Abstract
Background
The molecular mechanisms of esophageal squamous cell carcinoma (ESCC) remain poorly understood. Transmembrane emp24 trafficking protein 3 (TMED3) acts as an oncogene or tumor suppressor gene in different cancers. Our study was to explore the clinicopathological significance and functional roles of TMED3 in ESCC.
Methods
Immunohistochemistry, qPCR, and western blotting were used to analyze the expression of TMED3 in ESCC tissues and cells. Statistical analysis was performed to analyze the relationship between TMED3 expression and tumor characteristics in patients with ESCC. The role of TMED3 in vitro and in vivo was investigated by performing functional verification experiments and using a xenograft mouse model. Proteins that are functionally related to TMED3 were recognized by Affymetrix microarray and Ingenuity Pathway Analysis (IPA). Functional verification experiments were performed to analyze the role of FAM60A (a protein functionally related to TMED3) in vitro.
Results
We confirmed the overexpression of TMED3 was correlated with poor prognosis in ESCC patients. When TMED3 was knocked down, ESCC cell proliferation, migration, and invasion were inhibited whereas cell apoptosis was promoted in vitro, and tumorigenicity was inhibited in vivo. We further revealed significant changes in gene expression profile in TMED3 knockdown cells. Among these differentially expressed genes, FAM60A was overexpressed in ESCC tissues. Furthermore, knocking down FAM60A significantly weakened the proliferation ability of ESCC cells and reversed TMED3's tumorigenicity of ESCC cells.
Conclusion
Our study revealed an oncogenic role of TMED3 in ESCC.
Keywords: TMED3, FAM60A, Esophageal squamous cell carcinoma, Progression, Apoptosis
At a glance commentary
Scientific background on the subject
The molecular mechanisms of esophageal squamous cell carcinoma (ESCC) remain poorly understood. The transmembrane emp24 trafficking protein 3 (TMED3) acts as an oncogene or tumor suppressor gene in different cancers. It is of great significance to explore the clinicopathological significance and functional role of TMED3 in ESCC.
What this study adds to the field
The results of our study demonstrated that TMED3 overexpression plays an important role in ESCC through regulating FAM60A and inhibiting the apoptosis signaling pathway. These findings help us better understand the molecular mechanisms of ESCC and potentially may help the prognosis and treatment of ESCC in the future.
Esophageal carcinoma is the sixth leading cause of cancer-related death worldwide, with an estimated 572,034 new cases and 508,585 deaths each year [1]. Esophageal squamous cell carcinoma (ESCC) is the main pathological type in China, accounting for more than 90% of esophageal cancer cases [2]. Neoadjuvant chemoradiotherapy or chemotherapy followed by surgery and radical chemoradiotherapy are standard treatments for esophageal cancer [3]. Despite the rapid development of various treatment regimes in recent decades, the 5-year survival rate of patients with locally advanced ESCC is only 25%, and recurrence and metastasis are still the leading causes of death [4]. Therefore, identifying the molecular mechanisms of ESCC is urgent and highly needed to improve the clinical outcome of ESCC patients.
Transmembrane emp24 domain-containing protein (TMED) family has emerged as important protein transport regulators and is essential for embryonic development and homeostasis [5]. TMED proteins dimerize and interact with coatomer protein complexes to promote cargo selection and vesicle formation during anterograde and retrograde transport [6]. Abnormally controlled transport of proteins and expression of TMED proteins in the secretory pathway contribute to diseases such as cancer. TMED3 is one of TMED/p24 family members that share a similar structural organization, including a luminal Golgi-dynamics domain, a luminal coiled-coil domain, a transmembrane domain, and a short cytosolic tail [[7], [8], [9], [10]]. Many studies have shown that TMED3 plays a tumor suppressor or oncogenic role in different cell types, including prostate cancer, colon cancer, hepatocellular cancer, breast cancer, clear cell renal cell, and gastric cancer [[11], [12], [13], [14], [15], [16], [17]]. However, the role of TMED3 in ESCC is unknown.
In this study, we aimed to investigate the function, molecular mechanism, and clinicopathological significance of TMED3 in ESCC. Using human tissues, cell lines and a xenograft mouse model, our results demonstrated that TMED3 plays an oncogenic role in ESCC progression and may help the prognosis and treatment of ESCC in the future.
Materials and methods
Clinical tissue samples and patient information
One hundred sixty-six paraffin-embedded surgical specimens and clinical information from patients with ESCC at the Sun Yat-sen University Cancer Center, Guangzhou, China, were collected from January 2006 to December 2008. Clinicopathological classification and staging before treatment were based on the American Joint Committee on Cancer (AJCC) guidelines. All tissue samples were collected in cases without preoperative chemotherapy. All patients received platinum- or taxane-based postoperative chemotherapy, once every 3 weeks for 2–4 cycles. The use of clinical information and tissue samples in this study was approved by the Institutional Research Ethics Committee of our hospital and consented by the patients.
Immunohistochemistry (IHC)
IHC was performed on tissue sections as described [15]. After deparaffinization, the sections were stained using anti-TMED3 (ab223175, Abcam, Cambridge, MA), anti-FAM60A (NBP2-68734, Novus, Littleton, CO), anti-K67 (ab16667, Abcam) and anti-Bcl-2 (ab32124, Abcam). The primary antibody was replaced with normal goat IgG in negative controls. Stained sections were observed by two pathologists independently who were blinded to the clinicopathological information of the samples. Positive staining in the samples was scored as 0 (no positive staining), 1 (0–25% positively stained cancer cells), 2 (25–50%), 3 (50–75%), or 4 (>75%). Staining intensity was scored as 0 (no staining), 1 (weak staining, light brown), 2 (moderate staining, moderate brown), or 3 (strong staining, dark brown). IHC score was calculated by multiplying the positive staining score by the staining intensity score. IHC score of ≥6 was defined as high expression, and of <6 as low expression.
