Abstract
Aims
Fresh produce is often a vehicle for the transmission of foodborne pathogens such as human norovirus. Thus, it is recommended to wash the surface of produce before consumption, and one of the most common ways to wash produce is by rinsing under running tap water. This study determined the effectiveness of removal of human coronavirus‐OC43 (HCoV‐OC43), as a surrogate for severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) and murine norovirus‐1 (MNV‐1), as a surrogate for human norovirus, from contaminated lettuce, apples and cucumbers.
Methods and Results
The produce surfaces were artificially inoculated in conjunction with faecal material to represent natural contamination. Rinsing under tap water for 10 s at 40 ml/s removed 1.94 ± 0.44, 1.42 ± 0.00 and 1.42 ± 0.42 log of HCoV‐OC43 from apple, cucumber and lettuce respectively. The same washing technique removed 1.77 ± 0.17, 1.42 ± 0.07 and 1.79 ± 0.14 log of MNV‐1 from apple, cucumber and lettuce respectively. This washing technique was effective at reducing a significant amount of viral contamination, however, it was not enough to eliminate the entire contamination. There was no significant difference in the reduction of viral load between the two viruses, nor between the three surfaces tested in this study.
Conclusions
Our data suggest that washing under tap water would be an efficient way of reducing the risk of foodborne viral transmission only if the level of contamination is less than 2 log PFU.
Significance and Impact of Study
This study demonstrates that running tap water was effective at reducing the amount of infectious HCoV‐OC43 and MNV on produce surfaces, and washing produce continues to be an important task to perform prior to consumption to avoid infection by foodborne viruses, particularly for foods which are eaten raw.
Keywords: human coronavirus OC43, murine norovirus‐1, produce, tap water, wash
INTRODUCTION
The Coronaviridae family are positive‐sense single‐stranded RNA (+ssRNA) enveloped viruses and are divided into one of the four genera: alpha‐, beta‐, gamma‐ and delta‐coronaviruses (Chen, et al. 2020). Human coronaviruses (HCoV) are found only in the first two of these genera. Alpha‐coronavirus includes HCoV‐229E and HCoV‐NL63, both of which cause mild common cold‐like infections (Leao, et al. 2020). Beta‐coronavirus includes HCoV‐OC43 and HCoV‐HKU1, which also cause mild common cold‐like infections, but also includes the two pathogenic HCoVs, Middle‐East respiratory syndrome coronavirus (MERS‐CoV) and severe acute respiratory syndrome coronavirus‐1 (SARS‐CoV‐1), as well as the pandemic causing SARS‐CoV‐2 (Leao, et al. 2020).
The main routes of transmission for SARS‐CoV‐2 have widely been accepted to be direct person‐to‐person contact and via aerosolized respiratory droplets (Falahi and Kenarkoohi. 2020, Meyerowitz, et al. 2021). Secondary modes of transmission may exist, although are not as well established or proven. One of these secondary methods is via fomites (Kraay, et al. 2021). Fomite‐mediated transmission has been shown for other human viruses such as norovirus (Tuladhar, et al. 2013), rotavirus (Chia, et al. 2018) and enveloped respiratory viruses, notably influenza (Zhang and Li. 2018, Liu, et al. 2020). Nevertheless, fomite‐mediated transmission of SARS‐CoV‐2, where respiratory transmission could be ruled out, remains controversial (Goldman, 2020). To date, there are a few studies indicating the potential of fomite‐mediated transmission of COVID‐19. In one study, SARS‐CoV‐2 was able to transfer from surfaces to artificial skin (Behzadinasab, et al. 2021). Studies from China, based upon the epidemiological data, also predicted that cases of COVID‐19 were due to SARS‐CoV‐2 transmission from contaminated surfaces (Xie, et al. 2020, Cai, et al. 2020).
Various pieces of evidence have shown that SARS‐CoV‐2 is able to infect the gastrointestinal (GI) tract and be shed in faeces, and potentially even transmitted via a faecal‐oral route (Dergham, et al. 2021). Diarrhoea and vomiting, symptoms typically associated with GI infections are also common in COVID‐19 patients, potentially occurring in between 20 and 35% of all cases (Joshi, et al. 2021). As a result, levels of SARS‐CoV‐2 RNA in wastewater can be used as a way to predict increases in cases of COVID‐19 (Cheung, et al. 2020, Randazzo, et al. 2020). Faecal organic material has also been shown to aid in the transfer of HCoVs from artificially contaminated gloved hands to produce and surfaces (Dallner, et al. 2021). Once contaminated onto produce surfaces, HCoVs also demonstrated an ability to survive for up to 3 days (Blondin‐Brosseau et al. 2021).
