Significance
Autophagy defects are a risk factor for inflammatory bowel diseases (IBDs), but the mechanism remains unknown. We show here that conditional whole-body deletion of Atg5 or Fip200, but not Atg7, is lethal due to loss of ileum stem cells and barrier function likely caused by different kinetics of autophagy loss, which was rescued by slow deletion. Specific autophagy loss in PDGFRα+ mesenchymal cells (PMCs) resulted in loss of Wnt signaling responsible for failed stem cell renewal. We also observed depletion of aspartate and nucleotides throughout the ileum. Our results illustrate that autophagy is required for PMC metabolism and survival necessary to sustain intestinal stem cells and mouse survival, and failure to maintain PMCs through autophagy contributes to IBD.
Keywords: autophagy, intestine, stem cells, IBD, metabolism
Abstract
Autophagy defects are a risk factor for inflammatory bowel diseases (IBDs) through unknown mechanisms. Whole-body conditional deletion of autophagy-related gene (Atg) Atg7 in adult mice (Atg7Δ/Δ) causes tissue damage and death within 3 mo due to neurodegeneration without substantial effect on intestine. In contrast, we report here that whole-body conditional deletion of other essential Atg genes Atg5 or Fip200/Atg17 in adult mice (Atg5Δ/Δ or Fip200Δ/Δ) caused death within 5 d due to rapid autophagy inhibition, elimination of ileum stem cells, and loss of barrier function. Atg5Δ/Δ mice lost PDGFRα+ mesenchymal cells (PMCs) and Wnt signaling essential for stem cell renewal, which were partially rescued by exogenous Wnt. Matrix-assisted laser desorption ionization coupled to mass spectrometry imaging (MALDI-MSI) of Atg5Δ/Δ ileum revealed depletion of aspartate and nucleotides, consistent with metabolic insufficiency underlying PMC loss. The difference in the autophagy gene knockout phenotypes is likely due to distinct kinetics of autophagy loss, as deletion of Atg5 more gradually extended lifespan phenocopying deletion of Atg7 or Atg12. Thus, autophagy is required for PMC metabolism and ileum stem cell and mammalian survival. Failure to maintain PMCs through autophagy may therefore contribute to IBD.
The autophagy pathway directs the formation of double-membrane-bound vesicles called autophagosomes that capture cargo such as cytoplasmic proteins, organelles, and bacteria. Autophagosomes containing their cargo are trafficked to and fuse with lysosomes for degradation and recycling (1). Several major protein complexes are involved in autophagosome formation: the ULK1–FIP200–ATG13 autophagy initiation complex, the BECN1–VPS34–ATG14 complex, the ATG12–ATG5–ATG16 complex (formation requires ATG7 and ATG10), and the microtubule-associated protein 1 light chain 3 (LC3 or ATG8) complexes (formation requires ATG4, ATG7, and ATG3). The ATG12–ATG5–ATG16 complex is recruited to the autophagosome membrane and facilitates the lipidation of LC3, which is required for its membrane association and elongation and formation of autophagosomes (2). Therefore, both Atg5 and Atg7 are essential for the formation of functional autophagosomes and autophagy.
Autophagy serves as an important intracellular quality-control mechanism by removing damaged proteins, mitochondria, and invading bacteria (3). Selective autophagy requires autophagy receptors, such as the SQSTM1 (p62), that link these cargos to ATG8 family proteins on the autophagosome membrane, recruiting them to autophagosomes for degradation (4–6). This process is particularly important in the liver, muscle, and brain, likely because the buildup of damaged organelles, such as mitochondria, leads to oxidative stress and perturbation of metabolism (7). Thus, autophagy has tissue- and cargo-specific roles in maintaining homeostasis.
Autophagy plays important roles in survival during starvation and in maintaining metabolic homeostasis. In mammals, autophagy is dramatically induced under nutrient deprivation (8) and is required for metabolic homeostasis and survival (7, 9–11). Conditional, whole-body deletion of the essential autophagy gene Atg7 in adult mice (Atg7Δ/Δ) causes susceptibility to infection and liver damage, muscle wasting, and loss of white adipose tissue (WAT), with their lifespan decreased to 3 mo, primarily due to neurodegeneration, in part through activation of p53 (9, 12). Autophagy-deficient intestine functions normally, despite an increase in p53 and apoptosis in stem cells and some abnormalities in Paneth cells, which was surprising given the association of ATG16L1 mutations in inflammatory bowel diseases (IBDs) (13–15). Consistent with the role of autophagy in promoting survival in starvation, fasting Atg7Δ/Δ, but not wild-type, mice is lethal due to hypoglycemia and in part through activation of p53 (9, 12). Autophagy also plays a critical role in sustaining cancer metabolism in both a tumor cell-autonomous and -nonautonomous fashion (16). In tumor cells, autophagy provides amino acids, particularly glutamine to support tricarboxylic acid (TCA) cycle turning, aspartate, nucleotide synthesis, and pools (17). Thus, an essential function of autophagy is intracellular nutrient scavenging, particularly in response to starvation, to ensure an adequate substrate supply to support essential metabolic functions (18).
Since both Atg7 and Atg5 are essential autophagy genes, we generated a genetically engineered mouse model (GEMM) for conditional, whole-body Atg5 deletion in adult mice to address whether loss of Atg5 would have a similar phenotype to loss of Atg7 (9). In contrast to Atg7Δ/Δ mice that tolerate loss of ATG7 for up to 3 mo in the fed state, conditional whole-body deletion of Atg5 (Atg5Δ/Δ) caused ileum destruction with loss of barrier function, and mice survived less than 5 d. Conditional deletion of another essential autophagy gene, Fip200, also produced the same phenotype, indicating that it is not due to a function unique to Atg5. The rapid lethality of Atg5 deletion was due to rapid loss of autophagy associated with metabolic impairment in the Atg5Δ/Δ ileum, specifically in PMCs. Gradual whole-body deletion of Atg5 bypassed this lethality, resulting in death from neurodegeneration, similar to whole-body deletion of Atg7 and Atg12. Thus, Atg5 is required for PMC survival and intestinal homeostasis, the loss of function in which may contribute to the pathogenesis of IBDs.
Results
Conditional, Whole-Body Atg5 or Fip200 Deletion in Adult Mice Leads to Ileum Damage and Death.
To address Atg5 function in adult mice, we generated the inducible Ubc–CreERT2/+;Atg5flox/flox GEMM and treated these adult mice with five consecutive daily injections of tamoxifen (TAM), as we described previously to generate Ubc–Cre Atg7Δ/Δ (Atg7Δ/Δ) mice (9), to generate Ubc-Cre Atg5Δ/Δ (Atg5Δ/Δ) mice to compare with Ubc–Cre (wild-type control) mice (Fig. 1A). Atg7Δ/Δ mice accumulated damage in liver, brain, and several other tissues and died of neurodegeneration with a lifespan of 2 to 3 mo (9). Surprisingly, Atg5Δ/Δ mice showed a lifespan of 4 d post-TAM (Fig. 1B). To confirm that the phenotype is specifically caused by loss of Atg5, we performed TAM deletion in Cag–CreERT2/+; Atg5flox/flox mice, as well as Cag–CreERT2/+; Atg12flox/flox mice (SI Appendix, Fig. S1A). We observed a similar lifespan of 4 d using Cag–CreERT2/+ to delete Atg5, while Cag–Cre Atg12Δ/Δ mice died of neurodegeneration by 10 wk, similar to Atg7Δ/Δ mice (SI Appendix, Fig. S1A) (19). Thus, Atg7 or Atg12 conditional whole-body deletion in adult mice results in similar lifespan limitation of 3 mo, with death primarily due to neurodegeneration, while deleting Atg5 using two different Cre drivers was rapidly lethal. To obtain further clarity as to whether the rapid lethality of Atg5 conditional deletion was autophagy-function-related or not, we generated Ubc–Cre Fip200Δ/Δ mice. Conditional deletion of Fip200 in adult mice resulted in rapid lethality, similar to conditional deletion of Atg5 (SI Appendix, Fig. S1B). Thus, conditional deletion of essential autophagy genes in adult mice is either rapidly lethal (Atg5 and Fip200) or permits survival for 2 to 3 mo (Atg7 and Atg12). To determine why this was the case, we compared the phenotypes of Atg5Δ/Δ to Atg7Δ/Δ mice.
