Abstract
Despite several reports worldwide documenting the presence of Rickettsia asembonensis in samples derived from ectoparasites, animals and more recently humans, genomic information of these specimens remains scarce, and when available, is usually limited to small genomic fragments of limited value. We generated complete sequences for two conserved (17-kDa antigen gene and gltA) and three variable (sca4, ompB and ompA) genes in five R. asembonensis DNA samples detected in cat and dog fleas in Peru. Complete gene sequences were used to conduct multi-locus sequence typing and phylogenetic analyses to assess diversity and infer relationships among strains and other reference sequences. The 17-kDa antigen gene was highly conserved across Rickettsia species. Of the variable genes ompB was the most variable, but this diversity was not captured through phylogenetics alone even when efforts were made to maximize potential diversity in terms of flea species, animal host and location. Through a combination of de novo and reference-based genome assembly we identified a 75 bp insertion in ompA that encodes a 25 aa repetitive motif found in other Rickettsia species, but not present in the original prototype strain from Kenya. R. asembonensis has only recently been shown to be a bona-fide human pathogen. As such, and compounded by a lack of available genomic information, it remains understudied. Our work directly addresses the lack of genomic information available worldwide for the study of these novel Rickettsia species and specifically contributes to our understanding of the diversity and molecular epidemiology of R. asembonensis in Peru.
Keywords: Multilocus sequence typing, Rickettsia asembonensis, Peru
Introduction
Rickettsia asembonensis is a new Rickettsia species that belongs to the R. felis-like group of organisms (RFLOs) within the Transitional Group of Rickettsia (Maina et al. 2018). In recent years several new Rickettsia species, including R. asembonensis, have been identified in ectoparasites and described as emerging pathogens with world-wide distribution (Maina et al. 2018, Bitencourth et al. 2019, Blanton et al. 2019a, Kho et al. 2019, Moonga et al. 2019, Nziza et al. 2019, Schott et al. 2019, Betancourt-Ruiz et al. 2020, Eremeeva et al. 2020, Nataraj et al. 2020). The majority of these novel pathogens were initially identified in ectoparasites that are common transmission vectors for several human infectious diseases.
Given their ectoparasite hosts, and given a close phylogenetic relationship to other Rickettsia species known to infect humans, such as R. felis (Perez-Osorio et al. 2008), it was expected that several of these novel pathogens would also turn out to be bona-fide human pathogens. In the case of R. asembonensis, confirmation arrived late in 2018, when Peruvian researchers reported the presence of R. asembonensis in human samples derived from patients suffering from nonspecific acute febrile illness, between 2009 and 2012, in multiple Peruvian cities with historical evidence of rickettsial disease (Palacios-Salvatierra et al. 2018).
In that respect, it is likely that R. asembonensis is an emergent flea-borne pathogen in multiple areas of Peru, where rickettsial infections frequently go underdiagnosed and underreported, both because of an overlap in clinical symptoms caused by other emerging pathogens and because of a general lack of molecular tools for diagnosis and characterization of rickettsial strains (Blair et al. 2004a, Forshey et al. 2010, Kocher et al. 2017, Palacios-Salvatierra et al. 2017, Ricapa-Antay et al. 2018, Salmon-Mulanovich et al. 2019).
R. asembonensis was first isolated from fleas collected in Kenya and characterized by multilocus sequence typing (MLST) (Jiang et al. 2013). Since then, we have witnessed an explosion of reports that documented the presence of R. asembonensis in a variety of sample types around the world. In a few cases, these reports have been accompanied by genomic information that supports the existence of multiple highly similar strains (Maina et al. 2018). One such highly similar, yet distinct, RFLO is Rickettsia sp. strain RF2125, which was first detected in ectoparasites of domestic animals collected in Thailand between 2001 and 2002, and subsequently detected in a human sample from Malaysia in 2016 (Parola et al. 2003, Kho et al. 2016). Despite sporadic reports, complete sequence data for RFLOs, and for R. asembonensis in particular, have remained scarce.
We previously reported MLST characterization of five complete genes from the first Peruvian R. asembonensis strain, identified in an ectoparasite collected from a domestic animal in the city of Iquitos in the Peruvian Amazon (Loyola et al. 2018). Since then, we have identified multiple additional ectoparasites that have also tested positive for R. asembonensis in a separate study conducted in Puerto Maldonado, a different Amazonian city in Peru (Salmon-Mulanovich et al. 2019). In this study, we report complete sequences for five genes used in MLST characterization of five independent R. asembonensis DNA samples, and we assess their diversity among other Peruvian R. asembonensis.
Given that to date, only the original Kenyan and the first Peruvian DNA specimens have reported enough complete genomic information of the type needed to carry out full MLST characterizations and phylogenetic analyses based on complete genes, this report represents a significant contribution toward addressing the scarcity of genomic information available to study these novel pathogens. As additional complete Rickettsia DNA sequences become available from other parts of the world, we will be able to better understand the global evolutionary relationships among Rickettsia species in general, and in pathogenic species such as R. asembonensis in particular.