Cell culture, plasmid constructs, lentivirus production and transduction, and transfection
Human ESCC cell lines Eca-109 (CVCL_6898), TE-1 (CVCL_1759) were obtained commercially from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All human cell lines were authenticated using STR profiling within the last three years. All cell lines were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Bioind, Kibbutz Beit, Israel) in a humidified 5% CO2 atmosphere at 37 °C. Scanning electron microscopy was used to confirm that all cell lines tested negative for mycoplasma contamination. The design of shRNA, the construction and package of RNAi lentivirus and overexpression lentivirus, and lentiviral infection of target cells were serviced by Shanghai Biosciences (Shanghai, China). The knockdown efficiency at the mRNA and protein level of the target gene was determined by quantitative real-time PCR (q-PCR) and western blotting.
RNA extraction, reverse transcription, and q-PCR
Total RNA was extracted with Trizol according to the supplier's operating instructions (Sigma–Aldrich, St. Louis, MO). cDNA was obtained by reverse transcription using the HiScript II QRT SuperMix for qPCR (+gDNA WIPER) (Vazyme, Nanjing, China). Q-PCR was conducted using AceQ qPCR SYBR Green master mix (Vazyme). The relative expression level was calculated as 2-∆∆Ct, where Ct represented the threshold cycle of each transcript, and GAPDH was used to normalize gene expression. TMED3-specific primers were 5′-GGCGTGAAGTTCTCCCTGGATT-3′ (forward) and 5′-GCTGTCGTACTGCTTCTTCGTTTC-3′ (reverse). FAM60A-specific primers were 5′-CAAGCCAAAGATGTACCGAAGT-3′ (forward) and 5′-CATTGCAGATGTCTCCTGAACGA-3′ (reverse). GAPDH-specific primers were 5′-CGGATTTGGTCGTATTGGG-3′ (forward) and 5′-GATTTTGGAGGGATCTCGC-3′ (reverse).
Western blotting
Cultured cells were lysed with RIPA lysis buffer (Beyotime, Shanghai, China). BCA kit (Pierce, Rockford, IL) was used to measure protein concentrations. The Flag and HA sequences were inserted into plasmids by the primers containing specific restriction sites. The plasmids were transformed into competent cells, screened, and amplified with LB containing ampicillin. High-concentration recombinant Flag-TMED3 and HA-FAM60A were obtained. These two plasmids were verified by DNA sequencing. Endotoxin-free extraction was carried out for subsequent cell transfection [18]. The equivalent amounts of proteins were separated on 10% SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Millipore, Bedford, MA). PVDF membrane was blocked with a blocking buffer and was incubated with an anti-TMED3 (ab223175, Abcam), anti-FAM60A (ab167180, Abcam), anti-GAPDH (AP0063, Bioworld, Nanjing, China), anti-DYKDDDDK Tag (14,793, Cell Signaling, Danvers, MA), anti-HA (ab9110, Abcam), anti-GAPDH antibody (AP0063, Bioworld), and the corresponding secondary antibodies, goat anti-rabbit IgG (A0208, Beyotime) or goat anti-mouse IgG (A0216, Beyotime). Immobilon Western Chemiluminescent HRP Substrate kit (Millipore) was used for color development and the bands were detected using an Amersham Imager 600 (GE Healthcare Little Chalfont, Buckinghamshire, UK).
MTT assay
Cells were placed in a 96-well plate at a density of 2000/well and were incubated overnight. From the second day after plating, 20 μl of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-H-tetrazolium bromide (MTT; Genview, Beijing, China) reagent (5 mg/ml) was added into the designated wells. 100 μl DMSO was used to dissolve the MTT formazan precipitate. The absorbance at 490/570 nm was measured with a microplate reader (Tecan, Männedorf, Switzerland).
Colony formation
Clonogenic growth of Eca-109 and TE-1 cells was assessed by incubating 800 cells transfected with lentiviral-based shTMED3, shFAM60A, or control. Clonogenic cultures were performed in a 6-well plate in 1 ml DMEM (Corning, New York, NY). Colonies of more than 50 cells were photographed under a fluorescence microscope after fixing the cells with 4% paraformaldehyde for 30 min and washing with PBS. Clones were photographed under a fluorescence microscope (Olympus, Tokyo, Japan). Cells were stained by impurity-free GIEMSA (Shanghai Dingguo Biotechnology, Shanghai, China). Colonies of more than 50 cells were counted after being washed, dried, and photographed.
Wound healing assay
Cells were plated in 96 well plates which were slightly pushed up by the scratcher to form scratches and were washed with the serum-free medium to remove floating cells. Low-concentration serum medium (0.5% FBS) was added to the 96 well plates. The target cells were cultured in 5% CO2 incubator (Thermo, Waltham, USA) at 37 °C. Migrating cells were photographed under a fluorescence microscope (Olympus, Tokyo, Japan). Cell motility was quantified by measuring the distance between the boundaries of migrating cells.
Transwell assay
Transwell assay was performed in a 24-well Transwell chamber (Corning). The required quantity of chambers was placed in an empty 24-well plate and placed in the incubator for 1 h after adding 100 μl of serum-free medium. The medium in the lower chamber was replaced with 600 μl of medium with 30% FBS. After making cell suspension, 100 μl cell suspension (containing 100,000–200,000 cells) was added to each chamber. The chambers were transferred into the lower chamber containing a medium with 30% FBS and cultured for 4–24 h in the incubator. The cells were immersed in the staining solution. The cells on the underside of the membrane were stained and counted under a microscope after being washed and dried.
Flow cytometry
The cells were cultured in a 6-well plate after transfection with lentiviral-based shTMED3, shFAM60A, or control, until the confluence was about 70%, followed by being treated with 5 μl pancreatic enzymes and stained with 5 μl Annexin V-APC and 5 μl propidium iodide (PI). Cell cycle distribution was analyzed by FACS Calibur system (3550UV, BioRad, Hercules, CA). Viable cells were not stained with Annexin V, necrotic cells double-positive for Annexin V and PI, while apoptotic cells Annexin V-positive and PI-negative.
HCS cell proliferation assay
The target cells were cultured in a 96-well culture plate and then transfected by lentivirus for GFP expression for 2–3 days. The expression of GFP was observed under a fluorescence microscope. When the fluorescence rate reached 70–90%, the cells were cultured until the rate of cell fusion reached 70–90%, and then collected for the following experiment. GFP+ cells were recognized and pictures were taken by Celigo® Image Cytometer (Nexcelom Bioscience, Lawrence, MA). The image was analyzed and processed with the software, and the number of cells in different groups was calculated.
