Abstract
Atherosclerosis (AS) is a vascular disease with plaque formation. Unstable plaques can be expected to result in cardiovascular disease, such as myocardial infarction and stroke. Studies have verified that long noncoding RNAs (lncRNAs) play a critical role in atherosclerotic plaque formation (APF), including MALAT1, GAS5, and H19. A ceRNA network is a combination of these two interacting processes, which regulate the occurrence and progression of many diseases. However, lncRNA-associated ceRNA network in terms of APF is limited. This study sought to discover novel potential biomarkers and ceRNA network for APF. We designed a triple network based on the lncRNA-miRNA and mRNA-miRNA pairs obtained from lncRNASNP and starBase. Differentially expressed genes (DEGs) and lncRNAs in human vascular tissues derived from the Gene Expression Omnibus database (GSE43292, GSE97210) were systematically selected and analyzed. A ceRNA network was constructed by hypergeometric test, including 8 lncRNAs, 243 miRNAs, and 8 mRNAs. APF-related ceRNA structure was discovered for the first time by combining network analysis and statistical validation. Topological analysis determined the key lncRNAs with the highest centroid. GO and KEGG enrichment analysis indicated that the ceRNA network was primarily enriched in “regulation of platelet-derived growth factor receptor signaling pathway,” “negative regulation of leukocyte chemotaxis,” and “axonal fasciculation.” A functional lncRNA, HAND2-AS1, was identified in the ceRNA network, and the main miRNA (miRNA-570-3p) regulated by HAND2-AS1 was further screened. This present study elucidated the important function of lncRNA in the origination and progression of APF and indicated the potential use of these hub nodes as diagnostic biomarkers and therapeutic targets.
1. Introduction
Atherosclerotic cardiovascular disease is a major threat to human health and quality of life in modern society [1]. The formation of atherosclerotic plaques is one of the major causes of cardiovascular disease morbidity and mortality, such as myocardial infarction and stroke [2]. However, the underlying mechanism of atherosclerotic plaque formation (APF) is not entirely clear, so further exploration of the possible effective treatments has drawn substantial attention.
Noncoding RNAs (ncRNAs) refer to the functional RNAs that cannot encode proteins in transcriptome, including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). miRNAs are a class of highly conserved single-stranded noncoding small RNAs with posttranscriptional regulatory activity. Long noncoding RNAs (lncRNAs) are longer than 200 nucleotides with no protein coding capacity or limited protein coding potential [3–7]. Although ncRNAs cannot code proteins, they can exert their functions by regulating the coding RNAs. Studies have shown that functional ncRNA not only play an important role in cell proliferation, differentiation, and aging but also participate in the occurrence and development of many diseases, such as cancer, digestive system diseases, and cardiovascular diseases [8–13]. Atherosclerosis (AS) is a vascular disease with plaque formation. Unstable plaques can be expected to result in cardiovascular disease, such as myocardial infarction and stroke. Studies have verified that long noncoding RNAs (lncRNAs) play a crucial role in APF, including MALAT1, GAS5, and H19 [14–16].
Competitive endogenous RNA (ceRNA) is the basis of a novel regulatory mechanism between noncoding RNA and coding RNA. lncRNA can be used as ceRNA to competitively bind miRNA with target genes, thereby reducing the inhibitory effect of miRNAs on target genes [17]. Therefore, they can indirectly regulate the expression of target genes and thus participate in the development of diseases. For instance, Wang et al. found that long noncoding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma [18]. Li et al. discovered that the long noncoding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2 [19]. However, the mechanism of ceRNA associated with APF is still unclear. Therefore, this study was aimed at investigating the functional role of novel potential biomarkers associated with lncRNA in the progression of APF by lncRNA-associated ceRNA network analysis. The flow chart of this research is shown in Figure 1.
Figure 1.

Flowchart for an overview of the present analysis.
2. Materials and Methods
2.1. Construction of the Global Triple Network
In our study, lncRNA-miRNA interactions and miRNA-mRNA interactions were obtained from the lncRNASNP database (http://bioinfo.life.hust.edu.cn/lncRNASNP/) [20] and starBase V2.0 database (http://starbase.sysu.edu.cn/) [21], respectively. Then, we constructed the global triple network and visualized by Cytoscape software v3.9.0 (http://www.cytoscape.org/). Cytoscape is a popular bioinformatics package for biological network visualization and data integration [22]. We defined the lncRNA, miRNA, and mRNA as the nodes of the network in Cytoscape software. If they could interact with each other, we connected them with lines. Finally, a global triple ceRNA network was constructed.