Fig. 1.
Conditional, whole-body Atg5 deletion in adult mice leads to ileum damage and death. (A) Experimental design for generation of Atg7Δ/Δ mice and Atg5Δ/Δ mice. (B) Kaplan–Meier survival curve of TAM-treated wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. ****P < 0.0001 (log-rank test). (C) Representative ileum hematoxylin and eosin-stained (H&E) histology with villus length quantification at indicated times from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. (D) Body weight of wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice at 3 d post-TAM. (E and F) Blood glucose (E) and FITC–dextran (F) concentration measured in milligrams per deciliter from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice at indicated times. (G) Western blotting for ATG5, ATG7, and LC3 at indicated times with relative fold change of band intensity normalized to actin from the wild-type, Atg7Δ/Δ, and Atg5Δ/Δ ileum. All images represent one of three biological replicates. All quantification data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group. See also SI Appendix, Figs. S1 and S2.
To determine why Atg5Δ/Δ mice fail to survive, we compared the histology of wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mouse tissues. Atg5Δ/Δ mice displayed singularly highly specific damage to the ileum, with other tissues such as liver, muscle, and adipose tissue displaying normal histology (SI Appendix, Fig. S1C). The wild-type ileum showed normal villous architecture without inflammation or significant disruption of the epithelium. The Atg7Δ/Δ ileum maintained the normal villous architecture with little disruption to the glandular epithelium. In contrast, the Atg5Δ/Δ ileum demonstrated marked epithelial damage exemplified by the shortening of villi, cytoplasmic vacuolation, numerous apoptotic cells indicated by increased cleaved caspase 3 (CC3), and sloughing of the surface epithelium, especially at 3 d post-TAM, but no significant inflammatory response and cell infiltration (Fig. 1C and SI Appendix, Fig. S1D). Duodenum and jejunum did not display those phenotypes (SI Appendix, Fig. S1 E–H). The same phenotype was observed in the Cag–Cre Atg5Δ/Δ and Ubc–Cre Fip200Δ/Δ, but not Cag–Cre Atg12Δ/Δ, ileum (SI Appendix, Fig. S2 A–G). Atg5Δ/Δ mice also had decreased body weight at 3 d post-TAM compared to wild-type and Atg7Δ/Δ mice (Fig. 1D).
Atg5 Deletion Decreases Blood Glucose and Causes Loss of Intestinal Barrier Function.
As the ileum functions in dietary nutrient absorption, and fasting Atg7Δ/Δ mice causes lethal hypoglycemia, we sought to test if Atg5Δ/Δ mice were nutritionally compromised. Atg5Δ/Δ mice had decreased blood glucose levels compared to wild-type and Atg7Δ/Δ mice starting at 2 d post-TAM, which decreased further at 3 d (Fig. 1E). Supplementation of glucose restored blood glucose levels, but did not rescue Atg5Δ/Δ mice, suggesting that this was not solely responsible for death (SI Appendix, Fig. S3 A and B). Atg5Δ/Δ mice began to show increased fluorescein isothiocyanate (FITC)–dextran in the circulation at 2 d, which further increased at 3 d post-TAM compared to wild-type and Atg7Δ/Δ mice (Fig. 1F). These results suggest that Atg5Δ/Δ mice lost intestinal barrier function and likely efficient nutritional absorption, causing hypoglycemia, which led to death.
Deletion of Atg5 Causes More Rapid Loss of Autophagy in the Ileum than Deletion of Atg7.
Deletion of Atg5 or Atg7 in Atg5Δ/Δ and Atg7Δ/Δ mice, respectively, was detected by PCR, but Atg5 deletion occurred more rapidly and robustly compared to Atg7 (SI Appendix, Fig. S3 C and D). Loss of the ATG5–ATG12 complex was apparent in the ileum at 1 d post-TAM in Atg5Δ/Δ mice, whereas the ATG5–ATG12 complex persisted in Atg7Δ/Δ mice despite loss of ATG7 (Fig. 1G). This indicated that the ATG5–ATG12 complex may be functional longer in the absence of ATG7 than it is in the absence of ATG5. The ratio of the unprocessed form of LC3 (LC3-I) to the processed form of LC3 (LC3-II) was generally higher in the Atg5Δ/Δ ileum compared to the Atg7Δ/Δ ileum (Fig. 1G). These results were similar, but less robust, in duodenum and jejunum (SI Appendix, Fig. S3 E and F). Taken together, whole-body deletion of either Atg5 or Atg7 caused autophagy defects, but Atg5Δ/Δ mice lost autophagy function more rapidly than Atg7Δ/Δ mice due to the accelerated kinetics of complete loss of the ATG5–ATG12 complex in the ileum, which may explain the lethal phenotype of Atg5Δ/Δ mice.
Ileum Stem Cells Are Lost in Atg5Δ/Δ Mice.