While foodborne transmission of SARS‐CoV‐2 has not been shown, and is not predicted to be a likely cause of transmission (Rose‐Martel, et al. 2021), food surfaces and food packaging may be able to act as fomites (O'Brien, et al. 2020). In high‐activity areas where food is present, such as in grocery stores and restaurants, there is a possibility that SARS‐CoV‐2 from an infected individual or a food handler could be deposited onto food surfaces. Due to mask‐wearing mandates being present in many countries across the world at the height of the pandemic (Felter and Bussemaker. 2020), direct respiratory droplet contamination from the nose and mouth is likely to be reduced (Bandiera, et al. 2020). However, contaminated hands are another likely source of contamination. Hands can become contaminated by respiratory droplets or faecal material and deposit the virus upon touching surfaces (Kraay, et al. 2021). In fact, fomite‐mediated transmission is well‐established for several other human pathogens including human norovirus and rhinoviruses (Kraay, et al. 2018).
Faecal contamination of fresh produce is one of the main reasons for many foodborne illnesses including norovirus outbreaks (Verhaelen, et al. 2013, Nasheri, et al. 2019). Thus, interventions that kill or remove human pathogens on fresh produce have been recommended and employed. Washing produce by rinsing under running tap water remains the most common household pathogen reduction practice, and is recommended by the U.S. Food and Drug Administration (FDA) (FDA. 2018) as well as Health Canada (Health Canada 2021). Rinsing is preferred over soaking, because soaking can lead to the spread and internalization of certain foodborne pathogens on the produce (Gomez, et al. 2021).
This study aimed to address whether rinsing with water is sufficient in removing HCoVs, and norovirus from produce surfaces when contaminated in conjunction with faecal organic material, representative of faecal shedding. As working with SARS‐CoV‐2 requires a biosafety level 3 facility, this work was conducted using HCoV‐OC43, a surrogate for SARS‐CoV‐2, located in the same genera (Chen, et al. 2020), which was chosen due to similar physiochemical properties (Liu, et al. 2021, Warnes, et al. 2015), receptor binding proteins (Cueno and Imai. 2021), cross‐reactive antibodies (Patrick, et al. 2006) and genetic sequences. Murine norovirus‐1 (MNV‐1), which has similar physiochemical, pathogenic and genetic properties to human norovirus (Kniel. 2014), was also included in this study, as a surrogate for human norovirus, to compare enveloped viruses with nonenveloped viruses, and to include a virus that is known to spread faecal‐orally (Cannon, et al. 2006).
MATERIALS AND METHODS
Cell lines and viruses
The MRC‐5 cell line, human lung fibroblast cells (ATCC#CCL‐171), were obtained from the American Type Culture Collection (ATCC). These cells were grown in complete Eagle's minimum essential medium (MEM) (Gibco‐Invitrogen Co., Grand Island, NY) supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (FBS) (Gibco‐Invitrogen), 1% nonessential amino acids (Gibco‐Invitrogen), 1% GlutaMax‐1 (Gibco‐Invitrogen), 500 μg/ml penicillin/streptomycin (Gibco‐Invitrogen) and 0.22% (w/v) of sodium bicarbonate (Sigma Aldrich Canada). The BV‐2 cell line, mouse microglial cells, were obtained courtesy of Dr. Christiane Wobus (University of Michigan). These cells were maintained in complete Dulbecco's modified Eagle medium (DMEM) (Gibco‐Invitrogen) supplemented with 10% (w/v) heat‐inactivated FBS, 500 μg/ml penicillin/streptomycin and 0.22% (w/v) of sodium bicarbonate. Both cell lines were grown at 37°C and 5% CO2 and were split 1:2 or 1:3 every 2 days using 0.05% Trypsin–EDTA (Gibco‐Invitrogen).
Two viruses were used in this study; HCoV‐OC43 (ATCC#VR‐1558) obtained from ATCC and MNV‐1 obtained courtesy of Dr. Herbert Virgin (Washington University School of Medicine, St. Louis, MO). The initial stocks of virus obtained were used in subsequent infections of their respective host cells, MRC‐5 for HCoV‐OC43 and BV‐2 for MNV‐1, to establish a working stock of virus for further experimentation.
Surface preparation and wash treatment
Three produce types were used in this study. Gala apples, English cucumbers and romaine lettuce were obtained from local grocery stores in Ottawa, ON. Surfaces were prepared, six samples per each produce type, by cleaning with a Kim‐wipe to remove any dust, washing with tap water, drying and then disinfecting the surface with 70% ethanol before allowing the surface to air dry in a biosafety cabinet to remove any residual ethanol. A 5 × 5 cm2 surface was then demarcated on the produce surface using tape.