2.2. Screening of Differentially Expressed Genes (DEGs)
To identify the main DEGs between normal blood vessels and the blood vessels containing atherosclerotic plaques, microarray data GSE97210 and GSE43292 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). When we enter the keywords “lncRNA” and “atherosclerotic plaque” in the GEO dataset search box, the result shows that there is only the GSE97210 dataset. The GSE97210 lncRNA dataset was measured using GPL16956 Platforms (Agilent-045997 Arraystar human lncRNA microarray V3) and included data from 3 patients with APF and 3 healthy controls. The mRNA expression data in GSE43292 were from 32 patients with APF and 32 healthy controls and were based on GPL6244 Platforms (Affymetrix Human Gene 1.0 ST Array). Subsequently, data were imported into the R-studio and normalized with RMA algorithm [23]. DEGs were defined by the Bioconductor/R limma package as previously described [24]. Selected values of P < 0.05 and [logFC] > 1.5 were applied.
Since the differentially expressed lncRNAs that have been screened out were replaced by sequence numbers of probes, they were further reannotated through the BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Detecting Potential ceRNA Mechanism
A hypergeometric distribution is a discrete probability distribution in statistics. It was employed to detect the competing lncRNA-mRNA interactions and evaluated the significance of the same miRNA between each mRNA and lncRNA. In this study, we used this statistical method to detect potential ceRNA mechanism, which made the results more objective and scientific. The P value was calculated by the following equation:
| (1) |
where K represented the count of human genome miRNA, M represented the count of miRNA related to the mRNA, N represented the count of miRNA related to the lncRNA, and X represented the count of miRNA shared between lncRNA and mRNA.
Then, the original P values were corrected for false discovery rate (FDR) according to the Benjamini and Hochberg method [25]. We considered that FDR < 0.01 was statistically significant.
2.4. GO Enrichment and Signaling Pathway Analysis
DEGs were subjected to gene ontology (GO) enrichment using Cytoscape plugin ClueGO to explore their roles in various biological processes. After GO functional enrichment analysis, we considered biology process terms with P value < 0.05 to be statistically significant. With the KEGG option, the signaling pathways regulated by disease-related genes could be obtained. ClueGO network was constructed by using kappa statistics and showed the relationship between terms with similarly associated genes. These specific and common terms were automatically calculated [26].
2.5. Construction of ceRNA Network Related to APF
The differentially expressed lncRNAs and mRNAs were marked from the global triple network and found out the lncRNA-miRNA interactions and mRNA-miRNA interactions, respectively. Then, these interactions were combined by hypergeometric test and visualized through Cytoscape software. A new lncRNA-miRNA-mRNA network related to APF was constructed.
2.6. Definition of Topological Features for the ceRNA Network
Topological parameters mainly include degree, closeness, and betweenness centrality. These are the key criteria for measuring the importance of network nodes. The NetworkAnalyzer tool in Cytoscape was used to calculate and analyze the topological parameters of each node in the network, including degree, closeness, and betweenness centrality. The most commonly used and the most straightforward measurement of centrality in network analysis is the degree value. As reported, degree is defined as the quantity of links that a node connects with other nodes [27]. Closeness is defined as the average mean path from a node to all other nodes reachable from it [28]. Betweenness is an index evaluating the influence a node exerts over the spread of information through the network [29]. Finally, we screened the lncRNA with the highest value of 3 standards in the network; that is to say, this lncRNA is the major lncRNA that regulates APF.
It was found that the selected lncRNA and its downstream miRNAs and target genes constituted a new network from the global triple ceRNA network. lncRNAs with the highest degree were screened out of the new network. The major miRNA and its target genes regulated by lncRNA were constructed into a new small ceRNA network. Finally, functional analysis of the small regulatory network was performed to verify whether it was involved in the regulation of the APF-related ceRNA network.
3. Results
3.1. Construction of a Global Triple ceRNA Network
In order to construct a global triple network, 548195 lncRNA-miRNA interaction pairs were downloaded from the lncRNASNP database, and then, 606408 miRNA-mRNA interaction pairs were downloaded from the starBase V2.0 database. Then, we integrated all miRNA-mRNA interaction pairs and lncRNA-miRNA interaction pairs into a global triple ceRNA network through the Cytoscape software. Afterwards, we defined lncRNAs, miRNAs, and mRNAs as the nodes of the network. If they could interact with each other, we connected them with lines, completing a global triple ceRNA network (Figure 2).
Figure 2.

The global triple ceRNA network. The green nodes represent mRNAs, the red nodes represent lncRNAs, and the blue nodes represent miRNAs.
