inhibitor ARN-3236 produces strong antidepressant-like efficacy in mice via the hippocampal CRTC1-CREB-BDNF pathway. Front. Pharmacol. 11:624429. 10.3389/fphar.2020.624429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke A. E., Kahali B., Berndt S. I., Justice A. E., Pers T. H., Day F. R., et al. (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature 518 197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan R. W., McClung C. A. (2019). Rhythms of life: circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 20 49–65. 10.1038/s41583-018-0088-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze B. E., Ginty D. D. (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron 35 605–623. 10.1016/s0896-6273(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Lu Y., Day F. R., Gustafsson S., Buchkovich M. L., Na J., Bataille V., et al. (2016). New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 7:10495. 10.1038/ncomms10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra E. S. N. M., Lam M. P., Soares C. N., Munoz D. P., Milev R., De Felice F. G. (2019). Insulin resistance as a shared pathogenic mechanism between depression and type 2 diabetes. Front. Psychiatry 10:57. 10.3389/fpsyt.2019.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Zang T., Birnbaum S. G., Wang Z., Johnson J. E., Zhang C. L., et al. (2017). TrkB dependent adult hippocampal progenitor differentiation mediates sustained ketamine antidepressant response. Nat. Commun. 8:1668. 10.1038/s41467-017-01709-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazziti D., Rutigliano G., Baroni S., Landi P., Dell’osso L. (2014). Metabolic syndrome and major depression. CNS Spectr. 19 293–304. 10.1017/S1092852913000667 [DOI] [PubMed] [Google Scholar]
- Marosi K., Mattson M. P. (2014). BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 25 89–98. 10.1016/j.tem.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden W. N. (2013). Synaptic plasticity in depression: molecular, cellular and functional correlates. Prog. Neuropsychopharmacol. Biol. Psychiatry 43 168–184. 10.1016/j.pnpbp.2012.12.012 [DOI] [PubMed] [Google Scholar]
- Matsumura S., Ishikawa F., Sasaki T., Terkelsen M. K., Ravnskjaer K., Jinno T., et al. (2021). Loss of CREB coactivator CRTC1 in SF1 cells leads to hyperphagia and obesity by high-fat diet but not normal chow diet. Endocrinology 162:bqab076. 10.1210/endocr/bqab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B., Montminy M. (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2 599–609. 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- McAllan L., Maynard K. R., Kardian A. S., Stayton A. S., Fox S. L., Stephenson E. J., et al. (2018). Disruption of brain-derived neurotrophic factor production from individual promoters generates distinct body composition phenotypes in mice. Am. J. Physiol. Endocrinol. Metab. 315 E1168–E1184. 10.1152/ajpendo.00205.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E. M., Breuillaud L., Seredenina T., Magistretti P. J., Halfon O., Luthi-Carter R., et al. (2016a). Involvement of the agmatinergic system in the depressive-like phenotype of the Crtc1 knockout mouse model of depression. Transl. Psychiatry 6:e852. 10.1038/tp.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E. M., Halfon O., Magistretti P. J., Cardinaux J. R. (2016b). The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: possible relevance for treatment-resistant depression. Neuropharmacology 107 111–121. 10.1016/j.neuropharm.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi Y., Lamers F., Peyrot W. J., Baune B. T., Breen G., Dehghan A., et al. (2017). Genetic association of major depression with atypical features and obesity-related immunometabolic dysregulations. JAMA Psychiatry 74 1214–1225. 10.1001/jamapsychiatry.2017.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi Y., Simmons W. K., Van Rossum E. F. C., Penninx B. W. (2019). Depression and obesity: evidence of shared biological mechanisms. Mol. Psychiatry 24 18–33. 10.1038/s41380-018-0017-5 [DOI] [PubMed] [Google Scholar]
- Minokoshi Y., Alquier T., Furukawa N., Kim Y. B., Lee A., Xue B., et al. (2004). AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428 569–574. 10.1038/nature02440 [DOI] [PubMed] [Google Scholar]