A 1 × 106 median tissue culture infectious dose (TCID)50/ml stock of either HCoV‐OC43 or MNV‐1 was diluted to approximately 1 × 105 PFU/ml a 10% (w/v) mix of faecal material in dH2O, obtained from a healthy donor. The next step was to transfer 100 μl of this dilution onto the demarcated area of the produce surface, spread it using a pipette tip and allow it to dry for approximately 30 min. Afterwards, three of the six samples were washed under running tap water (Ottawa, ON) at 24°C for 10 s at a flow rate of 40 ml/s. The other three samples were unwashed controls, which were prepared on the same day. The ISO 15216‐1:2017 method for surfaces (ISO. 2017) was used for viral extraction following treatment. The demarcated area was then swabbed five times with a sterile cotton swab dipped into either: (1) 1000 μl of MEM maintenance media for HCoV‐OC43 (identical to MEM growth media but with 2% of FBS instead of 10%), releasing media back into the tube after each round of swabbing, then quantified using TCID50; or (2) 1000 μl of DMEM maintenance media for MNV‐1 (identical to DMEM growth media but with 0% FBS instead of 10%), then quantified using a plaque assay.
Quantification of infectious virus
The concentration of infectious viral particles for HCoV‐OC43 was determined by TCID50 using MRC‐5 cells as described previously (Harlow, et al. 2022). A negative control consisting of MEM maintenance media and a positive control consisting of diluted HCoV‐OC43 stock, were also included. The plates were then incubated at 33°C for 5 days and stained with 0.1% crystal violet. Wells were then inspected for the presence or absence of visual cytopathic effect (CPE), and the TCID50/ml was calculated using the Reed‐Muench method (Reed and Muench, 1938). TCID50/ml values were converted to PFU/ml by multiplying by 0.7, which is a constant value obtained based upon Poisson distribution (Wang, 2013).
The concentration of infectious viral particles for MNV‐1 was determined by plaque assay using BV‐2 cells as described previously (Nasheri, et al. 2021). A negative control consisting of just DMEM maintenance media and a positive control consisting of diluted viral stock, was also included. After the infection period was completed, the inocula were removed and the wells were washed once using phosphate‐buffered saline (PBS). The cells were then covered with 2 ml of overlay media, which consisted of a 50:50 mix of 2 × DMEM growth media and 2% agarose. Plates were incubated at 37°C and 5% CO2 for 2 days. Plates were then fixed using 3.7% paraformaldehyde for 2 to 4 h. The cells were then stained with 0.1% crystal violet for 20 min and then plaques were counted and PFU/ml determined.
Calculation of recovery rate
The recovery rate for each produce was determined as the ratio between the recovered viral titre (PFU/mL) from the unwashed produce to the inoculated viral titre (PFU/ml).
Statistical analysis
Statistical analysis was performed using GraphPad Prism v9.0 (GraphPad Software). Multiple unpaired t‐tests were used to determine significant differences between treatments.
RESULTS
Limit of detection (LOD) and recovery rate determination for swabbing of the produce surfaces
The LOD for the quantification methods used, namely swabbing using a sterile cotton swab followed by either determination of infectious viral particles by TCID50 or plaque assay, were determined in previous work by our research group (Blondin‐Brosseau, et al. 2021) for two of the surfaces, that is cucumber and apples. LOD on lettuce for both MNV‐1 and HCoV‐OC43 was determined in this study with identical methodology to previous work (Wang. 2013). The LOD for the quantification methods used in this study is determined and demonstrated in Table 1. As shown, the LOD range for HCoV‐OC43 was lower (5.9–31.6 PFU/ml) compared to MNV (26–88 PFU/ml) (Table 1).
TABLE 1.
Limit of detection in PFU/ml for the enumeration method used in this study, determined by swabbing with a sterile cotton swab followed by TCID50/ml or plaque assay
| Virus | Produce surface | LOD in PFU/ml |
|---|---|---|
| HCoV‐OC43 | English cucumber | 31.6 |
| Gala apple | 10.0 | |
| Romaine lettuce | 5.9 | |
| MNV‐1 | English cucumber | 63 |
| Gala apple | 26 | |
| Romaine lettuce | 88 |
The recovery rate was determined as the ratio between the extracted viral titre from each produce surface and the amount of virus used to inoculate that surface. The average recovery rate for each virus on the specific produce type is provided in Table 2. Generally, the recovery rates for HCoV‐OC43 (0.58%–0.84%) is considerably lower compared to the recovery rates obtained for MNV (9.46%–10.16%).
TABLE 2.
Average recovery rate in percentage for the viral extraction efficiency and is calculated as the ratio between the recovered viral titre (PFU/ml) from the unwashed produce surface and the inoculated viral titre
| Virus | Produce surface | Recovery rate (%) |
|---|---|---|
| HCoV‐OC43 | English cucumber | 0.84 |
| Gala apple | 0.58 | |
| Romaine lettuce | 0.68 | |
| MNV‐1 | English cucumber | 10.16 |
| Gala apple | 9.46 | |
| Romaine lettuce | 9.88 |
Effect of washing on the removal of HCoV‐OC43 from the produce surfaces
As demonstrated in Figure 1, HCoV‐OC43 artificially contaminated produce surfaces were washed with tap water to determine the effectiveness of this washing procedure in removing the virus from the surfaces. The infectious virus concentration was determined using TCID50, then converted to PFU/ml, and was expressed as a reduction in the infectious virus when compared to produce surfaces that were unwashed (Figure 2; Table 3). The washing reduced infectious virus concentration from 4.07 ± 0.52 to 2.12 ± 0.44 log PFU/ml (1.94 ± 0.44 log) on apples, from 4.26 ± 0.08 to 2.85 log PFU/ml (1.42 log) on cucumbers, and from 4.18 to 2.76 ± 0.42 log PFU/ml (1.42 ± 0.44 log) on lettuce.