3.2. Screening of Genes Related to APF
A total of two gene expression profiles (GSE97210, GSE43292) were downloaded from the GEO database, comparing the blood vessels between patients with APF and healthy controls. 39 differentially expressed lncRNAs (DELs) and 9 differentially expressed mRNAs (DEMs) were identified with [logFC] > 1.5 and adjusted P < 0.01 (Tables 1 and 2), including 30 genes upregulated and 18 genes downregulated in the APF group. Since these lncRNAs screened out are represented by the sequence of the probe, they are further reannotated by BLAST software. These overview boxplot, volcano plot, and heat map of the DEGs were obtained by GEO2R and R-studio (Figure 3).
Table 1.
Nine DEMs screened from GSE43292.
| Regulation | Gene symbol | logFC | P value |
|---|---|---|---|
| Upregulation | FABP4 | 2.454461 | 6.05E-08 |
| IBSP | 1.794982 | 1.03E-08 | |
| DPP4 | 1.610873 | 2.24E-07 | |
| Downregulation | chr4:174457998-174460620 | −1.50436 | 1.39E-07 |
| FHL5 | −1.56465 | 1.17E-08 | |
| MYOCD | −1.62845 | 6.08E-08 | |
| CASQ2 | −1.66766 | 2.16E-08 | |
| CNTN4 | −1.79233 | 3.57E-09 | |
| CNTN1 | −1.91103 | 3.00E-08 |
Table 2.
39 DELs screened from GSE97210.
| Regulation | Gene symbol | logFC | P value |
|---|---|---|---|
| Upregulation | PRDM16-DT | 7.58 | 2.39E-10 |
| PART1 | 7.44 | 3.27E-11 | |
| HAND2-AS1 | 6.83 | 7.03E-11 | |
| LINC01252 | 6.33 | 1.4E-10 | |
| PARD3-DT | 6.04 | 1.71E-10 | |
| LOC100507334 | 5.82 | 1.93E-10 | |
| TBX2-AS1 | 5.69 | 1.18E-09 | |
| SSTR5-AS1 | 5.48 | 7.59E-10 | |
| PSORS1C3 | 5.37 | 4.16E-10 | |
| LINC01139 | 5.08 | 2.41E-10 | |
| SAMMSON | 5.06 | 9.54E-10 | |
| LOC100129455 | 4.96 | 3.86E-10 | |
| LOC105369957 | 4.93 | 7.19E-10 | |
| LOC105379447 | 4.88 | 4.86E-10 | |
| LINC00165 | 4.86 | 4.01E-10 | |
| SPANXA2-OT1 | 4.83 | 7.52E-10 | |
| TMEM92-AS1 | 4.75 | 8.96E-10 | |
| GATA6-AS1 | 4.69 | 1.25E-09 | |
| KIF23-AS1 | 4.67 | 5.30E-10 | |
| LINC02718 | 4.64 | 6.24E-10 | |
| FSIP2-AS1 | 4.53 | 4.49E-10 | |
| HOXC-AS3 | 4.50 | 5.78E-10 | |
| CNN3-DT | 4.12 | 5.36E-10 | |
| LOC100506725 | 4.10 | 6.66E-10 | |
| LOC105374029 | 3.98 | 8.05E-10 | |
| LOC100507547 | 3.93 | 8.26E-10 | |
| LOC100130992 | 3.87 | 1.24E-09 | |
| Downregulation | SUGCT-AS1 | -4.60 | 4.10E-10 |
| LOC101927602 | -4.71 | 4.78E-10 | |
| VPS13A-AS1 | -4.73 | 4.39E-10 | |
| C1RL-AS1 | -4.76 | 5.67E-10 | |
| LOC105377323 | -4.89 | 3.50E-10 | |
| APOC1P1 | -5.34 | 3.49E-10 | |
| GAPLINC | -5.44 | 8.44E-10 | |
| LINC00900 | -5.84 | 2.27E-10 | |
| L3MBTL4-AS1 | -5.93 | 1.27E-10 | |
| LINC01827 | -6.27 | 8.47E-10 | |
| CNIH3-AS2 | -6.48 | 4.64E-10 | |
| MMP2-AS1 | -6.49 | 1.68E-10 |
Figure 3.
