Key words: Arginine methylation, post-translational modification, protein arginine methyltransferases, protozoan parasites
Abstract
Arginine methylation is a post-translational modification involved in gene transcription, signalling pathways, DNA repair, RNA metabolism and splicing, among others, mechanisms that in protozoa parasites may be involved in pathogenicity-related events. This modification is performed by protein arginine methyltransferases (PRMTs), which according to their products are divided into three main types: type I yields monomethylarginine (MMA) and asymmetric dimethylarginine; type II produces MMA and symmetric dimethylarginine; whereas type III catalyses MMA only. Nine PRMTs (PRMT1 to PRMT9) have been characterized in humans, whereas in protozoa parasites, except for Giardia intestinalis, three to eight PRMTs have been identified, where in each group there are at least two enzymes belonging to type I, the majority with higher similarity to human PRMT1, and one of type II, related to human PRMT5. However, the information on the role of most of these enzymes in the parasites biology is limited so far. Here, current knowledge of PRMTs in protozoan parasites is reviewed; these enzymes participate in the cell growth, stress response, stage transitions and virulence of these microorganisms. Thus, PRMTs are attractive targets for developing new therapeutic strategies against these pathogens.
Introduction
Post-translational modifications (PTMs) are biochemical changes of amino acid residues that alter the charge state, hydrophobicity, conformation or stability of the proteins. These modifications act as molecular switches regulating the interaction of proteins with nucleic acids, cofactors, lipids and other proteins (Venne et al., 2014). PTMs can be ‘read’ by specific effectors to regulate several cellular processes in response to extracellular and intracellular stimuli. To date, >450 unique protein modifications have been identified, including phosphorylation, acetylation, ubiquitination, SUMOylation and methylation, among others (Venne et al., 2014).
Protein methylation occurs at different amino acids and it has been estimated that from 0.6 to 1.6% of the eukaryote genomes encode methyltransferases, the enzymes that perform this PTM (Katz et al., 2003). Although initially the studies had been largely focused on lysine and arginine methylation of histones, which impact the transcription regulation (Jambhekar et al., 2019), nowadays it is known that methylation on non-histone proteins regulates other molecular mechanisms, including RNA stability, splicing and translation (Lanouette et al., 2014; Blanc and Richard, 2017; Al-Hamashi et al., 2020).
Arginine contains in its side chain an aliphatic sequence of three carbon atoms, ending with a guanidino group, which is protonated and positively charged at physiological pH, with five potential hydrogen-bonds donors to interact with other molecules (Bedford and Clarke, 2009). The addition of one or two methyl groups to arginine creates a bulkier and more hydrophobic residue that does not alter the global cationic charge of the amino acid, but disperses it towards the methyl groups (Bedford and Richard, 2005; Bedford and Clarke, 2009). Arginine methylation increases the affinity for specific proteins such as those containing aromatic cages, including the Tudor domain (Tripsianes et al., 2011), which interpret specific methylation marks to regulate several molecular events such as epigenetics, mRNA splicing, mRNA translation, DNA damage and stress response, among others (Blanc and Richard, 2017).
Infections with protozoan parasites are responsible for high morbidity and mortality in humans worldwide. The diseases produced by these microorganisms depend on the expression and activity of their virulence factors as well as the response of parasites to host immunity and the transition to different stages and hosts, events that require changes in gene expression and activation of specific signalling pathways that could be regulated by arginine methylation. Some advances have been made regarding the role of arginine methylation and the enzymes that perform this PTM, named as protein arginine methyltransferases (PRMTs), in the biology of certain protozoan parasites, mainly in Trypanosoma brucei; however, in others, such as Trypanosoma cruzi, many Leishmania and Plasmodium species as well as Trichomonas vaginalis, the arginine methylation and PRMTs have not yet been studied. Here, current knowledge on arginine methylation and PRMTs in protozoan parasites is reviewed.
Protein arginine methyltransferases (PRMTs)
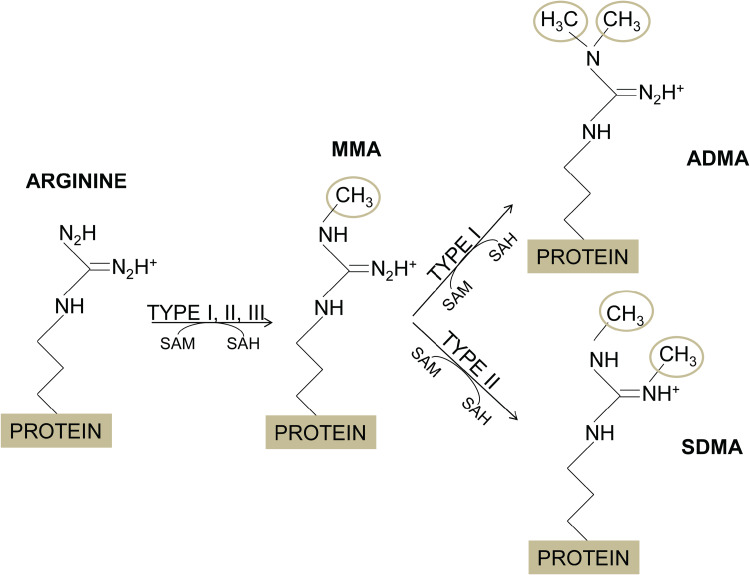
The addition of methyl groups to the guanidino nitrogen atoms of arginine residues is catalysed by the PRMTs, which use S-adenosyl-L-Methionine (AdoMet or SAM) as the donor of the methyl groups (Blanc and Richard, 2017). According to their activity, there are three main types of PRMTs, type I catalyses monomethylarginine (MMA) and asymmetric dimethylarginine (ADMA); type II yields MMA and symmetric dimethylarginine (SDMA); and type III produces MMA only (Fig. 1); additionally, yeasts have a type IV PRMT, which monomethylates the delta (δ) nitrogen atom of arginine (Bedford and Clarke, 2009). Most PRMTs substrates contain glycine- and arginine-rich (GAR) sequences that include multiple arginines in RGG or RXR contexts (Najbauer et al., 1993; Gary and Clarke, 1998; Feng et al., 2013).
Fig. 1.
Arginine methylation. The methylation of arginine residues is catalysed by the protein arginine methyltrnasferases (PRMTs), which transfer a methyl group from the S-adenosylmethionone (SAM or AdoMet), resulting in the arginine methylation and S-adenosylhomocysteine (SAH). PRMTs of type I, II and III produce monomethylarginine (MMA). In addition, PRMTs of type I add other methyl groups to the same nitrogen atom to yield asymmetric dimethylarginine (ADMA), while PRMTs of type II attach the second methyl group to the other N-terminal nitrogen of the arginine residue, forming symmetric dimethylarginine (SDMA).
