Abstract
RNA folding is hierarchical; therefore, predicting RNA secondary structure from sequence is an intermediate step in predicting tertiary structure. Secondary structure prediction is based on a nearest neighbor model using free energy minimization. To improve secondary structure prediction, all types of naturally occurring secondary structure motifs need to be thermodynamically characterized. However, not all secondary structure motifs are well characterized. Pentaloops, the second most abundant hairpin size, is one such uncharacterized motif. In fact, the current thermodynamic model used to predict the stability of pentaloops was derived from a small data set of pentaloops and from data for other hairpins of different sizes. Here, the most commonly occurring pentaloops were identified and optically melted. New experimental data for 22 pentaloop sequences were combined with previously published data for nine pentaloop sequences. Using linear regression, a pentaloop-specific model was derived. This new model is simpler and more accurate than the current model. The new experimental data and improved model can be incorporated into software that is used to predict RNA secondary structure from sequence.
Keywords: pentaloop, hairpin, secondary structure, RNA
INTRODUCTION
In addition to transcribing genetic information from DNA and translating it to protein, RNA can regulate gene expression (Tucker and Breaker 2005; Sun et al. 2018), catalyze reactions in the cell (Doudna and Cech 2002; Wilson and Lilley 2015), and act as a therapeutic target (Krützfeldt et al. 2005; Elmén et al. 2008; Esau 2008; Faghihi et al. 2008; Gupta et al. 2010; Gupta and Nandan 2017), to name a few. Due to a strong relationship between structure and function (Dyson et al. 1993; Lee et al. 1997; McCarthy 2005; Campagnola et al. 2015; Travers and Muskhelishvili 2015), determining RNA tertiary structure is the prime step in order to better understand RNA's diverse functionalities. An intermediate step in predicting tertiary structure from sequence is to predict secondary structure from sequence. The most common way to predict secondary structure from sequence is by free energy minimization using nearest neighbor parameters (Xia et al. 1998; Chen et al. 2012) derived for each secondary structure motif from optical melting experiments. However, there is still room for improving secondary structure prediction because not all secondary structure motifs have been sufficiently characterized thermodynamically.
One of the most common secondary structure motifs in RNA is a hairpin, formed when a strand of RNA folds back on itself to form a stem–loop structure. In Escherichia coli, ∼70% of rRNA nucleotides are involved in forming hairpins (Giese et al. 1998). Hairpins can act as nucleation sites for higher order folding (Uhlenbeck 1990) and recognition sites for other biomolecules (Legault et al. 1998; Wu et al. 2001; Koldobskaya et al. 2011; Koirala et al. 2018). One of the most common sizes of hairpins is a pentaloop, a hairpin containing 5 nt in the loop. In 16S rRNA of E. coli, pentaloops account for 13% of the total hairpins (Woese et al. 1990), and in large subunit rRNA, 24% of the hairpins are pentaloops (Gutell and Fox 1988). In addition to being prevalent, pentaloops can serve important biological functions. For example, pentaloops can play an important role in alternative splicing. In the transcript of the survival motor neuron, a temporary pentaloop is located at the junction of exon 7 and intron 7 and acts as a regulatory element for exon 7 inclusion (Singh et al. 2007, 2015). A pentaloop is present in the D3 and D5 domains of group II introns and facilitates tertiary interactions between domains (Jestin et al. 1997; Fedorova and Pyle 2005; Pechlaner et al. 2013). Pentaloops in spliceosomal U6 RNA can mediate tertiary folding (Cate et al. 1996) or act as a recognition site for proteins (Legault et al. 1998). In RNA crystallization experiments, pentaloops can serve as a recognition site for crystallization chaperones (Koldobskaya et al. 2011; Koirala et al. 2018). N protein of phages λ and P22 recognize boxB RNA by a pentaloop, which acts as a transcription antiterminator (Salstrom and Szybalski 1978; de Crombrugghe et al. 1979; Olson et al. 1982; Franklin 1985; Lazinski et al. 1989; Weisberg and Gottesman 1999). A pentaloop found in aptamer NEO1A can act as a recognition site for a wide range of aminoglycoside antibiotics (Ilgu et al. 2014) and can cause toxicity. In E. coli, the aminoacyl (A) site of 23S rRNA has a conserved pentaloop hairpin which is essential for the function of the peptidyltransferase center of rRNA (Samaha et al. 1995; Kim and Green 1999). Lastly, a pentaloop can mediate pre-mRNA modification; a highly conserved pentaloop at the R/G site of pre-mRNA in mammals and birds acts as a recognition site for adenosine deaminases that act on RNA (ADARs) (Aruscavage and Bass 2000; Stefl et al. 2006).