Xenograft nude mouse model
Four-week-old female nude mice (BALB/cSlcn/n) were purchased from SLAC Laboratory Animal Co, Ltd (Shanghai, China) and maintained under specific pathogen-free conditions. Mice were randomized into two groups (10 mice per group), control group, and TMED3KD group. Tumors were established by subcutaneously injecting Eca-109-TMED3KD cells or Eca-109-Control cells (1 × 107/ml; 200 μl per xenograft) into the right forelimb of BALB/cSlcn/n mice. The weight of the mice was measured every week after implantation. Mice were injected intraperitoneally with D-Luciferin (15 mg/ml) and anesthetized with 0.7% pentobarbital sodium. Bioluminescent images were measured 2 or 3 times a week after 14 days using IVIS Spectrum (Berthold Technologies, Wildbad, Germany). All mice were sacrificed after 21 days to collect the data of tumor weight. Animal experiments were performed in compliance with the Animal Ethical Committee of the Sun Yat-sen University Cancer Center (Guangzhou, China) after the approval of the local Animal Ethical Committee.
Gene array and enrichment analysis
The library for gene microarray was constructed with the TruSeq Stranded mRNA LT Sample Preparation Kit (Illumina, San Diego, CA) according to the standard protocols, and then scan it by Affymetrix Scanner 3000 (Affymetrix, Santa Clara, CA). Ingenuity Pathway Analysis (IPA) analysis was applied to identify the pathways that the differentially expressed genes (Fold Change≥1.5 and FDR<0.05) belong to. A Z-score>2 suggested significant activation of the pathway, and a Z-score < −2 significant inhibition.
Apoptosis antibody array
The expression profiles of apoptosis-related proteins were analyzed with a human apoptosis antibody array (ab134001, Abcam), according to the standard protocol. Place the capture and control antibodies in duplicate on the nitrocellulose membrane. Capture and control antibodies were placed in duplicate on the nitrocellulose membranes. Cellular extracts were diluted and incubated with the human apoptosis array overnight. The array was washed and then incubated with biotinylated detection antibodies.
Gene expression omnibus (GEO) data acquisition and analysis
We analyzed the expression of FAM60A in ESCC and normal tissue using the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GEO data sets (GSE23400, GSE20347, and GSE1735) were obtained from the GEO database. The data set included FAM60A mRNA expression in ESCC tissue and normal esophagus tissue. The FAM60A mRNA expression level in ESCC tissue and normal esophagus tissue was compared using t test. p < 0.05 indicated statistical significance.
Statistical analysis
Statistical analysis was performed by SPSS version 24.0 (SPSS, Chicago, IL) and GraphPad Prism 6.0 (GraphPad, La Jolla, CA). Mann–Whitney test and Kruskal–Wallis test were used for analyzing the relationship between TMED3 expression and tumor characteristics in patients with ESCC. T test, wilcoxon test and chi-square test were used in the data analysis. All data were presented as mean ± SD. Kaplan–Meier analysis was applied to assess survival probability, and a log-rank test was applied to compare differences in survival in univariate analysis.
Results
Upregulated TMED3 was significantly associated with poor clinical outcomes and prognosis of ESCC
To examine the expression of TMED3 in ESCC, we evaluated 166 paraffin-embedded ESCC specimens by IHC staining. We found that the expression of TMED3 in ESCC tissues was higher than that in adjacent normal tissues (Fig. 1A). In ESCC tissues, 1.0% had negative, 54.4% weak, 33.0% moderate, and 11.7% strong, staining of TMED3 protein. In adjacent normal tissues, 57.1% had negative staining, 42.9% weak, and none moderate or strong staining of TMED3 protein (Fig. 1B). 44.7% (46/103) of ESCC specimens were categorized as the high TMED3 group and the rest 55.3% (57/103) was the low TMED3 group. 100% (63/63) of adjacent normal tissues were categorized as the low TMED3 group and none as the high TMED3 group. The IHC score of TMED3 in ESCC tissues was significantly higher than that in adjacent normal tissues (p < 0.001; Fig. 1C).
Fig. 1.
TMED3 upregulation was related to poor clinical outcomes and prognosis in ESCC patients. (A) Representative examples of TMED3 staining on adjacent normal tissues and ESCC tissues. Scale bars = 100 μm or 200 μm. (B) Distribution of TMED3 staining intensity in ESCC and adjacent normal tissues. (C) IHC scores of TMED3 in the adjacent normal tissues (n = 63) and ESCC tissues (n = 103), which were analyzed by the Mann–Whitney U test. (D) Representative examples of TMED3 staining on ESCC tissues from patients with three lymph nodes or nine lymph nodes. (E) Representative examples of TMED3 staining on ESCC tissues from patients at AJCC Stage I, II, and III. The right panel of Fig. 1D and the right panel of Fig. 1E are from the same sample. (F) Kaplan–Meier analysis of overall survival for ESCC patients that were grouped according to TMED3 expression level (n = 103, log-rank test). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Statistical analysis was performed to analyze the relationship between TMED3 expression and tumor characteristics in ESCC patients. Analysis showed that the expression of TMED3 was significantly associated with the number of lymph nodes (p = 0.023), but not significantly related to other pathological variables, including age, gender, smoking history, drinking history, tumor size, grade, T stage, N stage and AJCC stage (Table 1). Combining the results of IHC staining, we further confirmed that patients with high TMED3 expression were more likely to have a higher rate of lymph node metastasis (Fig. 1D). We observed from the IHC results that the higher the AJCC stage, the higher the expression of TMED3, although the statistical analysis did not show that the expression of TMED3 was associated with the AJCC stage, which may be related to our insufficient sample size. Kaplan–Meier survival analysis showed that upregulated TMED3 was markedly associated with poor overall survival (p = 0.012; Fig. 1F). Taken together, upregulated TMED3 was related to poor clinical outcomes and prognosis in ESCC patients.
Table 1.
