Abstract
Two Gram-stain-negative, aerobic, non-motile and rod-shaped bacterial strains designated 3A5MI-3T and RSS-23T were isolated from the Dragon-shaped Wetland System in Beijing Olympic Park, PR China. Strain 3A5MI-3T grew at 15–45 °C, pH 5.0–9.0 and with 0–2 % NaCl (w/v), and strain RSS-23T grew at 15-40 oC, pH 5.5–9.0 and with 0–1 % NaCl (w/v). Phylogenetic analyses of 16S rRNA gene sequences revealed that strains 3A5MI-3T and RSS-23T were members of Bacteroidetes and Proteobacteria , respectively. Phylogenetically closest relatives of strains 3A5MI-3T and RSS-23T were Niabella pedocola R384T and Thermomonas aquatica SY21T, respectively. The cells of strain 3A5MI-3T contained menaquinone MK-7 and phosphatidylethanolamine, and the major cellular fatty acids were composed of iso-C15 : 0, iso-C15 : 1 ω6c and/or iso-C15 : 1 ω7c, iso-C17 : 0 3-OH, C16 : 0 and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). Strain RSS-23T contained ubiquinone Q-8 and diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, two unknown phospholipids and an unknown lipid, and its major cellular fatty acids were iso-C15 : 0, iso-C17 : 1 ω9c, iso-C11 : 0 3-OH and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). DNA sequencing resulted in 6.59 Mb for the strain 3A5MI-3T genome and 2.79 Mb for the strain RSS-23T genome. The calculated G+C molar contents for strains 3A5MI-3T and RSS-23T were 47.07 and 61.21 mol%, respectively. According to phenotypic and phylogenetic characteristics, strains 3A5MI-3T and RSS-23T represent novel species of the genera Niabella and Thermomonas for which the names Niabella beijingensis sp. nov. and Thermomonas beijingensis sp. nov. are proposed. The type strain for N. beijingensis sp. nov. is 3A5MI-3T (=CGMCC 1.17737T=KCTC 82817T). The type strain for T. beijingensis sp. nov. is RSS-23T (=CGMCC 1.17738T=KCTC 82820T).
Keywords: Niabella beijingensis sp. nov., Thermomonas beijingensis sp. nov., strain RSS-23T , strain 3A5MI-3T , CGMCC 1.17737T=KCTC 82817T , CGMCC 1.17738T=KCTC 82820T
Data Summary
The newly sequenced data included in this work are deposited under the nucleotide accession numbers: MZ919349 and MZ920050 and under the Bioproject accession numbers JAIQDI000000000, JAJNEC000000000 and JAIQDJ000000000 at a public domain server in the National Center for Biotechnology Information (NCBI) database.
All supporting data, code and protocols have been provided within the article or through supplementary data files. A supplementary table and further supplementary files can be found at: https://doi.org/10.6084/m9.figshare.18220907.v1 [1].
Although members of the genera Niabella and Thermomonas are often isolated from similar environmental samples [2–5], they belong to the Bacteroidetes and Proteobacteria , respectively. The genus Niabella is a member of the family Chitinophagaceae . Cells of Niabella species are Gram-stain-negative, aerobic, non-flagellated and short rods. Since the genus Niabella was firstly described by Kim et al. [6], 11 specific names have been validly published [2, 3, 6–13] (https://lpsn.dsmz.de/genus/niabella). The names of ‘ Niabella terrae ’ [14] and ‘ Niabella thaonhiensis ’ [15] were proposed but have not been validated at the time of this writing. The genus Thermomonas , a member of the family Xanthomonadaceae , was firstly described by Busse et al. [16]. At the time of writing, seven species have been validly published [4, 5, 16–19] (https://lpsn.dsmz.de/genus/thermomonas). The species of the genera Niabella and Thermomonas have been isolated from a wide range of habitats, and they share features such as cells that are Gram-strain-negative, aerobic and non-flagellated.
In this report, we describe two bacterial strains that were isolated from a constructed wetland system, by phylogenetic, physiological, biochemical, and genomic analyses.
The constructed wetland system, also called the Dragon-shaped Wetland, located in the central area of the Beijing Olympic Park area (40° 0.2′ N and 116° 22.28′ E), is the largest urban artificial water system in Asia. The water in the Dragon-shaped Water System comes mainly from the Beijing Qinghe Water Reclamation Plant, and the entire water system has a complete circulation system. Sludge samples from the Dragon-shaped Wetland water system were collected, and were serially diluted with 0.85 % NaCl (w/v) and plated onto Reasoner's 2A (R2A) agar (Difco). The isolates, designated as 3A5MI-3T and RSS-23T, were obtained after incubation for 3 days at 30 °C and were routinely stored at −80 °C as suspensions in R2A broth supplemented with 20 % (v/v) glycerol.
Genomic DNA was extracted with a commercial TIANamp Bacteria DNA Kit, and 16S rRNA genes were amplified by PCR with universal bacterial primers 27 F and 1492R [20], which was also used for sequencing the PCR product. The almost-complete sequence was compared with 16S rRNA gene sequences from GenBank. The 16S rRNA gene sequences were aligned using ClustalW [21]. Phylogenetic analyses were carried out using three phylogenetic algorithms: neighbour-joining [22], maximum-likelihood [23] and maximum-parsimony [24]. Phylogenetic trees were reconstructed and bootstrapped with 1000 replicates of each sequence using mega version 7.0 [25]. The CVTree method was used to reconstruct phylogenomic tree based on whole genomes [26].