Materials and Methods
Ectoparasite selection
We previously detected R. asembonensis in ectoparasites from domestic and peridomestic animals from Iquitos (3°44′S, 73°15′W) and Puerto Maldonado (12°36′S, 69°11′W) in Peru (Kocher et al. 2016, Salmon-Mulanovich et al. 2019). All samples described in this study were derived from those two studies, where ectoparasites had been previously confirmed as Rickettsia-positive by a quantitative real-time PCR (qPCR) assay (Rick17b) that targets the conserved 17-kDa surface antigen gene, and confirmed as R. asembonensis-positive by a species-specific qPCR assay (Rasemb) that targets a portion of the variable ompB gene (Jiang et al. 2012, 2013).
For this study, we selected five ectoparasite DNA samples that were Rick17b and Rasemb positive after considering and maximizing diversity for the available variables at play, which included geographical location (Iquitos and Puerto Maldonado), host type (cats and dogs), and ectoparasite type (Ctenocephalides felis and Ctenocephalides canis) (Table 1). All specimens selected for analysis were collected under studies approved by NAMRU-6's Institutional Review Board and Institutional Animal Care and Use Committee in compliance with all applicable regulations.
Table 1.
Source Characteristics of the Five Rickettsia asembonensis DNA Specimens Used for Multilocus Sequence Typing Characterization
| Specimen name | City | Host | Ectoparasite |
|---|---|---|---|
| 8294D3 | Iquitos | Canis lupus familiaris (Dog) | Ct. canis |
| 8556D1 | Canis lupus familiaris (Dog) | Ct. canis | |
| VGC2 | Felis catus (Cat) | Ct. felis | |
| LER197 | Puerto Maldonado | Canis lupus familiaris (Dog) | Ct. canis |
| LER205 | Felis catus (Cat) | Ct. felis |
NGS library preparation and sequencing
Genomic DNA was extracted individually from ectoparasites halves by mechanical disruption and used to prepare Ion Torrent and/or Ion Proton NGS libraries following the manufacturer's directions as previously described (Kocher et al. 2016, Loyola et al. 2018). Libraries were sequenced on PI v.2 chips using ion PI Hi-Q sequencing 200 kits (ThermoFisher, A26433). In some instances, libraries were sequenced more than once.
Bioinformatics processing
Before the analyses, raw shot-gun sequencing data from individual sample codes sequenced multiple times were first merged into a single file using SamTools (Li et al. 2009). Merged, sequencing data were then quality trimmed and filtered (<Q20) using PRINSEQ-lite v0.20.4 (Schmieder et al. 2011) and sequencing adapters were removed using Cutadapt v1.15 (Martin 2011). Preprocessed reads were initially de novo assembled into contigs with MEGAHIT V1.1.3 (Li et al. 2015).
Given that multiple contigs were generated during the de novo assembly process, these were subsequently reference-mapped with Bowtie2 v2.3.4.1 (Langmead et al. 2012) to the five open reading frames (ORFs) of interest: the conserved 17-kDa antigen gene, the conserved citrate synthase gene (gltA), the variable outer membrane protein A gene (ompA), the variable outer membrane protein B gene (ompB), and the variable surface cell antigen 4 gene (sca4). Raw reads were also reference-mapped to the five ORFs of interest and the resulting consensus sequences compared to those generated by de novo assembly. For reference mapping, we used reference sequences previously reported for the first R. asembonensis strain VGD7 isolated from an ectoparasite collected in Peru (Accession numbers KY650696-KY650700).
When discrepancies were identified between the consensus sequences generated by de novo assembly versus reference-assembly, as in the in/del reported for ompA, the consensus sequence generated via de novo assembly was given priority. All new consensus sequences generated as part of this study have been deposited in GenBank (Accession numbers MK923720-MK92374).
MLST and phylogenetic analysis
MLST analysis was independently performed at the nucleotide and amino acid levels for all five genes of interest from each R. asembonensis DNA against both the Peruvian R. asembonensis reference sample VGD7 and the Kenyan R. asembonensis reference strain NMRCii (JWSW01000078.1). Given the 75 nt in/del detected in ompA, this gene was further analyzed to explore functional domains along the full ORF (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Validated reference Rickettsia species of the transitional group (Gillespie et al. 2012) (R. akari, R. australis, R. felis, and members of the RFLO) were downloaded from GenBank (Table 2).
Table 2.
Reference Sequences of Rickettsia Species Used in the ompB Phylogenetic Analysis
| Rickettsia species | Nucleotides | Accession number |
|---|---|---|
| R. asembonensis type strain NMRCii | 4947 | JWSW01000078 |
| R. asembonensis strain VGD7 | 4947 | KY650699.1 |
| R. akari strain Hartford | 4977 | CP000847.1 |
| R. akari strain Kaplan | 4900 | AF123707.1 |
| R. australis strain Cutlack | 4935 | CP003338.1 |
| R. australis strain Phillips | 4903 | AF123709.1 |
| R. felis | 5225 | AF182279.1 |
| R. felis strain Ar3 | 5180 | GQ385243.1 |
| R. felis strain URRWXcal2 | 4965 | CP000053.1 |
| R. helvetica strain AS819 | 4848 | MF163037.1 |
| R. helvetica strain C9P9 | 4850 | AF123725.1 |
| R. helvetica strain Komi | 4833 | KP866151.1 |
| R. helvetica strain Novosibirsk-08-5 | 4850 | KU310591.1 |
| R. hoogstraalii strain RCCE3 | 5077 | EF629536.1 |
| R. senegalensis strain Cf_US_0036D | 4290 | KT304219.1 |
| R. senegalensis strain PU01-02 | 4858 | KF666470.1 |
Given that complete ORFs of the ompB gene were not available from public repositories for all the Rickettsia specimens of interest, we used partial ORFs (4263 nt, 86% of complete ORF) for phylogenetic analysis. Sequences were aligned using MUSCLE and trees were generated using the maximum likelihood algorithm with 2000 bootstrap replicates in Mega 10.0.5 (Edgar 2004, Kumar et al. 2018). The analysis, which included genetic distances, was calculated using a General Time Reversible Gamma Distributed model.