- Misztak P., Panczyszyn-Trzewik P., Sowa-Kucma M. (2018). Histone deacetylases (HDACs) as therapeutic target for depressive disorders. Pharmacol. Rep. 70 398–408. 10.1016/j.pharep.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Moda-Sava R. N., Murdock M. H., Parekh P. K., Fetcho R. N., Huang B. S., Huynh T. N., et al. (2019). Sustained rescue of prefrontal circuit dysfunction by antidepressant-induced spine formation. Science 364:eaat8078. 10.1126/science.aat8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. M., Christensen J. D., Lafer B., Fava M., Renshaw P. F. (1997). Lower levels of nucleoside triphosphate in the basal ganglia of depressed subjects: a phosphorous-31 magnetic resonance spectroscopy study. Am. J. Psychiatry 154 116–118. 10.1176/ajp.154.1.116 [DOI] [PubMed] [Google Scholar]
- Morhenn K., Quentin T., Wichmann H., Steinmetz M., Prondzynski M., Sohren K. D., et al. (2019). Mechanistic role of the CREB-regulated transcription coactivator 1 in cardiac hypertrophy. J. Mol. Cell Cardiol. 127 31–43. 10.1016/j.yjmcc.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Myers M. G., Jr., Munzberg H., Leinninger G. M., Leshan R. L. (2009). The geometry of leptin action in the brain: more complicated than a simple ARC. Cell Metab. 9 117–123. 10.1016/j.cmet.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neis V. B., Bettio L. B., Moretti M., Rosa P. B., Olescowicz G., Fraga D. B., et al. (2018). Single administration of agmatine reverses the depressive-like behavior induced by corticosterone in mice: comparison with ketamine and fluoxetine. Pharmacol. Biochem. Behav. 173 44–50. 10.1016/j.pbb.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Neis V. B., Bettio L. E. B., Moretti M., Rosa P. B., Ribeiro C. M., Freitas A. E., et al. (2016a). Acute agmatine administration, similar to ketamine, reverses depressive-like behavior induced by chronic unpredictable stress in mice. Pharmacol. Biochem. Behav. 150 108–114. 10.1016/j.pbb.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Neis V. B., Moretti M., Bettio L. E., Ribeiro C. M., Rosa P. B., Goncalves F. M., et al. (2016b). Agmatine produces antidepressant-like effects by activating AMPA receptors and mTOR signaling. Eur. Neuropsychopharmacol. 26 959–971. 10.1016/j.euroneuro.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Neis V. B., Rosa P. B., Olescowicz G., Rodrigues A. L. S. (2017). Therapeutic potential of agmatine for CNS disorders. Neurochem. Int. 108 318–331. 10.1016/j.neuint.2017.05.006 [DOI] [PubMed] [Google Scholar]
- Ni S., Huang H., He D., Chen H., Wang C., Zhao X., et al. (2019). Adeno-associated virus-mediated over-expression of CREB-regulated transcription coactivator 1 in the hippocampal dentate gyrus ameliorates lipopolysaccharide-induced depression-like behaviour in mice. J. Neurochem. 149 111–125. 10.1111/jnc.14670 [DOI] [PubMed] [Google Scholar]
- Nonaka M., Kim R., Fukushima H., Sasaki K., Suzuki K., Okamura M., et al. (2014). Region-specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron 84 92–106. 10.1016/j.neuron.2014.08.049 [DOI] [PubMed] [Google Scholar]
- Obrietan K., Impey S., Smith D., Athos J., Storm D. R. (1999). Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J. Biol. Chem. 274 17748–17756. 10.1074/jbc.274.25.17748 [DOI] [PubMed] [Google Scholar]
- Ostergaard L., Jorgensen M. B., Knudsen G. M. (2018). Low on energy? An energy supply-demand perspective on stress and depression. Neurosci. Biobehav. Rev. 94 248–270. 10.1016/j.neubiorev.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Pan W. W., Myers M. G., Jr. (2018). Leptin and the maintenance of elevated body weight. Nat. Rev. Neurosci. 19 95–105. 10.1038/nrn.2017.168 [DOI] [PubMed] [Google Scholar]
- Pan Y., He X., Li C., Li Y., Li W., Zhang H., et al. (2021). Neuronal activity recruits the CRTC1/CREB axis to drive transcription-dependent autophagy for maintaining late-phase LTD. Cell Rep. 36:109398. 10.1016/j.celrep.2021.109398 [DOI] [PubMed] [Google Scholar]