PRMTs contain a conserved core region of approximately 310 amino acids comprising the characteristic methyltransferase motif for the binding of the AdoMet cofactor, formed by a seven-strand twisted beta-sheet and the PRMT-specific ‘double E’ and ‘THW’ loops (Cheng et al., 2005). In addition, different studies demonstrated that the dimerization of PRMTs facilitates their AdoMet-binding and their catalytic activity (Zhang et al., 2000; Zhang and Cheng, 2003; Xu, 2004; Li et al., 2015; Zhou et al., 2015).
Nine PRMTs have been characterized in humans (PRMT1-9). PRMT1, 2, 3, 4 (also known as CARM1), 6 and 8 are type I, PRMT5 and 9 are type II, and PRMT7 is type III (Bedford and Clarke, 2009). Some of these proteins contain specific domains, PRMT2 has an SH3 domain, PRMT3 a Zn-finger domain (Frankel and Clarke, 2000) and PRMT8 is myristoylated at its N-terminus (Frankel et al., 2002). It has been demonstrated that most of type I PRMTs are active as homodimers (Zhang et al., 2000; Zhang and Cheng, 2003; Xu, 2004; Li et al., 2015; Zhou et al., 2015), while that PRMT5 forms a hetero-octameric complex (methylosome) with MEP50 (four subunits of each), in which MEP50 binds to the substrate and stimulates PRMT5 activity (Ho et al., 2013). PRMT1 is responsible for most of the total monomethylation and asymmetric dimethylation of arginine, whereas PRMT5 has the major type II activity (Bedford and Clarke, 2009).
Most of these enzymes are ubiquitously expressed, with exception of PRMT8, which is localized only in the central nervous system (Lee et al., 2005). In general, PRMTs are found in the nucleus and cytoplasm, but PRMT3 is located principally in the cytoplasm, PRMT6 is concentrated mainly in the nucleus and PRMT8 is linked to the plasma membrane (Frankel and Clarke, 2000; Frankel et al., 2002; Lee et al., 2005). Arginine methylation on histones is involved in the co-activation or co-repression of gene transcription and the effects on transcription of specific residues modified by different PRMTs have been determined (reviewed by Rakow et al., 2020). PRMT1 and PRMT4/CARM1 are the main co-activators (Bedford and Clarke, 2009), PRMT2 and PRMT7 function as co-repressors (Ganesh et al., 2006; Karkhanis et al., 2012), whereas PRMT5 and PRMT6 have been reported to have both functions (Zhao et al., 2009; Migliori et al., 2012; Casadio et al., 2013; Cheng et al., 2020). In addition, PRMTs also methylate several non-histone proteins such as transcription factors and protein involved in RNA metabolism, DNA damage repair and many signalling pathways (reviewed by Al-Hamashi et al., 2020).
PRMTs have also been identified in non-mammalian animals, filamentous fungi, yeasts, plants and protozoan parasites (McBride et al., 2007; Ahmad et al., 2011; Fisk and Read, 2011; Zhao et al., 2016; Hadjikyriacou and Clarke, 2017; Plett et al., 2017; Wang et al., 2017; Bauer et al., 2019). Enzymes similar to mammalian PRMT1, 3 and 5 are present in different eukaryotic organisms, whereas other PRMTs, such as PRMT2 and 8, are less conserved and it has been suggested that they could have evolved in the multicellular organisms as a requirement for tissue-specific functions (Bachand, 2007).
Arginine methylation and PRMTs in protozoan parasites
Protozoan parasites, except Giardia intestinalis, have several PRMTs that were named according to their similarity with the human enzymes [reviewed by Fisk and Read (2011)], although they have structural differences with their mammalian homologues (see below). All protozoan parasites have two or more PRMTs of type I and at least one of type II; interestingly, trypanosomatids also contain a type III enzyme (Fisk and Read, 2011). They also possess the canonical histones, suggesting that the arginine methylation observed in histones of other eukaryotes, as well as their effects in gene expression, are preserved in these microorganisms. In addition, arginine methylation in non-histone proteins may also be important in the biology of protozoan parasites. However, most of the non-histone protein targets in these pathogens remain to be identified.
Phylogenetic analysis shows that there is no strong relationship of PRMT3, PRMT4/CARM1 and PRMT6 of the parasites with their respective human enzymes (Fig. 2). Furthermore, this analysis clearly separates the PRMTs of parasites into four main clades, one of them grouped PRMTs of type I, most of them with a higher relationship with the human PRMTs 1, 3 and 8; the other clade comprises PRMTs of type II similar to human PRMT5; the third clade is present only in trypanosomatids and corresponds to type III (PRMT7); and the last clade includes the PRMT3 of T. brucei and the atypical PRMT of E. histolytica (Fig. 2).
Fig. 2.
Phylogenetic relationship of protozoan parasites PRMTs. The predicted amino acid sequences of PRMTs from human, protozoan parasites and Saccharomyces cerevisiae (types I and IV) were aligned by MUSCLE. Then, data were submitted to phylogenetic analysis by UPGMA using MEGA version 5.05. Accession numbers for the protein sequences of T. vaginalis are indicated. The other PRMTs used for this analysis were: Human: HsPRMT1 (NP_938074.2), Hs PRMT2 (NP_001526.2), HsPRMT3 (NP_005779.1), HsPRMT4/CARM1 (NP_954592.1), HsPRMT5 (NP_006100.2); HsPRMT6 (NP_060607.2), HsPRMT7 (NP_061896.1), HsPRMT8 (NP_062828.3). HsPRMT9 (NP_612373.2); S. cereviciae: ScRMT2 (NP_010753.1), ScHMT1 (NP_009590.1); T. brucei: TbPRMT1 (Tb927.1.4690), TbPRMT3 (Tb927.10.3560), TbPRMT5 (Tb927.10.640), TbPRMT6 (Tb927.5.3960), TbPRMT7 (Tb927.7.5490); P. falciparum: PfPRMT1 (PF14_0242), PfPRMT4/CARM1 (PF14_0242), PfPRMT5 (PF13_0323), T. gondii: TgPRMT1 (GT1_030400), Tg PRMT3 (GT1_001320), TgPRMT4/CARM1 (GT1_074560), TgPRMT5 (GT1_126490); E. histolytica: EhPRMT1a (EHI_105780), EhPRMT1b (EHI_152460), EhPRMT1c (EHI_202470), EhPRMTA (EHI_159780), EhPRMT5 (EHI_158560). The numbers at the nodes of the branches indicate the confidence percentages of the tree topology from the bootstrap analysis of 1000 replicates. Proteins are grouped into type I, II, III, IV and atypical.