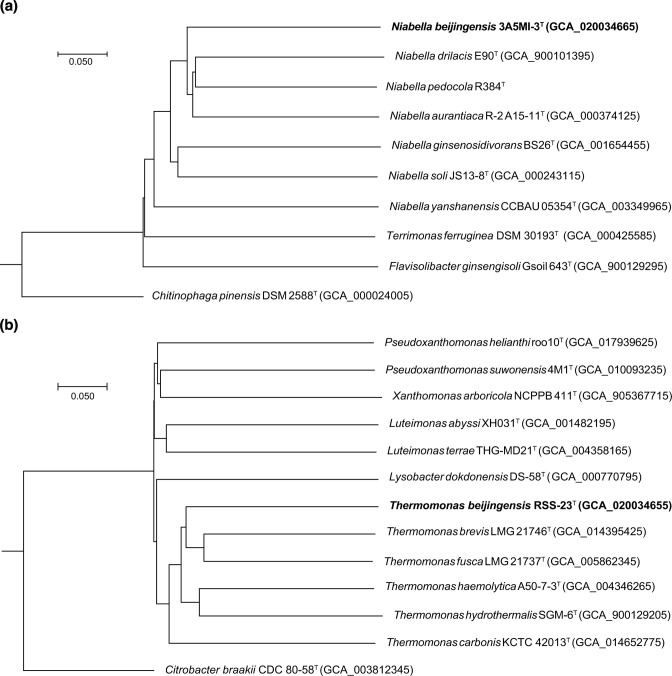
Analysis of 16S rRNA gene sequences in GenBank indicated that strain 3A5MI-3T was phylogenetically close to Niabella pedocola R384T (95.97 %), ‘ Niabella thaonhiensis ’ NHI-24T (95.68 %), Niabella aurantiaca R2A15-11T (95.61 %), Niabella tibetensis 15-4T (95.60 %), Niabella soli JS 13-8T (95.25 %), Niabella drilacis E90T (94.62 %), Niabella hirudinis CCM8411T (94.53 %), Niabella aquatica RP-2T (93.93 %), Niabella yanshanensis CCBAU 05354T (93.76 %), Niabella ginsenosidivorans BS26T (93.51%), ‘ Niabella terrae ’ ICM 1-15T (92.66 %), Niabella hibiscisoli THG-DN5.5T (92.52 %) and Niabella ginsengisoli GR 10-1T (92.45 %), as well as to Terrimonas rhizosphaerae CR94T (92.14 %) and was <92 % similar to some other species included in this analysis (Fig. 1a and S1, available in the online version of this article). Based on the results of phylogenetic analysis and 16S rRNA gene identity, N. pedocola JCM 31011T was selected as a reference strain for phenotypic tests (see following paragraph).
Fig. 1.
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences, showing the phylogenetic position of strains 3A5MI-3T (a) and RSS-23T (b) among closely related taxa. GenBank accession numbers are given in parentheses. Numbers at nodes indicate the percentage of 1000 bootstrap replicates yielding this topology; only values >50 % are shown. Filled circles indicate nodes recovered by all three treeing methods (neighbour-joining, maximum-likelihood and maximum-parsimony). Chitinophaga pinensis NBRC 18968T and Citrobacter bitternis SKKU-TP7T were used as outgroups.
Strain RSS-23T was phylogenetically close to Thermomonas aquatica SY21T (96.99 %), Thermomonas fusca LMG 21737T (96.40 %), Thermomonas brevis LMG 21746T (95.90 %), Thermomonas carbonis GZ436T (95.82 %), Thermomonas haemolytica A50-7-3T (95.75 %), Thermomonas koreensis NBRC 101155T (95.61 %) and Thermomonas hydrothermalis SGM-6T (95.44 %) (16S rRNA gene sequence identities >95 %). The phylogenetic tree showed that the strain grouped with seven members of the genus Thermomonas (Fig. 1b and S2). A phylogenomic tree based on genome sequences also showed the phylogenetic positions of strains 3A5MI-3T (Fig. 2a) and RSS-23T (Fig. 2b).
Fig. 2.
Phylogenomic trees of strains 3A5MI-3T (a) and RSS-23T (b). GenBank accession numbers are given in parentheses. We performed genome sequencing of Niabella pedocola JCM31011T and the sequence was deposited in the GenBank/EMBL/DDBJ with the accession number JAJNEC000000000. Chitinophaga pinensis DSM 2588T (=NBRC 18968T) and Citrobacter braakii CDC 80-58T (=SKKU-TP7T) were used as outgroups.
To analyse the genomic properties of strains 3A5MI-3T and RSS-23T, whole-genome sequencing was performed using the Illumina system. The genomic assembly was performed with the SPAdes software (version 3.9.0) using clean data [27]. Average nucleotide identity (ANI) values were calculated with ChunLab’s online ANI Calculator [28]. Digital DNA–DNA hybridization (dDDH) was performed by using the Genome-to-Genome Distance Calculator (3.0) [29].
