Abstract
Bovine fescue toxicosis (FT) is caused by grazing ergot alkaloid-producing endophyte (Epichloë coenophiala)-infected tall fescue. Endophyte’s effects on the animal’s microbiota and metabolism were investigated recently, but its effects in planta or on the plant–animal interactions have not been considered. We examined multi-compartment microbiota–metabolome perturbations using multi-‘omics (16S and ITS2 sequencing, plus untargeted metabolomics) in Angus steers grazing non-toxic (Max-Q) or toxic (E+) tall fescue for 28 days and in E+ plants. E+ altered the plant/animal microbiota, decreasing most ruminal fungi, with mixed effects on rumen bacteria and fecal microbiota. Metabolic perturbations occurred in all matrices, with some plant-animal overlap (e.g., Vitamin B6 metabolism). Integrative interactomics revealed unique E+ network constituents. Only E+ had ruminal solids OTUs within the network and fecal fungal OTUs in E+ had unique taxa (e.g., Anaeromyces). Three E+-unique urinary metabolites that could be potential biomarkers of FT and targeted therapeutically were identified.
Subject terms: Computational biology and bioinformatics, Microbiology, Physiology, Systems biology, Biomarkers
Introduction
Fescue toxicosis (FT) is a complex livestock disease that occurs when animals graze tall fescue, Lolium arundinaceum, infected with the endophyte Epichloë coenophiala1,2. While the plant-endophyte relationship is considered mutualistic because of beneficial agronomic attributes3, E. coenophiala produces ergot alkaloids implicated in FT etiology. Thus, wild-type tall fescue is referred to as toxic and leads to production deficits i.e., decreased weight gains1, resulting in over $1 billion in annual losses to the US beef industry4. Ergot alkaloids are promiscuous, interact with multiple monoamine receptors5–7, and elicit systemic pathophysiological responses. In the rumen, ergopeptine alkaloids (e.g., ergovaline) are metabolized to simpler metabolites, i.e., lysergic acid, that are able to cross gastric barriers8. This metabolism is likely driven by ruminal hyper-ammonia producing (HAB) and tryptophan-utilizing bacteria9, suggesting parent ergopeptine alkaloids may play a limited direct role in the systemic perturbations associated with FT. Considering this, the manner in which other plant-associated molecules and microbiota directly or indirectly affect the toxic (E+) fescue grazing animal, is of interest.
Evidence linking the bovine microbiota and animal productivity has spurred interest in microbiota’s contribution to FT. Altered bacterial ruminal liquid abundances of Ruminococcaceae, Coriobacteriaceae, and Erysipelotrichaceae were reported10). Also, decreased diversity and richness were associated with a lower tolerance to FT, while fecal Neocallimastigaceae was increased in high-tolerant steers11. Additionally, we found that: (i) E+ fescue grazing significantly perturbed certain fecal bacterial taxa, leading to a unique hindgut microbiota structure12; (ii) E+ exposure altered the bovine plasma and urine metabolomes13; and (iii) plasma and urinary biomarkers of a FT-specific hindgut microbiota were associated with decreased animal performance irrespective of additional external stressors e.g., hot and humid environmental conditions14. While these studies were performed using the hindgut microbiota, the foregut (rumen) microbiota is known to contain distinct populations15, with both solid and liquid fractions including multiple metabolically important microorganisms16,17. Although not evaluated in tall fescue, it is known the phyllosphere microbiota are of concern in grazing diseases like FT, and, besides grazing pressure, respond to factors in the soil, environment and other parasites18–22.
No study has yet evaluated microbial and metabolic changes induced in the tall fescue plant by the toxic endophyte, the plant and animal metabolomes/microbiomes simultaneously, or the microbiota–metabolome relationship in multiple biological matrices. To move beyond host–microbe interactions, we applied an integrative analysis of the plant and animal microbiome–metabolome networks as this is expected to provide deeper molecular insights into the FT integrome and help identify molecules, pathways and networks that are directly and/or indirectly responsible for FT and can be targeted therapeutically23.
Towards this end, using grazing beef steers, the specific goals of this study were to: (i) characterize the bacterial and fungal microbiota and metabolic differences between non-toxic (Max-Q) and toxic (E+) endophyte-infected tall fescue plant, (ii) assess changes in rumen solid/liquid and fecal microbial (bacterial and fungal) communities and of the rumen, plasma and urine metabolomes that result from E+ grazing, and, importantly (iii) assemble and evaluate the FT integrome by using systems biology approaches applied to the plant–animal microbiome–metabolome networks.
Materials and methods
Animals, pastures, and environmental conditions
All animal handling and sample collection were performed in accordance with all relevant guidelines and regulations. The study was carried out in compliance with the ARRIVE guidelines with experimental protocols approved in advance by the Institutional Animal Care and Use Committee of the University of Georgia. Post-weaning Angus steers (n = 12) were blocked by weight and randomly assigned to non-toxic (weight: 306.6 ± 12.2 kg [ ± SEM]; Max-Q; Jesup MaxQ with endophyte AR542; 3 paddocks; 2 steers per paddock) or toxic (weight: 312.2 ± 16.0 kg; E+; Jesup with wild-type endophyte; 3 paddocks; 2 steers per paddock) tall fescue pastures, as described previously13.
Sample collection and processing
Individual plant tillers were sampled from 15 random locations throughout the pastures and processed for ergot alkaloid analysis as described in12 in accordance with the most current guidelines for collection of leaf and plant tissue from cultivated pasture grasses by the College of Agricultural and Environmental Sciences of the University of Georgia, which are aligned with all relevant international guidelines. Temperature and humidity were recorded as in12. Steer body weights were recorded prior to (Pre) and at the end (28 days) of the study. Plasma, fecal, and urine samples were collected Pre (Day 0), 2, 7, 14, and 28 days post pasture placement, similar to12–14, at a working facility adjacent to the pastures. Voided urine was collected in clean collection cups via a free catch and plasma was harvested from blood collected via jugular blood draw. Ruminal samples on these dates were obtained with an ororuminal probe as in15. Rumen liquids and solids were separated with three layers of sterile cheesecloth into 3 mL (liquids) and ~ 5 g (solids) aliquots, frozen immediately on dry ice, then stored at − 80 °C.
Urinary ergot alkaloid analysis
Total urinary ergot alkaloid concentrations were determined for all samples as previously described13,24,25.
DNA extraction
Genomic DNA was extracted from all bovine matrices as previously described12,14. Tall fescue samples were stomached for 5 min in a sterile stomacher bag with TE extraction buffer. The supernatant was collected and subjected to the same extraction procedures as previously described12,14.
DNA amplification and sequencing
Sequencing of the bacterial 16S rRNA gene was performed as described12,14. For fungal sequencing, custom primers targeting the 5.8S-internal transcribed spacer 2 (ITS2) region (F-AGCCTCCGCTTATTGATATGCTTAART, R-AACTTTYRRCAAYGGATCWCT) were used, as in26. The primers also contained Illumina-specific sequencing adapters (F-AATGATACGGCGACCACCGAGATCTACAC; R- CAAGCAGAAGACGGCATACGAGAT). The following PCR cycling (40 cycles; 30 ng starting DNA) conditions were used: initial denaturation at 95 °C for 3 min; 39 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s; with the final extension set at 72 °C for 5 min. Controls and both 16S rRNA and ITS2 PCR products were treated as before and sequenced on an Illumina MiSeq using a v2 sequencing reagent kit (500-cycle)12,14.
NGS sequence processing and bioinformatics analysis
Raw 16S rRNA and ITS2 sequence files were processed using mothur v.1.41.3, as previously described12,14. After quality filtering, unique sequences were aligned to the SILVA version 119 reference alignment database27 and chimeras were removed using chimera.uchime (http://drive5.com/uchime). Bacterial sequences were aligned to the Greengenes database v13.8 (http://greengenes.secondgenome.com)28. Singletons were removed and the operational taxonomic units (OTUs) were normalized for sequence depth (i.e., each sample was normalized to the number of sequences in the smallest sample) and abundance filtered prior to statistical analysis. ITS2 sequences were aligned to the UNITE database v04.02.202029. Herein, sequences classified as belonging to the genus Neotyphodium in UNITE will be referred to its updated genus nomenclature Epichloë2.
High-resolution metabolomics (HRM)
Metabolomics sample processing for urine, plasma, and rumen liquids were performed as previously described for plasma and urine13. For tall fescue preparation, approximately 50 mg of plant material was added to vials with 100 μL acetonitrile, sonicated for 10 s and placed on ice for 30 min prior to centrifugation (10 min at 14,000 rpm). All HRM samples were analyzed with a Thermo Scientific linear triple quadrupole (LTQ) Orbitrap Velos with either hydrophilic liquid interaction chromatography (Waters Xbridge BEH Amide 2.5 μm, 2.1 × 100 mm) or C18 chromatography (Higgins Targa C18 5 μm, 2.1 × 100 mm) with 10 min gradient runs, positive ionization, and instrument settings at 120,000 resolving power, 5 min runs, and 10 μL injection. Detection of metabolomics features and QC was performed as previously described13. All metabolomics annotation presented herein was generated using xMSannotator30 and either the Human Metabolome Database HMDB31; or the T3DB toxic exposome database T3DB32. Pathway analysis was performed using mummichog33 and annotated with either the bovine or the thale cress KEGG databases for, respectively, bovine and tall fescue samples.