Despite the high frequency and functional importance of pentaloops, the thermodynamics of pentaloops are not well characterized. The current model (Vecenie and Serra 2004; Vecenie et al. 2006) used to predict the thermodynamic stability of pentaloops is based on a data set of experimental thermodynamic parameters from only nine pentaloop sequences (Serra et al. 1997; Giese et al. 1998) as well as data from different size hairpins. In the current model, two different equations are used based on the closing base pair of the hairpin (Vecenie and Serra 2004; Vecenie et al. 2006). The general hairpin equation for hairpins with a Watson–Crick closing base pair is:
| (1) |
For pentaloops, ΔG°37i(n) 5.7 kcal/mol. ΔG°37MM is the free energy of interaction between the first mismatch of the loop and the closing base pair (Freier et al. 1986; SantaLucia et al. 1991; Serra et al. 1994; Serra and Turner 1995; Giese et al. 1998; Vecenie and Serra 2004; Vecenie et al. 2006; Sheehy et al. 2010). For hairpins with a G-U or U-G closing base pair, the equation is:
| (2) |
Here, for pentaloops, ΔG°37i(n) = 5.0 kcal/mol. The bonus for G·A, U·U, and G·G first mismatches in Equations 1 and 2 was included based on the thermodynamics of hexaloops (Vecenie and Serra 2004; Vecenie et al. 2006). The model was also validated by predicting the stability of hexaloops (Vecenie and Serra 2004; Vecenie et al. 2006). Therefore, we hypothesized that a thermodynamic model derived from only pentaloop data will be more accurate at predicting pentaloop stability. Here, we report the thermodynamics of 22 pentaloops. We combine this data with the data for nine pentaloops from the literature (Serra et al. 1997; Giese et al. 1998) to derive a pentaloop-specific model. This new model can be incorporated into secondary structure prediction software to improve RNA secondary structure prediction from sequence.
RESULTS
Database searching
A total of 1589 pentaloops were found in the secondary structure database described in Materials and Methods, averaging about one pentaloop in every secondary structure. Almost all of the pentaloops were found within rRNA (∼75% were found in 23S RNA and ∼18% were found in 16S RNA). Group I introns (∼4%) and tRNA (∼1%) also contributed pentaloops, with all other types of RNA contributing <1% of the total pentaloops found. A summary of these pentaloops is shown in Table 1. Data set 1 shows frequency and percent occurrence of pentaloops when specifying the loop sequence and the closing base pair. Because previous studies have shown that the stability of hairpin loops depends on both the identity of the nucleotides in the loop and the closing base pair (Vecenie and Serra 2004; Vecenie et al. 2006; Sheehy et al. 2010; Thulasi et al. 2010), this categorization is most important. A total of 545 combinations of this type were found in the database. The top 18 most frequent pentaloops each account for 1%–4% of the total number of pentaloops, and together, they account for 36.8% of the total number of pentaloops. The remaining 527 pentaloops account for 63.2% of the total number of pentaloops, but each accounts for <1% of the total number of pentaloops. Data set 2 consists of pentaloops when specifying the loop nucleotides only (closing base pair is not included). A total of 358 pentaloops were found in the database. The top 19 most frequent pentaloops each account for 1.1%–6.4% of the total number of pentaloops, and together, they account for 46.5% of the total number of pentaloops. The remaining 339 pentaloops account for 53.1% of the total number of pentaloops, but each accounts for <1% of the total number of pentaloops. Data set 3 tallies the number of pentaloops with each of the six possible canonical closing base pairs. Lastly, data set 4 lists the pentaloop sequences when purine nucleotides are represented as “R” and pyrimidine nucleotides are represented as “Y.” While all 32 possible types of pentaloops were found in the database, the top 27 most frequent pentaloops each account for 1.0%–11.6% of the total number of pentaloops, and together, they account for 97.6% of the total number of pentaloops. The remaining five pentaloops account for 2.4% of the total number of pentaloops, with each accounting for <1% of the total number of pentaloops.
TABLE 1.