The genome of strain 3A5MI-3T contained three contigs, and the total length was 6.59 Mb, encoding 6358 genes. The calculated DNA G+C content of strain 3A5MI-3T was 47.07 mol%. The genome of strain 3A5MI-3T carries 49 ncRNA genes, including three rRNA operons (5S, 16S, 23S), 41 tRNA genes and five sRNA genes. In addition, there is a clustered regularly interspaced short palindromic repeat sequence (CRISPR) with a length of 2862 bp in this genome. The Kyoto Encyclopedia of Genes and Genomes (KEGG) functional category distribution revealed that large numbers of genes in the genome of strain 3A5MI-3T belonged to the metabolism (61 %), genetic information processing (16 %), environmental information processing (7 %) and cellular processes (7 %). Annotated with the RefSeq non-redundant proteins (NR) database, the genomes of strain 3A5MI-3T harboured nine genes encoding phosphohydrolase, nine genes encoding phosphatase, 11 genes encoding glucosidase, 51 genes encoding glycosyl hydrolase families, 36 genes encoding starch binding protein, six genes encoding polysaccharide lyase, seven genes encoding polysaccharide deacetylase, 28 genes encoding SusC/RagA family TonB-linked outer membrane protein, 13 genes encoding xylanase, one gene encoding cellulase and one gene encoding chondroitinase-B. We found genes for xylanase and cellulase in the strain 3A5MI-3T and N. pedocola JCM31011T genomes. APIZYM and carbon source utilization experiments confirmed that strain 3A5MI-3T and N. pedocola JCM31011T could utilize xylan and cellulose. Although strain 3A5MI-3T and N. pedocola JCM31011T both have the chitinase gene in their genome, neither of them can use chitin. Trypsin-like serine proteases were found in the strain 3A5MI-3T genome but not in strain N. pedocola JCM31011T, which corresponds to APIZYM trypsin activity for strain 3A5MI-3T but not for N. pedocola JCM31011T. Comparing the genomes of strain 3A5MI-3T and N. pedocola JCM 31011T, we found that the specific genes detected in strain 3A5MI-3T were as follows: four genes encoding lipopolysaccharide biosynthesis protein, three genes encoding biotin synthase, three genes encoding CRISPR-associated protein Cas1, two genes encoding 2-dehydropantoate 2-reductase, one gene encoding bifunctional NAD(P)H-nitrite reductase/anaerobic dehydrogenase, two genes encoding arabinogalactan endo-1,4-beta-galactosidase, three genes encoding TDP-4-oxo-6-deoxy-d-glucose aminotransferase, one gene encoding zinc-binding alcohol dehydrogenase family protein, three genes encoding 3-phytase, three genes encoding alpha-glucuronidase and one gene encoding Alg9-like mannosyltransferase family protein. The accession numbers of the sequences in the database relating to the specific or characteristic (marked in yellow) genes of strain 3A5MI-3T and N. pedocola JCM 31011T are provided as supplementary material. The ANI value between strains 3A5MI-3T and N. pedocola JCM 31011T was calculated using ChunLab’s online ANI Calculator [28]. We performed genome sequencing of N. pedocola JCM31011T and its GenBank/EMBL/DDBJ accession number is JAJNEC000000000. The ANI result was 77.48 %, below the 95 % inter-species threshold [30]. dDDH was performed with genome sequences of strains 3A5MI-3T and N. pedocola JCM 31011T using the Genome-to-Genome Distance Calculator 29]. The estimated dDDH value was 19.50 %, which was below the standard value generally recommended for species differentiation.
The genome of strain RSS-23T contained also three contigs, and the total length is 2.79 Mb, encoding 2575 genes. The calculated genomic DNA G+C content of strain RSS-23T was 61.21 mol%. The genome of strain RSS-23T carries 58 ncRNA genes, including six rRNA genes (5S, 16S, 23S), 49 tRNA genes and three sRNA genes. The KEGG functional category distribution revealed that large numbers of genes in the genome of strain RSS-23T belonged to the metabolism (53 %), genetic information processing (16 %), environmental information processing (14 %), cellular processes (8 %). Annotated with the RefSeq NR database, the genomes of strain RSS-23T harboured 61 genes encoding phosphatase, six genes encoding glucosidase, four genes encoding glycosyl hydrolase, 27 genes encoding glycosyl transferase and 11 genes encoding polysaccharide biosynthesis protein. APIZYM positive enzyme genes such as alkaline phosphatase, acid phosphatase and α-glucosidase were also found in the genome. Comparing the genomes of strains RSS-23T and T. fusca DSM 15424T, we found that the specific genes contained in strain RSS-23T are as follows: three genes encoding aminotransferase class I/II, six genes encoding chloride channel protein, three genes encoding penicillin acylase, three genes encoding UDP-glucose 4-epimerase, three genes encoding UDP-N-acetylglucosamine 2-epimerase and three genes encoding carbonate dehydratase. The accession numbers of the sequences on the database relating to the specific or characteristic (marked in yellow) genes of strain RSS-23T and T. fusca DSM 15424T are provided as supplementary files. The ANI values between strain RSS-23T and T. fusca DSM 15424T or T. aquatica SY21T were 78.10 and 76.11 %, respectively, which were below the 95 % inter-species threshold [30]. The estimated dDDH values of strain RSS-23T to T. fusca DSM 15424T and T. aquatica SY21T were 25.20 and 20.90 %, respectively, which were below the standard value generally recommended for species differentiation.
The morphology and size of cells grown on R2A agar at 30 °C for 2 days were observed by using transmission electron microscopy (JEM-1400, jeol), and motility was observed with light microscopy. Gram-staining was performed according to Hucker [31]. Growth was measured at temperatures of 16, 20, 30, 37,40, 45, 50, 60, 65 and 70 °C. Anaerobic growth was examined in R2A broth without oxygen, and using l-cysteine (1 g l−1) to consume residual oxygen and resazurin (1 mg l−1) as a redox indicator. Salt tolerance was investigated by supplementing NaCl concentrations of 0, 1, 2, 3, 4 and 5.0 % (w/v) to R2A broth. The effect of pH (at pH 5.0, 6.0, 7.0, 8.0 and 9.0) was tested in R2A broth using phosphate buffer (for pH 5.0–7.0) and Tris buffer (for pH 8.0–9.0). Catalase activity was determined using a 3 % (v/v) hydrogen peroxide solution. Oxidation of N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride was used to check oxidase activity [32]. Monosaccharide, disaccharide and polysaccharide utilization was tested with the following compounds and basic inorganic salt at concentration of 1 g 1−1: starch, cellulose, chitin, xylan, agar, fructose, galactose, mannose, xylose, arabinose, glucose, lactose, maltose, cellobiose, trehalose, sucrose and sorbose. Bacterial cells grown in R2A were collected and washed three times with basic inorganic salt medium, inoculated at 1 % (v/v), and cultivated for 2 days. Other physiological and biochemical analyses were carried out by using the API ZYM, API 20NE and Biolog GEN III systems, according to the manufacturers’ instructions. Cells of strains 3A5MI-3T and RSS-23T were Gram-stain-negative, aerobic, non-motile and short rod-shaped (Fig. S3). The distinctive physiological characteristics of strains 3A5MI-3T and RSS-23T are listed in Tables 1 and 2, respectively.
Table 1.
