Summary
Two-electrode voltage clamp (TEVC) combined with the Xenopus laevis oocytes heterologous expression system is a powerful electrophysiological tool widely used to study the properties of many transmembrane proteins. Here, we describe a protocol using this combined approach to identify the ligands of odorant receptors that form ligand-gated ion channels. We detail the procedures for site-directed mutagenesis, oocyte microinjection, and TEVC recording. This protocol can also be used to identify the key residues and illustrate the structure-function relationships of these proteins.
For complete details on the use and execution of this protocol, please refer to Cao et al. (2021).
Subject areas: Gene Expression, Model Organisms, Molecular Biology, Neuroscience
Graphical abstract

Highlights
-
•
A simple approach to identify the ligands of odorant receptors in Xenopus oocytes
-
•
Detailed procedures to perform TEVC recoding combined with site-directed mutagenesis
-
•
Identifying the key amino acid residues for ligand binding of odorant receptors
-
•
Applicable for studying other proteins encoding ligand-gated ion channels
Two-electrode voltage clamp (TEVC) combined with the Xenopus laevis oocytes heterologous expression system is a powerful electrophysiological tool widely used to study the properties of many transmembrane proteins. Here, we describe a protocol using this combined approach to identify the ligands of odorant receptors that form ligand-gated ion channels. We detail the procedures for site-directed mutagenesis, oocyte microinjection, and TEVC recording. This protocol can also be used to identify the key residues and illustrate the structure-function relationships of these proteins.
Before you begin
Primer design
Timing: 20 min
-
1.Design primers for preparing the mutants of target plasmids. The point mutation replacing the leucine with a valine at 321 position of HvirOR6 is introduced by site-directed mutagenesis using the following primers:
-
a.Forward: 5′-CAAGTGGCTGTCTCGTACTTCTCGAATG-3′
-
b.Reverse: 5′-CTTGGTGAAACGCGTAATATATGAAC-3′
-
a.
Note: The mutation codon is introduced with forward primer and marked with underline. The primers used to mutate target plasmid must be phosphorylated at the 5′ end to eliminate the need for a separate phosphorylation step before direct ligation. One or both primers can be designed with desired mutation(s), and each primer can have more than one mismatch, either separated by correctly matched nucleotides or present in consecutive nucleotides. Generally, the length of the correctly matched sequence in the mutagenic primers should be in average 24–30 nucleotides. The desired mutation should be in the middle of the primer with 10–15 perfectly matched nucleotides on each side. The primers are recommended to be purified with polyacrylamide gel electrophoresis (PAGE).
Reagent preparation
Timing: 2 h
-
2.Tetracycline stock solution (50 mg/mL)
-
a.Dissolve the 0.5 g tetracycline hydrochloride into 10 mL distilled water. Mix thoroughly and filter through 0.22 μm PES syringe filters to sterilize the solution and filter insoluble impurities. Store in 1 mL aliquots at −20°C for 1 year.
-
a.
-
3.Streptomycin stock solution (100 mg/mL)
-
a.Dissolve the 1 g streptomycin sulfate salt into 10 mL distilled water. Mix thoroughly and filter through 0.22 μm PES syringe filters. Store in 1 mL aliquots at −20°C for 1 year.
-
a.
