Abstract
AbstractEnhancer RNAs (eRNAs) can participate in enhancer regulation and target gene transcription, thus affecting the occurrence and development of tumors. In this study, we identified eRNAs closely related to bladder cancer (BLCA).
Gene expression profiles and clinical information from The Cancer Genome Atlas (TCGA) database were used in this study. The Atlas of Noncoding RNAs in Cancer (TANRIC) co-expression data was also studied to evaluate correlations between the inferred levels of eRNA and its predicted target genes. Moreover, we evaluated differences in tumor microenvironment between high and low ID2-AS1 expression groups, and predicted the response of high- and low-expression groups to immune checkpoint inhibitor (ICI) treatment. Finally, we analyzed the prognostic value of ID2-AS1 in different tumors.
ID2-AS1 and ID2 were identified as eRNAs and target genes related to the prognosis of BLCA. Low ID2-AS1 levels were associated with advanced age, low overall survival, high histological grade, and late BLCA staging. ID2-AS1 appeared to regulate epithelial mesenchymal transition, mitotic spindle assembly, and angiogenesis, thereby affecting BLCA progression. The ID2-AS1 high-expression group had better ICI treatment response. In addition, ID2-AS1 also had prognostic value in other cancers.
ID2-AS1 helps predict prognostic and immunotherapeutic effects in BLCA.
Keywords: bladder cancer, enhancer RNAs, ICI therapy, immunotherapeutic, prognosis, tumor microenvironment
1. Introduction
Bladder carcinoma (BLCA) is the second-most common urological malignancy worldwide, with an estimated 83,730 new cases and 17,200 deaths in 2021.[1] Ninety percent of BLCAs are caused by malignant transformation of urothelial cells. Approximately 25% of patients with BLCA have muscle-invasive bladder cancer (MIBC) or metastasis, and 75% have nonmuscle invasive bladder cancer (NMIBC).[2] If not diagnosed and treated early, NMIBC cells are likely to exhibit muscle invasion and metastasis. As a result, approximately 2/3 of advanced BLCA patients present with MIBC at their first clinical visit.[3] Half of these patients are reported to develop distant metastases within 2 years.[4] Once a tumor has developed to the metastatic stage, surgery combined with general chemotherapy are inadequate for treatment, resulting in a median survival of 14 months.[3,5] According to research, the response rate for novel immunotherapy and immune checkpoint inhibitors (ICIs) is 30% or less.[6] Therefore, there remains an urgent clinical demand for the discovery of novel molecular biomarkers by molecular profiling of BLCA cases.
With the development of high-throughput gene detection technology and the establishment of large-scale gene expression data sets, greater attention is being paid to long noncoding RNAs linked to BLCA.[7–9] The expression of lncRNAs in tissues and cell types has distinct specificity, making them potential cancer biomarkers.[7] Among these lncRNAs, enhancer RNAs (eRNAs) have attracted considerable attention because of their functions in mediating enhancer and gene transcription, and frequent overlap with noncoding risk gene loci associated with disease.[10–12] It has been shown in BLCA that downregulation of P2RY2e diminishes cancer promotion, which thus represents a potential therapeutic target.[13] Estrogen might promote the development of BLCA by inducing SMAD7e production.[14] Furthermore, high expression of MARC1 eRNA is associated with malignant cell behavior.[15] To the best of our knowledge, only 3 studies have been conducted regarding BLCA-associated eRNAs.