Results
We have characterized five R. asembonensis DNA specimens initially detected in ectoparasites collected from domestic dogs and cats in Iquitos and Puerto Maldonado in Peru (Table 1). We used genomic information derived from next-generation sequencing data to assemble complete ORFs for two conserved (17-kDa antigen gene [480 nt] and gltA [1314 nt]) and three variable (ompA [5151 nt], ompB [4947 nt], and sca4 [3033 nt]) genes from each specimen. MLST analyses revealed a high degree of conservation for most genes (Table 3). For the 17-kDa antigen gene, we detected no mutations at either the nucleotide or amino acid levels between the five strains characterized here and both the Peruvian (VGD7) and Kenyan (NMRCii) reference strains used in the analysis.
Table 3.
Multilocus Sequence Typing of Complete Genes of Five Rickettsia asembonensis Specimens from Iquitos and Puerto Maldonado in Peru
| Peruvian R. asembonensis strain VGD7 |
Kenyan R. asembonensis strain NMRCii |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Conserved genes |
Variable genes |
Conserved genes |
Variable genes |
||||||||
| 17-kDa | gltA | sca4 | ompB | ompA | 17-kDa | gltA | sca4 | ompB | ompA | ||
| 8294D3 | #changes: nt/aa | 0/0 | 0/0 | 0/0 | 3/2 | 75/25 | 0/0 | 3/1 | 5/4 | 3/2 | 80/29 |
| %id: nt/aa | 100/100 | 100/100 | 100/100 | 99.94/99.88 | 98.54/98.54 | 100/100 | 99.77/99.77 | 99.84/99.60 | 99.94/99.88 | 98.45/98.31 | |
| 8556D1 | #changes: nt/aa | 0/0 | 0/0 | 0/0 | 4/2 | 75/25 | 0/0 | 3/1 | 5/4 | 4/2 | 80/29 |
| %id: nt/aa | 100/100 | 100/100 | 100/100 | 99.92/99.88 | 98.54/98.54 | 100/100 | 99.77/99.77 | 99.84/99.60 | 99.92/99.88 | 98.45/98.31 | |
| VGC2 | #changes: nt/aa | 0/0 | 0/0 | 0/0 | 3/2 | 75/25 | 0/0 | 3/1 | 5/4 | 3/2 | 80/29 |
| %id: nt/aa | 100/100 | 100/100 | 100/100 | 99.94/99.88 | 98.54/98.54 | 100/100 | 99.77/99.77 | 99.84/99.60 | 99.94/99.88 | 98.45/98.31 | |
| LER197 | #changes: nt/aa | 0/0 | 0/0 | 0/0 | 3/2 | 75/25 | 0/0 | 3/1 | 5/4 | 3/2 | 80/29 |
| %id: nt/aa | 100/100 | 100/100 | 100/100 | 99.94/99.88 | 98.54/98.54 | 100/100 | 99.77/99.77 | 99.84/99.60 | 99.94/99.88 | 98.45/98.31 | |
| LER205 | #changes: nt/aa | 0/0 | 0/0 | 0/0 | 3/2 | 75/25 | 0/0 | 3/1 | 5/4 | 3/2 | 80/29 |
| %id: nt/aa | 100/100 | 100/100 | 100/100 | 99.94/99.88 | 98.54/98.54 | 100/100 | 99.77/99.77 | 99.84/99.60 | 99.94/99.88 | 98.45/98.31 | |
The MLST analysis is presented in number of changes and percentage of identity at the nucleotide (nt) and amino acid (aa) level. MLST results for the conserved 17 kDa gene evidenced 100.0% of identity at both levels for comparisons against R. asembonensis strains VGD7 and NMRCii.
MLST, multilocus sequence typing.
For gltA, we detected no mutations among Peruvian specimens, but three mutations between Peruvian and the Kenyan type strain; two silent mutations at nucleotide positions 138 and 537, and one missense mutation at position 868 encoding a conservative lysine-to-glutamic acid substitution. Similarly, for sca4, we detected no mutations among Peruvian specimens, but five mutations between Peruvian and Kenyan specimens; one silent mutation at nucleotide position 807 and four missense mutations at positions 383, 1824, 2260, and 2492, encoding one conservative substitution (glutamine-to-histidine at 1824) and three nonconservative substitutions (leucine-to-proline at 383, arginine-to-glycine at 2260, and isoleucine-to-threonine at 2492).
For ompB, we detected three mutations between the five DNA specimens characterized here and both the Peruvian and Kenyan specimens at positions 715, 1488, and 2791, with one additional mutation at position 24 in sample 8556D1 only. Of these, two were silent mutations and the other two encoded semiconservative amino acid substitutions; one glycine-to-serine change at position 715 and a serine-to-glycine change at position 2791.