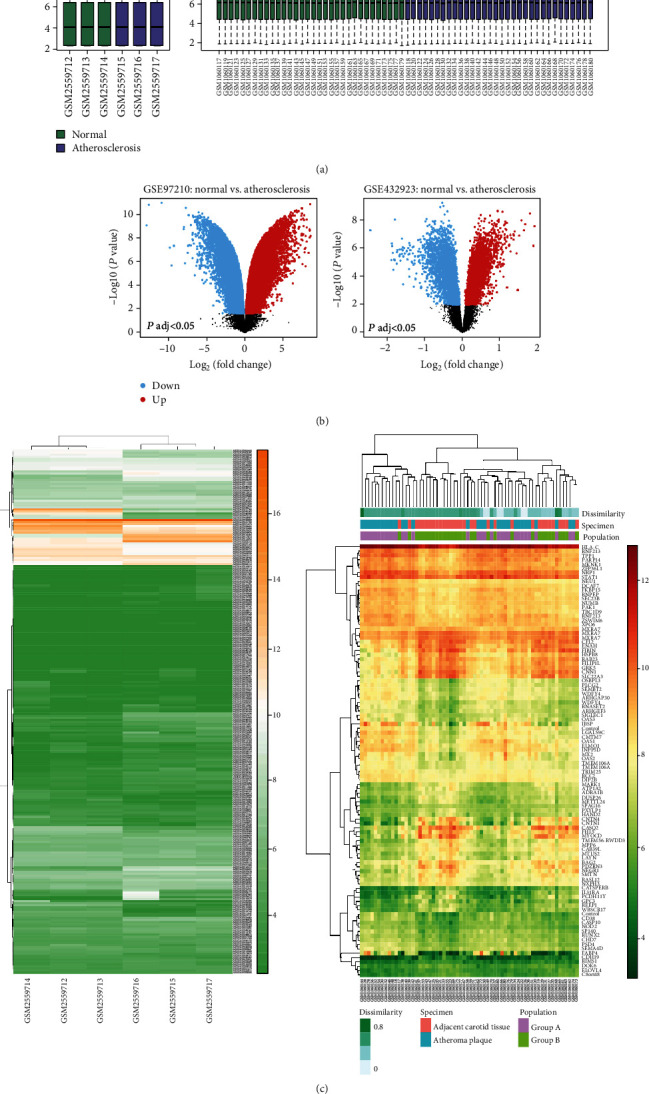
Identification of differentially expressed genes from the GSE97210 dataset and the GSE43292 dataset. (a) The boxplots of the sample expression level in GSE97210 and GSE43292. (b) The volcano plot of the differential expressed genes between the APF and control group in GSE97210 and GSE43292. (c) Heat map of the expression profiles including DEGs and lncRNAs between the APF and control group in GSE97210 and GSE43292.
3.3. Construction of the Network Related to APF from Global Triple ceRNA Network
First, the lncRNASNP database was used to predict differentially expressed miRNAs (DEMis) associated with DELs. The starBase V2.0 database was used to predict miRNAs that interact with DEGs. Then, a hypergeometric test was utilized to further detect the significance of these lncRNA-miRNA pairs and mRNA-miRNA pairs. Finally, we constructed a new lncRNA-miRNA-mRNA network related to APF in Cytoscape software (Figure 4). It consists of 8 lncRNA nodes, 243miRNA nodes, 8 mRNA nodes, and 1298 edges.
Figure 4.

Construction of the network related to APF by Cytoscape. The green nodes represent mRNAs, the red nodes represent lncRNAs, and the blue nodes represent miRNAs.
3.4. Functional Enrichment Analyses of APF-Related Network
Enrichment analysis of GO classification and KEGG pathway was performed on the DEGs in APF-related networks by employing ClueGO plugin in Cytoscape software. The results are shown in Figure 5. GO enrichment analysis has shown that 66 GO terms were enriched. KEGG pathway suggested that APF-related genes were mainly related with the “regulation of platelet-derived growth factor receptor signaling pathway.”
Figure 5.

Signaling pathway enrichment analysis for APF-related genes by Cytoscape. (a) A functionally grouped network of enriched categories for target genes. Every node is a disease-related GO term. (b) The proportion of each term.
3.5. Topological Analysis of ceRNA Network Related to APF
First, we used Cytoscape software to calculate the topology parameters of the nodes in the APF-related ceRNA network. Then, all of the nodes' topological parameters in the network were ranked (Table 3). It was previously reported that the degree value of a hub node should not be less than 5 [30]. A comprehensive examination revealed that three parameters of lncRNA HAND2-AS1 were significantly higher than those of other lncRNA nodes. Therefore, HAND2-AS1 was the hub node of this ceRNA network. In other words, HAND2-AS1 may be the key lncRNA regulating the development of APF. Further, we extracted miRNAs and mRNAs associated with the HAND2-AS1 from the APF-related ceRNA network and constructed a new ceRNA network of HAND2-AS1 by Cytoscape software (Figure 6(a)). Among the top 3 miRNAs (hsa-miRNA-466, hsa-miRNA-570-3p, and hsa-miRNA-4495), only hsa-miRNA-570-3p could directly interact with HAND2-AS1. The mRNAs that directly interact with hsa-miRNA-570-3p are CNTNI, MYOCD, CNTN4, and DPP4. KEGG functional enrichment analysis indicated that hsa-miRNA-570-3p may also act on the “regulation of platelet-derived growth factor receptor signaling pathway” (Figure 6(b)).