Phenotypic characteristics that differentiate strain 3A5MI-3T from the phylogenetically closely related strains of the genus Niabella
Strains: 1, 3A5MI-3T; 2, N. pedocola JCM 31011T [8]; 3, N. aquatica JCM 30952T [2]; 4, N. aurantiaca DSM 17617T [6]; 5, N. ginsengisoli JCM 15444T [3]; 6, N. ginsenosidivorans JCM 18199T [13]; 7, N. hibiscisoli KACC 18857T [10]; 8, N. hirudinis DSM 25812T [9]; 9, N. soli DSM 19437T [12]; 10, ‘ N. terrae ’ JCM 19502T [14]; 11, ‘ N. thaonhiensis ’ JCM 18864T [15]; 12, N. yanshanensis KACC 14980T [11]; 13, N. drilacis DSM25812T [9]; 14, N. tibetensis CCTCC AB 209167T [7]. +, Positive; −, negative; w, weakly positive. The data of strain 3A5MI-3T and N. pedocola JCM 31011T were taken from the current study, others were taken from the published literature.
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Source of isolation |
Sludge |
Soil |
Water |
Soil |
Soil |
Compost |
Soil |
Leeches |
Soil |
Soil |
Soil |
Soybean rhizosphere |
Leeches |
Soil |
|
pH range for growth |
5–9 |
5.5–10.5 |
5–9 |
5–8 |
6–8 |
4.5–10 |
5–8 |
4.5–10.5 |
5–8 |
5–8.5 |
6.5–11 |
6–10 |
4.5–10.5 |
5–8 |
|
Temperature range for growth (°C) |
15–45 |
15–40 |
10–37 |
10–35 |
5–35 |
18–42 |
4–40 |
15–30 |
15–35 |
15–35 |
15–37 |
15–35 |
15–30 |
15–34 |
|
Catalase/oxidase |
−/+ |
−/w |
+/− |
+/− |
+/− |
+/+ |
+/+ |
−/+ |
+/+ |
+/+ |
−/+ |
+/+ |
−/+ |
+/+ |
|
Gelatin hydrolysis |
+ |
− |
− |
+ |
− |
− |
+ |
− |
− |
+ |
+ |
+ |
− |
+ |
|
Assimilation tests (API 20NE and Biolog GENIII): |
||||||||||||||
|
d-Glucose |
− |
+ |
+ |
+ |
− |
+ |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
|
d-Mannitol |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
|
l-Fucose |
w |
− |
+ |
− |
+ |
+ |
+ |
− |
− |
+ |
− |
+ |
− |
− |
|
l-Serine |
+ |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
l-Alanine |
w |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
− |
+ |
− |
|
Enzyme activities (API ZYM): |
||||||||||||||
|
Cystine arylamidase |
w |
+ |
+ |
− |
+ |
+ |
+ |
+ |
− |
− |
w |
+ |
+ |
+ |
|
Trypsin |
w |
− |
− |
− |
+ |
− |
+ |
w |
− |
+ |
− |
+ |
w |
− |
Table 2.
Phenotypic characteristics that differentiate strain RSS-23T from the phylogenetically closely related species of the genus Thermomonas
Strains: 1, RSS-23T; 2, T. fusca DSM 15424T [18]; 3, T. aquatica KCTC 62191T [4]; 4, T. carbonis KCTC 42013T [17]; 5, T. brevis DSM 15422T [18]; 6, T. haemolytica DSM 13605T [16]; 7, T. koreensis KCTC 12540T [5]; 8, T. hydrothermalis DSM 14834T [19]. +, Positive; −, negative; w, weakly positive/sensitive. The data of strain RSS-23T and T. fusca DSM 15424T were taken from the current study, others were taken from the published literature.
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|---|---|---|---|---|---|---|---|---|
|
Colony colour on R2A agar |
Cream |
Beige |
Cream |
Yellow |
Beige |
Cream |
Beige |
Cream |
|
Cell shape |
Ovoid rod |
Rod |
Rod |
Ovoid rod |
Rod |
Rod |
Rod |
Rod |
|
Nitrate reduction |
+ |
+ |
− |
− |
− |
− |
+ |
+ |
|
Optimum growth temperature (°C) |
30 |
28–37 |
28 |
28 |
28–37 |
37–50 |
37 |
50 |
|
Motility |
− |
+ |
− |
− |
+ |
+ |
+ |
− |
|
Aesculin hydrolysis |
+ |
− |
− |
+ |
+ |
− |
+ |
+ |
|
Gelatin hydrolysis |
+ |
+ |
− |
− |
+ |
− |
+ |
+ |
|
Assimilation tests (API 20NE and Biolog): |
||||||||
|
d-Glucose |
+ |
− |
+ |
− |
+ |
− |
+ |
+ |
|
Maltose |
+ |
− |
+ |
w |
+ |
− |
+ |
+ |
|
d-Mannitol |
− |
− |
+ |
+ |
− |
− |
− |
− |
|
N-Acetylglucosamine |
+ |
− |
w |
+ |
+ |
− |
− |
− |
|
d-Mannose |
− |
− |
+ |
− |
+ |
− |
− |
− |
|
l-Proline |
+ |
+ |
+ |
+ |
+ |
− |
+ |
+ |
|
Enzyme activities (API ZYM): |
||||||||
|
Trypsin |
+ |
+ |
+ |
w |
+ |
− |
− |
− |
|
β-Glucosidase |
+ |
− |
+ |
− |
− |
− |
+ |
+ |
|
Cystine arylamidase |
+ |
+ |
+ |
w |
+ |
w |
+ |
− |
|
Chymotrypsin |
+ |
+ |
− |
w |
− |
+ |
− |
+ |
|
Lipase (C14) |
− |
− |
− |
− |
− |
+ |
− |
+ |
The cellular fatty acids were determined using cells grown on R2A agar for 2 days at 30 °C with the standard midi protocol (Sherlock Microbial Identification System, version 6.0B) and analysed with a gas chromatograph (GC6890, Agilent) [33]. The extraction, purification and analysis of quinones were carried out according to the methods of Collins et al. [34]. The total lipids were extracted using chloroform and methanol, followed by thin-plate biphasic chromatography to identify each lipid component [35].