For ompA, we detected five mutations between Peruvian and Kenyan specimens; one silent mutation at nucleotide position 828 and four missense mutations at positions 484, 828, 3913, 4435, and 4955, encoding one conservative substitution (leucine-to-valine at position 4435), one semiconservative substitution (valine-to-alanine at position 4955), and two nonconservative substitutions (tyrosine-to-aspartic acid at position 484 and arginine-to-glycine at position 3913).
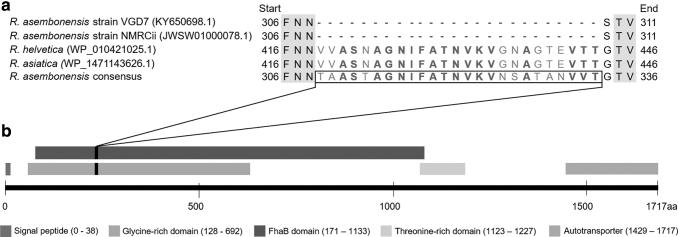
We also identified a 75-nucleotide insertion that encodes 25 amino acids not present in either the Peruvian (VGD7) or Kenyan (NMRCii) R. asembonensis reference strains. The insertion, which begins at position 926 and ends at position 1000, encodes a putative functional motif located in the FhaB region of the protein that is also present in other Rickettsia strains (Fig. 1). When translated BLASTp against all available Rickettsiaceae sequences, the query returns highly similar and significant (70.0–72.0%, e-value = 2e-6) matches to motifs repeated three times across the R. helvetica ompA gene (GenBank access: WP_010421025), and less similar matches to motifs found in two other R. asembonensis strains (GenBank access: WP_041079521 and AQZ41247, 62.5%, e-value = 5e-4) and one R. asiatica (58.0–70.0%, e-value = 2e-6, GenBank access: WP_147143626).
FIG. 1.
Schematic representation of the ompA gene, including the insertion/deletion, found in the FhaB domain of the protein at amino acid positions 309 to 333. (a) Zoom-in view of an alignment of five sequences over the FhaB domain region of ompA. The Rickettsia asembonensis consensus sequence represents strains 8294D3, 8556D1, VGC2, LER197, and LER205 described in this work, which share 100% identity at nucleotide and amino acid levels over the region represented. The insertion is not present in the NMRCii reference strain from Kenya, and it was missed in the Peruvian strain VGD7 when it was first reported. However, the reported insertion is present as a motif in the R. helvetica and R. asiatica ompA gene with a high degree of conservation (conserved amino acids shown in blue and variable amino acids shown in red). (b) Zoom-out view of the complete ompA gene, with various functional domains color coded according to the legend.
Presence of this insertion in all specimens characterized here but not in the original Peruvian reference strain (VGD7) was surprising, raising the possibility of an artifact introduced during genomic data assembly. However, the fact that we used a bioinformatics approach that combines de novo assembly followed by reference-based assembly limits the opportunities for error. Instead, we discovered that sequences of the original Peruvian R. asembonensis strain VGD7 (Loyola et al. 2018) were assembled using reference-based assembly only. That assembly was carried out with the Kenyan strain as reference, which does not contain the insertion, resulting in the omission of 75 nucleotides from the gene ORF. This was later verified through a reassembly of VGD7 raw data using the dual approach described here.
In summary, when assembled using de novo-based methods, followed by reference-based methods, all Peruvian R. asembonensis specimens return ORFs that contain the 75-nucleotide insertion.
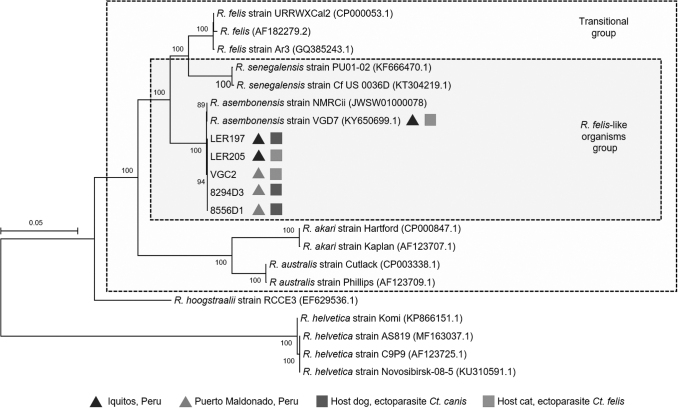
To further characterize the strains from Iquitos and Puerto Maldonado, we conducted phylogenetic analysis using the variable ompB gene, which has been previously used as a sequence-specific species detection target and to infer phylogenetic relationships among members of the RFLO group (Jiang et al. 2013, Mediannikov et al. 2015, Maina et al. 2018, Blanton et al. 2019b). Although reference sequences are available for multiple Rickettsiaceae, validated reference sequences are limited in number and length, particularly for the transitional group and for RFLOs (Table 3). Nevertheless, we were able to access enough references of sufficient length to conduct phylogenetic analysis using 86% (4263 nt) of the ompB ORF (Fig. 2). The analysis confirms that the five R. asembonensis specimens characterized here are closely related to the original Kenyan and Peruvian R. asembonensis type strains.
FIG. 2.
Phylogenetic analysis of R. asembonensis specimens using the variable ompB gene. The analysis included 4263 nucleotides (82.8%) of the complete open reading frame of the ompB gene. It was carried out using the maximum likelihood method based on the General Time Reversible Gamma distributed model. Scale bars represent the number of substitutions per site. R. hoogstraalii and R. helvetica strains were included as outgroups.