Relationship between TMED3 expression and tumor characteristics in patients with ESCC.
| Characteristics | No. | TMED3 expression |
p value | |
|---|---|---|---|---|
| Low | High | |||
| All | 103 | 57 | 46 | |
| Age (years) | 0.600 | |||
| ≤65 | 53 | 28 | 25 | |
| >65 | 50 | 29 | 21 | |
| Gender | 0.635 | |||
| Male | 76 | 41 | 35 | |
| Female | 27 | 16 | 11 | |
| Smoking history | 0.197 | |||
| Never | 21 | 9 | 12 | |
| Former or current | 82 | 48 | 34 | |
| Drinking history | 0.495 | |||
| Never | 35 | 21 | 14 | |
| Former or current | 68 | 36 | 32 | |
| Lymph node number | 0.023a | |||
| ≤6 | 47 | 31 | 16 | |
| >6 | 45 | 19 | 26 | |
| Tumor size | 0.744 | |||
| <5 cm | 41 | 20 | 21 | |
| ≥5 cm | 42 | 22 | 20 | |
| Grade | 0.405 | |||
| I | 8 | 6 | 2 | |
| II | 68 | 38 | 30 | |
| III | 27 | 13 | 14 | |
| AJCC Stage | 0.058 | |||
| I | 4 | 4 | 0 | |
| II | 43 | 26 | 17 | |
| III | 51 | 23 | 28 | |
| T stage | 0.074 | |||
| T1 | 4 | 4 | 0 | |
| T2 | 12 | 7 | 5 | |
| T3 | 81 | 44 | 37 | |
| T4 | 3 | 0 | 3 | |
| N stage | 0.327 | |||
| N0 | 46 | 28 | 18 | |
| N1 | 31 | 18 | 13 | |
| N2 | 19 | 8 | 11 | |
| N3 | 4 | 1 | 3 | |
Mann–Whitney test or Kruskal–Wallis test.
p < 0.05.
TMED3 knockdown (TMED3KD) inhibited the malignant behaviors of ESCC cells in vitro
To further explore the functional role of TMED3 in ESCC, we firstly detected TMED3 expression in Eca-109 and TE-1 cells. TMED3 was highly expressed in Eca-109 and TE-1 cells (Fig. 2A). We then successfully established two knockdown cells using shRNA, Eca-109-TMED3KD cells and TE-1-TMED3KD cells. The knockdown efficiency of TMED3 was validated at the mRNA level and protein level by q-PCR and western blotting (Fig. 2B). Then we conducted multiple experiments to assess the role of TMED3 in various malignant cellular behaviors of Eca-109 and TE-1 cells in vitro. Silencing TMED3 remarkably inhibited cell proliferation (Fig. 2C), colony formation (Fig. 2D), cell migration (Fig. 2E), cell invasion (Fig. 2F), but promoted apoptosis (Fig. 2G) of ESCC cells. Collectively, these results demonstrated that TMED3 promoted the malignant behaviors of ESCC cells in vitro.
Fig. 2.
TMED3 knockdown (TMED3KD) inhibited malignant behaviors of ESCC cells in vitro. (A) TMED3 mRNA expression in Eca-109 and TE-1 cells as detected by q-PCR. (B) The knockdown effect on TMED3 mRNA and protein in Eca-109 and TE-1 cells detected by q-PCR and western blotting. (C) Quantification of cell proliferation in TMED3KD and control Eca-109 and TE-1 cells using MTT assay (mean ± SD, triplicate experiment). (D) Representative images and quantification of colonies of TMED3KD and control Eca-109 and TE-1 cells (mean ± SD, triplicate experiment). (E) Representative images and quantification of cell migration of TMED3KD and control Eca-109 and TE-1 cells using wound-healing assay (mean ± SD, triplicate experiment). (F) Representative images and quantification of cell invasion in TMED3KD and control Eca-109 and TE-1 cells using transwell assay (mean ± SD, triplicate experiment). (G) Representative images of Annexin V-FITC/PI staining and quantification of apoptotic cells in TMED3KD and control Eca-109 and TE-1 cells (mean ± SD, triplicate experiment). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, student's t test.
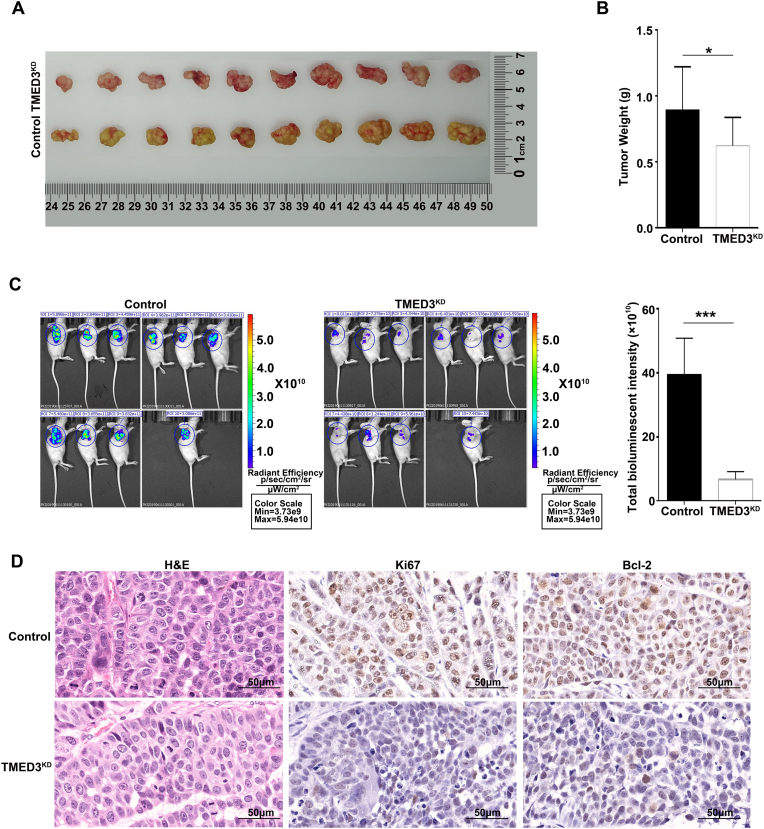
TMED3 knockdown (TMED3KD) inhibited tumor growth in vivo
To investigate the role of TMED3 in ESCC progression in vivo, we successfully established ESCC xenograft in mice. The stable Eca-109-TMED3KD cells were subcutaneously injected into nude mice (1 × 107/ml; 200 μl per xenograft) with Eca-109 cells as a control. On day 21, Eca-109-TMED3KD tumors were lighter in weight and smaller in size than the control tumors (Fig. 3A and B). The bioluminescent intensity of Eca-109-TMED3KD tumors was significantly lower than that of control tumors (Fig. 3C). Besides, H&E staining and IHC staining of a proliferation marker (Ki67) and an anti-apoptosis marker (Bcl-2) of the xenograft tumors also confirmed that Eca-109-TMED3KD cells were less crowded, less proliferative and expressed less Bcl2, than control tumors (Fig. 3D). These results revealed that TMED3 played an important role in ESCC progression in vivo.