Table 3.
Degree, closeness centrality, and betweenness centrality of DEGs in the APF-related ceRNA network.
(a).
| lncRNA | Degree | Closeness centrality | Betweenness centrality |
|---|---|---|---|
| HAND2-AS1 | 142 | 0.36317829457364337 | 0.20723471560902113 |
| LINC00900 | 72 | 0.3457564575645757 | 0.10401877331726586 |
| TBX2-AS1 | 12 | 0.20647862494490965 | 0.021243238559140364 |
| GATA6-AS1 | 12 | 0.24657894736842106 | 0.019403039645996233 |
| SPANXA2-OT1 | 6 | 0.22193273330175273 | 0.002378732808397449 |
| SSTR5-AS1 | 6 | 0.2113216057735679 | 0.006399994526995594 |
(b).
| miRNA | Degree | Closeness centrality | Betweenness centrality |
|---|---|---|---|
| Hsa-miRNA-570-3p | 5 | 0.41644444444444445 | 0.022132585913046563 |
| Hsa-miRNA-466 | 5 | 0.38783112582781454 | 0.014465726762748462 6 |
| Hsa-miRNA-4495 | 5 | 0.380275974025974 | 0.01051210577447764264 |
(c).
| mRNA | Degree | Closeness centrality | Betweenness centrality |
|---|---|---|---|
| CNTN1 | 289 | 0.4157054125998226 | 0.423378721015294453 |
| MYOCD | 226 | 0.3904166666666667 | 0.3305499570867205 |
| CNTN4 | 177 | 0.3781275221953188 | 0.2358729852351261564 |
| FHL5 | 108 | 0.3557327258921792 | 0.1310673691240780733 |
| DPP4 | 101 | 0.3538519637462236 | 0.1366076949975910749 |
| IBSP | 60 | 0.3322695035460993 | 0.061990908379872794 |
| CASQ2 | 56 | 0.33584229390681003 | 0.07356388148731316 |
| FABP4 | 26 | 0.30206318504190843 | 0.024157518050064344 |
Figure 6.
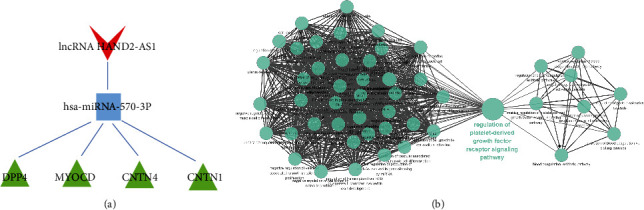
ceRNA regulatory network of the miRNA bound by HAND2-AS1 and the gene function of the network. (a) The ceRNA network of hsa-miRNA-570-3p sponged by lncRNA HAND2-AS1. (b) Gene function analysis of the ceRNA network.
4. Discussion
APF is a complex disease that has brought a heavy burden to people all over the world. Since the pathogenesis and process of this disorder are complex, it is necessary to solve the problem through a more comprehensive perspective. In recent years, with the rapid development of high-throughput genomics, noncoding RNA has received extensive attention. Noncoding RNA includes lncRNA and miRNA, which are involved in the regulation of multiple biological processes [31]. ceRNA is a common regulatory mechanism between lncRNA and miRNA, and it is an important method in bioinformatics analysis [32, 33]. However, the APF-related ceRNA regulation mechanism has not been systematically elucidated. In the current research, a synthetical bioinformatics method was applied to identify the key lncRNA involved in APF and further explore the molecular mechanism of APF.
First of all, construct a global triple network based on the ceRNA theory by using the interactive data of lncRNASNP and starBase [34]. Then, we collected the DEGs associated with APF by mining microarray data from the GEO database. A ceRNA network related to APF was extracted by mapping the DEGs into the triple global network. It contains 8 mRNA nodes, 243 miRNA nodes, 8 lncRNA nodes, and 1298 edges. Further, we applied the topological features and cluster analysis to the APF-related ceRNA network. Finally, with comprehensive consideration of degree, closeness, and betweenness centrality, HAND2-AS1 was considered to be a possible hub node. In other words, these results suggest that HAND2-AS1 might play an important role in the development of APF. By implementing the topological analysis to the downstream target of HAND2-AS1, it was found that only hsa-miRNA570-3p directly interacted with it. This also indicated that hsa-miRNA-570-3p may be the critical miRNA regulating APF.
APF has been shown to be closely related to inflammation and oxidative stress [35, 36]. Studies have reported that hsa-miRNA-570-3p is a novel regulator of inflammation [37]. It mainly participates in the occurrence and development of inflammation by regulating the expression of IL-6 and CXCL8, and these inflammatory factors have been confirmed to be closely associated with APF [38–40]. Moreover, a previous research reported the negative regulatory effect of has-miRNA-570-3p against MMP-9, which suggests this miRNA is closely related to the stability of the atherosclerotic plagues [37, 41].