Summary of the RNA pentaloops found in the secondary structure databasea
Thermodynamic parameters
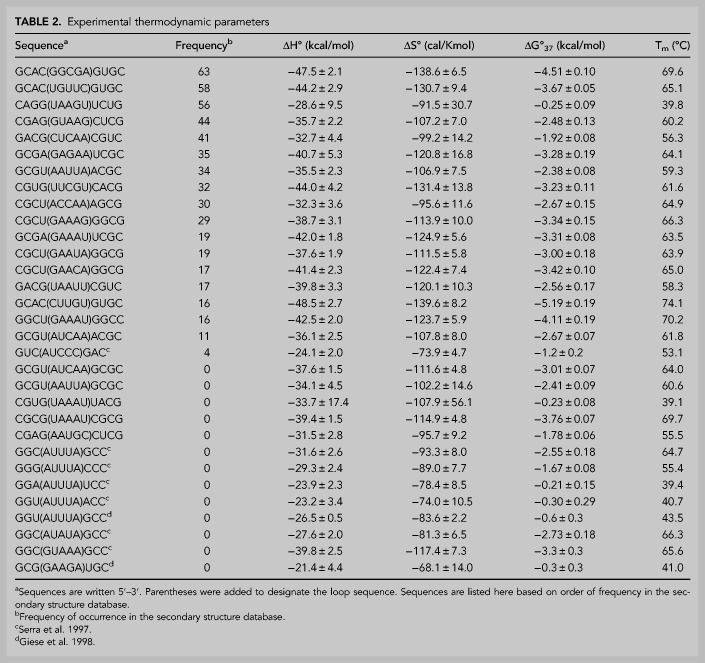
Seventeen of the most frequent pentaloops in the database were optically melted, and thermodynamic parameters were derived (Table 2). Due to possible competition from bimolecular association of strands, some frequent pentaloops were not studied here. In order to incorporate more sequence variability and to develop the most inclusive thermodynamic model, five additional pentaloop sequences that were not found in the database were also studied. These sequences were selected to fill in gaps (i.e., additional pentaloops with G-U and U-G closing pairs) in the data set and to see if frequency in the secondary structure database was related to stability.
TABLE 2.
Experimental thermodynamic parameters
Contribution of pentaloops to stem–loop free energy
Table 3 shows the thermodynamic contribution of pentaloops to stem–loop stability. In addition to the 22 pentaloops measured here, previously published data for nine additional pentaloops are included (Serra et al. 1997; Giese et al. 1998). The free energy contribution of pentaloops (ΔG°37,pentaloop) ranges from 2.58 to 5.74 kcal/mol.
TABLE 3.
Pentaloop contribution to stem–loop thermodynamics
Updated model for predicting the free energy of previously unmeasured pentaloops
To improve prediction of pentaloop stability and reduce the complexity of the current model, a new pentaloop-specific model was derived. Many different parameters were tested, but the following equation represents the best combination of parameters for a simple model with high accuracy:
| (3) |
Please note that the −1.00 kcal/mol bonus only applies to pentaloops closed by a C-G pair, with a C on the 5′ side of the loop (the bonus does not apply to G-C closing pairs), which is consistent with what was seen previously for triloops (Thulasi et al. 2010). Similarly, the 0.58 kcal/mol penalty only applies to pentaloops closed by a G-U pair, with a G on the 5′ side of the loop (the penalty does not apply to U-G closing pairs). Additional bonuses and/or additional penalties may be discovered with additional experiments. The standard error from the regression analysis for the pentaloop penalty, C-G closing pair bonus, G-U closing pair penalty, and A·A first mismatch penalty is 0.11, 0.18, 0.26, and 0.16 kcal/mol, respectively. For the new model, the average difference between the predicted and experimental free energy is 0.26 kcal/mol, with a range of 0.00–0.89 kcal/mol (Table 3). This can be compared to the current model (using Equations 1 and 2 for the same set of pentaloops) where the average difference between the predicted and experimental free energy is 0.42 kcal/mol, with a range of 0.04–2.32 kcal/mol (Table 3). Similar models without each of the individual parameters introduced in Equation 3 resulted in a significant decrease in accuracy.
A similar model was derived in order to predict the enthalpic contribution of pentaloops. Using the same parameters as Equation 3, the following equation resulted:
| (4) |
Experimental and predicted ΔH°pentaloop values can be found in Supplemental Table S2.