Fig. 3.
TMED3 knockdown (TMED3KD) suppressed tumor growth in vivo. (A) Xenograft tumors of TMED3KD and control Eca-109 cells. (B) Histogram of tumor weight (mean ± SD). (C) Bioluminescent images of ESCC xenograft tumors (mean ± SD). (D) Representative images of H&E staining, IHC staining of Ki67, and Bcl-2 of xenograft tumors. Scale bar = 50 μm ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Exploration of the molecular mechanism of TMED3 in ESCC cells
To further elucidate the molecular mechanism of TMED3 in ESCC, Eca-109-TMED3KD (n = 3, as an experimental group) and control cells (n = 3, as control) are utilized for Affymetrix array analysis. |Fold Change| value ≥ 1.5 and FDR value < 0.05 was used as the cutoff value for identifying the differentially expressed genes (DEGs). A total of 1180 DEGs were upregulated and 1318 DEGs down-regulated in the Eca-109-TMED3KD cells (Fig. 4A and B). We further explored the canonical signaling pathways and interaction network analysis to identify gene enrichment induced by TMED3 knockdown through performing IPA (Supplementary Fig. 1A and B), and verified that apoptosis signaling pathway was activated after TMED3 knockdown (TMED3KD) as shown in the IPA results (Supplementary Fig. 1C).
Fig. 4.
Exploration of the molecular mechanism of TMED3 in ESCC. (A) The volcano graph was drawn by referring to the fold change (≥1.5) between TMED3KD and control Eca-109 cells and the FDR (FDR<0.05) obtained from the significance test, Wilcoxon test. (B) The heatmap of differential gene expression between TMED3KD and control Eca-109 cells, wilcoxon test. (C) Differentially expressed mRNAs in TMED3KD and control Eca-109 cells detected by q-PCR, student's t test. (D) Differentially expressed proteins in TMED3KD and control Eca-109 cells detected by western blotting. (E) Celigo cell counting assay was performed to evaluate the effect of CDK6, FAM60A and GDAP1 knockdown on proliferation of Eca-109 cells, paired sample t-test. (F) Representative examples of FAM60A staining on adjacent normal tissues and ESCC tissues. Scale bar = 100 μm or 200 μm (left panel); IHC scores of FAM60A in the adjacent normal tissues (n = 63) and ESCC tissues (n = 103), which were analyzed by the Mann–Whitney U test (right panel). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We first identified DEGs through IPA analysis, and then screened the significantly down-regulated genes in the classical pathway downstream of TMED3 using the KEGG database, and combined with the results of survival analysis in the TCGA database to identify a total of 30 genes. Based on relevant literature we then further narrow down the list to 20 genes for qPCR analysis. We verified the expression of DEGs in the Eca-109-TMED3KD group by qPCR. Compared with the control cells, mRNA levels of TP53BP1, ACTA2, BAX, and ID2 were upregulated, while mRNA levels of TRIB3, CDK6, DDIT3, SMAD2, EGFR, INHBE, MAPK3, CLDN1, STC2, AJUBA, FAM60A, NEDD9, HSPA9, BIRC2, LAMP3, and GDAP1 down-regulated in Eca-109-TMED3KD cells (p < 0.05; Fig. 4C). Among these genes, the differential expression of BIRC2, CDK6, FAM60A, and GDAP1 at the protein level in Eca-109-TMED3KD cells were further validated by western blotting analysis (Fig. 4D). These results indicated that TMED3 regulated BIRC2, CDK6, FAM60A, and GDAP1 at the transcriptional level and post-transcriptional level. Among the four genes, CDK6, FAM60A, and GDAP1 were selected as candidates for further analysis, since the expression of these genes differs greatly between the Eca-109-TMED3KD cells and the control according to western blotting. Furthermore, lentivirus was utilized to deliver shRNAs into Eca-109 cells to knock down these genes (CDK6, FAM60A, and GDAP1), respectively. Then, we explored the role of these three genes in ESCC by performing celigo cell counting assay, and found that as compared to the control cells, Eca-109-CDK6 KD cells and Eca-109-GDAP1KD cells, the proliferation of Eca-109-FAM60AKD cells was most significantly inhibited (p < 0.001; Fig. 4E). Thus, FAM60A was selected for further studies. We hypothesized that TMED3 and FAM60A were functionally related. IHC analysis validated this hypothesis (Fig. 4F, left panel), indicating that FAM60A was highly expressed in ESCC tissues, compared with normal tissues (p < 0.001; Fig. 4F, right panel), which was also confirmed by the analysis of publicly available GEO datasets (GSE23400, GSE20347, and GSE17351) (Supplementary Fig. 2). Collectively, FAM60A was identified as a protein functionally related to TMED3 in Eca-109 cells.
Silencing FAM60A reversed the promoting effect of TMED3 overexpression in ESCC
We explored the role of TMED3 and FAM60A in ESCC cells by performing functional experiments. The expression of TMED3 and FAM60A in ESCC cell lines (Eca-109 and TE-1) at the mRNA and protein level was analyzed by q-PCR and western blotting. Compared with Eca-109 cells, TMED3 was highly expressed in TE-1 cells, and FAM60A was highly expressed in TE-1 cells (Supplementary Fig. 3A). With shRNAs acting on FAM60A at three different sites, termed FAM60AKD-1, FAM60AKD-2, FAM60AKD-3, respectively, we evaluated their knockdown efficiency by q-PCR. We chose FAM60AKD-3 which inhibited FAM60A expression by 68% for the following experiments (Supplementary Fig. 3B). We further established TMED3OE, FAM60AKD, and TMED3OE-FAM60AKD cells with parental Eca-109 cells, and confirmed gene overexpression and knockdown at the mRNA and protein levels by q-PCR and western blotting (Supplementary Fig. 3C).