Discussion
Since its initial discovery in 2013 in samples from Kenya, R. asembonensis has been reported worldwide and with increasing frequency (Maina et al. 2018). In South America, R. asembonensis has been documented in samples from Brazil (Silva et al. 2017, Bitencourth et al. 2019, Schott et al. 2019), Chile (Cevidanes et al. 2018), Colombia (Faccini-Martinez et al. 2016, Betancourt-Ruiz et al. 2020), Ecuador (Oteo et al. 2014), and Peru (Kocher et al. 2016, Palacios-Salvatierra et al. 2017, Salmon-Mulanovich et al. 2019). The first identification of R. asembonensis in Peru occurred 3 years after its discovery in Kenya, in ectoparasite samples collected in Iquitos (Kocher et al. 2016), and thereafter, it was also found in archived samples collected between 2010 and 2011 in Loreto and Madre de Dios (Salmon-Mulanovich et al. 2019).
In 2018, we published the first sequences of complete genes from a Peruvian R. asembonensis via MLST characterization of a single sample from Iquitos (Loyola et al. 2018). At that point, only the original Kenyan strain had been fully sequenced, so despite our contribution with one additional characterization, the availability of useful genomic information continued to be limited despite multiple detections worldwide. By the end of 2018, researchers at the National Institutes of Health of Peru, published a report describing R. asembonensis in human samples (Palacios-Salvatierra et al. 2018). In our view, this highlighted even further an urgent need to generate gene sequences to study these novel emerging pathogens.
Genomic information is an essential tool that enables us to develop appropriate diagnostics, to infer phylogenetic relationships, and to link potential genetic variations to pathogenicity. Genus-specific genes that are highly conserved, such as the 17-kDa antigen gene and gltA for example, are frequently used as targets for generic diagnostics based on PCR because they can easily identify a number of Rickettsia species with high specificity (Anderson et al. 1989, Roux et al. 1997, Jiang et al. 2012). Our MLST analysis confirmed that the 17-kDa antigen gene is conserved, as expected, as the sequences analyzed here were identical among the five specimens described herein and the Peruvian and Kenyan DNA specimens used as references. The gltA gene was also very highly conserved, although less so, as we detected three mutations between the Peruvian specimens and the Kenyan strain NMRCii.
Nevertheless, only one of these three mutations encoded an amino acid change, which was a conservative substitution. Based on these findings, we confirm that the 17-kDa antigen gene is a conserved gene useful for generic screenings of diverse Rickettsia species and that gltA is also highly conserved, although less so, and therefore potentially useful for the reconstruction of evolutionary relationship of diverse Rickettsia species, including novel ones such as R. asembonensis.
Genes such as sca4, ompA, and ompB, which encode cell surface antigens that are involved in a variety of cell processes, including adhesion, cell-to-cell spread, and induction of the adaptive immune response, tend to be more diverse, subject to positive selective pressure probably driven by hosts, and prone to recombination (Li et al. 1998, Diaz-Montero et al. 2001, Feng et al. 2004, Blanc et al. 2005, Jiggins 2006, Hillman et al., 2013, Sahni et al. 2013, Gillespie et al. 2015, and Lamason et al. 2016). As such, their sequences can be used to differentiate closely related Rickettsia species, and even strains, and infer their genetic relationships (Blanc et al. 2005, Maina et al. 2018).
Our MLST analysis revealed that sca4 was the most conserved of the three variable genes analyzed, with no mutations among Peruvian specimens, but five mutations between the Peruvian strains and the Kenyan reference strain. In the case of ompB, we detected differences even within Peruvian specimens only, as one of the four mutations detected was only identified in sample 8556D1. Given the variability observed in this gene, we used the five ompB sequences characterized here by MLST, together with other reference sequences, to carry out phylogenetic analysis that confirmed that the Peruvian strains are closely related, but distinct from the original Kenyan R. asembonensis strain, and that all of them belong to the RFLO group of organisms.
In the case of ompA, we detected the same five point mutations, previously reported for the VGD7 strain (Loyola et al. 2018), between the Peruvian strains characterized here and the original Kenyan reference strain. However, we also detected a 75 nucleotide insertion. This insertion is not present in the Kenyan reference strain, and was not reported for the first Peruvian (VGD7) strain characterized. Upon careful analysis of all the genomic data available for all Peruvian strains, including a reanalysis of the original VGD7 data, we realized that VGD7 sequence data had been assembled only by reference assembly to the Kenyan strain, which does not contain the 75 nucleotide insertion, resulting in omission of 25 amino acids from the coding sequence. This has since been corrected, through a combination of de novo assembly first, followed by reference-based assembly.
This correction was submitted to GenBank (Accession number: KY650698). With that approach, we have confirmed that the 75-nucleotide insertion is present in all Peruvian R. asembonensis specimens, but absent from the original Kenyan strain. One limitation of this study is a lack of additional corroboration of the insertion using an independent method such as Sanger sequencing or PCR. We would have liked to carry out such experiments, but we were unable to do so because the genomic DNA from the original samples was completely used up in the preparation of multiple NGS libraries. Others have reported that molecular techniques such as Sanger have an almost perfect agreement with NGS data for the identification and validation of in/dels (Beck et al. 2016, Arteche-López et al. 2021), suggesting that Sanger sequencing may not provide additional value in the validation of in/dels detected by NGS.