- Parra-Damas A., Chen M., Enriquez-Barreto L., Ortega L., Acosta S., Perna J. C., et al. (2017a). CRTC1 function during memory encoding is disrupted in neurodegeneration. Biol. Psychiatry 81 111–123. 10.1016/j.biopsych.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Parra-Damas A., Rubio-Ferrarons L., Shen J., Saura C. A. (2017b). CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters. Sci. Rep. 7:18004. 10.1038/s41598-017-18215-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Damas A., Saura C. A. (2019). Synapse-to-nucleus signaling in neurodegenerative and neuropsychiatric disorders. Biol. Psychiatry 86 87–96. 10.1016/j.biopsych.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Parra-Damas A., Valero J., Chen M., Espana J., Martin E., Ferrer I., et al. (2014). Crtc1 activates a transcriptional program deregulated at early Alzheimer’s disease-related stages. J. Neurosci. 34 5776–5787. 10.1523/JNEUROSCI.5288-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., Mcewen B. S., Epel E. S., Sandi C. (2018). An energetic view of stress: focus on mitochondria. Front. Neuroendocrinol. 49:72–85. 10.1016/j.yfrne.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piletz J. E., Aricioglu F., Cheng J. T., Fairbanks C. A., Gilad V. H., Haenisch B., et al. (2013). Agmatine: clinical applications after 100 years in translation. Drug Discov. Today 18 880–893. 10.1016/j.drudis.2013.05.017 [DOI] [PubMed] [Google Scholar]
- Pittenger C., Duman R. S. (2008). Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology 33 88–109. 10.1038/sj.npp.1301574 [DOI] [PubMed] [Google Scholar]
- Prosser R. A., Gillette M. U. (1991). Cyclic changes in cAMP concentration and phosphodiesterase activity in a mammalian circadian clock studied in vitro. Brain Res. 568 185–192. 10.1016/0006-8993(91)91396-i [DOI] [PubMed] [Google Scholar]
- Quteineh L., Preisig M., Rivera M., Milaneschi Y., Castelao E., Gholam-Rezaee M., et al. (2016). Association of CRTC1 polymorphisms with obesity markers in subjects from the general population with lifetime depression. J. Affect. Disord. 198 43–49. 10.1016/j.jad.2016.03.031 [DOI] [PubMed] [Google Scholar]
- Ragguett R. M., Tamura J. K., Mcintyre R. S. (2019). Keeping up with the clinical advances: depression. CNS Spectr. 24 25–37. 10.1017/S1092852919001159 [DOI] [PubMed] [Google Scholar]
- Rasgon N. L., McEwen B. S. (2016). Insulin resistance-a missing link no more. Mol. Psychiatry 21 1648–1652. 10.1038/mp.2016.162 [DOI] [PubMed] [Google Scholar]
- Reis D. J., Regunathan S. (2000). Is agmatine a novel neurotransmitter in brain? Trends Pharmacol. Sci. 21 187–193. 10.1016/s0165-6147(00)01460-7 [DOI] [PubMed] [Google Scholar]
- Rios M. (2013). BDNF and the central control of feeding: accidental bystander or essential player? Trends Neurosci. 36 83–90. 10.1016/j.tins.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M., Fan G., Fekete C., Kelly J., Bates B., Kuehn R., et al. (2001). Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol. Endocrinol. 15 1748–1757. 10.1210/mend.15.10.0706 [DOI] [PubMed] [Google Scholar]
- Rohde K., Keller M., La Cour Poulsen L., Ronningen T., Stumvoll M., Tonjes A., et al. (2019). (Epi)genetic regulation of CRTC1 in human eating behaviour and fat distribution. EBioMedicine 44 476–488. 10.1016/j.ebiom.2019.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti C., Sciarra D., Petit J. M., Eap C. B., Halfon O., Magistretti P. J., et al. (2017). Gender-specific alteration of energy balance and circadian locomotor activity in the Crtc1 knockout mouse model of depression. Transl. Psychiatry 7:1269. 10.1038/s41398-017-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic R. D., Mcnamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., et al. (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2:e377. 10.1371/journal.pbio.0020377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Norona F. E., Alzate-Correa D., Scarberry D., Hoyt K. R., Obrietan K. (2013). Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J. Neurosci. 33 9021–9027. 10.1523/JNEUROSCI.4202-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura C. A., Cardinaux J. R. (2017). Emerging roles of CREB-regulated transcription coactivators in brain physiology and pathology. Trends Neurosci. 40 720–733. 10.1016/j.tins.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Schumacher Y., Aparicio T., Ourabah S., Baraille F., Martin A., Wind P., et al. (2016). Dysregulated CRTC1 activity is a novel component of PGE2 signaling that contributes to colon cancer growth. Oncogene 35 2602–2614. 10.1038/onc.2015.283 [DOI] [PubMed] [Google Scholar]
- Schwartz M. W., Woods S. C., Porte D., Jr., Seeley R. J., Baskin D. G. (2000). Central nervous system control of food intake. Nature 404 661–671. 10.1038/35007534 [DOI] [PubMed] [Google Scholar]
- Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., et al. (2004). The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119 61–74. 10.1016/j.cell.2004.09.015 [DOI] [PubMed] [Google Scholar]
- Sekeres M. J., Mercaldo V., Richards B., Sargin D., Mahadevan V., Woodin M. A., et al. (2012). Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J. Neurosci. 32 17857–17868. 10.1523/JNEUROSCI.1419-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Liu P., Leitch B. (2011). Spatial learning-induced accumulation of agmatine and glutamate at hippocampal CA1 synaptic terminals. Neuroscience 192 28–36. 10.1016/j.neuroscience.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Sepanjnia K., Modabbernia A., Ashrafi M., Modabbernia M. J., Akhondzadeh S. (2012). Pioglitazone adjunctive therapy for moderate-to-severe major depressive disorder: randomized double-blind placebo-controlled trial. Neuropsychopharmacology 37 2093–2100. 10.1038/npp.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Wang B., Giribaldi M. G., Ayres J., Thomas J. B., Montminy M. (2016). Neuronal energy-sensing pathway promotes energy balance by modulating disease tolerance. Proc. Natl. Acad. Sci. U.S.A. 113 E3307–E3314. 10.1073/pnas.1606106113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara R., Aghajanian G. K., Abdallah C. G. (2021). Neurobiology of the rapid-acting antidepressant effects of ketamine: impact and opportunities. Biol. Psychiatry 90 85–95. 10.1016/j.biopsych.2020.12.006 [DOI] [PubMed] [Google Scholar]
- Shu H., Wang M., Song M., Sun Y., Shen X., Zhang J., et al. (2020). Acute nicotine treatment alleviates LPS-induced impairment of fear memory reconsolidation through AMPK activation and CRTC1 upregulation in hippocampus. Int. J. Neuropsychopharmacol. 23 687–699. 10.1093/ijnp/pyaa043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., Xue X., Liu S., Feng C., Zhang H., Zhang S., et al. (2021). CRTC1 signaling involvement in depression-like behavior of prenatally stressed offspring rat. Behav. Brain Res. 399:113000. 10.1016/j.bbr.2020.113000 [DOI] [PubMed] [Google Scholar]
- Skledar M., Nikolac M., Dodig-Curkovic K., Curkovic M., Borovecki F., Pivac N. (2012). Association between brain-derived neurotrophic factor Val66Met and obesity in children and adolescents. Prog. Neuropsychopharmacol. Biol. Psychiatry 36 136–140. 10.1016/j.pnpbp.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Song M., Martinowich K., Lee F. S. (2017). BDNF at the synapse: why location matters. Mol. Psychiatry 22 1370–1375. 10.1038/mp.2017.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Altarejos J., Goodarzi M. O., Inoue H., Guo X., Berdeaux R., et al. (2010). CRTC3 links catecholamine signalling to energy balance. Nature 468 933–939. 10.1038/nature09564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag T., Moresco J. J., Vaughan J. M., Matsumura S., Yates J. R., III, Montminy M. (2017). Analysis of a cAMP regulated coactivator family reveals an alternative phosphorylation motif for AMPK family members. PLoS One 12:e0173013. 10.1371/journal.pone.0173013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriya M., Maekawa F., Maejima Y., Onaka T., Fujiwara K., Nakagawa T., et al. (2010). Long-term infusion of brain-derived neurotrophic factor reduces food intake and body weight via a corticotrophin-releasing hormone pathway in the paraventricular nucleus of the hypothalamus. J. Neuroendocrinol. 22 987–995. 10.1111/j.1365-2826.2010.02039.x [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. U.S.A. 99 7728–7733. 10.1073/pnas.102075599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., Mcdearmon E. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308 1043–1045. 10.1126/science.1108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., Shumyatsky G. P. (2018). Synaptically localized transcriptional regulators in memory formation. Neuroscience 370 4–13. 10.1016/j.neuroscience.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., Teubner B. J., Hevi C., Hara K., Kobayashi A., Dave R. M., et al. (2017). CRTC1 nuclear translocation following learning modulates memory strength via exchange of chromatin remodeling complexes on the Fgf1 gene. Cell Rep. 18 352–366. 10.1016/j.celrep.2016.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., Yamagata H., Seki T., Watanabe Y. (2018). Epigenetic mechanisms of major depression: targeting neuronal plasticity. Psychiatry Clin. Neurosci. 72 212–227. 10.1111/pcn.12621 [DOI] [PubMed] [Google Scholar]