Strain 3A5MI-3T contained aliphatic saturated fatty acid (iso-C15 : 0) as its predominant cellular fatty acid that accounted for about half of total fatty acids, followed by iso-C15 : 1 ω6c and/or iso-C15 : 1 ω7c, iso-C17 : 0 3-OH and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). The composition of the fatty acids is shown in Table 3. The major cytoquinone was determined to be MK-7, as previously reported for described members of the genus Niabella . The polar lipid profile of strain 3A5MI-3T contained phosphatidylethanolamine and three unknown lipids (Fig. S4a).
Table 3.
Cellular fatty acid composition (% of total) of strain 3A5MI-3T and closely related species of the genus Niabella
Strains: 1, 3A5MI-3T; 2, N. pedocola JCM 31011T [8]; 3, N. aquatica JCM 30952T [2]; 4, N. aurantiaca DSM 17617T [6]; 5, N. ginsengisoli JCM 15444T [3]; 6, N. ginsenosidivorans JCM 18199T [13]; 7, N. hibiscisoli KACC 18857T [10]; 8, N. hirudinis DSM 25812T [9]; 9, N. soli DSM 19437T [12]; 10, ‘ N. terrae ’ JCM 19502T [14]; 11, ‘ N. thaonhiensis ’ JCM 18864T [15]; 12, N. yanshanensis KACC 14980T [11] ; 13, N. drilacis DSM25812T [9]; 14, N. tibetensis CCTCC AB 209167T [7]. −, Not detected or <0.1 %. The data of strain 3A5MI-3T and N. pedocola JCM 31011T were taken from the current study, others were taken from the published literature.
|
Fatty acid |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Saturated: |
||||||||||||||
|
iso-C15 : 0 |
40.6 |
40.9 |
40.4 |
33.7 |
32.8 |
42.0 |
27.5 |
52.4 |
29.2 |
38.9 |
36.7 |
39.4 |
40.8 |
41.3 |
|
anteiso-C15 : 0 |
1.9 |
1.2 |
0.9 |
1.6 |
– |
0.7 |
0.6 |
– |
1.2 |
3.8 |
0.9 |
– |
0.7 |
0.9 |
|
C16 : 0 |
5.5 |
5.9 |
2.2 |
3.5 |
7.9 |
2.1 |
13.3 |
2.9 |
6.8 |
2.6 |
3.6 |
4.3 |
3.2 |
3.4 |
|
Unsaturated: |
||||||||||||||
|
iso-C15 : 1 G* |
19.0 |
32.2 |
23.4 |
22.3 |
36.4 |
15.9 |
20.3 |
16.6 |
18.4 |
20.3 |
20.8 |
30.5 |
20.5 |
14.9 |
|
anteiso-C15 : 1 |
1.1 |
1.0 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
Hydroxy: |
||||||||||||||
|
iso-C15 : 0 3OH |
1.7 |
2.1 |
2.8 |
2.9 |
3.7 |
2.9 |
3.0 |
3.8 |
2.2 |
3.0 |
2.6 |
3.2 |
– |
2.4 |
|
iso-C16 : 0 3OH |
1.1 |
– |
– |
– |
– |
0.7 |
– |
– |
– |
2.2 |
– |
– |
– |
0.7 |
|
C16 : 0 3OH |
2.4 |
– |
1.8 |
2.4 |
– |
1.2 |
1.8 |
1.9 |
2.2 |
1.8 |
2.9 |
1.4 |
3.5 |
3.3 |
|
iso-C17 : 0 3OH |
11.4 |
7.6 |
13.9 |
15.5 |
9.5 |
16.4 |
10.7 |
9.7 |
11.8 |
12.9 |
9.1 |
7.7 |
9.2 |
– |
|
Summed feature 3† |
7.1 |
5.2 |
12.5 |
10.6 |
3.5 |
14.4 |
8.8 |
5.2 |
11.1 |
5.3 |
15 |
7.8 |
10.7 |
16.0 |
*iso-C15 : 1 G should correspond to either iso-C15 : 1 ω6c and/or iso-C15 : 1 ω7c. The double bond position indicated by a capital letter is unknown.
†Summed features are fatty acids that cannot be resolved reliably from another fatty acid using the chromatographic conditions chosen. The midi system groups these fatty acids together as one feature with a single percentage of the total. Summed feature 3 comprised C16 : 1 ω7c/C16 : 1 ω6c.
Strain RSS-23T also contained aliphatic saturated fatty acid (iso-C15 : 0) as its predominant cellular fatty acid, which accounted for about one third of total fatty acids. The fatty acid profiles are shown in Table 4. The major isoprenoid quinone was determined to be Q-8, as previously reported for members of the genus Thermomonas . The polar lipid profile contained diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, two unknown phospholipids and an unknown lipid (Fig. S4b).
Table 4.
Cellular fatty acid composition (% of total) of strain RSS-23T and closely related species of the genus Thermomonas
Strains: 1, RSS-23T; 2, T. fusca DSM 15424T [18]; 3, T. aquatica KCTC 62191T [4]; 4, T. carbonis KCTC 42013T [17]; 5, T. brevis DSM 15422T [18]; 6, T. haemolytica DSM 13605 [16]T [16]; 7, T. koreensis KCTC 12540T [5]; 8, T. hydrothermalis DSM 14834T [19]. −, Not detected or <0.1 %. The data of strain RSS-23T and T. fusca DSM 15424T were taken from the current study, others were taken from the published literature.