The result of celigo cell counting assay showed that as compared with control cells, TMED3OE cells exhibited a higher proliferation rate, while FAM60AKD cells and TMED3OE-FAM60AKD cells exhibited slower proliferation rates (Fig. 5A). Cell proliferation data was corroborated by the colony formation data (Fig. 5B). Wound-healing assay and transwell assay indicated that, as compared with control cells, the migration and invasion ability increased in the TMED3OE cells, and decreased in the FAM60AKD cells. However, the migration ability of TMED3OE-FAM60AKD cells was not significantly different from the control cells in the wound healing assay, while their invasion ability was significantly decreased in the transwell assay as compared with control cells (Fig. 5C and D). Moreover, flow cytometry demonstrated that cell apoptosis was significantly decreased in TMED3OE cells, and increased in FAM60AKD cells and TMED3OE-FAM60AKD cells (Fig. 5E). Collectively, these data proved that FAM60AKD rescued the promoting effect of TMED3 overexpression (TMED3OE) on the malignant behaviors of ESCC cells.
Fig. 5.
FAM60A knockdown reversed the oncogenic role of TMED3 in ESCC. (A) Representative images of cells expressing GFP and quantification of proliferating cells in TMED3OE, FAM60AKD, TMED3OE-FAM60AKD, and control Eca-109 cells using celigo cell counting assay (mean ± SD, triplicate experiment), paired t-test. (B) Representative images and quantification of colonies of TMED3OE, FAM60AKD, TMED3OE-FAM60AKD, and control Eca-109 cells using colony formation assay (mean ± SD, triplicate experiment), student's t test. (C) Representative images and quantification of cell migration in TMED3OE, FAM60AKD, TMED3OE-FAM60AKD, and control Eca-109 cells using wound-healing assay (mean ± SD, triplicate experiment), student's t test. (D) Representative images and quantification of cell invasion of TMED3OE, FAM60AKD, TMED3OE-FAM60AKD, and control Eca-109 cells using transwell assay (mean ± SD, triplicate experiment), Student's t test. (E) Representative images of Annexin V-FITC/PI staining and quantification of apoptotic cells in TMED3OE, FAM60AKD, TMED3OE-FAM60AKD, and control Eca-109 cells using colony formation assay (mean ± SD, triplicate experiment), Student's t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
This study is the first to show that TMED3 is overexpressed in ESCC and TMED3OE is significantly related to worse clinical outcomes and prognosis in ESCC patients. Furthermore, the effect of TMED3OE on the progression of ESCC was closely related to FAM60A.
For most eukaryotic cells, the secretory pathway plays a key role in the synthesis, transportation, and secretion of a variety of biologically active molecules involved in intercellular interactions. Members of the TMED/p24 family have been identified as important regulators of vesicular transport, which regulate the molecular transport between various membrane-bound compartments in cells and thus all major cellular activities [[19], [20], [21], [22], [23], [24], [25], [26]]. Pathological conditions in humans, such as cancer, liver disease, pancreatic disease, and immune system dysregulation [6]. Vainio et al. reported that TMED3 was overexpressed in prostate cancer and was related to oncogene expression [11]. Zheng et al. reported that upregulated TMED3 promoted hepatocellular carcinoma metastasis through the IL-11/STAT3 signaling pathway [14]. Pei et al. suggested that TMED3 upregulation promoted proliferation, migration, and invasion of breast cancer cells and was related to the poor prognosis of breast cancer patients [15]. In a retrospective multi-cohort analysis, Ha et al. demonstrated the carcinogenic effect of TMED3 in clear cell renal cell cancer [16]. Peng et al. found that TMED3, a direct target of mir-876–3p, enhanced cisplatin resistance, and strengthened stem cell-like features of gastric cancer cells [17]. Xie et al. revealed that TMED3 promoted the progression of lung squamous cell carcinoma by regulating EZR [27]. However, other than its carcinogenic effect, TMED3 has also been reported to have a tumor suppressor effect on human colon cancer through a whole-genome screening assay [12]. TMED3-mediated WNT signaling was shown to inhibit metastasis by down-regulated TMED9 in colon cancer [13]. These data suggested the role of TMED3 in human cancer depends on the cellular context. In this study, we confirmed the role of TMED3 in promoting ESCC from multiple aspects: 1) TMED3 was overexpressed in ESCC tissues and cells and its overexpression was positively correlated with poor prognosis; 2) TMED3 downregulation significantly inhibited cell proliferation, migration, and invasion, and promoted cell apoptosis in vitro; 3) TMED3 downregulation suppressed tumorigenicity in vivo. These data clearly demonstrated the oncogenic role of TMED3 in ESCC.
Our experimental data also showed that TMED3 inhibited the apoptosis of ESCC cells. Meanwhile, the result of IPA analysis declared that the apoptosis signaling pathway was activated after silencing TMED3. In addition, we found that the apoptosis induced by TMED3KD in Eca-109 cells was a combination of a series of apoptosis-related proteins. For example, anti-apoptosis proteins (Bcl-2, Bcl-w, HSP60, HSP70, IGF-I, IGF-1sR, Livin, Survivin, sTNF-R1, TRAILR-3, TRAILR-4, and XIAP) were significantly down-regulated in the TMED3KD cells. Many studies have revealed the role of anti-apoptotic proteins in ESCC. For example, it was reported that the expression of HSP60 and HSP70 in ESCC was higher than that of normal tissue, which may be related to the biological behavior of ESCC [28]. Moreover, Han et al. reported that a combination of a natural compound (Periplocin) and TRAIL contributed to the apoptosis of ESCC cells [29]. In addition, TMED3 is a component of the coated vesicles which are related to the transport of cargo molecules from the endoplasmic reticulum (ER) to the Golgi complex and serves as a receptor for specific secretory cargo [5]. It has been shown that protein denaturation and ER dysfunction induce a stress response leading to cancer cells apoptosis and targeting ER proteostasis and Golgi apparatus function has recently been proposed as a promising mechanism for cancer therapy [30,31]. Therefore, we concluded that knockdown of TMED3 induces ESCC cell apoptosis through regulating a series of apoptosis proteins. Moreover, based on the relationship between TMED3, ER and Golgi complex, targeting TMED3 may be a potential mechanism for treating cancer.