In this study, we screened differentially expressed eRNAs (DEEs) in normal and tumor tissues, and explored prognostic eRNAs. ID2-AS1 was found to be significantly associated with survival of patients with BLCA. High expression of ID2-AS1 resulted in elevated purity of tumor cells in the tumor microenvironment (TME), which suggested that ID2-AS1 affects the cellular composition of the TME and thus affects the progression and treatment of BLCA.[16,17]
2. Materials and methods
2.1. TCGA data analysis
We obtained RNA expression data for various cancers from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). At the same time, we also obtained clinical data (age, gender, pathological stage, etc.) and case survival information from TCGA database. To explore possible target genes regulated by eRNAs, we identified 1585 eRNAs and 2303 predicted target genes using the PreSTIGE algorithm. We also studied TANRIC co-expression data (https://bioinformatics.mdanderson.org/public-software/tanric) to assess correlations between the predicted levels of eRNA and predicted target gene expression. Among these eRNAs, only those that were significantly correlated with the overall survival (OS) rate (P < .05) and target gene expression levels (R > .4, P < .001) were considered as candidate key eRNAs in BLCA. Subsequently, we analyzed the relationship between the expression of eRNA and clinicopathological features of BLCA. In addition, we validated the prognostic value of the selected eRNA at the pan-cancer level in all 32 types of malignancies. All datasets in the present study were downloaded from public databases, thus ethical approval was not required.
2.2. Gene bioinformatics enrichment analysis
Gene set variation analysis (GSVA) uses standardized RNA-seq data from TCGA database. As an unsupervised gene set enrichment method, GSVA quantifies the pathway level through gene level quantification to analyze the difference in pathway levels between the ID2-AS1 low- expression group and ID2-AS1 high-expression cohort. We downloaded the “Hallmark” gene sets from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/index.jsp) for GSVA through the package “GSVA” in R. The “MCPcounter” package in R was used for analysis of microenvironment cell populations (MCPs) and quantification of immune cells from transcriptomic data. The results were delivered by the package “pheatmap” in R. Subsequently, the package “limma” in R was used to identify pathways with the most significant differences between patients in the signature group.
2.3. Evaluation of immune cell infiltration in TME and immune checkpoints Inhibitor responses
We evaluated the enrichment of immune and stromal cell gene markers. We used the ESTIMATE algorithm to calculate immunity and matrix scores. In addition, we chose a new TME scoring algorithm to explore differences between high- and low-risk groups based on Immune-related gene pairs. The CIBERSORT algorithm was used to infer the relative proportion of 22 infiltrating immune cells in each sample. In addition, the tumor immune dysfunction and rejection (TIDE) algorithm was used to evaluate ICI treatment responses.
2.4. Statistical analysis
All analyses in this study were performed using R software (v.3.6.3); P < .05 was considered statistically significant.
3. Results
3.1. Identification of DEEs and prognostic eRNAs
A total of 1585 eRNAs and 2303 predicted target genes were identified using the PreSTIGE algorithm.[18] We extracted eRNA expression data from TCGA-BLCA dataset and found 379 DEEs. Next, according to TCGA-BLCA patient clinical prognostic information, we filtered out 53 of 379 DEEs that were significantly correlated with overall survival (Table 1, Kaplan–Meier log-rank test, P < .05). ID2-AS1 was closely related to the prognosis of bladder cancer (P = .003), and positively correlated with ID2 (R = .71). In addition, ID2 has been shown to play an important role in a variety of tumors. So we chose ID2-AS1 in this study.
Table 1.
53 eRNAs and their predicted target genes.