Overlapping feature analysis
Sets of overlapping OTUs were determined using OTU tables that included only OTUs with greater than 10 sequences (50% presence) within a matrix and a treatment. For the metabolomics overlap, all features that were present in > 80% of samples within a matrix and a treatment were used. Venn Diagrams were generated using the Bioinformatics and Evolutionary Genomics online processor (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Targeted network analysis
Normalized bacterial and fungal OTUs detected in > 50% of samples in the E+ plant and rumen, specific to each analysis, were correlated to the Epichloë OTU using the Hmisc R package34; network analysis was performed on the significantly (P < 0.05) correlated features using qgraph35. The same approach was utilized for the rumen liquids metabolomics targeted network correlated to the putatively annotated ergot alkaloid metabolite ergovaline.
Integrative interactomics analysis
xMWAS v0.552(32,36; https://kuppal.shinyapps.io/xmwas/) was used for the integrative interactomics analyses with the following parameters: dataX = metabolomics; dataY = 16S OTU table; dataZ = ITS2 OTU table; RSD > 1; Maximum number of variables from dataX = 500, from dataY and dataZ = 250; integration methods = sparse partial least squares regression (sPLS) in canonical mode with optimization of sPLS components with 500 [dataX], 250 [dataY], and 250 [dataZ] variables selected by sPLS; the association analysis was set to (|r| > 0.5; P < 0.05); centrality analysis used eigenvector centrality; graphical options were default. For the fescue xMWAS, parameters were: maximum number of variables from dataX = 500, from dataY and dataZ = 250, correlation threshold (|r| > 0.3), 1000 [dataX], 100 [dataY], and 100 [dataZ] variables selected by sPLS. The rumen analysis included the same variables as the fescue plant analysis, but the correlation threshold was set to (|r| > 0.5). The output files were downloaded and differential networks were imported into Cytoscape37 for graphical visualization and analysis.
Targeted animal integrative interactomics analysis
Putative metabolites that were significantly affected by E+ grazing and annotated by mummichog using the bos taurus KEGG database38 into tryptophan metabolism, tyrosine metabolism, Vitamin B6 metabolism, steroid hormone biosynthesis, and primary bile acid metabolism were used to perform these integrative interactomics analysis by targeting metabolic pathways commonly perturbed by E+12,13. sPLS was performed using 290 metabolites and microbiota data from the untargeted analysis for both Max-Q and E+ data sets; differential network analysis were performed to identify metabolic and microbial nodes specific to E+ steers in Cytoscape.
Statistical analysis of weight gains and urinary ergot alkaloids
Analyses of weight gains and urinary ergot alkaloids was performed using Sigma Plot, v 12.5 (Systat Software, Inc., San Jose, CA; https://systatsoftware.com) using two-way ANOVA (within-subjects design) with Holm-Sidak post-hoc analysis performed where applicable as in12–14.
Transparency statement(s)
All DNA sequences are publicly available in the NCBI Sequence Read Archive and are accessible under BioProject accession number PRJNA817179. HRM feature intensity tables and metadata will be deposited in the Metabolomics Workbench (https://www.metabolomicsworkbench.org).
Results
Non-integrative results (physiological, microbiota, metabolomics) are presented and elaborated on in the supplemental files; some important outcomes are highlighted below.
Physiological results
E+ grazing significantly increased urinary ergot alkaloids and reduced cumulative and daily weight gains (Fig. S1), indicating presence of fescue toxicosis.
Sequencing results
After quality control measures, 16S sequencing resulted in 4,976,758 high-quality sequences and a total of 4911 operational taxonomic units (OTUs) for all samples. The post-QC ITS2 sequencing resulted in 4,518,088 high-quality sequences that clustered into a total of 3116 OTUs across all samples.
General influence of E+ on plant and animal bacterial and fungal microbiota profiles
For bacterial alpha diversity, Simpson’s diversity was increased by E+ in fescue plant and decreased by E+ in rumen liquids, with rumen liquids also exhibiting effect of time (Fig. S2). There were main effects of E+ and time for Chao1 richness in rumen liquids (increased in E+), rumen solids (increased in E+ after 14 days), and fecal matter (mixed effects; Table S1; Fig. S2). For ITS2 alpha diversity, there was a main effect of E+ (an increase) in rumen solids (Table S1; Fig. S2). Also, there were time effects in several matrices for both 16S and ITS2 diversity (Table S1; Fig. S2). Permutational analysis of variance (PERMANOVA) on bacterial and fungal microbiota revealed significant main effects of E+ and time for Bray–Curtis and Jaccard indices in all sample matrices, except ITS2 in fescue plant (Table S1). The only significant treatment by time interactions were for plant bacterial and fungal ruminal solids and liquids profiles (Table S1). These data indicate that microbiota of the fescue plant, rumen solids, rumen liquids and feces are all perturbed by E+.
Identification of E. coenophiala in the microbiota
After quality filtering, alignment, and normalization for sequencing depth, one OTU aligned to Epichloë remained and was detected only in E+ plant and rumen samples, with the greatest rumen abundance being after 14 days of grazing. This OTU was used for subsequent targeted network analysis in the E+ plant and rumen.
LEfSe results
Fescue plant
Most tall fescue bacteria (Fig. S3A) and fungi (Fig. S3B) that were altered by E+ were plant-specific. The most notable effect was increased Epichloë in E+ tall fescue. A full list of the significantly different bacteria and fungi between fescue cultivars tall fescue is in the Fig. S3 legend.
Rumen solids and liquids
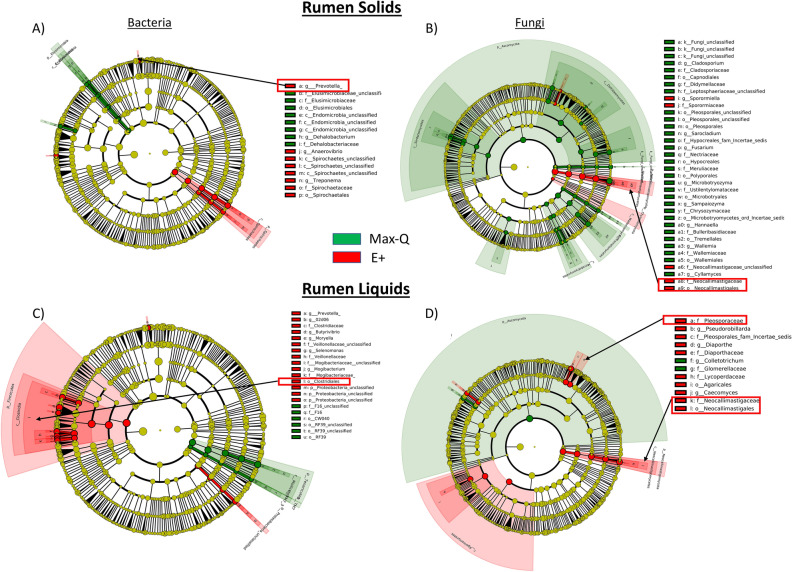
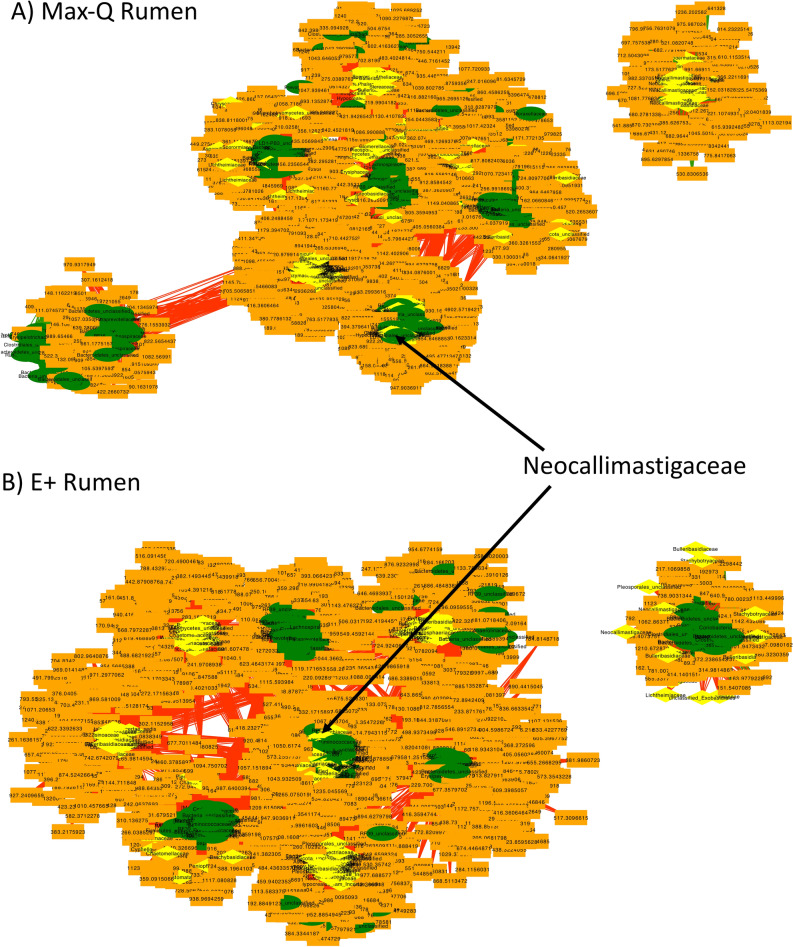
When considering Pre, Max-Q, and E+ steers, the rumen solids and liquids microbiota shifts post-pasture placement (Fig. S4). Focusing on E+ (Fig. 1A), g_Dehalobacterium. p_Spirochaetes (notably g_Treponema), and g_Prevotella were all increased. Rumen solids fungi were significantly more abundant for most taxa in Max-Q, but select fungi increased in E+ (Fig. 1B). Examples are g_Sporormiella, f_Neocallimastigaceae, and p_Chytridiomycota. Among E+ effects on bacteria in rumen liquids were increases of g_Mogibacteriu, and g_Prevotella (Fig. 1C); effects of E+ on rumen liquid fungal communities included increases of f_Neocallimastigaceae and f_Lycoperdaceae (Fig. 1D). Complete lists of affected bacteria and fungi are located in the legends of Fig. 1.