In the crypt of intestinal epithelium, the crypt base columnar (CBC) cells function as an active stem cell population and are interspersed with Paneth cells. CBC cells renew and differentiate into secretory or absorptive progenitor cells, which proliferate within the crypt and are called transit-amplifying (TA) cells (20). To investigate the cause of ileum damage, we examined which cell type was lost in the Atg5Δ/Δ ileum. Immunohistochemistry (IHC) confirmed the loss of ATG5 and ATG7 in the ileum, duodenum, and jejunum in Atg5Δ/Δ and Atg7Δ/Δ mice (SI Appendix, Fig. S3 G–L). The Atg5Δ/Δ intestine, however, accumulated more p62 aggregates compared to the Atg7Δ/Δ intestine, indicative of a more rapid functional autophagy loss (Fig. 2 A and B and SI Appendix, Fig. S4 A and B). There was also more p62 accumulation in the Cag–Cre Atg5Δ/Δ compared to the Cag–Cre Atg12Δ/Δ intestine (SI Appendix, Fig. S4 C–E). In contrast to the wild-type and Atg7Δ/Δ ileum, Alcian blue staining was completely lost at 3 d post-TAM in the Atg5Δ/Δ ileum, but not in the duodenum and jejunum, indicating loss of goblet cells (Fig. 2C and SI Appendix, Fig. S5 A and B), which was also observed in the Ubc–Cre Fip200Δ/Δ and Cag–Cre Atg5Δ/Δ ileum, but not the duodenum or jejunum (SI Appendix, Fig. S5 C–F). IHC for the TA cell marker Ki67 indicated significant loss of TA cells by 3 d post-TAM in the Atg5Δ/Δ (Fig. 2D and SI Appendix, Fig. S5 G–I), Cag–Cre Atg5Δ/Δ, and Ubc–Cre Fip200Δ/Δ ileum (SI Appendix, Fig. S5 J–M) compared to the wild-type and Atg7Δ/Δ ileum. IHC for the stem cell marker Olfactomedin 4 (OLFM4) revealed almost complete loss of stem cells by 3 d post-TAM in the Atg5Δ/Δ (Fig. 2E and SI Appendix, Fig. S6 A–C), Cag–Cre Atg5Δ/Δ, and Ubc–Cre Fip200Δ/Δ ileum (SI Appendix, Fig. S6 D–G) compared to the wild-type and Atg7Δ/Δ ileum. IHC for lysozyme showed similar staining in the wild-type, Atg7Δ/Δ, and Atg5Δ/Δ duodenum and jejunum (SI Appendix, Fig. S6 H–J); however, the Atg5Δ/Δ and Cag–Cre Atg5Δ/Δ ileum displayed a diffuse staining pattern by 3 d post-TAM (Fig. 2F and SI Appendix, Fig. S6 K–M). Immunofluorescence (IF) for OLFM4 and p62 showed increased p62 aggregates colocalized with OLFM4 in the Atg5Δ/Δ duodenum and jejunum compared to Atg7Δ/Δ at all times, which was greatest in the ileum; in the Atg5Δ/Δ ileum, p62 aggregates increased in comparison to the Atg7Δ/Δ ileum at 2 d, but not at 3 d, post-TAM, due to complete loss of stem cells (Fig. 2G and SI Appendix, Fig. S6 N and O). Combined IF for OLFM4 and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay displayed increased TUNEL colocalization with OLFM4 in the Atg5Δ/Δ ileum and the Cag–Cre Atg5Δ/Δ ileum compared to the wild-type and Atg7Δ/Δ ileum (Fig. 2H and SI Appendix, Fig. S6P). Taken together, these findings suggested that ileum stem cells undergo apoptosis due to acute loss of autophagy and that autophagy impairment is greater upon systemic deletion of Atg5 than Atg7.
Fig. 2.
Ileum stem cells are lost in Atg5Δ/Δ mice. (A–F) Representative ileum IHC staining of p62 (A) with quantification (B), Alcian blue staining (C), IHC staining of Ki67 (D), OLFM4 (E), and lysozyme (F) with quantification at indicated times from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. Red arrows indicate p62 aggregates. (G and H) Representative ileum IF costaining of p62 (G) and TUNEL (H) with OLFM4 and quantification at indicated times from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. White arrows indicate p62 or TUNEL colocalized with OLFM4. All images represent one of three biological replicates. All quantification data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group. See also SI Appendix, Figs. S3–S6.
In Vitro Deletion of Atg5 Leads to a Reduction in Stem Cells That Is Rescued by Wnt Ligands.
We hypothesized that loss of stem cells was the main cause of ileum damage and loss of barrier function. The canonical WNT signaling pathway is mediated by Wnt ligands Wnt3a or Wnt2b secreted by the Paneth cells in the crypt using β-catenin as the essential signal transducer, which supports the renewal of intestinal stem cells (21–23). It was previously shown that loss of autophagy in the epithelium layer caused a defect in Paneth cells (13, 14). To test if loss of stem cells upon deletion of Atg5 was due to WNT signaling failure in the epithelium layer, we isolated crypts from the ileum of Ubc–Cre Atg5flox/flox and Ubc–Cre Atg7flox/flox adult mice, deleted Atg5 or Atg7 in vitro once organoids were formed, and then cultured them in standard medium or medium with WNT ligands Wnt3a or Wnt2b (SI Appendix, Fig. S7A). The Atg5Δ/Δ organoids showed rapid and complete loss of the ATG5–ATG12 complex, while the Atg7Δ/Δ organoids showed loss of ATG7 protein, yet maintained the ATG5–ATG12 complex. Both the Atg5Δ/Δ and Atg7Δ/Δ organoids had a higher ratio of LC3-I/LC3-II post deletion (SI Appendix, Fig. S7B). These results indicated that ATG5 loss in ileum organoids produced a more rapid and complete autophagy defect than ATG7 loss, consistent with the ileum in vivo.
When passaged for three generations after deletion, the viability and budding number of Atg5Δ/Δ organoids was decreased compared to Atg7Δ/Δ organoids, which was rescued by supplementation with Wnt3a or Wnt2b (SI Appendix, Figs. S7 C and D and S8 A–D). Real-time PCR for the stem cell marker Olfm4, Lgr5, and the Wnt target genes Axin2 and Sox9 revealed decreased transcription level of these genes in the Atg5Δ/Δ compared to the Atg5flox/flox organoids, which was restored by supplementation with Wnt3a or Wnt2b across three passages (SI Appendix, Figs. S7E and S8 E and F). At passages two and three, Western blot of the Atg5Δ/Δ organoids showed loss of ATG5, high LC3-I, and no LC3-II, whereas the Atg7Δ/Δ organoids had ATG7 protein with low LC3-I and increased LC3-II, suggesting that undeleted outcompeted deleted stem cells in Atg7Δ/Δ organoids (SI Appendix, Fig. S8 G and H). These data indicated that stem cell depletion in Atg5Δ/Δ organoids is at least partially caused by insufficient Wnt signaling in the epithelium layer and that there was incomplete deletion and autophagy loss in the Atg7Δ/Δ organoids.
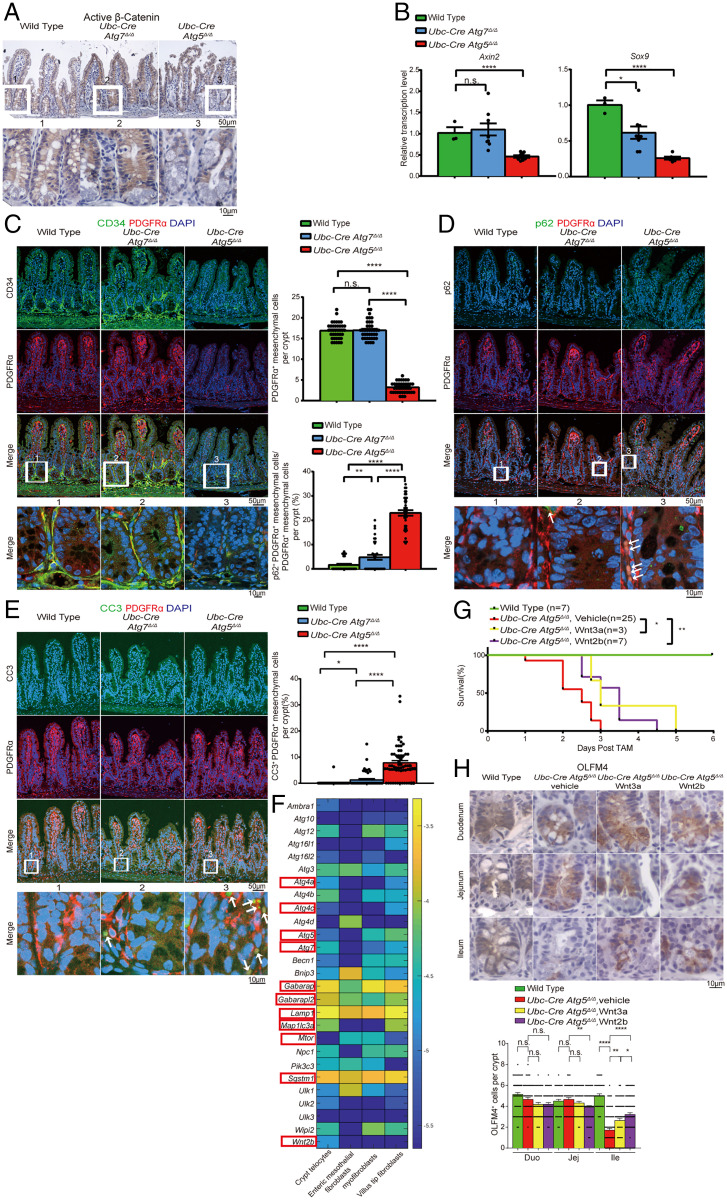
Atg5 Is Required to Maintain PMCs.
