Summary
Ripples are brief high-frequency electrographic events with important roles in episodic memory. However, the in vivo circuit mechanisms coordinating ripple-related activity among local and distant neuronal ensembles are not well understood. Here we define key characteristics of a long-distance projecting GABAergic cell group in the mouse hippocampus that selectively exhibits high-frequency firing during ripples while staying largely silent during theta-associated states when most other GABAergic cells are active. The high ripple-associated firing commenced before ripple onset and reached its maximum before ripple peak, with the signature theta-off, ripple-on firing pattern being preserved across awake and sleep states. Controlled by septal GABAergic, cholinergic, and CA3 glutamatergic inputs, these ripple-selective cells innervate parvalbumin and cholecystokinin-expressing local interneurons while also targeting a variety of extra-hippocampal regions. These results demonstrate the existence of a hippocampal GABAergic circuit element that is uniquely positioned to coordinate ripple-related neuronal dynamics across neuronal assemblies.
Keywords: GABA, inhibition, disinhibition, sharp wave ripple, hippocampus, muscarinic, medial septum, brain state, sleep
Graphical Abstract

eTOC blurb:
Szabo et al. identify a subset of hippocampal GABAergic cells with long-range projections and unique activity dynamics. Theta-OFF/Ripple-ON (TORO) cells are virtually silent during locomotion- and REM sleep theta but fire vigorously during sharp wave-ripples. TORO cells receive CA3 glutamatergic, septal GABAergic and inhibitory cholinergic inputs and target interneurons.
Introduction
Hippocampal sharp-wave ripples (SPW-Rs) are the most synchronous population patterns observable in local field potential (LFP) recordings from the mammalian brain, associated with a robust transient increase in excitability (Buzsáki, 1986; Csicsvari et al., 1999). Several lines of evidence support their prominent role in memory consolidation, including the replay of memory-related information both during specific awake and sleep states and in cognitive processes such as memory-guided decision-making (Diba and Buzsáki, 2007; Ego-Stengel and Wilson, 2010; Girardeau et al., 2009; Jadhav et al., 2012). However, the precise microcircuit mechanisms that regulate SPW-Rs in the hippocampus and coordinate SPW-R-related activity dynamics with extra-hippocampal regions are not well understood.
The sharp-wave component represents the dendritic depolarization evoked by the synchronous activity of subgroups of excitatory afferents from the CA3 region (Buzsáki, 1986; Buzsáki et al., 1983; Oliva et al., 2016; Sullivan et al., 2011; Suzuki and Smith, 1988), while the ripple oscillations (120-200 Hz) are thought to be generated by the microcircuit interactions between CA1 pyramidal cells (PCs) and local fast spiking interneurons whose activity is set into motion by the sharp wave-associated excitatory inputs (Schlingloff et al., 2014; Stark et al., 2014). Although it has been recognized that excitatory activity during SPW-Rs propagates from the hippocampus towards the subiculum and other downstream areas (Böhm et al., 2015; Chrobak and Buzsáki, 1994, 1996; Siapas and Wilson, 1998; Wierzynski et al., 2009), it is less clearly established if SPW-R-associated hippocampal inhibitory dynamics is similarly relayed to extra-hippocampal areas through long-distance GABAergic projections.
In this study, we aimed to define a GABAergic hippocampal circuit element that would be capable of providing inhibition to subsets of interneurons both locally and in downstream regions in a ripple-selective manner. A number of previous observations provided partial clues concerning the nature of such hypothetical ripple-selective GABAergic neurons. First, hippocampal interneurons display a variety of ripple-related spiking behaviors ranging from robust entrainment to the complete lack of activity (Ferraguti et al., 2005; Fuentealba et al., 2008; Jinno et al., 2007; Katona et al., 2017; Klausberger et al., 2004, 2005; Lasztóczi et al., 2011; Szabo et al., 2017; Varga et al., 2012, 2014; Viney et al., 2013), raising the possibility that the silent cells are actively inhibited during ripples. Second, there is at least one cell reported in the literature that discharged during ripples (albeit under urethane anesthesia) and preferentially innervated interneurons (Ferraguti et al., 2005). Third, several types of hippocampal CA1 GABAergic neurons give rise to long-range GABAergic projections to extra-hippocampal targets (Caputi et al., 2013; Gulyás et al., 2003; Jinno et al., 2007; Melzer et al., 2012; Takács et al., 2008; Toth et al., 1993), although some of these (e.g., VIP-expressing) cells did not seem to preferentially discharge during ripples (Francavilla et al., 2018; Kullander and Topolnik, 2021). Fourth, some interneurons in extra-hippocampal areas are suppressed during hippocampal SPW-Rs (Girardeau et al., 2017), suggesting the possibility that ripple-related GABAergic coordination between local hippocampal and distal circuits may indeed exist.
Using juxtacellular recordings, two-photon (2P) calcium imaging, and optogenetics in behaving mice, here we describe a unique GABAergic cell population in the CA1 that is highly active before and during ripples, but not in hippocampal states characterized by theta oscillations (locomotion and REM sleep). These cells are suppressed by medial septal inhibitory inputs and cholinergic activation of muscarinic type 2 receptors (M2Rs), and innervate other local GABAergic neurons while also robustly projecting to distant brain areas. These results outline a functional circuit element ideally positioned to regulate and broadcast ripple-related inhibitory activity both within- and beyond the hippocampus.
Results
Distinct activity patterns of SPW-R-activated non-pyramidal cells during theta oscillations in vivo
We carried out juxtacellular recordings of CA1 non-pyramidal neurons in awake, head-fixed mice, running or resting on a spherical treadmill. For each experiment, an electrode was placed in the CA1 pyramidal layer, allowing us to monitor hippocampal local field potentials (LFP), including theta oscillations and sharp-wave ripples. Cells that displayed robust spiking activity throughout ripples (ripple-ON) were filled with neurobiotin and processed for post-hoc anatomical reconstruction and neurochemical characterization (Szabo et al., 2017; Varga et al., 2012, 2014) (Figure 1). Similar to previously described interneurons (Lapray et al., 2012; Varga et al., 2012, 2014), a subset of ripple-ON cells displayed elevated activity during locomotion-associated theta oscillations compared to rest (theta-ON, Figures 1A-D, n=6). In contrast, we observed a subset of ripple-ON cells that had markedly reduced firing rates during theta (theta-OFF, Figures 1E-H, n=7). Strikingly, these theta-OFF, ripple-ON cells (or simply TORO cells) fired at distinctly high firing rates during ripples (Figure 1I), especially in contrast to the theta state (theta: 2 ± 2.5 Hz; ripple: 173.7 ± 26.0 Hz). Interestingly, the few TORO cell spikes that did occur during theta periods were strongly phase coupled to the oscillation in most cells (5/7 cells, Figure S1F). Moreover, the theta phase-preference of TORO cells (37±39.5°; ascending phase) was distinct from that of theta-ON cells (255±41.2°; descending phase) (Lapray et al., 2012; Varga et al., 2012, 2014), indicating their differential engagement in the circuit.
Figure 1. Distinct in vivo activity dynamics of theta-OFF, ripple-ON cells in the CA1 region of the mouse hippocampus assessed with juxtacellular recordings.
(A) Partial reconstruction of a representative theta-ON, ripple-ON, identified as parvalbumin-expressing basket cell (PVBC). s.o. – stratum oriens; s.p. – stratum pyramidale; s.r. – stratum radiatum.
(B) Immunopositivity for PV. Arrows point out positive boutons.
(C) Spiking activity of the cell shown in A. Note the robust firing during RUN and the spiking initiated at the onset of the ripple events.
(D) Average firing frequency during resting and running for all recorded PVBCs in these experiments (REST: 9.5 ± 6.8 Hz; RUN: 39.1 ± 18 Hz; paired sample t-test, t=−5.5, p=0.003, n=6). Individual values plus mean ± SD are shown.
(E) Partial reconstruction of an in-vivo juxtacellularly recorded and labeled representative theta-OFF, ripple-ON (“TORO”) cell. Inset (below reconstruction): schematic depicting the soma location of all juxtacellularly recorded and labeled CA1 TORO cells. s.l.-m. – stratum lacunosum-moleculare; alv. – alveus.
(F) Confocal micrograph showing somatostatin and M2R immunopositivity in the soma and in dendrites, respectively (arrows).
(G) Spiking activity of the cell shown in E during RUN and REST periods. Note the low level of activity during RUN, and the bouts of intense spiking initiated shortly before ripples.
(H) Average firing frequency during resting and running for all recorded CA1 TORO cells (REST: 11.8 ± 4.8 Hz; RUN: 2 ± 2.5 Hz paired sample t-test, t=6.9, p=0.0004, n=7). Data are presented as on D.
(I) Comparison of average spiking frequency outside of and during ripples for CA1 TORO cells vs. PVBCs. (TORO, ripple OUT: 8.9 ± 4.9 Hz, ripple IN: 173.7±26.0 Hz, t=−15.5, p<0.0001; PVBC, ripple OUT: 12.3 ± 7.3 Hz, ripple IN: 103.8 ± 42.1 Hz, paired sample t-test, t=−6.3, p=0.001). While both cell groups increased their firing during ripples, TORO cells reached higher firing frequencies during ripples than PVBCs (Welch’s t-test, t=3.5, p=0.008, n=13).
(J) Ripple center-triggered normalized spiking frequency curve of a representative PVBC and a TORO cell. Note the area under the curve (AUC) being larger before the center than after for the TORO cell.
(K) Comparison of the pre-ripple index (100-(AUCpost-ripple-center/AUCpre-ripple-center)*100) between CA1 TORO cells and PVBCs (Welch’s t-test, p=0.01). Bars indicate the means.
While all ripple-ON units by definition increased their firing rates during ripples, their firing patterns varied according to their theta response types (ON or OFF). During ripples, TORO cells fired at higher frequencies than theta-ON cells (Figure 1I). Moreover, TORO cells started firing at elevated rates even before the onset of ripple waves on the LFP (Figures 1G, S1C-D), and their activity was biased towards the early phase of ripple episodes (Figure 1J). In contrast, theta-ON cells started to increase their ripple-related firing later than TORO cells, reached a lower maximal firing frequency, and centered their maximal firing close to the ripple peak (Figures 1C, J, K, S1C-D). In addition, TORO cells had a relatively weak coupling to the late ascending phase of the individual ripple wavelets (144.6±34.5°), while theta-ON cells had strong coupling to the early ascending phase (56.9±14.7°, Figure S1G).
Morphological and neurochemical features were also distinct between theta-OFF and theta-ON groups of ripple active cells. While units with theta-ON activity were parvalbumin-expressing basket cells (PVBCs) located within or nearby the pyramidal layer (n=6, Figures 1A-D (Lapray et al., 2012; Varga et al., 2012, 2014)), neurons with TORO activity (n=7) were scattered across multiple layers within the CA1 (inset on Figure 1E) and none of the tested TORO cells expressed PV (0 of 4). Unlike PV cells, such as BCs and axo-axonic cells (AACs) that only express M2Rs at detectable levels presynaptically in their axon terminals (Hájos et al., 1997; Szabó et al., 2010), all tested TORO cells expressed robust immunoreactivity for M2Rs on their somata and dendrites (3 of 3, Fig 1F; for additional evidence on the high TORO cell M2R expression, see below). One of five TORO cells expressed calbindin (CB) and four of five expressed somatostatin (SOM) (see Discussion). Most TORO cells were found in the stratum oriens near the alveus with dendrites residing in the oriens (4/7) (inset on Figure 1E), while two cells were at the border of the strata radiatum and lacunosum-moleculare (LM), and one in the stratum radiatum with dendrites spanning from the stratum oriens to radiatum. TORO cell axons were located mostly in the stratum oriens (Figure 1E, Figure S1A, S6A) with collaterals in the stratum radiatum running along the radiatum-LM border, and in the subiculum. Axonal collaterals often formed characteristic short appendages called drumsticks (Figures S1E, S7B), previously described as a morphological feature of subsets of GABA cell axons (Hájos et al., 1997; Sik et al., 1994). In addition, 5 out of 7 TORO cells had thick axons in the alveus and projected to the subiculum. Two of these cells had axons up to ~1.6mm from the cell body towards the septal and caudal directions, including one cell with axons in the stria terminalis, internal capsule, and the splenium of the corpus callosum (Figures S1A-B, S6A-C), indicating that TORO cells may form long-range projections (for further analysis of such projections, see below). In contrast, PVBC axons were not observed leaving the hippocampus, in agreement with the lack of reports of extra-hippocampally projecting PV cells (Lapray et al., 2012; Varga et al., 2012, 2014) except for a single report of one PV-positive hippocampo-septal cell (Toth and Freund, 1992).
Taken together, the results above support the notion that TORO cells, defined by their unique ripple-selective activity, form a cell group with characteristic morphological and physiological features.
Classification criteria for unbiased TORO cell identification and characterization across datasets
Next, in order to move from the low-throughput juxtacellular method that relied on chance encounters with particular cells by the single recording electrode, we sought to identify TORO cells in larger datasets based on their characteristic behavior. Therefore, we identified criteria based on firing rates derived from our juxtacellular TORO cell recordings to provide suitable, easy to implement, and generalizable criteria for defining TORO cells across datasets. Firing rate cut-offs for defining TORO cells were set to within 3 standard deviations of the mean from juxtacellularly-recorded cells, such that firing rates must be less than 9 Hz during locomotion and greater than 50Hz during SPW-Rs, and we also required the SPW-R firing rate of a TORO cell to be at least 15 times greater than its locomotor firing rate. To validate our criteria, we analyzed LFP and spiking data recorded from 6 Neuropixel probes (Jun et al., 2017) spanning multiple areas of the mouse brain, including neocortex, hippocampus, thalamus, and midbrain, using the publicly available Allen Brain Observatory database (Siegle et al., 2021; n=13 mice). By plotting firing rates during locomotion vs. SPW-Rs for all recorded cells in the database (Figure 2A, n=9375), we observed a unique group of cells classified as TORO cells (Figure 2A, n=18). These units, which were recorded for an average of 2.6 ± 0.04 hours across a range of speeds and during an average of 2277 ± 320 SPW-Rs, gave us the opportunity to comprehensively characterize their firing rate dynamics.
Figure 2. Identification and characterization of hippocampal TORO cells in a publicly available dataset.
(A) Data from the Allen Brain Observatory (Siegle et al., 2021) functional connectivity dataset were analyzed to identify putative TORO units based on firing activity during ripples and locomotor activity. Scatter plot shows firing rate during locomotion and SPW-Rs for TORO (orange) and non-TORO (grey) units. Dashed orange lines indicate cut-offs for TORO cell identification (>50Hz for ripple, <9Hz for locomotion, >15:1 ripple:locomotion firing rate ratio).
(B) Firing rate during locomotion for a representative TORO cell located in the CA1 (top left). Top right panel shows average firing rate with respect to speed. Bottom panel shows firing rate changes during periods of locomotion and immobility.
(C) Brain areas that contain TORO cells (orange dot and line). Brain regions are color coded by neocortical, thalamic, hippocampal, and midbrain structures. The percentage of cells that are identified as TORO are displayed along with the # of TORO cells out of the total population per brain region. HPF (hippocampal formation) refers to a category that includes cells that were not assigned with high confidence to any subregion within the HPF. The TORO cell in this category was located in the white matter over the subiculum.
(D) Left shows histogram of speed vs. firing rate correlation values for TORO (orange) and non-TORO units (t9373=−8.47, p=2.8x10−17). Right panel (shared y-axis) shows TORO cell firing rate vs. speed correlation values for all data vs. when immobility data was withheld (t17=−9.29, p=4.5x10−8).
(E) Correlation values for TORO cell firing rates to ripple amplitude (t17=11.41, p=2.1x10−9), frequency (t17=8.35, p=4.5x10−8), and duration (t17=0.73, p=0.47).
(F) Scatter plot and histograms for TORO vs. CA1 theta-ON, ripple-ON cells comparing pre-ripple index (shared y-axis; same analysis as Fig. 1K; t102=−4.63, p=1.1x10−5) and burst index (shared x-axis; t102=−4.42, p=2.5x10−5). Pre-ripple index was calculated as in Figure 1K. Burst index, a score ranging from −1 to 1 (1 being most bursty), was calculated from interspike intervals (ISI, inset shows mean ISI density).
The firing rate of a representative TORO cell recorded from CA1 with respect to speed is shown in Figure 2B. The firing rate of this TORO cell (Figure 2B) and all other TORO cells were negatively correlated to speed and distinct from the overall non-TORO population (Figure 2D left). The firing rate dropped dramatically even at low speeds and remained similarly low across a range of increasing speeds (Figure 2B). This binary locomotor response, rather than a graded response across increasing speeds, was generally consistent across TORO cells, since all TORO cells were less negatively correlated to speed when immobility data was withheld (Figure 2D right). During SPW-Rs, the firing rates of TORO cells were positively correlated to the amplitude and frequency, but not the duration of the ripple oscillation (Fig. 2E). Thus, TORO cell firing activity varies as a function of ripple dynamics, further emphasizing their strong coupling to SPW-Rs.
We then determined if TORO cells are more broadly expressed in the brain or unique to CA1 where our previous experiments were focused. TORO cells were only found in the hippocampus, including the CA1 as expected, and no cells were found in the recorded neocortical, thalamic, and midbrain areas. Using this unbiased approach, however, we also observed one cell in the dentate gyrus, one cell in the CA3, and a relatively high proportion of cells in the subiculum (Figure 2C). Cells were often superficially located in the hippocampus near the alveus (Movie S1), similar to our TORO cells labeled by juxtacellular recordings (Fig. 1E).