|
Fatty acid |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|---|---|---|---|---|---|---|---|---|
|
Saturated: |
||||||||
|
iso-C11 : 0 |
4.9 |
4.3 |
7.8 |
8.8 |
11.0 |
9.7 |
8.7 |
9.5 |
|
iso-C13 : 0 |
1.3 |
2.5 |
– |
– |
– |
– |
2.9 |
– |
|
C14 : 0 |
4.8 |
3.4 |
4.3 |
3.2 |
1.3 |
– |
1.7 |
– |
|
iso-C14 : 0 |
4.3 |
13.8 |
– |
1.4 |
1.7 |
2.9 |
2.6 |
– |
|
iso-C15 : 0 |
31.1 |
21.9 |
27.9 |
35.8 |
44.3 |
48.7 |
47.4 |
52.3 |
|
anteiso-C15 : 0 |
2.9 |
1.8 |
1.9 |
– |
1.1 |
– |
– |
– |
|
C16 : 0 |
4.2 |
1.9 |
13.4 |
6.0 |
1.9 |
1.4 |
– |
2.0 |
|
iso-C16 : 0 |
5.0 |
11.1 |
2.3 |
2.7 |
3.0 |
10.4 |
1.5 |
1.2 |
|
iso-C17 : 0 |
1.0 |
– |
1.7 |
1.2 |
1.7 |
3.9 |
– |
7.6 |
|
Unsaturated: |
||||||||
|
iso-C15 : 1 F* |
5.7 |
3.0 |
1.3 |
3.1 |
1.8 |
1.0 |
8.3 |
1.2 |
|
iso-C17 : 1 ω9c |
15.7 |
12.5 |
– |
7.9 |
17.6 |
8.0 |
16.1 |
13.8 |
|
C16 : 1 ω9c |
1.5 |
1.0 |
– |
– |
– |
– |
– |
– |
|
Hydroxy: |
||||||||
|
iso-C11 : 0 3-OH |
6.3 |
5.9 |
10.3 |
7.8 |
8.6 |
10.6 |
5.9 |
8.0 |
|
Summed feature 3† |
7.9 |
10.3 |
– |
17.5 |
2.2 |
– |
1.5 |
– |
*iso-C15 : 1 F should correspond to either iso-C15 : 1 ω6c and/or iso-C15 : 1 ω5c. The double bond position is presumptive [36].
†Summed features are fatty acids that cannot be resolved reliably from another fatty acid using the chromatographic conditions chosen. The midi system groups these fatty acids together as one feature with a single percentage of the total. Summed feature 3 comprised C16 : 1 ω7c/C16 : 1 ω6c.
Based on the phylogenetic and phenotypic characteristics, we conclude that strains 3A5MI-3T and RSS-23T represent novel species of the genera Niabella and Thermomonas , respectively, for which the names Niabella beijingensis sp. nov. and Thermomonas beijingensis sp. nov. are proposed.
Description of Niabella beijingensis sp. nov.
Niabella beijingensis (bei.jing.en’sis. N.L. fem. adj. beijingensis, pertaining to Beijing, the geographical origin of the type strain).
Cells are Gram-stain-negative, strictly aerobic, non-motile and short rod-shaped (Fig. S3a). Colonies are convex, circular, smooth and edges were regular after incubation on R2A broth for 2 days at 30 °C. The strain can grow in a wide range of temperature (16–45 °C) and pH (5.0–9.0), and the range of NaCl tolerance is 0–2 % (w/v) NaCl. Optimal growth at 30 °C, pH 7.0 and with 1 % NaCl (w/v). Oxidase and catalase activities are positive and negative, respectively. According to the API 20NE system, nitrate cannot be reduced to nitrite. Positive for aesculin ferric citrate, gelatin, 4-nitrophenyl-β-d-galactopyranoside d-glucose, l-arabinose, d-mannose, N-acetyl-glucosamine and maltose, and negative for l-tryptophan, l-arginine, urea, d-mannitol, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate and phenylacetic acid (API 20NE). The following enzyme activities are positive: alkaline phosphatase, leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Esterase lipase (C8), cystine arylamidase and trypsin are weak; and esterase (C4), lipase (C14), α-chymotrypsin and β-glucuronidase are negative (API ZYM). Carbon substrates utilized include gentiobiose, sucrose, stachyose, maltose, trehalose, cellobiose, turanose, raffinose, lactose, melibiose, d-mannose, d-fructose, d-galactose, l-rhamnose, glycy-l-proline, l-aspartic acid, l-glutamic acid, l-serine, dextrin, pectin, Tween 40, acetoacetic acid, acetic acid, l-lactic acid, N-acetyl neuraminic acid, d-galacturonic acid, d-glucuronic acid, l-galactonic acid lactone, methyl β-d-glucoside, d-salicin, N-acetyl-d-glucosamine, N-acetyl-d-galactosamine, glucuronamide, gelatin, d-arabinose, d-sorbose, starch and agar. Weak utilization of l-fucose, d-glucose-6-PO4, l-alanine, l-histidine, d-gluconic acid, citric acid, glycerol, N-acetyl-β-d-mannosamine, methyl pyruvate, xylan and cellulose. No utilization of 3-methyl glucose, d-fucose, inosine, d-sorbitol, d-mannitol, d-arabitol, myo-inositol, d-fructose-6-PO4, d-aspartic acid, d-serine, l-arginine, l-pyroglutamic acid, mucic acid, quinic acid, d-saccharic acid, p-hydroxy-phenylacetic acid, d-lactic acid methyl ester, α-keto-glutaric acid, d-malic acid, l-malic acid, bromo-succinic acid, γ-amino-butryric acid, β-hydroxy-d,l-butyric acid, α-keto-butyric acid, propionic acid, formic acid and chitin (Biolog GENⅢ and cultivation examinations). The predominant cellular fatty acids are iso-C15 : 0, iso-C15 : 1 ω6c and/or iso-C15 : 1 ω7c, iso-C17 : 0 3-OH and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). Contains cytoquinone MK-7 as respiratory quinone. The polar lipid profile contains phosphatidylethanolamine and three unknown lipids. The G+C content of the type strain is 47.07 mol%.
The type strain, 3A5MI-3T (=CGMCC 1.17737T=KCTC 82817T), was isolated from constructed wetland sludge from Beijing, PR China.