| eRNA | KM | Target | cor | corPval |
|---|---|---|---|---|
| LINC01122 | 0.045473483 | FANCL | 0.40339467 | 1.63E-17 |
| AC004920.1 | 0.007582194 | GRB10 | 0.411626332 | 3.08E-18 |
| RSRP1 | 0.002617455 | RHD | 0.412420081 | 2.61E-18 |
| AC023632.2 | 0.005158216 | DPY19L4 | 0.414415151 | 0 |
| AC023794.2 | 0.002204621 | HOXC4 | 0.420125572 | 5.25E-19 |
| LINC01820 | 0.043439249 | EPAS1 | 0.431815996 | 4.25E-20 |
| AP004608.1 | 0.03042265 | GLB1L2 | 0.438955464 | 8.70E-21 |
| AL035587.1 | 0.029204762 | GNMT | 0.446748342 | 0 |
| AC023790.2 | 0.014139072 | HTR7P1 | 0.452870863 | 3.55E-22 |
| AC091849.2 | 0.046824814 | SDHAP3 | 0.46326598 | 2.95E-23 |
| AL139246.2 | 0.006990563 | TNFRSF14 | 0.465403499 | 1.75E-23 |
| AP000424.2 | 0.010317937 | RNF19A | 0.465827163 | 1.58E-23 |
| LINC01940 | 0.035013722 | TWIST2 | 0.471246138 | 4.13E-24 |
| ADCY10P1 | 0.009323816 | NFYA | 0.474857705 | 0 |
| STEAP1B | 0.036667699 | IL6 | 0.489882296 | 3.39E-26 |
| AC023632.2 | 0.005158216 | RAD54B | 0.493853437 | 0 |
| BLCAP | 0.005434537 | NNAT | 0.49918123 | 0 |
| LINC01357 | 0.025161781 | RHOC | 0.509990406 | 1.36E-28 |
| AL118511.1 | 0.004991953 | C1orf198 | 0.514862257 | 3.36E-29 |
| LINC01615 | 0.026431573 | THBS2 | 0.521304029 | 5.14E-30 |
| LILRP2 | 0.020981196 | LILRB1 | 0.522919934 | 3.19E-30 |
| LILRP2 | 0.020981196 | LILRB4 | 0.524310011 | 2.11E-30 |
| AP000424.1 | 0.014852208 | RNF19A | 0.527188751 | 8.93E-31 |
| LINC02615 | 0.043961018 | PGRMC2 | 0.527810686 | 0 |
| ADCY10P1 | 0.009323816 | UNC5CL | 0.544762173 | 0 |
| AP004608.1 | 0.03042265 | B3GAT1 | 0.547522101 | 1.60E-33 |
| AP000696.1 | 0.028988601 | SIM2 | 0.556052693 | 9.92E-35 |
| CROCCP2 | 0.007230662 | NBPF1 | 0.57597325 | 0 |
| LRRC3-DT | 0.015702475 | LRRC3 | 0.577134181 | 7.16E-38 |
| AC025539.1 | 0.043428858 | HS3ST1 | 0.578892178 | 3.83E-38 |
| PCBP1-AS1 | 9.10E-05 | TIA1 | 0.584023708 | 0 |
| AP001628.2 | 0.036292825 | PDE9A | 0.592521677 | 2.59E-40 |
| AC073316.2 | 0.03870831 | SDK1 | 0.599887858 | 1.58E-41 |
| BLCAP | 0.005434537 | SRC | 0.601001813 | 0 |
| LINC01357 | 0.025161781 | PPM1J | 0.608609553 | 5.22E-43 |
| LILRP2 | 0.020981196 | NCR1 | 0.623955584 | 9.95E-46 |
| AC019117.2 | 0.012389065 | AHR | 0.650786839 | 7.26E-51 |
| ZNF518A | 0.007141589 | CCNJ | 0.657033866 | 0 |
| AC087672.2 | 0.044452206 | LY96 | 0.661537466 | 4.50E-53 |
| ZFHX4-AS1 | 0.005136828 | ZFHX4 | 0.67490614 | 5.99E-56 |
| CDK6-AS1 | 0.004874166 | CDK6 | 0.686888248 | 1.17E-58 |
| LINC01357 | 0.025161781 | SLC16A1 | 0.695766686 | 9.41E-61 |
| ID2-AS1 | 0.003320138 | ID2 | 0.710186992 | 2.53E-64 |
| LEF1-AS1 | 0.033170247 | LEF1 | 0.710507193 | 2.10E-64 |
| TP53TG1 | 0.012233638 | CROT | 0.729915599 | 0 |