The functionality of lncRNA HAND2-AS1 has been well characterized in several types of human cancers [42, 43]. Furthermore, studies have confirmed that lncRNA HAND2-AS1 is positively related to insulin-like growth factor I (IGF-1) [44]. And IGF-1 has also been proven to reduce lipid oxidation and foam cell formation by downregulating 12/15-LOX and ultimately reduce APF [45]. A previous study revealed that lncRNA HAND2-AS1 could regulate the expression of hypoxia-inducible factor-1α (HIF-1α) [46]. HIF-1α has also been confirmed to promote macrophage necroptosis by regulating miRNA-210 and miRNA-383 [47]. However, there is no evidence to point out the regulatory role of lncRNA HAND2-AS1 in APF.
As far as we know, this research constructed a ceRNA network containing lncRNA HAND2-AS1, hsa-miRNA-570-3p, and APF-related genes in APF. Furthermore, KEGG analysis has shown that the “regulation of platelet-derived growth factor receptor signaling pathway” may be the main pathway resulting in APF. It was previously reported that platelet-derived growth factor receptor (PDGFR) signaling pathway could regulate smooth muscle cell (SMC) migration and proliferation in the vascular wall, leading to increased predisposition to cardiovascular disease [48]. It has been reported that patients with APF exhibit higher PDGFR expression in clinical study [49]. In vivo and in vitro experiments have also confirmed that PDGFR can promote the development of APF [50]. Therefore, understanding the PDGFR signaling pathway in the development of APF is helpful to find novel treatments.
5. Conclusions
In this study, a global triple network including lncRNAs, miRNAs, and mRNAs was constructed based on the ceRNA hypothesis, and then, a regulatory network of APF was screened. lncRNA HAND2-AS1, hsa-miRNA-570-3p, DPP4, MYOCD, CNTN4, and CNTN1 were identified as the key genes. These findings complemented our understanding of APF and may provide potential diagnostic biomarkers or therapeutic targets in APF.
Acknowledgments
This study was funded by President Foundation of The Fifth Affiliated Hospital of Southern Medical University (YZ2020QN02), and Basic and Applied Basic Research Project of Guangzhou Basic Research Plan in 2022 (2019KYQN12).
Abbreviations
- AS:
Atherosclerosis
- lncRNAs:
Long non-coding RNAs
- APF:
Atherosclerotic plaque formation
- DEGs:
Differentially expressed genes
- miRNAs:
MicroRNAs
- ceRNA:
Competitive endogenous RNA
- FDR:
False discovery rate
- GO:
Gene ontology
- DELs:
Differentially expressed lncRNAs
- DEMs:
Differentially expressed mRNAs
- DEMis:
Differentially expressed miRNAs.
Data Availability
All of the data was obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The lncRNA-miRNA and miRNA-mRNA interaction data were obtained from the lncRNASNP database (http://bioinfo.life.hust.edu.cn/lncRNASNP/) and starBase V2.0 database (http://starbase.sysu.edu.cn/).
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Yunci Ma, Jianhua Feng, and Weiqian Deng developed the method, carried out experiments, and drafted the manuscript. Danyu Li prepared and analyzed the data. All authors read and approved the final paper. Yunci Ma and Danyu Li are equal contributors.