DISCUSSION
Database searching
For triloops, G(GGG)C and GGG were the most common triloop sequences in data sets 1 and 2, respectively (Thulasi et al. 2010). For tetraloops, the most common tetraloop sequences in data sets 1 and 2 were C(GAAA)G and GAAA, respectively (Sheehy et al. 2010). Note that all four of these hairpins consist of all purine nucleotides. Common pentaloop sequences are a little different. For pentaloops, C(GGCGA)G and UGUUC were the most common pentaloop sequences in data sets 1 and 2, respectively. It was not until the seventh most common sequence for data set 1, A(GAGAA)U, and the sixth most common sequence for data set 2, GAAAG, did pentaloops exhibit all purine sequences. Only 52.3% of pentaloops were closed by C-G or G-C pairs, compared to 68.6% of triloops (Thulasi et al. 2010) and 69.3% of tetraloops (Sheehy et al. 2010). More specifically, pentaloops with C-G closing pairs (28.6%) were much fewer than triloops (42.0%) and tetraloops (49.2%) with C-G closing pairs. Similar to what was observed in data sets 1 and 2, data set 4 for triloops (Thulasi et al. 2010) and tetraloops (Sheehy et al. 2010) were dominated by all purine nucleotides, 25.2% and 34.6%, respectively. However, for pentaloops, all purine hairpins were only the third most abundant at 7.1%.
Thermodynamic contributions of pentaloops to motif stability
As was the case for triloops and tetraloops, it is clear that stability is not the only determinant of pentaloop frequency in nature. For example, the third most frequent pentaloop in the secondary structure database, G(UAAGU)U, is the least stable (ΔG°37,pentaloop = 5.74 kcal/mol) pentaloop measured in this study. On the other hand, the most stable pentaloop (2.58 kcal/mol) was only the 18th most frequent pentaloop in the secondary structure database, C(CUUGU)G. Additionally, there is very little difference in average stability between the pentaloops that were found frequently in the secondary structure database (4.3 ± 0.8 kcal/mol) and those that were not found in the secondary structure database (4.6 ± 0.5 kcal/mol).
Several trends emerged from the thermodynamic data which resulted in the updated model (Equation 3). Pentaloops with a C-G closing base pair (with the C on the 5′ side of the hairpin loop) were found to be more stable (average ΔG°37,pentaloop of 3.5 kcal/mol) than pentaloops with other closing base pairs (average ΔG°37,pentaloop of 4.7 kcal/mol). Pentaloops with a G-U closing base pair (with the G on the 5′ side of the hairpin loop) were found to be less stable (average ΔG°37,pentaloop of 5.0 kcal/mol) than pentaloops with other closing base pairs (average ΔG°37,pentaloop of 4.4 kcal/mol). Pentaloops with an A-A first mismatch were found to be less stable (average ΔG°37,pentaloop of 4.8 kcal/mol) than pentaloops with other first mismatches (average ΔG°37,pentaloop of 4.2 kcal/mol).
Updated model for predicting thermodynamics of pentaloops
Because we have collected thermodynamic data for 22 pentaloops that previously did not have experimental values, when predicting the free energy contributions of these pentaloops in an RNA stem–loop, the experimental values can be used. For pentaloops that still do not have experimental values, the predictive model (Equation 3) can be utilized.
The updated model for predicting thermodynamics of pentaloops is simple; the inclusion of bonuses and penalties (and the corresponding values) can be determined from sequence alone. Unlike the current model in Equations 1 and 2 (Vecenie and Serra 2004; Vecenie et al. 2006), the updated model does not require the calculated free energy change for the stacking of the first mismatch on the closing base pair. Not only is the new model simpler to use, but it also more accurately predicts the stability of the entire data set of pentaloops (0.42 vs. 0.26 kcal/mol average difference from experimental value). The higher accuracy is mostly likely due to the use of pentaloop data only, whereas the parameters of the current model were derived from several different sizes of hairpins and validated using a data set of hexaloops (Vecenie and Serra 2004; Vecenie et al. 2006). The new model is significantly better for certain subsets of the data. For example, ΔΔG°37 values (difference between predicted and measured ΔG°37 values) for pentaloops with a G·G first mismatch improved from 0.80 to 0.11 kcal/mol. Additional studies with more pentaloop sequences and adjacent base pairs may result in additional free energy bonuses/penalties. We recommend Equation 3 for predicting the stability of previously unmeasured pentaloops and the general Equations 1 and 2 for previously unmeasured hairpins of larger sizes.