While reduced stem cell gene expression and OLFM4 staining in the Atg5Δ/Δ organoid cultures indicated an epithelial cell-autonomous role for Atg5, the stem cell loss in vivo was more pronounced than in vitro, suggesting that additional cell populations likely depend upon Atg5 function to support ileum health. We first tested whether ATG5-dependent WNT signaling was required for stem cell maintenance in vivo. IHC revealed significant loss of stabilized β-catenin (not phosphorylated on Ser33/37/Thr41) in the crypt of the Atg5Δ/Δ ileum, consistent with decreased Axin2 and Sox9 transcription compared with the Atg7Δ/Δ ileum at 3 d post-TAM, suggesting WNT signaling reduction consistent with loss of stem cells (Fig. 3 A and B and SI Appendix, Fig. S9 A and B). The same phenotype was observed in the Cag–Cre Atg5Δ/Δ ileum (SI Appendix, Fig. S9 C and D). Wnt ligands come from Paneth cells in the epithelium (24); however, Atg5 deletion using epithelium-specific Villin–Cre does not cause ileum stem cell loss (13), suggesting that ATG5-dependent Wnts originate outside the epithelium. PMCs, including telocytes, CD81− stromal cells, and CD81+ trophocytes, surround the intestinal epithelium and are important in maintaining WNT and bone morphogenic protein gradients for intestinal homeostasis. Among these three cell types, the CD81+ trophocytes are sufficient for intestinal stem cell maintenance in vitro (25–28). Notably, targeted ablation of a subset of these PMCs in mice using a Grem1–CreERT2 driver specifically compromises ileal stem cell function, but not in the duodenum and jejunum (28), reminiscent of the Atg5Δ/Δ phenotype described above. Therefore, we tested if Atg5 was required for the function and survival of PMCs.
Fig. 3.
Atg5 is required to maintain PMCs. (A) Representative ileum IHC staining of stabilized β-catenin at 3 d post-TAM from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. (B) qRT-PCR of Axin2 and Sox9 for the ileum from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. Data represent mean ± SEM. *P < 0.05; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group and represents one of three biological replicates. (C–E) Representative ileum IF costaining of CD34 and PDGFRα at 3 d post-TAM (C), p62 and PDGFRα (D), and CC3 and PDGFRα at 2 d post-TAM (E) with quantification from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. White arrows indicate colocalization of p62 or CC3 with PDGFRα. (F) Single-cell RNA-sequencing expression map of essential autophagy genes. Color bar is the log10 of the fraction of cellular messenger RNA (averaged over all the sequenced cells from the indicated cluster). Selected autophagy genes and Wnt2b are marked with red rectangles. (G and H) Kaplan–Meier survival curve (G) and representative intestine IHC of OLFM4 at 3 d post-TAM with quantification (H) from Atg5Δ/Δ and Atg5Δ/Δ mice supplemented with Wnt3a or Wnt2b. Wnt3a or Wnt2b was administered to the mice by intraperitoneal injection with an amount of 50 mg/kg per mouse, two injections per day starting from the last day of TAM injection. *P < 0.05; **P < 0.01 (log-rank test) in G. All images represent one of three biological replicates. All quantification data except in G represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group. See also SI Appendix, Figs. S9 and S10. Duo, duodenum; Ile, ileum; Jej, jejunum.
IF for PMC markers PDGFRα and CD34 indicated that they were present at 1 and 2 d in Atg5Δ/Δ and Atg7Δ/Δ ileum (SI Appendix, Fig. S9 E and F), but nearly completely lost at 3 d post-TAM in the Atg5Δ/Δ, but not in the Atg7Δ/Δ, ileum (Fig. 3C). PMCs were similarly lost in the Cag–Cre Atg5Δ/Δ, and not in the Cag–Cre Atg12Δ/Δ, ileum (SI Appendix, Fig. S9 G and H). Prior to their loss, PMCs displayed increased p62 and CC3, indicative of apoptosis in the Atg5Δ/Δ compared to the Atg7Δ/Δ ileum at 2 d, which was lost on 3 d post-TAM due to PMC loss (Fig. 3 D and E and SI Appendix, Figs. S9 I and J and S10 A–D). p62 and CC3 similarly accumulated in PMCs of Cag–Cre Atg5Δ/Δ, which was less in the Cag–Cre Atg12Δ/Δ ileum (SI Appendix, Fig. S10 E–L). Greater p62 accumulation in the Atg5Δ/Δ ileum PMCs was consistent with more rapid and complete loss of autophagy function. Single-cell RNA sequencing of PMCs revealed considerable expression level of several essential autophagy genes and Wnt2b in the crypt and villus. These included genes involved in the autophagosome assembly (Atg4a, Atg4c, Atg5, and Atg7), cargo adapters and receptors (Gabarap and Gabarap2, Map1lc3a [LC3-I], and Sqstm1 [p62]), and lysosomes (Lamp1) indicative of active autophagy function in PMCs (Fig. 3F and SI Appendix, Table S1) (29, 30). Crypt PMCs showed higher expression level of Wnt2b compared to other types of PMCs, indicative of their potential role in Wnt signaling (Fig. 3F and SI Appendix, Table S1) (29, 30). These data suggested that PMCs may be autophagy-dependent.
We then tested if Wnt ligand supplementation could compensate for the loss of PMCs in the Atg5Δ/Δ ileum. Atg5Δ/Δ mice supplemented with Wnt3a or Wnt2b showed a 24-h extension of lifespan (Fig. 3G). There was increased OLFM4 in the Atg5Δ/Δ ileum with supplementation of Wnt3a and Wnt2b compared to the untreated Atg5Δ/Δ ileum, but not to the extent of in the wild-type ileum, suggesting that Wnt ligand supplementation partially rescued the stem cells, but cannot rescue survival of mice due to insufficient Wnt restoration or other potential defects caused by loss of autophagy (Fig. 3H and SI Appendix, Fig. S10M). These data indicated that PMCs require autophagy to survive and that they are essential for maintaining intestinal stem cells by providing Wnt ligands.