Finally, we compared TORO cell firing dynamics to cells in CA1 that had ripple firing rates exceeding the TORO cell cut-off (>50Hz) but were also active during locomotion (>20Hz, i.e. theta-ON, ripple-ON cells). Similar to Figure 1K, TORO cells in this dataset had a high pre-ripple firing index compared to theta-ON, ripple-ON cells (Figure 2F). We also found that TORO cells were distinctly and consistently more bursty compared to theta-ON cells (Figure 2F). Collectively, these data indicate that applying a functional definition for TORO cells, based only on firing rates measured during theta and ripple episodes in behaving mice, identifies a distinct group of hippocampal cells with unique properties that can be easily implemented on independent datasets.
TORO cells identified by two-photon imaging in vivo
Next, in order to analyze the behavior-dependent activity patterns of TORO cells in more detail, we carried out in vivo 2-photon imaging of virally expressed calcium indicators (GCaMP6f or jGCaMP7s; Chen et al., 2013; Dana et al., 2019) in head-fixed, behaving mice spontaneously running or resting on a linear treadmill, with simultaneous LFP recordings from the contralateral CA1 pyramidal layer (Buzsáki et al., 2003; Dudok et al., 2021; Geiller et al., 2020; Figures 3A-C, Figures S2A-B). In a subset of experiments, GABAergic neurons were selectively labeled using Dlx5/6-Cre transgenic mice and Cre-dependent viral constructs (Chen et al., 2013; Dana et al., 2019; Monory et al., 2006). Somatodendritic M2R expression of the imaged cells was assessed using post-hoc immunostaining (Figures 3E-F). In agreement with our data obtained using the juxtacellular method (Figure 1), we could identify ripple-ON cells without a theta-ON response in our calcium imaging data (Figures 3B-C). Relying on the stark contrast between TORO cell activity during ripple and theta states, we computed relative fluorescence changes in each pixel upon running and ripple onset and displayed these changes in contrasting color channels to visually identify TORO cells (see Methods). TORO cells in the resulting images (referred to as the “TORO image” below) appeared bright in the ripple-triggered channel but dark in the running-triggered channel, readily visualizing the somata and dendrites of TORO cells (Figure 3D; Figure S2D). As expected, overlaying TORO images with registered M2R immunostaining revealed that all tested TORO cells expressed high levels of somatodendritic M2Rs (15 of 15, Figure 3E-F, I), further validating our imaging approach to locate TORO cells in vivo even in the absence of a TORO-specific transgenic mouse line. In agreement with the higher firing rate of TORO cells compared to PVBCs during ripples (Figure 1I), calcium transients in TORO cells were greater than in any other GABAergic neurons recorded in the same experiments (Figures 3G-H, Figure S2C), underlining the unusually selective and exceptionally high activity of TORO cells during ripples.
Figure 3. Unique characteristics of TORO cell activity dynamics verified with 2-photon imaging in behaving mice.
(A) Experimental design.
(B) Two-photon in vivo calcium imaging trace of a representative TORO cell identified based on the “TORO image” (see text). Vertical magenta bars indicate ripple events detected in the contralateral CA1.
(C) Expanded view of the trace on B.
(D) Zoomed-in region from an average time projection 2-photon Ca-image from str. oriens (top) and the derived TORO image (bottom), where the average RUN response is shown on a cyan scale, while ripple response is shown on a magenta scale. TORO cells (arrows) were identified based on their low activity during RUN onsets but strong responses to ripple occurrences.
(E) Post-hoc confocal maximum intensity projection (MIP) image of the same cells shown on D (top). Cells #1 and #7 express M2R (bottom).
(F) Post-hoc confocal MIP image including several GABAergic neurons imaged during the same in vivo experiment as in panel D (note the same cells). The encircled cells are post hoc identified neurons expressing the respective markers. Image has been cropped and rotated to match the orientation of the cells shown on D-E, and the original boundaries of the confocal section are marked with a dashed line.
(G) Ripple-triggered average 2-photon calcium traces for individual GABAergic cells. Note that TORO cells #1 and #7 (same as on D-F) have larger ripple responses than any of the other non-TORO GABA cells (orange traces belong to TORO cells, grey traces are from non-TORO GABAergic cells).
(H) Comparison of the magnitude of ripple responses between 15 TORO cells and 109 non-TORO cells. TORO cell ripple response amplitude was greater than that of non-TORO cells (median[1st quartile, 3rd quartile]: TORO, 36.3[21.8, 43.9]; non-TORO, 4.1[1.4, 8.5], p <0.0001, n=124 cells from 5 mice, including 4 Dlx5/6-Cre mice (14 TORO cells) and 1 C57BL/6 mouse (1 TORO cell); Z=5.9; Mann-Whitney U test). Each cell was ranked according to their ripple response magnitude within their session (TORO, 100[100, 100]; nonTORO, 46.7[20.5, 70.9], p<0.0001, Z=6.26; Mann-Whitney U test on normalized ranks).
(I) All post-hoc identified TORO cells were found to be M2R-positive (n = 15).
Next, in order to gain insight into the molecular identity of the cells that express M2R in their somatodendritic axis, we quantified the fraction of M2R cells that expresses CB, SOM, vasoactive intestinal polypeptide (VIP) and/or nitric oxide synthase-1 (NOS1), which are neurochemical markers expressed in some long-projecting cells (Francavilla et al., 2018; Christenson Wick et al., 2019). This immunocytochemical assessment (performed on 1070 CA1 cells) revealed that these neurochemical markers are expressed heterogeneously in M2R cells (SOM: 19.7 ± 4.5%; CB: 51.9 ± 3.5%; NOS1: 2.5 ± 0.3%; VIP: 20.6 ± 7%, Figures S2E-I; for NOS1, see also Figure S6L).
Altogether, our juxtacellular recordings, analysis of large scale database of extracellular Neuropixel recordings, and our calcium imaging results indicated that M2R-expressing TORO cells were among the most active CA1 neurons during SPW-Rs, while they rarely discharged during theta. We next examined whether this unique activity pattern extends to analogous brain states during sleep.
TORO cells retain their brain state-specific activity patterns during sleep
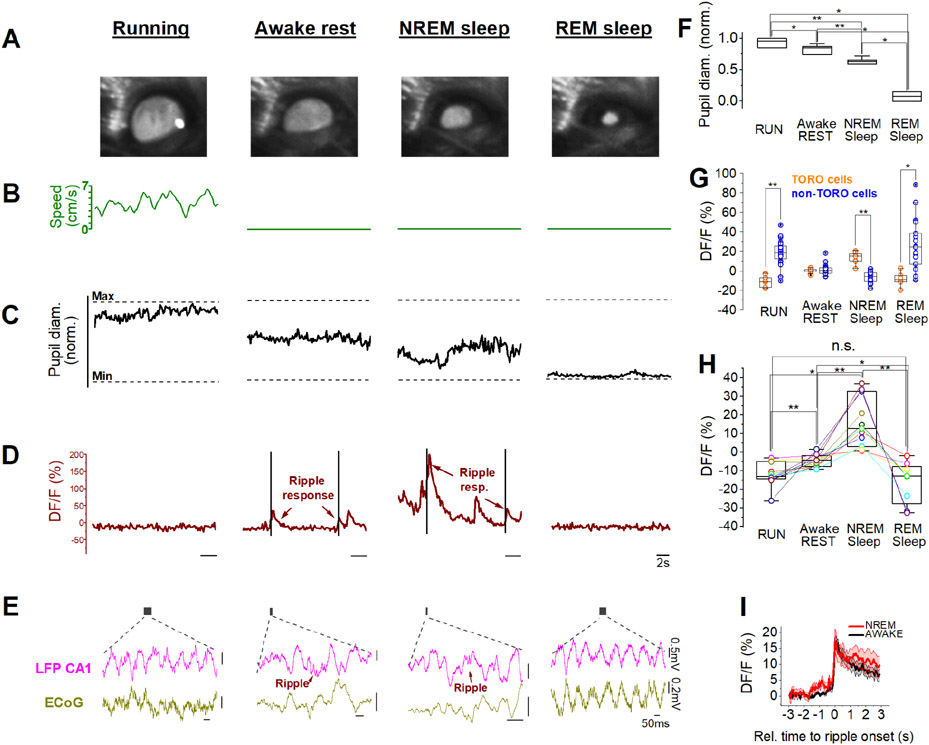
Theta oscillations can be observed during locomotion as well as during rapid eye movement (REM) sleep, while SPW-Rs occur during awake rest and non-REM (NREM) sleep (Buzsáki et al., 1992). Mice subjected to sleep deprivation can sleep in head-fixed condition with their eyelids open (Yüzgeç et al., 2018), allowing 2-photon calcium imaging while monitoring the pupil diameter, a known biomarker for the animal’s internal brain state (McGinley et al., 2015). Therefore, to determine TORO cell activity patterns during sleep, we imaged CA1 cells in the stratum oriens in sleep-deprived mice, with simultaneous electrocorticogram (ECoG), electromyogram (EMG), and LFP recordings (Figures 4A-E) to track sleep stages (see Methods). As expected, changes in pupil diameter were tightly correlated to the sleep scores obtained by conventional ECoG- and EMG analysis (Weber et al., 2015, 2018), indicating that pupil monitoring data can indeed be used as a reliable indicator of sleep stages in the head-fixed condition (Figures 4A, F; Figure S3A). During running, NREM or REM sleep stages, TORO cells were oppositely recruited compared to other neurons in the oriens layer (Figure 4G, for sleep transitions, see Figure S3B). Average TORO cell ΔF/F, likely indicative of overall activity in each brain state, was maximal during NREM sleep, and minimal during running and REM (Figure 4H). Despite the difference in average ΔF/F, the ripple response amplitudes of TORO cells were similar during ripples detected in awake or NREM stages (Figure 4I). Together, these experiments indicate that the characteristic theta-OFF, ripple-ON activity patterns are preserved during sleep, in spite of large differences in neuromodulatory dynamics between awake and sleep states (reviewed in Lee and Dan, 2012).
Figure 4. Preserved ripple-preference of TORO cells during sleep.
(A) Representative images of the pupil of a head-fixed mouse, corresponding to the different behavioral and brain states during 2-photon calcium imaging.
(B) Speed plots of locomotion activity of the mouse during running, awake rest, NREM and REM sleep stages.
(C) The corresponding pupil diameter measurements.
(D) Respective calcium imaging traces from a TORO cell. Black bars indicate ripples. Note the robust ripple-responses during awake rest and NREM sleep, and the lack of activity during running and REM sleep.
(E) LFP and electrocorticogram (ECoG) traces taken from the time windows (with respect to panels B-D) illustrated by the grey bars. Note the running- and REM-associated theta, and the presence of ripples during awake rest and NREM sleep.
(F) Pupil diameter is unique to each state (Kruskal-Wallis ANOVA completed with Mann-Whitney U test as pairs, X2(3)=25.3, p<0.001, n=4 mice; *p<0.05, **p<0.01 (M-W test)). Box plots show median±interquartile range (IQR), whiskers show range.
(G) TORO cell activity is distinct from non-TORO cells in 3 out of 4 states. Boxplots show median±IQR, whiskers show range, circles show individual values. (n=9 TORO and 24 non-TORO cells from 4 mice, Mann-Whitney U test; *p<0.05, **p<0.01).
(H) TORO cell activity changes from one state to another. Repeated measures ANOVA with Greenhouse-Geisser correction, F=12.4, p=0.0054; completed with pairwise comparisons with pair-sample t-test; n=11 cells, 6 mice. Note that TORO cell activity is similarly low during RUN and REM (pair-sample t-test; t=1.5, p=0.186 (RUN vs REM). Boxplots show median±IQR, whiskers show range, circles show individual values. (*p<0.05, **p<0.01)
(I) Ripple-triggered average 2-photon calcium traces for awake rest vs. NREM states do not differ significantly in TORO cells (t=0.7, p=0.48, pair sample t-test on ripple-response magnitudes, n=11 cells, 6 mice), indicating that TORO cells robustly increase their ripple-related activity during both awake and sleep states. MeanáSEM is shown.
TORO cells receive strong feed-forward excitation
Next, we explored the mechanisms that may contribute to TORO cell activity patterns. SPW-Rs are thought to be initiated by the synchronous bursting of a subpopulation of CA3 pyramidal cells which, in turn, drive CA1 pyramidal cells and interneurons via Schaffer collaterals (Buzsáki, 2015; Buzsáki et al., 1983; Sullivan et al., 2011). The robust and early recruitment of TORO cells compared to PVBCs during ripples (Figures 1I-K) may indicate that TORO cells are more strongly driven by feed-forward CA3 inputs. To test this, we expressed channelrhodopsin (ChR2) or non-opsin control (mCherry) in CA3 pyramidal cells and photostimulated their axons while imaging neuronal responses in CA1 oriens, which contains dense CA3 axonal innervation (Amaral and Witter, 1989; Schaffer, 1892; Takács et al., 2012; Figure 5A, Figure S4). TORO cells were identified based on TORO images (Figure 5B), as described above (Figure 3D). Photostimulation consistently evoked TORO cell calcium responses when CA3 pyramidal cells expressed ChR2, but not in controls (Figures 5C-D). Similar to the greater ripple response of TORO cells compared to other neurons (Figure 3G), TORO cells also displayed the greatest ΔF/F changes out of all imaged CA1 oriens neurons in response to optogenetically driving CA3 (Figures 5C-D). Altogether, these data indicate that TORO cells are particularly responsive to CA3 input, which may contribute to the robust and early firing of TORO cells during ripples.
Figure 5. TORO cells are strongly entrained by CA3 activation in vivo.
(A) Experimental design for driving CA3 inputs optogenetically (strategy 1; n=4 mice, strategy 2; n=2 mice).
(B) TORO image (RUN- and Ripple responses were overlaid) of a representative calcium movie taken from CA1 oriens. Cells #1&2 are TORO cells.
(C) While all cells shown on (B) responded positively to optogenetic activation of CA3-originating fibers, TORO cells #1&2 (orange traces) had more robust response than the rest of the cells (grey traces). Stimulation: 15ms light pulses at 15.6 Hz for the duration of 5 imaging frames or approximately 320ms. Note that the strongest ripple response (arrow) was also produced by the TORO cells.
(D) Average light-triggered response in all TORO cells vs. non-TORO cells (n=26 TORO cells from 4 mice, left) and the lack of response in mCherry controls (right, TORO cells shown, n=24 cells from 2 mice). TORO cells showed positive calcium responses upon light delivery compared to no-opsin controls (by 35±16% DF/F, X2(1)=5, p=0.028, likelihood ratio test, n=50 cells, 6 mice) and TORO cells had more robust light-response than non-TORO cells: see for example cells #1&2 on Figure 5B-C (effect size r=0.79, p=0.007, sign test, n=21 cells, 4 mice). Data are shown as mean±SEM.
Septal GABAergic inhibition may contribute to silencing TORO cells during theta
GABAergic input from the medial septum (MS) is known to inhibit hippocampal interneurons, and synchronize their firing during theta oscillations (Freund and Antal, 1988; Hangya et al., 2009; Kaifosh et al., 2013; Katona et al., 2020). The robust theta modulation of the sparse firing activity of TORO cells during locomotion (Figure S1F) is consistent with the possibility that these cells receive rhythmic inhibition during theta. Therefore, in order to determine whether TORO cells receive septal GABAergic inputs during theta, we first expressed mCherry in MS GABAergic cells in Dlx5/6-Cre mice (Figures 6A, S5A). Exploiting the specificity of somatodendritic M2R expression to TORO cells (Figures 1B, 3E-F; note that, as mentioned above, PVBCs do not show M2R immunoreactivity on their somata and dendrites), we quantified mCherry-positive boutons around M2R-immunopositive somata in the CA1 area. These immunocytochemical results showed the presence of MS GABAergic contacts on every (30/30) examined M2R-positive soma (Figures 6B-C), consistent with the possibility that MS inhibition may play a role in shaping the theta-OFF activity dynamics of TORO cells.
Figure 6. Septal GABAergic inhibition suppresses TORO cells during running.
(A) Labeling strategy for visualizing septal GABAergic fibers in the CA1.
(B) Confocal MIP images showing M2R-positive somata and dendrites contacted by septal GABAergic inputs.
(C) Quantification of septal GABAergic contacts. Box plot shows median±IQR, whiskers show range, circles show mean values for each animal; red dots are individual values for M2R cells.
(D) Labeling strategy for 2P imaging of septal GABAergic axon terminals while simultaneously imaging TORO cells.
(E) Representative example of simultaneous 2P imaging from a TORO cell (jRGECO, violet; defined based on TORO image, panels F-H) and from the GABAergic axons originating from the medial septum (Axon-GCaMP6s, green; yellow arrows).
(F) Ripple-triggered calcium response calculated from the jRGECO signal (4 post-ripple frames/10 pre-ripple frames), intensity is shown on a magenta scale. Note that this procedure also visualizes the otherwise faint dendritic segments of the TORO cell.
(G) RUN-triggered calcium response calculated on the Axon-GCaMP6s signal (RUN frames/no-RUN frames), intensity is shown on a cyan scale.
(H) Overlay of the two responses shown on panels (F) and (G). Note the septal GABAergic axons located in close juxtaposition to the TORO cell soma and dendrites.
(I) 2P calcium trace from the TORO cell and the surrounding septal axons shown on panel (H). Vertical black bars indicate ripples. Note the axonal activation during running.
(J) Run-triggered average for all TORO cells (violet) and the surrounding septal axons (green). Axon terminals surrounding TORO cells are primarily active during running (median[1st quartile, 3rd quartile]: pre-RUN: 5.0[0.1, 48.4] DF/F%, RUN: 176.1[142.5, 298.7] DF/F%, n=11 cells, 3 mice, p=0.0039, W=0, Wilcoxon signed ranks test). Data are shown as mean±SEM. See also Figure S5.