MATERIALS AND METHODS
Compiling and searching a database for RNA pentaloops
In order to determine which pentaloop sequences to characterize thermodynamically, sequences were selected based on frequency of occurrence in a database of secondary structures. A database of 1349 RNA secondary structures, consisting of 123 small subunit rRNAs (Gutell 1994), 223 large subunit rRNAs (Gutell et al. 1993; Schnare et al. 1996), 309 5S rRNAs (Szymanski et al. 1998), 484 tRNAs (Sprinzl et al. 1998), 91 signal recognition particles (Larsen et al. 1998), 16 RNase P RNAs (Brown 1998), 100 group I introns (Waring and Davies 1984; Damberger and Gutell 1994), and three group II introns (Michel et al. 1989), was searched for pentaloops. During this search, G-U base pairs were considered canonical base pairs. Pentaloops were required to have canonical closing pairs. The number of occurrences for each type of pentaloop were tallied.
Design of sequences for optical melting studies
To be consistent with the Watson–Crick thermodynamic parameters and parameters for most other RNA secondary structure motifs, the melting buffer used in this work contained 1 M NaCl. A major limitation of a thermodynamic analysis of RNA hairpins using a high salt concentration is the possible bimolecular association of RNA strands. To ensure that unimolecular pentaloop formation out-competed bimolecular association in a 1 M NaCl solution, the following equations, derived from the equilibrium equations and ΔG° = −RT ln K (see SI for derivation of Equation 5), were utilized:
| (5) |
| (6) |
| (7) |
Here, [H] is the concentration of hairpin, and %H is the percent hairpin in solution. [A]T is the total concentration of strand, and [D] is the concentration of duplex. KD and KH are the equilibrium constants for duplex and hairpin formation, respectively. KH and KD values were calculated at 37°C using ΔG°37 values predicted by RNAstructure (Reuter and Mathews 2010; Bellaousov et al. 2013) for hairpin and duplex formation, respectively. Calculations were done for [A]T = 1 µM and 0.1 mM, which is the typical concentration range for the melting experiments. Due to potential competition from duplex formation, some of the most frequently occurring pentaloops were not studied here; only those that were likely to form pentaloops were used. All of the sequences studied here had %[H] > 92% at [A]T = 0.1 mM and %[H] > 99% at [A]T = 1 µM.
Sequences of pentaloops and closing base pairs were designed to represent those found in the database described above. Each stem contained three Watson–Crick pairs in addition to the closing base pair. The terminal base pair was always a G-C pair in order to prevent end fraying of the duplex during melting. The duplexes were also designed to have a melting temperature between 40°C and 75°C. Care was taken to design the stem–loop sequences so that the pentaloop of interest would form, with little competition from other secondary structure motifs.
RNA synthesis and purification
The oligonucleotides were ordered from Integrated DNA Technologies, Inc. and purified by column chromatography and thin-layer chromatography as described previously (Davis and Znosko 2007; Wright et al. 2007; Christiansen and Znosko 2008).
Optical melting experiments and thermodynamics
Optical melting experiments were performed in a buffer containing 1 M NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA at pH 7.0 with a Beckman-Coulter DU800 spectrophotometer from 10°C–90°C at 260 and 280 nm. All stem–loops were melted at least nine times with a ∼50-fold concentration range. Each stem–loop melting curve resulted in a single transition, and all melts of a given sequence were concentration independent, suggesting stem–loop formation. Stem–loop thermodynamics were determined by averaging the thermodynamics derived from each individual curve fit using MeltWin 3.5 (McDowell and Turner 1996) in order to derive enthalpy (ΔH°), entropy (ΔS°), melting temperature (Tm), and free energy (ΔG°37) values. The free energy contribution of the pentaloop (ΔG°37,pentaloop) was calculated by subtracting the canonical pair contribution of the stem (Xia et al. 1998; Chen et al. 2012) from the measured ΔG°37 values for the stem–loops. Stem sequences in which the terminal pair of the stem (or the pentaloop closing base pair) is A-U or U-A utilize the 0.45 kcal/mol terminal A-U penalty (Xia et al. 1998) when calculating the contribution of the canonical pairs in the stem. The inclusion of this penalty here is likely the reason why no additional terminal pair bonuses or penalties emerged from the analysis of the pentaloop data.