| LNC-LBCS | 0.045061 | ID4 | 0.749029601 | 3.94E-75 |
| TM4SF1-AS1 | 0.000265242 | TM4SF1 | 0.763220081 | 1.37E-79 |
| LINC01833 | 0.026044281 | SIX3 | 0.771680093 | 2.11E-82 |
| SLC44A3-AS1 | 0.023976707 | SLC44A3 | 0.780605858 | 0 |
| NR2F1-AS1 | 0.003963551 | NR2F1 | 0.784468094 | 0 |
| AP001189.3 | 0.014542547 | LRRC32 | 0.784789791 | 5.25E-87 |
| AC105942.1 | 0.016728123 | CNN3 | 0.787381061 | 0 |
| AL162411.1 | 0.016024105 | GLDC | 0.79269484 | 6.09E-90 |
3.2. LncRNA ID2-AS1 is a key eRNA in BLCA
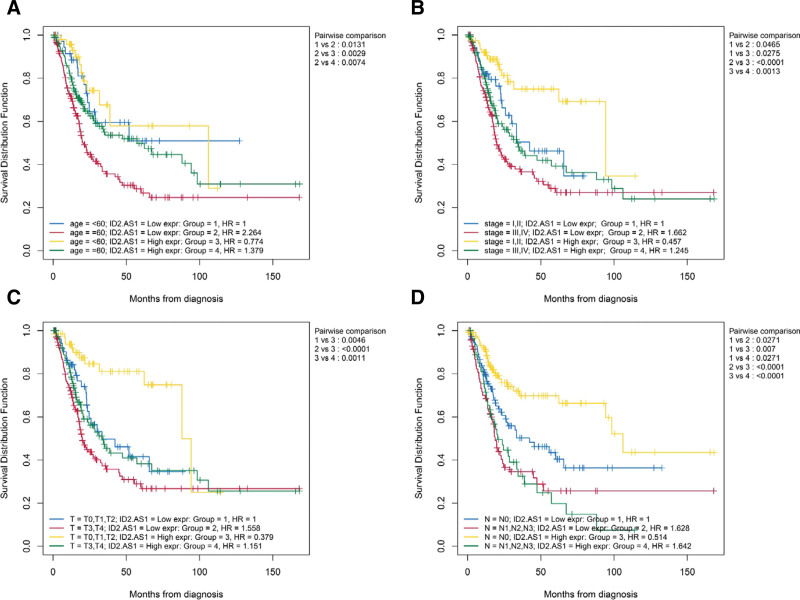
In tumors, the expression level of lncRNA ID2-AS1 was lower than that in normal tissues (Fig. 1A). In addition, in patients with BLCA, the ID2-AS1 high-expression group showed better overall survival than the low-expression group (Fig. 1B). There is a strong correlation between ID2 level and predicted target gene ID2-AS1 expression (Fig. 1C). Moreover, the summary of clinical characteristics of TCGA-BLCA was shown in Table 2. ID2-AS1 expression analysis was performed for different clinical feature subclasses (age, gender, WHO grade, American Joint Committee on Cancer (AJCC) stage, and TNM stage). We observed that patients with advanced BLCA (high WHO grade, high AJCC stage, and MIBC) had lower expression levels (Fig. 2A-G). Subsequently, OS curves using combinations of different clinical features and ID2-AS1 expression levels showed that lower expression levels were associated with poorer OS in patients aged > 60 years, AJCC-Stage I-II, T0-2 stage, and N0 stage subgroups (Figure 3A-D).
Figure 1.
Expression of ID2-AS1 in different tissues. (A) Expression of ID2-AS1 in normal and tumor tissues, (B) Kaplan–Meier curves of overall survival according to IRL-signature groups in the TCGA cohort. (C) Correlation analysis between ID2-AS1 and its prediction target ID2.
Table 2.
Summary of clinical characteristics of TCGA-BLCA patient data sets in the study.
| Characteristic | TCGA-BLCA data set (n = 406) |
|---|---|
| Vital status, n (%) | |
| Alive | 226 (55.7) |
| Dead | 180 (44.3) |
| Age, n (%) | |
| <60 | 86 (21.2) |
| ≥60 | 320 (78.8) |
| Grade, n (%) | |
| Low grade | 21 (5.2) |
| High grade | 382 (94.1) |
| Unknown | 3 (0.7) |
| Gender, n (%) | |
| Female | 106 (26.1) |
| Male | 300 (73.9) |
| WHO-Stage, n (%) | |
| I | 2 (0.5) |
| II | 129 (31.8) |
| III | 140 (34.5) |
| VI | 133 (32.7) |
| Unknown | 2 (0.5) |
| AJCC-T stage, n (%) | |
| T0 | 1 (0.2) |
| T1 | 3 (0.7) |
| T2 | 118 (29.1) |
| T3 | 193 (47.5) |
| T4 | 58 (14.3) |
| Unknown | 33 (8.1) |
| AJCC-N stage, n (%) | |
| N0 | 236 (58.1) |
| N1 | 45 (11.1) |
| N2 | 75 (18.5) |
| N3 | 8 (2.0) |
| Unknown | 42 (10.3) |
Figure 2.