Arginine methylation and PRMTs in trypanosomatids
The causative agents of African trypanosomiasis (T. brucei), Leishmaniasis (Leishmania spp.) and Chagas disease (T. cruzi) belong to the kinetoplastids group (trypanosomatids). The main mechanisms implicated in the regulation of gene expression in these parasites are the post-transcriptional processes, including RNA turnover, translation and editing, involving multiple RNA-binding proteins (RBPs), which are common targets of arginine methylation (Campagnaro et al., 2021). These microorganisms contain five PRMTs, which are homologues to the mammalian PRMT1, 3, 5, 6 and 7 (Fisk and Read, 2011); the studies related to these enzymes have been performed on the five enzymes of T. brucei and on the PRMT7 of Leishmania major, whereas there are no reports about the role of PRMTs in other Leishmania species or in T. cruzi.
Although the number of annual cases of African trypanosomiasis has been progressively decreasing, it has been estimated that approximately 1.8 million people live in high or very high-risk areas and 11.3 million people in moderate-risk zones (Simarro et al., 2015). T. brucei life cycle consists of bloodstream trypomastigotes into the vertebrate host and procyclic trypomastigotes, epimastigotes and metacyclic trypomastigotes into the tsetse fly (Glossina spp.).
A recent global proteomic analysis identified 1332 methylarginines in 676 proteins (approximately 10% of the proteome) (Lott et al., 2013), demonstrating that arginine methylation is a common PTM in this parasite. In addition, both type I and type II PRMT activities have been detected in cell extracts of T. brucei (Pelletier et al., 2002). TbPRMT1 catalyses the majority of the ADMA modification in vivo (Pelletier et al., 2005). It is expressed in all stages of the parasite life cycle and synthetic peptides containing GAR sequences are methylated by the recombinant protein (Pelletier et al., 2005). The TbPRMT1 knockdown does not produce defects in the in vitro growth of the procyclic or bloodstream forms, but its knockout downregulates enzymes involved in glycolysis and upregulates enzymes participating in proline degradation, reduces the cytoplasmic mRNA granules in the insect stage under starvation and decreases the virulence of T. brucei in mice (Pelletier et al., 2005; Kafková et al., 2017; Kafková et al., 2018). Interestingly, TbPRMT1 directly binds to RNA and to a lipin homologue that possesses activity of phosphatidic acid phosphatase (Pelletier et al., 2013; Kafková et al., 2018). All these results indicate that TbPRMT1 participates in carbohydrates metabolism, RNA biology, phospholipid biosynthesis and virulence (Table 1).
Table 1.
Functions of characterized PRMTs in protozoan parasites
| Parasite | PRMT | Type | Molecular and/or cellular function | Reference |
|---|---|---|---|---|
| Trypanosoma brucei | TbPRMT1 (Tb927.1.4690) | Type I | Forms active heterotetramers with TbPRMT3, RNA and carbohydrates metabolism, phospholipid biosynthesis, stress response, virulence, in vitro growth of bloodstream forms | Pelletier et al. (2013), Kafková et al. (2017, 2018) |
| TbPRMT3 (Tb927.10.3560) | Atypical | Inactive, but forms active heterotetramers with TbPRMT1 | Kafková et al. (2017) | |
| TbPRMT5 (Tb927.10.640) | Type II | RNA metabolism | Pasternack et al. (2007) | |
| TbPRMT6 (Tb927.5.3960) | Type I | Cell growth, morphology, together with PRMT7 is critical for proliferation of procyclic forms | Fisk et al. (2010), Lott et al. (2013) | |
| TbPRMT7 (Tb927.7.5490) | Type III | Together with TbPRMT6 is critical for proliferation of procyclic forms | Lott et al. (2013) | |
| Leishmania major | LmPRMT7 (LMJSD75_060015100) | Type III | RNA metabolism, life cycle progression, in vitro and in vivo virulence | Ferreira et al. (2014, 2020), Alcoforado Diniz et al. (2021) |
| Plasmodium falciparum | PfPRMT1 (PF14_0242) | Type I | RNA metabolism | Fan et al. (2009) |
| PfPRMT5 (PF13_0323) | Type II | RNA splicing | Hossain et al. (2013) | |
| Toxoplasma gondii | TgPRMT1 (GT1_030400) | Type I | Epigenetics, RNA metabolism and splicing, cell duplication | Musiyenko et al. (2012), El Bissati et al. (2016) |
| TgPRMT4/TgCARM 1 (GT1_074560) | Type I | Epigenetics, stage conversion | Saksouk et al. (2005) | |
| TgPRMT5 (GT1_126490) | Type II | Epigenetics, stage conversion | Liu et al. (2019) | |
| Entamoeba histolytica | EhPRMT1a (EHI_105780) | Type I | Epigenetics | Borbolla-Vázquez et al. (2015) |
| EhPRMTA (EHI_159780) | Atypical | RNA metabolism, cell growth, migration, stress response | Cázares-Apátiga et al. (2017), Medina-Gómez et al. (2021) |
Based on the homology to human PRMT3, TbPRMT3 was predicted to be a type I enzyme (Fisk and Read, 2011); however, most of the conserved amino acids involved in PRMT activity are absent in TbPRMT3, where both glutamate residues in the double E loop are changed to aspartate and tryptophan is the only conserved residue in the THW loop; concordantly, its recombinant protein has no enzymatic activity (Kafková et al., 2017). In fact, TbPRMT3 functions as a proenzyme that stabilizes the TbPRMT1 enzyme, producing an active heterotetramer formed by two enzyme-proenzyme pairs (TbPRMT1/TbPRMT3, also named TbPRMT1ENZ/TbPRMT1PRO) (Kafková et al., 2017). Our phylogenetic analysis suggested that TbPRMT3 is more related to the atypical enzyme of Entamoeba histolytica EhPRMTA than to other PRMT members (Fig. 2).
TbPRMT5 is constitutively expressed in procyclic and bloodstream forms, it is found mainly in the cytoplasm forming high-molecular-weight complexes that do not include to MEP50, but that contain a DEAD box protein and two kinetoplast-specific proteins involved in RNA editing (Pasternack et al., 2007). Interestingly, genes encoding MEP50 homologue have not been found in the genome database of T. brucei and recombinant TbPRMT5 displays enzymatic activity on broad substrates that includes the RNA-binding protein 16 (Pasternack et al., 2007). Thus, this enzyme participates in the processing or translation of RNA (Table 1).