-
4.Sodium pyruvate stock solution (275 mg/mL)
-
a.Dissolve 5.5 g sodium pyruvate into 20 mL distilled water. Mix thoroughly and filter through 0.22 μm PES syringe filters. Store in 1 mL aliquots at −20°C for 1 year.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Trans1-T1 Competent Cells | TransGEN | Cat#CD501-03 |
| Chemicals, peptides, and recombinant proteins | ||
| Phenol: Chloroform: Isoamyl Alcohol 25:24:1 | Sigma-Aldrich | Cat#P3803 |
| Ampicillin | TransGEN | Cat#GG101-01 |
| Tryptone | Thermo Fisher Scientific | Cat#LP0042B |
| Yeast Extract | Thermo Fisher Scientific | Cat#LP0021B |
| Agar | Coolaber | Cat#CA1331 |
| Tetracycline hydrochloride | Amresco | Cat#0422 |
| Streptomycin sulfate | Amresco | Cat#0382 |
| Sodium pyruvate | Sigma-Aldrich | Cat#P2256 |
| Horse serum | Thermo Fisher Scientific | Cat#26050-070 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#D4540; CAS: 67-68-5 |
| (Z)-9-tetradecenal (Z9-14:Ald) | Changzhou Nimrod Inc. | P5126; CAS: 53939-27-8 |
| (Z)-9-hexadecenal (Z9-16:Ald) | Changzhou Nimrod Inc. | P5111; CAS: 56219-04-6 |
| Mineral oil | Sigma-Aldrich | Cat#M8410 |
| Critical commercial assays | ||
| PhusionTM Site-Directed Mutagenesis Kit | Thermo Fisher Scientific | Cat#F541 |
| TIANprep Mini Plasmid Kit | TIANGEN | Cat#DP103-03 |
| mMESSAGE mMACHINE SP6 Kit | Thermo Fisher Scientific | Cat#AM1340 |
| Experimental models: Organisms/strains | ||
| Xenopus laevis oocytes | Xenopus Resource Center (Hangzhou, China) | N/A |
| Recombinant DNA | ||
| pSP64DV | Lu et al. (2007) | N/A |
| Oligonucleotides | ||
| Forward: 5′-CAAGTGGCTGT CTCGTACTTCTCGAATG-3′ |
Sangon Biotech | N/A |
| Reverse: 5′-CTTGGTGAAACG CGTAATATATGAAC-3′ |
Sangon Biotech | N/A |
| Software and algorithms | ||
| pCLAMP 10.2 | Molecular Device | N/A |
| GraphPad Prism 8.0 | GraphPad Software | https://www.graphpad.com |
| SAS v8 | SAS | https://www.sas.com/ |
| Other | ||
| 0.22 μm PES Syringe Filters | Any supplier | N/A |
| 50 mL centrifuge tubes | Sangon Biotech | N/A |
| Plastic Pasteur pipette | Any supplier | N/A |
| 90-mm Petri dish | Any supplier | N/A |
| 60-mm Petri dish | Corning Incorporated | Cat#430166 |
| 24-well plates | Corning Incorporated | Cat#3524 |
| Incubator | SANYO | MIR-154 |
| Stereo Microscopes | Olympus | SZX7 |
| Micropipette puller | Sutter Instrument | P-1000 |
| Glass capillaries | WPI | Cat#1B120F-3 |
| Microinjector | WPI | Nanoliter injector 2010 |
| Silver wires | Warner Instruments | Cat#AG25-10 |
| OC-725C Oocyte Clamp Amplifier | Warner Instruments | N/A |
| Digidata 1440A Data Acquisition System | Molecular Devices | N/A |
Materials and equipment
Preparation of LB media for bacterial culture
-
•
LB sterile liquid medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast Extract | 0.5 g/L | 5 g |
| NaCl | 10 g/L | 10 g |
| Ultra-pure water | To a final volume of 1,000 mL | n/a |
| Total | n/a | 1,000 mL |
Mix thoroughly with a magnetic stirrer and sterilize the medium in autoclave sterilizer appliance that generate temperatures around 130°C and store at 25°C for one month.
-
•
LB agar medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast Extract | 0.5 g/L | 5 g |
| NaCl | 10 g/L | 10 g |
| Agar | 15 g/L | 15 g |
| Ultra-pure water | To a final volume of 1,000 mL | n/a |
| Total | n/a | 1,000 mL |
Mix thoroughly with a magnetic stirrer and sterilize the medium in autoclave sterilizer appliance that generate temperatures around 130°C. After cooling around 40°C, add 500 μL Ampicillin, mix thoroughly and aliquot 15 mL into each 90-mm Petri dishes and store at 4°C for one month.
Preparation of materials/equipment for oocytes microinjection and culture
For preparing oocytes collection you will need:
90-mm Petri dish
60-mm Petri dish
Plastic Pasteur pipette to sort and transfer oocytes
24-well plates
Incubator to keep oocytes at 18°C
Solutions for oocytes microinjection and culture
-
•
10× Ringer’s buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 960 mM | 56.1 g |
| KCl | 20 mM | 1.5 g |
| MgCl2⋅6H2O | 50 mM | 10.2 g |
| HEPES | 50 mM | 11.9 g |
| Distilled water | To a final volume of 1,000 mL | n/a |
| Total | n/a | 1,000 mL |
Adjust pH to 7.6 with NaOH. Filter-sterilize the solution and store at 4°C for several months.