Analyze ID2-AS1 expression in different clinical features. Kaplan–Meier curves analyses of clinical subgroups in TCGA cohort: (A) Age < 60 years, (B) Gender, (C) WHO Grade: high, (D) AJCC Stage: Stage I, II, III, IV, (E) AJCC-T stage: T0-1, T2-4, (F) AJCC-N stage: N0, N1-3, (G) AJCC-M stage: M0, M1. AJCC = American Joint Committee on Cancer, WHO = World Health Organization;
Figure 3.
Evaluate whether lncRNA signature is an independent prognostic factor and relationship between risk score and clinical characteristics. Two factors Kaplan–Meier curves of overall survival for patients in TCGA cohort stratified by IRL-signature, age (A), AJCC Stage (B), AJCC-T stage (C), and AJCC-N stage (D).
3.3. Multiple aggressive, immunosuppressive, and angiogenic hallmarks related to low ID2-AS1 expression
To investigate differentially expressed features between ID2-AS1 high- and low- expression groups, we performed GSVA of the TCGA-BLCA cohort. Notably, gene sets related to tumor necrosis factor-α signaling via nuclear factor-κB, Myc targets, mTOR complex 1 signaling, epithelial mesenchymal transition, mitotic spindle, and angiogenesis signaling pathways were upregulated in the ID2-AS1 low-expression group (Fig. 4A-B).
Figure 4.
Biological function of 2 groups of patients. (A) Bar plot of “Hallmark” pathway score calculated by GSVA for 2 groups of patients. Heat map of GSVA analysis between 2 risk groups. (B) Heat map of most valuable pathway.
3.4. Differences in TME between ID2-AS1 high- and low-expression groups, and prediction of whether ID2-AS1 expression is correlated with ICI therapeutic responses
The ESTIMATE algorithm was used to evaluate tumor purity for each patient. There was a significant difference in tumor purity between the high- and low-expression groups (P <.001, Figure 5A). Furthermore, the TIDE algorithm, which was established to predict immunotherapy responses through transcriptomic data, was used to explore whether the expression level of ID2-AS1 could predict immunotherapeutic benefit in the TCGA-BLCA cohort. The results for each patient are shown in Figure 5B. These results revealed that the percentage of ICI therapy responders was significantly higher for the high-expression group (44%) compared to the low ID2-AS1 expression group (30%) (Chi-squared test, P =.00367; Figure 5B). ID2-AS1 expression was robustly positively correlated with ICI therapeutic response in BLCA patients.
Figure 5.
ID2-AS1 may affect the cellular composition of TME and thus affect the immunotherapy of BLCA. (A) ESTIMATE algorithm was used to evaluate tumor purity for each patient. Assess the difference in immune infiltration between the 2 groups. (B) IRL-signature was efficient in prediction the immunotherapeutic benefit in BLCA. (C) Differences of 10 cell abundances calculated by MCP-counter method between 2 groups of patients. (D) Box plot of ten gene sets calculated by MCP-counter. ns: P ≥ .05, *P < .05, **P < .01, ***P < .001, ****P < .0001.
To further characterize the levels of different immune cell subsets in the TME and chemotactic signals involved, we employed the microenvironment cell population-counter (MCP - counter) method, which allows for robust quantification of the absolute abundance of 8 immune and 2 stromal cell populations in heterogeneous tissues from transcriptomic data[19] (Figure 5C). As shown in Figure 5D, compared to the ID2-AS1 low-expression group, cytotoxic T-lymphocytes and T cells were upregulated in the high-expression group (P < .001 and P < .05). Strikingly, fibroblasts, a type of stromal cell, were downregulated in the high-expression group (P < .001).