(K) Labeling strategy for driving septal GABAergic inputs optogenetically while imaging TORO cells in the CA1.
(L) Example 2P imaging trace from a TORO cell suppressed by blue light. Note the lack of ripple responses when the light is delivered to the CA1. Stimulation pattern: 15ms pulses at 15.6 Hz for 220 frames (or approximately 14 sec).
(M) Summary plots of ChR2 (n=58 cells from 5 mice) and control (mCherry, n=20 cells from 2 mice) experiments. Blue light significantly reduced the magnitude of ripple responses in ChR2 expressing mice compared to no-opsin and no-light controls (by −5.9±2.1% DF/F, X2(1)=8, p=0.0006, likelihood ratio test, n=78 cells, 7 mice). Box plots show median±IQR, whiskers show range, square markers show mean.
Next, we carried out dual calcium imaging of MS GABAergic axons and CA1 GABAergic cells by employing the Cre-dependent expression of axon-targeted GCaMP6s (Broussard et al., 2018) in the septum and a red-shifted calcium indicator (jRGECO1a, Dana et al., 2016) in the CA1 of Dlx5/6-Cre mice (Figure 6D). MS GABAergic axon terminals surrounding CA1 TORO cells (identified based on the TORO images) (Figures 6E-H) were active during running, but importantly, not during ripples (Figures 6I-J, S5B-E). Thus, the activity of septal GABAergic contacts innervating TORO cells follows a pattern complementary to that of TORO cell activity.
Finally, to determine whether MS GABAergic axons were able to inhibit TORO cells in vivo, we expressed Cre-dependent ChR2 in the medial septum and calcium indicator in the CA1 of Dlx5/6-Cre mice (Figure 6K). TORO cells in ChR2-expressing mice remained relatively silent during photostimulation during running episodes (Figure 6L). However, TORO cell ripple responses during immobility were blunted during activation of septal inhibitory cells (Figures 6L-M).
Altogether, these data indicate that TORO cells are inhibited by MS GABAergic cells that are active during running, consistent with septal inhibition contributing to the suppressed activity of TORO cells during theta. Furthermore, the lack of MS GABAergic activity in axons innervating TORO cells during ripples may facilitate ripple-related firing.
Somatodendritic M2Rs are inhibitory
In addition to septal GABAergic inputs, cholinergic septohippocampal projections play an important role in modulating the activity of hippocampal neurons (Cai et al., 2012; Dannenberg et al., 2015; Mesulam et al., 1983; Moreau et al., 2008; Solari and Hangya, 2018). While the majority of CA1 GABAergic cells has been shown to depolarize in response to the activation of muscarinic receptors (Cea-del Rio et al., 2010; Lawrence et al., 2006; McQuiston and Madison, 1999; Parra et al., 1998; Szabó et al., 2010), the M2R that is enriched in TORO cells is an inhibitory G-protein coupled receptor (Lai et al., 1991; Peralta et al., 1988) that may instead cause hyperpolarization. To determine the physiological effect of postsynaptic M2R expression in TORO cells (Figure 1F, Figure 3E), we utilized the recently developed Chrm2-tdT-D reporter mice (D’Souza et al., 2019; Gămănuţ et al., 2018). Since this knocked-in gene encodes a fluorescent tag (tdTomato, tdT) fused to the endogenous M2R (tdTChrm2), we expected the distribution of the tdT signal to be similar to that of M2R. Indeed, we found that M2R immunostaining and tdT fluorescence completely overlapped (tdTChrm2, Figure S6D). Therefore, we carried out fluorescence-targeted whole-cell patch-clamp recordings of tdTChrm2 cells (i.e. putative TORO cells; see below for in vivo validation), followed by biocytin labeling and reconstruction. tdTChrm2 cells displayed a mostly linear injected current – firing rate (I-F) relationship, the presence of a prominent sag in response to hyperpolarizing inputs, large afterhyperpolarization following each action potential (AP), and their cellular reconstruction revealed morphologies similar to that of in-vivo labelled TORO cells (Figures 7A-B; Figures S6E-F; compare to Figure 1E). Consistent with the inhibitory nature of M2Rs (Egan and North, 1985; Pan and Williams, 1994), the cholinergic agonist carbachol resulted in a robust hyperpolarization of these cells by 5.5 ± 1.6 mV (Figure S6G), which could be blocked by the selective M2R antagonist AF-DX116 (0.7 ± 1.8 mV; Figure S6G) (Araujo et al., 1989; Giachetti et al., 1986).
Figure 7. TORO cells substantially contribute to extra-hippocampal GABAergic projections from the CA1.
(A) Firing characteristics of a representative tdTChrm2 cell in response to depolarizing (150, 300, 500, 800 pA) and hyperpolarizing (−100 pA) current steps.
(B) Intrinsic properties of in vitro recorded tdTChrm2-cells. Box plots show median±interquartile range (IQR), whiskers show range, blue dots are individual values per cell (n=40 from 8 mice). Adaptation ratio initially increased with current injection levels then decreased with more depolarized levels due to the smaller change of 1st ISI at higher frequencies (green trace).
(C) 2P image of the tdTChrm2 signal is shown from in vivo 2P calcium imaging experiments (C-E) carried out in a Chrm2-tdT-D mouse brain previously injected with an AAV to express GCaMP6f in neurons. Asterisk in panel C points out a tdTChrm2 cell, arrowheads indicate its dendrites. See also FigS6 H-I.
(D) GCaMP signal (average intensity across time) from the same imaging plane as on (C).
(E) TORO image of the GCaMP image from (D). The tdTChrm2 cell labeled with an asterisk in panels (C) and (D) is a TORO cell. See also Figures S6H-I for more examples.
(F) Labeling strategy for visualizing GABAergic cells used in the experiments illustrated in panels H-M (note that in order to avoid labeling cells in the cortex, brain tissue overlying the hippocampus was removed and a cannula window was fitted in place).
(G) Example confocal micrograph of a brain section including the hippocampus, part of the neighboring subiculum (SUB) and part of the retrosplenial cortex (RSC). Most GCaMP-expressing cell bodies were located in str. oriens, pyramidale and radiatum, and to a lesser extent at the CA1 radiatum-LM border in low numbers (arrowheads).
(H) High magnification MIP image from the SUB area indicated by a square in (G). Arrowheads and arrow point out tdTChrm2 positive and negative CA1-originating GABAergic axon terminals, respectively (same for (I) and (K))
(I) High magnification MIP image of the RSC area indicated by a rectangle in (G).
(J) Example confocal micrograph of a brain section including the lateral entorhinal cortex (EC lat.).
(K) High magnification MIP image from the EC lat. are indicated by a square in (J).
(L) Density of tdTChrm2 and GCaMP (i.e. CA1-originating) co-expressing boutons in six remote target regions (one-way ANOVA with Bonferroni correction, p<0.0001, F=11.4, n=5 mice). Data are presented as mean±SD.
(M) tdTChrm2-expressing fraction of the CA1-originating GABAergic axon terminals (p=0.022, F=3.29, one-way ANOVA with Bonferroni correction, n=5 mice). Data are presented as mean±SD.
M2R-expressing cells contribute a substantial fraction of CA1 long-range GABAergic projections
As noted above, juxtacellularly labeled M2R-expressing TORO cell axons appeared to give rise to long range projections (Figure S1A, S6C), therefore, we aimed to map their target brain areas. First, we carried out experiments to further validate that TORO cells were preferentially labeled in Chrm2-tdT-D mice (as suggested by Figures S6A&E) using in vivo calcium imaging. As expected, most cells with detectable somatic tdTChrm2 expression (15 of 18 or 83%, Figures 7C-E, Figure S6I-J) were TORO cells (as identified based on the TORO images). The remaining 3 cells were theta-OFF cells without a detectable ripple response (Figures S6H, J). Importantly, no tdTChrm2 cells were theta-ON cells (i.e., they were unlikely to be PV cells with axonal M2R expression, since all known PV cell classes increase their firing at theta onset (Varga et al., 2014)).
Based on the latter results indicating the utility of the Chrm2-tdT-D mice for putative TORO cell labeling, we next sought to identify putative CA1 TORO cell distal axons in Chrm2-tdT-D mice injected with GABAergic cell-specific Dlx-GCaMP6f virus (Dimidschstein et al., 2016) in the CA1. This viral injection strategy labeled GABAergic cells mostly in strata oriens, pyramidale and radiatum and to a lesser extent at the radiatum/lacunosum-moleculare border (Figure 7G). We detected CA1-originating tdTChrm2-expressing axons in the subiculum (SUB), dorsal lateral septal nucleus (LSD), lateral- and medial entorhinal cortex (EC), retrosplenial cortex (RSC), medial septum (MS) and anterior cingulate cortex (ACC) (Figures 7F-K). Although certain PV cells also express axonal M2R (Hájos et al., 1997; Szabó et al., 2010), long-distance extra-hippocampal PV cell projections from the CA1 to areas such as the LSD, EC, RSC, or ACC have not been reported (Klausberger et al., 2004; Lapray et al., 2012; Varga et al., 2012, 2014). Indeed, we did not detect PV immunopositivity in tdTChrm2-positive CA1-originating long-projecting axons in MS, EC and RSC (0 of 227, 0 of 141 and 0 of 60 boutons, respectively, from 2 mice; see also Figures S6K, S7G), and very few in the subiculum (35 of 1126 boutons or 3.1%, n = 2 mice). Thus, the vast majority of axons double positive for tdTChrm2 and GCaMP originated from CA1 M2R-expressing, non-PV GABAergic cells, i.e., putative TORO cells.
Next, to estimate the contribution of M2R-expressing cells to CA1 GABAergic long-range projections, we quantified the ratio of tdTChrm2-positive (i.e., M2R-expressing) boutons among all GCaMP-expressing (i.e., CA1-originating GABAergic) boutons in the six brain regions mentioned above. Surprisingly, in 5 out of 6 of these remote targets, we found that most CA1 GABAergic axon terminals were in fact originating from tdTChrm2 cells. Their fraction was the highest in SUB (92.1±7.0%) and the lowest in the MS (52.6±17.8%, Figure 7M). On average, in the above listed six brain areas, tdTChrm2-expressing putative TORO cells composed 77±14% of CA1 long-range inhibitory projections, indicating that TORO cells may robustly contribute to the GABAergic projections from the hippocampus to these brain regions.
TORO cells target other GABAergic neurons
Neurons that express M2R in their somatodendritic membranes have been reported to target other GABAergic cells (Francavilla et al., 2018; Katona et al., 2020). Therefore, we examined our samples containing individually labeled TORO cells, as well as genetically labeled tdTChrm2-expressing cells, and found that their labeled axons indeed appeared to be in close juxtaposition to and densely surrounding putative CA1 interneuron somata and dendrites (Figures S7A-B). In order to examine the innervation of GABAergic cells by TORO cells in more detail, we took advantage of insights from previous studies that showed that some PV-expressing or cholecystokinin (CCK)-expressing hippocampal interneurons are inhibited during SPW-Rs (Klausberger et al., 2005; Lasztóczi et al., 2011; Szabo et al., 2017; Varga et al., 2014). While the source of such inhibitory inputs has not been established, the high activity of TORO cells during SPW-Rs suggests that they may contribute to the reduced spiking of certain PV-expressing and CCK-expressing interneurons. Therefore, we quantified the number of tdTChrm2-positive, PV-negative putative TORO cell axon contacts around PV-positive or proCCK-positive somata using the dual labeling approach described above (Chrm2-tdT and Dlx-GCaMP, Figure 7F). PV-positive cell bodies were mostly located in the oriens and pyramidal cell layers and were frequently contacted by PV-negative tdTChrm2 GABAergic axons (Figures 8A&C). ProCCK-positive cell bodies were scattered across most CA1 layers, and each layer contained cells that received PV-negative tdTChrm2 GABAergic boutons in varying numbers, with the highest bouton number per cell found in the str. oriens and at the radiatum-LM border (Figures 8B&C). Moreover, we found that PV-positive or proCCK-positive somata in the EC were preferentially innervated by tdTChrm2 CA1 GABAergic axons in comparison to principal cells expressing calbindin or reelin (Varga et al., 2010; Figures S7G-J).
Figure 8. TORO cells preferentially innervate interneurons.
(A) Confocal MIP image of a representative CA1 oriens PV interneuron being contacted by several tdTChrm2 terminals (arrows), including virally labeled (i.e., originating from local cells) boutons from the CA1 (arrowheads). Note that PV expressing boutons were excluded from the analysis shown on panel C.
(B) Confocal MIP image of a representative CCK interneuron at the CA1 radiatum-LM border surrounded by several virally labeled tdTChrm2 boutons (arrowheads). Arrow points out an unlabeled (i.e., GCaMP6f-negative) tdTChrm2 bouton. Note that tdTChrm2 boutons are negative for PV at the radiatum-LM border.
(C) Quantification of tdTChrm2 expressing, PV negative boutons on PV and CCK cell bodies. PV negativity of the presynaptic tdTChrm2 axons was confirmed for all postsynaptic PV cells and for CCK cells where PV containing was available (40/56 CCK cells, see Methods). PV cell bodies were mostly located in the oriens and pyramidal cell layers where several of them received axon terminals from tdTChrm2-cells (median[1st quartile, 3rd quartile]: ORI: 2[0, 4], PYR:1[0, 3] boutons/cell body, n=80 cells, 7 mice). Within the CCK cell group, the average bouton number was not uniform across layers, with the highest bouton number per cell found in the oriens and radiatum-LM border (Kruskal-Wallis ANOVA completed with Mann-Whitney U test as pairs; X2(3)=10.9, p=0.012; (ORI: 3[2, 4], PYR: 0.5[0, 2], RAD: 1[0, 3], Radiatum-LM border: 3[2, 5] boutons/soma, n=56 cells, 6 mice). Box plots show median±IQR, whiskers show range, vertical ticks show individual values. (*p<0.05, **p<0.01, M-W test).
(D) Experimental design for paired recording of tdTChrm2 cells and pyramidal cells (PCs) or interneurons (INs).
(E) Connection probability obtained by paired recordings.
(F) Representative traces obtained from a synaptically connected tdTChrm2 cell – IN pair. Note the robust increase in IPSCs throughout the train of presynaptic APs. The IN is the axo-axonic cell (AAC) illustrated on panel H.
(G) Representative traces obtained from the single synaptically connected tdTChrm2 cell-PC pair. Asterisks indicate successful uIPSCs which are smaller and fewer compared to the uIPSCs in the pair shown on F. See also Figure S7 D-F.
(H) 2D reconstruction of the cell pair shown on F. Black: tdTChrm2 cell dendrite, orange: axon; olive: AAC dendrite, green: axon. Arrows point out synaptic contacts.
(I) Biocytin-labeled AAC axons target axon initial segments visualized by anti-p-IκBα. Arrows point out juxtapositions.
(J) PV immunopositivity of the postsynaptic AAC.
(K) Ten superimposed uIPSCs (thin grey lines) evoked by single presynaptic APs (orange) in the tdTChrm2 cell – AAC pair. The averaged uIPSC trace is shown in olive.
(L) Properties of uIPSCs obtained from the 4 tdTChrm2 cell – IN pairs. Note that properties were obtained from the responses followed by the 1st AP of the train. Peak amplitude: mean peak amplitude of uIPSC including failures; synaptic potency: mean peak amplitude excluding failures. (Note that similar metrics could not be obtained from the tdTChrm2 cell – PC pair because all of the first responses in the train were failures (100% failure probability – magenta); see also Figure S7E-F). Mean±SEM is shown.
(M) Representative trace showing that tdTChrm2 cells are able to evoke uIPSCs in IN-s even when being driven at 200Hz.
In order to explore the targeting preference of tdTChrm2 cells in the CA1 functionally, we performed paired recordings between tdTChrm2 cells and their potential interneuron and PC targets in the presence of AMPA and NMDA receptor antagonists (Figure 8D). Out of 21 simultaneously recorded tdTChrm2 cells and interneuron pairs, we found 4 monosynaptically connected pairs (19%), whereas monosynaptic connection was found in only one out 21 (4.7%) tested tdTChrm2 cell to PC pairs (Figure 8E). In each of the 4 tdTChrm2 cell to interneuron connected pairs, the presynaptic APs evoked large and fast (Figures 8K-L) IPSCs throughout the train of presynaptic spikes (Figure 8F; Figures S7C-F). In contrast, in the single tdTChrm2 cell to PC connected pair, most of the presynaptic APs resulted in failures (Figures 8G, L) and the unitary IPSCs (uIPSCs) were small (Figure 8G, asterisks; Figures S7E-F). Interestingly, the postsynaptic cell in one of the tdTChrm2 cell – interneuron pairs was identified as a PV-positive AAC in the stratum oriens (Figures 8H-J; for an example of a postsynaptic interneuron located in the stratum radiatum, see Figures S7C-D). Importantly, the presynaptic tdTChrm2 cells evoked uIPSCs in the postsynaptic interneurons in a sustained manner even when driven to fire at 200 Hz in all 3/3 pairs tested (Figure 8M).
Taken together, these immunocytochemical and paired recording data are consistent with the M2R-expressing TORO cells innervating local and distant interneurons.