Figure 1.
Linear discriminant analysis (LDA) effect size (LEfSe; Kruskal–Wallis [P < 0.05]; Pairwise Wilcoxon [P < 0.05]; logarithmic LDA score > 2.0) of the rumen solid (A) bacterial and (B) fungal and rumen liquid (C) bacterial and (D) fungal microbiota of Angus steers across a 28-day grazing trial after placement on either a non-toxic (Max-Q; n = 6) or toxic (E+; n = 6) endophyte-infected tall fescue. Green and red shading indicates greater abundance in Max-Q or E+ steers, respectively. Taxonomic rank labels are provided before microbe names: “p_; c_; o_; f_; g_” indicate phylum, class, order, family, and genus, respectively. Letters and numbers within the cladograms refer to respective bacterial or fungal names located in the keys to the right of each cladogram. Select taxa of interest are highlighted by boxes and arrows point to their position within a cladogram.
Feces
E+ increased c_Clostridia, g_Mogibacterium, and g_Clostridium (c_Clostridia); in the fecal fungal microbiota, it is notable that the c_Sordariomycetes and order (o_)Hypocreales were increased in E+ steers, but f_Clavicipitaceae and g_Epichloë were not (Fig. S5). Analysis, including samples prior to pasture placement (Pre), are presented as Fig. S6.
Metabolic effects of E+ exposure
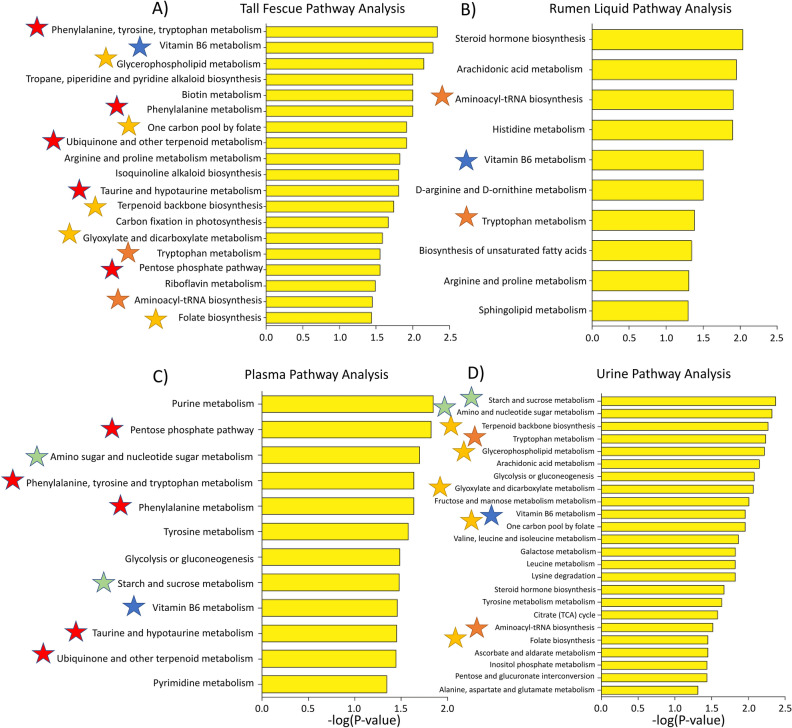
E. coenophiala-infection affected numerous metabolic pathways in the fescue plant, including phenylalanine, tyrosine and tryptophan metabolism, Vitamin B6 metabolism, and tropane, piperidine and pyridine alkaloid biosynthesis (Fig. 2A). E+ grazing perturbed steroid hormone biosynthesis, arachidonic acid, histidine, and Vitamin B6 metabolism in the rumen liquids (Fig. 2B); in the plasma, it did so on, among others, pentose phosphate pathway, phenylalanine, tyrosine, tryptophan, and Vitamin B6 metabolism (Fig. 2C). In the urine, E+ effects were on starch and sucrose metabolism, terpenoid backbone biosynthesis, tryptophan and arachidonic acid metabolism (Fig. 2D). Vitamin B6 metabolism was one of few metabolic pathways affected by E+ presence/grazing across all biological matrices; a full list of affected metabolic pathways is in Fig. 2.
Figure 2.
Metabolic pathway analysis performed on the (A) tall fescue plant, (B) rumen, (C) plasma and (D) urine high-resolution metabolomics features using mummichog. Putative metabolic pathways significantly (P < 0.05) affected by toxic tall fescue (E+) in the plant and animal throughout the 28-day grazing trial are presented. The negative log of the FDR- corrected P value for each metabolic pathway indicated on the y-axis is on the x-axis. Blue star signifies overlapping pathways between all biological matrices; orange is fescue grass, rumen liquid, and urine overlap; red is fescue grass and plasma overlap; yellow is fescue grass and urine overlap.
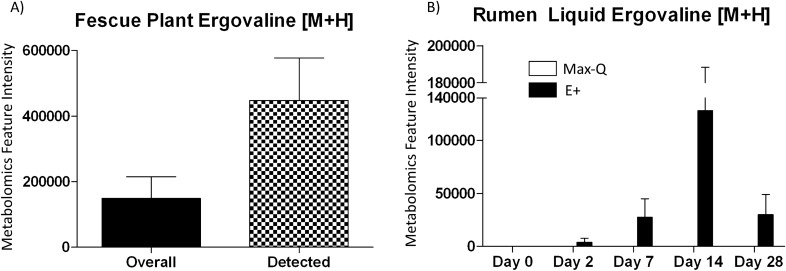
Putative ergovaline feature intensity
Using xMSannotator and the T3DB database, one metabolic feature (m/z 534.2708451, time 281.4581094; [M + H]) was putatively identified as ergovaline. Interestingly, this feature was only detected in the E+ plant and rumen liquid. In the E+ fescue, putative ergovaline was detected in multiple individual plant samples, with the average overall and positive sample intensities depicted in Fig. 3A. In the rumen liquid, ergovaline was not detected in any Max-Q or E+ samples before pasture placement; it was first detected in E+ steers on Day 2, peaked on Day 14, with Day 28 levels equaling Day 7 (Fig. 3B).
Figure 3.
Average putative ergovaline [M + H] feature intensity in (A) toxic (E+; n = 18) tall fescue plant for all samples (black) and only in samples where ergovaline was detected (checkered) and (B) the rumen liquids of Angus steers grazing either a non-toxic (Max-Q; n = 6) or toxic (E+; n = 6) tall fescue over the course of the 28-day grazing trial. Feature intensity data are presented as mean ± SEM.
Overlapping microbial and metabolic features
Plant and animal bacterial OTUs did not overlap, but an overlap between plant and rumen fungal OTUs (Max-Q 103 OTUs; E+ 115 OTUs) was present. Substantial overlap occurred between bacterial (332 Max-Q; 378 E+) OTUs in the rumen solids and rumen liquids regardless of fescue cultivar (Fig. 4A,B). Overlapping OTUs between the rumen solids and feces within E+ steers mapped to f_Coriobacteriaceae, f_Lachnospiraceae, and f_Ruminococcaceae families (Fig. 4B).