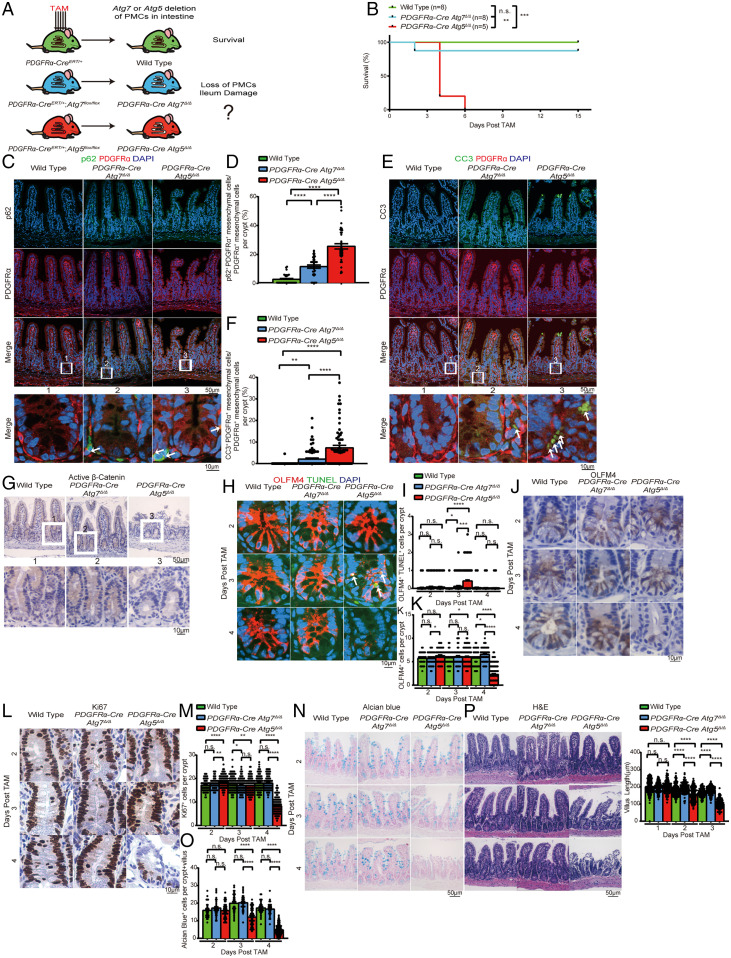
Deletion of Atg5 in PMCs Causes Loss of Ileum Stem Cells.
We then sought to investigate if loss of ileum function was specifically caused by loss of Atg5 in PMCs. To test this, we generated PDGFRα–CreERT/+;Atg7flox/flox and PDGFRα–CreERT/+;Atg5flox/flox mice and deleted Atg7 or Atg5 specifically in the PMCs (Fig. 4A). Most PDGFRα–Cre Atg5Δ/Δ mice died within 6 d post-TAM, while wild-type and PDGFRα–Cre Atg7Δ/Δ mice survived (Fig. 4B). Increased p62 aggregates with increased CC3 was first observed in PMCs from the PDGFRα–Cre Atg5Δ/Δ ileum at 3 d, preceding the complete loss of these cells at 4 d post-TAM, indicating that autophagy blockage caused loss of PMCs in the PDGFRα–Cre Atg5Δ/Δ ileum via apoptosis (Fig. 4 C–F and SI Appendix, Fig. S11 A–H). Combined IF for OLFM4 and TUNEL assay in the PDGFRα–Cre Atg5Δ/Δ ileum revealed increased TUNEL in stem cells at 3 d, following loss of active β-catenin and OLFM4 at day 4, suggesting loss of WNT signaling and stem cells via apoptosis (Fig. 4 G–K and SI Appendix, Fig. S11I). Ki67 staining was largely decreased in the PDGFRα–Cre Atg5Δ/Δ ileum at 4 d post-TAM, suggesting loss of TA cells (Fig. 4 L and M and SI Appendix, Fig. S11J). Alcian blue staining of the PDGFRα–Cre Atg5Δ/Δ ileum was completely lost at 4 d post-TAM, indicating loss of goblet cells (Fig. 4 N and O). IHC for lysozyme revealed normal and no diffuse staining pattern in the PDGFRα–Cre Atg5Δ/Δ ileum, suggesting unaffected Paneth cells (SI Appendix, Fig. S11K). The PDGFRα–Cre Atg5Δ/Δ ileum also showed similar epithelial damage as the Atg5Δ/Δ ileum at 4 d post-TAM, with no effect on the duodenum and jejunum (Fig. 4P and SI Appendix, Fig. S11 L–N). These data indicated that the ileum-damage phenotype in Atg5Δ/Δ mice can largely be attributed to loss of PMCs.
Fig. 4.
Deletion of Atg5 in PMCs causes loss of ileum stem cells. (A) Experimental design for generation of PDGFRα–CreERT/+ Atg5Δ/Δ and Atg7Δ/Δ mice. (B) Kaplan–Meier survival curve of wild-type, PDGFRα–Cre Atg7Δ/Δ, and PDGFRα–Cre Atg5Δ/Δ mice. **P< 0.01, ***P < 0.001; n.s., not significant (log-rank test). (C–F) Representative ileum IF costaining of p62 and PDGFRα (C and D) and costaining of CC3 and PDGFRα (E and F) with quantification at 3 d post-TAM from wild-type, PDGFRα–Cre Atg7Δ/Δ, and PDGFRα–Cre Atg5Δ/Δ mice. White arrows indicate colocalization of p62 or CC3 with PDGFRα. (G–P) Representative ileum IHC staining of stabilized β-catenin (G) at 4 d post-TAM, costaining of OLFM4 and TUNEL (H and I), OLFM4 (J and K), Ki67 (L and M), Alcian blue staining (N and O), and H&E histology with villus length quantification (P) at the indicated times from wild-type, PDGFRα–Cre Atg7Δ/Δ, and PDGFRα–Cre Atg5Δ/Δ mice. All images represent one of three biological replicates. All quantification data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group. See also SI Appendix, Fig. S11.
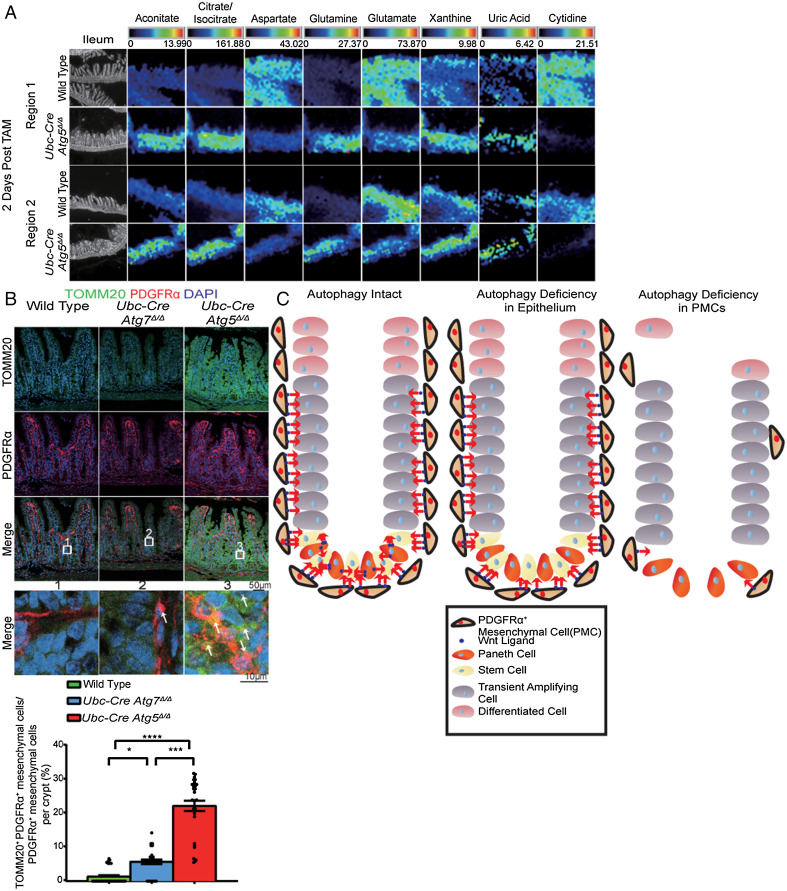
Autophagy Maintains Ileum Aspartate and Nucleotide Pools.