Discussion
TORO cells as a circuit element with ripple-selective activity dynamics
In this study, we identified a GABAergic circuit element whose activity dynamics is closely associated with hippocampal ripples, with connectivity properties consistent with a potential role in relaying ripple-related activity to intra- and extra-hippocampal areas. Key characteristics of the firing pattern of these TORO cells included the early onset of firing before ripples, the significantly higher frequency of ripple-related spikes (even compared to the intensely firing fast-spiking PVBCs), and the low-frequency but strongly phase-modulated firing exhibited during theta and locomotion. Since SPW-Rs are relatively short in duration and do not occur very often (juxtacellular recordings: 0.28±0.1 Hz; calcium imaging experiments: 0.29±0.2 Hz), the overall activity of TORO cells across all brain states is low, which may explain why these cells have not been recognized in most prior in vivo studies. Our easy to implement set of quantitative criteria for TORO cell identification from spike data should enable the efficient and objective search for TORO cells in existing databases and future experiments. Indeed, when these criteria were applied to the Allen Brain Observatory database, TORO cells were found to form a distinct group of neurons located uniquely in the hippocampal formation whose activity is gated by locomotion in an on-off manner rather than inversely scaling with speed.
The signature theta-OFF, ripple-ON spiking dynamics of TORO cells also enable the investigations of these cells with in vivo functional calcium imaging, overcoming the fundamental problem that the low overall activity of TORO cells makes it extremely challenging to identify them using conventional image segmentation strategies. Cells identified as TORO cells showed the highest calcium response to ripples among all other hippocampal neurons, in agreement with the juxtacellular data, and all cells identified based on their TORO images were subsequently confirmed to be M2R immunopositive. In contrast to the uniformly high somato-dendritic expression of M2R in TORO cells, other potential markers (e.g., calbindin, SOM, VIP) appeared to label variable proportions of functionally identified TORO cells or M2R expressing neurons (note that recent RNA-seq data also support the existence of multiple M2R clusters (Harris et al., 2018)).
The access to TORO cells provided by these 2-photon imaging results also made it possible to study them for prolonged periods during sleep. In spite of the robust brain state differences between the awake and sleep conditions overall, TORO cells preserved the defining features of their activity dynamics (intense activity during ripples in NREM sleep and virtually no activity during theta oscillations in REM) across states. It is interesting in this regard that acetylcholine levels are particularly high during REM compared to NREM states (Marrosu et al., 1995; Zhang et al., 2021), which is consistent with the possibility that M2R-mediated inhibition may suppress TORO cell activity during theta states both during awake and sleep conditions.
As expected from intensely ripple-activated cells possessing dendrites in the Schaffer collateral termination zones, TORO cells showed the strongest activation among hippocampal neurons in response to CA3 pyramidal cell stimulation (note that such intense activation may be mediated by both direct and indirect excitation from pyramidal cells in the CA3 and CA1, respectively). In contrast, medial septal GABAergic afferents that targeted TORO cells were only active during theta and activating such afferents suppressed the activity of TORO cells, suggesting their potential contribution to the theta-OFF nature of TORO cells (note that ripple-active septal GABAergic axons that target different interneuronal populations may also exist (Unal et al., 2018; Viney et al., 2013)). Therefore, TORO cells are under the control of specific inhibitory septal afferents, including theta-ON GABAergic inputs and cholinergic afferents hyperpolarizing TORO cells through M2Rs. Taken together, these results indicate the likely influence of glutamatergic, GABAergic, and cholinergic systems on TORO cells, and additional inputs (for example, locomotion-activated VIP expressing cells (Turi et al., 2019)) may also contribute to the emergence of the uniquely ripple-selective discharge patterns of these neurons.
TORO cells may constitute a substantial fraction of GABAergic projections to extra-hippocampal areas
CA1 glutamatergic pathways convey ripple-related information to distant brain areas, however, the existence of parallel ripple-modulated GABAergic pathways has not been previously demonstrated. We took advantage of the high proportion of TORO cell-like activity patterns of cells labeled in the Chrm2-tdT-D transgenic line to show that TORO cells likely provide hippocampal GABAergic inputs to a variety of brain areas, including the SUB, LSD, MS, lateral and medial EC, RSC and the ACC. Our results showed that 77% of the long-range projections from CA1 were tdTChrm2-expressing, PV-negative axons. The latter data, combined with the observation that 83% of the tdTChrm2 cells were TORO cells in the oriens layer, indicate that TORO cells likely contribute a sizable and perhaps even the dominant portion of GABAergic projections from the CA1 to extra-hippocampal areas. It should be noted, however, that our viral labeling strategy most likely did not label all CA1 layers evenly, potentially leaving some long-projecting GABAergic cells (e.g., Yamawaki et al., 2019) underrepresented.
In agreement with prior reports that GABAergic long-distance projections from cortical areas often preferentially innervate other GABAergic cells (Francavilla et al., 2018; Katona et al., 2020; Melzer et al., 2012; Toth et al., 1993; Yamawaki et al., 2019), our immunohistochemical investigations performed on in vivo juxtacellularly labeled TORO cell axons and tdTChrm2-expressing, GABAergic axons originating from the CA1 suggested that TORO cells innervate PV and CCK cells both within the CA1 as well as in extra-hippocampal target areas. Regarding PV cells, our data from a tdTChrm2 cell - AAC connected pair provide direct evidence that AACs can receive inhibition from M2R cells (Figures 8F&H-M), suggesting that TORO cells may contribute to the suppression of some AACs during SPW-Rs (Dudok et al., 2021, Szabo et al., 2017). Many PV-positive cells increase their firing during ripples in vivo (Lapray et al., 2012), likely contributing to the overall increased inhibition that CA1 PCs receive during these events (Gan et al., 2017; Valero et al., 2022). However, each of the three PV interneuronal classes targeting different postsynaptic domains (AACs, PVBCs, bistratified cells) exhibits considerable heterogeneity in their ripple-related firing patterns (Varga et al., 2014). In general agreement with the latter results, the number of boutons that PV cells received from M2R cells varied considerably (Fig. 8C), raising the possibility that the contribution of TORO cells to the regulation of ripple-related firing of PV cells may be cell-type specific.
Both our paired recordings and immunohistochemical experiments suggested that tdTChrm2 cells preferred interneurons over principal cells. On the other hand, we considered only cell bodies as potential targets in our immunocytochemical experiments, and a differential target preference for interneurons over PCs by dendrite-targeting long-range GABAergic cells (Yamawaki et al., 2019; Francavilla et al., 2018) cannot be excluded. Nevertheless, based on our paired recording and immunocytochemical results, it seems reasonable to suggest that TORO cells may convey a disinhibitory signal prior to and during SPW-Rs, perhaps selectively gating local and distant neuronal circuits to facilitate computations associated with the ripple-related information transmitted by CA1 PC axons. Since unusually thick myelin sheath has been demonstrated for some long-range GABAergic cells (Jinno et al., 2007), it is possible that TORO cells also exhibit this feature which would allow rapid transmission of ripple-related activity patterns. While the suppression of certain hippocampal and extra-hippocampal interneurons during ripples (Fuentealba et al., 2008; Girardeau et al., 2017; Klausberger et al., 2005; Szabo et al., 2017; Varga et al., 2014) is generally consistent with potential ripple-associated disinhibition, future work will be required to identify the details of the circuit computations that may be influenced by TORO cells, including the replay of memory-related information during SPW-Rs (Jadhav et al., 2012).
Relationship of TORO cells to other GABAergic projection cell types
TORO cells showed remarkable functional homogeneity in their activity dynamics, as represented not only by their signature theta-OFF, ripple-ON firing patterns, but also by their high firing frequency during ripples, high pre-ripple index, robust ripple preference in both awake and sleep states, and strong responses to CA3 activation. Theta-OFF units have been previously recognized in the hippocampus (Buzsáki et al., 1983; Colom and Bland, 1987; Mizumori et al., 1990) and the cellular identity of one theta-OFF cell population have been recently identified as VIP/M2R-co-expressing GABAergic projection cells (Francavilla et al., 2018). However, in contrast to TORO cells, VIP/M2R-co-expressing theta-OFF cells lacked elevated activity during ripples (Francavilla et al., 2018). In addition, a characteristic feature of VIP/M2R long-range projecting cells is the presence of spiny dendrites (Francavilla et al., 2018), which we did not observe on TORO cells. Therefore, it is unlikely that TORO cells overlap with the VIP/M2R CA1 GABAergic projection cells. However, it is interesting to note that these cells may have been among the 17% of tdTChrm2 cells that were identified as theta-OFF and ripple-OFF cells in our calcium imaging experiments.
Other previously reported GABAergic long-projecting CA1 neurons may also share some similarities to TORO cells, although either the lack of in vivo functional data, or differences in experimental conditions (e.g., presence or absence of anesthesia), species, developmental stages, or low sample sizes can make such comparisons challenging. First, early anatomical studies reported long-range GABAergic neurons with morphological characteristics similar to that of TORO cells (Hájos et al., 1997; Katona et al., 1999), suggesting a possible overlap between these neuronal populations. However, the in vivo activity patterns of the anatomically-identified GABAergic long-range projecting neurons were not investigated in these studies. Second, a cell population was recently reported to express NOS1 but little M2R (Christenson Wick et al., 2019). Our immunocytochemical results also indicated little overlap between the NOS1 and M2R cell populations (Figure S2I) and we found no evidence for NOS1 expression in our CA1-originating tdTChrm2 axons (Figure S6L). Third, trilaminar GABAergic cells (Ferraguti et al., 2005; Sik et al., 1995, Katona et al., 2020) express M2R, but only limited information is available about these cells during theta and ripples from animals without anesthesia. For example, based on a single cell available from awake animals (Katona et al., 2020), trilaminar cells may fire at a considerably higher frequency during running relative to the TORO cells. Therefore, it is not currently possible to definitively establish the nature of the relationship of trilaminar and TORO cells. Fourth, the long-range GABAergic neurons reported by Jinno et al., (2007) share some similarities with TORO cells. However, the ripple-related firing frequency of these former cells (between 10-69Hz, in anesthetized rats) may be well below that of TORO cells (174Hz; note, however, that a recent study in freely moving rats reported two SOM-positive long-projecting cells that displayed higher ripple-related firing (Katona et al., 2017)). Finally, early-born, GABAergic projection cells also often express M2R, and these so-called hub cells fire before and during hippocampal bursts in early postnatal development (Bonifazi et al., 2009; Picardo et al., 2011; Villette et al., 2016). Whether such hub cells include TORO cells will need to be established, although recent evidence indicated that early-born GABAergic hub cells were recruited during running when examined in adult animals (Bocchio et al., 2020).
Taken together, our results suggest that the TORO cell activity dynamics described in this paper constitute a useful, simple and clear definition to guide future electrophysiological and cellular resolution functional imaging studies aimed at understanding the role of long-distance GABAergic projections in ripple-associated computations in the brain.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Gergely G. Szabo (szergely@stanford.edu).
Materials availability
This study did not generate new unique mouse lines or reagents.
Data and code availability
The datasets generated during the current study are available from the Lead Contact on reasonable request.
Custom scripts used in this study have been deposited to a publicly accessible repository. The URL is listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-DsRed | Takara | #632496, RRID: AB_10013483 |
| goat anti-GFP | abcam | ab5450, RRID:AB_304897 |
| chicken anti-GFP | Aves | #GFP-1020, RRID: AB_10000240 |
| rat anti-muscarinic acetylcholine receptor type 2 | Millipore | MAB367, RRID:AB_2152546 |
| rat anti-somatostatin | Millipore | MAB354, RRID:AB_2255365 |
| rabbit-anti-somatostatin | Peninsula | Cat# T-4103.0050 RRID:AB_518614 |
| rabbit anti calbindin | Swant | CB38, RRID:AB_10000340 |
| rabbit anti-parvalbumin | Swant | PV27, RRID:AB_2631173 |
| mouse anti- parvalbumin | Sigma | #P3088, RRID: AB_477329 |
| rabbit anti-proCCK | Frontiers Neuroscience | #AF350, RRID: AB_2571674 |
| rabbit anti-neuronal nitric oxide synthase | Cayman Chemical | 160870, RRID:AB_10080041 |
| mouse anti-GABA | Sigma | A0310, RRID:AB_476667 |
| goat anti-RFP | Rockland | 200-101-379 RRID:AB_2744552 |
| rabbit anti-VIP | Immunostar | RRID:AB_10730725 |
| mouse anti-reelin | Millipore | RRID: AB_2285132 |
| anti-phospho-IκBα | Cell Signaling Technology | Cat# 2859; RRID:AB_561111 |
| Bacterial and virus strains | ||
| AAV1-Syn-GCaMP6f-WPRE-SV40 | Penn Vector Core | N/A |
| AAV1-Syn-Flex-GCaMP6f-WPRE-SV40 | Penn Vector Core | N/A |
| AAV1-Syn-Flex-jGCaMP7s-WPRE | Dana et al., 2019 doi: 10.1038/s41592-019-0435-6 | Addgene #104491-AAV1, RRID: Addgene_104491 |
| AAV1-Syn-Flex-NES-jRGECO1a-WPRE-SV40 | Dana et al., 2016 doi:10.7554/eLife.12727 | Addgene #100853-AAV1, RRID: Addgene_100853 |
| AAV-DJ-Ef1a-fDIO GCaMP6f | Stanford Neuroscience Gene Vector and Virus Core | #167 |
| AAV5-EF1a-DIO-hChR2(H134R)-mCherry-WPRE-pA | UNC Vector Core | N/A |
| AAV5-EF1a-DIO-mCherry-WPRE-pA | UNC Vector Core | N/A |
| AAV5-hSyn-hChR2(H134R)-mCherry-WPRE | UNC Vector Core | N/A |
| pAAV-hSynapsin1-FLEx-axon-GCaMP6s (AAV9) | Broussard et al., 2018 doi: 10.1038/s41593-018-0211-4 | Addgene #112010-AAV9, RRID: Addgene_112010 |
| pAAV-mDlx-GCaMP6f | Dimidschstein et al., 2016 doi: 10.1038/nn.4430 | Addgene #83899, RRID: Addgene_83899 |
| Chemicals, peptides, and recombinant proteins | ||
| NBQX | Tocris | Cat. No. 1044 |
| D-APV | Cayman Chemical | #14539 |
| PTX | Tocris | Cat. No. 1128 |
| CGP35348 | Tocris | Cat. No. 1245 |
| AF-DX116 | Tocris | Cat. No. 1105 |
| carbachol | Tocris | Cat. No. 2810 |
| neurobiotin | Vector Laboratories | SP1120 |
| biotin-ethylenediamine (N-(2-Aminoethyl)Biotinamide, HBr | Thermofisher Scientific | A1593 |
| Experimental models: Organisms/strains | ||
| Dlx5/6-Cre transgenic (Tg(dlx5a-cre)1Mekk/J) | JAX | JAX Stock No. 008199 |
| C57BL/6J | JAX | JAX Stock No. 000664 |
| Chrm2-tdT-D knock-in | JAX | JAX Stock No. 030330 |
| Tg(Grik4-cre)G32-4Stl/J | JAX | JAX Stock No.006474 |
| ChAT-IRES –Cre | JAX | JAX Stock No.006410 |
| Npy-IRES2-FlpO-D | JAX | JAX Stock No.030211 |
| Tg(Amigo2-cre)1Sieg/J | JAX | JAX Stock No.030215 |
| Software and algorithms | ||
| Custom scripts | Present study; Zenodo | https://zenodo.org/record/6402500#.YkXUQbhlDdo |
| MATLAB | Mathworks | R2019b |
| Scanbox | Neurolabware | v4.6 |
| ZEN | Zeiss | 2012 SP5 FP1 v14 |
| Python | Python.org | v3.6 |
| sima (Python) | Kaifosh et al., 2014 doi: 10.3389/fninf.2014.00080 | https://github.com/losonczylab/sima PMID: 25295002 |
| pClamp | Molecular Devices | v10.7.0.3 |
| Clampfit | Molecular Devices | v10.7.0.3 |
| OriginPro | Originlab | 2019b |
| Allen Software Development Kit | Allen Institute | https://allensdk.readthedocs.io/en/latest/ |
| QuickNII | Puchades et al., 2019 doi: 10.1371/journal.pone.0216796 | RRID: SCR_016854, https://github.com/HumanBrainProject/QuickNII |
| Brainrender GUI | Claudi et al. 2021 doi: 10.7554/eLife.65751 | https://github.com/brainglobe/brainrender |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal procedures were performed in accordance with NIH guidelines and with the approval of the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University or the Institutional Animal Care and Animal Use Committee (IACUC) of the University of California, Irvine. For juxtacellular recordings adult (4- to 14-month-old) C57BL/6 male mice were used (Jackson laboratory stock # 000664). For two-photon imaging a variety of transgenic heterozygous adult (4- to 15-month-old) mice of both sexes, on a C57BL/6J background, were used. Dlx5/6-Cre transgenic mice having Cre recombinase expression directed by the regulatory sequences of the zebrafish dlx5a/dlx6a genes were used to genetically access GABAergic cells (JAX Stock No. 008199). Chrm2-tdT-D knock-in mice are designed to have expression of a Chrm2/tdTomato fusion protein directed by the endogenous M2 muscarinic acetylcholine receptor promoter/enhancer sequences (JAX Stock No.030330). Validation of this mouse line for selectivity for M2R expressing cells in the CA1 region as well as electrophysiological characterization of tdT cells is provided in Figure S5 of the current study and also by others (D’Souza et al., 2019; Gămănuţ et al., 2018). Tg(Grik4-cre)G32-4Stl/J mouse line (JAX Stock No.006474) was used for optogenetic targeting of CA3 pyramidal cells. Additional TORO cells were recorded in imaging experiments using C57BL/6, ChAT-IRES –Cre (JAX Stock No.006410), Tg(Amigo2-cre)1Sieg/J (JAX Stock No.0302150) and Npy-IRES2-FlpO-D knock-in (JAX Stock No.030211) mice. For the immunhistochemical quantitative assessment, 3 C57BL/6 mice (of both sexes, 2- to 6-month-old) were used. Chrm2-tdT-D knock-in mice of both sexes were used in all the TORO-output mapping experiments (injected at 3- to 5-month) as well as in all in vitro recordings (7- to 12-week old). Mice were housed in groups of 2-5 animals on a standard laboratory diet and 12/12 light dark cycle.