Figure 4.
Top, Middle: Venn diagrams representing specific bacterial (16S; A, B) and fungal (ITS2; C, D) OTUs that overlapped between biological matrices in steers grazing a novel, non-toxic (Max-Q; n = 6; left) or a toxic (E+; n = 6; right) tall fescue over the course of a 28 day grazing trial. Only OTUs with sequence counts (nseq > 10) were included in the analysis. Red arrows indicate specific microbes of interest with overlapping OTUs. Bottom: Venn diagrams representing specific metabolic features with exact mass-to-charge ratios (m/z’s) that overlapped between biological matrices in steers grazing (E) a non-toxic (Max-Q; n = 6) or (G) a toxic (E+; n = 6) tall fescue over the course of a 28 day grazing trial. (F) Represents shared or distinct features between Max-Q and E+ that overlapped between all four biological matrices in each respective cultivar. Only metabolic features present in > 80% of samples within a treatment and matrix were included in the analysis.
For the fungi, more OTUs overlapped between the grass and rumen liquids than grass and rumen solids in both Max-Q and E+ steers (Fig. 4C,D, respectively). In E+ steers, one OTU aligned to the Epichloë genus, alongside multiple Phaeosphaeriaceae OTUs, overlapped between these biological matrices. Most OTUs overlapping between the fescue grass, rumen solids, and rumen liquids were cultivar specific, including Cryptococcus aureus and Cryptococcus dimennae in E+ (Fig. 4D). Multiple f_Neocallimastigaceae OTUs overlapped between all animal matrices irrespective of treatment. Sub-family overlap was largely cultivar-specific, and the full list of overlapping bacterial and fungal OTUs is in File S1.
Similar metabolite (m/z) overlap was observed for Max-Q and E+ steers. 533 and 543 metabolic features overlapped between all biological matrices in Max-Q (Fig. 4E) and E+ steers (Fig. 4G), respectively. Of these, 526 were shared, and 7 and 17 were distinct to Max-Q and E+, respectively (Fig. 4F). Among the distinct E+ features were metabolites putatively annotated as: 11-dimethoxydecane (m/z 102.1042 [M + H]), urea (m/z 121.0718 [2M + H]), L-kynurenine (m/z 209.0921 [M + H]), and (R)-Pterosin B (m/z 219.1379), plus several unannotated metabolites.
Targeted Epichloë and ergovaline correlation network analysis
E. (coenophiala) targeted network analysis in the fescue plant revealed one highly interconnected network of 29 fungal and 31 bacterial OTUs (Fig. 5A). Of note, the E. (coenophiala) OTU had the third highest centrality measure, preceded only by one Periconia and one Nocardioidaceae OTU (Fig. 5B). The classification of most OTUs in the network were unique (i.e., 1 OTU per family/genus/species). The only classified taxa with more than 1 OTU were Lichtheimia ramose, Neocallimastigaceae, Comamonadaceae, Dyadobacter, and Methylobacterium adhaesivum.
Figure 5.
Targeted correlation-based network analysis of significantly correlated features with (A, B) fescue plant Epichloë (coenophiala) OTU, (C, D) rumen liquid E.(coenophiala) OTU, and (E, F) ergovaline. (A) Network of toxic tall fescue plant (E+; P < 0.05) bacterial and fungal OTUs (B) respective centrality measurements with E. (coenophiala) marked in red; (C) network of toxic tall fescue grazing steers (E+; P < 0.05) ruminal bacterial and fungal OTUs with (D) respective centrality measurements with Epichloë marked in red; (E) focused network of toxic tall fescue grazing steers (E+; |r| > 0.6; P < 0.05) ruminal metabolic features significantly correlated with ruminal ergovaline and (F) respective centrality measurements with (ergovaline) marked in red. Blue and red nodes in (A, B, C, D) represent fungal and bacterial nodes, respectively. Yellow, green, blue, and white nodes in (E, F) indicate, respectively, ergovaline, metabolites involved in primary bile acid biosynthesis, metabolites involved in steroid hormone biosynthesis, and metabolites from unannotated pathways. E. (coenophiala) presence in the network is highlighted by arrows. Green and red lines indicate positive and negative correlations, respectively.
For the rumen, targeted analysis was performed using the E. (coenophiala) OTU in the rumen liquids; 180 total (27 fungi [8 solids, 19 liquids], 153 bacteria [61 solids, 92 liquids]) OTUs were significantly correlated with the Epichloë OTU (Fig. 5C). Of these, 4 liquid and 2 solid Orpinomyces sp, 2 solid and 1 liquid Cyllamyces aberensis, and 2 Neocallimastigaceae OTUs were the most prominent. Aspergillus cristatus and Piromyces sp were the other fungi in the rumen network that had an OTU from both rumen solids and rumen liquids. From the bacteria, Prevotella and Lachnospiraceae both had 14 liquid and 13 solid OTUs in the network. Other notable bacteria included BS11 (7 liquid, 4 solid OTUs), candidate CF231 (6 liquid OTUs), Butyrivibrio (6 liquid, 3 solid OTUs), and Ruminococcaceae (5 liquid OTUs). The Epichloë OTU had median centrality within the network (Fig. 5D).
Finally, E+ rumen targeted metabolite network using the putative E. coenophiala-derived metabolite ergovaline was performed with (|r| > 0.4; P < 0.05). 497 metabolic features (255 c18, 242 HILIC) were correlated with the ergovaline feature intensity profile (Fig. S7). Ergovaline-associated metabolites from the rumen liquids were involved in steroid hormone biosynthesis, tryptophan, tyrosine, amino and nucleotide sugar, glucose and energy, and Vitamin B6 metabolism. Considering the large size of this network, the analysis was further restricted (|r| > 0.6; P < 0.05). The newly generated network (Fig. 5E) had 43 metabolites, where ergovaline was at the center of the network and had the highest centrality measure (Fig. 5F). Pathway analysis revealed that most metabolites in the focused network were involved in steroid hormone, tryptophan, and tyrosine metabolism.
Differential integrative interactomics
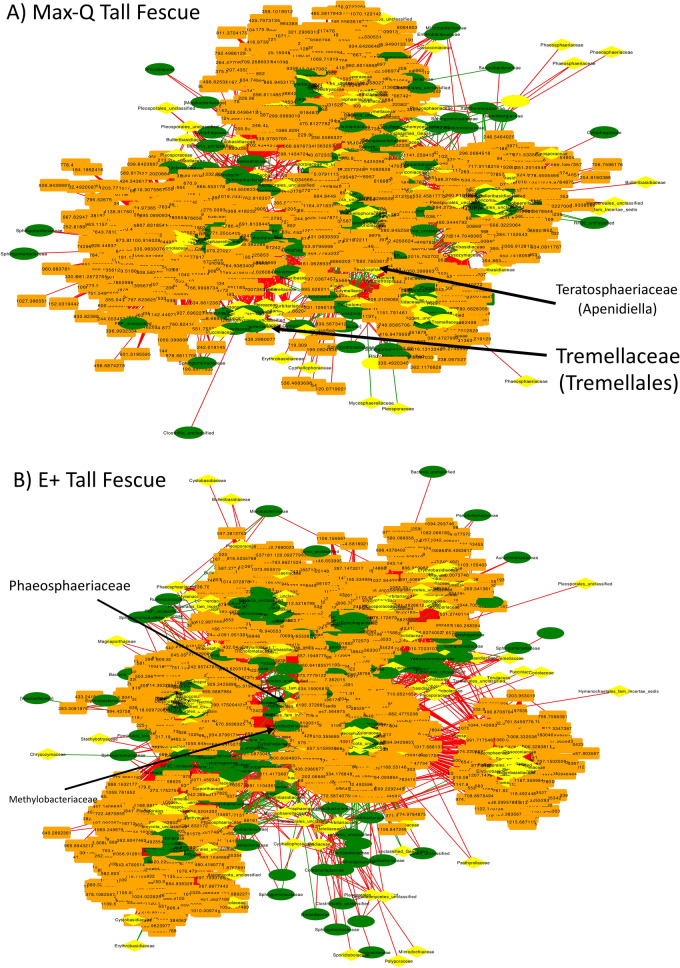
Fescue plant integrative interactomics
In both Max-Q and E+ tall fescue networks, bacterial and fungal OTUs were the central nodes in most clusters and were surrounded by associated metabolites (Fig. 6). The Max-Q network consisted of five clusters, whereas the E+ network had seven (Fig. 6). Of the nodes that had centrality measurements > 0.5, 71 were metabolites, 30 were bacteria, and 27 were fungi in the Max-Q network (File S2A); those in the E+ network consisted of 0 metabolites, 22 bacteria, and 34 fungi (File S2B). Cluster 5 of the E+ network (File S2B) had most nodes with high centrality measurements. Notably, in the E+ tall fescue network (Fig. 6; File S2B), no OTU aligning to E. (coenophiala), or higher taxonomic levels (e.g., Clavicipitaceae), was identified. Numerous OTUs were present only in the E+ tall fescue network, such as Clostridium (Ruminococcaceae), Cladosporium, and Mogibacteriaceae (File S2B). Interestingly, the steroid biosynthesis, nucleic acid, and glucosinolate biosynthesis pathways appeared in both Max-Q and E+ networks when querying nodes with centrality measurements above (> 0.2). Metabolic pathways singular to E+ include, among others, ubiquinone and other terpenoid-quinone biosynthesis, tyrosine metabolism, brassinosteroid biosynthesis, and valine, leucine and isoleucine metabolism.