FIGURE 1.
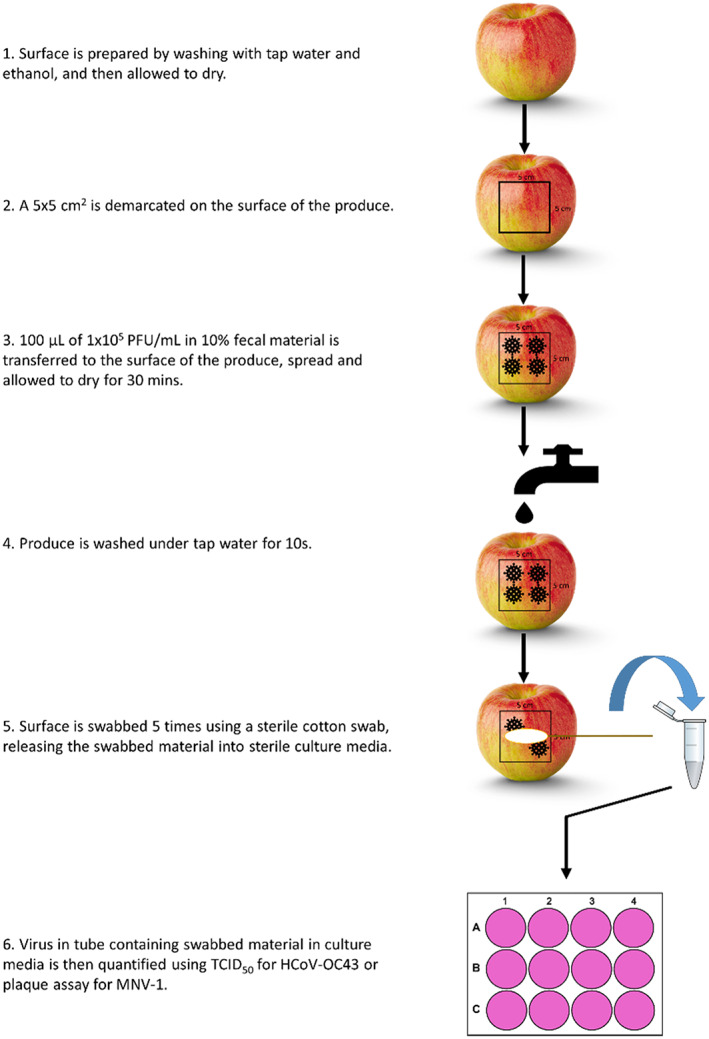
Schematic representation of the experimental procedures in this study.
FIGURE 2.
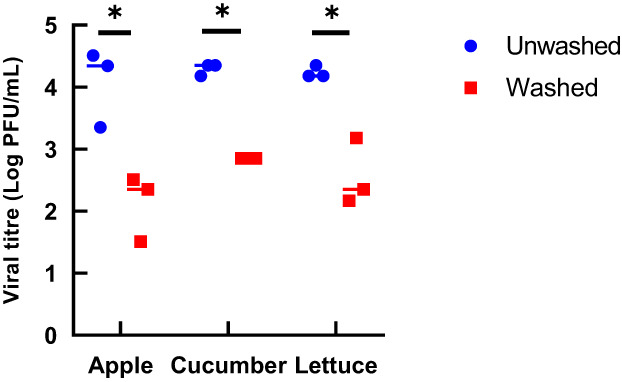
Concentration of infectious HCoV‐OC43 before and after rinsing with tap water for 10 s at a rate of 40 ml/s for the three produce surfaces tested. TCID50/ml values were converted to PFU/ml by multiplying by 0.7 (Wang. 2013). The data are from three independent experiments. *p < 0.05, calculated by t‐test.
TABLE 3.