In this work the original samples were made into individual sequencing libraries and individually sequenced multiple times, which is essentially the equivalent of carrying out independent verification experiments. The insertion was detected in every case provided that the analysis included a de novo assembly step. In/dels and transversions have been reported in ompA in both R. rickettsia (Clark et al. 2015) and R. felis (Zavala-Castro et al. 2005) using comparative analyses of Ricketssia strains based on NGS data alone as we have done here. Furthermore, given that the insertion we report was consistently found in all ompA genes from Peruvian R. asembonensis samples, including in the reanalysis of the original VGD7 strain, and given that the insertion is a triplet-encoding sequence highly similar to motifs found in other Rickettsia species, suggests that the insertion is real and not the product of a sequencing artifact.
The insertion is located within the FhaB domain of the ompA protein, and corresponds to a repetitive motif found in R. helvetica and R. asiatica. The ompA gene, or remnants of such in the form of a pseudogene, is present in all recognized Rickettsia species of the spotted fever and transitional groups, including R. felis and other RFLOs, and may have evolved under positive selection (Li et al. 1998, Jiggins 2006, Gillespie et al. 2015, Lamason et al. 2016). Evidence of major insertion/deletion events and recombination in Rickettsia species is rare in comparison to evidence of point mutations (Fuerst et al. 1990, Jiggins 2006, Gillespie et al. 2012, Clark et al. 2015, Akter et al. 2017).
However, some of these events have been reported, including a deletion in the ompA gene that occurred after several passages of a well-characterized R. rickettsii strain maintained in culture (Matsumoto et al. 1996), an insertion of 891 bp in the ompA gene or R. rickettsii (Clark et al. 2015), and various in/dels and transversions in the ompA gene of R. felis (Zavala-Castro et al. 2005). We hypothesize that these types of changes could be used to study the selective pressures that promote genetic insertion/deletion events, as well as to carefully infer evolution of Rickettsia species between geographically distant regions.
Conclusions
In Peru, rickettsial infections remain a neglected problem because they are underdiagnosed and underreported despite being endemic to the region. A number of Rickettsia species have been identified locally, including R. asembonensis, R. felis, “Candidatus R. andeanae,” Rickettsia bellii, and Rickettsia parkeri (Blair et al. 2004b, Jiang et al. 2005, Ogrzewalska et al. 2012, Flores-Mendoza et al. 2013, Palacios-Salvatierra et al. 2017, Loyola et al. 2018), and yet, their distribution remains poorly understood. Complete genomic information on these diverse Rickettsia species is required to address their evolution and molecular epidemiology, particularly for recently discovered pathogenic strains such as R. asembonensis.
The MLST characterization described here, which includes complete genes from five R. asembonensis DNA samples, derived from distinct ectoparasites collected in different regions of Peru at different times, contributes valuable genomic information that is currently available in very limited supply, and that is required to expand our understanding of the molecular epidemiology of these pathogens in the region and around the world.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the National Institutes of Health, the Department of the Navy, Department of Defense, nor the U.S. Government.
Copyright Statement
Several authors are employees of the U.S. government. This work was prepared as part of their official duties. Title 17, U.S.C., §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military Service member or employee of the U.S. Government as part of that person's official duties.
Ethical Statement
The study protocol was approved by NAMRU-6's IACUC in compliance with all applicable Federal regulations. The experiments reported herein were conducted in compliance with the Animal Welfare Act and in accordance with principles set forth in the “Guide for the Care and Use of Laboratory Animals,” Institute of Laboratory Animals Resources, National Research Council, National Academy Press, 2011.
Author Disclosure Statement
No conflicting financial interests exist.
Funding Information
This work was supported by the Global Emerging Infections Surveillance (GEIS) Branch of the Armed Forces Health Surveillance Division (https://health.mil/Military-Health-Topics/Combat-Support/Armed-Forces-Health-Surveillance-Branch/Global-Emerging-Infections-Surveillance-and-Response) work unit number No. 800000.82000.25GB.B0016, Promis ID# MLeguia-PO166-14 to M.L. M.L. has additional support from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health under award U01AI151814. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
References
- Akter A, Ooka T, Gotoh Y, Yamamoto S, et al. Extremely low genomic diversity of Rickettsia japonica distributed in Japan. Genome Biol Evol 2017; 9:124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BE, Tzianabos T. Comparative sequence analysis of a genus-common rickettsial antigen gene. J Bacteriol 1989; 171:5199–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteche-López A, Ávila-Fernández A, Romero R, Riveiro-Álvarez R, et al. Sanger sequencing is no longer always necessary based on a single-center validation of 1109 NGS variants in 825 clinical exomes. Sci Rep 2021; 11:5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck TF, Mullikin JC; NISC Comparative Sequencing Program, Biesecker LG. Systematic evaluation of sanger validation of next-generation sequencing variants. Clin Chem 2016; 62:647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt-Ruiz P, Martinez-Diaz HC, Gil-Mora J, Ospina C, et al. Candidatus Rickettsia senegalensis in cat fleas (Siphonaptera: Pulicidae) collected from dogs and cats in Cauca, Colombia. J Med Entomol 2020; 57:382–387. [DOI] [PubMed] [Google Scholar]