Alternatives: 1× Ringer’s buffer can also be prepared by adding 10× stock solutions of each ingredient. Before that, prepare 10× stock solutions of each component by dissolving each chemical compound into 1,000 mL distilled water, adjust pH of HEPES stock, filter sterilize and keep at 4°C. Prepare 1× Ringer's buffer fresh prior to use.
-
•
100× CaCl2 solution
| Reagent | Final concentration | Amount |
|---|---|---|
| CaCl2⋅2H2O | 60 mM | 4.41 g |
| Distilled water | To a final volume of 500 mL | n/a |
| Total | 60 mM | 500 mL |
Filter-sterilize the solution and store at 4°C for several months.
-
•
Incubation medium
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Ringer’s buffer | n/a | 100 mL |
| Sodium pyruvate stock solution | 550 μg/mL | 2 mL |
| Tetracycline stock solution | 50 μg/mL | 1 mL |
| Streptomycin stock solution | 100 μg/mL | 1 mL |
| Dialyzed horse serum | n/a | 50 mL |
| Distilled water | To a final volume of 1,000 mL | n/a |
| Total | n/a | 1,000 mL |
Filter-sterilize the solution. Prepare on the day of frog dissection.
Reagents and solutions for electrophysiological recordings
-
•
KCl solution
| Reagent | Final concentration | Amount |
|---|---|---|
| KCl | 3 M | 11.175 g |
| Ultra-pure water | To a final volume of 1,000 mL | n/a |
| Total | n/a | 50 mL |
Filter-sterilize the solution and store at room temperature (15°C–25°C) for several months.
-
•
Working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Ringer’s buffer | n/a | 200 mL |
| 100× CaCl2 stock solution | 600 μM | 20 mL |
| Ultra-pure water | To a final volume of 2,000 mL | n/a |
| Total | n/a | 2,000 mL |
Mix thoroughly and store at room temperature for several months.
-
•
1 M candidate ligand Z9-14:Ald stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Z9-14:Ald | 1 M | 125.36 μL |
| DMSO | To a final volume of 500 μL | n/a |
| Total | 1 M | 500 μL |
Mix thoroughly and store at −20°C for several months.
-
•
1 M candidate ligand Z9-16:Ald stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Z9-16:Ald | 1 M | 141.91 μL |
| DMSO | To a final volume of 500 μL | n/a |
| Total | 1 M | 500 μL |
Mix thoroughly and store at −20°C for several months.
Experimental model and subject details
The female sexually mature Xenopus frogs used in this study are purchased from the Xenopus Resource Center (Hangzhou, China). Frogs are maintained in a recirculating tank system in a room at a temperature of 18°C–20°C. All experiments involving frogs are approved by the Institutional Animal Care and Use Committee of Chinese Academy of Agricultural Sciences, Beijing, China.
Step-by-step method details
Site-directed mutagenesis of target plasmid
Timing: 1 day for plasmid mutation and 20 min for sequence analysis
The protocol is performed using a PhusionTM Site-Directed Mutagenesis Kit (https://www.thermofisher.cn/order/catalog/product/F541) based on the manufacturer’s instructions. The protocol can also be used to introduce insertions and deletions in any type of plasmid DNA (Römer et al., 2009; Briant et al., 2017). The main procedures are shown in Figure 1.
-
1.PCR to amplify the target plasmid using the above-mentioned primers (Primer design).
-
a.Prepare the mix below:
Reagent Amount 5× Phusion HF Buffer 10 μL 10 mM dNTPs 1 μL Forward primer (10 μM) 2 μL Reverse primer (10 μM) 2 μL HvirOR6-pSP64DV plasmid 0.5 μL Phusion Hot Start DNA Polymerase (2 U/μL) 0.5 μL dd H2O Fill to 50 μL -
b.Use the following thermocycler parameters for the PCR.
PCR cycling conditions
Steps Temperature Time Cycles Initial Denaturation 98°C 1 min 1 Denaturation 98°C 10 s 25 cycles Annealing 67°C 30 s Extension 72°C 2 min Final Extension 72°C 5 min 1 Hold 4°C Forever
-
a.