Linear regression and pentaloop thermodynamic parameters
Experimental data for the 22 hairpins measured here were combined with data for nine previously published pentaloops (Serra et al. 1997; Giese et al. 1998), which were also melted in 1 M NaCl. A new predictive model for pentaloops was derived using the LINEST function (linear regression) in Microsoft Excel. The calculated experimental contribution of the pentaloop to stem–loop stability was used as a constant when doing linear regression. Many combinations of variables, including a parameter for a U·U, G·G, G·A, pyrimidine·pyrimidine, and purine·purine first mismatch and a parameter for pyrimidine or purine as the middle nucleotide in the loop, were tested, with the best combination of variables resulting in the simple, highly accurate model described in the “Results” section. The robustness of the predictive model was tested by removing individual pentaloops and small sets of pentaloops that were not predicted well by the original model and rederiving a new model. In the cases that we tested, removing data resulted in ΔΔG°37 values that improved by <0.1 kcal/mol (data not shown). As a result, the predictive model using all of the data is presented here.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work has been supported by National Institutes of Health (NIH) grant number R15GM085699-04.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078915.121.
MEET THE FIRST AUTHOR
Md. Sharear Saon.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Sharear Saon is the first author of this paper, “Thermodynamic characterization of naturally occurring RNA pentaloops.” Sharear is a fifth-year graduate student working in Dr. Znosko's laboratory in the Department of Chemistry at Saint Louis University. His research projects are focused on the study of RNA structure and stability.
What are the major results described in your paper and how do they impact this branch of the field?
In this work, thermodynamic parameters for 22 naturally occurring sequences are reported. For previously unmeasured pentaloop sequences, the model developed here can be used. The new model can predict the stability of pentaloops more accurately than the current model proposed in Vecenie et al. The average difference between experimental and predicted free energy of pentaloop formation was 0.42 and 0.26 kcal/mol for current and new model, respectively. This improved model can be incorporated into software used to predict secondary structure from sequence.
What led you to study RNA or this aspect of RNA science?
I began studying RNA because I was fascinated with the diverse functionalities of RNA. This fascination convinced me to pursue research investigating RNA structure and stability in the hopes that this research can help explain the functionalities of RNA.
If you were able to give one piece of advice to your younger self, what would that be?
I would advise myself to read more articles from fields outside my research interest. This would broaden my knowledge and give me different perspectives.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
I first learned about John Nash from the movie, A Beautiful Mind. The most important lesson I have learned from John Nash is that, regardless of the obstacles, dedication and focus can help us achieve our dreams, or in some cases, achieve beyond our dreams.
What are your subsequent near- or long-term career plans?
Right after my defense, I want to explore more in the field of RNA structure and function. Long term, through my own research group, I want to develop tools to perform predictive analyses on RNA structure and function.
REFERENCES
- Aruscavage PJ, Bass BL. 2000. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA 6: 257–269. 10.1017/S1355838200991921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaousov S, Reuter JS, Seetin MG, Mathews DH. 2013. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res 41: W471–W474. 10.1093/nar/gkt290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW. 1998. The ribonuclease P database. Nucleic Acids Res 26: 351–352. 10.1093/nar/26.1.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnola G, McDonald S, Beaucourt S, Vignuzzi M, Peersen OB. 2015. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J Virol 89: 275–286. 10.1128/JVI.01574-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. 1996. RNA tertiary structure mediation by adenosine platforms. Science 273: 1696–1699. 10.1126/science.273.5282.1696 [DOI] [PubMed] [Google Scholar]
- Chen JL, Dishler AL, Kennedy SD, Yildirim I, Liu B, Turner DH, Serra MJ. 2012. Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters. Biochemistry 51: 3508–3522. 10.1021/bi3002709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen ME, Znosko BM. 2008. Thermodynamic characterization of the complete set of sequence symmetric tandem mismatches in RNA and an improved model for predicting the free energy contribution of sequence asymmetric tandem mismatches. Biochemistry 47: 4329–4336. 10.1021/bi7020876 [DOI] [PubMed] [Google Scholar]