TbPRMT6 was named based on its higher homology to human PRMT6 than to another type I enzymes, but in the parasite, protein lacks the N-terminal nuclear localization signal detected in the human enzyme and its chrystal structure revealed several features that are distinct from previously characterized type I PRMTs, including four stretches of insertion, the absence of the β15 strand and a unique dimerization arm (Fisk et al., 2010; Wang et al., 2014a). It methylates bovine H4R3, components of the nuclear pore complex and flagellar proteins, but it does not methylate several parasite proteins that contain GAR sequences (Fisk et al., 2010; Wang et al., 2014a). Remarkably, the TbPRMT6 knockdown reduces the growth rate of procyclic and bloodstream stages and yields aberrant morphologies in both forms (Fisk et al., 2010). Therefore, this enzyme participates in epigenetics, cell growth and cell morphology of T. brucei (Table 1).
TbPRMT7 is expressed in the bloodstream and procyclic stages, but is apparently dispensable (Fisk et al., 2009). Unlike human PRMT7, the trypanosomatid homologue is localized only in the cytoplasm and lacks the second AdoMet binding-like domain, which is required for the activity of mammalian PRMT7; however, the parasite enzyme showed a robust monomethylation activity on several targets (Fisk et al., 2009). Homodimerization of TbPRMT7 is due to the interaction between a dimerization arm of one subunit with the AdoMet-binding domain of the other and consequently, mutations on the dimerization arm remove dimerization and significantly reduce the methyltransferase activity (Wang et al., 2014b; Debler et al., 2016). In addition, the change of a glutamate residue to aspartate in the double E loop (E181D) confers to TbPRMT7 the ability to perform ADMA, whereas an additional change of glutamine residue to alanine in the THW loop (E181D/Q329A double mutant) generated SDMA (Jain et al., 2016), indicating that specific residues in the double E and THW loops are responsible for the product formed by different PRMTs.
An study analysing single and double TbPRMTs knockdowns (Lott et al., 2014) showed that: (i) the reduction of TbPRMT1 induces a decrease in TbPRMT3 protein levels and similarly, the TbPRMT3 knockdown reduced the TbPRMT1 expression; (ii) the knockdown of TbPRMT1 or TbPRMT3 produced a decrease in ADMA with a concomitant increase in MMA, confirming that TbPRMT1 and TbPRMT3 form functional hetero-oligomers; (iii) in TbPRMT7 knockdown, a very slight reduction of ADMA and unchangeable levels of MMA were observed, but the double TbPRMT1/7 knockdown dramatically reduced ADMA and MMA, proposing that both enzymes can compete for the arginine monomethylation on the same substrates; (iv) in single TbPRMT6 and double TbPRMT6/7 knockdowns no significant changes were observed in ADMA, confirming that TbPRMT6 methylates a relatively narrow group of substrates, but double knockdowns showed a more dramatic growth defect in the procyclic form that single repression of TbPRMT6, indicating that patterns of ADMA and MMA are critical to proliferation of procyclic forms (Lott et al., 2014). All these results indicate that TbPRMTs display a functional interplay at multiple levels.
Leishmaniasis is a group of tropical diseases that affect 12 million people worldwide with 1.5 million new cases diagnosed each year (Alvar et al., 2012). Leishmania spp. has two forms: promastigote in the sand fly vector and amastigote into the infected macrophages of the vertebrate host. The presence of arginine-methylated proteins in Leishmania sp. was demonstrated in 1986 and some of them have been identified (Paolantonacci et al., 1986). Leshmania sp. has five PRMT homologues to those of T. brucei, but studies showed certain differences, for example, the L. major PRMT3 has an intact double E loop, suggesting that it could be an active enzyme (Campagnaro et al., 2021). The expression of LmPRMT7 is high in the promastigote logarithmic growth phase and intracellular amastigotes, but low in the promastigote stationary phase (Ferreira et al., 2014). Interestingly, the LmPRMT7 knockout augmented infectivity in vitro and in vivo; and in concordance, the overexpression of this enzyme leads to a reduced progression of the lesion; moreover, the LmPRMT7 deletion rescues the pathogenic phenotype of an attenuated strain of L. major (Ferreira et al., 2014; Alcoforado Diniz et al., 2021). The absence of LmPRMT7 also impairs parasite development within the sand fly vector Phlebotomus duboscqi (Alcoforado Diniz et al., 2021). The proteome comparison of wild-type and Δprmt7 parasites allowed the identification of 40 hypomethylated proteins in the mutant cells, including 17 RBPs (Ferreira et al., 2020). Interestingly, the half-life of the RBP16 protein was reduced in the stationary phase of Δprmt7 cells compared to wild-type parasites, whereas the half-life of Alba3 was similar between both populations, although the absence of PRMT7 diminished the binding of Alba3 to the mRNA of the virulence-factor δ-amastin, affecting its stability (Ferreira et al., 2020), suggesting that LmPRMT7 may regulate the protein stability or the affinity of RBPs for their mRNA targets, impacting in the life cycle progression and virulence of L. major (Table 1).
Arginine methylation and PRMTs in Plasmodium falciparum
Parasites of the genus Plasmodium are the causative agents of malaria. The World Health Organization estimated about 229 million malaria cases and 409 000 malaria-related deaths in 2019 (World Health Organization, 2020). The major species infecting humans are P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi, although the majority of the malaria cases are produced by P. falciparum, which is associated with the most severe morbidity and mortality in Africa, whereas P. vivax is the specie more widely distributed (Jong et al., 2020). Parasites of the genus Plasmodium have several stages in the life cycle: gametes, zygotes, ookynetes, oocysts and sporozoites in the insect vector (females mosquitoes of the genus Anopheles) and schizonts, merozoites, ring, trophozoites and gametocytes in the vertebrate hosts. In addition, in P. vivax and P. ovale infections, some parasites become in dormant forms called ‘hypnozoites’.
Immunoprecipitation assays in blood stages of P. falciparum using anti-methyl arginine antibodies showed the presence of 843 proteins with this PTM involved in several functions such as protein synthesis, chromatin organization and haemoglobin metabolic processes, among others, indicating that arginine methylation may play an important regulatory role in the parasite biology, including host–parasite interactions (Zeeshan et al., 2017). P. falciparum encodes three PRMTs: PfPRMT1, PfPRMT4/CARM1 and PfPRMT5 (Fisk and Read, 2011). Recombinant PfPRMT1 methylates histones H4 and H2A and several conserved substrates involved in RNA metabolism, such as fibrillarin, poly(A)-binding protein II, ribosomal protein S2 and a splicing factor; in concordance, this enzyme is localized in both the cytoplasm and the nucleus (Fan et al., 2009). In summary, these studies indicate that PfPRMT1 may play important roles in epigenetics and RNA metabolism (Table 1).