3.5. Pan-cancer perspective of ID2-AS1 expression profiles and diagnostic/prognostic potential
Differences in expression of ID2-AS1 were analyzed in 24 different cancer types in TCGA (Fig. 6). We found that the most prevalent gene expression changes involved downregulation, as opposed to upregulation, in tumor tissue, such as in BLCA (P < .001), invasive breast carcinoma (BRCA, P < .0001), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, P < .05), colon adenocarcinoma (COAD, P < .0001), glioblastoma multiforme (GBM, P < .05), kidney chromophobe (KICH, P < .0001), lung adenocarcinoma (LUAD, P < .0001), lung squamous cell carcinoma (LUSC, P < .0001), prostate adenocarcinoma (PRAD, P < .0001), rectal adenocarcinoma (READ, P < .0001), thymoma (THYM, P < .0001), and uterine corpus endometrial carcinoma (UCEC, P < .01) compared to normal tissue. In addition, the expression of ID2-AS1 was independently analyzed for its ability to predict OS in 32 different cancer types using tumor sample data from TCGA. Each cancer type was considered to be independent when examining ability of ID2-AS1 expression to predict OS. Hazard ratios and P-values for the 32 cancer types are shown in Figure 7A. Of these, significant survival differences were detected for adrenocortical carcinoma (ACC, P =.008), BLCA (P =.003), mesothelioma (MESO, P = .023), brain lower grade glioma (LGG, P < .001), uterine carcinosarcoma (UCS, P = .007), and pancreatic adenocarcinoma (PAAD, P =.013). Better OS was associated with high ID2-AS1 expression in BLCA, MESO, LGG, UCS, and PAAD (Figure 7B).
Figure 6.
Gene expression differences for ID2-AS1 were analyzed in 24 different cancer types. ns: P ≥ .05, *P < .05, **P < .01, ***P < .001, ****P < .0001.
Figure 7.
Identification the prognostic effect of ID2-AS1 in pan-cancer. (A) Correlation of ID2-AS1 expression and Hazard ratio in pan-cancer. (B) ID2-AS1 expression downregulation was significantly correlated with poorer overall survival in BLCA, MESO, LGG, UCS and PAAD. However, ID2-AS1 expression downregulation was significantly correlated with better overall survival in ACC.
4. Discussion
In recent years, studies have found that eRNAs regulate gene expression by promoting the enhancer promoter chain, and knockout of eRNAs is usually accompanied by a decrease in target gene expression. A large body of evidence shows that abnormal expression of eRNAs in various cancers is closely related to tumorigenesis. In various types of cancer, eRNAs may modulate the occurrence and progression of tumor cells by regulating certain signaling pathways. However, few researchers have examined BLCA prognosis-related eRNAs. Our study provides insights into the potential role of ID2-AS1 in tumor immunology and its use as a new BLCA-related molecular biomarker.
In our study, we searched for eRNAs related to BLCA prognosis. Through a series of comprehensive analyses, we found that ID2-AS1 and its corresponding target gene, ID2, had a significant impact on survival of BLCA patients. Therefore, we regarded ID2-AS1 as a primary eRNA in BLCA. The results showed that low expression of ID2-AS1 represented an independent prognostic factor in patients with BLCA. We further evaluated the clinical characteristics and relationships between ID2-AS1 expression and BLCA. Interestingly, low expression of ID2-AS1 predicted advanced tumor staging better, and the same results were also found in different clinical subgroup analyses. As an important member of the DNA-binding inhibitor family, ID2 has been shown in previous studies to play key roles in the growth and invasion of various tumor cells.[20,21] Upregulation of ID2 indicates poor prognosis of BRCA patients after breast-conserving surgery.[21] In head and neck squamous cell carcinoma, ID2 is a new molecule involved in the regulation of tumor cell invasion and lymph node metastasis.[20] These findings coincide with the results of our pan-cancer analysis.