- Damberger SH, Gutell RR. 1994. A comparative database of group I intron structures. Nucleic Acids Res 22: 3508–3510. 10.1093/nar/22.17.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Znosko BM. 2007. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry 46: 13425–13436. 10.1021/bi701311c [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Mudryj M, Dilauro R, Gottesman M. 1979. Specificity of the bacteriophage lambda N gene product (pN): nut sequences are necessary and sufficient for antitermination by pN. Cell 18: 1145–1151. 10.1016/0092-8674(79)90227-7 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. 2002. The chemical repertoire of natural ribozymes. Nature 418: 222–228. 10.1038/418222a [DOI] [PubMed] [Google Scholar]
- Dyson MR, Mandal N, RajBhandary UL. 1993. Relationship between the structure and function of Escherichia coli initiator tRNA. Biochimie 75: 1051–1060. 10.1016/0300-9084(93)90004-C [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899. 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- Esau CC. 2008. Inhibition of microRNA with antisense oligonucleotides. Methods 44: 55–60. 10.1016/j.ymeth.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C. 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med 14: 723–730. 10.1038/nm1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM. 2005. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. EMBO J 24: 3906–3916. 10.1038/sj.emboj.7600852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin NC. 1985. Conservation of genome form but not sequence in the transcription antitermination determinants of bacteriophages λ, φ21 and P22. J Mol Biol 181: 75–84. 10.1016/0022-2836(85)90325-0 [DOI] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Caruthers MH, Neilson T, Turner DH. 1986. Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry 25: 3209–3213. 10.1021/bi00359a019 [DOI] [PubMed] [Google Scholar]
- Giese MR, Betschart K, Dale T, Riley CK, Rowan C, Sprouse KJ, Serra MJ. 1998. Stability of RNA hairpins closed by wobble base pairs. Biochemistry 37: 1094–1100. 10.1021/bi972050v [DOI] [PubMed] [Google Scholar]
- Gupta SC, Nandan YT. 2017. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer 140: 1955–1967. 10.1002/ijc.30546 [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR. 1994. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res 22: 3502–3507. 10.1093/nar/22.17.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Fox GE. 1988. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res 16: r175–r269. 10.1093/nar/16.suppl.r175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. 1993. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res 21: 3055–3074. 10.1093/nar/21.13.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgu M, Fulton DB, Yennamalli RM, Lamm MH, Sen TZ, Nilsen-Hamilton M. 2014. An adaptable pentaloop defines a robust neomycin-B RNA aptamer with conditional ligand-bound structures. RNA 20: 815–824. 10.1261/rna.041145.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jestin JL, Dème E, Jacquier A. 1997. Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO J 16: 2945–2954. 10.1093/emboj/16.10.2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DF, Green R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4: 859–864. 10.1016/S1097-2765(00)80395-0 [DOI] [PubMed] [Google Scholar]
- Koirala D, Shelke SA, Dupont M, Ruiz S, DasGupta S, Bailey LJ, Benner SA, Piccirilli JA. 2018. Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res 46: 2624–2635. 10.1093/nar/gkx1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldobskaya Y, Duguid EM, Shechner DM, Suslov NB, Ye J, Sidhu SS, Bartel DP, Koide S, Kossiakoff AA, Piccirilli JA. 2011. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol 18: 100–106. 10.1038/nsmb.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689. 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Larsen N, Samuelsson T, Zwieb C. 1998. The Signal Recognition Particle Database (SRPDB). Nucleic Acids Res 26: 177–178. 10.1093/nar/26.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazinski D, Grzadzielska E, Das A. 1989. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59: 207–218. 10.1016/0092-8674(89)90882-9 [DOI] [PubMed] [Google Scholar]
- Lee K, Varma S, SantaLucia J, Cunningham PR. 1997. In vivo determination of RNA structure-function relationships: analysis of the 790 loop in ribosomal RNA. J Mol Biol 269: 732–743. 10.1006/jmbi.1997.1092 [DOI] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. 1998. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell 93: 289–299. 10.1016/S0092-8674(00)81579-2 [DOI] [PubMed] [Google Scholar]
- McCarthy N. 2005. Form and function. Nat Rev Cancer 5: 669. 10.1038/nrc1702 [DOI] [Google Scholar]
- McDowell JA, Turner DH. 1996. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry 35: 14077–14089. 10.1021/bi9615710 [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30. 10.1016/0378-1119(89)90026-7 [DOI] [PubMed] [Google Scholar]