A study about the organization and assembly of the Sm core complex in the P. falciparum spliceosome suggested that its proteins form a heptameric ring, in which the methylation of PfSmD1, catalysed by PfPRMT5, allows the interaction of this Sm protein with two proteins containing the Tudor domain: PfSMN (survival of motor neurons) and PfTSN (Tudor Staphylococcal nuclease) (Hossain et al., 2013), indicating that PfPRMT5 has an important role in splicing (Table 1).
At the moment, neither the role of PfPRMT4/CARM1 in the biology of P. falciparum nor the function of PRMTs in the other Plasmodium species has been investigated.
Arginine methylation and PRMTs in Toxoplasma gondii
Toxoplasma gondii is an obligate intracellular parasite that produces a disease called toxoplasmosis. It has been estimated that this microorganism infects 16–40% of the population in the USA and up to 80% of the population in other countries (Hill and Dubey, 2002). Members of the cat family are its final hosts, whereas various warm-blooded animals including humans, are intermediate hosts (Montoya and Liesenfeld, 2004). The T. gondii life cycle consists of two stages, the replicating tachyzoite and the latent bradyzoite. Bradyzoites can remain in host tissues for its lifetime without health consequences, but when host immunity is attenuated, bradyzoites can differentiate into actively replicating tachyzoites, causing life-long chronic infection.
This parasite has four PRMTs similar to PRMT1, PRMT3, PRMT4/CARM1 and PRMT5 (Fisk and Read, 2011). Approximately 50% of the MMA-proteins found in wild-type parasites do not exhibit this modification in TgPRMT1 knockout cells, indicating that they are putative substrates for this enzyme (Yakubu et al., 2017). TgPRMT1 substrates include the H4 histone, two ApiAP2 transcription factors, several RBPs, a lysine methyltransferase containing a SET domain, myosin E and SPM2 (a component of the subpellicular microtubules that regulate cell division) and, interestingly, most of them are also susceptible to phosphorylation, suggesting a relationship between both PTMs (Saksouk et al., 2005; Yakubu et al., 2017). The fact that myosin E and SPM2 are putative substrates of TgPRMT1 could explain why this enzyme is detected concentrated in the centrosome and in a pericentriolar material during the replication of tachyzoites (El Bissati et al., 2016). Another target of TgPRMT1 is the Argonaute protein (TgAgo), a component of the RNA-induced silencing complex (RISC) that in T. gondii displays low RNA slicer activity, but its methylation by TgPRMT1 induces the recruitment of the Tudor Staphylococcal Nuclease (TgTSN), making it a second potent RNA slicer for RISC (Musiyenko et al., 2012). Parasites lacking TgPRMT1 grow slower than wild-type ones and show abnormal daughter buds, perturbed centrosome stoichiometry and loss of synchronous replication (Musiyenko et al., 2012; El Bissati et al., 2016). Thus, TgPRMT1 participates in epigenetics, RNA metabolism and splicing, mechanisms that may control cell duplication (Table 1).
TgPRMT4/CARM1 catalyses the methylation of H3R17, an epigenetic mark for gene activation that is present in genes specifically expressed by tachyzoites; concordantly, inhibition of TgPRMT4/CARM1 induces conversion to bradyzoites, as well as a decrease of H3R17 methylation (Saksouk et al., 2005). On the other hand, TgPRMT5 produces MMA and SMDA on H3R26 and H4R3 and is expressed in both tachyzoites and bradyzoites, but it has different localization in both stages, in tachyzoites, it is found in the cytoplasm, while in bradyzoites it is located mainly in the nucleus (Liu et al., 2019). Therefore, TgPRMT4/CARM1 and TgPRMT5 participate in epigenetics, which in turn regulates the stage conversion (Table 1).
Arginine methylation and PRMTs in Entamoeba histolytica
Entamoeba histolytica is the microorganism that causes human amebiasis. This parasite infects up to 50 million people worldwide each year, producing intestinal and extra-intestinal diseases that cause between 40 000 and 100 000 deaths annually (Walsh, 1986). Its life cycle consists of two stages, the infective cysts and the invasive trophozoites.
The E. histolytica chromatin is organized in nucleosomes, but the distance of the DNA linker is irregular (Torres-Guerrero et al., 1991). Moreover, its histone H4 is longer than the consensus due to the insertion at the N-terminus of three amino acid segments, one of them adds three lysine residues that could be susceptible to being modified by acetylation and/or methylation (Binder et al., 1995; Lozano-Amado et al., 2016). Regarding the arginine methylation of histones, the antibodies against MMA- and ADMA-H4R3 recognized nuclear proteins of amoeba, indicating the presence of this epigenetic mark in E. histolytica, although the alignment of the amino acid sequence of its H4 histone with the eukaryotic consensus sequence, showed that the modified arginine corresponds to position 8 (H4R8) (Borbolla-Vázquez et al., 2015; Lozano-Amado et al., 2016).
This parasite contains five PRMTs, four of them show similarity to type I enzymes, whereas the other is related to the PRMT5 family (type II) (Fisk and Read, 2011) (Fig. 2). The amino acid sequences of the four type I PRMTs display higher similarity to human PRMT1 and do not contain the specific signatures for other type I PRMTs; therefore, they were initially classified as PRMT1 homologues (Fisk and Read, 2011). However, our phylogenetic analysis suggested that one of these EhPRMTs does not show significant homology with any PRMT family (Borbolla-Vázquez et al., 2015) (Fig. 2), therefore, we named it as atypical PRMT (EhPRMTA), whereas the other three type I enzymes were termed EhPRMT1a, EhPRMT1b and EhPRMT1c (Borbolla-Vázquez et al., 2015). An antibody against human PRMT1 recognized these EhPRMT1 proteins and they were localized in the nucleus and the cytoplasm, suggesting that they could methylate both histones and non-histone proteins (Borbolla-Vázquez et al., 2015). In fact, the recombinant protein of EhPRMT1a catalysed the asymmetric dimethylation of H4R3 in commercial histones (Borbolla-Vázquez et al., 2015), indicating that it may be involved in epigenetics (Table 1); however, until now it is unknown whether the three EhPRMT1 proteins have redundant or different functional roles.
EhPRMTA is the type I enzyme that shows the lowest similarity to PRMT1 (Fisk and Read, 2011). In concordance, this protein was not recognized by an antibody against human PRMT1 (Borbolla-Vázquez et al., 2015). In addition, it does not contain several of the canonical amino acid residues of type I PRMTs (Medina-Gómez et al., 2021) and a phylogenetic analysis grouped this protein with the inactive PRMT3 of T. brucei (Fig. 1), although the EhPRMTA recombinant protein displayed enzymatic activity on commercial histones (Medina-Gómez et al., 2021). On the other hand, EhPRMTA interacts with the EhTSN protein, which is a transcription factor of E. histolytica (Cázares-Apátiga et al., 2017) and its homologues in other protozoan parasites participate in RNA metabolism (Hossain et al., 2008; Musiyenko et al., 2012), suggesting that this EhPRMT is involved in transcription regulation and in RNA metabolism; moreover, the EhPRMTA-knockdown increased cell growth and phagocytosis, but decreased cell migration and survival of trophozoites submitted to heat shock (Medina-Gómez et al., 2021), indicating that this PRMT participates in these events (Table 1), although the identification of its protein targets remains to be determined.