-
2.Digest the PCR product with the enzyme DpnI to digest the Dam-methylated plasmid DNA template.
-
a.Add 1 μL of FastDigest DpnI enzyme (included in kit) to the PCR product. Mix thoroughly by pipetting.
-
b.Incubate at 37°C for 30 min.
-
a.
-
3.Prepare 10 μL ligation reaction mix.
-
a.Prepare the mix below:
Regent Amount PCR product 4 μL 5× Rapid Ligation Buffer 2 μL T4 DNA Ligase 0.5 μL dd H2O Fill to 10 μL -
b.Centrifuge briefly and incubate at 25°C for 30 min, then chill on ice.
-
a.
-
4.Perform transformation reaction.
-
a.5 μL of the product from the ligation reaction is used to transform Trans1-T1 Escherichia coli competent cells.
-
b.Cultivate the transformants on Ampicillin (working concentration 50 μg/mL) LB agar plates at 37°C for 12–14 h.
-
a.
-
5.
In the next day, 3–5 individual colonies are cultured in 500 μL of liquid Ampicillin (working concentration 50 μg/mL) LB medium at 37°C for 4–6 h with shaking at 220 rpm. The cultured medium containing each colony is sent to sequence to check for the right mutations.
Figure 1.
Schematic workflow of site-directed mutagenesis of target plasmid
Preparation of cRNA
Timing: 1 day
The protocol is performed using a mMESSAGE mMACHINE SP6 Kit according to the manufacturer’s instructions (https://www.thermofisher.cn/order/catalog/product/AM1340).
-
6.
Culture the colonies of wild type HvirOR6 and its mutant, HvirOR6-L321V, in liquid Ampicillin LB at 37°C for 14 h in an incubator with shaking at 220 rpm.
-
7.
Extract the plasmid DNA using a Plasmid DNA extraction kit (TIANprep Mini Plasmid Kit).
-
8.
Plasmid DNA is linearized with restriction enzyme SmaI and then purified with Phenol: Chloroform: Isoamyl alcohol (25:24:1), and resuspended in 7 μL nuclease-free water.
-
9.
The quantity and integrity of linearized plasmid DNA are determined using a Nanodrop ND-1000 spectrophotometer (Nano-Drop products, Wilmington, USA) and gel electrophoresis analysis.
CRITICAL: The plasmid DNA must be completely digested before the transcription reaction, because even a small amount of circular template can adversely affect the transcription output.
-
10.Prepare 20 μL in vitro transcription reaction mix.
-
a.Prepare the mix below:
Regent Amount 2× CAP/NTP 10 μL 10× Reaction Buffer 2 μL Linearized plasmid template 6 μL Enzyme Mix 2 μL -
b.Pipette the mixture gently, and briefly centrifuge. Incubate at 37°C for 2 h.
-
a.
-
11.
Stop the reaction and precipitate the RNA by adding 30 μL nuclease-free water and 30 μL LiCl Precipitation Solution (included in kit). Chill for 1 h at −20°C.
-
12.
Centrifuge at 13,523 g at 4°C for 30 min to pellet the RNA.
-
13.
Wash the RNA twice with 1 mL 70% ethanol. The synthesized cRNA is diluted into nuclease-free water to final concentration of 2 μg/μL and stored in 1 μL aliquots at −70°C until use.
Oocytes microinjection
Timing: 3 h
-
14.
Choose mature and healthy oocytes (stage V-VII) under a stereomicroscope and transfer them into a new 60 mm Petri dishes (Figure 2A).
-
15.
Mix 1 μL cRNA of HvirOR6 or 1 μL cRNA of its mutant, HvirOR6-L321V with 1 μL cRNA of Orco (HvirOrco), respectively.
-
16.
Prepare injection needles by pulling glass capillaries with a micropipette puller. The tip of a needle is cut with a blade to a 20 μm opening in diameter.
-
17.
Fill the glass needle with mineral oil and install it onto the dispenser of a microinjector.
-
18.
Transfer the 2 μL cRNA of each mixture onto a piece of parafilm and draw it into the glass needle under the control of the microinjector manipulator.
CRITICAL: Make sure no air has been drawn into the glass needle, which will greatly affect the injection of oocytes.