We then sought to test why autophagy was required for the maintenance of PMCs. Autophagy deficiency limits tumor growth in part by activating p53 and apoptosis in lung, breast, and pancreatic cancer, and deletion of p53 partly restores tumor growth (16, 18). Moreover, p53 deficiency delays death from neurodegeneration and allows survival to fasting in Atg7Δ/Δ mice (12). To test if p53 activation was responsible for ileum damage, we generated Ubc–CreERT2/+;Atg5flox/flox;Trp53flox/flox mice, and TAM was administered to generate Atg5Δ/Δp53Δ/Δ mice (SI Appendix, Fig. S12A). Whole-body codeletion of p53 with Atg5 did not rescue the survival of Atg5Δ/Δ mice, indicating that p53 activation did not cause ileal PMC or stem cell loss (SI Appendix, Fig. S12B). Autophagy is also required for cellular metabolism and mitochondrial function essential for cell and mammalian survival (9, 17, 31). Thus, we tested whether autophagy is required to maintain PMC metabolism.
To assess the metabolic consequences of ATG5 loss in PMCs and the ileum, we performed matrix-assisted laser desorption ionization (MALDI) coupled to mass spectrometry imaging (MALDI-MSI) to distinguish between a specific metabolic defect in PMCs and an effect on the entire ileum. PMCs were localized by IF, and the metabolic profile of the ileum was overlaid. Nine out of 164 putative metabolites (hereafter metabolites) showed dramatic differences in relative concentrations between the wild-type and Atg5Δ/Δ ileum. TCA-cycle intermediates aconitate and citrate/isocitrate were elevated in Atg5Δ/Δ compared to wild type, suggesting alteration of the TCA cycle. The Atg5Δ/Δ ileum displayed decreased aspartate and glutamate, but elevated glutamine, which are all TCA-cycle-derived amino acids, suggesting dysfunction of mitochondria as previously reported (17, 32). The Atg5Δ/Δ ileum showed increased xanthine and uric acid and decreased cytidine compared to wild type, indicative of increased purine and pyrimidine degradation (Fig. 5A and SI Appendix, Fig. S12 C and D) (33). The Atg5Δ/Δ ileum also had elevated glucose-1 phosphate, suggesting higher glycogen consumption (SI Appendix, Fig. S12E) (33). These metabolite changes were reflected broadly in the ileum and were not PMC-specific, suggesting a global effect on the ileum that may be related to mitochondria dysfunction caused by autophagy deficiency (17). Elevated mitochondria numbers indicated by increased TOMM20 staining in PMCs in the Atg5Δ/Δ ileum may also contribute to altered levels of mitochondrial metabolites (Fig. 5B and SI Appendix, Fig. S12F). Thus, autophagy is critical for the metabolism of the ileum generally, and PMCs may be selectively sensitive to these metabolic alterations, which may compromise PMC survival and thereby WNT signaling required for stem cell survival and intestinal homeostasis (Fig. 5C).
Fig. 5.
Autophagy maintains ileum aspartate and nucleotide pools. (A) MALDI-MSI data from the wild-type and Atg5Δ/Δ ileum at 2 d post-TAM from two different regions. The scale bar on the top of each image set represents the intensity range of ions detected for each metabolite. The white pixels represent the PMCs. (B) Representative ileum IF costaining of TOMM20 and PDGFRα with quantification at 2 d post-TAM from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. All images represent one of three biological replicates. (C) Mechanism by which autophagy is required for PMC survival to maintain intestinal homeostasis. All images represent one of three biological replicates. All quantification data were a combination of three biological replicates and represent mean ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n > 3 mice per group. See also SI Appendix, Fig. S12.
Gradual Atg5 Deletion Rescues Ileum Function, Prolongs Survival, and Causes Death from Neurodegeneration.
Systemic deletion of Atg5 causes more rapid and complete loss of autophagy in the ileum than that of Atg7. This raised the possibility that gradual loss of autophagy may allow adaptation and survival. We tested if administering TAM to Ubc–Cre Atg5flox/flox mice once per week for 4 wk (slow deletion) instead of for five consecutive days (fast deletion) to delete Atg5 more gradually would allow adaptation and rescue the lethality of Atg5Δ/Δ mice (Fig. 6A). Atg5Δ/Δ mice generated by slow deletion survived 3 mo post-TAM, more comparable to Atg7Δ/Δ mice generated by slow or fast deletion (Fig. 6B). Both Atg5Δ/Δ and Atg7Δ/Δ mice generated by slow deletion revealed limited tissue damage at 3 and 14 d post-TAM; had liver damage, muscle wasting, and loss of WAT, pyramidal neurons, and Purkinje cells at late times; and they died of neurodegeneration, similar to Atg7Δ/Δ mice generated by fast deletion (SI Appendix, Fig. S13 A–D) (9). Loss of Atg5 or Atg7 in the intestine was efficient following slow deletion, but did not affect the normal intestine architecture (Fig. 6C and SI Appendix, Fig. S13 E–G). Loss of the ATG5–ATG12 complex was evident at 3 d post-TAM in the Atg5Δ/Δ intestine by slow deletion, which was persistent in the Atg7Δ/Δ intestine. The Atg5Δ/Δ intestine displayed accumulation of LC3-I, decreased levels of LC3-II, and increased p62 aggregates compared to the Atg7Δ/Δ intestine generated by slow deletion, indicating more rapid loss of autophagy (Fig. 6 D and E and SI Appendix, Fig. S13 H and I). Ileum stem cells and PMCs were retained in Atg5Δ/Δ mice generated by slow deletion (Fig. 6 F–H). There was no decrease in the relative transcriptional level of Axin2 and less than twofold change of Sox9, suggesting intact WNT signaling (SI Appendix, Fig. S13J). Decreased p62 aggregates were observed in the Atg5Δ/Δ and Atg7Δ/Δ PMCs generated by slow deletion compared to the Atg5Δ/Δ PMCs generated by fast deletion, indicating intact autophagy in most of the PMCs (Figs. 3D and 6 I and J). Taken together, Atg5Δ/Δ mice generated by slow deletion overcame the deleterious ileum phenotype and resembled Atg7Δ/Δ mice due to more gradual loss and retention of some autophagy function.
Fig. 6.
Gradual Atg5 deletion rescues ileum function, prolongs survival, and causes death from neurodegeneration. (A) Experimental design for generation of Atg7Δ/Δ and Atg5Δ/Δ mice by slow deletion. (B) Kaplan–Meier survival curve of TAM-treated wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice. *P < 0.05; ****P < 0.0001 (log-rank test). (C) Representative ileum H&E histology from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice by slow deletion at the indicated times. (D–F) Western blotting for ATG5, ATG7, and LC3 (D) and representative ileum IHC staining of p62 (E) and OLFM4 (F) with quantification at 3 d post-TAM from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice by slow deletion. Red arrows indicate p62 aggregates. (G–J) Representative ileum IF costaining of CD34 and PDGFRα with quantification (G and H) and costaining of p62 and PDGFRα with quantification (I and J) at 3 d post-TAM from wild-type, Atg7Δ/Δ, and Atg5Δ/Δ mice by slow deletion. White arrows indicate colocalization of p62 with PDGFRα, and we magnified the crypt region to focus on PMCs, which are adjacent to stem cells. All images represent one of three biological replicates. All quantification data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired t test). n ≥ 3 mice per group. All the time points were counted from the fourth injection of TAM (day 0). See also SI Appendix, Fig. S13.