Figure 6.
Global fescue plant integrative interactomics networks of relationships between bacterial (green, oval) and fungal (yellow, diamond) OTUs and metabolites (orange, rectangle) in the tall fescue plant within non-toxic (A; Max-Q; n = 6) or toxic (B; E+; n = 6) endophyte-infected plants. Green and red edges indicate positive and negative correlations, respectively. Select nodes of interest are highlighted by arrows and text.
Rumen integrative interactomics
Analysis of the rumen revealed one main cohort of clusters with a single cluster unattached to the main cohort in both Max-Q and E+ (Fig. 7). Additionally, in Max-Q (File S2C) and E+ (File S2D) networks, fungal OTUs had highest centrality measurements. While some fungi with high centrality measurements were present in both (e.g., Neocallimastigaceae), others were distinct. Fungi specific to the E+ network included Aspergillus, Leptospora, and Fusarium (Fig. 7; File S2D). The features with the next highest centrality measurements were bacterial OTUs and many were similar in Max-Q and E+ networks; however, Pyramidobacter and Treponema were unique to E+ (File S2C). E+ network-specific metabolic pathways included glycosphingolipid biosynthesis, metabolism of xenobiotics by cytochrome P450, and folate biosynthesis (Fig. 7).
Figure 7.
Global rumen integrative interactomics networks demonstrating relationships between bacterial (green, oval) and fungal (yellow, diamond) OTUs and metabolites (orange, rectangle) of non-toxic (A; Max-Q; n = 6) or toxic (B; E+; n = 6) grazing beef steers. Green and red edges indicate positive and negative correlations, respectively. Select nodes of interest are highlighted by arrows and text.
Global animal integrative interactomics
Global (i.e., rumen, plasma, urine, feces; microbiota and metabolomes) xMWAS resulted in two unique networks between Max-Q and E+ steers.
The nodes with the highest centrality measurements in the E+ network were from the fecal fungal, rumen solids/liquids bacterial, and fecal bacterial features (File S2F; eigenvector centrality > 0.7; rest of nodes centrality was less than 0.3); in the Max-Q network, those nodes were fecal fungi, rumen liquids bacteria, and fecal bacteria (File S2E; centrality > 0.84; rest less than 0.3). The fecal fungal OTUs within the Max-Q and E+ networks were quite distinct, with the E+ network having two Neocallimastigaceae Anaeromyces, one Aspergillus, one Acremonium brachypenium, and one Meyerozyma OTU (File S2F). Although ruminal liquid OTUs had some overlap between the two treatments (i.e., Lachnospiraceae), most were distinct. The OTUs solely in the E+ network aligned to unclassified Betaproteobacteria and Bacteroidetes, one from candidates BS11 and LD1-PB3, and one Ruminococcaceae and Mogibacteriaceae (File S2F). The only fecal bacterial OTUs unique to the E+ network were one Erysipelotrichaceae and one Anaerolinaceae OTU (File S2F). The rumen solid bacterial OTUs in the global E+ network were diverse (File S2F).
Interestingly, we sought to identify metabolites with high centrality (> 0.2) that were different between the Max-Q and E+ global networks (Fig. 8). Most node metabolites were urinary metabolites, with rumen liquid and plasma metabolites making up a smaller portion. Plasma metabolites unique to the E+ network were mainly involved in fatty acid and riboflavin metabolism. The rumen metabolic features unique to the E+ networks were primarily involved in steroid biosynthesis, folate biosynthesis, and metabolism of xenobiotics by cytochrome P450. The E+-specific urinary metabolites were associated with steroid hormone biosynthesis, purine, arachidonic acid, pentose phosphate pathway, and tryptophan and tyrosine metabolism. Urinary steroid hormone biosynthesis as a pathway appeared in both networks, but the metabolic features annotated within this pathway were network-specific. The Max-Q plot had three stand-alone clusters apart from the main network structure (Fig. 8A), which was not seen in the E+ network (Fig. 8B). Full node tables for the fescue plant, rumen, and global xMWAS analyses can be found in File S2.
Figure 8.
Global whole animal integrative interactomics networks of the relationships between bacterial (oval) and fungal (diamond) OTUs and metabolites (rectangle) in the rumen solid (green), rumen liquid (blue), plasma (orange), urine (yellow), and feces (brown) of either (A) non-toxic (Max-Q; n = 6) or (B) toxic (E+; n = 6) endophyte-infected tall fescue grazing beef steers. Green and red edges indicate positive and negative correlations, respectively.
Targeted animal integrative interactomics analysis
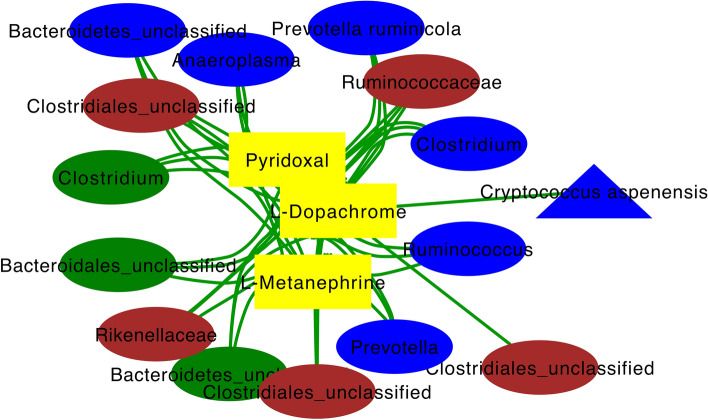
Targeted analysis assessed the multi-compartment relationship between metabolites and microbes that were significantly affected by E+ and present solely in a targeted E+ integrome network. One resultant cluster specific to E+ was centered on three urinary metabolic features that were highly associated (|r| > 0.7) with bacterial and fungal OTUs from every animal biological matrix (Fig. 9). The three metabolites central to this network are l-metanephrine, l-dopachrome, and pyridoxal, which are involved in tyrosine and Vitamin B6 metabolism, respectively (Fig. 9). OTUs associated with all three metabolites in the network include: Anaeroplasma, Prevotella ruminicola, Clostridium, Ruminococcus, and Prevotella (rumen liquids), Clostridium (rumen solids), Ruminococcaceae, Rikenellaceae (feces), other unclassified OTUs in all matrices (Fig. 9). Notably, one unclassified fecal bacterial OTU and one rumen liquid Cryptococcus aspenensis OTU were associated solely with urinary L-dopachrome in the targeted integrome network (Fig. 9).
Figure 9.
Targeted whole animal integrative interactomics networks of the relationships between bacterial (oval) and fungal (triangle) OTUs and metabolites (rectangle) in the rumen solid (green), rumen liquid (blue), urine (yellow), and feces (brown) of steers on toxic (E+; n = 6) endophyte-infected tall fescue. Metabolic pathways targeted in this analysis include tryptophan, tyrosine, Vitamin B6, steroid hormone, and bile acid metabolism. Green edges indicate positive correlations.
Discussion
Herein, is the first fescue toxicosis analysis using an integrative multi-‘omics approach that includes both the plant and animal. E. coenophiala infection and exposure significantly altered both the plant and animal multi-compartment microbiota and metabolomes, and led to unique integrome structure. Further, while there is little overlap between the plant and animal microbiota, some E+-associated metabolic changes were common to all biological matrices.
E. coenophiala infection altered the tall fescue phyllosphere microbiota, including increases in numerous plant-specific bacteria and fungi, g_Epichloë included. Plant microbiota is influenced by plant/environmental factors, including by endophytes such as E. coenophiala18–20. Most bacteria/fungi affected by E+ in the plant and animal were distinct, but it is significant that the Epichloë genus was increased in the E+ rumen liquids. While it is unlikely that aerobic microbes, fungi included, are able to thrive in the anaerobic ruminal environment, one study has found that anaerobic fungi within the GI tract of cattle have one life cycle stage that provides increased aerobic tolerance39, suggesting some tolerance flexibility. Considering this and reports of aerobic fungi being viable in bovine feces40, one explanation for the number of fungal OTUs that overlapped between all biological matrices is that the complex life cycle and sporulation of fungi could allow them to persist in adverse environments39,40.