Description of Thermomonas beijingensis sp. nov.
Thermomonas beijingensis (bei.jing.en’sis. n.l. fem. adj. beijingensis, pertaining to Beijing, the geographical origin of the type strain).
Cells are Gram-stain-negative, aerobic, non-motile and rod-shaped. Colonies are convex, circular, smooth and edges are regular after incubation on R2A broth for 2 days at 30 °C. The strain can grow in a wide range of temperature (16–45 °C) and pH (5.0–9.0). The range of NaCl tolerance is 0–1 % NaCl (w/v). Optimal growth at 30 °C, pH 7.0 and with 1 % NaCl (w/v). Oxidase and catalase activities are positive. According to the API 20NE system, nitrate can be reduced to nitrite. Positive for aesculin ferric citrate, gelatine, d-glucose, N-acetyl-glucosamine and maltose, and negative for l-tryptophan, l-arginine, urea, 4-nitrophenyl-β-d-galactopyranoside, l-arabinose, d-mannose, d-mannitol, potassium gluconate, caprate, adipate, malate, citrate and phenylacetate (API 20NE). The following enzyme activities of alkaline phosphatase, leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, esterase lipase (C8), cystine arylamidase, trypsin, esterase (C4) and α-chymotrypsin are positive; α-mannosidase. α-fucosidase, lipase (C14), α-galactosidase, β-galactosidase and β-glucuronidase are negative (API ZYM). Carbon sources utilized include dextrin, maltose, cellobiose, gentiobiose, N-acetyl-d-glucosamine, N-acetyl-d-galactosamine, gelatin, glycy-l-proline, l-alanine, l-arginine, l-aspartic acid, l-glutamic acid, l-serine, methyl pyruvate, Tween 40, α-hydroxy-butyric acid, β-hydroxy-d,l-butyric acid, α-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, d-arabinose, d-sorbose, starch, xylan and agar. Weak utilization of turanose, N-acetyl-β-d-mannosamine, d-fructose, d-glucose-6-PO4, d-aspartic acid and pectin. No growth occurs on trehalose, sucrose, stachyose, raffinose, lactose, melibiose, methyl β-d-glucoside, d-salicin, N-acetyl neuraminic acid, α-d-glucose, d-mannose, d-galactose, 3-methyl glucose, d-fucose, l-fucose, l-rhamnose, inosine, d-sorbitol, d-mannitol, d-arabitol, myo-inositol, glycerol, d-serine, l-histidine, l-pyroglutamic acid, d-galacturonic acid, l-galactonic acid lactone, d-gluconic acid, d-glucuronic acid, glucuronamide, mucic acid, quinic acid, d-saccharic acid, p-hydroxy-phenylacetic acid, d-lactic acid methyl ester, l-lactic acid, citric acid, α-keto-glutaric acid, d-malic acid, l-malic acid, bromo-succinic acid, γ-amino-butryric acid, formic acid, cellulose and chitin (Biolog GENⅢ and cultivation examinations). The predominant cellular fatty acids are iso-C15 : 0, iso-C17 : 1 ω9c, iso-C11 : 0 3-OH and summed feature 3 (C16 : 1 ω7c/C16 : 1 ω6c). The major isoprenoid quinone is Q-8. The polar lipid profile contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, two unknown phospholipids and an unknown lipid.
The type strain, RSS-23T (=CGMCC 1.17738T=KCTC 82820T), was isolated from soil of the Dragon-shaped Wetland System in Beijing Olympic Park, PR China. The DNA G+C content of the type strain is 61.21 mol%.
Funding information
This work was financially supported by National Natural Science Foundation of China (Grant No. 31861133002).
Acknowledgements
We thank Prof. Yuguang Zhou at Institute of Microbiology, Chinese Academy of Sciences (CAS) for coordination of deposits of type strains.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; dDDH, digital DNA–DNA hybridization; KEGG, Kyoto Encyclopedia of Genes and Genomes; R2A, Reasoner's 2A.
References
- 1.Guo S-Z, Wu T, Zhu H-Z, Yan L, Liu Z, et al. Niabella beijingensis sp. nov. and Thermomonas beijingensis sp. nov., two bacteria from constructed wetland. Figshare. 2022. 10.6084/m9.figshare.18220907.v1 doi: 10.1099/ijsem.0.005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqi MZ, Im WT. Niabella aquatica sp. nov., isolated from lake water. Int J Syst Evol Microbiol. 2016;66:2774–2779. doi: 10.1099/ijsem.0.001053. [DOI] [PubMed] [Google Scholar]
- 3.Weon H-Y, Yoo S-H, Kim B-Y, Son J-A, Kim Y-J, et al. Niabella ginsengisoli sp. nov., isolated from soil cultivated with Korean ginseng. Int J Syst Evol Microbiol. 2009;59:1282–1285. doi: 10.1099/ijs.0.004333-0. [DOI] [PubMed] [Google Scholar]
- 4.Ju J-H, Kim J-S, Lee D-H, Jeon JH, Heo S-Y, et al. Thermomonas aquatica sp. nov., isolated from an industrial wastewater treatment plant. Int J Syst Evol Microbiol. 2019;69:3399–3404. doi: 10.1099/ijsem.0.003630. [DOI] [PubMed] [Google Scholar]
- 5.Kim MK, Im WT, In JG, Kim SH, Yang DC. Thermomonas koreensis sp. nov., a mesophilic bacterium isolated from a ginseng field. Int J Syst Evol Microbiol. 2006;56:1615–1619. doi: 10.1099/ijs.0.64049-0. [DOI] [PubMed] [Google Scholar]
- 6.Kim B-Y, Weon H-Y, Yoo S-H, Hong S-B, Kwon S-W, et al. Niabella aurantiaca gen. nov., sp. nov., isolated from a greenhouse soil in Korea. Int J Syst Evol Microbiol. 2007;57:538–541. doi: 10.1099/ijs.0.64614-0. [DOI] [PubMed] [Google Scholar]