- Bitencourth K, Amorim M, de Oliveira SV, Voloch C, et al. Genetic diversity, population structure and rickettsias in Amblyomma ovale in areas of epidemiological interest for spotted fever in Brazil. Med Vet Entomol 2019; 33:256–268. [DOI] [PubMed] [Google Scholar]
- Blair PJ, Schoeler GB, Moron C, Anaya E, et al. Evidence of rickettsial and leptospira infections in Andean northern Peru. Am J Trop Med Hyg 2004a; 70:357–363. [PubMed] [Google Scholar]
- Blair PJ, Jiang J, Schoeler GB, Moron C, et al. Characterization of spotted fever group rickettsiae in flea and tick specimens from northern Peru. J Clin Microbiol 2004b; 42:4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Ngwamidiba M, Ogata H, Fournier PE, et al. Molecular evolution of rickettsia surface antigens: Evidence of positive selection. Mol Biol Evol 2005; 22:2073–2083. [DOI] [PubMed] [Google Scholar]
- Blanton LS, Quade BR, Bouyer DH. Differentiation of Rickettsia felis and Rickettsia felis-like organisms via restriction fragment length polymorphism analysis. Vector Borne Zoonotic Dis 2019b; 19:637–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LS, Vohra RF, Fistein L, Quade B, et al. Rickettsiae within the fleas of feral cats in Galveston, Texas. Vector Borne Zoonotic Dis 2019a; 19:647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevidanes A, DiCataldo S, Vera F, Lillo P, et al. Molecular detection of vector-borne pathogens in rural dogs and associated Ctenocephalides felis fleas (Siphonaptera: Pulicidae) in Easter Island (Chile). J Med Entomol 2018; 55:1659–1663. [DOI] [PubMed] [Google Scholar]
- Clark TR, Noriea NF, Bublitz DC, Ellison DW, et al. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun 2015; 83:1568–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Montero CM, Feng HM, Crocquet-Valdes PA, Walker DH. Identification of protective components of two major outer membrane proteins of spotted fever group Rickettsiae. Am J Trop Med Hyg 2001; 65:371–378. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremeeva ME, Capps D, McBride CL, Williams-Newkirk AJ, et al. Detection of Rickettsia asembonensis in fleas (Siphonaptera: Pulicidae, Ceratophyllidae) collected in five counties in Georgia, United States. J Med Entomol 2020; 57:1246–1253. [DOI] [PubMed] [Google Scholar]
- Faccini-Martinez AA, Ramirez-Hernandez A, Forero-Becerra E, Cortes-Vecino JA, et al. Molecular evidence of different Rickettsia species in Villeta, Colombia. Vector Borne Zoonotic Dis 2016; 16:85–87. [DOI] [PubMed] [Google Scholar]
- Feng HM, Whitworth T, Olano JP, Popov VL, et al. Fc-dependent polyclonal antibodies and antibodies to outer membrane proteins A and B, but not to lipopolysaccharide, protect SCID mice against fatal Rickettsia conorii infection. Infect Immun 2004; 72:2222–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mendoza C, Florin D, Felices V, Pozo EJ, et al. Detection of Rickettsia parkeri from within Piura, Peru, and the first reported presence of Candidatus Rickettsia andeanae in the tick Rhipicephalus sanguineus. Vector Borne Zoonotic Dis 2013; 13:505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, Stewart A, Morrison AC, Galvez H, et al. Epidemiology of spotted fever group and typhus group rickettsial infection in the Amazon basin of Peru. Am J Trop Med Hyg 2010; 82:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PA, Poetter KP, Pretzman C, Perlman PS. Molecular genetics of populations of intracellular bacteria: The spotted fever group rickettsiae. Ann N Y Acad Sci 1990; 590:430–438. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Joardar V, Williams KP, Driscoll T, et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol 2012; 194:376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, et al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 2015; 39:47–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman RD Jr., Baktash YM, Martinez JJ.. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with alpha2beta1 integrin. Cell Microbiol 2013; 15:727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Blair PJ, Felices V, Moron C, et al. Phylogenetic analysis of a novel molecular isolate of spotted fever group Rickettsiae from northern Peru: Candidatus Rickettsia andeanae. Ann N Y Acad Sci 2005; 1063:337–342. [DOI] [PubMed] [Google Scholar]
- Jiang J, Maina AN, Knobel DL, Cleaveland S, et al. Molecular detection of Rickettsia felis and Candidatus Rickettsia asemboensis in fleas from human habitats, Asembo, Kenya. Vector Borne Zoonotic Dis 2013; 13:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Stromdahl EY, Richards AL. Detection of Rickettsia parkeri and Candidatus Rickettsia andeanae in Amblyomma maculatum Gulf Coast ticks collected from humans in the United States. Vector Borne Zoonotic Dis 2012; 12:175–182. [DOI] [PubMed] [Google Scholar]