- Olson ER, Flamm EL, Friedman DI. 1982. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell 31: 61–70. 10.1016/0092-8674(82)90405-6 [DOI] [PubMed] [Google Scholar]
- Pechlaner M, Sigel RKO, van Gunsteren WF, Dolenc J. 2013. Structure and conformational dynamics of the domain 5 RNA hairpin of a bacterial group II intron revealed by solution nuclear magnetic resonance and molecular dynamics simulations. Biochemistry 52: 7099–7113. 10.1021/bi400784r [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salstrom JS, Szybalski W. 1978. Coliphage λnutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol 124: 195–221. 10.1016/0022-2836(78)90156-0 [DOI] [PubMed] [Google Scholar]
- Samaha RR, Green R, Noller HF. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377: 309–314. 10.1038/377309a0 [DOI] [PubMed] [Google Scholar]
- SantaLucia J, Kierzek R, Turner DH. 1991. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: evidence for stable hydrogen-bonded U.U and C.C+ pairs. Biochemistry 30: 8242–8251. 10.1021/bi00247a021 [DOI] [PubMed] [Google Scholar]
- Schnare MN, Damberger SH, Gray MW, Gutell RR. 1996. Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23 S-like) ribosomal RNA. J Mol Biol 256: 701–719. 10.1006/jmbi.1996.0119 [DOI] [PubMed] [Google Scholar]
- Serra MJ, Turner DH. 1995. Predicting thermodynamic properties of RNA. Methods Enzymol 259: 242–261. 10.1016/0076-6879(95)59047-1 [DOI] [PubMed] [Google Scholar]
- Serra MJ, Axenson TJ, Turner DH. 1994. A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry 33: 14289–14296. 10.1021/bi00251a042 [DOI] [PubMed] [Google Scholar]
- Serra MJ, Barnes TW, Betschart K, Gutierrez MJ, Sprouse KJ, Riley CK, Stewart L, Temel RE. 1997. Improved parameters for the prediction of RNA hairpin stability. Biochemistry 36: 4844–4851. 10.1021/bi962608j [DOI] [PubMed] [Google Scholar]
- Sheehy JP, Davis AR, Znosko BM. 2010. Thermodynamic characterization of naturally occurring RNA tetraloops. RNA 16: 417–429. 10.1261/rna.1773110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Singh RN, Androphy EJ. 2007. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res 35: 371–389. 10.1093/nar/gkl1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Lee BM, Singh RN. 2015. Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions. Ann N Y Acad Sci 1341: 176–187. 10.1111/nyas.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153. 10.1093/nar/26.1.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FHT. 2006. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure 14: 345–355. 10.1016/j.str.2005.11.013 [DOI] [PubMed] [Google Scholar]
- Sun Q, Hao Q, Prasanth KV. 2018. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 34: 142–157. 10.1016/j.tig.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Specht T, Barciszewska MZ, Barciszewski J, Erdmann VA. 1998. 5S rRNA data bank. Nucleic Acids Res 26: 156–159. 10.1093/nar/26.1.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasi P, Pandya LK, Znosko BM. 2010. Thermodynamic characterization of RNA triloops. Biochemistry 49: 9058–9062. 10.1021/bi101164s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Muskhelishvili G. 2015. DNA structure and function. FEBS J 282: 2279–2295. 10.1111/febs.13307 [DOI] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. 2005. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15: 342–348. 10.1016/j.sbi.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC. 1990. Tetraloops and RNA folding. Nature 346: 613–614. 10.1038/346613a0 [DOI] [PubMed] [Google Scholar]
- Vecenie CJ, Serra MJ. 2004. Stability of RNA hairpin loops closed by AU base pairs. Biochemistry 43: 11813–11817. 10.1021/bi049954i [DOI] [PubMed] [Google Scholar]
- Vecenie CJ, Morrow CV, Zyra A, Serra MJ. 2006. Sequence dependence of the stability of RNA hairpin molecules with six nucleotide loops. Biochemistry 45: 1400–1407. 10.1021/bi051750u [DOI] [PubMed] [Google Scholar]
- Waring RB, Davies RW. 1984. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing: a review. Gene 28: 277–291. 10.1016/0378-1119(84)90145-8 [DOI] [PubMed] [Google Scholar]
- Weisberg RA, Gottesman ME. 1999. Processive antitermination. J Bacteriol 181: 359–367. 10.1128/JB.181.2.359-367.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Lilley DMJ. 2015. RNA catalysis—is that it? RNA 21: 534–537. 10.1261/rna.049874.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR. 1990. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci 87: 8467–8471. 10.1073/pnas.87.21.8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DJ, Rice JL, Yanker DM, Znosko BM. 2007. Nearest neighbor parameters for inosine·uridine pairs in RNA duplexes. Biochemistry 46: 4625–4634. 10.1021/bi0616910 [DOI] [PubMed] [Google Scholar]
- Wu H, Yang PK, Butcher SE, Kang S, Chanfreau G, Feigon J. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J 20: 7240–7249. 10.1093/emboj/20.24.7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson−Crick base pairs. Biochemistry 37: 14719–14735. 10.1021/bi9809425 [DOI] [PubMed] [Google Scholar]