- 7.Dai J, Jiang F, Wang Y, Yu B, Qi H, et al. Niabella tibetensis sp. nov., isolated from soil, and emended description of the genus Niabella . Int J Syst Evol Microbiol. 2011;61:1201–1205. doi: 10.1099/ijs.0.022103-0. [DOI] [PubMed] [Google Scholar]
- 8.Dahal RH, Kim J. Niabellapedocola sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2016;66:2650–2656. doi: 10.1099/ijsem.0.001099. [DOI] [PubMed] [Google Scholar]
- 9.Glaeser SP, Galatis H, Martin K, Kämpfer P. Niabella hirudinis and Niabella drilacis sp. nov., isolated from the medicinal leech Hirudo verbana . Int J Syst Evol Microbiol. 2013;63:3487–3493. doi: 10.1099/ijs.0.050823-0. [DOI] [PubMed] [Google Scholar]
- 10.Ngo HTT, Trinh H, Yan Z-F, Moya G, Kook M, et al. Niabella hibiscisoli sp. nov., isolated from soil of a rose of Sharon garden. Int J Syst Evol Microbiol. 2017;67:784–788. doi: 10.1099/ijsem.0.001595. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Zhang YZ, Man CX, Chen WF, Sui XH, et al. Niabella yanshanensis sp. nov., isolated from the soybean rhizosphere. Int J Syst Evol Microbiol. 2009;59:2854–2856. doi: 10.1099/ijs.0.010447-0. [DOI] [PubMed] [Google Scholar]
- 12.Weon H-Y, Kim B-Y, Joa J-H, Kwon S-W, Kim W-G, et al. Niabella soli sp. nov., isolated from soil from Jeju Island, Korea. Int J Syst Evol Microbiol. 2008;58:467–469. doi: 10.1099/ijs.0.65304-0. [DOI] [PubMed] [Google Scholar]
- 13.Yi KJ, Im WT, Kim DW, Liu QM, Kim SK. Niabella ginsenosidivorans sp. nov., isolated from compost. J Microbiol. 2015;53:762–766. doi: 10.1007/s12275-015-5463-z. [DOI] [PubMed] [Google Scholar]
- 14.Ahn J-H, Jo E-H, Kim B-Y, Song J, Kwon S-W, et al. Niabella terrae sp. nov. isolated from greenhouse soil. J Microbiol. 2013;51:731–735. doi: 10.1007/s12275-013-3507-9. [DOI] [PubMed] [Google Scholar]
- 15.Pham VHT, Kim J. Niabella thaonhiensis sp. nov., isolated from the forest soil of Kyonggi University in Korea. Curr Microbiol. 2014;69:176–181. doi: 10.1007/s00284-014-0565-0. [DOI] [PubMed] [Google Scholar]
- 16.Busse HJ, Kämpfer P, Moore ERB, Nuutinen J, Tsitko IV, et al. Thermomonas haemolytica gen. nov., sp. nov., a gamma-proteobacterium from kaolin slurry. Int J Syst Evol Microbiol. 2002;52:473–483. doi: 10.1099/00207713-52-2-473. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Zheng S, Wang D, Wang L, Wang G. Thermomonas carbonis sp. nov., isolated from the soil of a coal mine. Int J Syst Evol Microbiol. 2014;64:3631–3635. doi: 10.1099/ijs.0.063800-0. [DOI] [PubMed] [Google Scholar]
- 18.Mergaert J, Cnockaert MC, Swings J. Thermomonas fusca sp. nov. and Thermomonas brevis sp. nov., two mesophilic species isolated from a denitrification reactor with poly(epsilon-caprolactone) plastic granules as fixed bed, and emended description of the genus Thermomonas . Int J Syst Evol Microbiol. 2003;53:1961–1966. doi: 10.1099/ijs.0.02684-0. [DOI] [PubMed] [Google Scholar]
- 19.Alves MP, Rainey FA, Nobre MF, da Costa MS. Thermomonas hydrothermalis sp. nov., a new slightly thermophilic gamma-proteobacterium isolated from a hot spring in central Portugal. Syst Appl Microbiol. 2003;26:70–75. doi: 10.1078/072320203322337335. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto M, Hayashi H, Benno Y. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol Immunol. 2003;47:133–142. doi: 10.1111/j.1348-0421.2003.tb02796.x. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 24.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406. doi: 10.2307/2412116. [DOI] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo G. CVTree: A parallel alignment-free phylogeny and taxonomy tool based on composition vectors of gParallel Alignment-free Phylogeny and Taxonomy Tool based on Composition Vectors of Genomes. Genomics Proteomics Bioinformatics. 2021;19:1–6. doi: 10.1016/j.gpb.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie vVan Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 29.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, et al. Microbial genomic taxonomy. BMC Genomics. 2013;14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hucker GJ. A new modification and application of the Gram stain. J Bacteriol. 1921;6:395–397. doi: 10.1128/jb.6.4.395-397.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacs N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956;178:703. doi: 10.1038/178703a0. [DOI] [PubMed] [Google Scholar]
- 33.Sasser M. MIDI; 1990. MIDI Technical Note 101. Identification of bacteria by gas chromatography of cellular fatty acids. [Google Scholar]
- 34.Collins MD, Jones D, Goodfellow M, Minnikin DE. Isoprenoid quinone composition as a guide to the classification of Listeria, Brochothrix, Erysipelothrix and Caryophanon . J Gen Microbiol. 1979;111:453–457. doi: 10.1099/00221287-111-2-453. [DOI] [PubMed] [Google Scholar]
- 35.Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- 36.Yassin AF, Chen W-M, Hupfer H, Siering C, Kroppenstedt RM, et al. Lysobacter defluvii sp. nov., isolated from municipal solid waste. Int J Syst Evol Microbiol. 2007;57:1131–1136. doi: 10.1099/ijs.0.64966-0. [DOI] [PubMed] [Google Scholar]