Discussion
Despite the known association of autophagy dysfunction with IBDs, mice with Atg5, Atg16L1, or Atg7 deficiency specifically in the intestine epithelium induced by Villin–Cre have a granule abnormality in Paneth cells, but normal intestine structure, and mice survive (13, 14). These findings indicate that autophagy in the epithelial compartment is not essential for ileum function. Moreover, conditional deletion of Atg7 or Atg12 does not cause loss of ileum function, as seen with conditional deletion of Atg5 or Fip200. There are several possible, nonmutually exclusive explanations to reconcile these findings. First, loss of Atg5 or Fip200 may act differently from loss of Atg7 or Atg12; second, some tissues may have a capacity to adapt to autophagy loss; and third, autophagy in a compartment outside of the epithelium may be essential for ileum stem cell survival.
First, why does conditional loss of Atg5 or Fip200 act differently from loss of Atg7 or Atg12? While Atg5 may have an autophagy-independent function, this is unlikely, as deletion of another Atg gene, Fip200, produces a similar phenotype. Alternatively, ileum stem cell and PMC loss in Atg5Δ/Δ, but not Atg7Δ/Δ or Atg12Δ/Δ, mice by fast deletion may instead be due to different kinetics of autophagy loss. Atg5Δ/Δ mice displayed more rapid loss of the ATG5–ATG12 complex in the ileum and greater p62 accumulation in the ileum and ileum PMCs compared to Atg7Δ/Δ and Atg12Δ/Δ mice, indicating more rapid loss of autophagy. Upon slow deletion, however, Atg5Δ/Δ PMCs had similar p62 accumulation compared to Atg7Δ/Δ PMCs. Therefore, slow deletion might allow activation of a protective mechanism in PMCs to compensate for loss of autophagy. Similarly, Atg5−/− mice with transgenic ATG5 expression in neurons to rescue neonatal lethality displayed shorter and wider villi and deeper crypts that did not affect mouse survival where adaptation may occur (34). One important compensatory protective adaptation mechanism to autophagy loss is activation of the master regulator of antioxidant defense nuclear factor erythroid 2-related factor 2 (NRF2). NRF2 is activated by autophagy deficiency, as the resulting accumulation of p62 inhibits the NRF2 inhibitor Kelch-like ECH-associated protein 1, thereby activating NRF2 and antioxidant defense (35–38). Turning on NRF2 enables survival of autophagy-deficient cell lines in vitro (38), and Atg7 deficiency is synthetically lethal in the absence of Nrf2 in vivo due to intestine damage (12). With more rapid loss of autophagy upon deletion of Atg5, but not Atg7, there may be insufficient time to activate NRF2 to protect PMCs from cell death.
Another compensatory mechanism is repopulation of stem cells with rare undeleted cells. The slower autophagy loss upon deletion of Atg7 may enable compensatory competition of rare undeleted stem cells in Atg7Δ/Δ organoids and mice, which is not possible with Atg5 deletion. Indeed, Atg7-deleted organoids showed more undeleted cells than Atg5-deleted organoids, explaining their improved growth. Slow deletion of Atg5 may permit establishment of these adaptation mechanisms that occur with deletion of Atg7. Although it is still possible that Atg5 has an autophagy-independent function that distinguishes it from Atg7, Atg5Δ/Δ mice generated by slow deletion allow PMC survival, and the mice later die of neurodegeneration, similar to Atg7Δ/Δ mice, indicating that they likely both are functioning in a canonical autophagy pathway.
As to whether autophagy in a compartment outside of the epithelium may be essential for ileum stem cell survival, it is clear that autophagy deficiency specifically in the intestine epithelial layer using Villin–Cre in mice is not lethal (13, 15, 39, 40), unlike the conditional whole-body Atg5 or Fip200 deletion reported here. By blocking autophagy function systemically beyond the epithelium, notably in PMCs that are not affected in the Villin–Cre mouse models, we revealed the critical role for autophagy in PMCs in maintaining ileum homeostasis. In Villin–Cre mouse models, the intestinal stem cells can still receive Wnt ligands from PMCs with intact autophagy, maintaining their renewal and allowing mice to survive without exogenous stress (39). Deleting Atg5 specifically in PMCs rapidly eliminated them along with ileum stem cells, which was independent of p53. As such, intestinal homeostasis cannot be maintained, which causes mice to die of hypoglycemia and loss of intestinal barrier function. Therefore, Atg5 is essential for PMC-dependent Wnt signaling and the survival of ileum stem cells, and the epithelial and mesenchymal layer were equally important in providing Wnt ligands to the intestinal stem cells (Fig. 5C). Interestingly, we did not see these phenotypes in the duodenum and jejunum, which may be explained by the more rapid loss of autophagy in the ileum in comparison to the duodenum and jejunum. The ileum appears more sensitive to loss of PMCs, which was also observed in Grem1Δ/Δ PMC-deficient GEMMs (28).
Autophagy is important for maintaining cellular metabolism and the pool size of TCA-cycle intermediates, amino acids, and nucleotides (9, 17, 41). As demonstrated by [U13 C5]-glutamine tracing in cancer cells in vitro, autophagy-mediated recycling also provides substrates to sustain TCA-cycle turning, enabling de novo aspartate and nucleotide synthesis (17). Using MALDI-MSI, we identified nine metabolites significantly different between the wild-type and Atg5Δ/Δ ileum in vivo, and aspartate may be the key metabolite for PMC survival. Aspartate is a TCA-cycle-derived nonessential amino acid required for cell proliferation and growth (42, 43). Accumulation of mitochondria in Atg5Δ/Δ PMCs suggested that mitochondrial dysfunction may cause decreased aspartate. Aspartate donates its amino group to inosine monophosphate to make adenosine monophosphate for purine synthesis and is a direct component of the pyrimidine ring for pyrimidine synthesis. The entire Atg5Δ/Δ ileum, including PMCs, has a marked deficit in aspartate, consistent with decreased nucleotide pools. Moreover, concurrent energy crisis due to impaired mitochondrial function may force degradation of nucleotides reflected by increased xanthine and uric acid, compounding the decrease in nucleotide pools and further compromising survival, similar to what occurs in autophagy-deficient cancer cell lines (17).