We also determined FAM60A as a protein functionally related to TMED3 by a genome-wide expression profiling analysis and IPA. FAM60A was initially reported as a new subunit of the SIN3A-HDAC complex by two independent studies [32,33]. SIN3A affects the developmental processes (e.g., stem cell function) and pathological processes (e.g., cancer), and controls cellular metabolism, cell cycle, and cell survival [34,35]. Consistent with our data, FAM60A has been reported to be overexpressed in ESCC tissues. Besides, FAM60AKD reduced the percentage of G1 phase ESCC cells, arrested ESCC cells in the G2/M phase [36]. Our study uncovered that silencing FAM60A alleviated the promoting effect of TMED3OE on ESCC, which suggested that TMED3 may promote the progression of ESCC by regulating FAM60A. Furthermore, our results found that TMED3 upregulated the expression of FAM60A at the transcription and translation level. It was reported that FAM60A was regulated on the transcription level by the E2F and core pluripotency transcription factor networks [37]. E2F, a large family of transcription factors that play various biological roles, including cell cycle control [38], is often overexpressed in tumors and plays an important role in cell proliferation [39,40]. Considering the function of TMED3 in transporting secretory cargo, we speculated that TMED3 may upregulate FAM60A expression by accelerating intracellular transport of transcription factor E2F. However, how TMED3 affects the expression of FAM60A at the transcription and translation level requires further research.
Conclusions
The results of our study demonstrated that TMED3OE plays an important role in ESCC through regulating FAM60A and inhibiting the apoptosis signaling pathway. These findings help us better understand the molecular mechanisms of ESCC and potentially may help the prognosis and treatment of ESCC in the future.
Funding
This work was supported by Natural Science Foundation of China (81874220), Fundamental Research Funds for the Central Universities (19ykpy176), and Natural Science Foundation of Guangdong Province (2019A1515011420, 2020A1515010030).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of interest
All the authors declare that they have no competing interests.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2022.03.013.
Contributor Information
Mian Xi, Email: ximian@sysucc.org.cn.
Lei Zhao, Email: zhaolei@sysucc.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Results of IPA analysis. (A) Significant enrichment of differentially expressed genes in several pathways examined by IPA analysis, Chi-square test. (B) Interaction network analysis between TMED3 and the genes in the above pathways. (C) Significantly down-regulated proteins on the apoptosis signaling pathway in TE-1-TMED3KD cells verified by apoptosis antibody array, Student's t test.
Fig. S2.
FAM60A mRNA expression in the normal and ESCC tissues using original data from three GEO datasets. GSE23400, GSE20347, and GSE1735, which were analyzed by the paired t-test.
Fig. S3.
TMED3 and FAM60A expression in human ESCC cell lines. (A) TMED3 and FAM60A mRNA expression as detected by q-PCR. (B) Downregulation of FAM60A mRNA expression by shRNAs as detected by q-PCR. (C) Modulation of mRNA and protein expression by knockdown or overexpression as detected by q-PCR and Western blotting. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, Student's t test.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro J., van Lanschot J.J.B, Hulshof M.C.C.M, van Hagen P., van Berge Henegouwen M.I., Wijnhoven B.P.L., et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 2015;16(9):1090–1098. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- 4.Herskovic A., Russell W., Liptay M., Fidler M.J., Al-Sarraf M. Esophageal carcinoma advances in treatment results for locally advanced disease: review. Ann Oncol. 2012;23(5):1095–1103. doi: 10.1093/annonc/mdr433. [DOI] [PubMed] [Google Scholar]
- 5.Anantharaman V., Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3(5) doi: 10.1186/gb-2002-3-5-research0023. research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aber R., Chan W., Mugisha S., Jerome-Majewska L.A. Transmembrane emp24 domain proteins in development and disease. Genet Res(Camb) 2019;101:e14. doi: 10.1017/S0016672319000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strating J.R., Hafmans T.G., Martens G.J. Functional diversity among p24 subfamily members. Biol Cell. 2009;101(4):207–219. doi: 10.1042/BC20080075. [DOI] [PubMed] [Google Scholar]
- 8.Nagae M., Hirata T., Morita-Matsumoto K., Theiler R., Fujita M., Kinoshita T., et al. 3D structure and interaction of p24β and p24δ Golgi dynamics domains: implication for p24 complex formation and cargo transport. J Mol Biol. 2016;428(20):4087–4099. doi: 10.1016/j.jmb.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Nagae M., Liebschner D., Yamada Y., Morita-Matsumoto K., Matsugaki N., Senda T., et al. Crystallographic analysis of murine p24γ2 Golgi dynamics domain. Proteins. 2017;85(4):764–770. doi: 10.1002/prot.25242. [DOI] [PubMed] [Google Scholar]
- 10.Pastor-Cantizano N., Montesinos J.C., Bernat-Silvestre C., Marcote M.J., Aniento F. p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma. 2016;253(4):967–985. doi: 10.1007/s00709-015-0858-6. [DOI] [PubMed] [Google Scholar]