METHOD DETAILS
Juxtacellular recordings
Anesthesia and surgical procedures were performed as described previously (Szabo et al., 2017; Varga et al., 2012, 2014). Briefly, stainless steel head bars were affixed to the skull during deep isoflurane anesthesia. On the day of the experiment, a small (1 mm) craniotomy was made over the right or left hippocampus (−2 mm antero-posterior and 2 mm lateral) under isoflurane anesthesia. Mice were then placed onto an 8-inch spherical treadmill, and recording began after the animals had recovered, approximately 30 mins after the termination of anesthesia, when the animal became completely alert. Two borosilicate recording electrodes at 15° from the vertical plane were lowered into the brain, one was placed into the CA1 str. pyramidale (LFP channel, positioned where the ripple amplitude was the highest), and the other (juxtacellular pipette) was used for the unit recording. Juxtacellular recordings and field recordings were performed using an ELC-03XS universal amplifier (NPI Electronics) and a Neurodata IR283 (Cygnus Technology) amplifier, respectively. The LFP electrode was filled with 1M NaCl solution, juxtacellular electrode was filled with 0.5M NaCl or Ringer solution containing 135mM NaCl, 5.4mM KCl, 5mM HEPES, 1.8mM CaCl 2 , and 1mM MgCl 2 (pH was adjusted to 7.2 by adding NaOH; target osmolality was 290mmol). The juxtacellular electrode (12-20 MΩ) also contained 1.5-2% (wt/vol) Neurobiotin tracer (SP1120; Vector Laboratories) or biotin-ethylenediamine (N-(2-Aminoethyl)Biotinamide, HBr) (A1593; Thermofisher Scientific) as a tracer for juxtacellular labelling. Both channels were low pass (Bessel) filtered at 5 kHz and digitized at 20 kHz using NIDAQ data acquisition cards (National Instruments). Data acquisition was carried out using custom-written routines in MATLAB that also operated continuous video recording of the animal synchronized to the recording session. Nonpyramidal cells were tentatively identified based on unit firing, by narrow spike width (peak to trough duration<0.6 ms) and by the lack of complex spike bursts characteristic of pyramidal cells. After 10-15 min of recording the juxtacellular pipette was advanced toward the cell and positive 200ms long current pulses (1-5nA) were delivered using a PG4000 Stimulator (Neurodata) so that the cell fired As for 10-40 min.
Neuronal identification and antibody staining after juxtacellular labeling
Experiments were terminated and the mice were perfused transcardially within two hours after the experiment. The mice were placed under deep anesthesia using ketamine (100 mg/kg) and Xylazine (10 mg/kg) in saline (0.9% NaCl) and were perfused using saline followed by a fixative containing 4% paraformaldehyde (PFA) and 0.2% picric acid dissolved in 0.1 M phosphate buffer (PB). Following overnight fixation, the brains were sectioned 60 μm thickness using a Vibratome (Leica VT1200). Sections were first incubated in Alexa Fluor 594- or 488-conjugated streptavidin (1:1000, Molecular Probes) then sections containing the labelled cell were selected for further immunohistochemical analysis. The following primary antibodies were used: rat anti-muscarinic acetylcholine receptor type 2 (MAB354; 1:1000; Millipore, Temecula, CA); rabbit anti-parvalbumin (PV27; 1:2500; Swant, Switzerland); rat anti-somatostatin (MAB354; 1:1000; Millipore, Temecula, CA); rabbit anti-neuronal nitric oxide synthase (160870; 1:1000; Cayman Chemical, MI); rabbit anti calbindin (CB38; 1:1000; Swant, Switzerland); mouse anti-GABA (1:1000, A0310, Sigma Aldrich). Primary antibodies were diluted in 0.1 M PB containing 0.25% Triton X100 and sections were incubated overnight at 4°C in the antibody cocktails. Primary antibodies were detected using secondary antibodies raised in goat against rabbit/rat/guinea pig conjugated to Alexa Fluor 488/594 (11005, 11008, 11007, 11001, 11076, 11006; Invitrogen) or raised in donkey against rabbit and conjugated to Alexa Fluor 647 (711-605-152; Jackson Laboratories). Immunoreactivity was documented using a CCD camera attached to a Zeiss Axioscope microscope or a Zeiss LSM 710 confocal microscope. After the immunochemical characterization, all cells were further visualized by 3,3’-diaminobenzidine tetrahydrochloride (DAB) to generate high resolution Z-stack images using a Zeiss bright-field microscope. Dendrites and axons of selected cells were then traced using the pencil tool in Adobe Photoshop after loading the Z-stack images into the program as individual layers. Cells were identified based on their protein expression and/or targets.
In vitro electrophysiology
Mice were deeply anesthetized by isoflurane, decapitated, then coronal or sagittal 300μm thick slices were cut using a vibratome (VT 1200S, Leica Biosystems) in ice-cold cutting solution containing (in mM): 85 NaCl, 75 sucrose, 25 glucose, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2 and 24 NaHCO3. Brain slices were then incubated in 35°C cutting solution for ~12 minutes then slices were left in the chamber to cool down to room temperature. Intracellular recordings were performed in a submerged chamber perfused with oxygenated aCSF consisting of (in mM): 126 NaCl, 26 NaHCO3, 10 glucose, 2.5 KCl, 2 MgCl2, 2 CaCl2, 1.25 NaH2PO4. Flow rate was 2.5 ml/min and temperature were maintained at 33°C by a chamber heater (BadController V, Luigs and Neumann). Hippocampal neurons were visualized using DIC illumination on an Olympus BX61WI microscope (Olympus Microscopy) with an sCMOS camera (Flash 4.0 LT+, Hamamatsu). All solutions were equilibrated with 95% O2/5% CO2. Recording pipettes were pulled from thin-walled borosilicate capillary glass (King Precision Glass) using a P97 puller (Sutter Instruments) and were filled with (in mM): 126 K-gluconate, 10 HEPES, 4 KCl, 4 ATP-Mg, 0.3 GTP-Na, 10 phosphocreatine (pH-adjusted to 7.3 with KOH, osmolarity 290 mOsm), as well as 0.2% biocytin. Pipettes had a 3-5 MΩ tip resistance. Whole cell recordings were performed on td-Tomato-positive neurons in the CA1 dorsal hippocampus. Data were acquired in pClamp software (Molecular Devices) using a Multiclamp 700B amplifier (Molecular Devices), low-pass filtered at 2 kHz, and digitized at 10 kHz (Digidata 1440A, Molecular Devices). After recording, brain slices were transferred into a fixative solution containing 4% paraformaldehyde and 0.2% picric acid in 0.1 M phosphate buffer for 24 hrs at 4°C. Brain slices were then embedded in 2% agarose and 60 μm coronal sections were prepared using a vibratome for fluorescent immunostaining and confocal imaging. Biocytin was labeled with the addition of AF488-strepavidin (1:1000, Life Technologies) simultaneously with the other secondary antibodies during immunostaining.
Cholinergic experiments were assessed in the presence of a “cocktail” to block synaptic transmission, consisting of 20 μM D-APV (to inhibit NMDA receptors), 5 μM NBQX (to inhibit AMPA receptors), 400 μM PTX (to block GABAA receptors) and 1.7 μM CGP35348 (to block GABAB receptors). Both carbachol and AF-DX116 were applied in 5μM concentrations.
All paired recordings were performed in the presence of glutamatergic transmission antagonists using 20 μM D-APV and 5 μM NBQX. Intrapipette solution for the presynaptic cell was the same as described above, while the postsynaptic solution contained (in mm) 80 CsCl2, 60 Cs-gluconate, 1 MgCl2, 2 Mg-ATP, 3 NaCl, 10 HEPES and 5 QX-314 [2(triethylamino)- N-(2,6-dimethylphenyl) acetamine], and was adjusted to pH 7.3 and to an osmolarity of 295 mOsm/L. Both the presynaptic and postsynaptic solutions contained 0.2-0.4% biocytin. Presynaptic tdTChrm2 cells were held in current-clamp mode around a membrane potential of - 65 mV, and stimulated by brief current pulses (2 ms, 2-4 nA). Postsynaptic pyramidal cells and interneurons were clamped at a holding potential of −65 mV. Series resistance was frequently monitored and compensated by 65% using the Rs-compensation function of the amplifier. Cells that changed 25% during recording were discarded from further analysis. Potential postsynaptic cells localized in the str. oriens and str. radiatum layers were considered to be putative interneurons, whereas cells located within the pyramidal layer, with soma shapes and sizes similar to the other cells in the layer, were considered putative pyramidal cells. Importantly, postsynaptic cells at all the synaptically connected interneuron to interneuron pairs were confirmed to be interneurons posthoc based on biocytin labeling. Connections between cells were tested using 10 consecutive trains of action potentials (AP-s), each train was 3s long and contained 10 AP-s delivered at 20 Hz or 25 APs delivered at 50 Hz.
Neuronal identification and antibody staining after in vitro electrophysiology
To visualize biocytin labeled neurons slices were first incubated in Alexa Fluor 488-conjugated streptavidin (1:1000, Molecular Probes) and z-stack images of the labeled cell(s) were taken using a Zeiss LSM 710 confocal microscope or a Nikon A1 confocal microscope using 20x 0.8 NA or 63x 1.4 NA objectives. Then slices were embedded in an agarose block and resectioned to 60 μm thickness using a vibratome (Leica VT1200S) for further immunohistochemical analysis. Rabbit anti-DsRed antibody was used to amplify tdT signal (1:1000, Takara #632496, RRID: AB_10013483). For demonstrational purposes cells were reconstructed by tracing the dendrites and axons using the pencil tool in Adobe Photoshop after loading the Z-stack images into the software as individual layers. The AAC was identified after visualizing the axon-initial segments of the pyramidal cells using rabbit anti-phospho-IκBα (1:1000, Cell Signaling Technology).
Viral injections
For calcium imaging experiments, right hemisphere CA1 was injected (2.3 posterior (P), 1.5 lateral (L), 1.45-1.35 ventral (V) to Bregma, units in mm) using a Hamilton syringe as previously described (Bui et al., 2018), with one of the following viruses (300-400 nl each, full titer): AAV1-Syn-GCaMP6f-WPRE-SV40, AAV1-Syn-Flex-GCaMP6f-WPRE-SV40, AAV1-syn-FLEX-jGCaMP7s-WPRE, AAV1-Syn-Flex-NES-jRGECO1a-WPRE-SV40, AAV-DJ-Ef1a-fDIO GCaMP 6f (in one NPY-Flp mouse).
For experiments with optogenetics, besides injecting the respective virus into the CA1 for Ca-imaging, CA3 or medial septum were injected with 200 to 400 nL of AAV5-EF1a-DIO-hChR2(H134R)-mCherry-WPRE-pA, AAV5-EF1a-DIO-mCherry-WPRE-pA or AAV5-CamKIIa-hChR2(H134R)-mCherry. For axonal imaging, medial septum was injected with pAAV-hSynapsin1-FLEx-axon-GCaMP6s (AAV9) in combination with AAV1-syn-FLEX-jGCaMP7s-WPRE injected into the CA1. The needle tip was left in position for 10 minutes to allow diffusion. Coordinates to target the CA3 and medial septum were −2.3P, 2.8L, 2.7V and +0.8P, 0.2L, 4.5V relative to bregma, respectively. Accuracy of injections were post-hoc verified in each animal.
Injections with the pAAV-mDlx-GCaMP6f virus (200 to 300 nL to CA1) were immediately followed by aspirating the brain tissue overlying the hippocampus to avoid viral labelling in the cortex. A cannula window was implanted in place of the aspirated brain tissue similarly to the procedure for two-photon microscopy. Stock AAV virus preparations were obtained from the Penn Vector Core (AAV1-Syn-GCaMP6f-WPRE-SV40, AAV1.Syn.Flex.GCaMP6f.WPRE.SV40), the UNC Vector Core (AAV5-EF1a-DIO-hChR2(H134R)-mCherry-WPRE-pA, AAV5-EF1a-DIO-mCherry-WPRE-pA, AAV5-CamKIIa-hChR2(H134R)-mCherry, from the Stanford Neuroscience Gene Vector and Virus Core (AAV-DJ-Ef1a-fDIO-GCaMP6f) or from Addgene (AAV1-syn-FLEX-jGCaMP7s-WPRE, AAV1-Syn-Flex-NES-jRGECO1a-WPRE-SV40, pAAV-hSynapsin1-FLEx-axon-GCaMP6s (AAV9). pAAV-mDlx-GCaMP6f was kindly provided by Jordane Dimidschstein and Gord Fishell and is deposited to Addgene (catalog # 83899).
In vivo two-photon microscopy and related surgeries
After the experimental mice recovered from virus injection, mice were anaesthetized with isoflurane and secured into a stereotaxic frame and an imaging window was surgically implanted over the hippocampus similarly to that previously described (Danielson et al., 2016). Briefly, following induction of anesthesia (Isoflurane: 3-4% induction, 1-2% maintenance; 1.0 L/min O2) and administration of analgesia (using bupivacaine (1-2mg/kg, SQ) and buprenorphine (0.03-1 mg/kg, SQ)), the scalp was removed, and a ~3.0 mm diameter craniotomy centered over the dorsal hippocampus (−2.3P, 1.5L) was performed, using a fine-tipped dental drill. The cortical brain tissue overlying the injection site was aspirated while flushing the brain with ice-cold cortex buffer (Holtmaat et al., 2009), to minimize bleeding. A stainless steel cannula with attached coverglass was then implanted over the hippocampus (Kaifosh et al., 2013; Lovett-Barron et al., 2014) followed by the fixation of a stainless steel headbar onto the skull. On the following days mice were frequently monitored for signs of distress and pain and appropriate analgesic treatments were applied accordingly (Carprofen (5mg/kg, SQ) once a day for the first three days). The same procedure was done for each subsequent surgery.
After recovery, typically on the day before the experiment, mice were implanted with a chronic LFP electrode in the contralateral CA1 pyramidal layer while awake on the Styrofoam ball. In a few initial experiments, to keep LFP recording circumstances similar to juxtacellular recordings, a custom-made glass capillary previously bent to the required angle was lowered into the hippocampus and fixed at the same location where the LFP electrode at the juxtacellular experiments was placed. In subsequent experiments, a bipolar wire electrode (tungsten, 0.002,” 0.5 mm tip separation, A-M systems) was used instead of the glass capillary, which we found to be more stable long-term. Implantation of either electrode type was performed while recording to accurately target the CA1 pyramidal cell layer based on the presence of high amplitude ripple events. Electrodes were then cemented in place for chronic recordings.
Before two-photon imaging experiments, mice were acclimated to handling for approximately 20 mins. Then, mice were placed on a 2m long treadmill. In some experiments, mice were trained to run for a water reward, while in most experiments mice were not specifically trained to run. Experiments were typically performed within 2 hrs and then returned to the homecage.
In vivo two photon images were obtained using a resonant scanner (8 kHz) 2-photon microscope (Neurolabware), equipped with a pulsed IR laser (Mai Tai, Spectra-Physics), gated GaAsP PMT detectors (H11706P-40, Hamamatsu), and a 16x water immersion objective (0.8 NA, 3.0 mm WD; WI, Nikon). Excitation wavelength was 920 nm for most of the imaging experiments. For axonal-imaging, where axonal-GCaMP6s and jRGECO signals were imaged simultaneously excitation wavelength was 1000 nm. Identification of tdT-expressing cells were performed with the laser tuned to 1040 nm. Two-photon image acquisition was controlled by a Scanbox (Neurolabware) system, which also synchronized video acquisition (Mako, Allied Vision), treadmill speed monitoring and field potential recording via a DAC (National Instruments) to the imaging frames. Speed was determined based on signal of a rotary encoder attached to the treadmill, negative speed means the mouse is backing up. Glass LFP capillary electrode was connected to an amplifier (ELC-03XS, NPI), and referenced against the immersion bath. LFP signal from the bipolar electrodes was recorded using a differential amplifier (Model 1700, A-M Systems).
Imaging experiments began with scanning most of the CA1 region that was visible through the imaging window at low magnification, focusing on the oriens, resulting in 6-7 separate, at least 5-min long movies including both RUN and REST states. Ripples were detected from the correlated LFP recordings from the contralateral hippocampus as described above (see juxtacellular recordings). Then, the TORO image was generated on each movie and the putative TORO cells were re-imaged at higher magnification.
For optogenetic stimulation, 470 nm light from a 400 mW LED (M470-L3, ThorLabs) was delivered through the imaging objective at ¾-maximal intensity (~19 mW/mm2 before entering the brain). Optogenetic stimuli consisted of 15 ms pulses at the beginning of each imaging frame acquisition while the PMT detectors were gated for the duration of the stimulus. Data acquisition was not started for an additional 15 ms within each frame to prevent artefact due to the light emitted from the LED after de-energized. Data were then recorded following the pulse in the remainder of each frame, at normal frame rate. For the septal optogenetic stimulation experiments, light stimulation lasted for 15, 40 or 220 frames (~1s, ~1.4s or ~14s, respectively). Data from these experiments were pooled as all photostimulation protocols were similarly effective. For the CA3 opto-stimulation experiments light delivery was applied for 5 frames (~0.3 s) and LED intensity was adjusted to the minimum level where the cells response to light first became apparent, typically at ½-maximal intensity.