-
19.
Inject 27.6 nL cRNA mixture into each oocyte under a stereomicroscope (Figure 2B). Oocytes microinjected with 27.6 nL RNase-free water are used as the negative control. After injection, transfer the injected oocytes into 24-well plates (2–3 oocytes in each well) filled with incubation medium. Keep the oocytes in the incubator for 3–5 days for heterologous expression at 18°C (Figure 2C).
Figure 2.
The main steps of oocytes microinjection and TEVC recording
(A) Select mature and healthy oocytes.
(B) Microinject oocytes under a stereomicroscope.
(C) Culture injected oocytes into 24-well plates in an incubator at 18°C.
(D) Perform TEVC recording experiment.
Electrophysiological recording with TEVC
Timing: 2 days
The TEVC experiment is performed with the equipment shown in Figure 2D. The main operation steps are as follows:
-
20.Prepare working solutions of Z9-14:Ald and Z9-16:Ald.
-
a.Dilute 5 μL of 1 M Z9-14:Ald or Z9-16:Ald stock solution into 50 mL working solution in a 50 mL tube to prepare working solutions of 10-4 M for each chemical. And 3 × 10-4 M working solution of each chemical can be prepared by adding 15 μL of 1 M chemical working solution into 50 mL working solution.
-
b.10-5 M and 3 × 10-5 M working solutions of each chemical are prepared by adding 5 mL of 10-4 M and 3 × 10-4 M chemical working solution into 45 mL working solution, respectively. Similarly, chemical solutions of other concentrations including 10-6 M, 10-7 M and 3 × 10-6 M are prepared.
-
a.
CRITICAL: The chemical working solutions should be freshly prepared.
-
21.
Prepare silver wires for recording. The silver wires used in voltage electrode, current electrode and bath probe electrodes must be chlorinated prior to assembly and use. Briefly polish silver wires with sandpaper, and then immerse them in NaClO solution to half length, until the immersed part becomes uniformly black.
-
22.
Load the working solution and chemical working solutions in each perfusion tubes, and adjust the height of tubes to keep the flow rate at about 5 mL/min.
-
23.
Prepare glass electrodes. Pull glass capillaries to a fine taper using a micropipette puller with proper parameters.
CRITICAL: The parameters are set according to the RAMP value for each batch of glass capillaries. Test the RAMP value of one glass capillary each time a new batch of glass capillaries is to be used.
-
24.
Fill 3 M KCl into two glass electrodes (about 2/3 volume of the glass electrode). Insert an oxidized silver wire and a glass electrode into each holder, and then install the holder on micromanipulators.
-
25.Measure the resistance of the electrodes.
-
a.Turn on OC-725C Oocyte Clamp Amplifier and Digidata 1440A Data Acquisition System, and open the Clampex 10.2 software.
-
b.Move the electrodes into bath solution with the micromanipulators and turn “Vm OFFSET” and “Ve OFFSET” to zero.
-
c.Press buttons “Vm Electrode Test” and “Ve Electrode Test” respectively to display the resistance of each electrode. The electrode resistance should be in the range of 0.1–1 MΩ.
-
a.
-
26.
Transfer an oocyte expressing the target gene into the hole at the middle of bath chamber with a pipette.
-
27.
Move the voltage electrode to gently impale the oocyte by operating the micromanipulator, and then the current electrode. Generally, the resting membrane potential of the oocyte is displayed in the range of −10 to −50 mV.
-
28.
Turn the buttons “CLAMP” to “SLOW” and “GAIN” to “MAX” on the Digidata 1440A Data Acquisition System, and click the button “Record” on the interface of Clampex 10.2 software to start recording.
CRITICAL: Chemical-induced currents are recorded with the TEVC at holding potential of -80 mV. Signals are amplified using an OC-725C amplifier with lowpass filtered at 50 Hz and digitized at 1 kHz. Data acquisition is carried out with Digidata 1440A Data Acquisition System and pCLAMP 10.2 software.
-
29.Stimulate oocytes with different concentrations of chemical solutions for desired purposes.
-
a.For the purpose to identify ligands of HvirOR6 and its mutant. In this experiment, the oocytes expressing HvirOR6 or its mutant are challenged with 10-4 M of Z9-14:Ald and Z9-16:Ald.