Discussion
Because there is a correlation between the expression level of some PRMTs and cancer (Yang and Bedford, 2013), investigation in both academic laboratories and the pharmaceutical industry has been leading to the discovery of several small molecules targeting different PRMTs that at micromolar and submicromolar concentrations display anti-tumour effects in cellular assays (Hu et al., 2016; Zhang and Zheng, 2016; Guccione and Richard, 2019). In fact, three specific inhibitors for PRMT5 (GSK3325695, JNJ-64619178 and EPZ015938) and one for type I PRMTs (GSK3368715) have been analysed in phase I clinical trials (Rakow et al., 2020). As PRMTs of protozoan parasites play important roles in their cell growth, virulence, conversion stages and response to stress conditions (Table 1), anti-PRMT drugs could also be effective for the treatment of diseases produced by those pathogenic microorganisms. In fact, several small molecules that inhibit the activity of PfPRMT1 also restrict the P. falciparum growth (Fan et al., 2009). Moreover, a study analysing the antiplasmodial activity of several compounds that inhibit epigenetic modifiers in cancer cells, showed that sinefungin, a pan-inhibitor of AdoMet-dependent methyltransferases, affected the asexual stage of P. falciparum, whereas the ellagic acid, a PRMT inhibitor, was 10-fold more potent against the asexual stage and early gametocytes than against mammalian cells (Coetzee et al., 2020). In addition, the structural differences shown by the PRMTs of the parasites with respect to their human homologues could be used to design specific anti-parasitic drugs.
Conclusions
Methylation of arginine residues is involved in the regulation of several molecular processes, such as transcription, RNA processing and signal transduction pathways, which in parasites may control pathogenesis-related events. Although most PRMTs from protozoan parasites have not been characterized, current research on some of these enzymes shows that they participate in cell growth, metabolism, stress response, stage transitions and virulence. Thus, the PRMTs of protozoan parasites are attractive targets to develop new therapeutic strategies that could help in the control of the public health problems generated by these pathogens. Actually, the considerable advances in the identification of clinically relevant PRMT inhibitors for cancer therapy could be extended to find effective drugs against the protozoan parasites.
Financial support
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) Grant FORDECYT-PRONACES/194163/2020.
Ethical standards
Not applicable.
Conflict of interest
The author declares there are no conflicts of interest.
References
- Ahmad A, Dong Y and Cao X (2011) Characterization of the PRMT gene family in rice reveals conservation of arginine methylation. PLoS One 6, e22664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hamashi AA, Diaz K and Huang R (2020) Non-histone arginine methylation by protein arginine methyltransferases. Current Protein & Peptide Science 21, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoforado Diniz J, Chaves MM, Vaselek S, Miserani Magalhães RD, Ricci-Azevedo R, de Carvalho RVH, Lorenzon LB, Ferreira TR, Zamboni D, Walrad PB, Volf P, Sacks DL and Cruz AK (2021) Protein methyltransferase 7 deficiency in Leishmania major increases neutrophil associated pathology in murine model. PLoS Neglected Tropical Diseases 15, e0009230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J and Boer Md (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7, e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand F (2007) Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryotic Cell 6, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer I, Lechner L, Pidroni A, Petrone A-M, Merschak P, Lindner H, Kremser L, Graessle S, Golderer G, Allipour S and Brosch G (2019) Type I and II PRMTs regulate catabolic as well as detoxifying processes in Aspergillus nidulans. Fungal Genetics and Biology 129, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT and Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Molecular Cell 33, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT and Richard S (2005) Arginine methylation. Molecular Cell 18, 263–272. [DOI] [PubMed] [Google Scholar]
- Binder M, Ortner S, Plaimauer B, Födinger M, Wiedermann G, Scheiner O and Duchêne M (1995) Sequence and organization of an unusual histone H4 gene in the human parasite Entamoeba histolytica. Molecular and Biochemical Parasitology 71, 243–247. [DOI] [PubMed] [Google Scholar]
- Blanc RS and Richard S (2017) Arginine methylation: the coming of age. Molecular Cell 65, 8–24. [DOI] [PubMed] [Google Scholar]
- Borbolla-Vázquez J, Orozco E, Betanzos A and Rodríguez MA (2015) Entamoeba histolytica: protein arginine transferase 1a methylates arginine residues and potentially modify the H4 histone. Parasites & Vectors 8, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnaro GD, Nay E, Plevin MJ, Cruz AK and Walrad PB (2021) Arginine methyltransferases as regulators of RNA-binding protein activities in pathogenic kinetoplastids. Frontiers in Molecular Biosciences 8, 692668. doi: 10.3389/fmolb.2021.692668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio F, Lu X, Pollock SB, LeRoy G, Garcia BA, Muir TW, Roeder RG and Allis CD (2013) H3R42me2a is a histone modification with positive transcriptional effects. Proceedings of the National Academy of Sciences 110, 14894–14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cázares-Apátiga J, Medina-Gómez C, Chávez-Munguía B, Calixto-Gálvez M, Orozco E, Vázquez-Calzada C, Martínez-Higuera A and Rodríguez MA (2017) The Tudor staphylococcal nuclease protein of Entamoeba histolytica participates in transcription regulation and stress response. Frontiers in Cellular and Infection Microbiology 7, 52. doi: 10.3389/fcimb.2017.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Collins RE and Zhang X (2005) Structural and sequence motifs of protein (histone) methylation enzymes. Annual Review of Biophysics and Biomolecular Structure 34, 267–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Gao G, Di Lorenzo A, Jayne S, Hottiger MO, Richard S and Bedford MT (2020) Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6. Journal of Biological Chemistry 295, 17060–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee N, von Grüning H, Opperman D, van der Watt M, Reader J and Birkholtz L-M (2020) Epigenetic inhibitors target multiple stages of Plasmodium falciparum parasites. Scientific Reports 10, 2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Jain K, Warmack RA, Feng Y, Clarke SG, Blobel G and Stavropoulos P (2016) A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proceedings of the National Academy of Sciences 113, 2068–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K, Suvorova ES, Xiao H, Lucas O, Upadhya R, Ma Y, Hogue Angeletti R, White MW, Weiss LM and Kim K (2016) Toxoplasma gondii arginine methyltransferase 1 (PRMT1) is necessary for centrosome dynamics during tachyzoite cell division. mBio 7, e02094-15. doi: 10.1128/mBio.02094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Miao J, Cui L and Cui L (2009) Characterization of PRMT1 from Plasmodium falciparum. Biochemical Journal 421, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson J-Y and Clarke SG (2013) Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. Journal of Biological Chemistry 288, 37010–37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira TR, Alves-Ferreira EVC, Defina TPA, Walrad P, Papadopoulou B and Cruz AK (2014) Altered expression of an RBP associated arginine methyltransferase 7 in Leishmania major affects parasite infection. Molecular Microbiology 94, 1085–1102. [DOI] [PubMed] [Google Scholar]