- Umemori J., Winkel F., Didio G., Llach Pou M., Castren E. (2018). iPlasticity: induced juvenile-like plasticity in the adult brain as a mechanism of antidepressants. Psychiatry Clin. Neurosci. 72 633–653. 10.1111/pcn.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T. J., Calderon G. A., Bradley L. C., Sena-Esteves M., Rios M. (2007). Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. J. Neurosci. 27 14265–14274. 10.1523/JNEUROSCI.3308-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbay T. I. (2012). The pharmacological importance of agmatine in the brain. Neurosci. Biobehav. Rev. 36 502–519. 10.1016/j.neubiorev.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Valverde A. P., Camargo A., Rodrigues A. L. S. (2021). Agmatine as a novel candidate for rapid-onset antidepressant response. World J. Psychiatry 11 981–996. 10.5498/wjp.v11.i11.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooij M. A., Jene T., Treccani G., Miederer I., Hasch A., Voelxen N., et al. (2018). Chronic social stress-induced hyperglycemia in mice couples individual stress susceptibility to impaired spatial memory. Proc. Natl. Acad. Sci. U.S.A. 115 E10187–E10196. 10.1073/pnas.1804412115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V., Feng J., Robison A. J., Nestler E. J. (2013). Epigenetic mechanisms of depression and antidepressant action. Annu. Rev. Pharmacol. Toxicol. 53 59–87. 10.1146/annurev-pharmtox-010611-134540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videbech P. (2000). PET measurements of brain glucose metabolism and blood flow in major depressive disorder: a critical review. Acta Psychiatr. Scand. 101 11–20. 10.1034/j.1600-0447.2000.101001011.x [DOI] [PubMed] [Google Scholar]
- Wang C., Wang M., Ma J. (2021). Analysis of genome-wide DNA methylation patterns in obesity. Endocr. J. 68 1439–1453. 10.1507/endocrj.EJ20-0734 [DOI] [PubMed] [Google Scholar]
- Wang Y. J., Liu L., Wang Y., Wang J. L., Gao T. T., Wang H., et al. (2020). Imipramine exerts antidepressant-like effects in chronic stress models of depression by promoting CRTC1 expression in the mPFC. Brain Res. Bull. 164 257–268. 10.1016/j.brainresbull.2020.08.028 [DOI] [PubMed] [Google Scholar]
- Watson K., Nasca C., Aasly L., Mcewen B., Rasgon N. (2018). Insulin resistance, an unmasked culprit in depressive disorders: promises for interventions. Neuropharmacology 136 327–334. 10.1016/j.neuropharm.2017.11.038 [DOI] [PubMed] [Google Scholar]
- Watts A. G., Sanchez-Watts G., Liu Y., Aguilera G. (2011). The distribution of messenger RNAs encoding the three isoforms of the transducer of regulated cAMP responsive element binding protein activity in the rat forebrain. J. Neuroendocrinol. 23 754–766. 10.1111/j.1365-2826.2011.02178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D., Pfaffenseller B., Wollenhaupt-Aguiar B., Paul Gea L., Cardoso T. A., Kapczinski F. (2019). Agmatine as a potential therapeutic intervention in bipolar depression: the preclinical landscape. Expert. Opin. Ther. Targets 23 327–339. 10.1080/14728222.2019.1581764 [DOI] [PubMed] [Google Scholar]
- Weiss T., Bernard R., Bernstein H. G., Veh R. W., Laube G. (2018). Agmatine modulates spontaneous activity in neurons of the rat medial habenular complex-a relevant mechanism in the pathophysiology and treatment of depression? Transl. Psychiatry 8:201. 10.1038/s41398-018-0254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S. T., Sanacora G. (2019). A new generation of antidepressants: an update on the pharmaceutical pipeline for novel and rapid-acting therapeutics in mood disorders based on glutamate/GABA neurotransmitter systems. Drug Discov. Today 24 606–615. 10.1016/j.drudis.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. W., Elmquist J. K. (2012). From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat. Neurosci. 15 1350–1355. 10.1038/nn.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfer M., Kasties V., Kahlfuss S., Walter M. (2019). The role of depressive subtypes within the neuroinflammation hypothesis of major depressive disorder. Neuroscience 403 93–110. 10.1016/j.neuroscience.2018.03.034 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2017). Depression and Other Common Mental Disorders. WHO Reference Number: WHO/MSD/MER/2017.2. Geneva: World Health Organization. [Google Scholar]