-
b.For the purpose to study the response sensitivity of HvirOR6 or its mutant to chemicals. In this stage, the oocytes expressing HvirOR6 or its mutant are treated with increasing concentrations (3 × 10-4 M to 10-7 M) of chemical solutions.
-
a.
CRITICAL: During the recording, chemical solutions are supplied for about 15 s at a flow rate of 5 mL/min. After each stimulus, the oocytes are washed with working solution until the current returns to the baseline.
-
30.
Stop the recording. After challenged with all stimuli, click the button “Stop” to end the recording, and then turn the “GAIN” to “0”, and “CLAMP” to “OFF” in turn.
-
31.
Withdraw the two electrodes carefully and remove the tested oocyte. Put another oocyte in the chamber and start a new recording as described above.
-
32.
Finish the recording. Close the Clampex 10.2 software, and turn off the machines. Empty the remaining salt solutions in the perfusion system by washing with clean water.
Expected outcomes
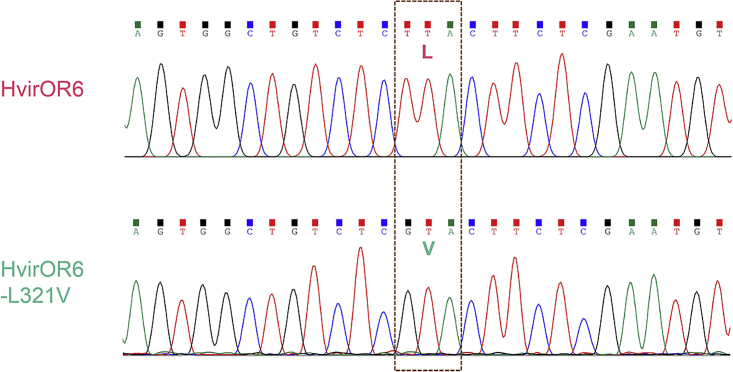
Examples of the expected outcomes in this protocol are given in Figures 3, 4, and 5. By using the site-directed mutagenesis protocol, we can easily obtain a mutated plasmid with desired mutation site(s). Figure 3 shows an example of a mutation at 321 amino acid site of HvirOR6 in vector pSP64DV, where the nucleotide thymine mutated into guanine and resulting in a mutation of the amino acid leucine (L) into valine (V).
Figure 3.
The sequencing chromatograms of HvirOR6 and its mutant, HvirOR6-L321V
Figure 4.
The electrophysiological responses of HvirOR6 and HvirOR6-L321V to 10-4 M chemical solutions
(A) Inward current responses of oocytes expressing HvirOR6 to Z9-14:Ald and Z9-16:Ald.
(B) Inward current responses of oocytes expressing HvirOR6-L321V to Z9-14:Ald and Z9-16:Ald.
(C) Response profiles of oocytes expressing HvirOR6 and its mutant to Z9-14:Ald and Z9-16:Ald. Error bars indicate SEM (n = 16–22).
Figure adapted from Cao et al. (2021).
Figure 5.
The response sensitivity of HvirOR6 and HvirOR6-L321V to Z9-14:Ald and Z9-16:Ald
(A) HvirOR6 expressing oocytes stimulated with a range of Z9-14:Ald concentrations.
(B) Dose-response curve of HvirOR6 expressing oocytes treated with Z9-14:Ald. EC50 = 6.196 × 10-6 M (n = 10).
(C) HvirOR6 and HvirOR6-L321V expressing oocytes stimulated with a range of Z9-16:Ald concentrations.
(D) Dose-response curve of HvirOR6 and HvirOR6-L321V expressing oocytes treated with Z9-16:Ald. EC50 = 2.476 × 10-5 M and 4.083 × 10-5 M, respectively (n = 7).
Figure adapted from Cao et al. (2021).
The example shown in Figure 4 reveals the binding profiles of HvirOR6 and its mutant to candidate ligands, Z9-14:Ald and Z9-16:Ald. Figures 4A and 4B show the electrophysiological responses of HvirOR6 and HvirOR6-L321V to 10-4 M tested chemical solutions, which indicates that HvirOR6 and its mutant both can be activated by Z9-14:Ald and Z9-16:Ald but with different binding affinities. Figure 4C shows that when mutating the amino acid L at 321 position of HvirOR6 to V, the mutant almost loses its response to Z9-14:Ald (t = 13.3, P < 0.001), but the response to Z9-16:Ald does not change significantly (t = -0.06, P = 0.9532) at 10-4 M concentration. The results of Figure 4 imply that L321 is a critical amino acid for the ligands binding of HvirOR6.