- 11.Vainio P., Mpindi J.P., Kohonen P., Fey V., Mirtti T., Alanen K.A., et al. High-throughput transcriptomic and RNAi analysis identifies AIM1, ERGIC1, TMED3 and TPX2 as potential drug targets in prostate cancer. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duquet A., Melotti A., Mishra S., Malerba M., Seth C., Conod A., et al. A novel genome-wide in vivo screen for metastatic suppressors in human colon cancer identifies the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol Med. 2014;6(7):882–901. doi: 10.15252/emmm.201303799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra S., Bernal C., Silvano M., Anand S., Ruiz I., Altaba A. The protein secretion modulator TMED9 drives CNIH4/TGFα/GLI signaling opposing TMED3-WNT-TCF to promote colon cancer metastases. Oncogene. 2019;38(29):5817–5837. doi: 10.1038/s41388-019-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng H., Yang Y., Han J., Jiang W.H., Chen C., Wang M.C., et al. TMED3 promotes hepatocellular carcinoma progression via IL-11/STAT3 signaling. Sci Rep. 2016;6:37070. doi: 10.1038/srep37070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei J., Zhang J., Yang X., Wu Z., Sun C., Wang Z., et al. TMED3 promotes cell proliferation and motility in breast cancer and is negatively modulated by miR-188-3p. Cancer Cell Int. 2019;19:75. doi: 10.1186/s12935-019-0791-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Ha M., Moon H., Choi D., Kang W., Kim J.H., Lee K.J., et al. Prognostic role of TMED3 in clear cell renal cell carcinoma: a retrospective multi-cohort analysis. Front Genet. 2019;10:355. doi: 10.3389/fgene.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng C., Huang K., Liu G., Li Y., Yu C. MiR-876-3p regulates cisplatin resistance and stem cell-like properties of gastric cancer cells by targeting TMED3. J Gastroenterol Hepatol. 2019;34(10):1711–1719. doi: 10.1111/jgh.14649. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Li Y., Luan D., Kang J., He R., Zhang Y., et al. Dynamic replacement of H3.3 affects nuclear reprogramming in early bovine SCNT embryos. Theriogenology. 2020;154:43–52. doi: 10.1016/j.theriogenology.2020.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Carney G.E., Bowen N.J. p24 proteins, intracellular trafficking, and behavior: Drosophila melanogaster provides insights and opportunities. Biol Cell. 2004;96(4):271–278. doi: 10.1016/j.biolcel.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Dancourt J., Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 21.Jerome-Majewska L.A., Achkar T., Luo L., Lupu F., Lacy E. The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Dev Biol. 2010;341(1):154–166. doi: 10.1016/j.ydbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakariyah A., Hou W., Slim R., Jerome-Majewska L. TMED2/p24β1 is expressed in all gestational stages of human placentas and in choriocarcinoma cell lines. Placenta. 2012;33(3):214–219. doi: 10.1016/j.placenta.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Viotti C. ER to golgi-dependent protein secretion: the conventional pathway. Methods Mol Biol. 2016;1459:3–29. doi: 10.1007/978-1-4939-3804-9_1. [DOI] [PubMed] [Google Scholar]
- 24.Wada Y., Sun-Wada G.H., Kawamura N., Yasukawa J. Membrane dynamics in mammalian embryogenesis: implication in signal regulation. Birth Defects Res C Embryo Today. 2016;108(1):33–44. doi: 10.1002/bdrc.21124. [DOI] [PubMed] [Google Scholar]
- 25.Hou W., Gupta S., Beauchamp M.C., Yuan L., Jerome-Majewska L.A. Non-alcoholic fatty liver disease in mice with heterozygous mutation in TMED2. PLoS One. 2017;12(8):e0182995. doi: 10.1371/journal.pone.0182995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou W., Jerome-Majewska L.A. TMED2/emp24 is required in both the chorion and the allantois for placental labyrinth layer development. Dev Biol. 2018;444(1):20–32. doi: 10.1016/j.ydbio.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Xie A., Xu X., Kuang P., Zhang L., Yu F. TMED3 promotes the progression and development of lung squamous cell carcinoma by regulating EZR. Cell Death Dis. 2021;12(9):804. doi: 10.1038/s41419-021-04086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J.H., Chen L.M., Xu L.Y., Wu M.Y., Shen Z.Y. [Expression and significance of heat shock proteins in esophageal squamous cell carcinoma] Zhonghua zhong liu za zhi. 2006;28(10):758–761. Chinese. [PubMed] [Google Scholar]
- 29.Han L., Dai S., Li Z., Zhang C., Wei S., Zhao R., et al. Combination of the natural compound Periplocin and TRAIL induce esophageal squamous cell carcinoma apoptosis in vitro and in vivo: implication in anticancer therapy. J Exp Clin Cancer Res. 2019;38(1):501. doi: 10.1186/s13046-019-1498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Ye Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Res. 2011;21(6):867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin M., Vandenbroeck K. The endoplasmic reticulum protein folding factory and its chaperones: new targets for drug discovery? Br J Pharmacol. 2011;162(2):328–345. doi: 10.1111/j.1476-5381.2010.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muñoz I.M., MacArtney T., Sanchez-Pulido L., Ponting C.P., Rocha S., Rouse J. Family with sequence similarity 60A (FAM60A) protein is a cell cycle-fluctuating regulator of the SIN3-HDAC1 histone deacetylase complex. J Biol Chem. 2012;287(39):32346–32353. doi: 10.1074/jbc.M112.382499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith K.T., Sardiu M.E., Martin-Brown S.A., Seidel C., Mushegian A., Egidy R., et al. Human family with sequence similarity 60 member A (FAM60A) protein: a new subunit of the Sin3 deacetylase complex. Mol Cell Proteomics. 2012;11(12):1815–1828. doi: 10.1074/mcp.M112.020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders A., Huang X., Fidalgo M., Reimer M.H., Jr., Faiola F., Ding J., et al. The SIN3A/HDAC corepressor complex functionally cooperates with NANOG to promote pluripotency. Cell Rep. 2017;18(7):1713–1726. doi: 10.1016/j.celrep.2017.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadamb R., Mittal S., Bansal N., Batra H., Saluja D. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol. 2013;92(8-9):237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Dong G., Mao Q., Yu D., Zhang Y., Qiu M., Dong G., et al. Integrative analysis of copy number and transcriptional expression profiles in esophageal cancer to identify a novel driver gene for therapy. Sci Rep. 2017;7:42060. doi: 10.1038/srep42060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streubel G., Fitzpatrick D.J., Oliviero G., Scelfo A., Moran B., Das S., et al. Fam60a defines a variant Sin3a-Hdac complex in embryonic stem cells required for self-renewal. EMBO J. 2017;36(24):2216–2232. doi: 10.15252/embj.201696307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attwooll C., Lazzerini Denchi E., Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park S.M., Choi E.Y., Bae D.H., Sohn H.A., Kim S.Y., Kim Y.J. The LncRNA EPEL promotes lung cancer cell proliferation through E2F target activation. Cell Physiol Biochem. 2018;45(3):1270–1283. doi: 10.1159/000487460. [DOI] [PubMed] [Google Scholar]
- 40.Bracken A.P., Ciro M., Cocito A., Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.