- Wu Z., Huang X., Feng Y., Handschin C., Gullicksen P. S., Bare O., et al. (2006). Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. U.S.A. 103 14379–14384. 10.1073/pnas.0606714103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zhang C., Grigoroiu-Serbanescu M., Wang L., Li L., Zhou D., et al. (2018). The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol. Psychiatry 23 1957–1967. 10.1038/mp.2017.243 [DOI] [PubMed] [Google Scholar]
- Xie Y., Tang Q., Chen G., Xie M., Yu S., Zhao J., et al. (2019). New insights into the circadian rhythm and its related diseases. Front. Physiol. 10:682. 10.3389/fphys.2019.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Goulding E. H., Zang K., Cepoi D., Cone R. D., Jones K. R., et al. (2003). Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat. Neurosci. 6 736–742. 10.1038/nn1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Xie X. (2016). Neurotrophic factor control of satiety and body weight. Nat. Rev. Neurosci. 17 282–292. 10.1038/nrn.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Anderson D., Lurie-Beck J. (2011). The relationship between abdominal obesity and depression in the general population: a systematic review and meta-analysis. Obes. Res. Clin. Pract. 5 e267–e360. 10.1016/j.orcp.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Yan P., Xue Z., Li D., Ni S., Wang C., Jin X., et al. (2021). Dysregulated CRTC1-BDNF signaling pathway in the hippocampus contributes to Abeta oligomer-induced long-term synaptic plasticity and memory impairment. Exp. Neurol. 345:113812. 10.1016/j.expneurol.2021.113812 [DOI] [PubMed] [Google Scholar]
- Yang H., Li W., Meng P., Liu Z., Liu J., Wang Y. (2018). Chronic unpredictable mild stress aggravates mood disorder, cognitive impairment, and brain insulin resistance in diabetic rat. Evid. Based Compl. Alternat. Med. 2018:2901863. 10.1155/2018/2901863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. C., Reis D. J. (1999). Agmatine selectively blocks the N-methyl-D-aspartate subclass of glutamate receptor channels in rat hippocampal neurons. J. Pharmacol. Exp. Ther. 288 544–549. [PubMed] [Google Scholar]
- Yeo G. S., Connie Hung C. C., Rochford J., Keogh J., Gray J., Sivaramakrishnan S., et al. (2004). A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 7 1187–1189. 10.1038/nn1336 [DOI] [PubMed] [Google Scholar]
- Yeo G. S., Heisler L. K. (2012). Unraveling the brain regulation of appetite: lessons from genetics. Nat. Neurosci. 15 1343–1349. 10.1038/nn.3211 [DOI] [PubMed] [Google Scholar]
- Zanos P., Gould T. D. (2018). Mechanisms of ketamine action as an antidepressant. Mol. Psychiatry 23 801–811. 10.1038/mp.2017.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanos P., Thompson S. M., Duman R. S., Zarate C. A., Jr., Gould T. D. (2018). Convergent mechanisms underlying rapid antidepressant action. CNS Drugs 32 197–227. 10.1007/s40263-018-0492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shen X., Dong J., Liu W. C., Song M., Sun Y., et al. (2019). Inhibition of reactive astrocytes with fluorocitrate ameliorates learning and memory impairment through upregulating crtc1 and synaptophysin in ischemic stroke rats. Cell Mol. Neurobiol. 39 1151–1163. 10.1007/s10571-019-00709-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Wu X., Yan S., Xie X., Fan Y., Zhang J., et al. (2016). The antidepressant-like effects of pioglitazone in a chronic mild stress mouse model are associated with PPARgamma-mediated alteration of microglial activation phenotypes. J Neuroinflammation 13:259. 10.1186/s12974-016-0728-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wu H., Li S., Chen Q., Cheng X. W., Zheng J., et al. (2006). Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One 1:e16. 10.1371/journal.pone.0000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomkowski A. D., Hammes L., Lin J., Calixto J. B., Santos A. R., Rodrigues A. L. (2002). Agmatine produces antidepressant-like effects in two models of depression in mice. Neuroreport 13 387–391. 10.1097/00001756-200203250-00005 [DOI] [PubMed] [Google Scholar]