To further clarify the effect of L321 to the function of HvirOR6, we perform dose-response experiments to compare the binding ability of HvirOR6 and its mutant to their ligands. Figures 5A and 5B show the dose-response curve of HvirOR6 to Z9-14:Ald and the EC50 value is 6.196 × 10-6 M. Figures 5C and 5D show the dose-response curves of HvirOR6 and its mutant, HvirOR6-L321V, to Z9-16:Ald, and their EC50 values are 2.476 × 10-5 M and 4.629 × 10-5 M, respectively. These results indicate that the mutation of one site, 321, causes a functional impact of HvirOR6, demonstrating the critical and essential role of this amino acid site.
CRITICAL: To fit data to produce a dose-response curve, we can perform the following steps: a. Create an XY table, and enter the data. Fill the concentrations into “X” column, ligand names into “Group A, B, C” et al. and replicate values into corresponding tables. b. Analyze the data. Click “Analyze” to transform the concentrations using function X = Log (X). c. Fit a dose-response curve. Click “Fit a curve with nonlinear regression” in “Analyze”, choose “Nonlinear regression (curve fit)” in “XY analyses”, then click “log(agonist) vs. response – Variable slope (four parameters)” in “Dose-response-Stimulation”. d. Beautify and export the figure. To fit a dose-response curve and calculate ligand's EC50 values, usually 5 or more groups of data are needed.
Quantification and statistical analysis
For statistical analysis of the response intensities of HvirOR6 and its mutant to ligands, the mean response value of HvirOR6 to Z9-14:Ald was normalized as 1, and all response values were normalized by dividing the mean response value. The normalized response values were used to compare with the response value of HvirOR6 to the two chemical compounds individually using Student’s t-test with SAS v8 for Windows. Dose-response curves and EC50 value were fitted and calculated with GraphPad Prism 8.0 software.
Limitations
The quality of oocytes can affect the electrophysiological responses of oocytes expressing target genes to ligands, both on the response intensities and sensitivities. Thus, only mature and healthy oocytes should be selected and used in the experiment. Besides, it is hard to perform the dose-response experiment for a target protein with small response to ligand. And the EC50 value is not accurate, either. Because during recording, the baseline may slightly fluctuate due to some background noise or/and changes in liquid height. The response values are obtained by calculating the difference value between the baseline and the trough, but their locations are artificially determined. If the response of a receptor to a ligand is very small, the response value is easily influenced by multiple factors, and it is easier to cause some errors.
Troubleshooting
Problem 1
The quality of oocytes affects the results of TEVC experiments (steps 24 and 29).
Potential solution
Considering the possible impacts caused by the state of oocytes on identifying ligands and response sensitivities of target proteins, we recommend using good quality oocytes at least from 3 different frogs in the experiments. In addition, at least 3 oocytes from each frog should be used to verify the results. For the purpose of identifying key amino acid residues, the response intensities and sensitivities of target proteins and their mutants to candidate ligands should be compared with oocytes from a same frog. Furthermore, it is better to perform the electrophysiological recordings at around the same time after RNA injection to eliminate possible differences on the expression time or level of proteins in different times.
Problem 2
Hard to find out key amino acid residues involved in ligand binding (steps 1–5).
Potential solution
First, find out if there are any orthologous genes that have different response profiles or show divergent sensitivity to a same ligand. After sequences alignment, their diverse amino acid residues are potential candidates (Pellegrino et al., 2011; Leary et al., 2012; Cao et al., 2021). Second, divide the amino acid sequence into several regions and construct chimeric genes. After functional comparison of original sequence and chimeric genes, find out the key regions (Yang et al., 2017; Cao et al., 2021). Third, previous studies imply that the amino acids at the transmembrane domains and extracellular loops (ECL) affect the function of proteins (Nichols and Luetje, 2010; Pellegrino et al., 2011; Leary et al., 2012; Hughes et al., 2014; Rahman and Luetje, 2017; Yang et al., 2017; Cao et al., 2021).