IBDs are complex diseases that start with chronic relapsing and remitting intestinal inflammation, culminating in tissue fibrosis. IBDs are usually driven by a combination of genetic, environmental, and microbiota factors in the intestine or colon (44, 45). Several genetic variants related to autophagy defects have been identified by genome-wide association studies to be linked to IBD onset. The most outstanding one is Atg16L1, which is a direct component important for autophagosome assembly. The T300A polymorphism in Atg16L1 is a risk allele in Crohn’s disease patients, which display granule abnormality in Paneth cells, as in Villin–Cre Atg16L1-deficient mice (13). This polymorphism also causes morphological defects in goblet cells and increased production of cytokine IL-1β (46). Atg16L1 polymorphism causes failure of intestinal sensing of protective signals from the microbiome, which is a critical gene-environment etiology for development of IBD (47). Another IBD susceptibility gene is Nod2, which works together with Atg16L1 as a pattern-recognition receptor involved in the integrity of intestinal immunity (48–50). Moreover, the autophagy initiator ULK1 can directly phosphorylate wild-type ATG16L1 to promote xenophagy upon infection, which is destabilized by the ATG16L1 T300A polymorphism (51). Importantly, PMCs are significantly decreased as IBD progresses to an advanced stage in patient samples from both Crohn’s disease and ulcerative colitis (52, 53). Since we identified a critical role for autophagy in PMC survival, this raises the possibility that decreased autophagy in PMCs in IBD results in failure of WNT signaling and stem cell survival, contributing to the manifestation of disease. As such, our findings represent a possible mechanism for IBD development, and restoration of autophagy, PMCs, or activation of WNT signaling in IBD patients might be potential therapies.
Materials and Methods
See SI Appendix for details.
All animal care and treatments were carried out in compliance with Rutgers University and University of California, San Francisco Institutional Animal Care and Use Committee guidelines.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 CA130893, CA188096, and CA163591 and the Ludwig Princeton Branch of the Ludwig Institute for Cancer Research at Princeton University (to E.W.) and NIH F31 Fellowship F31CA217015 (to T.M.). Services, results, and/or products in support of the research project were generated by the Rutgers Cancer Institute of New Jersey Biospecimen Repository and Histopathology Service Shared Resource, supported, in part, with funding from National Cancer Institute (NCI) Cancer Center Support Grant (CCSG) P30CA072720-5919. Similarly, the Metabolomics Shared Research received support from NCI CCSG P30CA072720-5923. S.D. was supported by a New Investigator Award (NCI CCSG P30CA072720-5931). M.G. was supported by NCI T32 Grant CA257957. We also thank Dr. Shalev Itzkovitz (Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel) for contributing the single-cell RNA-sequencing data.
Footnotes
Reviewers: K.R., Cancer Research UK Beatson Institute; and A.T., ANSCHUTZ Cancer Center.
Competing interest statement: E.W. owns stock in Forma Therapeutics and is a founder of Vescor LLC, neither of which has anything to do with this manuscript. Note that K.R. was a coauthor on a 2021 Autophagy Guidelines paper with hundreds of others in the field, including several coauthors here.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202016119/-/DCSupplemental.
Data Availability
All raw data will be made available upon publication. All the MALDI-MSI data were uploaded to the Proteomics Identifications Database (https://www.ebi.ac.uk/pride/, accession code PXD023428). Previously published data used for this work are in the Gene Expression Omnibus database (accession code GSE134479). All other relevant data are included in the main text and/or SI Appendix.
References
- 1.Kaur J., Debnath J., Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Johansen T., Lamark T., Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto K., Organellophagy: Eliminating cellular building blocks via selective autophagy. J. Cell Biol. 205, 435–445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wild P., et al. , Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabinowitz J. D., White E., Autophagy and metabolism. Science 330, 1344–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N., Kuma A., Autophagosomes in GFP-LC3 transgenic mice. Methods Mol. Biol. 445, 119–124 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Karsli-Uzunbas G., et al. , Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara T., et al. , Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Komatsu M., et al. , Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y., et al. , Autophagy promotes mammalian survival by suppressing oxidative stress and p53. Genes Dev. 34, 688–700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadwell K., et al. , A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittkopf N., et al. , Lack of intestinal epithelial Atg7 affects Paneth cell granule formation but does not compromise immune homeostasis in the gut. Clin. Dev. Immunol. 2012, 278059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trentesaux C., et al. , Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc. Natl. Acad. Sci. U.S.A. 117, 11136–11146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poillet-Perez L., White E., Role of tumor and host autophagy in cancer metabolism. Genes Dev. 33, 610–619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J. Y., et al. , Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 30, 1704–1717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimmelman A. C., White E., Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhotra R., Warne J. P., Salas E., Xu A. W., Debnath J., Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11, 145–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barker N., Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Valenta T., et al. , Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Clevers H., Loh K. M., Nusse R., Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Fevr T., Robine S., Louvard D., Huelsken J., Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farin H. F., Van Es J. H., Clevers H., Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Shoshkes-Carmel M., et al. , Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stzepourginski I., et al. , CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. U.S.A. 114, E506–E513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greicius G., et al. , PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 115, E3173–E3181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy N., et al. , Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 26, 391–402.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halpern K. B., et al., Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat. Commun. 11, 1936 (2020). [DOI] [PMC free article] [PubMed]
- 30.Halpern K. B., et al., Lgr5+ telocytes are a signaling hub at the intestinal villus tip. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134479. Deposited 5 February 2020. [Google Scholar]
- 31.Guo J. Y., et al. , Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 27, 1447–1461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain I. H., et al. , Leigh syndrome mouse model can be rescued by interventions that normalize brain hyperoxia, but not HIF activation. Cell Metab. 30, 824–832.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., White E., Autophagy in PDGFRα+ Mesenchymal Cells is Essential for Intestinal Stem Cell Survival: MALDI data, Proteomics Identifications Database (https://www.ebi.ac.uk/pride/) with accession code PXD023428, Deposited 6 January 2021. [Google Scholar]
- 34.Yoshii S. R., et al. , Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev. Cell 39, 116–130 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Lau A., et al. , A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol. Cell. Biol. 30, 3275–3285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T., et al. , p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 7, 12030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strohecker A. M., et al. , Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3, 1272–1285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towers C. G., et al. , Cancer cells upregulate NRF2 signaling to adapt to autophagy inhibition. Dev. Cell 50, 690–703.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asano J., et al. , Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep. 20, 1050–1060 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Matsuzawa-Ishimoto Y., et al. , Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214, 3687–3705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poillet-Perez L., et al. , Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birsoy K., et al. , An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan L. B., et al. , Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieder F., Fiocchi C., Intestinal fibrosis in inflammatory bowel disease—Current knowledge and future perspectives. J. Crohn’s Colitis 2, 279–290 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Graham D. B., Xavier R. J., Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lassen K. G., et al. , Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl. Acad. Sci. U.S.A. 111, 7741–7746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu H., et al. , Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooney R., et al. , NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Travassos L. H., et al. , Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Homer C. R., Richmond A. L., Rebert N. A., Achkar J. P., McDonald C., ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 139, 1630–1641, 1641.e1-2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alsaadi R. M., et al. , ULK1-mediated phosphorylation of ATG16L1 promotes xenophagy, but destabilizes the ATG16L1 Crohn’s mutant. EMBO Rep. 20, e46885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milia A. F., et al. , Telocytes in Crohn’s disease. J. Cell. Mol. Med. 17, 1525–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manetti M., Rosa I., Messerini L., Ibba-Manneschi L., Telocytes are reduced during fibrotic remodelling of the colonic wall in ulcerative colitis. J. Cell. Mol. Med. 19, 62–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data will be made available upon publication. All the MALDI-MSI data were uploaded to the Proteomics Identifications Database (https://www.ebi.ac.uk/pride/, accession code PXD023428). Previously published data used for this work are in the Gene Expression Omnibus database (accession code GSE134479). All other relevant data are included in the main text and/or SI Appendix.