- Ferreira TR, Dowle AA, Parry E, Alves-Ferreira EVC, Hogg K, Kolokousi F, Larson TR, Plevin MJ, Cruz AK andWalrad PB (2020) PRMT7 regulates RNA-binding capacity and protein stability in Leishmania parasites. Nucleic Acids Research 48, 5511–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC and Read LK (2011) Protein arginine methylation in parasitic protozoa. Eukaryotic Cell 10, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Sayegh J, Zurita-Lopez C, Menon S, Presnyak V, Clarke SG and Read LK (2009) A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. Journal of Biological Chemistry 284, 11590–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Zurita-Lopez C, Sayegh J, Tomasello DL, Clarke SG and Read LK (2010) TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryotic Cell 9, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A and Clarke S (2000) PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Journal of Biological Chemistry 275, 32974–32982. [DOI] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S and Bedford MT (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. Journal of Biological Chemistry 277, 3537–3543. [DOI] [PubMed] [Google Scholar]
- Ganesh L, Yoshimoto T, Moorthy NC, Akahata W, Boehm M, Nabel EG and Nabel GJ (2006) Protein methyltransferase 2 inhibits NF-κB function and promotes apoptosis. Molecular and Cellular Biology 26, 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD and Clarke S (1998) RNA and protein interactions modulated by protein arginine methylation. Progress in Nucleic Acid Research and Molecular Biology 61, 65–131. [DOI] [PubMed] [Google Scholar]
- Guccione E and Richard S (2019) The regulation, functions and clinical relevance of arginine methylation. Nature Reviews Molecular Cell Biology 20, 642–657. [DOI] [PubMed] [Google Scholar]
- Hadjikyriacou A and Clarke SG (2017) Caenorhabditis elegans PRMT-7 and PRMT-9 are evolutionarily conserved protein arginine methyltransferases with distinct substrate specificities. Biochemistry 56, 2612–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D and Dubey JP (2002) Toxoplasma gondii: transmission, diagnosis and prevention. Clinical Microbiology and Infection 8, 634–640. [DOI] [PubMed] [Google Scholar]
- Ho M-C, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, Carter LG, Onikubo T, Kumar PR, Chan MK, Brenowitz M, Cheng RH, Reimer U, Almo SC and Shechter D (2013) Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One 8, e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MJ, Korde R, Singh S, Mohmmed A, Dasaradhi PVN, Chauhan VS and Malhotra P (2008) Tudor domain proteins in protozoan parasites and characterization of Plasmodium falciparum Tudor staphylococcal nuclease. International Journal for Parasitology 38, 513–526. [DOI] [PubMed] [Google Scholar]
- Hossain M, Sharma S, Korde R, Kanodia S, Chugh M, Rawat K and Malhotra P (2013) Organization of Plasmodium falciparum spliceosomal core complex and role of arginine methylation in its assembly. Malaria Journal 12, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Qian K, Ho M-C and Zheng YG (2016) Small molecule inhibitors of protein arginine methyltransferases. Expert Opinion on Investigational Drugs 25, 335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Warmack RA, Debler EW, Hadjikyriacou A, Stavropoulos P and Clarke SG (2016) Protein arginine methyltransferase product specificity is mediated by distinct active-site architectures. Journal of Biological Chemistry 291, 18299–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Dhall A and Shi Y (2019) Roles and regulation of histone methylation in animal development. Nature Reviews Molecular Cell Biology 20, 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong RM, Tebeje SK, Meerstein-Kessel L, Tadesse FG, Jore MM, Stone W and Bousema T (2020) Immunity against sexual stage Plasmodium falciparum and Plasmodium vivax parasites. Immunological Reviews 293, 190–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafková L, Debler EW, Fisk JC, Jain K, Clarke SG and Read LK (2017) The major protein arginine methyltransferase in Trypanosoma brucei functions as an enzyme-prozyme complex. Journal of Biological Chemistry 292, 2089–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafková L, Tu C, Pazzo KL, Smith KP, Debler EW, Paul KS, Qu J and Read LK (2018) Trypanosoma brucei PRMT1 is a nucleic acid-binding protein with a role in energy metabolism and the starvation stress response. mBio 9. doi: 10.1128/mBio.02430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis V, Wang L, Tae S, Hu Y-J, Imbalzano AN and Sif S (2012) Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. Journal of Biological Chemistry 287, 29801–29814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JE, Dlakić M and Clarke S (2003) Automated identification of putative methyltransferases from genomic open reading frames. Molecular & Cellular Proteomics 2, 525–540. [DOI] [PubMed] [Google Scholar]
- Lanouette S, Mongeon V, Figeys D and Couture J (2014) The functional diversity of protein lysine methylation. Molecular Systems Biology 10, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S and Bedford MT (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. Journal of Biological Chemistry 280, 32890–32896. [DOI] [PubMed] [Google Scholar]
- Li H-T, Gong T, Zhou Z, Liu Y-T, Cao X, He Y, Chen CD and Zhou J-Q (2015) Yeast Hmt1 catalyses asymmetric dimethylation of histone H3 arginine 2 in vitro. Biochemical Journal 467, 507–515. [DOI] [PubMed] [Google Scholar]
- Liu M, Li F-X, Li C-Y, Li X-C, Chen L-F, Wu K, Yang P-L, Lai Z-F, Liu T, Sullivan WJ, Cui L and Chen X-G (2019) Characterization of protein arginine methyltransferase of TgPRMT5 in Toxoplasma gondii. Parasites & Vectors 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Li J, Fisk JC, Wang H, Aletta JM, Qu J and Read LK (2013) Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular process impacted by arginine methylation. Journal of Proteomics 91, 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Zhu L, Fisk JC, Tomasello DL and Read LK (2014) Functional interplay between protein arginine methyltransferases in Trypanosoma brucei. MicrobiologyOpen 3, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Amado D, Herrera-Solorio AM, Valdés J, Alemán-Lazarini L, Almaraz-Barrera MdJ, Luna-Rivera E, Vargas M and Hernández-Rivas R (2016) Identification of repressive and active epigenetic marks and nuclear bodies in Entamoeba histolytica. Parasites & Vectors 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AE, Zurita-Lopez C, Regis A, Blum E, Conboy A, Elf S and Clarke S (2007) Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryotic Cell 6, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gómez C, Bolaños J, Borbolla-Vázquez J, Munguía-Robledo S, Orozco E and Rodríguez MA (2021) The atypical protein arginine methyltrasferase of Entamoeba histolytica (EhPRMTA) is involved in cell proliferation, heat shock response and in vitro virulence. Experimental Parasitology 222, 108077. [DOI] [PubMed] [Google Scholar]
- Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M and Guccione E (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nature Structural & Molecular Biology 19, 136–144. [DOI] [PubMed] [Google Scholar]
- Montoya J and Liesenfeld O (2004) Toxoplasmosis. The Lancet 363, 1965–1976. [DOI] [PubMed] [Google Scholar]
- Musiyenko A, Majumdar T, Andrews J, Adams B and Barik S (2012) PRMT1 methylates the single Argonaute of Toxoplasma gondii and is important for the recruitment of Tudor nuclease for target RNA cleavage by antisense guide RNA. Cellular Microbiology 14, 882–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najbauer J, Johnson BA, Young AL and Aswad DW (1993) Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. The Journal of Biological Chemistry 268, 10501–10509. [PubMed] [Google Scholar]
- Paolantonacci P, Lawrence F, Lederer F and Robert-Gero M (1986) Protein methylation and protein methylases in Leishmania donovani and Leishmania tropica promastigotes. Molecular and Biochemical Parasitology 21, 47–54. [DOI] [PubMed] [Google Scholar]
- Pasternack DA, Sayegh J, Clarke S and Read LK (2007) Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryotic Cell 6, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Xu Y, Wang X, Zahariev S, Pongor S, Aletta J and Read LK (2002) Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Molecular and Biochemical Parasitology 18, 49–59. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Pasternack DA and Read LK (2005) In vitro and in vivo analysis of the major type I arginine methyltransferase from Trypanosoma brucei. Molecular and Biochemical Parasitology 144, 206–217. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Frainier AS, Munini DN, Wiemer JM, Karpie AR and Sattora JJ (2013) Identification of a novel lipin homologue from the parasitic protozoan Trypanosoma brucei. BMC Microbiology 13, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett KL, Raposo AE, Bullivant S, Anderson IC, Piller SC and Plett JM (2017) Root morphogenic pathways in Eucalyptus grandis are modified by the activity of protein arginine methyltransferases. BMC Plant Biology 17, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakow S, Pullamsetti SS, Bauer U-M and Bouchard C (2020) Assaying epigenome functions of PRMTs and their substrates. Methods (San Diego, Calif) 175, 53–65. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Cesbron-Delauw M-F and Hakimi M-A (2005) Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Molecular and Cellular Biology 25, 10301–10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro PP, Cecchi G, Franco JR, Paone M, Diarra A, Priotto G, Mattioli RC and Jannin JG (2015) Monitoring the progress towards the elimination of gambiense human African trypanosomiasis. PLoS Neglected Tropical Diseases 9, e0003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Guerrero H, Peattie DA and Meza I (1991) Chromatin organization in Entamoeba histolytica. Molecular and Biochemical Parasitology 45, 121–130. [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM and Sattler M (2011) Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nature Structural & Molecular Biology 18, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Venne AS, Kollipara L and Zahedi RP (2014) The next level of complexity: crosstalk of posttranslational modifications. Proteomics 14, 513–524. [DOI] [PubMed] [Google Scholar]
- Walsh JA (1986) Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of morbidity and mortality. Clinical Infectious Diseases 8, 228–238. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhu Y, Chen J, Li X, Peng J, Chen J, Zou Y, Zhang Z, Jin H, Yang P, Wu J, Niu L, Gong Q, Teng M and Yunyu S (2014a) Chrystal structure of arginine methyltransferase 6 from Trypanosoma brucei. PLoS One 9, e87267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhu Y, Caceres TB, Liu L, Peng J, Wang J, Chen J, Chen X, Zhang Z, Zuo X, Gong Q, Teng M, Hevel JM, Wu J and Shi Y (2014b) Structural determinants for the strict monomethylation activity by Trypanosoma brucei protein arginine methyltransferase 7. Structure (London, England: 1993) 22, 756–768. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Wang C-W, Lin W-C, Tsai Y-J, Chang C-P, Lee Y-J, Lin M-J and Li C (2017) Identification, chromosomal arrangements and expression analyses of the evolutionarily conserved prmt1 gene in chicken in comparison with its vertebrate paralogue prmt8. PLoS One 12, e0185042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020) World Malaria Report 2020: 20 Years of Global Progress and Challenges. Geneva: World Health Organization. [Google Scholar]
- Xu W (2004) A methylation-mediator complex in hormone signaling. Genes & Development 18, 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubu RR, Simon de Moterri NC, Nieves E, Kim K and Weiss LM (2017) Comparative monomethylarginine proteomics suggests that protein arginine methyltransferase 1 (PRMT1) is a significant contributor to arginine monomethylation in Toxoplasma gondii. Molecular & Cellular Proteomics 16, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y and Bedford MT (2013) Protein arginine methyltransferases and cancer. Nature Reviews Cancer 13, 37–50. [DOI] [PubMed] [Google Scholar]
- Zeeshan M, Kaur I, Joy J, Saini E, Paul G, Kaushik A, Dabral S, Mohmmed A, Gupta D and Malhotra P (2017) Proteomic identification and analysis of arginine-methylated proteins of Plasmodium falciparum at asexual blood stages. Journal of Proteome Research 16, 368–383. [DOI] [PubMed] [Google Scholar]
- Zhang X and Cheng X (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure (London, England: 1993) 11, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J and Zheng YG (2016) SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chemical Biology 11, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou L and Cheng X (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. The EMBO Journal 19, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM and Jane SM (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature Structural & Molecular Biology 16, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X-X, Zhang Y-B, Ni P-L, Wu Z-L, Yan Y-C and Li Y-P (2016) Protein arginine methyltransferase 6 (Prmt6) is essential for early zebrafish development through the direct suppression of gadd45αa stress sensor gene. Journal of Biological Chemistry 291, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Xie Y, Hu H, Hu G, Patel VS, Zhang J, Yu K, Huang Y, Jiang H, Liang Z, Zheng YG and Luo C (2015) Molecular mechanism underlying PRMT1 dimerization for SAM binding and methylase activity. Journal of Chemical Information and Modeling 55, 2623–2632. [DOI] [PubMed] [Google Scholar]