Problem 3
How to identify and select mature and healthy oocytes (step 14).
Potential solution
Stage V-VII oocytes are selected and used in the next microinjection and electrophysiological recording experiments. These oocytes are characterized by their size (1.0–1.3 mm in diameter) and appearance (good contrast between the dark pigmented animal hemisphere and the yellowish vegetal hemisphere). The healthy oocytes are usually round with uniform color in each side.
Problem 4
Hard to cut pipet tips to a suitable size for oocyte microinjection (step 16).
Potential solution
The appropriate size of pipets is important for oocyte microinjection, because a pipet with smaller size will be easily blocked and is hard for injection, while a pipet with bigger size will cause any damage to oocytes. Thus, it is necessary to produce some pipets with suitable size. We usually cut the pipet tips under a stereomicroscope with the maximum power, and then measure the opening diameter using a stereomicroscope with a graduated scale. We can adjust the cutting site according to the opening size. When the size is suitable for microinjection, other pipets can also be cut under a stereomicroscope with the maximum power by comparing with the well prepared one. It is easy to cut pipet tips with good opening size for someone experienced.
Problem 5
Hard to regulate the external interference to electrophysiological signal during recording (steps 21–29).
Potential solution
First, maintain the machine well grounded. Second, chlorinate the silver wires uniformly used in electrodes. Third, keep a steady flow rate of chemical solutions and working solution. Fourth, prepare a pair of glass electrodes in good shapes. Adjust the parameters to produce electrodes with short but sharp tips, and test the resistances in a proper range (step 25). Test the RAMP value of one glass capillary each time a new batch of glass capillaries is to be used. Besides, it is recommended to purchase and use capillaries of a same brand.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guirong Wang (wangguirong@caas.cn).
Materials availability
No materials were generated in this study.
Acknowledgments
This work was supported by National Natural Science Foundation of China (31725023 to G.W. and 32072509 to Y.L.) and Shenzhen Science and Technology Program (KQTD20180411143628272 to G.W.).
Author contributions
Conceptualization, S.C., Y.L., and G.W.; methodology, S.C. and Y.L.; investigation, S.C. and Y.L.; writing – original draft, S.C. and Y.L.; writing – review & editing, S.C., Y.L., and G.W.; funding acquisition, Y.L. and G.R.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Song Cao, Email: csong_onnz@163.com.
Yang Liu, Email: yangliu@ippcaas.cn.
Guirong Wang, Email: wangguirong@caas.cn.
Data and code availability
No data or code was generated in this study.
References
- Briant K., Johnson N., Swanton E. Transmembrane domain quality control systems operate at the endoplasmic reticulum and Golgi apparatus. PLoS One. 2017;12:e0173924. doi: 10.1371/journal.pone.0173924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Liu Y., Wang B., Wang G. A single point mutation causes one-way alteration of pheromone receptor function in two Heliothis species. iScience. 2021;24:102981. doi: 10.1016/j.isci.2021.102981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D.T., Wang G., Zwiebel L.J., Luetje C.W. A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem. Senses. 2014;39:761–769. doi: 10.1093/chemse/bju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary G.P., Allen J.E., Bunger P.L., Luginbill J.B., Jr., Linn C.E., Macallister I.E., Kavanaugh M.P., Wanner K.W. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Qiu Y., Wang G., Kwon J.Y., Rutzler M., Kwon H., Pitts R.J., van Loon J.J.A., Takken W., Carlson J.R., Zwiebel L.J. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A.S., Luetje C.W. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J. Biol. Chem. 2010;285:11854–11862. doi: 10.1074/jbc.M109.058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M., Steinbach N., Stensmyr M.C., Hansson B.S., Vosshall L.B. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature. 2011;478:511–514. doi: 10.1038/nature10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., Luetje C.W. Mutant cycle analysis identifies a ligand interaction site in an odorant receptor of the malaria vector Anopheles gambiae. J. Biol. Chem. 2017;292:18916–18923. doi: 10.1074/jbc.M117.810374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P., Recht S., Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Huang L., Ning C., Wang C. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife. 2017;6:e29100. doi: 10.7554/eLife.29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data or code was generated in this study.