- Jiggins FM. Adaptive evolution and recombination of Rickettsia antigens. J Mol Evol 2006; 62:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho KL, Koh FX, Singh HK, Zan HA, et al. Spotted fever group rickettsioses and murine typhus in a Malaysian Teaching Hospital. Am J Trop Med Hyg 2016; 95:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho KL, Tay ST. Identification of rickettsial infections (Rickettsia sp. TH2014) in Ctenocephalides orientis fleas (Siphonaptera: Pulicidae). J Med Entomol 2019; 56:526–532. [DOI] [PubMed] [Google Scholar]
- Kocher C, Jiang J, Morrison AC, Castillo R, et al. Serologic evidence of scrub typhus in the peruvian Amazon. Emerg Infect Dis 2017; 23:1389–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher C, Morrison AC, Leguia M, Loyola S, et al. Rickettsial disease in the Peruvian Amazon Basin. PLoS Negl Trop Dis 2016; 10:e0004843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Bastounis E, Kafai NM, Serrano R, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell 2016; 167:670–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, et al. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015; 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog 1998; 24:289–298. [DOI] [PubMed] [Google Scholar]
- Loyola S, Flores-Mendoza C, Torre A, Kocher C, et al. Rickettsia asembonensis characterization by multilocus sequence typing of complete genes, Peru. Emerg Infect Dis 2018; 24:931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina AN, Jiang J, Luce-Fedrow A, St John HK, et al. Worldwide presence and features of flea-borne Rickettsia asembonensis. Front Vet Sci 2018; 5:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17:10–12. [Google Scholar]
- Matsumoto M, Tange Y, Okada T, Inoue Y, et al. Deletion in the 190 kDa antigen gene repeat region of Rickettsia rickettsii. Microb Pathog 1996; 20:57–62. [DOI] [PubMed] [Google Scholar]
- Mediannikov O, Aubadie-Ladrix M, Raoult D. Candidatus “Rickettsia senegalensis” in cat fleas in Senegal. New Microbes New Infect 2015; 3:24–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonga LC, Hayashida K, Nakao R, Lisulo M, et al. Molecular detection of Rickettsia felis in dogs, rodents and cat fleas in Zambia. Parasit Vectors 2019; 12:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataraj N, Muthuraman K, Sundaram D, Ayyanar E, et al. Molecular detection of Candidatus Rickettsia asembonensis in fleas collected from pets and domestic animals in Puducherry, India. Med Vet Entomol 2020; 34:498–502. [DOI] [PubMed] [Google Scholar]
- Nziza J, Tumushime JC, Cranfield M, Ntwari AE, et al. Fleas from domestic dogs and rodents in Rwanda carry Rickettsia asembonensis and Bartonella tribocorum. Med Vet Entomol 2019; 33:177–184. [DOI] [PubMed] [Google Scholar]
- Ogrzewalska M, Literak I, Cardenas-Callirgos JM, Capek M, et al. Rickettsia bellii in ticks Amblyomma varium Koch, 1844, from birds in Peru. Ticks Tick Borne Dis 2012; 3:254–256. [DOI] [PubMed] [Google Scholar]
- Oteo JA, Portillo A, Portero F, Zavala-Castro J, et al. “Candidatus Rickettsia asemboensis” and Wolbachia spp. in Ctenocephalides felis and Pulex irritans fleas removed from dogs in Ecuador. Parasit Vectors 2014; 7:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Salvatierra R, Anaya-Ramirez E, Juscamayta-Lopez J, Caceres-Rey O, et al. Epidemiological and molecular profile of Rickettsiosis in peruvian border locations. Rev Peru Med Exp Salud Publica 2017; 34:76–84. [DOI] [PubMed] [Google Scholar]
- Palacios-Salvatierra R, Caceres-Rey O, Vasquez-Dominguez A, Mosquera-Visaloth P, et al. Rickettsial species in human cases with non-specific acute febrile syndrome in Peru. Rev Peru Med Exp Salud Publica 2018; 35:630–635. [DOI] [PubMed] [Google Scholar]
- Parola P, Sanogo OY, Lerdthusnee K, Zeaiter Z, et al. Identification of Rickettsia spp. and Bartonella spp. in ffrom the Thai-Myanmar border. Ann N Y Acad Sci 2003; 990:173–181. [DOI] [PubMed] [Google Scholar]
- Perez-Osorio CE, Zavala-Velazquez JE, Arias-Leon JJ, Zavala-Castro JE. Rickettsia felis as emergent global threat for humans. Emerg Infect Dis 2008; 14:1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricapa-Antay F, Diaz-Melon K, Silva-Caso W, Del Valle LJ, et al. Molecular detection and clinical characteristics of Bartonella bacilliformis, Leptospira spp., and Rickettsia spp. in the Southeastern Peruvian Amazon basin. BMC Infect Dis 2018; 18:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the Rickettsiae. Int J Syst Bacteriol 1997; 47:252–261. [DOI] [PubMed] [Google Scholar]
- Sahni SK, Narra HP, Sahni A, Walker DH. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol 2013; 8:1265–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Mulanovich G, Simons MP, Flores-Mendoza C, Loyola S, et al. Seroprevalence and risk factors for rickettsia and leptospira infection in Four Ecologically Distinct Regions of Peru. Am J Trop Med Hyg 2019; 100:1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D, Souza UA, Dall'Agnol B, Webster A, et al. Detection of Rickettsia spp. and Bartonella spp. in Ctenocephalides felis fleas from free-ranging crab-eating foxes (Cerdocyon thous). Med Vet Entomol 2019; 33:536–540. [DOI] [PubMed] [Google Scholar]
- Silva AB, Vizzoni VF, Costa AP, Costa FB, et al. First report of a Rickettsia asembonensis related infecting fleas in Brazil. Acta Trop 2017; 172:44–49. [DOI] [PubMed] [Google Scholar]
- Zavala-Castro JE, Small M, Keng C, Bouyer DH, et al. Transcription of the Rickettsia felis ompA gene in naturally infected fleas. Am J Trop Med Hyg 2005; 73:662–666. [PMC free article] [PubMed] [Google Scholar]