Across the grazing trial, E+ exposure reduced most ruminal fungal taxa in the solid and liquid fractions. Fungi digest cellulose in the rumen; mycelium penetration of feed particles breaks fibers apart, increasing surface area for better degradation41. Their ability to degrade fibrous particles of feedstuffs is an important part of ruminant nutrient extraction41. Increased rumen fill, unexplained by increased dry matter intake, has been reported in FT6,42–44 and might reflect decreased ruminal passage rates. In this study, we noticed, but did not quantify, that E+ steers had greater rumen solid contents fraction. Considering the role ruminal fungi play in feed degradation, the relationship between fungal shifts in response to E+ exposure and alterations in rumen fill/passage rates should be evaluated, especially considering that specific fungal microbiota may provide tolerance to E+ exposure11.
While plant–animal carryover effects were limited to the microbiota, several metabolic pathways were affected by E+ in both the plant and animal. Some (e.g., tryptophan) pathways align with our earlier E+ grazing studies12–14, but we also identified other important pathways, such as Vitamin B6 metabolism, as significantly perturbed in all biological matrices. We previously reported altered urinary Vitamin B6 metabolism in fescue toxicosis14. Here, we found it altered in all biological matrices. Multiple forms of Vitamin B6 exist45 and it is an essential vitamin in humans and animals46. While plants synthesize it de novo, animals must obtain it through diet47,48. Alterations in Vitamin B6 metabolism in the plant and rumen could perturb downstream amino acid metabolism through microbial means49. Also, multiple enzymes in tryptophan metabolic pathways (e.g., kynureninase) use Vitamin B6 as an essential co-factor and its reduction decreases tryptophan metabolites in mice50,51. Notably, l-kynurenine was an E+-specific metabolite we found overlapping between all biological matrices (plant and animal). Vitamin B6 is also a co-factor for transaminases52 that are induced by glucocorticoids following stress, indicating possible widespread effects of Vitamin B6 alterations. Overall, plant and animal alterations in Vitamin B6 metabolism could influence subsequent amino acid metabolism that is consistently altered in E+ grazing studies13,14. Folate (Vitamin B9) biosynthesis was a metabolic pathway that appeared only in the E+ rumen and global integrative networks. Previously, it was demonstrated that thiamin (Vitamin B1) supplementation provided benefits to E+ grazing steers53. Thus, the relationship between vitamins in the B family with FT pathophysiology and, perhaps, broader vitamin supplementation/monitoring as a FT therapeutic is worthy of further investigation.
Interestingly, this study putatively identified ergovaline, the most prevalent toxic fescue ergopeptine7,54, via untargeted metabolomics. This is novel and confirming this metabolic feature by targeted means is important. The pattern of putative ergovaline detection aligns with what would be expected from the literature. No ergovaline was detected in biological matrices where he non-toxic Max-Q endophyte was used, which is expected since this endophyte was created to not produce ergot alkaloids while providing other agronomic benefits55–57. In E+, putative ergovaline was found in the fescue plant and rumen. In the plant, one-third of E+ samples had this m/z. Whether this is due to the untargeted metabolomics-based, non-ergot alkaloid specific58 extraction method, varied level of endophyte infection, small sample size, or other factors, is unknown. Given the small variability of stem and leaf ergovaline levels measured by targeted analysis59, small sample amount in combination with the broad, untargeted metabolomics extraction method is likely responsible for the non-uniform detection.
In the rumen of E+ steers, putative ergovaline was undetected before pasture placement, with levels increasing until peaking at Day 14. Urinary ergot alkaloids in this study indicate these steers were not recently exposed to toxic tall fescue pastures, so it was not expected to detect ergovaline on Day 0. The pattern of ergovaline in the rumen follows a similar pattern to total urinary ergot alkaloids, a sensitive biomarker of ergot alkaloid exposure24, until Day 14 of the grazing trial. Nominal decrease in ruminal ergovaline on Day 28 of the grazing trial could have multiple origins. We have found that urinary ergot alkaloid levels plateau or decrease and differences between the E+ and Max-Q microbiota become stable after 14 days of grazing E+ fescue in the fall, spring, and early summer12–14. This plateau, including herein, across seasons may indicate an adaptive response to E+ after 14 days on pasture and/or that steady state metabolism has been reached. If the animal microbiota shifted and/or cytochrome P450 levels increased60,61 to adapt to ergovaline and other ergot alkaloids, accelerated metabolism and/or biotransformation into less toxic metabolites would take place and result in decreased ruminal ergovaline towards the end of the study as observed here.
Targeted plant and rumen network analysis revealed most OTUs associated with the E. (coenophiala) OTU were distinct between the plant and animal. The bacterial family Lachnospiraceae, a family commonly affected by E+ in the grazing animal12–14, and the Orpinomyces genus were the only microbes that had OTUs significantly associated with E. (coenophiala) in both matrices. Notably, none of the Lachnospiraceae or Orpinomyces OTUs in the plant and the animal overlapped, indicating that sub-genus differences exist between what is present in the plant and the animal. The only fungus that was associated with E. (coenophiala) and was increased in the plant was one Leptospora sp OTU. A member of the Phaeosphaeriaceae family, which contains economically costly plant pathogens, some Leptospora sp relatives, have been identified as endophytes in monocotyledons plants62, like tall fescue. The genera Sphingomonas and Methylobacterium were associated with E. (coenophiala), but were decreased in E+ tall fescue. The relationship between them and E. (coenophiala) is unclear, but it seems possible that they could be competing for resources with Epichloë and/or be influenced by Epichloë secondary metabolites.
Within the rumen, most correlating OTUs were bacteria, but some notable fungi associated with E. (coenophiala) OTU too. The Neocallimastigaceae family, namely the Orpinomyces genus, had the majority of fungal Epichloë-associated fungal OTUs. Notably, two Orpinomyces OTUs were correlated with Epichloë in both the rumen solids and rumen liquids, one positively and one negatively. Of the 15 rumen liquid Prevotella OTUs, 10 were negatively correlated and 5 were positively correlated; of the 13 rumen solid OTUs, 3 were negatively correlated and 10 were positively correlated. Overall, for most taxa with multiple correlations, there was a mix of positively and negatively correlated OTUs, indicating that sub-genus targeted analysis of these fungi/bacteria might help understanding the complex rumen microbiota relationship in FT. Such work, in a different context, has outlined how different strains of active dry yeast influence ruminal acidosis and methane production63 and found strain-specific carbohydrate-utilization patterns in ruminal bacteria64. These data highlight the feasibility of a sub-genus, targeted microbiota analyses in FT context.
Targeted ergovaline network analysis revealed highly interconnected network between rumen metabolites. The majority of the metabolites in this network were involved in steroid hormone biosynthesis, tryptophan, tyrosine, and Vitamin B6 metabolism. This is interesting considering these metabolites are components of metabolic pathways most significantly affected by E+ grazing. Previously, we found that E+ altered tryptophan and tyrosine metabolism13,14. So, while putative ergovaline did not have the highest centrality in the full network, the metabolic pathways it was associated with were the same ones identified by broader metabolomics methods. In the focused networks, the only unique affected pathways were steroid hormone and primary bile acid biosynthesis and ergovaline had the highest centrality of all metabolic features. It has been previously shown that ergot alkaloids can influence systemic hormonal homeostasis65, but current data indicate ergovaline and E+ tall fescue grazing may begin to induce hormonal imbalances presystemically, i.e., in the rumen; however, this will require further investigation.
Integrative analysis revealed the general structures of the global Max-Q and E+ networks were similar (i.e., microbial nodes as anchors with peripherally associated metabolites), but the constituents of the networks were distinct. Notably, in both networks, fecal fungal OTUs had the highest centrality measurements, but the fungal classifications were mostly network-specific. One of the E+-unique genera was fecal Neocallimastigaceae Anaeromyces. Although Anaeromyces was not a genus reported, it was previously found that the Neocallimastigaceae family was increased in steers with greater tolerance to E+ exposure, indicating this family may be important in the structure of the E+ integrome and play a modulatory role in the severity of FT11. Considering that E+ reduced the abundance of most fungal taxa, yet ruminal solid fungal OTUs appear only in the E+ integrome, further exploration of the specific influence of E+ on ruminal fungi homeostasis is warranted.
Previously, urinary ergot alkaloids have been proposed as a sensitive biomarker of exposure12–14,24,25 and, potentially, a biomarker of effect24 for E+ in beef cattle. Although these provide great utility for producers and scientists alike, additional biomarkers that encapsulate the molecular mechanisms of E+-induced decreased weight gains may be therapeutically valuable. In our search for subsequent/supplemental biomarkers, we identified a ruminal Epichloë OTU only in E+ animals that was most abundant after 14 days of grazing. As this E+ specific OTU did not track well with pasture alkaloids, urinary ergot alkaloids, or weight gains in E+ steers, its identification is interesting, likely consequential, but not an ideal biomarker as its presence might be related to endophyte breakdown together with the plant material in the rumen.