References
- 1.Fuster V. Global burden of cardiovascular disease: time to implement feasible strategies and to monitor results. Journal of the American College of Cardiology . 2014;64(5):520–522. doi: 10.1016/j.jacc.2014.06.1151. [DOI] [PubMed] [Google Scholar]
- 2.Fowkes F. G., Rudan D., Rudan I., et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet . 2013;382(9901):1329–1340. doi: 10.1016/S0140-6736(13)61249-0. [DOI] [PubMed] [Google Scholar]
- 3.Qiu G. Z., Tian W., Fu H. T., Li C. P., Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochemical and Biophysical Research Communications . 2016;471(1):135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W., Zhang D., Xu B., et al. Long non-coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomedicine & Pharmacotherapy . 2015;69:90–95. doi: 10.1016/j.biopha.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Wei S., Xie W., Shang Q., Bing S. The long noncoding RNA MEG3 is downregulated and inversely associated with VEGF levels in osteoarthritis. BioMed Research International . 2015;2015:5. doi: 10.1155/2015/356893.356893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naemura M., Murasaki C., Inoue Y., Okamoto H., Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Research . 2015;35(10):5377–5382. [PubMed] [Google Scholar]
- 7.Aguilo F., Di Cecilia S., Walsh M. J. Long non-coding RNA ANRIL and Polycomb in human cancers and cardiovascular disease. Current Topics in Microbiology and Immunology . 2016;394:29–39. doi: 10.1007/82_2015_455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ounzain S., Burdet F., Ibberson M., Pedrazzini T. Discovery and functional characterization of cardiovascular long noncoding RNAs. Journal of Molecular and Cellular Cardiology . 2015;89:17–26. doi: 10.1016/j.yjmcc.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Holdt L. M., Beutner F., Scholz M., et al. ANRILexpression is associated with atherosclerosis risk at chromosome 9p21. Arteriosclerosis, Thrombosis, and Vascular Biology . 2010;30(3):620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 10.Wu G., Cai J., Han Y., et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation . 2014;130(17):1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Shen Q., Zhang X., et al. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cellular Physiology & Biochemistry International Journal of Experimental Cellular Physiology Biochemistry & Pharmacology . 2017;41(6):2221–2229. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 12.Chen D. L., Chen L. Z., Lu Y. X., et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial- mesenchymal transition in colorectal cancer. Cell Death & Disease . 2017;8(8):p. e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y., Wang L., Li Y., Chen M., He W., Qi L. The long non-coding RNA XIST interacted with miR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR) Cellular Physiology & Biochemistry International Journal of Experimental Cellular Physiology Biochemistry & Pharmacology . 2017;43(1):405–418. doi: 10.1159/000480419. [DOI] [PubMed] [Google Scholar]
- 14.Han Y., Qiu H., Pei X., Fan Y., Tian H., Geng J. Low-dose sinapic acid abates the pyroptosis of macrophages by downregulation of lncRNA-MALAT1 in rats with diabetic atherosclerosis. Journal of Cardiovascular Pharmacology . 2018;71(2):104–112. doi: 10.1097/FJC.0000000000000550. [DOI] [PubMed] [Google Scholar]
- 15.Chen L., Yang W., Guo Y., et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One . 2017;12(9):p. e185406. doi: 10.1371/journal.pone.0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Gao W., Long Q. Q., et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Scientific Reports . 2017;7(1):p. 7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilyugin M., Irminger-Finger I. Long non-coding RNA and microRNAs might act in regulating the expression of BARD1 mRNAs. International Journal of Biochemistry & Cell Biology . 2014;54:356–367. doi: 10.1016/j.biocel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Cao L., Wu J., Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. International Journal of Oncology . 2018;52(1):77–88. doi: 10.3892/ijo.2017.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D. X., Fei X. R., Dong Y. F., Cheng C. D., Niu C. S. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget . 2017;8(50):88163–88178. doi: 10.18632/oncotarget.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ya-Ru M., Wei L., Qiong Z., An-Yuan G. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Research . 2018;46(D1):D276–D280. doi: 10.1093/nar/gkx1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J. H., Liu S., Hui Z., Qu L. H., Yang J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research . 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics . 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbron C., Chang K. M., South M. C. RefPlus: an R package extending the RMA algorithm. Bioinformatics . 2007;23(18):2493–2494. doi: 10.1093/bioinformatics/btm357. [DOI] [PubMed] [Google Scholar]
- 24.Cao C., Chen J., Chengqi L., et al. Bioinformatics analysis of the effects of tobacco smoke on gene expression. PLoS One . 2015;10(12):p. e0143377. doi: 10.1371/journal.pone.0143377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J., Zhang S. A Bayesian extension of the hypergeometric test for functional enrichment analysis. Biometrics . 2014;70(1):84–94. doi: 10.1111/biom.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabriela B., Bernhard M., Hubert H., et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics . 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otte E., Rousseau R. Social network analysis: a powerful strategy, also for the information sciences. Journal of Information Science . 2002;28(6):441–453. doi: 10.1177/016555150202800601. [DOI] [Google Scholar]