Log reduction in infectious HCoV‐OC43 after rinsing for the three produce surfaces tested. The results are the mean of three independent experiments ± standard deviation
| Produce surface | Log reduction |
|---|---|
| Gala apple | 1.94 ± 0.44 |
| English cucumber | 1.42 ± 0.00 |
| Romaine lettuce | 1.42 ± 0.42 |
Effect of washing on the removal of MNV‐1 from the produce surfaces
MNV‐1 artificially contaminated produce surfaces were washed with tap water to determine the effectiveness of this washing procedure in removing the virus from the surfaces. The infectious virus concentration was determined using plaque assay and was expressed as a reduction of infectious virus when compared to produce surfaces that were unwashed (Figure 3; Table 4). The washing procedure reduced infectious virus concentration from 2.85 ± 0.15 to 1.06 ± 0.17 log PFU/ml (1.77 ± 0.17 log reduction) for apples, 3.21 ± 0.07 to 1.79 ± 0.07 log PFU/ml (1.42 ± 0.07 log reduction) for cucumbers, and 2.84 ± 0.02 to 1.15 ± 0.06 log PFU/ml (1.79 ± 0.14 log reduction) for lettuce.
FIGURE 3.
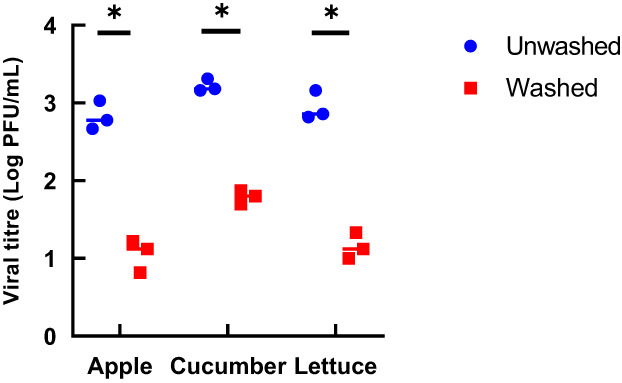
Concentration of infectious MNV‐1 before and after rinsing with tap water for 10 s at a rate of 40 ml/s for the three produce surfaces tested. The data are from three independent experiments. *p < 0.05, calculated by t‐test.
TABLE 4.
Log reduction in infectious MNV‐1 after rinsing for the three produce surfaces tested. The results are the mean of three independent experiments ± standard deviation
| Produce surface | Log reduction |
|---|---|
| Gala apple | 1.77 ± 0.17 |
| English cucumber | 1.42 ± 0.07 |
| Romaine lettuce | 1.79 ± 0.14 |
DISCUSSION
Many human pathogens that transmit faecal‐orally, such as norovirus, utilize food, often fresh produce, as a vehicle for transmission (Nasheri, et al. 2019, Chatziprodromidou, et al. 2018, Bennett, et al. 2018). To date, there is no conclusive evidence regarding the faecal‐oral transmission of HCoVs, including SARS‐CoV‐2, and foodborne transmission has not been definitively shown (Britton, et al. 2021). The risk of fomite‐mediated transmission of coronaviruses is considered low (Sobolik, et al. 2022, Butot, et al. 2022, Dallner, et al. 2021). However, various pieces of evidence have supported the possibility of faecal‐oral transmission of SARS‐CoV‐2, including: (1) the presence of viable virus particles being shed in faeces (Joshi, et al. 2021); (2) the presence of cellular receptors in the gut epithelium of humans (Lamers, et al. 2020); (3) the ability of animal models to support oral transmission of SARS‐CoV‐2 (Chak‐Yiu Lee, et al. 2020); (4) the presence of faecal material aiding in the transfer of HCoVs via fomites (Dallner, et al. 2021) and (5) the survival of HCoVs on produce surfaces (Blondin‐Brosseau, et al. 2021).
Faecal‐oral transmission presents both challenges and opportunities for infection control. Opportunities arise due to the implementation of possible intervention strategies that could result in pathogen reduction on surfaces. One of the main intervention strategies that is commonly employed in households for fresh produce is washing by rinsing under running tap water (Kilonzo‐Nthenge, et al. 2006). This study aimed to address the effectiveness of at‐home, consumer‐friendly washing with tap water in the removal of HCoV‐OC43 and MNV‐1 from produce surfaces. Many studies investigating the effectiveness of sanitizing agents and washing procedures do not consider the effect of organic matter on disinfection efficacy (Dawley, et al. 2021). Herein, the surfaces were artificially contaminated with the virus in the presence of faecal material to mimic contamination of produce by faecal material containing the virus. The faecal material used in this study was not examined for its microflora composition and to date, little is known about the effect of the composition of faecal microflora on viral stability and survival. Three produce surfaces were tested: apples, cucumbers and lettuce. For both viruses and all three surfaces, the amount of infectious virus removed by washing was between 1.42 and 1.94 Log. It is interesting to note that there were no drastic differences between the removal of the nonenveloped MNV‐1 and the enveloped HCoV‐OC43, as enveloped viruses are less resistant to environmental conditions than nonenveloped viruses (Firquet, et al. 2015). However, the recovery rates for HCoV‐OC43 are over 10 times lower compared to MNV, which might suggest that the extraction method for the enveloped coronavirus was not optimum. There were also no significant differences in infectious virus reduction among the three surfaces tested. For both viruses, rinsing apples had a higher reduction in infectious virus concentration than cucumber (1.94 ± 0.44 vs. 1.42 ± 0.00 log reduction for HCoV‐OC43 and 1.77 ± 0.17 vs. 1.42 ± 0.07 log reduction for MNV‐1), possibly due to the former having a smoother surface where water is able to access easier. Lettuce had more varied results between the two viruses, 1.42 ± 0.42 log reduction for HCoV‐OC43 and 1.79 ± 0.14 log reduction for MNV‐1, possibly due to the uneven nature of the surface of lettuce.