Subsequently, the results of GSVA revealed that there were significant differences in biological behavior between the ID2-AS1 high-expression group and the ID2-AS1 low-expression group. Compared to the high-expression group, the low-expression cohort scored higher in epithelial mesenchymal transformation, mitotic spindle formation, and angiogenesis signaling pathways. There is evidence that these signals are closely related to the occurrence and development of tumors. In a study by Zhou et al, ID2-AS1 and ID2 inhibited twist-induced epithelial mesenchymal transformation in hepatocellular carcinoma cells to inhibit tumor metastasis,[22] which is consistent with our results.
Tumor tissues are mainly composed of tumor cells, stromal cells, and immune cells. Tumor purity represents the proportion of tumor cells in the tumor tissue. Interestingly, in studies of glioma, gastric cancer, and colon cancer, patients with low tumor purity tended to have higher malignancy and poorer prognosis.[23–25] In our study, the ID2-AS1 high expression group had a higher tumor density and better OS. TME immune cell infiltration is considered an important and unpredictable phenomenon to predict the prognosis of various cancers and immunotherapeutic responses. The difference analysis of immune cell infiltration showed that the infiltration abundance of CTLs and T cells in the high expression group was significantly higher, and the infiltration abundance of fibroblasts in the low expression group was higher, which is consistent with the results of GSVA to a certain extent. We used the TIDE algorithm to predict the response of patients with BLCA to immunotherapy. It was encouraging that patients in the high expression group are more likely to benefit from immunotherapy.
The significant advantage of our research is the comprehensive analysis conducted, which revealed that ID2-AS1 is an independent prognostic factor for BLCA. We also predicted the relationship between high expression of ID2-AS1 and positive ICI treatment, and found that it is associated with OS in many tumors. Our research has potential value in future tumor prognosis and immunotherapy. However, we did not verify the prognostic ability of ID2-AS1 in the external database; therefore, the predictive value of ID2-AS1 in terms of immunotherapeutic responses needs to be verified in future experiments. Other researchers can use this study as a cornerstone to further reveal the cellular mechanisms involving ID2-AS1 in the occurrence and development of various tumors.
5. Conclusions
In conclusion, ID2-AS1 is an independent protective factor in BLCA, and may become a potentially powerful tool to improve the diagnosis and prognosis of BLCA patients. Hence, ID2-AS1 is a potential promising immune-related therapeutic target for the precise treatment of a variety of malignant tumors in the future.
Author contributions
Conceptualization: Jie Liu, Lei Zhang.
Data curation: Jie Liu.
Formal analysis: Jie Liu.
Investigation: Jie Liu.
Methodology: Jie Liu, Lei Zhang.
Resources: Junfeng Liu.
Software: Junfeng Liu.
Supervision: Jianjun Liu, Degang Ding.
Validation: Ning Wang.
Writing - original draft: Lei Zhang.
Junfeng Liu, Ning Wang, Jie Liu.
Writing - review & editing: Lei Zhang, Junfeng Liu, Ning Wang, Jie Liu.
Abbreviations:
- ACC =
- adrenocortical carcinoma
- AJCC =
- American Joint Committee on Cancer
- BRCA =
- invasive breast carcinoma
- BLCA =
- bladder cancer
- CESC =
- cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD =
- colon adenocarcinoma
- DEEs =
- differentially expressed eRNAs
- eRNAs =
- enhancer RNAs
- GBM =
- glioblastoma multiforme
- GSVA =
- gene set variation analysis
- ICI =
- immune checkpoint inhibitor
- KICH =
- kidney chromophobe
- LUAD =
- lung adenocarcinoma
- LUSC =
- lung squamous cell carcinoma
- LGG =
- brain lower grade glioma
- MIBC =
- muscle-invasive bladder cancer
- OS =
- overall survival
- PAAD =
- pancreatic adenocarcinoma
- PRAD =
- prostate adenocarcinoma
- READ =
- rectal adenocarcinoma
- TCGA =
- The Cancer Genome Atlas
- TIDE =
- tumor immune dysfunction and rejection
- TME =
- tumor microenvironment
- UCEC =
- uterine corpus endometrial carcinoma
- UCS =
- uterine carcinosarcoma
Funding: Science and Technique Program of Henan Province, China (No. 202102310100) supported this study.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article : Zhang L, Ding D, Liu J, Liu J, Wang N, Liu J. Identification of prognostic and immunotherapy-related eRNA ID2-AS1 in bladder cancerMedicine 2022;101:26(e29759).