Post-hoc immunohistochemical characterization of imaged cells, M2R-cells and long range targets
After the last imaging session mice were perfused transcardially using saline followed by a fixative containing 4% PFA and 0.2% picric acid dissolved in 0.1 M phosphate buffer (PB) under deep anesthesia. Then the brain was removed from the skull, the two hemispheres were separated and the ventral basis of the right hemisphere was cut off at an angle such that the cut surface was parallel with the plane of previous in vivo imaging (8°). Then the brain was sectioned to 60 μm thickness and sections containing the imaged brain region were mounted. Confocal Z-stack images were taken of the GCaMP-expressing cells and the imaged neurons were identified based on morphology and the relative position of the cells. For further neurochemical identification and for the amplification of the GCaMP signal the following primary antibodies were used: rat anti-muscarinic acetylcholine receptor type 2 (MAB367; 1:1000; Millipore, Temecula, CA); rat anti-somatostatin (MAB354; 1:1000; Millipore, Temecula, CA); chicken anti-GFP (1:3000, Aves #GFP-1020, RRID: AB_10000240); goat anti-GFP 1:5000, abcam, ab5450); rabbit anti-DsRed (1:1000, Takara #632496, RRID: AB_10013483); rabbit anti-calbindin (CB38; 1:1000; Swant, Switzerland). Primary antibodies were diluted in 0.1 M PB containing 0.25% Triton X100 and sections were incubated overnight at 4°C in the antibody cocktails. Primary antibodies were detected using secondary antibodies as described at the juxtacellular recordings section.
For the quantitative assessment of other neurochemical markers in M2R cells, animals used for VIP staining were transcardially perfused with sucrose ACSF containing (in mM) 2 KCl, 1.25 NaH2PO4, 7 MgSO4, 26 NaHCO3, 10 glucose, 219 sucrose and 0.5 CaCl2. This was followed by perfusion with 4% PFA (Francavilla et al., 2018). For the rest of the experiments (SOM, NOS1 and calbindin) brain sections were used both from normally perfused (with saline and PFA) and sucrose-perfused mice. For each marker, four 60 μm thick randomly chosen coronal brain sections containing the dorsal hippocampus (from both hemispheres) were selected from each animal and were then completely imaged along the z-axis using a Nikon A1 confocal microscope with 20 x 0.8 NA objective. Immunohistochemical characterization of the M2R cells was performed offline using the z-stack-tile images. The following antibodies were used: rat anti-muscarinic acetylcholine receptor type 2 (MAB367; 1:1000; Millipore, Temecula, CA); rabbit anti-VIP (Immunostar, Id.: 20077); rabbit anti-Somatostatin (Peninsula, T-4103), rabbit anti-calbindin (CB38; 1:1000; Swant, Switzerland); rabbit anti-neuronal nitric oxide synthase (160870; 1:1000; Cayman Chemical, MI).
For the output mapping of tdTChrm2 cells, mice were perfused and coronal or sagittal sections were cut using a vibratome 3-4 weeks after the injection. Apart from the antibodies listed above, to further characterize the long range tdTChrm2 axons and their targets the following primary antibodies were used: rabbit anti-parvalbumin (PV27; 1:2500; Swant, Switzerland); mouse anti-PV (1:1000, Sigma #P3088, RRID: AB_477329); goat-anti RFP (1:3000, Rockland (200-101-379, RRID:AB_2744552); rabbit anti-neuronal nitric oxide synthase (160870; 1:1000; Cayman Chemical, MI); rabbit anti-proCCK (1:1000, Frontiers Neuroscience #AF350, RRID: AB_2571674) and mouse anti-reelin (MAB5366, Millipore). Primary antibodies were diluted in 0.1 M PB containing 0.25% Triton X100 and sections were incubated overnight at 4°C in the antibody cocktails. Primary antibodies were detected using secondary antibodies raised in goat and conjugated to Alexa Fluor 488/594 (Invitrogen), raised in donkey and conjugated to Alexa Fluor 647 (711-605-152; Jackson Laboratories) or with CF405S, CF488A, CF568 or Dy-Light647, highly cross-absorbed, by Biotium or Thermo Fisher).
Sleep experiments
Mice used for sleep experiments were implanted with an imaging window and a bipolar LFP electrode as described at two-photon imaging. In addition, mice were implanted with an ECoG electrode and an EMG electrode. ECoG signal was obtained using custom made Teflon coated tungsten wire electrodes placed over the primary motor cortex in the left hemisphere (1.5 mm lateral and 1.5 mm rostral from bregma) and referenced to the signal of a similar electrode placed over the cerebellum (midline, 6 mm caudal). For EMG recordings two coated tungsten wires twisted together were used as a bipolar electrode and was placed in the neck muscle using a 24G needle as guidance. A small section of insulation was removed on each wire about 1 mm apart thus forming the recording sites (Pearson et al., 2005). Output wires of LFP, ECoG and EMG electrodes were soldered to connector pins fixed to the skull.
After recovery, mice underwent a total sleep deprivation procedure. Prior to head-fixed experiments animals were individually housed in a 10-14 inch diameter circular arena equipped with a low-torque overhead sweeper bar that rotated at around 1 revolution per minute. Flexible arms of the sweeper bar promoted the animal to regularly relocate within the arena, but lacked sufficient force to inflict physical harm upon a stationary animal. Animals were allowed at least 2 days of undisrupted sleep in their home cage after every session of sleep deprivation. Animals had ad libitum access to food and water while in the sleep deprivation arena. Recording usually took place in the morning after the mice spent the previous night in the sleep-deprivation setup. Imaging session was not longer than 3 hrs and if the mouse did not spend sufficient amount of time in sleep the sleep deprivation procedure was started over on a different day. Mice were not exposed to sleep deprivation longer than 24 hrs at once.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of in vivo recordings
Recordings were analyzed using custom-written routines in MATLAB using the Signal Processing Toolbox. Traces were zero-phase filtered using finite impulse–response filters. The juxtacellularly recorded signal was high-pass filtered at 400 Hz, and spikes were detected after crossing a threshold manually set by the experimenter (typically between 200 and 400 μV). Superimposing all detected spikes revealed single units in all cases. Average firing frequency during running and rest was calculated by dividing the number of action potentials by the time spent in these states, determined using time-stamps based on the video recording of the mouse. Ripple detection was carried out during non-running periods. For ripple detection, the LFP was filtered between 90-200 Hz. The program examined the envelope on the absolute values of the signals, and when the envelope crossed the threshold of 5 standard deviations, the event was considered a ripple. The start and end points of ripples were specified as the time points where the signal reached a threshold of 2 standard deviations. All detected ripple events were inspected manually and artifacts were discarded. To determine if a cell was significantly modulated by ripples or the APs during ripples arose by chance, simulations were performed either by randomizing the spike location (Varga et al., 2012) or by shuffling the ripple location (Lasztóczi et al., 2011). Additionally, spike frequency plots for each cell triggered by ripple center were visually inspected for verification. The average firing frequency during ripples was obtained by dividing the number of action potentials within a ripple by the duration of the ripple. For comparisons of firing frequencies inside and outside ripples the spiking frequency was calculated using a 50ms-long sliding window. For calculating pre-ripple index, a ripple center-triggered average spiking frequency plot was generated for each cell. After normalization, area under the curve (AUC) values were then determined both for the negative (pre-ripple center) and for the positive time values (post-ripple center). The degree of precession was then expressed according to the following equation: pre-ripple index=100-(AUCpost-RIPPLE/AUCpre-RIPPLE)*100.
The running-associated theta oscillation was detected from the LFP signal using a band-pass filter (5–10 Hz). Preferred firing phase and strength of modulation during oscillations of individual cells were calculated as described earlier (Varga et al., 2012, 2014) and expressed as orientation and length of vectors calculated by summing the spike phases normalized by the number of spikes. Depending on the orientations of the different phase angles, the length of the normalized vector (r) ranged from 0 (no phase preference) to 1 (all phase angles identical). This length of the vector was used to measure the magnitude of phase modulation, and the direction of r indicated the mean phase angle of the cell (Varga et al., 2012, 2014). Differences between cell classes (TOROs vs. PVBCs) in terms of phase locking were evaluated using Watson–Williams circular test (Senior et al., 2008) ran in MATLAB. Directional data were presented as mean ± SD using the CircStat toolbox in Matlab (Berens, 2009). Additional statistical comparisons were obtained and figures were generated using Origin Pro 2019b.
Analysis of TORO cell activity from Allen Brain Observatory database
Sessions with complete behavioral data from the functional connectivity dataset, which include Neuropixel recordings from male (n=10) and female (n=3) adult mice (P108-P136), were used for these experiments. All recorded cells were included in our analyses to avoid potential biases from quality control filters and data was analyzed in Python. Session names, probe identification numbers, and channels used to detect ripples are included in a documented Jupyter notebook. SPW-Rs were detected as outlined in Analysis of in vivo recordings by bandpass filtering, computing a Hilbert transformation, and thresholding the envelope using the same cut-offs. Only ripples lasting at least 30 milliseconds were included in this analysis. Ripple amplitude, duration, and average frequency during the event were computed for each identified ripple using the Hilbert transformation. Firing rates during ripples were calculated from the number of spikes occurring during a ripple divided by the duration of the ripple and averaged across all ripples within the session. For each cell, Pearson correlation coefficients were calculated for ripple firing rate vs. ripple amplitude, duration, and frequency.
For calculating firing rate across the entire session, spikes were grouped into 100 millisecond bins and smoothed with a 1-dimensional gaussian filter (sigma=5). Speed data was resampled to align with session firing rate data and the Pearson correlation coefficient was calculated for each cell across the entire session for all data points or data points where immobility data was withheld (speed < 1 cm/s). Locomotor firing rate for each unit was calculated as the average firing rate of this trace for speeds greater than 2.5 cm/s. Cut-offs for defining TORO cells were derived from juxtacellularly-identified TORO cell (n=10, including 3 additional TORO cells in the CA3 reported in Szabo at al. 2017) locomotor firing rates and ripple firing rates, using 3 standard deviations from the mean rounded to the nearest integer value (>50Hz for ripples, and <9Hz for locomotion). This inclusive threshold was further refined by the requirement that TORO cells ripple firing rates must be 15 times greater than the locomotor firing rate. Identified TORO cell locations within the Allen Mouse Brain Common Coordinate Framework (Wang et al., 2020) were plotted in 2-dimensional coronal sections using QuickNII (Puchades et al., 2019) and plotted in 3 dimensions using the Brainrender graphical user interface (Claudi et al., 2021).
Pre-ripple index was calculated as outlined in Analysis of in vivo recordings. Burst index was calculated similar to Royer et al., 2012. Interspike interval (ISI) histograms (1 millisecond bins) were generated from the list of spike times and the burst index was computed as a number ranging from −1 to 1 (increasing in burstiness). The mean number of ISI between 40 and 50 milliseconds is subtracted from the mean number of ISI less than 7 milliseconds. Positive values were normalized to the mean number of ISI less than 7 milliseconds whereas negative values were normalized to the mean number of ISI between 40 and 50 milliseconds. Theta-ON, ripple-ON cells were selected for comparison to TORO cells if their ripple firing rates were >50Hz, locomotor firing rates were >20Hz, and were located in CA1.
Analysis of in vitro recordings
Firing properties were assessed during current injection steps (from −100 up to 800 pA, in 25 pA increments, for 1s) in neurons where the holding current was adjusted to maintain a holding membrane potential close to −65 mV. Rheobase was taken as the minimum depolarizing current that evoked at least one spike. Steady-state membrane potential was estimated as the mean membrane potential over the last 100 ms of hyperpolarizing current injection of −100 pA, while input resistance was taken as the difference between steady-state and holding potentials divided by 100 pA. Sag ratio was calculated as the ratio of the difference between the peak hyperpolarized deflection and the holding voltage to the difference between the steady-state and holding potentials, in response to −100 pA current injection. All AP properties were estimated for the first spike following +100 pA current injection. AP threshold was the voltage where the dV/dt prior to a detected event first exceeded 3 times the standard deviation while AP peak was the highest depolarized potential achieved. AP height was calculated as the difference between AP peak and AP threshold. Afterhyperpolarization potential was the largest hyperpolarized deflection from AP threshold. Spike width was the time an AP, resampled at 100 kHz, exceeded the half-height between threshold and peak voltages. Firing rate was taken as the number of spikes generated divided by stimulation time, while adaptation ratio was calculated as the ratio of the last ISI to the first ISI in response to depolarizing current injections, for traces that had at least three spikes. Data analysis was performed using Clampfit (Molecular Devices) or custom written Python scripts.
Properties of uIPSCs were analyzed using Clampfit and Origin Pro 2019b. For tdTChrm2 cell to IN pairs peak amplitude was calculated as the mean peak amplitude of the first IPSC during the train including failures, while synaptic potency was calculated as the mean peak amplitude of the first IPSC in the train excluding failures. Synaptic latency was calculated as the distance between the presynaptic AP peak and the postsynaptic IPSC onset. Statistics on paired recording experiments were done in Origin Pro 2019b.
Processing of 2-photon calcium imaging data
Calcium imaging data were processed and analyzed using Python scripts. Motion correction was performed using the HiddenMarkov2D function of SIMA (Kaifosh et al., 2014). This was followed by generating the “TORO image”, i.e. RUN-response (all RUN frames/no-RUN frames) and ripple-response (4 post-ripple frames were taken for each ripple and all averaged, then this was divided by the average of 10 ripple-preceding frames for each ripple) were visualized on two different color scales and overlaid on each other. RUN frames were defined as frames recorded while locomotion (based on the movement of the belt) was detected. The TORO image-identified TORO cells showed robust individual Ca peaks at ripple occurrences, which was not observed at other IN-s that otherwise had a positive response to ripples on average (Figure 3C; G). TORO images revealed cell bodies (i.e. TORO cells) that were otherwise barely visible on the average intensity projection images. Therefore, to segment interneuron cell bodies, binary regions of interest (ROIs) were drawn manually, and any cell visible either in the average intensity projection or the TORO image was included. Next, the fluorescence intensity traces were extracted for each ROI by averaging the included pixel intensities within each frame. DF/F traces were obtained as described previously (Dudok et al., 2021). In a subset of experiments, brief (typically 5 min) movies were taken at low magnification to localize TORO cells. This was followed by reimaging the cells of interest at higher magnification, resulting in multiple 2P images of the same or overlapping sets of cells recorded with different durations. TORO images done on these multiple movies always resulted in similar TORO images, i.e. the same cells appeared as ripple-active and RUN-inactive across sessions.
Event-triggered averages were calculated as described previously (Dudok et al., 2021). The process of event detection and averaging was fully automated to remove investigator bias. LFP traces were automatically processed to detect ripple events as described at the juxtacellular section. Detected events were then manually curated in a blinded manner by displaying events (1 s-long snippets of LFP) in the order of decreasing ripple-band power, with no context of behavioral or calcium imaging data.
The video recordings were analyzed in Matlab utilizing the built-in Matlab function regionprops. Frames first were cropped isolating the eye of the mouse. For each video frame an estimate of the diameter and the coordinates of the pupil was obtained. For the frames that were unable to produce a reliable estimate (e.g. rapid eye movement during REM sleep) the pupil diameter and coordinates were estimated manually.
ECoG/EMG- and pupil-based brain state classification
ECoG and EMG data were recorded at 10 kHz, then down-sampled to 500 Hz and filtered (ECoG: 1-20 Hz, EMG: 20-200). For ECoG/EMG-based scoring fast Fourier transform (FFT) power spectrum analysis was performed on the filtered ECoG and EMG signals using 4 s sliding windows shifted in 2 s increments with 0.25 Hz resolution. Next, theta and delta powers were computed by summing the ECoG power ranging from 1 to 4 Hz and from 6 to 9 Hz, as well as the EMG power from 20 to 200 Hz. Furthermore, we employed theta versus delta (theta/delta) power ratio and delta power threshold (separating the typically bimodal distribution of the delta power into a lower and higher range, (Weber et al., 2015, 2018) that have been used to measure brain states in rodent.
The brain state was determined using an automated threshold algorithm based on: 1. normalized EMG power; 2. theta/delta power ratio; 3. delta power threshold. A state was classified as RUN if the EMG power was higher than two standard deviations above the mean. A state was classified as NREM if the theta/delta ratio was lower than one standard deviation below the mean. A state was classified as REM if the theta/delta ratio was higher than two standard deviations above the mean and EMG power was lower than its mean plus one standard deviation. The remaining states were classified as AWAKEREST. In addition, if a single AWAKEREST epoch was preceded and followed by a NREM epoch, it was re-scored as NREM. Similarly, single NREM epochs preceded and followed by AWAKEREST epochs were re-scored as AWAKEREST. Finally, automatic classification was verified manually. State transitions were identified manually when EEG/EMG criterion changes were predominant for more than 50% of the epoch duration (Herrera et al., 2016).
Besides the conventional ECoG scoring method above all mice were re-scored based on LFP, pupil diameter and movement data. Since the two approaches resulted in very similar outcomes (n=4 mice), two additional mice with no ECoG signal were added to the analysis of TORO cell behavior across brain states. Classification criteria for brain states in these cases were the following: RUN: movement of the belt detected; REM: 1) no sign of movement, 2) high power theta on LFP similar to running, 3) rapid-eye-movement appears, 4) pupil diameter below 50% of maximum; NREM: 1) no sign of movement, 2) large irregular activity with frequent ripple occurrence on LFP, 3) pupil diameter between 40-70% of maximum; AWAKEREST: all the rest of the epochs (Figure 4F; Figure S3A; see also (Yüzgeç et al., 2018)).