In a similar study, it was shown that rinsing lettuce, artificially inoculated with human norovirus, for 30 s under running water led to 1.15 log reduction, as determined by RNA copy number (Bae, et al. 2011). This reduction is slightly lower than what was observed in the present study but it is notable that a proportion of quantified viral RNA does not belong to infectious particles and thus the reduction in viral RNA is significantly lower than the decrease in viral infectivity (Nasheri, et al. 2021). Rinsing blueberries under running water for 1 min resulted in approximately 1.5 log reduction in infectious MNV‐1 and HAV (Leblanc, et al. 2021).
Although rinsing did reduce the infectious virus concentration of both viruses by between 1.42 and 1.94 log, a small, concentration of virus was left on the produce surfaces. For all three produce types, there were greater than 2 and 1 log of HCoV and MNV‐1, respectively, remaining on the surface. The aim of this study was to represent a real‐life scenario of how a consumer would wash their produce, that is, under running tap water at 40 ml/s for 10 s. As some virus was still left on the produce surface, modifications to this washing technique could help to reduce the infectious concentration further, even without the use of detergents and sanitizers, which are less readily available to everyday consumers. This could include increasing the flow rate of the tap water, increasing or reducing water temperature and increasing the rinse time. Scrubbing of the produce surface could also aid in removing the virus.
CONFLICT OF INTEREST
None to be declared.
ACKNOWLEDGEMENTS
This study is financially supported by the Bureau of Microbial Hazards, Health Canada. The authors thank Dr. Sean Li and Dr. Brent Dixon from the Health Products and Food Branch, Health Canada, for thorough review of the manuscript.
Dallner, M. , Harlow, J. & Nasheri, N. (2022) Efficacy of washing produce in removing human coronavirus OC43 and murine norovirus. Journal of Applied Microbiology, 133, 1800–1807. Available from: 10.1111/jam.15667
REFERENCES
- Bae, J.Y. , Lee, J.S. , Shin, M.H. , Lee, S.H. & Hwang, I.G. (2011) Effect of wash treatments on reducing human norovirus on iceberg lettuce and perilla leaf. Journal of Food Protection, 74, 1908–1911. [DOI] [PubMed] [Google Scholar]
- Bandiera, L. , Pavar, G. , Pisetta, G. , Otomo, S. , Mangano, E. , Seckl, J.R. et al. (2020) Face coverings and respiratory tract droplet dispersion. Royal Society Open Science, 7, 201663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadinasab, S. , Chin, A. W. H. , Hosseini, M. , Poon, L. L. M. and Ducker, W. A. 2021. SARS‐CoV‐2 virus transfers to skin through contact with contaminated solids. medRxiv, 2021.04.24.21256044. [DOI] [PMC free article] [PubMed]
- Bennett, S.D. , Sodha, S.V. , Ayers, T.L. , Lynch, M.F. , Gould, L.H. & Tauxe, R.V. (2018) Produce‐associated foodborne disease outbreaks, USA, 1998‐2013. Epidemiology and Infection, 146, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin‐Brosseau, M. , Harlow, J. , Doctor, T. & Nasheri, N. (2021) Examining the persistence of human coronavirus 229E on fresh produce. Food Microbiology, 98, 103780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, G.J. , Chen‐Liaw, A. , Cossarini, F. , Livanos, A.E. , Spindler, M.P. , Plitt, T. et al. (2021) Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS‐ CoV‐2‐specific IgA in patients with acute COVID‐19. Scientific Reports, 11, 13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butot, S. , Zuber, S. , Moser, M. & Baert, L. (2022) Data on transfer of human coronavirus SARS‐CoV‐2 from foods and packaging materials to gloves indicate that fomite transmission is of minor importance. Applied and Environmental Microbiology, 88, e0233821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, J. , Sun, W. , Huang, J. , Gamber, M. , Wu, J. & He, G. (2020) Indirect virus transmission in cluster of COVID‐19 cases, Wenzhou, China, 2020. Emerging Infectious Diseases, 26, 1343–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, J.L. , Papafragkou, E. , Park, G.W. , Osborne, J. , Jaykus, L.A. & Vinje, J. (2006) Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. Journal of Food Protection, 69, 2761–2765. [DOI] [PubMed] [Google Scholar]