Akin to what we found for plasma/urinary metabolites having utility as a biomarker of a decreased productivity-associated hindgut microbiota12, targeted global integrative analyses performed herein revealed three urinary metabolic features (l-metanephrine, l-dopachrome, and pyridoxal) positively associated with the E+ microbiota in multiple animal matrices. All three were urinary metabolites, not plasma or rumen liquid, indicating urine can be discriminatory between Max-Q and E+ steers and ideal for easy-to-access biomarkers of FT. Urinary metanephrines have shown equal utility as plasma metanephrines as a diagnostic biomarker of pheochromocytoma66 and urinary dopachrome tautomerase protein has been suggested as a potentially sensitive biomarker of drug-induced liver injury67. We reported that several urinary catecholamines in E+ steers after 28 days on pasture are altered13. This, together with the current results, suggests pathways associated with urinary catecholamines are consistently perturbed in FT. The positive association of metanephrine and dopachrome with microbiota from multiple compartments hints that these urinary metabolites may be useful biomarkers for FT from a therapeutics perspective. Urinary pyridoxal was the other metabolite appearing in this focused network, which is notable, considering Vitamin B6 metabolism was perturbed in all plant and animal matrices tested. Finally, these data provide foundational evidence that show the previous perturbations we have identified12–14 are snapshots of systemic perturbations that occur in E+ grazing steers; understanding the multi-level, multi-compartment, integrome will provide more actionable insights. Although the relationship between urinary l-dopachrome and Cryptococcus aspenensis in FT context is unclear, it is interesting that l-dopachrome was the only feature associated with this fungal OTU.
In this novel study, effects of E. coenophiala infection on the plant and animal microbiota, metabolome, and the multi-compartment, multi-‘omics integration are presented. The data suggest the majority of the microbiota profiles, and the effects of E+, are distinct between the plant and the animal, but effects of E+ on the plant and multi-compartment animal metabolome shared some similarities. Herein, is the first overview of the complex interactions between the bovine multi-compartment microbiota, both bacterial and fungal, and metabolome; these relationships are endophyte-specific. We found that only the E+ integrome had rumen solid OTUs, indicating these may be an important microbial point-of-origin for E+ pathophysiology. Overall, these data align with our previous work showing E+ disrupts plasma and urine metabolic and fecal microbiota homeostasis12–14. Additionally, our current finding that E+ begins altering the microbiota/metabolome in planta and in the rumen of toxic fescue grazing steers and these changes associate with plasma/urine metabolic and fecal microbiota changes is novel; it suggests a complex, systemic pathophysiological response in FT. Future therapeutic- or management-based intervention strategies, as well as detailed evaluation of adaptive vs pathophysiological responses to E+ and in the context of other complex diseases, should take advantage of such integrome-based integrative analyses.
Supplementary Information
Acknowledgements
This research was funded from the National Institute of Food and Agriculture (NIFA) Agriculture and Food Research Initiative (AFRI) Grants # 67030-25004 and 67015-31301 to NMF. We would also like to thank the Interdisciplinary Toxicology Program, the Department of Physiology and Pharmacology, and the Graduate School of the University of Georgia for partial support to RSM. Help with research, animal handling and care, and other assistance from the skillful personnel at the J. Phil Campbell Natural Resources Conservation Center of the University of Georgia (Watkinsville, GA) is greatly appreciated.
Author contributions
N.M.F. conceived and designed the study. R.S.M., N.S.H., J.M.C., J.M.L., T.R.C., and N.M.F. assisted in study design, execution, sample collection and storage and some lab work. J.H.S. and G.S. performed the next-generation sequencing of all samples. V.T.T., M.R.S., and D.P.J. contributed the high-resolution metabolomics analysis and feature extraction and quantification for all samples. R.S.M. performed the bioinformatics analysis and wrote the draft of the manuscript with input from N.M.F., J.H.S., and G.S. R.S.M. and N.M.F. edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08540-2.
References
- 1.Paterson J, Forcherio C, Larson B, Samford M, Kerley M. The effects of fescue toxicosis on beef cattle productivity. J. Anim. Sci. 1995;73:889–898. doi: 10.2527/1995.733889x. [DOI] [PubMed] [Google Scholar]
- 2.Leuchtmann A, Bacon CW, Schardl CL, White JF, Jr, Tadych M. Nomenclatural realignment of Neotyphodium species with genus Epichole. Mycologia. 2014;106:202–215. doi: 10.3852/13-251. [DOI] [PubMed] [Google Scholar]
- 3.Clay K, Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 2002;160(Suppl 4):S99–S127. doi: 10.1086/342161. [DOI] [PubMed] [Google Scholar]
- 4.Thompson FN, Stuedemann JA. Pathophysiology of fescue toxicosis. Agr. Ecosyst. Environ. 1993;44:263–281. doi: 10.1016/0167-8809(93)90050-y. [DOI] [Google Scholar]
- 5.Foote AP, Harmon DL, Strickland JR, Bush LP, Klotz JL. Effect of ergot alkaloids on contractility of bovine right ruminal artery and vein. J. Anim. Sci. 2011;89:2944–2949. doi: 10.2527/jas.2010-3626. [DOI] [PubMed] [Google Scholar]
- 6.Foote AP, et al. Ergot alkaloids from endophyte-infected tall fescue decrease reticuloruminal epithelial blood flow and volatile fatty acid absorption from the washed reticulorumen. J. Anim. Sci. 2013;91:5366–5378. doi: 10.2527/jas.2013-6517. [DOI] [PubMed] [Google Scholar]
- 7.Klotz JL. Activities and effects of ergot alkaloids on livestock physiology and production. Toxins (Basel) 2015;7:2801–2821. doi: 10.3390/toxins7082801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayers AW, et al. Ruminal metabolism and transport of tall fescue ergot alkaloids. Crop Sci. 2009;49:2309–2316. doi: 10.2135/cropsci2009.01.0018. [DOI] [Google Scholar]
- 9.Harlow BE, et al. Ruminal tryptophan-utilizing bacteria degrade ergovaline from tall fescue seed extract. J. Anim. Sci. 2017;95:980–988. doi: 10.2527/jas.2016.1128. [DOI] [PubMed] [Google Scholar]
- 10.Melchior EA, et al. Effects of red clover isoflavones on tall fescue seed fermentation and microbial populations in vitro. PLoS ONE. 2018;13:e0201866. doi: 10.1371/journal.pone.0201866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koester L, Poole D, Serão N, Schmitz-Esser S. Beef cattle that respond differently to fescue toxicosis have distinct gastrointestinal tract microbiota. PLoS ONE. 2020;15:e0229192. doi: 10.1371/journal.pone.0229192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mote RS, et al. Response of beef cattle fecal microbiota to grazing on toxic tall fescue. Appl. Environ. Microbiol. 2019;85:e00032–00019. doi: 10.1128/AEM.00032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mote RS, et al. Metabolomics of fescue toxicosis in grazing beef steers. Food Chem. Toxicol. 2017;105:285–299. doi: 10.1016/j.fct.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Mote RS, et al. Toxic tall fescue grazing increases susceptibility of the Angus steer fecal microbiota and plasma/urine metabolome to environmental effects. Sci. Rep. 2020;10:2497. doi: 10.1038/s41598-020-59104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lourenco J, et al. Comparison of the ruminal and fecal microbiotas in beef calves supplemented or not with concentrate. PLoS ONE. 2020;15:e0231533. doi: 10.1371/journal.pone.0231533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson G, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackmann TJ, Spain JN. Invited review: Ruminant ecology and evolution: Perspectives useful to ruminant livestock research and production. J. Dairy Sci. 2010;93:1320–1334. doi: 10.3168/jds.2009-2071. [DOI] [PubMed] [Google Scholar]
- 18.Wallace JG, Kremling KA, Kovar LL, Buckler ES. Quantitative genetics of the maize leaf microbiome. Phytobiomes J. 2018;2:208–224. doi: 10.1094/pbiomes-02-18-0008-r. [DOI] [Google Scholar]
- 19.Zarraonaindia I, et al. The soil microbiome influences grapevine-associated microbiota. mBio. 2015;6:e02527–e2614. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorholt JA. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 21.Fierer N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 22.Hardoim PR, et al. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mote RS, Filipov NM. Use of integrative interactomics for improvement of farm animal health and welfare: An example with fescue toxicosis. Toxins (Basel) 2020;12:633. doi: 10.3390/toxins12100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill NS, Thompson FN, Stuedemann JA, Dawe DL, Hiatt EE., 3rd Urinary alkaloid excretion as a diagnostic tool for fescue toxicosis in cattle. J. Vet. Diagn. Investig. 2000;12:210–217. doi: 10.1177/104063870001200303. [DOI] [PubMed] [Google Scholar]
- 25.Stuedemann JA, et al. Urinary and biliary excretion of ergot alkaloids from steers that grazed endophyte-infected tall fescue. J. Anim. Sci. 1998;76:2146–2154. doi: 10.2527/1998.7682146x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor DL, et al. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for illumina amplicon sequencing. Appl. Environ. Microbiol. 2016;82:7217. doi: 10.1128/AEM.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson RH, et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uppal K, Walker DI, Jones DP. xMSannotator: An R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017;89:1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart D, et al. T3DB: The toxic exposome database. Nucleic Acids Res. 2015;43:D928–D934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hmisc: Harrell Miscellaneous. https://cran.r-project.org/web/packages/Hmisc/index.html (2018).