Analyzing confocal images for output mapping
We defined TORO cell distal axons in Chrm2-tdT-D mice injected with interneuron-specific Dlx-GCaMP6f virus (Dimidschstein et al., 2016) in the CA1, by the combination of 3 criteria: 1) expression of tdT, to identify M2R-expressing cells; 2) expression of GCaMP, to restrict the axons to those originating from CA1 interneurons; and 3) the absence of PV immunolabeling. However, as we found no CA1-originating M2-expressing axons to be PV-positive (Figure S6K; Figure S7G), and, with the exception of one report about PV interneurons giving rise to commissural projections between the two hippocampi (Christenson Wick et al., 2017), there are no known distally projecting, PV-expressing cell types. Thus, we did not stain all samples for PV.
We first established that there are six target regions that consistently receive tdTChrm2 and GCaMP co-expressing axon terminals based on surveying the entire anterior-posterior axis in sagittal sections. For quantification, high resolution confocal Z-stack images (63x 1.4 NA objective of the Zeiss LSM 710 imaging system) were taken from each of the target areas using each experimental mouse in which the GCaMP expression was restricted to the CA1. Next, using the ZEN software, the GCaMP-expressing boutons were counted within a 50 μm3 volume to which optical sections corresponding to an estimated 50 μm physical section thickness (pre-mounting) were included. Counting was performed such that the analyzing person was blinded to the tdT channel. This was followed by counting tdTChrm2-positive boutons within the same volume. The same procedure was repeated at 3 randomly selected locations within each target region and for each brain. The marker immunostaining channels were not displayed when the 3 locations of the volume were selected to avoid bias. The mean bouton number from the 3 random locations were used as density for a given target region from a given brain.
Putative synaptic contacts around M2R-, PV- and CCK cell bodies were determined by 3D inspection of optical sections through z-stacks. For the quantification of GCaMP-expressing septal GABAergic boutons around M2R cells, the center of the cell body was placed into the middle of a 50 μm2 square and putative contacts were counted throughout the entire 60 μm thick section within the square. To quantify tdTChrm2- and GCaMP-expressing boutons targeting PV and CCK cells, immunolabeled cell bodies were selected such that the tdT channel was not displayed to avoid bias. All boutons that targeted PV cell bodies as well as most CCK cell-targeting boutons were also tested for PV and the positive ones were not taken into account. In agreement with earlier reports (Hájos et al., 1997), we did not observe PV and M2R-positive axons at the radiatum-LM border or in the radiatum layer. Therefore, a minority of CCK cells located at radiatum-LM and in the radiatum were analyzed in samples not immunostained for PV. For the quantification of the cellular targeting of tdTChrm2- and GCaMP-expressing boutons in the EC a 100 μm2 or a 50 μm2 square was selected on low magnification Z-stack images (20x) for interneuronal (PV and pro-CCK) or principal cell markers (calbindin and reelin), respectively. Three squares containing tdTChrm2- and GCaMP-expressing axons were selected for each marker and from each animal while the marker immunostaining channels were not displayed to avoid bias. Then, contacts around cell bodies positive for the respective marker were counted throughout the entire 60 μm thick (physical) section within each square on high magnification (63x) z-stack images. Figures showing microscopy images were prepared using the ZEN software and Adobe Photoshop. Maximum intensity projections were created using the orthogonal function of ZEN and exported as 8-bit RGB images. Pseudocoloring and adjustments of gamma were performed for each channel using the Hue and Curves commands in Photoshop. All adjustments were made adhering to common image integrity standards, and were applied uniformly to the entire image.
Statistical analysis
Statistical analysis was performed using Origin Pro 2019b, Python and R. Plots were generated using Origin Pro 2019b and Python. Appropriate statistical tests were selected based on study design and data was checked for meeting the assumptions of the applied tests. Normality of the data was tested using a Shapiro-Wilk test and further parametric or nonparametric tests were used accordingly, unless specified otherwise. Reported measures of central tendency and error are mean ± SD, unless stated differently. All tests are 2-sided. For experiments with optogenetics, linear mixed effects models were used. For likelihood ratio tests, a mixed model was constructed using the lmer function of lme4 in R. The reported effect sizes are the fixed effect ± standard error as returned by the test model fitted with the REML approach. The number of cells per animal was not pre-determined (all available cells in each animal were included). Statistical parameters are reported in the Results or in the Figure legends.
Supplementary Material
Movie S1 related to Figure 2. 3-Dimensional TORO cell locations in the brain TORO cells are represented by orange dots, whereas all other recorded cells are represented by partially transparent black dots. Hippocampal structures, including the CA1, CA3, DG, and subiculum, are filled to show cell locations relative to structural boundaries.
Highlights:
Theta-OFF/Ripple-ON (TORO) cells are hippocampal GABAergic neurons.
TORO cells rarely spike during theta states but burst during sharp wave-ripples.
TORO cells express inhibitory muscarinic type-2 receptors.
TORO cells preferentially innervate interneurons and project extrahippocampally.
Acknowledgements:
Research reported in this publication was supported by the National Institute of Mental Health (NIMH) and the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH) under Award Numbers U19NS104590 (to I.S.), NINDSR01NS99457 (to I.S.), K99NS117795 (to BD), and by the Lundbeck Foundation R273-2018-212 (to M.C. and I.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank the Stanford Neuroscience Gene Vector and Virus Core for providing AAV vector services, Andrew Olson and the Stanford Neuroscience Microscopy Service for microscopy support, Mikko Oijala, Peter Klein and Ernie Hwaun for help with data analysis techniques, Lisa Topolnik for the VIP immunostaining protocol, as well as Sylwia Felong, Theresa Nguyen, Pardis Zarifkar and Sandra Linder for technical and administrative support.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaral DG, and Witter MP (1989). The three-dimensional organization of the hippocampal formation: A review of anatomical data. Neuroscience 31, 571–591. [DOI] [PubMed] [Google Scholar]
- Araujo DM, Lapchak PA, Regenold W, and Quirion R (1989). Characterization of [3H]AF-DX 116 binding sites in the rat brain: Evidence for heterogeneity of muscarinic-M2 receptor sites. Synapse 4, 106–114. [DOI] [PubMed] [Google Scholar]
- Berens P (2009). CircStat : A MATLAB Toolbox for Circular Statistics. J. Stat. Softw 31, 10.18637/jss.v031.i10. [DOI] [Google Scholar]
- Bocchio M, Gouny C, Angulo-Garcia D, Toulat T, Tressard T, Quiroli E, Baude A, and Cossart R (2020). Hippocampal hub neurons maintain distinct connectivity throughout their lifetime. Nat. Commun 11, 4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm C, Peng Y, Maier N, Winterer J, Poulet JFA, Geiger JRP, and Schmitz D (2015). Functional diversity of subicular principal cells during hippocampal ripples. J. Neurosci 35, 13608–13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifazi P, Goldin M, Picardo MA, Jorquera I, Cattani A, Bianconi G, Represa A, Ben-Ari Y, and Cossart R (2009). GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science 326, 1419–1424. [DOI] [PubMed] [Google Scholar]
- Broussard GJ, Liang Y, Fridman M, Unger EK, Meng G, Xiao X, Ji N, Petreanu L, and Tian L (2018). In vivo measurement of afferent activity with axon-specific calcium imaging. Nat. Neurosci 21, 1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui AD, Nguyen TM, Limouse C, Kim HK, Szabo GG, Felong S, Maroso M, and Soltesz I (2018). Dentate gyrus mossy cells control spontaneous convulsive seizures and spatial memory. Science 359, 787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G (1986). Hippocampal sharp waves: Their origin and significance. Brain Res. 398, 242–252. [DOI] [PubMed] [Google Scholar]
- Buzsáki G (2015). Hippocampal sharp wave-ripple: A cognitive biomarker for episodic memory and planning. Hippocampus 25, 1073–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Lai-Wo S,L, and Vanderwolf CH (1983). Cellular bases of hippocampal EEG in the behaving rat. Brain Res. Rev 6, 10.1016/0165-0173(83)90037-1. [DOI] [PubMed] [Google Scholar]
- Buzsáki G, Horváth Z, Urioste R, Hetke J, and Wise K (1992). High-frequency network oscillation in the hippocampus. Science 256, 1025–1027. [DOI] [PubMed] [Google Scholar]
- Buzsáki G, Buhl DL, Harris KD, Csicsvari J, Czéh B, and Morozov A (2003). Hippocampal network patterns of activity in the mouse. Neuroscience 116, 201–211. [DOI] [PubMed] [Google Scholar]
- Cai L, Gibbs RB, and Johnson DA (2012). Recognition of novel objects and their location in rats with selective cholinergic lesion of the medial septum. Neurosci. Lett 506, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi A, Melzer S, Michael M, and Monyer H (2013). The long and short of GABAergic neurons. Curr. Opin. Neurobiol 23, 179–186. [DOI] [PubMed] [Google Scholar]
- Cea-del Rio CA, Lawrence JJ, Tricoire L, Erdelyi F, Szabo G, and McBain CJ (2010). M3 muscarinic acetylcholine receptor expression confers differential cholinergic modulation to neurochemically distinct hippocampal basket cell subtypes. J. Neurosci 30, 6011–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson Wick Z, Leintz CH, Xamonthiene C, Huang BH, and Krook-Magnuson E (2017). Axonal sprouting in commissurally projecting parvalbumin-expressing interneurons. J. Neurosci. Res 95, 2336–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrobak JJ, and Buzsáki G (1994). Selective activation of deep layer (V-VI) retrohippocampal cortical neurons during hippocampal sharp waves in the behaving rat. J. Neurosci 14, 6160–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrobak JJ, and Buzsáki G (1996). High-frequency oscillations in the output networks of the hippocampal-entorhinal axis of the freely behaving rat. J. Neurosci 16, 3056–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudi F, Tyson AL, Petrucco L, Margrie TW, Portugues R, and Branco T (2021). Visualizing anatomically registered data with brainrender. Elife 10, DOI: 10.7554/eLife.65751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom LV, and Bland BH (1987). State-dependent spike train dynamics of hippocampal formation neurons: evidence for theta-on and theta-off cells. Brain Res. 422, 277–286. [DOI] [PubMed] [Google Scholar]
- Csicsvari J, Hirase H, Czurkó A, Mamiya A, and Buzsáki G (1999). Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving rat. J. Neurosci 19, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza RD, Bista P, Meier AM, Ji W, and Burkhalter A (2019). Spatial Clustering of Inhibition in Mouse Primary Visual Cortex. Neuron 104, 588–600.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, et al. (2016). Sensitive red protein calcium indicators for imaging neural activity. Elife 5, eLife 2016;5:e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657. [DOI] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M, Losonczy A, 2016. Sublayer-Specific Coding Dynamics during Spatial Navigation and Learning in Hippocampal Area CA1. Neuron 91, 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg H, Pabst M, Braganza O, Schoch S, Niediek J, Bayraktar M, Mormann F, and Beck H (2015). Synergy of direct and indirect cholinergic septo-hippocampal pathways coordinates firing in hippocampal networks. J. Neurosci 35, 8394–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diba K, and Buzsáki G (2007). Forward and reverse hippocampal place-cell sequences during ripples. Nat. Neurosci 10, 1241–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci 19, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudok B, Klein PM, Hwaun E, Lee BR, Yao Z, Fong O, Bowler JC, Terada S, Sparks FT, Szabo GG, et al. (2021). Alternating sources of perisomatic inhibition during behavior. Neuron 109, 997–1012.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan TM, and North RA (1985). Acetylcholine acts on m2-muscarinic receptors to excite rat locus coeruleus neurones. Br. J. Pharmacol 85, 733–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ego-Stengel V, and Wilson MA (2010). Disruption of ripple-associated hippocampal activity during rest impairs spatial learning in the rat. Hippocampus 20, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti F, Klausberger T, Cobden P, Baude A, Roberts JDB, Szucs P, Kinoshita A, Shigemoto R, Somogyi P, and Dalezios Y (2005). Metabotropic glutamate receptor 8-expressing nerve terminals target subsets of GABAergic neurons in the hippocampus. J. Neurosci 25, 10520–10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla R, Villette V, Luo X, Chamberland S, Muñoz-Pino E, Camiré O, Wagner K, Kis V, Somogyi P, and Topolnik L (2018). Connectivity and network state-dependent recruitment of long-range VIP-GABAergic neurons in the mouse hippocampus. Nat. Commun 9, 5043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF, and Antal M (1988). GABA-containing neurons in the septum control inhibitory interneurons in the hippocampus. Nature 336, 170–173. [DOI] [PubMed] [Google Scholar]
- Fuentealba P, Tomioka R, Dalezios Y, Márton LF, Studer M, Rockland K, Klausberger T, and Somogyi P (2008). Rhythmically active enkephalin-expressing GABAergic cells in the CA1 Area of the hippocampus project to the subiculum and preferentially innervate interneurons. J. Neurosci 28, 10017–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gămănuţ R, Kennedy H, Toroczkai Z, Ercsey-Ravasz M, Van Essen DC, Knoblauch K, and Burkhalter A (2018). The Mouse Cortical Connectome, Characterized by an Ultra-Dense Cortical Graph, Maintains Specificity by Distinct Connectivity Profiles. Neuron 97, 698–715.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, ming Weng S, Pernía-Andrade AJ, Csicsvari J, and Jonas P (2017). Phase-Locked Inhibition, but Not Excitation, Underlies Hippocampal Ripple Oscillations in Awake Mice In Vivo. Neuron 93, 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiller T, Vancura B, Terada S, Troullinou E, Chavlis S, Tsagkatakis G, Tsakalides P, Ócsai K, Poirazi P, Rózsa BJ, et al. (2020). Large-Scale 3D Two-Photon Imaging of Molecularly Identified CA1 Interneuron Dynamics in Behaving Mice. Neuron 108, 968–983.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachetti A, Micheletti R, and Montagna E (1986). Cardioselective profile of AF-DX 116, a muscarine M2 receptor antagonist. Life Sci. 38, 1663–1672. [DOI] [PubMed] [Google Scholar]
- Girardeau G, Benchenane K, Wiener SI, Buzsáki G, and Zugaro MB (2009). Selective suppression of hippocampal ripples impairs spatial memory. Nat. Neurosci 12, 1222–1223. [DOI] [PubMed] [Google Scholar]
- Girardeau G, Inema I, and Buzsáki G (2017). Reactivations of emotional memory in the hippocampus-amygdala system during sleep. Nat. Neurosci 20, 1634–1642. [DOI] [PubMed] [Google Scholar]
- Gulyás AI, Hájos N, Katona I, and Freund TF (2003). Interneurons are the local targets of hippocampal inhibitory cells which project to the medial septum. Eur. J. Neurosci 17, 1861–1872. [DOI] [PubMed] [Google Scholar]
- Hájos N, Papp EC, Acsády L, Levey AI, and Freund TF (1997). Distinct interneuron types express m2 muscarinic receptor immunoreactivity on their dendrites or axon terminals in the hippocampus. Neuroscience 82, 355–376. [DOI] [PubMed] [Google Scholar]
- Hangya B, Borhegyi Z, Szilágyi N, Freund TF, and Varga V (2009). GABAergic neurons of the medial septum lead the hippocampal network during theta activity. J. Neurosci 29, 8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Hochgerner H, Skene NG, Magno L, Katona L, Bengtsson Gonzales C, Somogyi P, Kessaris N, Linnarsson S, and Hjerling-Leffler J (2018). Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics. PLoS Biol. 16, e2006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CG, Cadavieco MC, Jego S, Ponomarenko A, Korotkova T, and Adamantidis A (2016). Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat. Neurosci 19, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee WCA, et al. (2009). Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc 4, 1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav SP, Kemere C, German PW, and Frank LM (2012). Awake hippocampal sharp-wave ripples support spatial memory. Science 336, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S, Klausberger T, Marton LF, Dalezios Y, Roberts JDB, Fuentealba P, Bushong EA, Henze D, Buzsáki G, and Somogyi P (2007). Neuronal diversity in GABAergic long-range projections from the hippocampus. J. Neurosci 27, 8790–8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydin Ç, et al. (2017). Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR, and Losonczy A (2013). Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat. Neurosci 16, 1182–1184. [DOI] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, and Losonczy A (2014). SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona I, Acsády L, and Freund TF (1999). Postsynaptic targets of somatostatinimmunoreactive interneurons in the rat hippocampus. Neuroscience 88, 37–55. [DOI] [PubMed] [Google Scholar]