- Chak‐Yiu Lee, A. , Zhang, A.J. , Fuk‐Woo Chan, J. , Li, C. , Fan, Z. , Liu, F. et al. (2020) Oral SARS‐CoV‐2 inoculation establishes subclinical respiratory infection with virus shedding in golden Syrian hamsters. Cell Reports Medicine, 1, 100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziprodromidou, I.P. , Bellou, M. , Vantarakis, G. & Vantarakis, A. (2018) Viral outbreaks linked to fresh produce consumption: a systematic review. Journal of Applied Microbiology, 124, 932–942. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Liu, Q. & Guo, D. (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. Journal of Medical Virology, 92, 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, K.S. , Hung, I.F. , Chan, P.P. , Lung, K.C. , Tso, E. , Liu, R. et al. (2020) Gastrointestinal manifestations of SARS‐CoV‐2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta‐analysis. Gastroenterology, 159, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, G. , Ho, H.J. , Ng, C.G. , Neo, F.J. , Win, M.K. , Cui, L. et al. (2018) An unusual outbreak of rotavirus G8P[8] gastroenteritis in adults in an urban community, Singapore, 2016. Journal of Clinical Virology, 105, 57–63. [DOI] [PubMed] [Google Scholar]
- Cueno, M.E. & Imai, K. (2021) Structural comparison of the SARS CoV 2 spike protein relative to other human‐infecting coronaviruses. Frontiers in Medicine, 7, 594439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallner, M. , Harlow, J. & Nasheri, N. (2021) Human coronaviruses do not transfer efficiently between surfaces in the absence of organic materials. Viruses, 13, 1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawley, C.R. , Lee, J.A. & Gibson, K.E. (2021) Reduction of norovirus surrogates alone and in association with bacteria on leaf lettuce and tomatoes during application of aqueous ozone. Food and Environmental Virology, 13, 390–400. [DOI] [PubMed] [Google Scholar]
- Dergham, J. , Delerce, J. , Bedotto, M. , La Scola, B. & Moal, V. (2021) Isolation of viable SARS‐CoV‐2 virus from feces of an immunocompromised patient suggesting a possible fecal mode of transmission. Journal of Clinical Medicine, 10, 2696. 10.3390/jcm10122696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahi, S. & Kenarkoohi, A. (2020) Transmission routes for SARS‐CoV‐2 infection: review of evidence. New Microbes and New Infections, 38, 100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2018. Selecting and serving produce safely. 2021.
- Felter, C. and Bussemaker, N. 2020. Which countries are requiring face masks? https://www.cfr.org/in‐brief/which‐countries‐are‐requiring‐face‐masks [Google Scholar]
- Firquet, S. , Beaujard, S. , Lobert, P.E. , Sane, F. , Caloone, D. , Izard, D. et al. (2015) Survival of enveloped and non‐enveloped viruses on inanimate surfaces. Microbes and Environments, 30, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, E. (2020) Exaggerated risk of transmission of COVID‐19 by fomites. The Lancet Infectious Diseases, 20, 892–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, C.B. , Ryser, E.T. & Marks, B. (2021) Kitchen‐scale treatments for reduction of listeria monocytogenes in prepared produce. Journal of Food Protection, 84(9), 1603–1609. [DOI] [PubMed] [Google Scholar]
- Harlow, J. , Dallner, M. & Nasheri, N. (2022) Protective effect of food against inactivation of human coronavirus OC43 by gastrointestinal fluids. Journal of Food and Environmental Virology, 23, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada . 2021. Produce safety. 2021.
- Joshi, T. , Ahmed, A. & Cholankeril, G. (2021) Gastrointestinal manifestations of coronavirus disease 2019. Current Opinion in Infectious Diseases, 34, 471–476. [DOI] [PubMed] [Google Scholar]
- Kilonzo‐Nthenge, A. , Chen, F.C. & Godwin, S.L. (2006) Efficacy of home washing methods in controlling surface microbial contamination on fresh produce. Journal of Food Protection, 69, 330–334. [DOI] [PubMed] [Google Scholar]
- Kniel, K.E. (2014) The makings of a good human norovirus surrogate. Current Opinion in Virology, 4, 85–90. [DOI] [PubMed] [Google Scholar]
- Kraay, A.N.M. , Hayashi, M.A.L. , Berendes, D.M. , Sobolik, J.S. , Leon, J.S. & Lopman, B.A. (2021) Risk for fomite‐mediated transmission of SARS‐CoV‐2 in child daycares, schools, nursing homes, and offices. Emerging Infectious Diseases, 27, 1229–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraay, A.N.M. , Hayashi, M.A.L. , Hernandez‐Ceron, N. , Spicknall, I.H. , Eisenberg, M.C. , Meza, R. et al. (2018) Fomite‐mediated transmission as a sufficient pathway: a comparative analysis across three viral pathogens. BMC Infectious Diseases, 18, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers, M.M. , Beumer, J. , van der Vaart, J. , Knoops, K. , Puschhof, J. , Breugem, T.I. et al. (2020) SARS‐CoV‐2 productively infects human gut enterocytes. Science, 369, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao, J.C. , Gusmao, T.P.L. , Zarzar, A.M. , Leao Filho, J.C. , Barkokebas Santos de Faria, A. , Morais Silva, I.H. et al. (2020) Coronaviridae‐old friends, new enemy! Oral Diseases, 1, 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc, D. , Gagné, M. & Brassard, J. (2021) Effectiveness of water and sanitizer washing solutions for removing enteric viruses from blueberries. Food Control, 126, 108043. [Google Scholar]