- 28.Wolfe A. W. Social network analysis: methods and applications. Contemporary Sociology . 1995;91(435):219–220. [Google Scholar]
- 29.Wang C., Jiang W., Li W., et al. Topological properties of the drug targets regulated by microRNA in human protein-protein interaction network. Journal of Drug Targeting . 2011;19(5):354–364. doi: 10.3109/1061186X.2010.504261. [DOI] [PubMed] [Google Scholar]
- 30.Jdj H., Bertin N., Hao T., et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature . 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 31.Wang K. C., Chang H. Y. Molecular mechanisms of long noncoding RNAs. Molecular Cell . 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulitsky I., Bartel D. lincRNAs: genomics, evolution, and mechanisms. Cell . 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. A _ceRNA_ hypothesis: the Rosetta Stone of a hidden RNA language? Cell . 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song C., Zhang J., Liu Y., Pan H., Li C. Q. Construction and analysis of cardiac hypertrophy-associated lncRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in cardiac hypertrophy. Oncotarget . 2016;7(10):10827–10840. doi: 10.18632/oncotarget.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchio P., Guerra-Ojeda S., Vila J. M., Aldasoro M., Mauricio M. D. Targeting early atherosclerosis: a focus on oxidative stress and inflammation. Oxidative Medicine and Cellular Longevity . 2019;2019:32. doi: 10.1155/2019/8563845.8563845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B., Li W., Li X., Hong Z. Inflammation: a novel therapeutic target/direction in atherosclerosis. Current Pharmaceutical Design . 2016;23(8) doi: 10.2174/1381612822666161230142931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker J. R., Vuppusetty C., Colley T., et al. MicroRNA-570 is a novel regulator of cellular senescence and inflammaging. The FASEB Journal . 2019;33(2):1605–1616. doi: 10.1096/fj.201800965R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomohisa H., Mika I., Satoshi A., et al. SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS One . 2015;10(1):p. e116480. doi: 10.1371/journal.pone.0116480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavin B., Phinikaridou A., Andia M. E., Potter M., Botnar R. M. Sustained focal vascular inflammation accelerates atherosclerosis in remote arteries. Arteriosclerosis Thrombosis and Vascular Biology . 2020;40(9) doi: 10.1161/ATVBAHA.120.314387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber C. Inflammatory Chemokines in Atherosclerosis. Cells . 2021;10(2):p. 226. doi: 10.3390/cells10020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T., Li X., Feng Y., Dong G., Yang J. The role of matrix metalloproteinase-9 in atherosclerotic plaque instability. Mediators of Inflammation . 2020;2020:13. doi: 10.1155/2020/3872367.3872367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Lin J., Hui Z., Zhu F., Xie R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochemical & Biophysical Research Communications . 2018;503(3):1848–1853. doi: 10.1016/j.bbrc.2018.07.125. [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Wang C. C., Lee W., Trovik J., Chung T., Kwong J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Letters . 2018;413:23–34. doi: 10.1016/j.canlet.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 44.Cheng X., Jiang H. Long non-coding RNA HAND2-AS1 downregulation predicts poor survival of patients with end-stage dilated cardiomyopathy. The Journal of International Medical Research . 2019;47(8):3690–3698. doi: 10.1177/0300060519849295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sukhanov S., Snarski P., Vaughn C., et al. Insulin-like growth factor I reduces lipid oxidation and foam cell formation via downregulation of 12/15-lipoxygenase. Atherosclerosis . 2015;238(2):313–320. doi: 10.1016/j.atherosclerosis.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang Y., Zhu X., Xu Y., et al. Energy stress-induced lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits osteosarcoma progression. American Journal of Cancer Research . 2018;8(3):p. 526. [PMC free article] [PubMed] [Google Scholar]
- 47.Karshovska E., Wei Y., Subramanian P., Mohibullah R., Schober A. HIF-1α (hypoxia-inducible factor-1α) promotes macrophage necroptosis by regulating miR-210 and miR-383. Arteriosclerosis Thrombosis and Vascular Biology . 2020;40(3) doi: 10.1161/ATVBAHA.119.313290. [DOI] [PubMed] [Google Scholar]
- 48.Vindis C., Escargueil-Blanc I., Uchida K., Elbaz M., Salvayre R., Negre-Salvayre A. Lipid oxidation products and oxidized low-density lipoproteins impair platelet-derived growth factor receptor activity in smooth muscle cells: implication in atherosclerosis. Redox Report . 2007;12(1-2):96–100. doi: 10.1179/135100007X162248. [DOI] [PubMed] [Google Scholar]
- 49.Irvine C. D., George S. J., Sheffield E., Johnson J. L., Lamont P. M. The association of platelet-derived growth factor receptor expression, plaque morphology and histological features with symptoms in carotid atherosclerosis1. Cardiovascular Surgery . 2000;8(2):121–129. doi: 10.1016/S0967-2109(99)00090-3. [DOI] [PubMed] [Google Scholar]
- 50.Pontén A., Folestad E. B., Pietras K., Eriksson U. Platelet-derived growth factor D induces cardiac fibrosis and proliferation of vascular smooth muscle cells in heart-specific transgenic mice. Circulation Research . 2005;97(10):1036–1045. doi: 10.1161/01.RES.0000190590.31545.d4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data was obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The lncRNA-miRNA and miRNA-mRNA interaction data were obtained from the lncRNASNP database (http://bioinfo.life.hust.edu.cn/lncRNASNP/) and starBase V2.0 database (http://starbase.sysu.edu.cn/).