- Katona L, Micklem B, Borhegyi Z, Swiejkowski DA, Valenti O, Viney TJ, Kotzadimitriou D, Klausberger T, and Somogyi P (2017). Behavior-dependent activity patterns of GABAergic long-range projecting neurons in the rat hippocampus. Hippocampus 27, 359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona L, Hartwich K, Tomioka R, Somogyi J, Roberts JDB, Wagner K, Joshi A, Klausberger T, Rockland KS, and Somogyi P (2020). Synaptic organisation and behaviour-dependent activity of mGluR8a-innervated GABAergic trilaminar cells projecting from the hippocampus to the subiculum. Brain Struct. Funct 225, 705–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Márton LF, Baude A, Roberts JDB, Magill PJ, and Somogyi P (2004). Spike timing of dendrite-targeting bistratified cells during hippocampal network oscillations in vivo. Nat. Neurosci 7, 41–47. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Marton LF, O’Neill J, Huck JHJ, Dalezios Y, Fuentealba P, Suen WY, Papp E, Kaneko T, Watanabe M, et al. (2005). Complementary roles of cholecystokinin- and parvalbumin-expressing GABAergic neurons in hippocampal network oscillations. J. Neurosci 25, 9782–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullander K, and Topolnik L (2021). Cortical disinhibitory circuits: cell types, connectivity and function. Trends Neurosci. 44, 643–657. [DOI] [PubMed] [Google Scholar]
- Lai J, Waite SL, Bloom JW, Yamamura HI, and Roeske WR (1991). The m2 muscarinic acetylcholine receptors are coupled to multiple signaling pathways via pertussis toxin-sensitive guanine nucleotide regulatory proteins. J. Pharmacol. Exp. Ther 258, 938–944. [PubMed] [Google Scholar]
- Lapray D, Lasztoczi B, Lagler M, Viney TJ, Katona L, Valenti O, Hartwich K, Borhegyi Z, Somogyi P, and Klausberger T (2012). Behavior-dependent specialization of identified hippocampal interneurons. Nat. Neurosci 15, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasztóczi B, Tukker JJ, Somogyi P, and Klausberger T (2011). Terminal field and firing selectivity of cholecystokinin-expressing interneurons in the Hippocampal CA3 Area. J. Neurosci 31, 18073–18093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JJ, Statland JM, Grinspan ZM, and McBain CJ (2006). Cell type-specific dependence of muscarinic signalling in mouse hippocampal stratum oriens interneurones. J. Physiol 570, 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, and Dan Y (2012). Neuromodulation of Brain States. Neuron 76, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV, and Losonczy A (2014). Dendritic inhibition in the hippocampus supports fear learning. Science 343, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrosu F, Portas C, Mascia MS, Casu MA, Fà M, Giagheddu M, Imperato A, and Gessa GL (1995). Microdialysis measurement of cortical and hippocampal acetylcholine release during sleep-wake cycle in freely moving cats. Brain Res. 671, 329–332. [DOI] [PubMed] [Google Scholar]
- McGinley MJ, David SV, and McCormick DA (2015). Cortical Membrane Potential Signature of Optimal States for Sensory Signal Detection. Neuron 87, 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuiston AR, and Madison DV (1999). Muscarinic receptor activity has multiple effects on the resting membrane potentials of CA1 hippocampal interneurons. J. Neurosci 19, 5693–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Michael M, Caputi A, Eliava M, Fuchs EC, Whittington MA, and Monyer H (2012). Long-range-projecting gabaergic neurons modulate inhibition in hippocampus and entorhinal cortex. Science 335, 1506–1510. [DOI] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Wainer BH, and Levey AI (1983). Central cholinergic pathways in the rat: An overview based on an alternative nomenclature (Ch1-Ch6). Neuroscience 10, 1185–1201. [DOI] [PubMed] [Google Scholar]
- Mizumori SJY, Barnes CA, and McNaughton BL (1990). Behavioral correlates of theta-on and theta-off cells recorded from hippocampal formation of mature young and aged rats. Exp. Brain Res 80, 365–373. [DOI] [PubMed] [Google Scholar]
- Monory K, Massa F, Egertová M, Eder M, Blaudzun H, Westenbroek R, Kelsch W, Jacob W, Marsch R, Ekker M, et al. (2006). The Endocannabinoid System Controls Key Epileptogenic Circuits in the Hippocampus. Neuron 51, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau PH, Cosquer B, Jeltsch H, Cassel JC, and Mathis C (2008). Neuroanatomical and behavioral effects of a novel version of the cholinergic immunotoxin mu p75-saporin in mice. Hippocampus 18, 610–622. [DOI] [PubMed] [Google Scholar]
- Oliva A, Fernández-Ruiz A, Buzsáki G, and Berényi A (2016). Role of Hippocampal CA2 Region in Triggering Sharp-Wave Ripples. Neuron 91, 1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZZ, and Williams JT (1994). Muscarine hyperpolarizes a subpopulation of neurons by activating an M2 muscarinic receptor in rat nucleus raphe magnus in vitro. J. Neurosci 14, 1332–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra P, Gulyás AI, and Miles R (1998). How many subtypes of inhibitory cells in the hippocampus? Neuron 20, 983–993. [DOI] [PubMed] [Google Scholar]
- Pearson KG, Acharya H, and Fouad K (2005). A new electrode configuration for recording electromyographic activity in behaving mice. J. Neurosci. Methods 148, 36–42. [DOI] [PubMed] [Google Scholar]
- Peralta EG, Ashkenazi A, Winslow JW, Ramachandran J, and Capon DJ (1988). Differential regulation of PI hydrolysis and adenylyl cyclase by muscarinic receptor subtypes. Nature 334, 434–437. [DOI] [PubMed] [Google Scholar]
- Picardo MA, Guigue P, Bonifazi P, Batista-Brito R, Allene C, Ribas A, Fishell G, Baude A, and Cossart R (2011). Pioneer GABA cells comprise a subpopulation of hub neurons in the developing hippocampus. Neuron 71, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades MA, Csucs G, Ledergerber D, Leergaard TB, and Bjaalie JG (2019). Spatial registration of serial microscopic brain images to three-dimensional reference atlases with the QuickNII tool. PLoS One 14, e0216796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, and Buzsáki G (2012). Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat. Neurosci 15, 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer K (1892). No TitleBeitrag zur histologie der Amnionshornformation. Arch Mikrosk Anat 39, 611–632. [Google Scholar]
- Schlingloff D, Káli S, Freund TF, Hájos N, and Gulyás AI (2014). Mechanisms of sharp wave initiation and ripple generation. J. Neurosci 34, 11385–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior TJ, Huxter JR, Allen K, O’Neill J, and Csicsvari J (2008). Gamma oscillatory firing reveals distinct populations of pyramidal cells in the CA1 region of the hippocampus. J. Neurosci 28, 2274–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siapas AG, and Wilson MA (1998). Coordinated interactions between hippocampal ripples and cortical spindles during slow-wave sleep. Neuron 21, 1123–1128. [DOI] [PubMed] [Google Scholar]
- Siegle JH, Jia X, Durand S, Gale S, Bennett C, Graddis N, Heller G, Ramirez TK, Choi H, Luviano JA, et al. (2021). Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sik A, Ylinen A, Penttonen M, and Buzsáki G (1994). Inhibitory CA1-CA3-hilar region feedback in the hippocampus. Science 265, 1722–1724. [DOI] [PubMed] [Google Scholar]
- Sik A, Penttonen M, Ylinen A, and Buzsáki G (1995). Hippocampal CA1 interneurons: An in vivo intracellular labeling study. J. Neurosci 15, 6651–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari N, and Hangya B (2018). Cholinergic modulation of spatial learning, memory and navigation. Eur. J. Neurosci 48, 2199–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Roux L, Eichler R, Senzai Y, Royer S, and Buzsáki G (2014). Pyramidal cell-interneuron interactions underlie hippocampal ripple oscillations. Neuron 83, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D, Csicsvari J, Mizuseki K, Montgomery S, Diba K, and Buzsáki G (2011). Relationships between hippocampal sharp waves, ripples, and fast gamma oscillation: Influence of dentate and entorhinal cortical activity. J. Neurosci 31, 8605–8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SS, and Smith GK (1988). Spontaneous EEG spikes in the normal hippocampus II. Relations to synchronous burst discharges. Electroencephalogr. Clin. Neurophysiol 69, 532–540. [DOI] [PubMed] [Google Scholar]
- Szabo GG, Du X, Oijala M, Varga C, Parent JM, and Soltesz I (2017). Extended Interneuronal Network of the Dentate Gyrus. Cell Rep. 20, 1262–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó GG, Holderith N, Gulyás AI, Freund TF, and Hájos N (2010). Distinct synaptic properties of perisomatic inhibitory cell types and their different modulation by cholinergic receptor activation in the CA3 region of the mouse hippocampus. Eur. J. Neurosci 31, 2234–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takács VT, Freund TF, and Gulyás AI (2008). Types and synaptic connections of hippocampal inhibitory neurons reciprocally connected with the medial septum. Eur. J. Neurosci 28, 148–164. [DOI] [PubMed] [Google Scholar]
- Takács VT, Klausberger T, Somogyi P, Freund TF, and Gulyás AI (2012). Extrinsic and local glutamatergic inputs of the rat hippocampal CA1 area differentially innervate pyramidal cells and interneurons. Hippocampus 22, 1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth K, Borhegyi Z, and Freund TF (1993). Postsynaptic targets of GABAergic hippocampal neurons in the medial septum-diagonal band of Broca complex. J. Neurosci 13, 3712–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, and Freund TF (1992). Calbindin D28k-containing nonpyramidal cells in the rat hippocampus: Their immunoreactivity for GABA and projection to the medial septum. Neuroscience 49, 793–805. [DOI] [PubMed] [Google Scholar]
- Turi GF, Li WK, Chavlis S, Pandi I, O’Hare J, Priestley JB, Grosmark AD, Liao Z, Ladow M, Zhang JF, et al. (2019). Vasoactive Intestinal Polypeptide-Expressing Interneurons in the Hippocampus Support Goal-Oriented Spatial Learning. Neuron 101, 1150–1165.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal G, Crump MG, Viney TJ, Katona L, Klausberger T, Somogyi P, 2018. Spatio-temporal specialization of GABAergic septo-hippocampal neurons for rhythmic network activity. Brain Struct. Funct 223, 2409–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero M, Zutshi I, Yoon E, and Buzsáki G (2022). Probing subthreshold dynamics of hippocampal neurons by pulsed optogenetics. Science 375, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C, Lee SY, and Soltesz I (2010). Target-selective GABAergic control of entorhinal cortex output. Nat. Neurosci 13, 822–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C, Golshani P, and Soltesz I (2012). Frequency-invariant temporal ordering of interneuronal discharges during hippocampal oscillations in awake mice. Proc. Natl. Acad. Sci. U. S. A 109, E2726–E2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C, Oijala M, Lish J, Szabo GG, Bezaire M, Marchionni I, Golshani P, and Soltesz I (2014). Functional fission of parvalbumin interneuron classes during fast network events. Elife 3, e04006 DOI: 10.7554/eLife.04006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V, Guigue P, Picardo MA, Sousa VH, Leprince E, Lachamp P, Malvache A, Tressard T, Cossart R, and Baude A (2016). Development of early-born γ-Aminobutyric acid hub neurons in mouse hippocampus from embryogenesis to adulthood. J. Comp. Neurol 524, 2440–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney TJ, Lasztoczi B, Katona L, Crump MG, Tukker JJ, Klausberger T, and Somogyi P (2013). Network state-dependent inhibition of identified hippocampal CA3 axo-axonic cells in vivo. Nat. Neurosci 16, 1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ding SL, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, et al. (2020). The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Chung S, Beier KT, Xu M, Luo L, and Dan Y (2015). Control of REM sleep by ventral medulla GABAergic neurons. Nature 526, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Hoang Do JP, Chung S, Beier KT, Bikov M, Saffari Doost M, and Dan Y (2018). Regulation of REM and Non-REM Sleep by Periaqueductal GABAergic Neurons. Nat. Commun 9, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick ZC, Tetzlaff MR, and Krook-Magnuson E (2019). Novel long-range inhibitory nNOS-expressing hippocampal cells. Elite 8, eLife 2019;8:e46816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzynski CM, Lubenov EV, Gu M, and Siapas AG (2009). State-Dependent Spike-Timing Relationships between Hippocampal and Prefrontal Circuits during Sleep. Neuron 61, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki N, Li X, Lambot L, Ren LY, Radulovic J, and Shepherd GMG (2019). Long-range inhibitory intersection of a retrosplenial thalamocortical circuit by apical tuft-targeting CA1 neurons. Nat. Neurosci 22, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüzgeç Ö, Prsa M, Zimmermann R, and Huber D (2018). Pupil Size Coupling to Cortical States Protects the Stability of Deep Sleep via Parasympathetic Modulation. Curr. Biol 28, 392–400.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao L, Varga V, Jing M, Karadas M, Li Y, and Buzsáki G (2021). Cholinergic suppression of hippocampal sharp-wave ripples impairs working memory. Proc. Natl. Acad. Sci. U. S. A 118, e2016432118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 related to Figure 2. 3-Dimensional TORO cell locations in the brain TORO cells are represented by orange dots, whereas all other recorded cells are represented by partially transparent black dots. Hippocampal structures, including the CA1, CA3, DG, and subiculum, are filled to show cell locations relative to structural boundaries.
Data Availability Statement
The datasets generated during the current study are available from the Lead Contact on reasonable request.
Custom scripts used in this study have been deposited to a publicly accessible repository. The URL is listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-DsRed | Takara | #632496, RRID: AB_10013483 |
| goat anti-GFP | abcam | ab5450, RRID:AB_304897 |
| chicken anti-GFP | Aves | #GFP-1020, RRID: AB_10000240 |
| rat anti-muscarinic acetylcholine receptor type 2 | Millipore | MAB367, RRID:AB_2152546 |
| rat anti-somatostatin | Millipore | MAB354, RRID:AB_2255365 |
| rabbit-anti-somatostatin | Peninsula | Cat# T-4103.0050 RRID:AB_518614 |
| rabbit anti calbindin | Swant | CB38, RRID:AB_10000340 |
| rabbit anti-parvalbumin | Swant | PV27, RRID:AB_2631173 |
| mouse anti- parvalbumin | Sigma | #P3088, RRID: AB_477329 |
| rabbit anti-proCCK | Frontiers Neuroscience | #AF350, RRID: AB_2571674 |
| rabbit anti-neuronal nitric oxide synthase | Cayman Chemical | 160870, RRID:AB_10080041 |
| mouse anti-GABA | Sigma | A0310, RRID:AB_476667 |
| goat anti-RFP | Rockland | 200-101-379 RRID:AB_2744552 |
| rabbit anti-VIP | Immunostar | RRID:AB_10730725 |
| mouse anti-reelin | Millipore | RRID: AB_2285132 |
| anti-phospho-IκBα | Cell Signaling Technology | Cat# 2859; RRID:AB_561111 |
| Bacterial and virus strains | ||
| AAV1-Syn-GCaMP6f-WPRE-SV40 | Penn Vector Core | N/A |
| AAV1-Syn-Flex-GCaMP6f-WPRE-SV40 | Penn Vector Core | N/A |
| AAV1-Syn-Flex-jGCaMP7s-WPRE | Dana et al., 2019 doi: 10.1038/s41592-019-0435-6 | Addgene #104491-AAV1, RRID: Addgene_104491 |
| AAV1-Syn-Flex-NES-jRGECO1a-WPRE-SV40 | Dana et al., 2016 doi:10.7554/eLife.12727 | Addgene #100853-AAV1, RRID: Addgene_100853 |
| AAV-DJ-Ef1a-fDIO GCaMP6f | Stanford Neuroscience Gene Vector and Virus Core | #167 |
| AAV5-EF1a-DIO-hChR2(H134R)-mCherry-WPRE-pA | UNC Vector Core | N/A |
| AAV5-EF1a-DIO-mCherry-WPRE-pA | UNC Vector Core | N/A |
| AAV5-hSyn-hChR2(H134R)-mCherry-WPRE | UNC Vector Core | N/A |
| pAAV-hSynapsin1-FLEx-axon-GCaMP6s (AAV9) | Broussard et al., 2018 doi: 10.1038/s41593-018-0211-4 | Addgene #112010-AAV9, RRID: Addgene_112010 |
| pAAV-mDlx-GCaMP6f | Dimidschstein et al., 2016 doi: 10.1038/nn.4430 | Addgene #83899, RRID: Addgene_83899 |
| Chemicals, peptides, and recombinant proteins | ||
| NBQX | Tocris | Cat. No. 1044 |
| D-APV | Cayman Chemical | #14539 |
| PTX | Tocris | Cat. No. 1128 |
| CGP35348 | Tocris | Cat. No. 1245 |
| AF-DX116 | Tocris | Cat. No. 1105 |
| carbachol | Tocris | Cat. No. 2810 |
| neurobiotin | Vector Laboratories | SP1120 |
| biotin-ethylenediamine (N-(2-Aminoethyl)Biotinamide, HBr | Thermofisher Scientific | A1593 |
| Experimental models: Organisms/strains | ||
| Dlx5/6-Cre transgenic (Tg(dlx5a-cre)1Mekk/J) | JAX | JAX Stock No. 008199 |
| C57BL/6J | JAX | JAX Stock No. 000664 |
| Chrm2-tdT-D knock-in | JAX | JAX Stock No. 030330 |
| Tg(Grik4-cre)G32-4Stl/J | JAX | JAX Stock No.006474 |
| ChAT-IRES –Cre | JAX | JAX Stock No.006410 |
| Npy-IRES2-FlpO-D | JAX | JAX Stock No.030211 |
| Tg(Amigo2-cre)1Sieg/J | JAX | JAX Stock No.030215 |
| Software and algorithms | ||
| Custom scripts | Present study; Zenodo | https://zenodo.org/record/6402500#.YkXUQbhlDdo |
| MATLAB | Mathworks | R2019b |
| Scanbox | Neurolabware | v4.6 |
| ZEN | Zeiss | 2012 SP5 FP1 v14 |
| Python | Python.org | v3.6 |
| sima (Python) | Kaifosh et al., 2014 doi: 10.3389/fninf.2014.00080 | https://github.com/losonczylab/sima PMID: 25295002 |
| pClamp | Molecular Devices | v10.7.0.3 |
| Clampfit | Molecular Devices | v10.7.0.3 |
| OriginPro | Originlab | 2019b |
| Allen Software Development Kit | Allen Institute | https://allensdk.readthedocs.io/en/latest/ |
| QuickNII | Puchades et al., 2019 doi: 10.1371/journal.pone.0216796 | RRID: SCR_016854, https://github.com/HumanBrainProject/QuickNII |
| Brainrender GUI | Claudi et al. 2021 doi: 10.7554/eLife.65751 | https://github.com/brainglobe/brainrender |