- Liu, D.X. , Liang, J.Q. & Fung, T.S. (2021) Human coronavirus‐229E, ‐OC43, ‐NL63, and ‐HKU1 (Coronaviridae). In: Bamford, D.H. & Zuckerman, M. (Eds.) Encyclopedia of virology, Fourth edition. Oxford: Academic Press, pp. 428–440. [Google Scholar]
- Liu, Q. , Brookbank, L. , Ho, A. , Coffey, J. , Brennan, A.B. & Jones, C.J. (2020) Surface texture limits transfer of S. aureus, T4 bacteriophage, influenza B virus and human coronavirus. PLoS One, 15, e0244518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz, E.A. , Richterman, A. , Gandhi, R.T. & Sax, P.E. (2021) Transmission of SARS‐CoV‐2. Annals of Internal Medicine, 174, 1037. [DOI] [PubMed] [Google Scholar]
- Nasheri, N. , Harlow, J. , Chen, A. , Corneau, N. & Bidawid, S. (2021) Survival and inactivation by advanced oxidative process of foodborne viruses in model low‐moisture foods. Food and Environmental Virology, 13, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasheri, N. , Vester, A. & Petronella, N. (2019) Foodborne viral outbreaks associated with frozen produce. Epidemiology and Infection, 147, e291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, B. , Goodridge, L. , Ronholm, J. & Nasheri, N. (2020) Exploring the potential of foodborne transmission of respiratory viruses. Food Microbiology, 95, 103709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, D.M. , Petric, M. , Skowronski, D.M. , Guasparini, R. , Booth, T.F. , Krajden, M. et al. (2006) An outbreak of human coronavirus OC43 infection and serological cross‐reactivity with SARS coronavirus. Canadian Journal of Infectious Diseases and Medical Microbiology, 17, 330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, W. , Cuevas‐Ferrando, E. , Sanjuan, R. , Domingo‐Calap, P. & Sanchez, G. (2020) Metropolitan wastewater analysis for COVID‐19 epidemiological surveillance. International Journal of Hygiene and Environmental Health, 230, 113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, L. & Muench, H. (1938) A simple method of estimating fifty per cent endpoints. American Journal of Hygiene, 27, 493–497. [Google Scholar]
- Rose‐Martel, M. , Tompkins, E. , Rutley, R. , Romero‐Barrios, P. & Buenaventura, E. (2021) Exposure profile of severe acute respiratory syndrome coronavirus 2 in Canadian food sources. Journal of Food Protection, 84, 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolik, J.S. , Sajewski, E.T. , Jaykus, L.A. , Cooper, D.K. , Lopman, B.A. , Kraay, A.N.M. et al. (2022) Decontamination of SARS‐CoV‐2 from cold‐chain food packaging provides no marginal benefit in risk reduction to food workers. Food Control, 136, 108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuladhar, E. , Hazeleger, W.C. , Koopmans, M. , Zwietering, M.H. , Duizer, E. & Beumer, R.R. (2013) Transfer of noroviruses between fingers and fomites and food products. International Journal of Food Microbiology, 167, 346–352. [DOI] [PubMed] [Google Scholar]
- Verhaelen, K. , Bouwknegt, M. , Carratala, A. , Lodder‐Verschoor, F. , Diez‐Valcarce, M. , Rodriguez‐Lazaro, D. et al. (2013) Virus transfer proportions between gloved fingertips, soft berries, and lettuce, and associated health risks. International Journal of Food Microbiology, 166, 419–425. [DOI] [PubMed] [Google Scholar]
- Wang, Y. (2013) Comparative study of different detection method on the virus titers. Journal of Applied Virology, 2, 6–13. [Google Scholar]
- Warnes, S.L. , Little, Z.R. & Keevil, C.W. (2015) Human coronavirus 229E remains infectious on common touch surface materials. mBio, 6, e01697–e01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, C. , Zhao, H. , Li, K. , Zhang, Z. , Lu, X. , Peng, H. et al. (2020) The evidence of indirect transmission of SARS‐CoV‐2 reported in Guangzhou, China. BMC Public Health, 20, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. & Li, Y. (2018) Transmission of influenza a in a student office based on realistic person‐to‐person contact and surface touch behaviour. International Journal of Environmental Research and Public Health, 15, 1699. 10.3390/ijerph15081699 [DOI] [PMC free article] [PubMed] [Google Scholar]