The authors have no conflicts of interest to disclose.
Contributor Information
Lei Zhang, Email: zhanglei10475@163.com.
Degang Ding, Email: 1350384819@163.com.
Jianjun Liu, Email: syurol@163.com.
Junfeng Liu, Email: syurol@163.com.
Ning Wang, Email: 15000031838@163.com.
References
- [1].Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- [2].Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388:2796–810. [DOI] [PubMed] [Google Scholar]
- [3].Godwin JL, Hoffman-Censits J, Plimack E. Recent developments in the treatment of advanced bladder cancer. Urol Oncol. 2018;36:109–14. [DOI] [PubMed] [Google Scholar]
- [4].Lokeshwar VB, Morera DS, Hasanali SL, et al. A novel splice variant of HYAL-4 drives malignant transformation and predicts outcome in patients with bladder cancer. Clin Cancer Res. 2020;26:3455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Merseburger AS, Apolo AB, Chowdhury S, et al. SIU-ICUD recommendations on bladder cancer: systemic therapy for metastatic bladder cancer. World J Urol. 2019;37:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Koshkin VS, Grivas P. Emerging role of immunotherapy in advanced urothelial carcinoma. Curr Oncol Rep. 2018;20:48. [DOI] [PubMed] [Google Scholar]
- [7].Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24:257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhan Y, Zhang L, Yu S, et al. Long non-coding RNA CASC9 promotes tumor growth and metastasis via modulating FZD6/Wnt/beta-catenin signaling pathway in bladder cancer. J Exp Clin Cancer Res. 2020;39:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhan Y, Chen Z, He S, et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer. 2020;19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. [DOI] [PubMed] [Google Scholar]
- [11].Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–23. [DOI] [PubMed] [Google Scholar]
- [13].Ding M, Zhan H, Liao X, et al. Enhancer RNA - P2RY2e induced by estrogen promotes malignant behaviors of bladder cancer. Int J Biol Sci. 2018;14:1268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Che W, Ye S, Cai A, et al. CRISPR-Cas13a targeting the enhancer RNA-SMAD7e inhibits bladder cancer development both in vitro and in vivo. Front Mol Biosci. 2020;7:607740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Y, Ding M, Liao X, et al. High expression of enhancer RNA MARC1 or its activation by DHT is associated with the malignant behavior in bladder cancer. Exp Cell Res. 2018;370:303–311. [DOI] [PubMed] [Google Scholar]
- [16].Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rhee JK, Jung YC, Kim KR, et al. Impact of tumor purity on immune gene expression and clustering analyses across multiple cancer types. Cancer Immunol Res. 2018;6:87–97. [DOI] [PubMed] [Google Scholar]
- [18].Vucicevic D, Corradin O, Ntini E, et al. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle. 2015;14:253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biology. 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bae WJ, Koo BS, Lee SH, et al. Inhibitor of DNA binding 2 is a novel therapeutic target for stemness of head and neck squamous cell carcinoma. Br J Cancer. 2017;117:1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu Y, Pandey PR, Sharma S, et al. ID2 and GJB2 promote early-stage breast cancer progression by regulating cancer stemness. Breast Cancer Res Treat. 2019;175:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yz A, Lin HA, Yw A, et al. LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett. 2020;469:399–409. [DOI] [PubMed] [Google Scholar]
- [23].Mao Y, Feng Q, Zheng P, et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag Res. 2018;10:3569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang CB, Cheng W, Ren X, et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23:6279–6291. [DOI] [PubMed] [Google Scholar]
- [25].Gong Z, Zhang J, Guo W. Tumor purity as a prognosis and immunotherapy relevant feature in gastric cancer. Cancer Med. 2020;9:9052–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]