- 35.Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: Network visualizations of relationships in psychometric data. J. Stat. Softw. 2012;1(4):1–18. doi: 10.18637/jss.v048.i04. [DOI] [Google Scholar]
- 36.Uppal K, Ma C, Go YM, Jones DP, Wren J. xMWAS: A data-driven integration and differential network analysis tool. Bioinformatics. 2018;34:701–702. doi: 10.1093/bioinformatics/btx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes CT, et al. Cytoscape Web: An interactive web-based network browser. Bioinformatics (Oxford, England) 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies D, Theodorou M, Lawrence M, Trinci A. Distribution of anaerobic fungi in the digestive tract of cattle and their survival in faeces. J. Gen. Microbiol. 1993;139:1395–1400. doi: 10.1099/00221287-139-6-1395. [DOI] [PubMed] [Google Scholar]
- 40.McGranaghan P, Davies JC, Griffith GW, Davies DR, Theodorou MK. The survival of anaerobic fungi in cattle faeces. FEMS Microbiol. Ecol. 1999;29:293–300. doi: 10.1111/j.1574-6941.1999.tb00620.x. [DOI] [Google Scholar]
- 41.Russell JB. Rumen Microbiology and Its Role in Ruminant Nutrition. James B. Russell; 2002. [Google Scholar]
- 42.Koontz AF, Kim DH, McLeod KR, Klotz JL, Harmon DL. Effect of fescue toxicosis on whole body energy and nitrogen balance, in situ degradation and ruminal passage rates in Holstein steers. Anim. Prod. Sci. 2015;55:988. doi: 10.1071/an14037. [DOI] [Google Scholar]
- 43.Koontz AF, et al. Evaluation of a ruminally dosed tall fescue seed extract as a model for fescue toxicosis in steers. J. Anim. Sci. 2012;90:914–921. doi: 10.2527/jas.2011-4292. [DOI] [PubMed] [Google Scholar]
- 44.Koontz AF, et al. Alteration of fasting heat production during fescue toxicosis in Holstein steers. J. Anim. Sci. 2013;91:3881–3888. doi: 10.2527/jas.2013-6232. [DOI] [PubMed] [Google Scholar]
- 45.Rosenberg I. A history of the isolation and identification of vitamin B (6) Ann. Nutr. Metab. 2012;61:236–238. doi: 10.1159/000343113. [DOI] [PubMed] [Google Scholar]
- 46.Percudani R, Peracchi A. The B6 database: A tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinform. 2009;10:1–8. doi: 10.1186/1471-2105-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fitzpatrick, T. B. in Advances in Botanical Research Vol. 59 (eds Fabrice Rébeillé & Roland Douce) 1–38 (Academic Press, 2011).
- 48.Tambasco-Studart M, et al. Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13687. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohammed N, Onodera R, Itabashi H, Lila ZA. Effects of ionophores, vitamin B6 and distiller's grains on in vitro tryptophan biosynthesis from indolepyruvic acid, and production of other related compounds by ruminal bacteria and protozoa. Anim. Feed Sci. Technol. 2004;116:301–311. doi: 10.1016/j.anifeedsci.2004.07.017. [DOI] [Google Scholar]
- 50.Bender DA, Njagi EN, Danielian PS. Tryptophan metabolism in vitamin B6-deficient mice. Br. J. Nutr. 1990;63:27–36. doi: 10.1079/bjn19900089. [DOI] [PubMed] [Google Scholar]
- 51.da Silva VR, et al. Metabolite profile analysis reveals functional effects of 28-day vitamin B-6 restriction on one-carbon metabolism and tryptophan catabolic pathways in healthy men and women. J. Nutr. 2013;143:1719–1727. doi: 10.3945/jn.113.180588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romo AJ, Liu H-W. Mechanisms and structures of vitamin B(6)-dependent enzymes involved in deoxy sugar biosynthesis. Biochem. Biophys. Acta. 2011;1814:1534–1547. doi: 10.1016/j.bbapap.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lauriault L, Dougherty C, Bradley N, Cornelius P. Thiamin supplementation and the ingestive behavior of beef cattle grazing endophyte-infected tall fescue. J. Anim. Sci. 1990;68:1245–1253. doi: 10.2527/1990.6851245x. [DOI] [PubMed] [Google Scholar]
- 54.Klotz JL, Nicol AM. Ergovaline, an endophytic alkaloid. 1. Animal physiology and metabolism. Anim. Prod. Sci. 2016;56:1761–1774. doi: 10.1071/an14962. [DOI] [Google Scholar]
- 55.Bouton JH, et al. Reinfection of tall fescue cultivars with non-ergot alkaloid-producing endophytes. Agron. J. 2002;94:567–574. doi: 10.2134/agronj2002.5670. [DOI] [Google Scholar]
- 56.Parish JA, et al. Use of nonergot alkaloid-producing endophytes for alleviating tall fescue toxicosis in stocker cattle1,2. J. Anim. Sci. 2003;81:2856–2868. doi: 10.2527/2003.81112856x. [DOI] [PubMed] [Google Scholar]
- 57.Gunter SA, Beck PA. Novel endophyte-infected tall fescue for growing beef cattle1. J. Anim. Sci. 2004;82:E75–E82. doi: 10.2527/2004.8213_supplE75x. [DOI] [PubMed] [Google Scholar]
- 58.Patel RM, et al. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion. 2015;55:544–552. doi: 10.1111/trf.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rottinghaus GE, Garner GB, Cornell CN, Ellis JL. HPLC method for quantitating ergovaline in endophyte-infested tall fescue: Seasonal variation of ergovaline levels in stems with leaf sheaths, leaf blades, and seed heads. J. Agric. Food Chem. 1991;39:112–115. doi: 10.1021/jf00001a022. [DOI] [Google Scholar]
- 60.Settivari RS, Evans TJ, Rucker E, Rottinghaus GE, Spiers DE. Effect of ergot alkaloids associated with fescue toxicosis on hepatic cytochrome P450 and antioxidant proteins. Toxicol. Appl. Pharmacol. 2008;227:347–356. doi: 10.1016/j.taap.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Moubarak AS, Rosenkrans CF., Jr Hepatic metabolism of ergot alkaloids in beef cattle by cytochrome P450. Biochem. Biophys. Res. Commun. 2000;274:746–749. doi: 10.1006/bbrc.2000.3210. [DOI] [PubMed] [Google Scholar]
- 62.Phookamsak R, et al. Revision of phaeosphaeriaceae. Fungal Divers. 2014;68:159–238. doi: 10.1007/s13225-014-0308-3. [DOI] [Google Scholar]
- 63.Chung YH, Walker ND, McGinn SM, Beauchemin KA. Differing effects of 2 active dried yeast (Saccharomyces cerevisiae) strains on ruminal acidosis and methane production in nonlactating dairy cows. J. Dairy Sci. 2011;94:2431–2439. doi: 10.3168/jds.2010-3277. [DOI] [PubMed] [Google Scholar]
- 64.Klassen L, et al. Quantifying fluorescent glycan uptake to elucidate strain-level variability in foraging behaviors of rumen bacteria. Microbiome. 2021;9:23. doi: 10.1186/s40168-020-00975-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliver, J. W. in Neotyphodium in Cool-Season Grasses. (eds C.A. Roberts, C.P. West, & D.E. Spiers) 291–304 (Blackwell Publ., 2005).
- 66.Grouzmann E, et al. Diagnostic accuracy of free and total metanephrines in plasma and fractionated metanephrines in urine of patients with pheochromocytoma. Eur. J. Endocrinol. 2010;162:951–960. doi: 10.1530/EJE-09-0996. [DOI] [PubMed] [Google Scholar]
- 67.van Swelm RPL, et al. Identification of novel translational urinary biomarkers for acetaminophen-induced acute liver injury using proteomic profiling in mice. PLoS ONE. 2012;7:e49524. doi: 10.1371/journal.pone.0049524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.